This work examines the chromosomal positional history of all genes in Arabidopsis thaliana through evolutionary time in the rosid superorder, finding that mobile gene families are under different selection pressure than syntenic gene families.
Abstract
Certain types of gene families, such as those encoding most families of transcription factors, maintain their chromosomal syntenic positions throughout angiosperm evolutionary time. Other nonsyntenic gene families are prone to deletion, tandem duplication, and transposition. Here, we describe the chromosomal positional history of all genes in Arabidopsis thaliana throughout the rosid superorder. We introduce a public database where researchers can look up the positional history of their favorite A. thaliana gene or gene family. Finally, we show that specific gene families transposed at specific points in evolutionary time, particularly after whole-genome duplication events in the Brassicales, and suggest that genes in mobile gene families are under different selection pressure than syntenic genes.
INTRODUCTION
As more and more genomes are sequenced and made available to the public, our ability to study phylogenetic relationships among taxa continues to expand beyond what researchers had envisioned even a decade ago. In plants, for instance, many monocot and eudicot genomes have been sequenced and released for research, and even more are in the pipeline. These sequenced genomes permit us to compare gene collinearity among species, which allows us to ask what sorts of gene families carry genes that tend to be syntenically retained over time and what genes, and, thus, gene families, tend to be deleted entirely, duplicated by some mechanism, or have transposed in certain lineages only.
Some genes shared between related species are syntenic or colocalized on corresponding chromosomes. They can also be collinear, meaning they remain in similar chromosomal orders over time (Coghlan et al., 2005) (Figure 1). Retained genes are defined as having been preserved collinearly after a whole-genome duplication (WGD) event. In plants, WGDs occur quite frequently over evolutionary time (Blanc and Wolfe, 2004) (Figure 2A), and certain gene families are consistently retained after WGDs, going back to before the monocot-eudicot split (Jiao et al., 2011; reviewed in Van de Peer, 2011). This retention is likely due to functional buffering (Chapman et al., 2006) or gene dosage (Veitia, 2004; Birchler and Veitia, 2007; Conant and Wolfe, 2008). It is thought that these genome duplications may present an opportunity for novelty (Fawcett et al., 2009). While syntenic genes have been studied extensively, little attention has been paid to the types of genes that have tended to transpose over evolutionary time. Recent work in Drosophila melanogaster and plants has demonstrated that many functional, nontransposable element genes are mobile, either via DNA- or RNA-mediated transposition (Wang et al., 2006; Freeling et al., 2008; Yang et al., 2008; Zhu et al., 2009; Wicker et al., 2010; Woodhouse et al., 2010). One mode of DNA-mediated transposition is through intrachromosomal recombination (Yang et al., 2008; Woodhouse et al., 2010). Recombination frequency has been observed to increase after a polyploidization event in Arabidopsis thaliana (Pecinka et al., 2011), and other sorts of rearrangements, such as duplications and translocations, are known to happen after a polyploidy event (reviewed in Gaeta and Pires, 2010). It is possible that in plants, a WGD may induce gene transposition via recombination or some other likely mechanism.
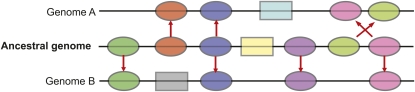
Figure 1.
Using Comparative Genomics to Define Synteny and Collinearity.
Some genes shared between related species are syntenic or colocalized on corresponding chromosomes. They can also be collinear, meaning they remain in similar chromosomal orders over time. In comparison to the ancestral genome, genomes A and B share some genes that are syntenic and collinear, though not always the same ones. Rectangular genes represent nonsyntenic genes. Notice that a gene can be syntenic without necessarily being collinear.
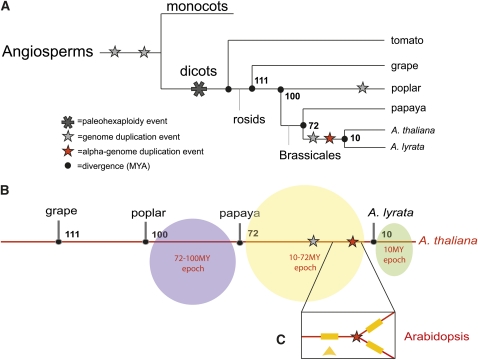
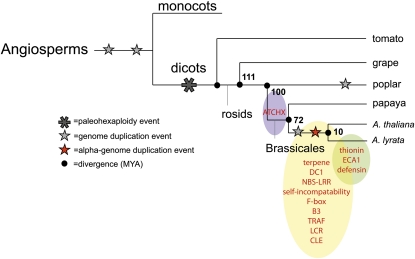
Figure 2.
Cladogram of the Key Species Used in This Study: A. thaliana and A. lyrata, Papaya, Poplar, and Grape.
(A) The genus Arabidopsis belongs in the order Brassicales, as does papaya. A. thaliana and A. lyrata diverged from each other ~10 MYA. Papaya and A. thaliana diverged from each other ~72 MYA. A. thaliana and poplar diverged ~100 MYA. Grape is the most distantly related outgroup from A. thaliana and diverged from A. thaliana ~111 MYA. The red star represents the α-duplication event that occurred 20 to 60 MYA (Paterson et al., 2010); the gray star represents an earlier genome duplication event from the α; the larger star is a paleohexaploidy. Citations are provided by Jiao et al. (2011).
(B) The positional history timeline and the epochs in which each existing A. thaliana gene had transposed throughout the A. thaliana lineage. Three time points are represented: the A. thaliana ancestral genes that transposed after poplar split from the A. thaliana lineage 100 MYA (72 to 100 MYA epoch), genes that had transposed after papaya split from the A. thaliana lineage 72 MYA (10 to 72 MYA), and genes that had transposed after A. lyrata had split from A. thaliana (10 MYA).
(C) Genes that had transposed prior to the α-duplication event in the A. thaliana lineage. The insertion event is represented by the yellow arrow; the inserted gene is represented by the yellow rectangle. After the α-duplication event (red star), the inserted gene is now duplicated in the A. thaliana lineage. Both copies of the transposed gene are unlikely to be retained.
Our lab has found that certain gene families in the order Brassicales, in particular F-box genes, nucleotide binding site–Leu-rich repeat (NBS-LRR) disease resistance genes, and AGAMOUS-like (AGL) genes, transpose more than others (Freeling et al., 2008; Woodhouse et al., 2010). The relative nonsynteny of certain gene families may be informative regarding the role of gene transposition in conferring novel functions within a gene family. Gene families that are particularly rich in genes that underwent transposition, such as F-box genes and NBS-LRR disease resistance genes, are also those families containing genes subject to tandem duplication (Freeling et al., 2008; Woodhouse et al., 2010). It has been argued from differential expression data that local gene duplication in these families and others permits subfunctionalization among the duplicated copies (Cannon et al., 2004; Leister, 2004; Rizzon et al., 2006), where a pair of once-identical genes lose nonessential components of their cis-elements such that only both genes together encode the full information from the ancestor; these duplicates should now be permanent (Force et al., 1999). Following this logic, the transposition of a gene to a new locus may confer changes in gene regulation that could lead to novel expression patterns even without taking position effects into account; for example, in addition to the possibility of landing in a position with novel cis-regulatory elements, the gene might transpose without some of its enhancer/repressor elements. However, the specific relationship between gene movement and subfunctionalization has not been tested rigorously.
Here, we report the positional history of all annotated A. thaliana genes in the rosid superorder, using Arabidopsis lyrata, papaya (Carica papaya), poplar (Populus trichocarpa), and French grape (Vitis vinifera) as outgroups. We built a public database that allows researchers to find the positional history of their gene or gene family of interest using the species that are now available to us, and the database is designed to be updated as new eudicot genomes are released. This database is linked to The Arabidopsis Information Resource (TAIR), so that plant geneticists may easily transition from TAIR to our site. What makes our databases different from others, such as PLAZA (Proost et al., 2009), is that ours specifically defines genes as being nonsyntenic or transposed as well as syntenous. The positional history for each A. thaliana gene published here may be retrieved on this website, along with a link to a multigenome comparison tool so that our data preloaded for each gene along with their genomic neighborhood can be visualized and proofed. Furthermore, we demonstrate that different types of gene families appear to have transposed at different times within the rosids, specifically during or after the WGD events in the Brassicales, and we discuss what role gene transposition might have in the diversification of gene families. Our data are preliminary in the sense that all positional data will improve as more and deeper eudicot genomes are sequenced and released.
