We used maize translocations to identify rough endosperm3 (rgh3) as a locus required in the endosperm to promote embryo development. The rgh3 mutant impairs or delays endosperm cell differentiation and encodes a hypomorphic allele of URP, a core RNA splicing factor. A subset of alternatively spliced transcripts shows differences in splicing patterns in wild-type and rgh3 tissues.
Abstract
Endosperm and embryo development are coordinated via epigenetic regulation and signaling between these tissues. In maize (Zea mays), the endosperm–embryo signals are not known, but endosperm cellularization is a key event for embryos to form shoots and roots. We screened seed mutants for nonautonomous functions in endosperm and embryo development with genetically nonconcordant seeds and identified the recessive mutant rough endosperm3 (rgh3). The wild-type Rgh3 allele is required in the endosperm for embryos to develop and has an autonomous role in embryo and seedling development. Endosperm cell differentiation is defective in rgh3. Results from endosperm cell culture indicate that rgh3 mutants remain in a proliferative state through mid-seed development. Rgh3 encodes the maize U2AF35 Related Protein (URP), an RNA splicing factor involved in both U2 and U12 splicing. The Rgh3 allele produces at least 19 alternative splice variants with only one isoform encoding a full-length ortholog to URP. The full-length RGH3α isoform localizes to the nucleolus and displays a speckled pattern within the nucleoplasm, and RGH3α colocalizes with U2AF65. A survey of alternatively spliced transcripts found that, in the rgh3 mutant, a fraction of noncanonical splicing events are altered. Our findings suggest that differentiation of maize endosperm cell types is necessary for embryos to develop. The molecular cloning of Rgh3 suggests that alternative RNA splicing is needed for cell differentiation, development, and plant viability.
INTRODUCTION
Angiosperm seeds develop an embryo and endosperm from double fertilization of the egg and central cell of the megagametophyte (De Smet et al., 2010; Linkies et al., 2010). The endosperm is a nutritive tissue for the embryo, and in grasses, it is a persistent storage tissue that is used upon germination (Sabelli and Larkins, 2009). The evolutionary conservation of the endosperm in angiosperms and the coordinated development of endosperm and embryo suggest that these tissues have interacting developmental pathways. In Arabidopsis thaliana, some aspects of embryo development are controlled nonautonomously. Endosperm-to-embryo signaling is supported by two endosperm-specific genes, ABNORMAL LEAF SHAPE1 (ALE1) and RETARDED GROWTH OF EMBRYO1/ZHOUPI (RGE1), which are required for embryos to develop a cuticle and separate from the endosperm (Tanaka et al., 2001; Kondou et al., 2008; Yang et al., 2008). Embryo patterning is not affected in ale1 and rge1 mutants, but ablation of the endosperm with tissue-specific expression of diphtheria toxin causes embryo patterning defects and seed abortion (Weijers et al., 2003).
Early endosperm development is controlled through imprinted genes and genome-wide epigenetic modification of maternal and paternal genomes (Berger and Chaudhury, 2009; Springer, 2009). Maternal and paternal genome dosage is important for seed development. In Arabidopsis interploidy crosses, increased paternal genome copies lead to greater endosperm growth and larger final seed size (Scott et al., 1998). The haiku2 and miniseed3 mutations reduce endosperm growth and seed size by inducing precocious endosperm cellularization (Luo et al., 2005). Thus, endosperm–embryo growth interactions can be uncoupled from interactions influencing patterning. In maize (Zea mays), epigenetic control of the endosperm has a stronger influence on embryo viability with an endosperm requirement for a 2:1 ratio of maternal to paternal genomes for both tissues to develop normally (Lin, 1984).
Genetically nonconcordant seeds, in which the endosperm and embryo have different genotypes, provide a powerful tool to identify nonautonomous genes underlying endosperm–embryo interactions. In maize, nonconcordant endosperm and embryo genotypes can be generated with B-A translocations between the supernumerary B chromosome and standard A chromosomes (Beckett, 1994a). B centromeres undergo nondisjunction during the second pollen mitosis, producing gametophytes with mixed sperm cells (Roman, 1947). When crossed as a male, B-A translocations generate seeds carrying a deletion of the paternal A chromosome arm in either the embryo or endosperm. B-A translocation crosses will uncover (reveal) recessive defective kernel (dek) mutant phenotypes when the dek maps to the translocated segment. These nonconcordant uncovered kernels can determine if the dek has a tissue autonomous function in growth or development (Sheridan and Neuffer, 1982; Neuffer et al., 1986; Chang and Neuffer, 1994; Scanlon and Myers, 1998; Costa et al., 2003).
We surveyed published primary data for B-A translocation phenotypes and found the vast majority of dek mutants show developmental autonomy (i.e., developmental patterning of the tissue is in accordance to the genotype) (see Supplemental Table 1 online). In these studies, two types of nonautonomous developmental interactions were observed. First, dek2, dek12, dek13, or dek32 mutant embryos are rescued by wild-type endosperm (Chang and Neuffer, 1994). Second, globby1 (glo1), discolored1 (dsc1), or dek26 mutant endosperm inhibits wild-type embryo development (Chang and Neuffer, 1994; Scanlon and Myers, 1998; Costa et al., 2003). Both glo1 and dsc1 cause defects in endosperm cellularization, suggesting a cellular endosperm is needed for embryo development in maize.
Here, we report a maize mutant, rough endosperm3 (rgh3), with a nonautonomous endosperm function that promotes embryo development. We show Rgh3 is required in the endosperm to switch from cellular proliferation to differentiation. These data suggest a unique mechanism by which endosperm cell differentiation influences embryo patterning and development. Molecular cloning of Rgh3 implicates alternative RNA splicing as an essential process to endosperm cell differentiation as well as embryo and plant development.
RESULTS
rgh Mutants Show Endosperm–Embryo Developmental Interactions
To identify seed mutants affecting endosperm–embryo developmental interactions, we crossed B-A translocation stocks for chromosome arms 5L, 6L, and 9L onto 140 rgh mutants. These mutants have a characteristic rough surface to the endosperm that is also termed etched or pitted (Scanlon et al., 1994; Neuffer et al., 1997). We also completed crosses for a small number of rgh mutants using 19 translocations that uncover all chromosome arms except 8S. These experiments uncovered 27 isolates distributed over five chromosome arms (see Supplemental Table 2 online). The uncovered rgh kernels were analyzed with hand sections to identify isolates with endosperm–embryo interactions (Figure 1). Mutants were considered not to have an interaction when they showed autonomous defective endosperm or embryo phenotypes in the two classes of nonconcordant kernels (Figure 1B). An endosperm–embryo interaction was inferred when nonconcordant kernels with both defective endosperm and defective embryo tissues were present. Among the uncovered rgh isolates, 40% (11/27) showed nonautonomous development (see Supplemental Table 2 online). By contrast, we estimate <20% (7/42) of defective kernels described in the literature have nonautonomous endosperm and embryo phenotypes (see Supplemental Table 1 online). These data suggest rgh mutants are enriched for loci required in endosperm–embryo developmental interactions.
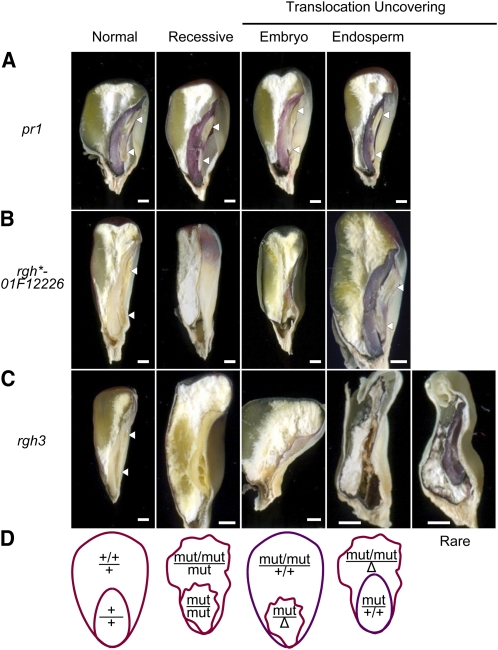
Figure 1.
B-A Translocation Crosses Reveal a Nonautonomous Role for rgh3 in Seed Development.
(A) to (C) Sagittal hand sections comparing recessive mutant and TB-5La translocation-uncovering kernel phenotypes. Arrowheads indicate normal embryo shoots and roots. The translocation is indicated by purple aleurone or embryo tissues in the uncovering panels. Bars = 1 mm.
(A) pr1 anthocyanin marker.
(B) rgh*-01F12226 in a pr1/pr1 background.
(C) rgh3 in a pr1/pr1 background.
(D) Schematic of expected anthocyanin and developmental phenotypes of autonomous endosperm and embryo defective mutants. Δ represents deletion of the long arm of chromosome 5.
We also inferred the direction of the endosperm–embryo interaction from the phenotypes of nonconcordant kernel classes in uncovering crosses. For example, uncovering crosses with the rgh3 mutant produced an embryo defective class with a normal endosperm as well as a defective kernel class with both endosperm and embryo defects. The embryo-defective class suggests wild-type endosperm can develop in conjunction with a mutant embryo. Thus, the defective endosperm and embryo phenotype suggests rgh3 endosperm negatively impacts wild-type embryo development. To confirm this inference, we developed marked nonconcordant kernels by crossing rgh3 with the linked pr1 marker. The B-A translocation carries the dominant Pr1 allele, which confers purple color to the aleurone or embryo (Figure 1A). Figure 1B shows marked uncovering crosses for an autonomous isolate, rgh*-01F12226, in which mutant, uncovered tissues are red. In rgh3 uncovering crosses, the embryo-defective kernel class has a purple aleurone, indicating the endosperm has the translocation and the embryo was uncovered (Figure 1C). In the nonautonomous kernels, the aleurone is red, indicating the endosperm was uncovered. Rarely (8/82 kernels sectioned), the nonautonomous kernels would develop sufficient embryo tissue to score anthocyanin color, and these embryos are purple, confirming a wild-type embryo with two copies of the normal Rgh3 allele fails to develop in conjunction with rgh3 mutant endosperm.
Inheritance of rgh3
Imprinted loci have multiple roles in seed development, and some imprinted genes have biparental expression at later stages of seed development (Danilevskaya et al., 2003; Gutiérrez-Marcos et al., 2004; Raissig et al., 2011). It is formally possible to explain the rgh3 seed uncovering phenotypes if Rgh3 is an imprinted locus required specifically for early embryo development. Based on this hypothesis, we would expect rgh3 to show transmission bias due to the parent of origin. In self-pollinations of rgh3/+ plants, we observe a 3:1 ratio of normal to rgh3 endosperm phenotypes, suggesting that rgh3 is a recessive allele for endosperm defects (see Supplemental Table 3 online). We did not observe a kernel class with a normal endosperm and a defective embryo in these self-pollinations. To test male and female transmission, we made reciprocal crosses with the reference UniformMu inbred W22 and self-pollinated the F1 progeny to score ears segregating for rgh3. We found ratios close to 1:1 of normal to rgh3 segregating ears regardless of the direction of the cross (see Supplemental Table 3 online). These results indicate rgh3 transmits equally through both gametes and is unlikely to have a parent-of-origin effect on embryo development. Combined with the marked B-A translocation phenotypes, we conclude that rgh3 mutant endosperm has a negative effect on wild-type embryo development. To understand the developmental basis of this interaction, we characterized rgh3 in more detail with a focus on the mutant endosperm.