RESULTS
The Positional History of All Genes in A. thaliana: A Searchable Database at http://biocon.berkeley.edu/athaliana
The outgroups examined in this study span the rosid eudicot superorder (Figure 2). A. thaliana diverged from A. lyrata ~10 million years ago (MYA; Hu et al., 2011), and from papaya ~72 MYA (Ming et al., 2008). The outgroup poplar (Tuskan et al., 2006) diverged from the Arabidopsis genus ~100 MYA (confidence interval 102 to 114 MYA) (Wang et al., 2009). Grape (Jaillon et al., 2007) is the outgroup most distantly related to A. thaliana in this study, diverging from A. thaliana ~111 MYA (confidence interval 109 to 115 MYA) (Wang et al., 2009). All divergence time estimation criteria are described in the literature cited above.
The positional history of each gene in A. thaliana (TAIR9) was found using parameters described in Methods. In short, we automated the initial chromosomal synteny search and classification, but later used manual proofing to check the correctness of the automated results and apply corrections when necessary. At the heart of our methods is a special algorithm particularly useful in sorting out runs of orthologous genes between genomes. Briefly, a 40-gene window was centered on every query A. thaliana (TAIR9) gene to check for a syntenic region in each target genome. LASTZ (default parameters) were used to define anchors and required that the syntenic region have at least four collinear anchors (out of 40 possible anchors). The nearest anchors on both sides of genes were identified to define a tight syntenic location. For each A. thaliana gene and its chromosomal neighborhood, we attempt to find the one (or two, as is the case in poplar because of a poplar-specific duplication; Tuskan et al., 2006) orthologous chromosomal segment in each outgroup genome. Depending on whether an ortholog of the query gene is found in the expected syntenic location or not, the query gene was determined to be either syntenic (S) or not syntenic (indicating a potential gene loss or transposition event).
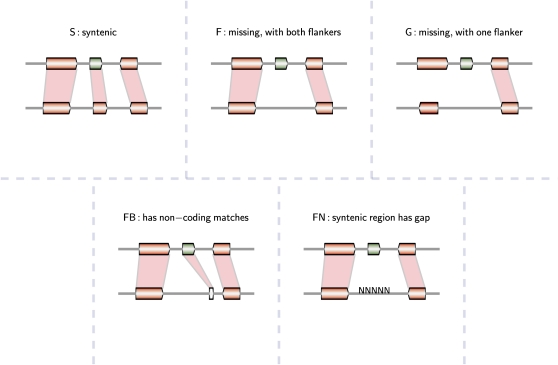
To facilitate downstream study of gene transposition events and rule out artifacts, nonsyntenic genes were further subdivided into those having one flanking gene in the 40-gene interval (G) or two flanking genes in the interval (F) (Figure 2). Genes that had no flankers (for example, genes in the pericentromeric regions in the genome) in the outgroup were denoted with a “−.” Genes that fall into the F category were characterized as potentially transposed, pending further analysis. If an F gene had a BLAST hit to noncoding sequences in the interval in the outgroup, it was denoted as FB. F genes that had assembly gaps (Ns) (i.e., missing sequences) between the flankers in the outgroup were denoted as FN. These careful labels of pipeline output are to minimize the effects of annotation and assembly artifacts on our essential classification of a gene as syntenic or nonsyntenic.
From our Positional History homepage, simply enter the gene LOC ID or TAIR gene description keyword (for example, “MADS box”) to determine the positional history of your favorite gene or group, select a previously TAIR-annotated gene family from the pull-down menu to observe the positional history of a particular gene family, or choose the pattern of synteny/nonsynteny per outgroup that you wish to study. Since we go to TAIR for gene categories and gene descriptions, please refer to TAIR for citations. Currently the TAIR gene descriptions are for TAIR9.
Syntenic Genes among the Rosids
An A. thaliana query gene is considered syntenic throughout the rosids if it has been found to be syntenic (S) in at least grape, as grape is the most distantly related outgroup from A. thaliana. There are 13,350 A. thaliana query genes that are categorized as syntenic in the rosids, or ~40% of all genes in A. thaliana. Excluding transposons, unknown genes, and pseudogenes, we found that 10,770 of all characterized genes are syntenous within rosids, or 49% (Table 1). The full set of characterized genes has also been uploaded to the Dryad data repository (http://datadryad.org/). Presumably, the genes that make up the remaining nonsyntenic half are either newly arisen, transposed, or have evolved so rapidly that their deeper rosid orthologs are not detected in the expected syntenous location.
Table 1.
Gene Stability versus Homoeolog Retention and CNSs
| Category | Genome | Syntenic | Nonsyntenic | Transposed | Syntenic (%) | Nonsyntenic (%) | Transposed (%) | χ2 Syntenic/Nonsyntenic | χ2 Transposed |
| All | 21,665 | 10,770 | 10,895 | 4,575 | 49.70% | 50.30% | 21.00% | n/a | n/a |
| Homoeolog | 5,193 | 3,502 | 1,689 | 383 | 67.50% | 32.50% | 7.00% | <0.0001 | <0.0001 |
| Single-copy | 6,076 | 3,177 | 2,899 | 970 | 52.30% | 47.70% | 16.00% | 0.0046 | <0.0001 |
| Total CNS | 4,502 | 3,239 | 1,261 | 190 | 71.90% | 28.00% | 4.20% | <0.0001 | <0.0001 |
| Total 5′ CNS | 2,032 | 1,450 | 582 | 88 | 71.40% | 28.60% | 4.30% | <0.0001 | 0.0003 |
| Total 3′ CNS | 799 | 562 | 235 | 42 | 70.30% | 29.40% | 5.30% | <0.0001 | 0.175 |
| Total intronic CNS | 1,671 | 1,227 | 444 | 56 | 73.40% | 26.60% | 3.40% | <0.0001 | <0.0001 |
| >5 CNSs | 825 | 605 | 218 | 15 | 73.30% | 26.40% | 1.80% | <0.0001 | <0.0001 |
| <5 CNSs | 2,735 | 1,893 | 848 | 125 | 69.20% | 31.00% | 4.60% | <0.0001 | 0.0002 |
The number of genes that are syntenic, nonsyntenic, and transposed in the genome, among genes with a homoeolog, among single-copy genes, and among genes with CNSs. χ2 for transposed homoeologs is based on the number of genes that have transposed in the genome. χ2 for total CNSs is based on the number of transposed genes that had a homoeolog. n/a, not applicable.
Genes That Have Putatively Transposed in the Rosids
The study of transposition over time is a function of synteny in the nearest outgroups versus nonsynteny in the more distant outgroups. Our criteria for a putatively transposed gene are described in Methods and illustrated in Figure 3. It should be noted that a significant limitation arises in the frequent occurrence of unsequenced DNA between flankers in the outgroups (for example, the papaya genome has nearly 100 Mb of missing sequences throughout its genome). The incompleteness of the genome sequence requires us to state that, in many cases described below, transposition is inferred to have most likely happened in one or more outgroup lineages but that this transposition is not proved because the syntenic gene might actually exist in an unsequenced gap (this scenario is captured in our FN class).
Figure 3.
Illustration of Different Scenarios When Classifying the Positional History for a Gene in Question (Colored in Green).
A 40-gene window was centered on every query A. thaliana (TAIR9) gene to check for a syntenic region in each target genome. LASTZ (default parameters) were used to define anchors and required that the syntenic region to have at least four collinear anchors (out of 40 possible anchors) in the interval. Each query gene is categorized based on the flank anchors and more sensitive search on the tight interval as follows: gene match in the interval, syntenic (S) or not syntenic but have both flankers (F) or one flanker (G). Genes labeled as F are further validated as follows: BLAST matches (e.g., to noncoding sequences) in the interval (FB) and contains assembly gaps (Ns) in the interval (FN). Because the region between flankers is unsequenced (FN), we cannot determine whether or not there is a gene in that space that could be syntenic with the query gene.
In our study of putatively transposed genes in A. thaliana, we characterized the time points when these genes had transposed in the A. thaliana ancestor in terms of three epochs based on the divergence times of the outgroups from A. thaliana (Figure 2B). An A. thaliana gene that had transposed since the divergence of A. thaliana and A. lyrata belongs in the <10 MYA epoch; an A. thaliana gene that had transposed before the divergence of A. thaliana and A. lyrata but after the divergence of papaya from what became the A. thaliana lineage falls within the 10 to 72 MYA epoch; and an ancestral A. thaliana gene that had transposed before the divergence of papaya from the A. thaliana lineage but after the divergence of poplar belongs in the 72 to 100 MYA epoch (Figure 2B). In the latter case, it is possible that the gene in question was lost in both the poplar and grape lineages, but we suggest that the likelihood of a loss at both loci is a less parsimonious inference than a single ancient loss pre-Brassicales divergence. In total, there are 4575 of these genes that meet our criteria for a putatively transposed gene in the A. thaliana lineage (independent of epoch), ~21% of the A. thaliana genome (Table 1). All 4575 of these transposed genes were further manually proofed.
Genes with Homoeologs (Posttetraploidy Pairs) Retained from the Most Recent A. thaliana Lineage Genome Duplication Event Tend to Be Syntenic throughout the Rosids
WGD events have occurred repetitively throughout plant evolutionary history (Paterson et al., 2010). After a genome-wide duplication event, certain types of genes tend to retain their duplicate copy, or homoeolog (Maere et al., 2005), such as many transcription factor genes. In the two Arabidopsis species, the most recent WGD event, known as the α duplication event, occurred ~20 to 60 MYA (Paterson et al., 2010) (Figure 2). It is thought that these genes retain their homoeolog because their protein products are sensitive to the dosage of the protein complexes in which they interact, so loss of the homoeolog would have caused an unfit haploinsufficiency syndrome (Veitia, 2002; Birchler and Veitia, 2007). Our definition of an α-retained homoeolog is based on Thomas and coworkers’ revision (Thomas et al., 2006) of the Bowers and coworkers’ original pairs list (Bowers et al., 2003). We asked if genes that retain their homoeolog also tended to remain syntenic throughout the rosids, going back to the grape common ancestor 111 MYA. We found that 1751 retained A. thaliana gene pairs were syntenic in grape, representing 3502 individual genes (Table 1). We next asked if the ancestor of any α-retained genes had transposed before the WGD event; the order of events would have been (1) a gene transposed into a new location in the A. thaliana lineage, (2) the genome duplicated, and (3) both copies of the transposed gene were retained after the WGD (Figure 2C). These genes would have transposed after poplar diverged from the A. thaliana lineage (100 MYA) but before the α-duplication event (20 to 60 MYA), which predated the divergence of A. thaliana and A. lyrata (10 MYA). In other words, the gene had to have transposed in the 10 to 72 MYA epoch (Figure 2B). Such a gene would be syntenic in A. lyrata and A. thaliana and would also have a homoeolog in A. thaliana. We found only 192 gene pairs that meet these criteria, representing 192 single-gene, pre-WGD insertion events (Table 1). These data demonstrate that retained genes are more likely to have been syntenic than to have transposed in earlier epochs.
Because recent WGD gene duplicates tend to be syntenic throughout the rosids, we asked if single-copy genes also demonstrated a bias in synteny. Single-copy genes are those that appear to always lose their homoeologs after a genome duplication event (Duarte et al., 2010). However, we found that the difference in frequency of syntenic genes among single-copy genes and the frequency of syntenic genes within the genome is not statistically significant (Table 1). Interestingly, while the frequency of transposed genes among single-copy genes is less than the frequency of transposed genes found in the genome (16% versus 21%, χ2 test <0.0001), it is greater than the frequency of transposed genes with a retained homoeolog (16% versus 7%, χ2 test <0.0001). Clearly, the syntenic genes would be more useful for systematic analyses, and our work demonstrates which ones these are.
A Gene’s Positional Stability Is Correlated with the Presence, Size, and Location of Conserved Noncoding Sequences
Conserved noncoding sequences (CNSs) are sequences outside of the coding region that have been retained over evolutionary time and have been shown, in general, to have some function (Inada et al., 2003; Freeling and Subramaniam, 2009). This function is inferred by the very fact that they are retained as CNS pairs under conditions where functionless DNA would be expected to drift by base mutations to undetectability, assuming that the functionless sequence avoided outright deletion. It is thought that CNSs play a part in gene regulation, most likely as cis-acting protein binding sites (Freeling and Subramaniam, 2009). In A. thaliana, CNSs are in part characterized by their retention as homoeologous pairs after the most recent tetraploidy (Thomas et al., 2007). Because of this, we expected that CNSs would not be associated with transposed genes but would be associated with syntenic genes. We found that 72% of CNS-rich genes are syntenic versus 67% of homoeologous genes that are syntenic, a difference that is statistically significant (χ2 test <0.0001). Conversely, the number of pre-α-duplication CNS-containing genes that had transposed is quite low, even lower than the number of transposed genes that have a homoeolog: 190 CNS-containing transposed genes (4.2%) representing 95 pairs versus 384 transposed homoeologous genes (7%) representing 192 pairs (χ2 test <0.0001).
Tracking CNSs and gene mobility allowed us to answer specific questions as to the unit of transposition: Does a transposed gene tend to take with it its full complement of associated regulatory DNA? If not, of course, the gene becomes a candidate neomorphic mutant in the sense that it might pick up, perhaps by necessity, regulatory information from its new chromosomal position. Previous research has suggested that one way genes can transpose is by a DNA-based copy-paste mechanism, where, after duplication, the original parent gene remains in situ after the daughter copy transposes to a new site (Woodhouse et al., 2010). Using BLASTN (word size 11, E-value cutoff ≤ 0.001), we identified the best hits outside of the homoeolog for each of our CNS-carrying transposed pairs and asked whether these hits contained sequence that corresponded to the described CNSs in these pairs. We found only 12 separate transposed pairs whose best hits were either a syntenic, CNS-containing gene (the potential parent gene) or another transposed gene where at least one of the CNSs was present (a potential daughter or sibling gene) (Table 2; see Supplemental Figure 1 online). This further confirms that transposed genes do not tend to arise from genes with CNSs nor take with them their suite of regulatory sequence.
Table 2.