Seed and Seedling Phenotype of rgh3
Mature rgh3 seeds show a rough, etched, or pitted endosperm surface as well as a reduced seed size compared with normal siblings (Figure 2A). Within individual segregating ears, rgh3 seeds show variable severity, ranging from almost normal grain fill to a nearly empty pericarp phenotype (Figure 2B). rgh3 kernels are reduced in weight but have an overlapping distribution with normal siblings (Figure 2C). The severity of rgh3 embryo defects is correlated to total seed weight with the least severe mutants developing near normal embryos. The most severe class of rgh3 mutants have similar endosperm and embryo phenotypes to rgh3 uncoverings (cf. Figures 1C to 2B), suggesting that a hypoploid endosperm or embryo has a more severe phenotype than the recessive mutant.
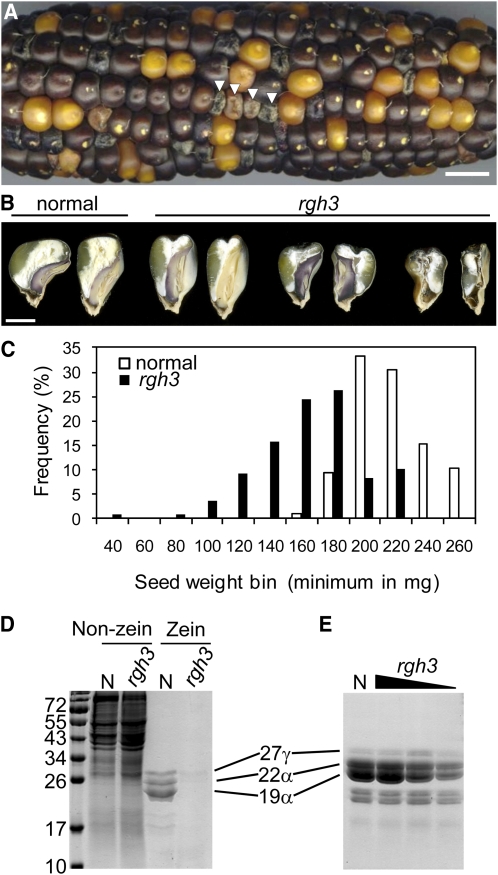
Figure 2.
Range of rgh3 Kernel Phenotypes.
(A) Self-pollinated ear segregating for rgh3. Arrowheads indicate rgh3 kernels. Bar = 1 cm.
(B) Sagittal hand sections of mature kernels from a single ear. Bar = 3 mm.
(C) Histogram of seed weight comparing normal and rgh3 seed from a single self-pollinated rgh3/+ ear.
(D) SDS-PAGE of kernel proteins showing 17-DAP extracts of non-zein and zein fractions.
(E) Zein fractions from mature kernels of normal and rgh3 mutants. Lanes 2 to 4 represent extracts from the following mutant weight classes: >200 mg, 140 to 200 mg, and <140 mg. All zein fractions were extracted from 20 μg total seed protein and loaded on an equal total seed protein basis.
[See online article for color version of this figure.]
The rgh3 seed phenotype becomes visible at 12 d after pollination (DAP), concomitant with visible differentiation of distinct endosperm cellular features (Lopes and Larkins, 1993). Accumulation of major seed storage carbohydrates and proteins occurs between 10 and 45 DAP (Kodrzycki et al., 1989). To determine if rgh3 impacts seed storage protein accumulation, zein and non-zein protein fractions were analyzed from 17-DAP kernels (Figure 2D). In comparison to normal siblings, rgh3 has reduced zein proteins relative to cellular proteins, suggesting rgh3 delays zein accumulation. In mature rgh3 kernels, zein accumulation is correlated to total seed weight with less severe kernels accumulating near normal levels (Figure 2E). Mature rgh3 kernel composition was determined from average weight mutants with standard analytical methods (see Supplemental Table 4 online). Only a slight reduction in kernel fat content was observed, which would be consistent with reduced embryo development of typical rgh3 kernels. These data suggest that rgh3 does not specifically limit protein, starch, or oil biosynthesis.
The range of rgh3 mutant embryo phenotypes led us to test whether rgh3 plants are able to develop. Less than 50% of rgh3 seeds germinate in soil, and the seedlings are smaller and stunted when compared with normal siblings 10 d after planting (Figure 3). All rgh3 seedlings exhibit aberrant development with adherent, narrow leaves resulting in hooked seedlings that died 15 to 18 d after planting. Lethal seedling phenotypes were also observed in F2 progeny after rgh3 was crossed to the B73, Mo17, and A636 inbreds (see Supplemental Figure 1 online). These lethal phenotypes suggest rgh3 impacts multiple developmental processes or Rgh3 is required for cell viability.
Figure 3.
Abnormal rgh3 Seedling Phenotype.
(A) Normal and rgh3 seedlings 10 d after planting. Bar = 2 cm.
(B) Higher magnification of the rgh3 seedling. Bar = 1 cm.
[See online article for color version of this figure.]
Rgh3 Regulates Endosperm Cell Proliferation in Culture
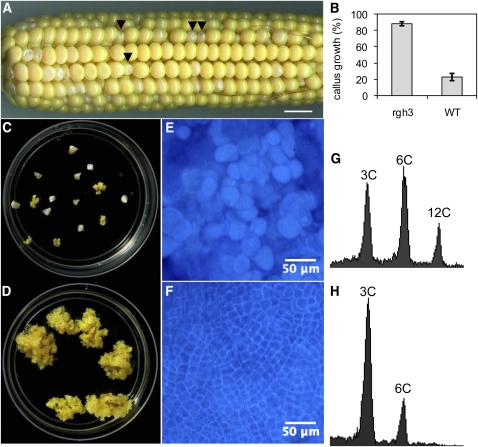
To differentiate between these hypotheses, we used cell culture to test rgh3 mutants for a general role in cellular viability. Shannon and Batey (1973) developed an endosperm culture system that produces immortalized, callus-like endosperm cells. Endosperm culture growth is best with the A636 inbred. We crossed rgh3 into A636 and initiated cultures from self-pollinations of the BC1 and BC2 generations. Mutant and normal seeds could be scored readily at 12 DAP due to a pale color in rgh3 seeds (Figure 4A). Growth in culture was scored with individual endosperm tissues. More than 87% of rgh3 endosperms grew in vitro compared with only 22% of normal siblings (Figure 4B). After 35 d in culture, rgh3 shows massive growth compared with normal siblings (Figures 4C and 4D). Calcofluor white staining of mutant and normal calli was used to visualize cell walls and estimate cell size (Figures 4E and 4F). The mutant cells were smaller than normal siblings, suggesting the excess growth is due to greater cell division in rgh3. Flow cytometry of nuclei extracted from normal endosperm cultures had 3C, 6C, and 12C DNA content, indicating the wild-type Rgh3 genotype undergoes endoreduplication in culture (Figure 4G). By contrast, rgh3 nuclei were mostly 3C with some 6C DNA content, consistent with mitotic cycling (Figure 4H). We then tested the endosperm culture system using 6-, 13-, and 18-DAP ears (see Supplemental Figure 2 online). In these assays, wild-type endosperm tissue readily initiates culture at 6 DAP, while rgh3 homozygous tissue grows readily even if harvested at 18 DAP. These data show Rgh3 inhibits the mitotic cell cycle, although this may be an indirect effect of the mutant. We conclude Rgh3 has roles in endosperm, embryo, and seedling development.
Figure 4.
Increased Cell Proliferation in rgh3 Endosperm Cultures.
(A) Self-pollinated 12-DAP ear segregating for rgh3. Arrowheads indicate rgh3 kernels with reduced size and pale color. Bar = 1 cm.
(B) Callus induction of rgh3 and normal sibling endosperms scored at 35 d in culture. Error bars are se of 16 rgh3 and 18 normal biological replicates. WT, wild type.
(C) and (D) Normal (C) and rgh3 (D) endosperm culture plates at 35 d.
(E) and (F) Calcofluor white–stained endosperm cultures from normal (E) and rgh3 (F). Bars = 50 μm.
(G) and (H) Flow cytometry of nuclei from normal (G) and rgh3 (H) endosperm culture cells.
[See online article for color version of this figure.]
rgh3 Shows Multiple Endosperm Differentiation Defects
The proliferative state of rgh3 endosperm cells in tissue culture is not simple to reconcile with the reduced endosperm of rgh3 kernels. Developing maize kernels transport nutrients from the maternal plant through the basal endosperm transfer cell layer (BETL), and prior reports of reduced grain fill mutations suggest that defects in transfer cells are associated with reduced seed size (Lowe and Nelson, 1946; Brink and Cooper, 1947; Maitz et al., 2000; Costa et al., 2003; Gutiérrez-Marcos et al., 2007). We analyzed sections stained with toluidine blue for BETL morphological defects. A developmental time course of wax-embedded sections from 12 to 16 DAP compares mutant and normal differentiation of the BETL region (see Supplemental Figure 3 online). Figures 5A and 5B show higher resolution semithin sections of BETL regions at 15 DAP. The BETL can be clearly identified in normal kernels as one or two layers of elongated transfer cells with extensive secondary cell wall ingrowths. By contrast, the rgh3 BETL fails to develop elongated cells and basal cells have very few cell wall ingrowths. Both the cells in the BETL region and overlying starchy endosperm cells are smaller in rgh3 when compared with normal siblings.
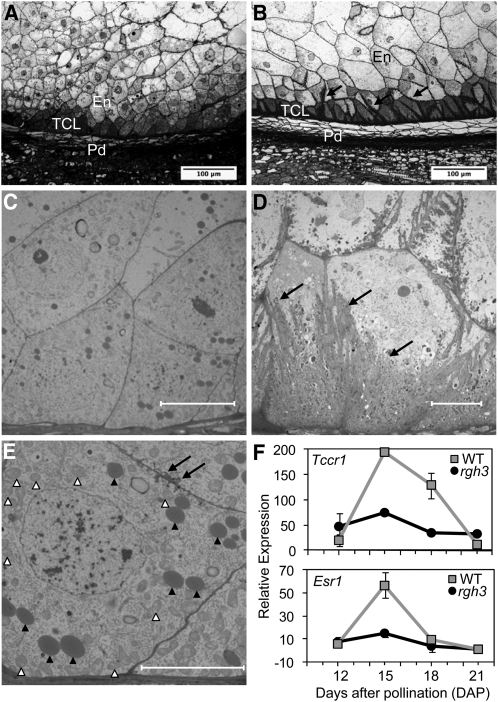
Figure 5.
BETL Defects in rgh3 Kernels.
(A) and (B) Toluidine blue–stained semithin sections of rgh3 (A) and normal (B) BETL region at 15 DAP. Black arrows point to cell wall ingrowths of normal kernels. En, inner endosperm; Pd, pedicel; TCL, transfer cell layer. Bars = 100 μm.
(C) to (E) TEM of rgh3 ([C] and [E]) and normal (D) BETL region at 15 DAP. Black arrows mark cell wall ingrowths. White arrowheads indicate mitochondria, and black arrowheads point to protein bodies. Bars = 10 μm.
(F) Quantitative RT-PCR of total RNA extracted from the lower half of normal (wild-type [WT]) and rgh3 developing kernels. Error bars show se from two biological replicates.
Transmission electron microscopy (TEM) of the BETL region at 15 DAP confirmed the reduced cell wall ingrowths in rgh3 (Figures 5C to 5E). Mitochondria within rgh3 BETL cells are distributed throughout the cytosol (Figure 5E, white arrowheads) as opposed to normal, polar distribution along on the basal cell wall of differentiating BETL cells (Kang et al., 2009). Moreover, rgh3 cells in the BETL region have a cuboidal cell shape and contain electron-dense bodies similar to protein storage bodies in aleurone cells (Figure 5E). These observations suggest rgh3 BETL differentiation is impaired or delayed.