Genes That Have Transposed with Their CNSs
| Query TAIR | No. of CNSs | 5′ CNSs | 3′ CNSs | Intronic CNSs | Homoeolog of Query TAIR | Transposed Best Hit TAIR | No. of CNSs Retained by Transposed | Transposed CNS Position | Transposed CNS Distance |
| AT3G05660 | 1 | 1 | AT5G27060 | AT2G15080 | 1 | 5′ | <50 bp | ||
| AT3G28890 | 1 | 5′ | <50 bp | ||||||
| AT3G11010 | 1 | 5′ | <50 bp | ||||||
| AT4G13820 | 1 | 5′ | 6 kb (separated by two genes) | ||||||
| AT4G13880 | 1 | 5′ | <50 bp | ||||||
| AT5G07210 | 1 | 1 | AT5G62110 | AT2G27070 | 1 | 5′ | <50 bp | ||
| AT1G64000 | 2 | 2 | AT5G41570 | AT2G46130 | 1 | 5′ | <50 bp | ||
| AT1G05570 | 11 | 1 | 10 | AT2G31960 | AT5G36870 | 7 | Intronic | Intronic | |
| AT1G21140 | 3 | 2 | 1 | AT1G76800 | AT3G43660 | 1 | 5′ | <50 bp | |
| AT3G25190 | 1 | 5′ | <50 bp | ||||||
| AT1G21160 | 3 | 3 | AT1G76825 | AT2G27700 | 1 | Intronic | Intronic | ||
| AT1G76720 | 2 | Intronic | Intronic | ||||||
| AT1G29470 | 1 | 1 | AT2G34300 | AT5G27800 | 1 | Intronic | Intronic | ||
| AT1G63400 | 1 | 1 | AT5G41170 | AT1G12620 | 1 | 5′ | 5 kb (separated by a gene) | ||
| AT4G31650 | 5 | 2 | 3 | AT2G24650 | AT2G13990 | 1 | 5′ | <50 bp | |
| AT4G00260 | 1 | 5′ | 1 kb | ||||||
| AT2G24748 | 1 | 5′ | 800 bp | ||||||
| AT1G26680 | 1 | 5′ | 1 kb | ||||||
| AT4G37260 | 12 | 10 | 2 | AT2G23290 | AT3G50060 | 2 | 5′ | 300 bp, 100 bp | |
| AT3G55730 | 1 | 5′ | 600 bp | ||||||
| AT5G18080 | 11 | 10 | 1 | AT3G03820 | near AT3G13820 | 1 | 5′ | 500 bp | |
| AT1G11803 | 1 | 5′ | 500 bp | ||||||
| AT4G38850 | 1 | 5′ | 500 bp | ||||||
| AT2G21200 | 1 | 3′ | 100 bp | ||||||
| AT5G66940 | 4 | 4 | AT3G50410 | AT4G38000 | 1 | 5′ | <50 bp |
The TAIR9 ID of the CNS-containing gene’s query sequence, its description, the numbers and types of CNSs it has, the TAIR ID of its homoeolog, the TAIR ID of its best hit, and the number, position, and distance of the best hit’s sequences corresponding to the query sequence’s CNSs. Most transposition events where a gene took with it a CNS from the donor site have taken or retained only one CNS. Most CNSs transposed are proximal to the 5′ start site. There are cases where one query gene has many possible best hits; the empty cells are placeholders.
Table 2 demonstrates that number, distance, and placement of CNSs are all limiting factors when it comes to a transposed gene carrying with it, then afterward retaining, CNSs from its parent site. Indeed, gene size itself may play a role in limiting gene mobility: When we compared the functional gene space (comprising the gene itself along with all known regulatory features upstream and downstream from the start-stop sites, including CNSs) of syntenous and transposed genes, we found that the percentage of syntenic genes over 3 kb was more than twice that of mobile genes (18% versus 7%, respectively), while 36% of mobile genes were under 1 kb in length, as opposed to only 6% of syntenic genes (Table 3), and 19% of mobile genes were under 500 bp, in comparison to only 1% of syntenic genes. The unit of mobility seems to be single gene: Upon manually proofing all 4575 transposed genes, we found <1% of cases where more than one gene transposed, and in those situations, it was usually one and a half genes (data not shown). The likelihood of having introns is also correlated with synteny: 11.6% of all syntenic genes lacked introns, whereas ~30% of all transposed genes lacked introns (Table 3). This lack of introns is expected, as it has been demonstrated in previous work that transposed genes often originate from families that do not tend to have introns (Woodhouse et al., 2010), though the mechanism of gene transposition via RNA-mediated retroposition (which gives rise to transposed genes without introns) may also play a role (Zhang et al., 2005) in addition to DNA-mediated transposition (Wicker et al., 2010; Woodhouse et al., 2010).
Table 3.
Transposed Genes Have a Smaller Functional Gene Space Than Stable Genes and Fewer Introns
| Functional Gene Space (bp) |
No. of Introns | ||||||||||
| Position | >3,000 | 3,000 | 2,000 | 1,000 | <1,000 | >500 | <500 | Total | ≥1 Intron | 0 Introns | Total |
| Syntenous | 1,968 | 1,805 | 3,313 | 3,052 | 624 | 547 | 77 | 10,764 | 9,396 | 1,228 | 10,624 |
| % Syntenous | 18% | 17% | 31% | 28% | 6% | 5% | 1% | – | 88.40% | 11.60% | – |
| Transposed | 322 | 321 | 830 | 1,457 | 1,633 | 768 | 865 | 4,563 | 2,470 | 1,068 | 3,538 |
| % Transposed | 7% | 7% | 18% | 32% | 36% | 17% | 19% | – | 69.80% | 30.20% | – |
Functional gene space is defined as the gene itself as well as all regulatory regions as inferred from CNS positions upstream and downstream of the gene. Units are in base pairs. Based on TAIR9 exon annotation data, we found that 30% of transposed genes lack introns, in comparison to only 11.6% of stable genes that lack introns.
Certain Classes of Genes Have Transposed at Specific Points in Time
We next examined gene classest to determine if any tended to transpose at specific points in evolutionary time, or epochs. This would also allow us to ask if recent genome duplication events might have played a role in expansion of gene transposition within certain clades and thus have given rise to novelty, as discussed by Fawcett et al. (2009). We found that, as expected F-box, NBS-LRR, defensin, and AGL genes were overrepresented for transposition within the <10 MYA epoch (Woodhouse et al., 2010), and most also transposed to a significant degree within the 10 to 72 MYA epoch (Freeling et al., 2008), which is the epoch that includes two genome duplication events (Figure 2). Previously overlooked gene categories also demonstrated a bias for gene transposition in the 10 to 72 MYA epoch (Figure 4; see Supplemental Table 1 and Supplemental Table 2 online). These include the B3-domain-containing genes, Locus Control Region (LCR)/self-incompatibility protein-related genes, the CLE (for CLAVATA3/ESR-related) developmental genes, and the meprin and TRAF homology domain proteins associated with developmental and pathological processes. By contrast, only one family, the Cation/H+ Exchanger (CHX) family of ttransporter genes, appears to have undergone a transposition radiation prior to the 10 to 72 MYA epoch.
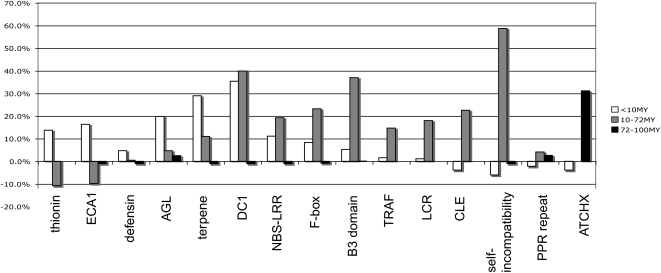
Figure 4.
The Epoch Specificities of the Major Gene Families That Tend to Transpose in the Rosids.
This chart describes the percentage over or under (y axis) expected incidence of transposition for each gene family based on the data from Supplemental Table 2. Expect values are based on frequency of transposition for each epoch per genome. Genes encoding ECA1s, thionins, and defensins transpose within the <10 MYA epoch, though their relative undetectability in poplar does not preclude their having transposed in earlier epochs. Genes encoding AGL and terpene synthases transposed primarily in the <10 MYA epoch, but some transposition occurred in the 10 to 72 MYA epoch. DC1 genes transposed almost equally within the <10 and 10 to 72 MYA epochs. NBS-LRRs and F-box genes mostly transposed in the 10 to 72 MYA epoch but also transposed more recently. Genes encoding B3, self-incompatibility, CLE, meprin, TRAF, and LCR proteins transposed exclusively within the 10 to 72 MYA epoch. PPR genes transposed in both the 10 to 72 and 72 to 100 MYA epoch. ATCHX genes transposed almost exclusively in the 72 to 100 MYA epoch.