To test this hypothesis, we analyzed expression of BETL specific markers by quantitative RT-PCR (Figure 5F) and gel analysis of PCR products (see Supplemental Figure 4 online). Aberrant expression was observed for three BETL markers tested. Figure 5F shows that the BETL marker Tcrr1 is expressed at a basal level in rgh3 kernels at 15 and 18 DAP, which is the normal peak of expression. The same pattern of reduced expression was observed using an embryo surrounding region (ESR) marker, ESR1 (Figure 5F). ESR is an endosperm cell type that is physically associated with the developing embryo and expresses secreted peptides (Opsahl-Ferstad et al., 1997; Bonello et al., 2002; Balandín et al., 2005). These cell-type marker data indicate that rgh3 has defects in both the BETL and ESR.
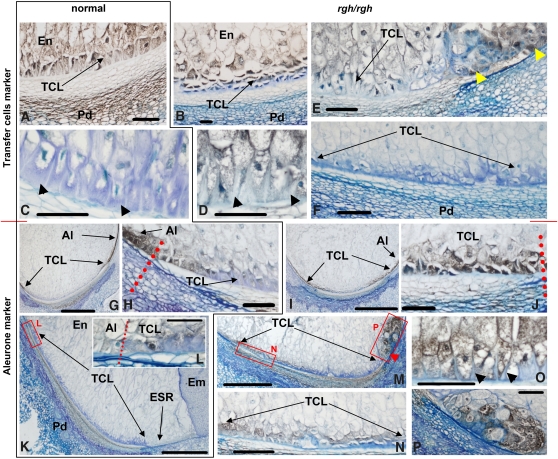
We extended cellular marker studies to immunolocalization of BETL and aleurone marker proteins at 10 and 20 DAP (Figure 6). At 10 DAP, normal and rgh3 kernels cannot be distinguished prior to sectioning, but microscopy readily identified kernels with altered endosperm differentiation (Figures 6A and 6B). The αBETL2 antibody (Serna et al., 2001) labels transfer cells, and BETL2 accumulation reaches a maximum at early developmental stages in normal kernels. Normal sibling kernels had elongated transfer cells accumulating sufficient BETL2 to exclude the blue, azure B, counterstain for secondary cell walls (Figure 6A, BETL signal in brown to black/gray). By contrast, the presumably rgh3 kernels had flat BETL cells with few cell wall ingrowths and showed greatly reduced BETL2 signal and enhanced azure B counterstaining (Figure 6B).
Figure 6.
Immunolocalization of Transfer and Aleurone Cell Markers.
(A) to (F) Kernel sections decorated with antibodies against BETL2.
(G) to (P) Kernel sections decorated with antibodies against an aleurone protease.
Normal Rgh3 kernels are grouped in the left panels and are outlined with a black border ([A], [C], [G], [H], [K], and [L]). Mutant rgh3/rgh3 kernels are shown in the right panels ([B], [D] to [F], [I], [J], and [M] to [P]). Kernel sections are organized chronologically with (A), (B), and (G) to (J) from 10-DAP kernels. (C) to (F) and (K) to (P) are from 20-DAP kernels. Black arrowheads point to cell wall ingrowths in (C), (D), and (O). Yellow arrowheads in (E) point to a mass of cells in the ESR decorated with αBETL2. Red arrowhead in (M) points to a mass of cells, at the position of the embryo suspensor, decorated with the aleurone marker. Red dotted lines mark the border between aleurone and transfer cells ([H], [J], and [L]). Red boxes labeled L, N, and P frame areas shown at higher magnification in adjacent panels (L), (N), and (P). Bar = 1 mm in (G), (I), and (K) and 100 μm in all other panels. Positive immunostaining appears gray, black, or dark brown against blue counterstaining.
(A) Normal Rgh3 kernels, 10 DAP, anti-BETL2 antibodies.
(B) Mutant rgh3/rgh3 kernels, 10 DAP, anti-BETL2 antibodies.
(C) Normal Rgh3 kernels, 20 DAP, anti-BETL2 antibodies.
(D) Mutant rgh3/rgh3 kernels, 20 DAP, anti-BETL2 antibodies.
(E) Mutant rgh3/rgh3 kernels, 20 DAP, anti-BETL2 antibodies.
(F) Mutant rgh3/rgh3 kernels, 20 DAP, anti-BETL2 antibodies.
(G) Normal Rgh3 kernels, 10 DAP, antialeurone protease antibodies.
(H) Normal Rgh3 kernels, 10 DAP, antialeurone protease antibodies.
(I) Mutant rgh3/rgh3 kernels, 10 DAP, antialeurone protease antibodies.
(J) Mutant rgh3/rgh3 kernels, 10 DAP, antialeurone protease antibodies.
(K) Normal Rgh3 kernels, 20 DAP, antialeurone protease antibodies.
(L) Normal Rgh3 kernels, 20 DAP, antialeurone protease antibodies.
(M) Mutant rgh3/rgh3 kernels, 20 DAP, antialeurone protease antibodies.
(N) Mutant rgh3/rgh3 kernels, 20 DAP, antialeurone protease antibodies.
(O) Mutant rgh3/rgh3 kernels, 20 DAP, antialeurone protease antibodies.
(P) Mutant rgh3/rgh3 kernels, 20 DAP, antialeurone protease antibodies.
Al, aleurone; Em, embryo; En, inner endosperm; Pd, pedicel; TCL, transfer cell layer.
To investigate the possible causes of rgh3 variable phenotypic severity, we sorted 20-DAP rgh3 into classes of phenotypic severity and compared with normal sibling kernels. At 20 DAP, αBETL2 signal is very weak in normal, fully developed transfer cells (Figure 6C, gray/black color), which appear densely covered by cell wall ingrowths (arrowheads, blue stain). Mild rgh3 kernels at 20 DAP had apparently younger transfer cells, less densely covered by cell wall ingrowths and accumulating significant amounts of BETL2 (Figures 6D and 6E). Occasionally cell masses outside the BETL region, in the position of ESR cells, were decorated with αBETL2 (Figure 6E, yellow arrowheads). The most severe rgh3 kernels showed no αBETL2 signal, almost no cell wall ingrowths, and only a slightly thickened cell wall (Figure 6F). These results suggest BETL development is delayed in mild rgh3 mutants and is not specified correctly when the kernel phenotype is severe.
The rgh3 mutants were also examined with an antibody directed against a protease expressed in immature aleurone (G. Hueros, unpublished data). In normal 10 DAP kernels, the aleurone-specific antibody decorated, in addition to the aleurone layer, a few cells at the border between the transfer cell layer and aleurone (Figures 6G and 6H). In mutant 10-DAP kernels, the aleurone marker was found throughout most of the transfer cell layer (Figures 6I and 6J). Similar to 10-DAP kernels, normal 20-DAP kernels had weak signal at the abgerminal edge of the BETL that extends for several cells into the transfer cell area (Figures 6K and 6L). No other signal is detected in normal transfer cells by this antibody at any developmental stage. By contrast, the aleurone marker showed significant signal in the BETL region of severe rgh3 mutants (Figures 6M and 6N). Figures 6F, 6M, and 6N are sections from the same mutant kernel. Thus, endosperm regions that lack expression of BETL2 express an immature aleurone marker. Consistent with reduced expression of the Esr1 marker (Figure 5F), internal endosperm cell masses, in the position of ESR cells, expressed the aleurone marker in severe rgh3 mutants (Figure 6M, red arrowhead, and Figure 6P). In mild rgh3 kernels, the aleurone marker labeled only the abgerminal edge of the BETL in a pattern similar to normal kernels except that the labeling was more intense (Figure 6O). These results suggest that aleurone cells are developmentally delayed in mild rgh3 mutants and that the BETL, ESR, and other endosperm cell types are incorrectly specified when the kernel phenotype is severe.
Rgh3 Encodes a Predicted RNA Splicing Factor
We mapped rgh3 to a 0.7-centimorgan interval between mmc0081 and AC207400-2 using an F2 population from crosses of rgh3 with the B73 inbred as well as two-point linkage data with pr1 (Figure 7A). Based on the Genetic 2005 map at MaizeGDB (Sen et al., 2009), two seed mutant loci map near pr1: dek33 and pitted rough germless1 (prg1). Three other seed mutant loci, dek9, dek26, and dek27, were mapped to 5L with B-A translocations. All five mutants are seed lethal and were tested for genetic complementation with rgh3 by crossing normal progeny from segregating ears. Results from these tests suggest all five loci are not alleles of rgh3 (see Supplemental Table 5 online).
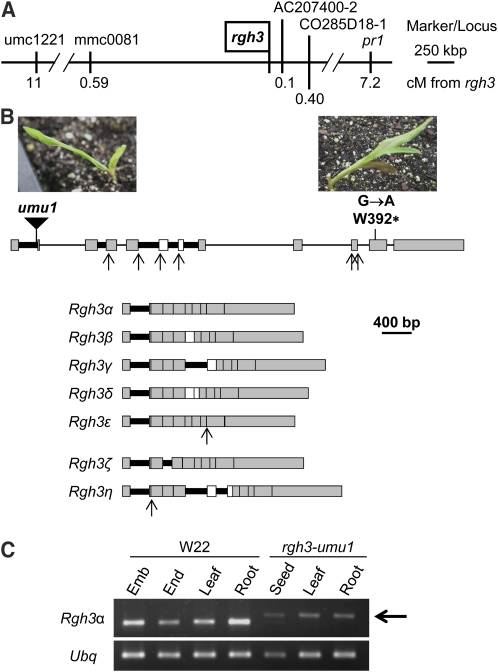
Figure 7.
Cloning of rgh3.
(A) Integrated physical-genetic map for rgh3. Molecular markers are positioned relative to the B73 AGPv2 sequence. Genetic distances are from F2 mapping populations or for pr1 are based on two-point linkage analysis. cM, centimorgan.
(B) Schematic of the rgh3 locus with the location and photographs of rgh3 mutant alleles. Alternative splicing gives rise to at least 19 splice variants, signified by Greek letters. Gray boxes indicate exons required to code the RGH3α protein, and open boxes are skipped exons for this isoform. Thick black lines indicate introns that are retained in one or more splice variants. Arrows indicate alternative 5′ and 3′ splice sites. Examples of splicing patterns that code for seven RGH3 protein variants are shown at the bottom.
(C) RT-PCR of the Rgh3α transcript variant in W22 and rgh3-umu1 tissues. Arrow indicates the rgh3-umu1 splice variant that contains part of the Mu transposon sequence. Ubiquitin (Ubq) was amplified as a loading control.
[See online article for color version of this figure.]
DNA gel blots identified a Mu1 insertion linked to the rgh3 phenotype (see Supplemental Figure 5 online). Robertson's Mutator insertion site thermal asymmetric interlaced PCR was used to amplify and clone the linked Mu1 insertion site (Settles et al., 2004), and linkage analysis was expanded to 1020 meiotic products without any recombinants between the Mu1 element and rgh3 observed. This Mutator-induced rgh3 reference allele was then named rgh3-umu1. We identified a second allele of rgh3 using chemical mutagenesis tagging crosses. Ethyl methanesulfonate (EMS)–mutagenized W22 pollen was crossed onto rgh3-umu1/+ plants. Among the F1 progeny, we identified four rgh3/rgh3 seedlings that included three homozygous rgh3-umu1 self-contaminants (Figure 7B). The remaining rgh3-umu1/rgh3* seedling was analyzed by PCR amplification of the genomic locus and CelI digest to identify two point mutations in the Rgh3 gene (see Supplemental Figure 6 online). One mutation within intron 6 is predicted to be silent. The second mutation was a G-to-A transition resulting in a premature stop codon (Figure 7B, right side). Combined, these data indicate that we identified the Rgh3 locus.