Many Mobile Gene Families Have Primarily Transposed during or Shortly after the Brassicales WGD Events
As noted, many of the transposition events occurred in the 10 to 72 MYA epoch, which is the epoch in which both the α- (~25 MYA) and the earlier β- (~50 MYA) WGD events (Jiao et al., 2011) had taken place in the Brassicales (Figure 2). We asked if we could more closely pinpoint the times in which these transpositions occurred. By acquiring the (Anisimova and Gascuel, 2006) gene IDs and descriptions in grape (Jaillon et al., 2007), poplar (Tuskan et al., 2006), papaya (Ming et al., 2008), and A. lyrata (Hu et al., 2011) from the sequenced genome database Phytozome (http://www.phytozome.net/), we looked at all the known, annotated genes in each of these species for the CHX, CLE, TRAF, B3, and LCR families of genes to ask how many genes there were in each family per species. From this information, we can deduce more precisely when the transposition events took place in the A. thaliana lineage (Figure 5; see Supplemental Table 3 online). We found that the 10 to 72 MYA genes (B3, TRAF, and LCR) expanded specifically in the A. thaliana lineage sometime after the A. thaliana ancestor diverged from the papaya ancestor but before A. lyrata diverged from A. thaliana, as these gene families had fewer representatives in papaya but contained many more in the A. thaliana species, which were primarily syntenic between A. lyrata and A. thaliana, but not syntenic in papaya (see Supplemental Table 3 online). This supports the hypothesis that these gene transposition events occurred during or soon after the WGD events in the Brassicales. When we examined the CHX genes, known to have expanded in eudicots after the eudicots had diverged from the monocots (Sze et al., 2004), we found that this gene family had expanded after the grape ancestor diverged from the rosids but not since (Figure 5; see Supplemental Table 2 online). Because this discrepancy might have been accounted for by gene movement after the poplar genome duplication event that took place after the A. thaliana lineage diverged from the poplar ancestor (represented in Figure 1), we examined the peach genome (also within the rosid 1 subclade) (http://www.phytozome.net/dataUsagePolicy.php?org=Org_Ppersica) for the presence of these transposed CHX genes and found they gave us a similar result regarding the number of genes in the A. thaliana lineage that had transposed in the 72 to 100 MYA epoch. In comparing the A. thaliana and poplar CHX protein sequences phylogenetically (see Methods), we find that the poplar genes not syntenic in grape (as deduced by SynMap; see Methods) and the nonsyntenic and transposed A. thaliana CHX genes tend to cluster (see Supplemental Figure 2 and Supplemental Data Set 1 online), suggesting that the similarity of these genes in different species are due to a single expansion event sometime before the poplar and A. thaliana ancestors diverged. Conversely, the nonsyntenic and transposed genes in the A. thaliana B3-domain family do not cluster around any poplar B3-domain genes; rather, the nonsyntenous poplar B3-domain genes tend to cluster with each other, and syntenous poplar B3-domain genes tended to cluster with syntenous A. thaliana genes (see Supplemental Figure 3 and Supplemental Data Set 2 online), as would be expected from a radiation that occurred after the poplar ancestor diverged from the A. thaliana lineage.
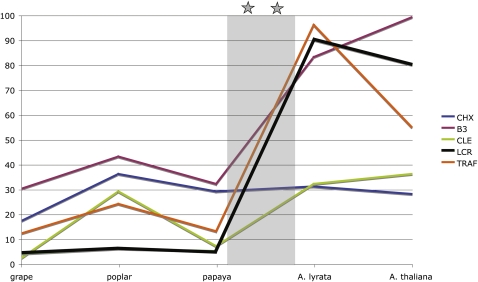
Figure 5.
Gene Expansion in the 10 to 72 MYA Epoch Occurred during or after the WGD Events in A. thaliana.
The increase in genes for most of these gene families occurred after the papaya ancestor diverged from the A. thaliana lineage but before A. lyrata diverged from A. thaliana, sometime during one or both of the WGDs (represented by the shaded area and the two stars). The exception is the CHX family of genes, which shows an increase in numbers of genes in poplar but not after. Most of these new genes in the A. thaliana lineage were not syntenic with papaya (see Supplemental Table 4 online).
Subclasses of the B3 and CHX Genes Tended to Transpose Relative to Other Subclasses
We asked if, within each epoch, certain subclasses of specific gene families had transposed and whether they were related to a particular function. In this study, we examined the B3-domain family of transcription factors (10 to 72 MYA) and the CHX family of transporters (72 to 100 MYA). These two families were chosen because their mobility had not been previously examined, they are overrepresented for transposition within their particular epoch, and they are well characterized. Table 4 describes the types of genes within each family that tend to transpose. Of the 99 B3 genes in our study, two major types were represented: The B3 genes associated with auxin response factor domain proteins and those associated with REM domain proteins, which are thought to be involved in floral organ development (Franco-Zorrilla et al., 2002; Swaminathan et al., 2008). Based on described functions for these genes, we found that a significant proportion (31/51) of transposed genes were of the REM subclass in comparison to the number of syntenic REM genes (7/51). For the CHX genes, we considered the patterns of tissue-specific expression as described by Sze et al. (2004) and Song et al. (2009). ATCHX genes code for cation:anion antiporters, most members of which are expressed in the male gametophyte in A. thaliana and are hypothesized to be involved in pollen tube growth (Sze et al., 2004; Song et al., 2009). Nine of the 10 transposed genes for which there is expression data are strongly expressed in pollen (Sze et al., 2004) (Table 4; see Supplemental Data Set 3 online). Additionally, the transposed pollen-associated CHX genes tended to cluster in phylogenetic tests, as noted by Sze et al. (2004).
Table 4.
The Subclass Specificity of Gene Transposition within Three Representative Gene Families
| Gene Family | Total | Syntenic | Nonsyntenic | Transposed |
| B3 | 91 | 34 | 14 | 43 |
| ARF | 21 | 11 | 1 | 9 |
| REM | 51 | 7 | 12 | 32 |
| Other | 19 | 16 | 1 | 2 |
| ATCHX | 28 | 7 | 11 | 10 |
| Pollen | 21 | 3 | 9 | 10 |
| Roots | 8 | 5 | 1 | 2 |
| Leaves | 11 | 5 | 3 | 3 |
Shown are the different classes within each gene family and the number of genes total, the number of syntenic genes, the number of nonsyntenic genes, and the number of transposed genes for each class for which there was available data. In B3 genes, REM-type genes disproportionately transpose (Swaminathan et al., 2008); for ATCHX genes, all transposed genes are associated with strong pollen expression. Most syntenic genes are associated with leaf and root expression (Sze et al., 2004).
These results are preliminary; our Arabidopsis Gene Positional History resource should facilitate continuing, improved positional histories. The goal of positional history of plant genes is a work in progress. The sequenced genomes we use as outgroups are being revised and updated now, and many new eudicot outgroup genome sequences are in progress.
DISCUSSION
The ever-increasing number of sequenced genomes made available to the scientific community has permitted us to trace the positional history of each A. thaliana gene as it and its descendents manifest in rosid genomes. By comparing the location of each A. thaliana gene to the outgroups A. lyrata, papaya, poplar, and grape (each placed further away from A. thaliana in evolutionary time), we have been able to deduce which genes have remained syntenic throughout the rosids versus those genes that have transposed or are otherwise nonsyntenic or are positioned within rearrangement-prone chromosomal regions. This resource is available in a searchable public database at http://biocon.berkeley.edu/athaliana. We intend to keep this website updated as new, relevant genomes become publicly available. Also, all data as of November, 2011 have been uploaded to the Dryad database (http://datadryad.org/); see accession numbers below.
Several characteristics can be used to predict a gene’s propensity for mobility. One is gene size, and another is the number and complexity of a gene’s putative regulatory regions. We demonstrated that transposed genes tend to have a smaller gene space than syntenic genes. We have also shown that transposed genes only rarely have CNS associated with them. Based on the small functional gene space and the lack of CNSs associated with mobile genes, we can infer that genes that tend to transpose are those that are not under the usual forms of regulation and where some function is not absolutely dependent upon regulatory sites far away from the transcriptional unit. This is further supported by studies of retroposed genes, whose relocation generally gives rise to different expression patterns, although these types of mobile genes are less abundant than DNA-based transposed genes (Kaessmann et al., 2009). Transposed genes seem likely candidates for genes encoding novel function (neomorphs).
Given the propensity for mobile genes to be small, it is not surprising that many highly mobile gene families, such as defensins, thionins, LCR, and ECA1 genes, encode small, secreted, Cys-rich proteins. These genes also are observed to transpose exclusively in the <10 MYA epoch (Figure 6), with the exception of LCR genes, which transpose mostly within the 10 to 72 MYA epoch. As more and more ecotypes of A. thaliana are sequenced (Schneeberger et al., 2011), it will be interesting to study recent transposition frequency and copy number of these gene families.
Figure 6.
The Major Transposition Events Studied.
This figure places the transposition events per epoch at their appropriate points over evolutionary time. Most of the transposition events occurred after papaya diverged from the A. thaliana ancestor. This may be due to the genome duplication events giving rise to an increase in gene transposition generally.
It is possible that the epoch-specific mobility we observe among the above gene families is a function of small gene size and high birth-and-death rates; these genes are simply not detectable in poplar (see Supplemental Table 2 online). Consequently, differentiating between deletion and transposition in an earlier epoch is simply not possible experimentally. By contrast, most F-box and NBS-LRR genes, which are observed to have transposed within both the <10 and 10 to 72 MYA epochs (Figure 3) but not the 72 to 100 MYA epoch, are detectable within the poplar outgroup (see Supplemental Table 2 online), suggesting that they did not transpose in the interval 72 to 100 MYA. In comparison, B3, self-incompatibility, CLE, meprin, and TRAF homology domain genes have not transposed within the <10 MYA epoch, but, rather, transposed almost exclusively within the 10 to 72 MYA epoch. CLE, TRAF, and self-incompatibility genes are relatively small genes (<1000 bp), yet they have been detected within the poplar outgroup. Therefore, their mobility within the 10 to 72 MYA epoch suggests truly time-specific transposition events. Notably, these gene classes tend to transpose within the time frame of not one but two WGD events in the Brassicales (Figure 6). We hypothesize that WGD events and gene mobility may be correlated in plants; this is plausible, as recombination and other types of rearrangements are known to occur after plant genome duplication events (Gaeta and Pires, 2010). The lone, pre-WGD epoch-specific radiation is the CHX family of antiporter genes, whose transposition is relegated almost exclusively to the rosid I clade and suggests a role in pollen development and fitness in the later-evolving rosid species. It will be interesting to see if gene transposition is correlated with other WGDs in other species, such as soybean (Glycine max) and the newly sequenced Brassica rapa genome that had undergone a recent hexaploidy event (Wang et al., 2011).
Within epoch-specific transposed gene families, certain subfamilies of genes have transposed and others have not (Table 4). A subfamily of B3-domain genes, the REM genes, are overrepresented for mobility. CHX genes encode cation:proton antiporters that have undergone expansion within the eudicots; many of the CHX genes that are unique to the eudicots are those whose expression is localized to the male gametophyte (Sze et al., 2004). All transposed CHX genes showed strong pollen expression, while a few of them showed expression in roots and leaves. Interestingly, we observed that the majority of CHX genes are not syntenic. In fact, only seven of the 28 known CHX genes have been syntenic throughout the rosids.
Duplication of A. thaliana genes via retention following its most recent tetraploidy has produced cases of subfunctionalization (Haberer et al., 2004). In theory (Sémon and Wolfe, 2007), all duplicate genes are subject to diversification by the neutral processes of subfunctionalization or cis-acting gene component loss (fractionation) or by the acquisition of some new function requiring positive selection (neofunctionalization), perhaps as a consequence of a new chromosomal position. Our findings demonstrate that, within the A. thaliana lineage, gene expansion via transposition in certain families has happened at specific points in time, in particular during or after a WGD event. These findings lead one to ask if different gene families have transposed at wholly different times in different species (e.g., grape or rice) and, if so, how might these expansions correlate with fitness in a particular clade.
Our Arabidopsis Gene Positional History database provides a useful platform from which to study gene synteny throughout the eudicots. The data from all eudicot genomes tested are anchored on orthologous genes or regions where such orthologous genes are expected, and a graphic of the BLASTZ alignment output of all orthologus outgroups is available via a link to GEvo (http://synteny.cnr.berkeley.edu/CoGe/GEvo.pl), the alignment viewer within the CoGe toolbox (Lyons et al., 2008). This link permits visual proofing of our results and facilitates on-the-fly research among orthologs. Our database is directly linked from TAIR, so that plant geneticists have easy access to it. We hope that our results serve as a foundation on which more profound hypotheses might be put to the test and that our Arabidopsis Gene Positional History Web tool might facilitate such research.
METHODS
The Positional History Whole Annotated Genome Pipeline (Positional History Pipeline)
A pipeline is a series of scripts that, given the input of annotated whole genomes, automatically labels genes that would provide useful results with minimal manual intervention. A 40-gene window was centered on every query Arabidopsis thaliana (TAIR9) gene to check for a syntenic region in each target genome. (B)LASTZ (default parameters: word size 8, gap start penalty 400, gap extend penalty 30, score threshold 300) was used to define anchors and required that the syntenic region to have at least four collinear anchors (out of 40 possible anchors). The nearest anchors on both sides of genes were identified to define a tight syntenic location.
We implemented a careful labeling pipeline for the positional history of genes described below and illustrated in Figure 3. Each query gene is categorized for each outgroup, based on the flank anchors and more sensitive search on the tight interval as follows: gene match in the interval, syntenic (S), not syntenic, have both flankers (F), or one flanker (G). For genes labeled as F, further validation is as follows: BLAST matches (e.g., to noncoding sequences) in the interval between flankers (FB) and assembly gaps (Ns) in the interval between flankers (FN). Because the region between flankers is unsequenced (FN), we cannot determine whether or not there is a gene in that space that could be syntenic with the query gene. To err on the side of caution, we denote it as FN instead of F. For Arabidopsis lyrata, a second test was done for S genes to ensure they weren’t truly F but had been denoted as S simply because they were within the 40-gene window. This was necessary because the A. thaliana and A. lyrata genomes are so similar. However, this test was not required for papaya (Carica papaya), poplar (Populus trichocarpa), and grape (Vitis vinifera). Poplar has a lineage-specific genome duplication (Tuskan et al., 2006), so, in most cases, each A. thaliana gene has two orthologs. Sometimes the codes can be conflicting in the case of poplar. For example, one copy is syntenic in position (S) and another has transposed (F). In this case, our rule is to report the gene in poplar as syntenic (i.e., S takes priority over F among homoeologs). This is a parsimonious explanation because S (synteny) is clearly the ancestral state; a change from S to F is much more probable than changes from F to S. The priorities for our codes are S, F, FB, FN, and G, in this order. This pipeline is available in Python format at https://github.com/tanghaibao/positional-history.
Rules for Being Labeled a Transposed Gene in Three Distinct Epochs
A transposed gene is defined as one that is not syntenic in any outgroup originating at a time older than the taxon carrying the gene in question. Such a gene is denoted F in at least one of the remaining outgroups. For instance, a gene would be considered as having transposed within the <10 MYA epoch if it is not syntenic (S) in any of the outgroups and is F in at least one of the outgroups. The criteria for a gene to be considered as having transposed within the 10 to 72 MYA epoch are that it must be syntenic (S) in A. lyrata and F in at least one of the other outgroups and not syntenic in the remainder. A gene is considered as having transposed within the 72 to 100 MYA epoch if it is syntenic (S) in A. lyrata and papaya and F in either poplar or grape. Of statistical necessity, our criteria for transposed genes must include genes that are FB or FN in some of the outgroups. However, as FN represents missing sequence between flankers in the outgroup and as FB represents the presence of small segments of noncoding sequence corresponding to the query gene in the outgroup interval, neither of these situations can guarantee a perfect test for whether or not the query gene had been deleted or partially lost in the outgroup, rather than having transposed. While our calls of synteny are solid, our calls of transposed certainly contain error that, if it is not due to incomplete sequencing, can be improved by manual proofing. We include GEvo links to all expected syntenic positions in all outgroup genomes (see http://datadryad.org/ or http://biocon.berkeley.edu/athaliana) to facilitate manual research using the enhanced visualization software of the GEvo sequence alignment tool in the CoGe suite of comparative genomics tools, http://synteny.cnr.berkeley.edu/CoGe/.