The Rgh3 locus is not correctly assembled in the current B73 AGPv2 reference, and we cloned and sequenced the locus in two overlapping fragments from the maize BAC ZMMBBc497J22. The predicted RGH3 protein is homologous to human U2AF35 Related Protein (URP). URP is involved in RNA splicing of both U2 and U12 introns in mammals (Tronchère et al., 1997; Shen et al., 2010). We amplified, cloned, and sequenced 45 independent Rgh3 RT-PCR products. These clones identified 19 Rgh3 splice variants with the potential to encode seven protein variants. The Rgh3 transcript variants result from variable intron retention, exon skipping, and alternative 5′ and 3′ splice sites. Figure 7B shows schematics for seven examples of Rgh3 isoforms that code for the different protein sequences. The Rgh3α isoform encodes a predicted 86.1-kD protein that is orthologous to URP. Splicing of intron 1 produces a likely noncoding message that deletes the start codon but still contains an uninterrupted URP reading frame that could initiate translation within the UHM domain. The other 17 isoforms are predicted to encode partial proteins ranging from 20 to 41.1 kD (see Supplemental Figure 7 online). There were four Rgh3 transcript variants that are predicted to code for the RGH3β protein and nine transcript variants predicted to code for the RGH3γ protein. Only a single clone was recovered for each of the four other predicted RGH3 protein variants.
To estimate the relative levels of transcripts coding for the RGH3α protein, we designed a quantitative RT-PCR assay for the exon skipping RNA splicing event required to code for the full-length protein (skipping both exons 6 and 7). We compared the relative abundance of the exon 6/7 skip to three splicing events that cause truncation of the open reading frame (ORF) (retaining intron 6, skipping exon 7, and splicing intron 5 to include exon 6). The levels of these Rgh3 splicing events are regulated in a tissue- and developmental-specific manner (see Supplemental Figure 8 online). The exon 6/7 skip event accumulates at its highest levels in 3-cm immature ears and late in seed development, post 30 DAP. The exon 6/7 skip is also found at high levels in early seed development from 6 to 16 DAP as well as in the ligule and midvein of adult leaves. The usage of Rgh3 alternative splice sites is regulated with the exon 6/7 skip, representing 8.1 to 26.7% of the total Rgh3 transcripts assayed (see Supplemental Figure 8C online). Rgh3α-like transcripts were most efficiently spliced at 20-, 34-, and 45-DAP in seeds.
Surprisingly, Rgh3α-specific primers that flanked the Mu1 insertion were able to amplify a cDNA fragment from rgh3-umu1 mutant tissues with only a small size shift relative to wild-type products (Figure 7C). Sequencing of eight rgh3-umu1 cDNA clones identified 141 bp from the Mu1 terminal inverted repeat, indicating Mu1 is partially spliced from mutant transcripts. The spliced Mu1 sequence deletes 12 amino acids of the wild-type protein and inserts 47 new amino acids, suggesting the mutation is likely to be a hypomorph (see Supplemental Figure 7 online).
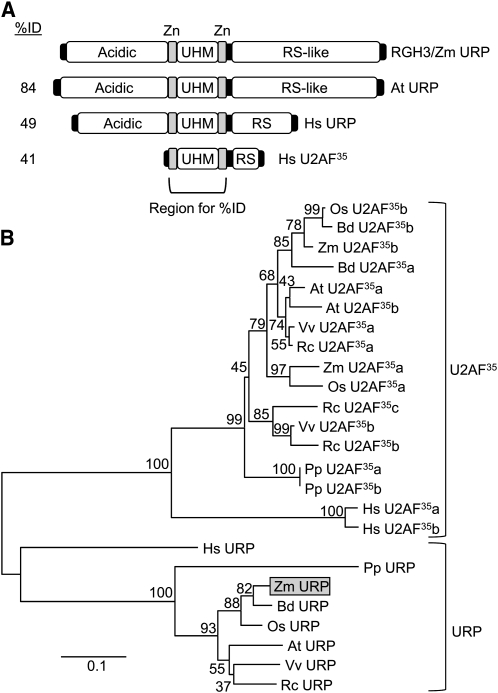
The full-length RGH3α protein encodes a 755–amino acid peptide with a domain structure similar to URP (Figure 8A). URP has an N-terminal acidic domain, a central U2AF Homology Motif (UHM) flanked by zinc-finger motifs, and a C-terminal Ser/Arg-rich region (RS domain). The UHM is related to the RNA Recognition Motif but is responsible for protein–protein interactions rather than binding RNA (Kielkopf et al., 2004). The zinc-finger and UHM region of the protein is highly conserved between maize, Arabidopsis, and humans, whereas the N- and C-terminal sequences are variable with only sequence composition being conserved (see Supplemental Figure 9 online). The U2AF35 protein lacks the acidic N-terminal domain. Phylogenetic analysis of the zinc-finger/UHM region indicates that URP is a distinct clade from U2AF35 proteins and is a single-copy gene in maize, Arabidopsis, and rice (Oryza sativa; Figure 8B). The Rgh3 transcript variants coding for RGH3ζ and RGH3η truncate the ORF such that the conserved zinc fingers, UHM, and RS domains are out of frame with the N-terminal acidic domain (see Supplemental Figure 7 online). RGH3β, RGH3γ, and RGH3δ include the N-terminal zinc finger in the ORF. RGH3ε is predicted to code for protein that includes the acidic, one zinc finger, and the UHM domain, while the EMS allele only truncates the RS domain.
Figure 8.
The Predicted RGH3 Protein Is an Ortholog of URP.
(A) Schematic of protein domains of URP and U2AF35 homologs. Sequence identity to the RGH3 zinc-finger-UHM-zinc-finger region is indicated. Zn, zinc finger.
(B) Phylogenetic tree of the zinc-finger-UHM-zinc-finger domain of URP and U2AF35 homologs. At, Arabidopsis thaliana; Bd, Brachypodium distachyon; Hs, Homo sapiens; Os, Oryza sativa; Pp, Physcomitrella patens; Rc, Ricinus communis; Vv, Vitis vinifera; Zm, Zea mays.
RGH3 Is Localized to Subnuclear Compartments
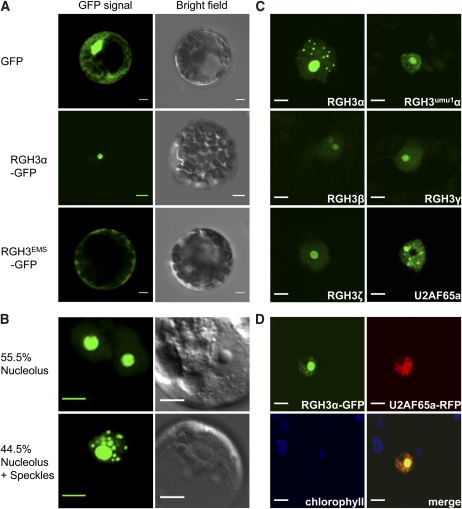
Transient expression of an RGH3α-green fluorescent protein (GFP) fusion in Arabidopsis mesophyll protoplasts localized to compartments of varying size within the nucleus (Figure 9). Analysis of 200 transformed protoplasts gave an equal number of cells either with RGH3α-GFP localized to the nucleolus or with a speckled pattern within the nucleoplasm plus localization to the nucleolus (Figure 9B). A similar pattern of localization to the nucleolus or nuclear speckles was observed when URP, the Arabidopsis ortholog of RGH3, was fused to GFP and expressed in Arabidopsis protoplasts (see Supplemental Figure 10A online). These data suggest the maize localization pattern is not an artifact of heterologous expression. Coexpression of RGH3α-GFP with an established nucleolus marker, Arabidopsis PRH75 (Lorković et al., 2004), confirmed RGH3α localization to the nucleolus (see Supplemental Figure 10B online). RNA processing proteins, such as RNA splicing factors, are typically localized to nuclear speckles and can be localized to the nucleolus in response to cellular stress (Tillemans et al., 2005; Koroleva et al., 2009). We then expressed a GFP fusion with the EMS-truncated reading frame, RGH3EMSα-GFP, which resulted in cytoplasmic localization (Figure 9A). Using an N-terminal GFP fusion expressed in Nicotiana benthamiana, we found GFP-RGH3α localized to the nucleolus and nuclear speckles as in Arabidopsis protoplasts (Figure 9C). We then used this expression system to localize RGH3umu1α as well as three additional wild-type RGH3 isoforms: β, γ, and ζ. These fusions expressed at low levels and localized to the nucleolus as well as diffusely throughout the nucleoplasm. These data suggest RGH3 localization, and potentially protein stability, result from signals or interacting domains within the acidic, UHM, and RS domains. Both the Mu element insertion and EMS alleles alter RGH3α protein localization, suggesting the alleles alter protein function.
Figure 9.
Subcellular Localization of GFP-Tagged RGH3 Proteins.
(A) Transient expression of GFP alone (top panels), RGH3α with a C-terminal GFP fusion (middle panels), and the RGH3EMS allele with a C-terminal GFP fusion in Arabidopsis mesophyll protoplasts (bottom panels).
(B) Transient expression of the RGH3α-GFP fusion in Arabidopsis suspension cell culture protoplasts shows dual targeting in subnuclear compartments. Percentages are the frequency of GFP localization patterns from 200 nuclei in two biological replicates.
(C) Transient expression of N-terminal GFP fusion proteins in N. benthamiana. RGH3α is the full-length RGH3 protein. RGH3umu1α is the full-length protein with the 47–amino acid indel polymorphism caused by the rgh3-umu1 allele. Additional RGH3 variants are truncated forms encoding the N-terminal acidic domain (ζ) or the acidic domain and one zinc finger (β, γ). U2AF65a shows subnuclear localization to speckles in the absence of RGH3α.
(D) Transient coexpression of C-terminal fusions of RGH3α-GFP and U2AF65a-RFP in N. benthamiana.
Bar = 5 μm in all panels.
To confirm that the localization patterns of GFP fusions with RGH3α represent RNA splicing complexes, we coexpressed RGH3α-GFP with a core splicing factor. We fused the ORF of the maize U2 Auxiliary Factor 65-kD subunit locus a (U2AF65a) to red fluorescent protein (RFP). Figure 9D shows that RGH3α-GFP expression overlaps with U2AF65a-RFP expression in both the nucleolus and in speckles. These results suggest the speckled pattern in RGH3α fusion proteins is likely to represent spliceosomes. Interestingly, expression of GFP-U2AF65a resulted in a speckled pattern, with U2AF65a being excluded from the nucleolus. Thus, high levels of RGH3α expression influence U2AF65a subnuclear localization. These data are consistent with the known protein–protein interaction of human URP and U2AF65 (Tronchère et al., 1997).
Rgh3 Is Required for a Subset of RNA Splicing Events
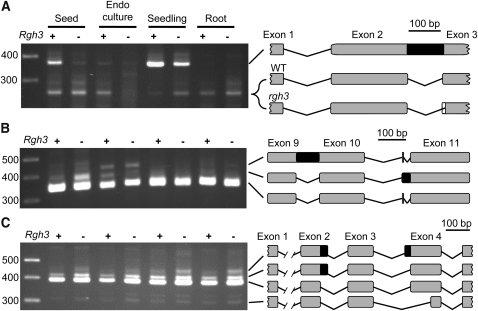
In human cells, URP participates in both U2 and U12 splicing and is essential for cell culture survival (Tronchère et al., 1997; Shen et al., 2010). The viability of rgh3 endosperm cells in culture suggests the rgh3-umu1 hypomorph allele does not disrupt core splicing functions. To assay rgh3 mutants for altered splicing, we selected 21 maize genes with evidence for alternative splicing in maize, Arabidopsis, and rice (Wang and Brendel, 2006; see Supplemental Table 6 online). Only three genes had differences in isoform usage in rgh3 and wild-type tissues. GRMZM2G165901 encodes a Gly-rich RNA binding protein with two major splice variants (Figure 10A). Sequencing of these isoforms found a noncanonical intron retention with GU-CG dinucleotides at the 5′ and 3′ splice sites instead of GU-AG. The rgh3 mutation reduces the level of intron retention in seed and seedling tissue, suggesting noncanonical splicing is more efficient in the mutant. The smaller variant contained different splice junctions in rgh3 and the wild type with rgh3 shifting the splice acceptor site by three bases.