Gene Detectability
To study the status of gene transposition during the <10, 10 to 72, and 72 to 100 MYA epochs, we performed BLAST on all putatively transposed genes in the <10 MYA epoch to A. lyrata and the 10 to 72 and 72 to 100 MYA epochs to poplar (TBLASTX for genes with CDSs and BLASTn for non-CDS genes, such as tRNA genes and RNA genes) using a loose cutoff of e-value 0.001 and a bit score of 45 (see Supplemental Table 1 online). To differentiate between a newly arisen gene and a gene that had actually transposed into its new location, we looked for a best hit in each outgroup in question for every putatively transposed A. thaliana gene, and those with a best hit not syntenic to the query gene are considered transposed. Those genes that did not have a best hit outside themselves in the representative genomes were discounted. When we compare putatively transposed A. thaliana genes to orthologous expected positions in outgroups that have anciently diverged from A. thaliana, such as poplar, some have no believable BLAST hits anywhere in the outgroup’s genome. Finding no hit between flankers in an outgroup may be the result of rapid divergence of that gene or high base substitution frequencies (drift), such that our algorithm cannot detect it. For instance, many genes, such as tRNAs, small nucleolar RNAs, and MIR genes, are denoted F, or putatively transposed, in our pipeline, but these genes have no best hit in the outgroup. In particular, MIR genes tend to be undetected because of high base substitution frequencies (Howell et al., 2007). Other gene families that tend to have short coding sequences, such as defensins, thionins, and other small, Cys-rich genes, are automatically suspected of being difficult to detect. Such genes and gene families cannot be proven to have transposed, but transposition can still be inferred if we can document recent transposition events between A. thaliana and A. lyrata.
Determining Transposed Genes for Gene Transposition per Epoch
Each gene in each epoch in this experiment had a believable BLAST hit within the representative outgroup. For instance, all genes that have transposed within the <10 MYA epoch had a believable blast hit in A. lyrata, and all transposed genes within the 10 to 72 MYA and the 72 to 100 MYA epochs had a believable blast hit in poplar. Note that few defensins, thionins, or other Cys-rich genes are represented within the 10 to 72 MYA epoch or before; this is only because no believable BLAST hit was found within the representative outgroup for these genes. This does not mean that transposition within these families did not take place.
Phylogenetic Analysis of the B3-Domain and CHX Genes
We deduced which B3-domain genes in poplar were syntenous by comparing the poplar genome to grape using our program SynMap, which generates a whole-genome dot plot analysis. Once we obtained this data, we aligned the B3 protein sequence in both poplar and A. thaliana and the CHX genes, respectively, using the alignment program MUSCLE (Edgar, 2004) using default parameters, then constructed our phylogenetic tree using the program Phylogeny.fr (http://www.phylogeny.fr; Dereeper et al., 2008). Branch lengths were calculated using the criteria described by Guindon and Gascuel (2003) and Anisimova and Gascuel (2006).
Accession Numbers
Sequence data from this article have been deposited in the Dryad Repository (http://dx.doi.org/10.5061/dryad.275kv81m).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Some Transposed Genes Retain CNSs.
Supplemental Figure 2. The CHX Family of Genes Expanded before the Divergence of the Poplar Ancestor and the A. thaliana Lineage.
Supplemental Figure 3. The B3-Domain Family of Genes Underwent a Transposition Expansion after the Poplar Ancestor Diverged from the A. thaliana Lineage.
Supplemental Table 1. Total Transposed Genes per Family versus Number of Transposed Genes per Family with a Best Hit in the Outgroup.
Supplemental Table 2. Genes That Transposed per Epoch.
Supplemental Table 3. Genes That Transposed in the 10 to 72 MYA Epoch Did So during or after the Two A. thaliana Genome Duplication Events.
Supplemental Data Set 1. Text File of PHYLIP Alignment of CHX Genes.
Supplemental Data Set 2. Text File of PHYLIP Alignment of B3-Domain Genes.
Supplemental Data Set 3. Subclasses within Gene Families Are Associated with Epoch-Specific Transposition.
Acknowledgments
This work was funded by National Science Foundation Grant MCB0820821 to M.F., an Arabidopsis 2010 grant (http://www.nsf.gov/). The funder had no role in study design, data collection, and analysis, decision to publish, or preparation of the manuscript. We thank Yuheng Huang of Project SEED for assisting with the proofing of our data sets. Sequence data for the peach genome were produced by the U.S. Department of Energy Joint Genome Institute (http://www.jgi.doe.gov/) in collaboration with the user community.
AUTHOR CONTRIBUTIONS
M.R.W. and M.F. designed the research. H.T. contributed computational tools, designed the positional history whole annotated genome pipeline, and designed and implemented the Arabidopsis Gene Positional History database. M.R.W. performed the research, analyzed the data, and wrote the article.
References
- Anisimova M., Gascuel O. (2006). Approximate likelihood-ratio test for branches: A fast, accurate, and powerful alternative. Syst. Biol. 55: 539–552 [DOI] [PubMed] [Google Scholar]
- Birchler J.A., Veitia R.A. (2007). The gene balance hypothesis: From classical genetics to modern genomics. Plant Cell 19: 395–402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanc G., Wolfe K.H. (2004). Widespread paleopolyploidy in model plant species inferred from age distributions of duplicate genes. Plant Cell 16: 1667–1678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowers J.E., Chapman B.A., Rong J., Paterson A.H. (2003). Unravelling angiosperm genome evolution by phylogenetic analysis of chromosomal duplication events. Nature 422: 433–438 [DOI] [PubMed] [Google Scholar]
- Cannon S.B., Mitra A., Baumgarten A., Young N.D., May G. (2004). The roles of segmental and tandem gene duplication in the evolution of large gene families in Arabidopsis thaliana. BMC Plant Biol. 14: 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman B.A., Bowers J.E., Feltus F.A., Paterson A.H. (2006). Buffering of crucial functions by paleologous duplicated genes may contribute cyclicality to angiosperm genome duplication. Proc. Natl. Acad. Sci. USA 103: 2730–2735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coghlan A., Eichler E.E., Oliver S.G., Paterson A.H., Stein L. (2005). Chromosome evolution in eukaryotes: A multi-kingdom perspective. Trends Genet. 21: 673–682 [DOI] [PubMed] [Google Scholar]
- Conant G.C., Wolfe K.H. (2008). Probabilistic cross-species inference of orthologous genomic regions created by whole-genome duplication in yeast. Genetics 179: 1681–1692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dereeper A., Guignon V., Blanc G., Audic S., Buffet S., Chevenet F., Dufayard J.F., Guindon S., Lefort V., Lescot M., Claverie J.M., Gascuel O. (2008). Phylogeny.fr: Robust phylogenetic analysis for the non-specialist. Nucleic Acids Res. 36(Web Server issue): W465–W469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duarte J., Wall P.K., Edger P.P., Landherr L.L., Ma H., Pires J.C., Leebens-Mack J., dePamphilis C.W. (2010). Identification of shared single copy nuclear genes in Arabidopsis, Populus, Vitis and Oryza and their phylogenetic utility across various taxonomic levels. BMC Evol. Biol. 10: 61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar R.C. (2004). MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32: 1792–1797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fawcett J.A., Maere S., Van de Peer Y. (2009). Plants with double genomes might have had a better chance to survive the Cretaceous-Tertiary extinction event. Proc. Natl. Acad. Sci. USA 106: 5737–5742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Force A., Lynch M., Pickett F.B., Amores A., Yan Y.L., Postlethwait J. (1999). Preservation of duplicate genes by complementary, degenerative mutations. Genetics 151: 1531–1545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franco-Zorrilla J.M., Cubas P., Jarillo J.A., Fernández-Calvín B., Salinas J., Martínez-Zapater J.M. (2002). AtREM1, a member of a new family of B3 domain-containing genes, is preferentially expressed in reproductive meristems. Plant Physiol. 128: 418–427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freeling M., Lyons E., Pedersen B., Alam M., Ming R., Lisch D. (2008). Many or most genes in Arabidopsis transposed after the origin of the order Brassicales. Genome Res. 18: 1924–1937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freeling M., Subramaniam S. (2009). Conserved noncoding sequences (CNSs) in higher plants. Curr. Opin. Plant Biol. 12: 126–132 [DOI] [PubMed] [Google Scholar]