Figure 10.
RNA Splicing Defects in rgh3 Detected with RT-PCR.
Schematics show intron-exon structures as determined by cloning and sequencing of the RT-PCR products. Exons are numbered according to the B73 annotation of release 5b.60. Black boxes indicate retained intron sequences.
(A) GRMZM2G165901 has a novel splice acceptor site in rgh3 with three additional bases included in the mutant intron (white box).
(B) GRMZM2G051276 has a four-base exon between annotated exons 10 and 11. The retained segment of annotated intron 10 also shifts the splice acceptor sequence by two bases relative to the four-base exon.
(C) GRMZM2G081642 has an alternative donor site at the 3′ end of exon 2 and an alternative acceptor site at the 5′ end of exon 4.
GRMZM2G051276 encodes a putative inositol monophosphatase with three splice variants (Figure 10B). Sequencing of these variants found complex alternative splicing within annotated intron 10. First, a four-base exon is found that results from splicing of two noncanonical introns with UU-AA and AU-UU dinucleotides. Second, an alternative variant contains only a single intron between the annotated exons 10 and 11. Assuming the same donor site is used in the alternative variant, the dinucleotides for the predicted intron are UU-UU, indicating this intron is also noncanonical. Relative to the wild type, the alternative, retained intron variant accumulates to higher levels in rgh3 seed tissue but to lower levels in endosperm culture.
GRMZM2G081642 encodes a protein of unknown function, which is conserved within plants and algal species. Sequencing of four variants identified an alternative donor site for intron 2 in which the shorter variant is a GC-AG intron. The longer variant splices with a canonical GU-AG intron, and the rgh3 tissues have higher levels of this canonical donor site. Intron 3 has three alternative acceptor sites that all have canonical dinucleotides. Combined, these data suggest that RGH3umu1 modulates splice site selection for a subset of noncanonical introns.
DISCUSSION
A Role for Endosperm Cell Differentiation in Endosperm–Embryo Developmental Interactions
The endosperm is thought to have a relatively minor role in embryo development but important roles in embryo and seed growth by providing nutrients and acting as a regulator of seed size (De Smet et al., 2010). Maize dek B-A translocation uncoverings generally support this theory with most types of reduced or defective endosperm tissues being tolerated by genetically wild-type embryos (Neuffer and Sheridan, 1980; Neuffer et al., 1986; Chang and Neuffer, 1994). For a few cases, mutant embryo growth benefits from the presence of a wild-type endosperm, supporting the model in which the endosperm nurtures the embryo (see Supplemental Table 1 online). By contrast, we found the rgh mutant class to be enriched for nonautonomous endosperm–embryo developmental interactions (see Supplemental Table 2 online). Most of these interactions are consistent with mutant endosperms causing wild-type embryos to abort or develop abnormally. For rgh3, we confirmed the endosperm to embryo interaction by marking the nonconcordant genotypes using the pr1 anthocyanin locus. These data suggest the rgh mutant class is likely to reveal mechanisms by which the endosperm promotes embryo development. Our characterization of the rgh3 phenotype illustrates a unique developmental mechanism for endosperm–embryo interactions.
As far as we are aware, glo1 is the only other dek mutant with a similar endosperm–embryo developmental interaction for which the mutant endosperm defects have been characterized at a cellular level (Costa et al., 2003). Cellularization and cytokinesis are affected in glo1, and Glo1 is likely to be essential for cell viability. The dsc1 mutation has a similar endosperm to embryo developmental interaction. At a macroscopic level, dsc1 endosperm fails to solidify by 16 DAP, suggesting defects in endosperm cellularization (Scanlon and Myers, 1998). The dsc1 and glo1 defects imply endosperm cellularization is critical for embryo development. By contrast, Arabidopsis mutants in the SPÄTZLE and ENDOSPERM DEFECTIVE1 (EDE1) loci can produce homozygous viable embryos even though the endosperm never cellularizes, suggesting endosperm cellularization is not an absolute requirement for embryo development (Sørensen et al., 2002; Pignocchi et al., 2009).
The rgh3 mutant further argues that cellularization is not the key endosperm event required for embryo development. The rgh3-umu1 allele does not impact endosperm cell viability or cellularization. Bulk kernel composition analysis suggests there are no major differences in the ability of rgh3 cells to accumulate seed storage molecules (see Supplemental Table 4 online). Instead, the cellular phenotypes of rgh3 indicate the gene is necessary for cells to switch from a proliferative to differentiating state. Multiple lines of evidence argue for this interpretation. The in vitro rgh3 cell cultures have smaller cell size and do not undergo endoreduplication (Figures 4E to 4H), while developing endosperm cells in rgh3 kernels are reduced in size (Figures 5A and 5B). Morphological, molecular, and protein markers indicate that rgh3 cells have endosperm cell differentiation defects. The starchy endosperm has reduced storage protein levels (Figures 2D and 2E); the BETL and ESR do not express cell type markers at wild-type levels (Figure 5F); an aleurone marker is delayed in expression and expressed in inappropriate cell positions (Figures 6G to 6P); and rgh3 endosperm retains the ability to grow in vitro 12 d later in development than the wild type (see Supplemental Figure 2 online). These data argue that one or more aspects of endosperm cell differentiation are needed for the embryo to develop. We suggest mutants affecting endosperm cellularization are likely upstream of the cell differentiation pathways affected in rgh3.
Is There a Key Endosperm Cell Type Required for Embryo Development?
A common endosperm phenotype in both glo1 and rgh3 is a defective BETL. It is tempting to conclude that loss of BETL function would limit the sink strength of a developing kernel and lead to defective embryo development. However, multiple dek mutants disrupt BETL differentiation without inhibiting embryo development. Mutants in defective endosperm17, minature1, reduced grain-filling1, and baseless1 (bsl1) produce viable embryos but have morphological BETL defects correlated with reduced grain fill (Lowe and Nelson, 1946; Brink and Cooper, 1947; Maitz et al., 2000; Gutiérrez-Marcos et al., 2006; Kang et al., 2009). Similar to rgh3, bsl1 shows a range of seed phenotypes, including an embryo-lethal phenotype in seeds with severely affected endosperm tissues (Gutiérrez-Marcos et al., 2006). Other seed mutations, such as empty pericarp4, have defective endosperm development, including BETL defects, but excised embryos develop to reproductive maturity (Gutiérrez-Marcos et al., 2007). These mutants suggest reduced sink strength from altered BETL function primarily affects grain fill. The fact that many seed mutants with reduced BETL function produce viable embryos and plants indicates the BETL is unlikely to be responsible for nonautonomous endosperm roles in embryo development. Thus, rgh3 BETL defects can explain nutrient-limited reduced grain fill on the cob even though rgh3 cells overproliferate in culture, when nutrients are not limiting. The lethal seedling phenotype of mild rgh3 kernels suggests the locus has a more global effect on plant development, and rgh3 seedling lethality could have a similar developmental mechanism to the reduced grain fill phenotype, in which critical cell types have differentiation defects.
The ESR also has cell differentiation defects in rgh3, and there is more direct evidence for ESR involvement in endosperm–embryo developmental interactions. The maize ESR produces secreted peptides known as ZmESRs and are hypothesized to have roles in embryo defense or to encode CLAVATA3-like signaling peptides (Opsahl-Ferstad et al., 1997; Bonello et al., 2002; Balandín et al., 2005). Our results show rgh3 kernels ectopically express aleurone and BETL cell markers in ESR-like cells and reduce ESR marker gene expression (Figures 5F, 6C, and 6H). In Arabidopsis, ALE1 and RGE1 are expressed specifically in the ESR and disrupt embryo cuticle development (Tanaka et al., 2001; Kondou et al., 2008; Yang et al., 2008). RGE1 encodes a basic helix-loop-helix transcription factor required for expression of ALE1, which encodes a secreted subtilisin-like Ser protease. The nonautonomous effects of RGE1 and ALE1 provide strong evidence for endosperm-to-embryo signaling during development, but the RGE1/ALE1 pathway does not affect ESR specification or embryo transition (Yang et al., 2008). It may also be significant that the ede1 endosperm cellularization mutant has a subset of endosperm nuclei tightly associated with developing embryos similar to an ESR (Pignocchi et al., 2009). The ede1 mutant develops viable embryos, and the polarized ede1 syncytium is consistent with a requirement for an ESR-like endosperm function to support embryo development.
Weak Alleles of rgh3 Reveal Developmental Roles for Maize URP
The Rgh3 gene is a predicted core RNA splicing factor. In plants, a variety of RNA splicing factors have been shown to have specific roles in abiotic stress tolerance and flowering time (reviewed in Lorković, 2009). Mutants and misexpression transgenics of both U2 and U12 splicing factors can have pleiotropic developmental defects (Lopato et al., 1999; Kalyna et al., 2003; Ali et al., 2007; Chung et al., 2007; Coury et al., 2007; Kim et al., 2010). Most plant genes contain introns that must be removed for protein expression; therefore, RNA splicing should be an essential function for cell viability. Consistent with this reasoning, loss-of-function alleles of core splicing factors are gametophyte or seed lethal (Liu et al., 2009; Wang and Okamoto, 2009; Kim et al., 2010).
The RGH3/ZmURP homologs in animals have only been studied to a limited extent. Human URP is a required core factor for both U2 and U12 splicing in vitro (Tronchère et al., 1997; Shen et al., 2010). URP proteins have a similar domain structure to U2AF35 and can interact with U2AF65 (Tronchère et al., 1997). We found RGH3α to colocalize with maize U2AF65a and redirect U2AF65a into the nucleolus supporting a similar model for URP protein–protein interactions in plants (Figure 9D). URP contacts the 3′ splice site in both U2 and U12 splicing reactions but has different roles for each type of intron with U2AF65 absent from U12 spliceosomes (Shen et al., 2010). Small interfering RNA knockdowns of URP and U2AF35 show that the human genes are essential for cell viability (Shen et al., 2010). Although the rgh3-umu1 mutation is seed and seedling lethal, RNA splicing is not broadly affected in rgh3 mutants, and rgh3 endosperm is completely viable in cell culture. These data are consistent with the recessive inheritance and molecular nature of rgh3-umu1, suggesting the allele is hypomorphic. The Mu insertion disrupts the coding sequence of the locus, but the transposon contains a cryptic splice site that creates an insertion-deletion mutation in the acidic domain of the RGH3α protein (Figure 7; see Supplemental Figure 7 online). The primary amino acid sequence of the acidic and RS domains of URP homologs is not highly conserved. Instead, these domains have conserved amino acid composition. The acidic domain of the RGH3umu1α protein has a pI of 5.64, which is identical to the wild-type protein. B-A translocation uncovering of rgh3 further supports the hypothesis that rgh3-umu1 encodes a weak allele. Uncovered embryo and endosperm phenotypes are more severe than recessive rgh3-umu1 mutant seeds (cf. Figures 1 and 2). In uncovered kernels, the entire 5L chromosome arm is deleted. It is possible there are rgh3 modifier loci on 5L that are haploinsufficient. However, we think it more likely that a single dose of RGH3umu1α is insufficient to promote embryo or endosperm development. The EMS-induced rgh3* allele may also be hypomorphic. The EMS allele supported embryo development and retains the conserved domains predicted to be involved in protein–protein interactions with U2AF65.
GFP fusions with RGH3 isoforms and mutant alleles provide insight on the function of RGH3 protein domains. For RGH3α, we observed exclusive localization to the nucleolus as well as a distribution between nucleolar and speckled localization. The speckled localization is likely to represent splicing complexes because RGH3α colocalizes with U2AF65a in the nucleoplasm. The distribution of nucleolar and speckle localization of RGH3α is not typical for plant pre-mRNA splicing factors (Docquier et al., 2004; Fang et al., 2004; Lorković et al., 2008; Tanabe et al., 2009). A few splicing factors show dynamic localization between the nucleoplasm and the nucleolus (Tillemans et al., 2005; Koroleva et al., 2009). The targeting of these splicing factors to the nucleolus is influenced by cellular stress, such as hypoxia, and levels of protein phosphorylation. RGH3α may show similar dynamic regulation of its localization. It is also possible that overexpression of RGH3α fusion proteins may stress the cell sufficiently to cause nucleolar localization.
Nuclear speckle localization is lost in RGH3umu1α and in RGH3 splice variants lacking the UHM, one zinc-finger, and the RS domains. The RGH3umu1α protein suggests that the acidic domain is important for the protein to be incorporated into nuclear speckles, while the RGH3β, γ, and ζ proteins implicate the UHM, C-terminal zinc-finger, or RS domain of the protein for nuclear speckle localization. In Arabidopsis, deletion of the RS domain from RSZp22 results in a similar loss of speckle localization and directs the splicing factor to the nucleolus (Tillemans et al., 2005). However, the rgh3* EMS allele truncates only the RS domain and shows cytoplasmic localization. The RS domain may be required to unmask the nuclear localization signal in the N-terminal acidic domain. Alternatively, a nuclear export signal may reside in the C-terminal region of the UHM domain or the C-terminal zinc finger. Combined, the GFP fusion data suggest RGH3α contains multiple targeting signals that can direct nuclear versus cytoplasmic localization as well as subnuclear localization to the nucleolus or to speckles.
Alternative RNA Splicing in Endosperm Cell Differentiation
In animals, alternative splicing is associated with cell-type and tissue-specific expression patterns (Blencowe, 2006). Multiple genetic diseases have been identified that show alternative RNA splicing is essential for different cell types to function correctly (Ferreira et al., 2010; Twine et al., 2011). However, only a few proteins with KH-type RNA binding domains have been shown to act as cell differentiation regulators (Ule et al., 2005; Volk et al., 2008). Endosperm cell differentiation is clearly defective in rgh3 mutants. We observe delayed or misexpression of all endosperm cell types, including zein storage proteins in the starchy endosperm, a protease in the aleurone, multiple BETL markers, and Esr1. Moreover, rgh3 endosperm retains the ability to grow in culture up to the latest time point tested, 18 DAP, while wild-type endosperm only grows at similar efficiency at 6 DAP. These data indicate a role for RNA splicing in endosperm cell differentiation. Our survey of splicing defects in rgh3 suggests that the RGH3umu1α protein alters splicing in a subset of noncanonical introns. Genome-wide-level analysis of rgh3 splicing defects would help identify the specific genes required to promote normal endosperm cell differentiation. Similar differentiation defects could be the cause of the rgh3 lethal seedling phenotype, but we did not investigate the rgh3 seedling phenotype in detail in this study.
In conclusion, we have shown that endosperm cell differentiation is required for wild-type embryo development. By comparing rgh3 cellular defects with other maize seed mutants, we speculate that the ESR is a critical cell type to promote embryo development. The cloning of rgh3 identifies alternative RNA splicing as a key process required for endosperm cell differentiation. The hypomorphic rgh3-umu1 allele suggests the acidic domain of URP has a role in alternative splicing in maize.
METHODS
B-A Translocation Genetics
All rgh mutants were heritable isolates from the UniformMu population (McCarty et al., 2005). Plants segregating for rgh/+ genotypes were crossed as females with hyperploid or heterozygous B-A translocations (Birchler and Alfenito, 1993; Beckett, 1994a, 1994b). For each translocation, 10 to 15 crosses were attempted with multiple B-A plants. A mutant was considered uncovered when multiple F1 crosses showed rgh phenotypes specific to a single chromosome arm. To identify endosperm–embryo interactions, sagittal sections of mature kernels were cut with a utility knife. To generate marked uncovering crosses, rgh3/+ plants carrying dominant alleles for anthocyanin genes were crossed onto a red aleurone (pr1) stock in the W22 inbred. F2 pr1/pr1 kernels were self-pollinated to identify rgh3 recombinants. Normal kernels from segregating F3 ears were used as females for B-A crosses.
Molecular and Genetic Mapping
F2 progeny from B73 × rgh3/+ crosses were scored for rgh3/rgh3 seedling phenotypes. DNA was extracted from seedling leaves as described by Settles et al. (2004). Simple sequence repeat markers from chromosome 5 were amplified and scored for recombination, and Haldane map distances were calculated. PCR conditions were optimized for each marker based on initial conditions from Martin et al. (2010). Primer sequences for the markers are in Supplemental Table 7 online.
Genetic complementation tests used reciprocal crosses between plants segregating for rgh3, dek9, dek26, dek27, dek33, and prg1 heterozygotes. The first ears of male parents in the crosses were self-pollinated to determine plant genotypes. The second ears of female parents in each cross were self-pollinated when possible. Crosses were screened visually, and seed phenotypes were expected to approximate segregation ratios of self-pollinated parents.
The rgh3 Mu1 insertion site was cloned from a PstI digest of genomic DNA from a rgh3/rgh3 mutant seedling at the BC3S1 generation of a Mu-inactive introgression into W22. The DNA fraction from 3.5 to 4.2 kb was gel purified and used as template for Robertson's Mutator insertion site thermal asymmetric interlaced PCR (Settles et al., 2004). PCR products were cloned and sequenced as described (Settles et al., 2004). The majority of the Rgh3 locus was subcloned from a BamHI digest of the B73 BAC, ZMMBBc497J22. A 6-kb overlapping 3′ region was amplified from the BAC using TaKaRa LA Taq DNA Polymerase and cloned using a pTOPO vector (Invitrogen). Full-length Rgh3 cDNA was amplified using 5′ and 3′ rapid amplification of cDNA ends PCR with the GeneRacer kit (Invitrogen), and splice variants were amplified using Phusion high fidelity polymerase (Finnzymes). cDNA products were cloned with the Zero Blunt TOPO PCR cloning kit (Invitrogen).
The rgh3* allele was generated by pollinating 150 rgh3-umu1/+ plants with EMS-mutagenized W22 pollen as described (Neuffer, 1993; Douglas et al., 2010). Approximately 4000 progeny were screened for the rgh3 seedling phenotype. Point mutations in the Rgh3 locus were identified using the Surveyor mutation detection kit (Transgenomics).
Quantitative Kernel Phenotypes
Normal and rgh kernels from 10 rgh3 segregating ears were separated. Seed weights were collected from an automated microbalance as described by Spielbauer et al. (2009). Kernels were ground to a fine meal using a mortar and pestle. Proteins were extracted from 100 mg of flour in 2 mL of extraction buffer (10% glycerol, 2% SDS, 100 mM DTT, 50 mM Tris-HCl, pH 6.8, and 1× Complete protease inhibitor cocktail [Roche Applied Science]) at 37°C with agitation for 2 h. Extracts were centrifuged at 12,000g for 10 min and supernatants processed for zein purification as described by Gibbon et al. (2003). Zein and non-zein protein fractions were analyzed on a fresh weight basis with 12.5% SDS-PAGE. Bulk composition analysis was completed by Waters Agricultural Laboratories using 15 g of meal from normal and average weight rgh3 kernels (140 to 200 mg) with Association of Analytical Communities methods.
Endosperm Culture
Self-pollinations from A636 × rgh3/+ BC1 and BC2 generations were used for endosperm cultures as described by Shannon (1994). Briefly, 6-, 12-, 13-, or 18-DAP ears were surface sterilized, and individual endosperm tissues were used to initiate cultures on Murashige and Skoog media, pH 5.7, with 3% Suc, 4 ppm thiamine, 0.2% Asn, and 2% phytagel. Cultures were incubated at 30°C in the dark for 35 d. Growth was assessed for over 250 cultured endosperms for both rgh3 and normal siblings. Fragments of calli were stained with calcofluor-white (0.01% in 1× PBS), mounted on glass slides, and observed using a Nikon Diaphot inverted microscope with epifluorescence illumination and a UV filter. Representative pictures were taken with a Leica DC300 digital camera.
Flow Cytometry
Approximately 150 mg of endosperm culture was chopped in 1.5 mL of ice-cold chopping buffer (20 mM MOPS, pH 7.2, 45 mM MgCl2, 30 mM Na3Citrate, 0.1% Triton X-100, and 0.01% RNase A) using a razor blade. Homogenates were gravity filtered through four layers of cheesecloth and then 100-μm nylon mesh. Samples were centrifuged for 2 min at 600g. Pelleted nuclei were resuspended in chopping buffer with 0.05% propidium iodide. BD FACSort (BD Biosciences) with “no gate” for the setting parameter was used for flow cytometry. Flow cytometry plots are representative of 10 replicates of 5000 events counted.
Microscopy
Purple kernels from crosses of Rgh3 pr1/Rgh3 pr1 × rgh3 Pr1/Rgh3 pr1 were selected to enrich for ~93% rgh3/+ plants. Developing ears were harvested from 10 to 20 DAP. Kernels were processed for semithin (500 nm) and thin (80 nm) sections as described by Kang et al. (2009). Representative pictures of semithin, toluidine blue (0.05%) stained sections were taken using an Olympus BH2 microscope and Retiga 200R digital camera (QImaging). Transmission electron micrographs of thin sections were captured with a Hitachi H-7000 microscope operated at 80 kV. For the developmental time course and immunolocalizations, kernels were fixed at 4°C overnight in FAA solution (3.7% formaldehyde, 5% glacial acetic acid, and 45% ethanol). Kernels were dehydrated in an ethanol series, embedded in paraffin or ParaPlast plus embedding medium (Sigma-Aldrich), and cut into 8- to 10-μm sections. The sections in the developmental time course were stained with toluidine blue. Immunolocalization used each preserum-serum pair at equivalent dilutions. Slides were rehydrated in an ethanol series, and endogenous peroxidase activity was deactivated in 0.3% hydrogen peroxide. The sections were blocked with 2% normal donkey serum and incubated with antisera or presera. The slides were washed twice in 140 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4, 1.8mM KH2PO4, and 0.1% Tween-20, pH 7.3, and incubated in biotin-conjugated α-rabbit goat antibody at 1/750 (Sigma-Aldrich). After two washes, an Extravidin-peroxidase (Sigma-Aldrich) solution at 1/800 was incubated with each slide. Following two washes, SIGMAFAST DAB with Metal Enhancer (Sigma-Aldrich) was incubated until the gray, black, or brown precipitate was visible on sera slides. Reactions were stopped by washing in water and counterstained for 3 min in azure B, pH 4.5. Micrographs were taken using a Zeiss Axiophot microscope.