- Gaeta R.T., Pires J.C. (2010). Homoeologous recombination in allopolyploids: The polyploid ratchet. New Phytol. 186: 18–28 [DOI] [PubMed] [Google Scholar]
- Guindon S., Gascuel O. (2003). A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst. Biol. 52: 696–704 [DOI] [PubMed] [Google Scholar]
- Haberer G., Hindemitt T., Meyers B.C., Mayer K.F. (2004). Transcriptional similarities, dissimilarities, and conservation of cis-elements in duplicated genes of Arabidopsis. Plant Physiol. 136: 3009–3022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howell M.D., Fahlgren N., Chapman E.J., Cumbie J.S., Sullivan C.M., Givan S.A., Kasschau K.D., Carrington J.C. (2007). Genome-wide analysis of the RNA-DEPENDENT RNA POLYMERASE6/DICER-LIKE4 pathway in Arabidopsis reveals dependency on miRNA- and tasiRNA-directed targeting. Plant Cell 19: 926–942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu T.T., et al. (2011). The Arabidopsis lyrata genome sequence and the basis of rapid genome size change. Nat. Genet. 43: 476–481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inada D.C., Bashir A., Lee C., Thomas B.C., Ko C., Goff S.A., Freeling M. (2003). Conserved noncoding sequences in the grasses. Genome Res. 13: 2030–2041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaillon O., et al. ; French-Italian Public Consortium for Grapevine Genome Characterization (2007). The grapevine genome sequence suggests ancestral hexaploidization in major angiosperm phyla. Nature 449: 463–467 [DOI] [PubMed] [Google Scholar]
- Jiao Y., et al. (2011). Ancestral polyploidy in seed plants and angiosperms. Nature 473: 97–100 [DOI] [PubMed] [Google Scholar]
- Kaessmann H., Vinckenbosch N., Long M. (2009). RNA-based gene duplication: Mechanistic and evolutionary insights. Nat. Rev. Genet. 10: 19–31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leister D. (2004). Tandem and segmental gene duplication and recombination in the evolution of plant disease resistance gene. Trends Genet. 20: 116–122 [DOI] [PubMed] [Google Scholar]
- Lyons E., Pedersen B., Kane J., Alam M., Ming R., Tang H., Wang X., Bowers J., Paterson A., Lisch D., Freeling M. (2008). Finding and comparing syntenic regions among Arabidopsis and the outgroups papaya, poplar, and grape: CoGe with rosids. Plant Physiol. 148: 1772–1781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maere S., De Bodt S., Raes J., Casneuf T., Van Montagu M., Kuiper M., Van de Peer Y. (2005). Modeling gene and genome duplications in eukaryotes. Proc. Natl. Acad. Sci. USA 102: 5454–5459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ming R., et al. (2008). The draft genome of the transgenic tropical fruit tree papaya (Carica papaya Linnaeus). Nature 452: 991–996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paterson A.H., Freeling M., Tang H., Wang X. (2010). Insights from the comparison of plant genome sequences. Annu. Rev. Plant Biol. 61: 349–372 [DOI] [PubMed] [Google Scholar]
- Pecinka A., Fang W., Rehmsmeier M., Levy A.A., Mittelsten Scheid O. (2011). Polyploidization increases meiotic recombination frequency in Arabidopsis. BMC Biol. 9: 24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Proost S., Van Bel M., Sterck L., Billiau K., Van Parys T., Van de Peer Y., Vandepoele K. (2009). PLAZA: A comparative genomics resource to study gene and genome evolution in plants. Plant Cell 21: 3718–3731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rizzon C., Ponger L., Gaut B.S. (2006). Striking similarities in the genomic distribution of tandemly arrayed genes in Arabidopsis and rice. PLoS Comput. Biol. 2: e115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneeberger K., et al. (2011). Reference-guided assembly of four diverse Arabidopsis thaliana genomes. Proc. Natl. Acad. Sci. USA 108: 10249–10254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sémon M., Wolfe K.H. (2007). Consequences of genome duplication. Curr. Opin. Genet. Dev. 17: 505–512 [DOI] [PubMed] [Google Scholar]
- Song L.F., Zou J.J., Zhang W.Z., Wu W.H., Wang Y. (2009). Ion transporters involved in pollen germination and pollen tube tip-growth. Plant Signal. Behav. 4: 1193–1195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swaminathan K., Peterson K., Jack T. (2008). The plant B3 superfamily. Trends Plant Sci. 13: 647–655 [DOI] [PubMed] [Google Scholar]
- Sze H., Padmanaban S., Cellier F., Honys D., Cheng N.H., Bock K.W., Conéjéro G., Li X., Twell D., Ward J.M., Hirschi K.D. (2004). Expression patterns of a novel AtCHX gene family highlight potential roles in osmotic adjustment and K+ homeostasis in pollen development. Plant Physiol. 136: 2532–2547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas B.C., Pedersen B., Freeling M. (2006). Following tetraploidy in an Arabidopsis ancestor, genes were removed preferentially from one homeolog leaving clusters enriched in dose-sensitive genes. Genome Res. 16: 934–946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas B.C., Rapaka L., Lyons E., Pedersen B., Freeling M. (2007). Intragenomic conserved noncoding sequences in Arabidopsis. Proc. Natl. Acad. Sci. USA 104: 3348–3353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuskan G.A., et al. (2006). The genome of black cottonwood, Populus trichocarpa (Torr. & Gray). Science 313: 1596–1604 [DOI] [PubMed] [Google Scholar]
- Van de Peer Y. (2011). A mystery unveiled. Genome Biol. 12: 113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veitia R.A. (2002). Exploring the etiology of haploinsufficiency. Bioessays 24: 175–184 [DOI] [PubMed] [Google Scholar]
- Veitia R.A. (2004). Gene dosage balance in cellular pathways: Implications for dominance and gene duplicability. Genetics 168: 569–574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H., Moore M.J., Soltis P.S., Bell C.D., Brockington S.F., Alexandre R., Davis C.C., Latvis M., Manchester S.R., Soltis D.E. (2009). Rosid radiation and the rapid rise of angiosperm-dominated forests. Proc. Natl. Acad. Sci. USA 106: 3853–3858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W., et al. (2006). High rate of chimeric gene origination by retroposition in plant genomes. Plant Cell 18: 1791–1802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., et al. (2011). The genome of the mesopolyploid crop species Brassica rapa. Nat. Genet. 43: 1035–1039 [DOI] [PubMed] [Google Scholar]
- Wicker T., Buchmann J.P., Keller B. (2010). Patching gaps in plant genomes results in gene movement and erosion of colinearity. Genome Res. 20: 1229–1237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodhouse M.R., Pedersen B., Freeling M. (2010). Transposed genes in Arabidopsis are often associated with flanking repeats. PLoS Genet. 6: e1000949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang S.A., Arguello J.R., Li X., Ding Y., Zhou Q., Chen Y., Zhang Y., Zhao R.P., Brunet F., Peng L.X., Long M.Y., Wang W. (2008). Repetitive element-mediated recombination as a mechanism for new gene origination in Drosophila. PLoS Genet. 4: e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Wu Y., Liu Y., Han B. (2005). Computational identification of 69 retroposons in Arabidopsis. Plant Physiol. 138: 935–948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Z., Zhang Y., Long M. (2009). Extensive structural renovation of retrogenes in the evolution of the Populus genome. Plant Physiol. 151: 1943–1951 [DOI] [PMC free article] [PubMed] [Google Scholar]