Subcellular Localization
N- and C-terminal GFP fusion proteins were constructed by cloning cDNAs for RGH3 isoforms and the coding Arabidopsis thaliana URP isoform into pDONR221 or pENTR (Invitrogen) according to the manufacturer’s instructions. The ORFs were subcloned into p2GWF7 for C-terminal GFP fusions or pB7-WGF2 for N-terminal GFP fusions (Karimi et al., 2002, 2007) by LR recombination (Invitrogen). Transient expression experiments in Arabidopsis were performed with protoplasts isolated from leaves or cell suspension culture (Yoo et al., 2007). Transient expression experiments in Nicotiana benthamiana were completed essentially as described by Kapila et al. (1996) with the following modifications. Binary vectors were transformed into Agrobacterium tumefaciens strain ABi by a freeze-thaw method (Wise et al., 2006). MES was not included in the Agrobacterium growing media, and N. benthamiana infiltration was completed with a 10-mL needleless syringe on 4- to 5-week-old plants grown in growth chamber at 22 to 24°C with 16/8-h day/night. For colocalization experiments, Agrobacterium strains carrying individual plasmids were mixed in a 1:1 ratio prior to infiltration. Fusion protein expression was visualized 24 to 48 h after transient transformation, and representative pictures of subcellular localization were obtained using a Zeiss Pascal LSM5 confocal laser scanning microscope as previously described (Pribat et al., 2010). GFP/RFP colocalization images were obtained with a Leica TCS SP2 confocal laser scanning microscope (Leica Microsystems). GFP was excited at 488 nm and detected with an emission band of 500 to 545nm. Monomeric RFP was excited at 594 nm and detected with an emission band of 605 to 645 nm.
Expression Analysis
Total RNA was extracted from maize (Zea mays) tissues as described by Reid et al. (2006) using 10 mL of extraction buffer/g of fresh weight. For molecular markers, maize kernels were cut in half with a transverse section as described by Gómez et al. (2009), and RNA was extracted from the basal sections. RNA samples were treated with RNase-free DNase I (New England Biolabs), extracted with phenol-chloroform, and ethanol precipitated. RT-PCR was performed on first-strand cDNA and synthesized by Superscript III (Invitrogen) using gene-specific primer sets from Supplemental Tables 6 and 7 online. PCR cycles were optimized for each gene to identify the linear amplification range. Representative results from two biological replicates are shown. Quantitative RT-PCR used first-strand oligo(dT)-primed cDNA, 500 nM of gene-specific primer sets from published cell-type markers or from Rgh3 isoform specific primers, and 1× SYBR Green Master Mix (Applied Biosystems). PCR was conducted on a StepOnePlus real-time PCR machine (Applied Biosystems). All quantifications were normalized against the Ubq control gene (Bommert et al., 2005), and data were expressed as 2−cycle threshold (Livak and Schmittgen, 2001). Primer specificity was confirmed by agarose gel electrophoresis and melting curve analysis.
Phylogenetic Analysis
Predicted protein sequences of URP and U2AF35 were identified through BLASTP and TBLASTN searches in the National Center for Biotechnology Information databases (http://www.ncbi.nlm.nih.gov/), the Joint Genome Institute (www.jgi.doe.gov) servers, and the plaza plant comparative genomics resource (Proost et al., 2009) using default settings at each server. Conserved domains were identified using Prosite scans at the European Bioinformatics Institute server. Multiple sequence alignments and phylogenetic tree of the zinc-finger-UHM-zinc finger region of the proteins were generated with MEGA4 (Tamura et al., 2007). The multiple sequence alignment (see Supplemental Data Set 1 online) was completed with the ClustalW module within MEGA4 using default parameters: gap opening penalty = 10, gap extension penalty = 0.2, protein weight matrix = Gonnet with residue-specific and hydrophylic penalties, gap separation distance = 4, and a 30% delay divergent cutoff. A rooted phylogenetic tree was constructed using the neighbor-joining algorithm within MEGA4 according to a Poisson model with uniform rates among sites and complete deletion of gaps and missing data. Bootstrap values were calculated from 1000 iterations. Protein sequence alignments in Supplemental Figures 7 and 9 online were completed with ClustalW2 at the European Bioinformatics Institute server using default parameters: Gonnet protein weight matrix, gap open = 10, gap extension = 0.2, gap distances = 5, no end gaps = no, iteration = none, numiter = 1, and clustering = NJ.
Accession Numbers
Sequence data for the maize Rgh3 genomic locus and cDNA clones can be found in the GenBank/EMBL data libraries under accession numbers JN692485, JF681144, and JN791417 to JN791437. The accession number for Arabidopsis URP is At1g10320.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Inbred Background Effects on the Phenotype of rgh3 Seedlings 10 d after Planting.
Supplemental Figure 2. Endosperm Tissue Culture Response at Multiple Developmental Time Points.
Supplemental Figure 3. Developmental Series of the BETL Region in rgh3 and Normal Siblings.
Supplemental Figure 4. RT-PCR of Endosperm Cell-Type Markers.
Supplemental Figure 5. Mu1 Element Linked to rgh3-umu1.
Supplemental Figure 6. Point Mutations in the EMS-Tagged rgh3* Allele.
Supplemental Figure 7. Multiple Sequence Alignment of RGH3 Protein Isoforms and Mutant Alleles.
Supplemental Figure 8. Relative Expression of Rgh3 Alternative Splicing Events.
Supplemental Figure 9. Multiple Sequence Alignment of the Conserved Zinc-Finger-UHM-Zinc-Finger Region of URP and U2AF35 Proteins.
Supplemental Figure 10. Transient Expression of AtURP-GFP in Arabidopsis Mesophyll Protoplasts and Coexpression of RGH3α-GFP with AtPRH75-RFP.
Supplemental Table 1. Summary of Inferred Seed Developmental Phenotypes from Published B-A Translocation Uncovering Crosses.
Supplemental Table 2. Inferred Developmental Phenotypes from B-A Translocation Uncovering Crosses of Uniform Mu rgh Mutants.
Supplemental Table 3. Transmission of rgh3.
Supplemental Table 4. Bulk Seed Composition Analysis.
Supplemental Table 5. Complementation Tests for rgh3 with Seed Loci on Chromosome Arm 5L.
Supplemental Table 6. Maize Genes Surveyed for Splicing Defects in rgh3 Mutants.
Supplemental Table 7. Primers Used in This Study.
Supplemental Data Set 1. Text File of the Sequences and Alignment Used for the Phylogenetic Analysis Shown in Figure 8B.
Acknowledgments
We thank Martha James (Iowa State University), Jim Birchler (University of Missouri-Columbia), Don Auger (South Dakota State University), and the Maize Genetics Cooperation Stock Center (USDA-ARS) for providing seed stocks as well as Byung-Ho Kang and Steve McClellan at the University of Florida Interdisciplinary Center for Biotechnology Research for technical support in microscopy and flow cytometry experiments. We thank Tariq Akhtar (University of Florida) for providing the Arabidopsis cell culture and Zdravko Lorković (Medical University of Vienna, Austria) for the AtPRH75-RFP marker. This work was supported by the National Research Initiative of the USDA Cooperative State Research, Education, and Extension Service (Grant 2007-35304-18439), the National Science Foundation Plant Genome Research Program (Grant 0076700), the Vasil-Monsanto Endowment, and the Spanish Ministerio de Ciencia e Innovación (Grant BIO2009-11856).
AUTHOR CONTRIBUTIONS
R.F., F.M., D.S.F., C.M.G., E.G., C.-W.T., T.P., G.H., and A.M.S. designed and performed the research. R.F., F.M., D.S.F., C.M.G., E.G., G.H., and A.M.S. analyzed data. R.F., G.H., and A.M.S. wrote the article.
References
- Ali G.S., Palusa S.G., Golovkin M., Prasad J., Manley J.L., Reddy A.S. (2007). Regulation of plant developmental processes by a novel splicing factor. PLoS ONE 2: e471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balandín M., Royo J., Gómez E., Muniz L.M., Molina A., Hueros G. (2005). A protective role for the embryo surrounding region of the maize endosperm, as evidenced by the characterisation of ZmESR-6, a defensin gene specifically expressed in this region. Plant Mol. Biol. 58: 269–282 [DOI] [PubMed] [Google Scholar]
- Beckett J.B. (1994a). Comprehensive list of B-A translocations in maize. The Maize Handbook, Freeling M., Walbot V., (New York: Springer-Verlag; ), pp. 336–341 [Google Scholar]
- Beckett J.B. (1994b) Locating recessive genes to chromosome arms with B-A translocations. The Maize Handbook, Freeling M., Walbot V., (New York: Springer-Verlag; ), pp. 313–327 [Google Scholar]
- Berger F., Chaudhury A. (2009). Parental memories shape seeds. Trends Plant Sci. 14: 550–556 [DOI] [PubMed] [Google Scholar]
- Birchler J.A., Alfenito M.R. (1993). Marker systems for B-A translocations in maize. J. Hered. 84: 135–138 [Google Scholar]
- Blencowe B.J. (2006). Alternative splicing: new insights from global analyses. Cell 126: 37–47 [DOI] [PubMed] [Google Scholar]
- Bommert P., Lunde C., Nardmann J., Vollbrecht E., Running M., Jackson D., Hake S., Werr W. (2005). thick tassel dwarf1 encodes a putative maize ortholog of the Arabidopsis CLAVATA1 leucine-rich repeat receptor-like kinase. Development 132: 1235–1245 [DOI] [PubMed] [Google Scholar]
- Bonello J.F., Sevilla-Lecoq S., Berne A., Risueño M.C., Dumas C., Rogowsky P.M. (2002). Esr proteins are secreted by the cells of the embryo surrounding region. J. Exp. Bot. 53: 1559–1568 [DOI] [PubMed] [Google Scholar]
- Brink R.A., Cooper D.C. (1947). Effect of the De17 allele on development of the maize caryopsis. Genetics 32: 350–368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang M.T., Neuffer M.G. (1994). Endosperm - Embryo interaction in maize. Maydica 39: 9–18 [Google Scholar]
- Chung T., Kim C.S., Nguyen H.N., Meeley R.B., Larkins B.A. (2007). The maize zmsmu2 gene encodes a putative RNA-splicing factor that affects protein synthesis and RNA processing during endosperm development. Plant Physiol. 144: 821–835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa L.M., Gutierrez-Marcos J.F., Brutnell T.P., Greenland A.J., Dickinson H.G. (2003). The globby1-1 (glo1-1) mutation disrupts nuclear and cell division in the developing maize seed causing alterations in endosperm cell fate and tissue differentiation. Development 130: 5009–5017 [DOI] [PubMed] [Google Scholar]
- Coury D.A., Zhang C., Ko A., Skaggs M.I., Christensen C.A., Drews G.N., Feldmann K.A., Yadegari R. (2007). Segregation distortion in Arabidopsis gametophytic factor 1 (gfa1) mutants is caused by a deficiency of an essential RNA splicing factor. Sex. Plant Reprod. 20: 87–97 [Google Scholar]
- Danilevskaya O.N., Hermon P., Hantke S., Muszynski M.G., Kollipara K., Ananiev E.V. (2003). Duplicated fie genes in maize: Expression pattern and imprinting suggest distinct functions. Plant Cell 15: 425–438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Smet I., Lau S., Mayer U., Jürgens G. (2010). Embryogenesis - The humble beginnings of plant life. Plant J. 61: 959–970 [DOI] [PubMed] [Google Scholar]
- Docquier S., Tillemans V., Deltour R., Motte P. (2004). Nuclear bodies and compartmentalization of pre-mRNA splicing factors in higher plants. Chromosoma 112: 255–266 [DOI] [PubMed] [Google Scholar]
- Douglas R.N., Wiley D., Sarkar A., Springer N., Timmermans M.C.P., Scanlon M.J. (2010). ragged seedling2 Encodes an ARGONAUTE7-like protein required for mediolateral expansion, but not dorsiventrality, of maize leaves. Plant Cell 22: 1441–1451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang Y., Hearn S., Spector D.L. (2004). Tissue-specific expression and dynamic organization of SR splicing factors in Arabidopsis. Mol. Biol. Cell 15: 2664–2673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferreira E.N., Rangel M.C., Galante P.F., de Souza J.E., Molina G.C., de Souza S.J., Carraro D.M. (2010). Alternative splicing enriched cDNA libraries identify breast cancer-associated transcripts. BMC Genomics 11 (Suppl. 5): S4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibbon B.C., Wang X., Larkins B.A. (2003). Altered starch structure is associated with endosperm modification in Quality Protein Maize. Proc. Natl. Acad. Sci. USA 100: 15329–15334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gómez E., Royo J., Muñiz L.M., Sellam O., Paul W., Gerentes D., Barrero C., López M., Perez P., Hueros G. (2009). The maize transcription factor myb-related protein-1 is a key regulator of the differentiation of transfer cells. Plant Cell 21: 2022–2035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutiérrez-Marcos J.F., Costa L.M., Biderre-Petit C., Khbaya B., O’Sullivan D.M., Wormald M., Perez P., Dickinson H.G. (2004). maternally expressed gene1 Is a novel maize endosperm transfer cell-specific gene with a maternal parent-of-origin pattern of expression. Plant Cell 16: 1288–1301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutiérrez-Marcos J.F., Costa L.M., Evans M.M. (2006). Maternal gametophytic baseless1 is required for development of the central cell and early endosperm patterning in maize (Zea mays). Genetics 174: 317–329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutiérrez-Marcos J.F., Dal Prà M., Giulini A., Costa L.M., Gavazzi G., Cordelier S., Sellam O., Tatout C., Paul W., Perez P., Dickinson H.G., Consonni G. (2007). empty pericarp4 encodes a mitochondrion-targeted pentatricopeptide repeat protein necessary for seed development and plant growth in maize. Plant Cell 19: 196–210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalyna M., Lopato S., Barta A. (2003). Ectopic expression of atRSZ33 reveals its function in splicing and causes pleiotropic changes in development. Mol. Biol. Cell 14: 3565–3577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang B.H., Xiong Y., Williams D.S., Pozueta-Romero D., Chourey P.S. (2009). Miniature1-encoded cell wall invertase is essential for assembly and function of wall-in-growth in the maize endosperm transfer cell. Plant Physiol. 151: 1366–1376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapila J., de Rycke R., van Montagu M., Angenon G. (1996). An Agrobacterium-mediated transient gene expression system for intact leaves. Plant Sci. 122: 101–108 [Google Scholar]
- Karimi M., Depicker A., Hilson P. (2007). Recombinational cloning with plant gateway vectors. Plant Physiol. 145: 1144–1154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karimi M., Inzé D., Depicker A. (2002). GATEWAY vectors for Agrobacterium-mediated plant transformation. Trends Plant Sci. 7: 193–195 [DOI] [PubMed] [Google Scholar]
- Kielkopf C.L., Lücke S., Green M.R. (2004). U2AF homology motifs: Protein recognition in the RRM world. Genes Dev. 18: 1513–1526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim W.Y., Jung H.J., Kwak K.J., Kim M.K., Oh S.H., Han Y.S., Kang H. (2010). The Arabidopsis U12-type spliceosomal protein U11/U12-31K is involved in U12 intron splicing via RNA chaperone activity and affects plant development. Plant Cell 22: 3951–3962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kodrzycki R., Boston R.S., Larkins B.A. (1989). The opaque-2 mutation of maize differentially reduces zein gene transcription. Plant Cell 1: 105–114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondou Y., Nakazawa M., Kawashima M., Ichikawa T., Yoshizumi T., Suzuki K., Ishikawa A., Koshi T., Matsui R., Muto S., Matsui M. (2008). RETARDED GROWTH OF EMBRYO1, a new basic helix-loop-helix protein, expresses in endosperm to control embryo growth. Plant Physiol. 147: 1924–1935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koroleva O.A., Calder G., Pendle A.F., Kim S.H., Lewandowska D., Simpson C.G., Jones I.M., Brown J.W., Shaw P.J. (2009). Dynamic behavior of Arabidopsis eIF4A-III, putative core protein of exon junction complex: fast relocation to nucleolus and splicing speckles under hypoxia. Plant Cell 21: 1592–1606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin B.Y. (1984). Ploidy barrier to endosperm development in maize. Genetics 107: 103–115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linkies A., Graeber K., Knight C., Leubner-Metzger G. (2010). The evolution of seeds. New Phytol. 186: 817–831 [DOI] [PubMed] [Google Scholar]
- Liu M., Yuan L., Liu N.Y., Shi D.Q., Liu J., Yang W.C. (2009). GAMETOPHYTIC FACTOR 1, involved in pre-mRNA splicing, is essential for megagametogenesis and embryogenesis in Arabidopsis. J. Integr. Plant Biol. 51: 261–271 [DOI] [PubMed] [Google Scholar]
- Livak K.J., Schmittgen T.D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 25: 402–408 [DOI] [PubMed] [Google Scholar]
- Lopato S., Kalyna M., Dorner S., Kobayashi R., Krainer A.R., Barta A. (1999). atSRp30, one of two SF2/ASF-like proteins from Arabidopsis thaliana, regulates splicing of specific plant genes. Genes Dev. 13: 987–1001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopes M.A., Larkins B.A. (1993). Endosperm origin, development, and function. Plant Cell 5: 1383–1399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorković Z.J. (2009). Role of plant RNA-binding proteins in development, stress response and genome organization. Trends Plant Sci. 14: 229–236 [DOI] [PubMed] [Google Scholar]
- Lorković Z.J., Hilscher J., Barta A. (2004). Use of fluorescent protein tags to study nuclear organization of the spliceosomal machinery in transiently transformed living plant cells. Mol. Biol. Cell 15: 3233–3243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorković Z.J., Hilscher J., Barta A. (2008). Co-localisation studies of Arabidopsis SR splicing factors reveal different types of speckles in plant cell nuclei. Exp. Cell Res. 314: 3175–3186 [DOI] [PubMed] [Google Scholar]
- Lowe J., Nelson O.E. (1946). Miniature seed-A study in the development of a defective caryopsis in maize. Genetics 31: 525–533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo M., Dennis E.S., Berger F., Peacock W.J., Chaudhury A. (2005). MINISEED3 (MINI3), a WRKY family gene, and HAIKU2 (IKU2), a leucine-rich repeat (LRR) KINASE gene, are regulators of seed size in Arabidopsis. Proc. Natl. Acad. Sci. USA 102: 17531–17536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maitz M., Santandrea G., Zhang Z., Lal S., Hannah L.C., Salamini F., Thompson R.D. (2000). rgf1, a mutation reducing grain filling in maize through effects on basal endosperm and pedicel development. Plant J. 23: 29–42 [DOI] [PubMed] [Google Scholar]
- Martin F., Dailey S., Settles A.M. (2010). Distributed simple sequence repeat markers for efficient mapping from maize public mutagenesis populations. Theor. Appl. Genet. 121: 697–704 [DOI] [PubMed] [Google Scholar]
- McCarty D.R., et al. (2005). Steady-state transposon mutagenesis in inbred maize. Plant J. 44: 52–61 [DOI] [PubMed] [Google Scholar]
- Neuffer M.G. (1993). Mutagenesis. The Maize Handbook, Freeling M., Walbot V., (New York: Springer-Verlag; ), pp. 212–219 [Google Scholar]
- Neuffer M.G., Chang M.T., Clark J.K., Sheridan W.F. (1986). The genetic control of maize kernel development. In Regulation of Carbon and Nitrogen Reduction and Utilization in Maize, J.C. Shannon, D.P. Knievel, and C.D. Boyer, eds (Rockville, MD: American Society of Plant Physiologists), pp. 35–50 [Google Scholar]
- Neuffer M.G., Coe E.H., Wessler S.R. (1997). Mutants of Maize. (Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press) [Google Scholar]
- Neuffer M.G., Sheridan W.F. (1980). Defective kernel mutants of maize. I. Genetic and lethality studies. Genetics 95: 929–944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Opsahl-Ferstad H.G., Le Deunff E., Dumas C., Rogowsky P.M. (1997). ZmEsr, a novel endosperm-specific gene expressed in a restricted region around the maize embryo. Plant J. 12: 235–246 [DOI] [PubMed] [Google Scholar]
- Pignocchi C., Minns G.E., Nesi N., Koumproglou R., Kitsios G., Benning C., Lloyd C.W., Doonan J.H., Hills M.J. (2009). ENDOSPERM DEFECTIVE1 is a novel microtubule-associated protein essential for seed development in Arabidopsis. Plant Cell 21: 90–105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pribat A., et al. (2010). Nonflowering plants possess a unique folate-dependent phenylalanine hydroxylase that is localized in chloroplasts. Plant Cell 22: 3410–3422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Proost S., Van Bel M., Sterck L., Billiau K., Van Parys T., Van de Peer Y., Vandepoele K. (2009). PLAZA: A comparative genomics resource to study gene and genome evolution in plants. Plant Cell 21: 3718–3731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raissig M.T., Baroux C., Grossniklaus U. (2011). Regulation and flexibility of genomic imprinting during seed development. Plant Cell 23: 16–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reid K.E., Olsson N., Schlosser J., Peng F., Lund S.T. (2006). An optimized grapevine RNA isolation procedure and statistical determination of reference genes for real-time RT-PCR during berry development. BMC Plant Biol. 6: 27–37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roman H. (1947). Mitotic nondisjunction in the case of interchanges involving the B-type chromosome in maize. Genetics 32: 391–409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabelli P.A., Larkins B.A. (2009). The contribution of cell cycle regulation to endosperm development. Sex. Plant Reprod. 22: 207–219 [DOI] [PubMed] [Google Scholar]
- Scanlon M.J., Myers A.M. (1998). Phenotypic analysis and molecular cloning of discolored-1 (dsc1), a maize gene required for early kernel development. Plant Mol. Biol. 37: 483–493 [DOI] [PubMed] [Google Scholar]
- Scanlon M.J., Stinard P.S., James M.G., Myers A.M., Robertson D.S. (1994). Genetic analysis of 63 mutations affecting maize kernel development isolated from Mutator stocks. Genetics 136: 281–294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott R.J., Spielman M., Bailey J., Dickinson H.G. (1998). Parent-of-origin effects on seed development in Arabidopsis thaliana. Development 125: 3329–3341 [DOI] [PubMed] [Google Scholar]
- Sen T.Z., Andorf C.M., Schaeffer M.L., Harper L.C., Sparks M.E., Duvick J., Brendel V.P., Cannon E., Campbell D.A., Lawrence C.J. (2009). MaizeGDB becomes ‘sequence-centric’. Database (Oxford) 2009: bap020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serna A., Maitz M., O’Connell T., Santandrea G., Thevissen K., Tienens K., Hueros G., Faleri C., Cai G., Lottspeich F., Thompson R.D. (2001). Maize endosperm secretes a novel antifungal protein into adjacent maternal tissue. Plant J. 25: 687–698 [DOI] [PubMed] [Google Scholar]
- Settles A.M., Latshaw S., McCarty D.R. (2004). Molecular analysis of high-copy insertion sites in maize. Nucleic Acids Res. 32: e54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shannon J.C. (1994). Establishment and culture of maize endosperm. In The Maize Handbook, M. Freeling and V. Walbot, eds (New York: Springer-Verlag), pp. 719–722 [Google Scholar]
- Shannon J.C., Batey J.W. (1973). Inbred and hybrid effects on establishment of in vitro cultures of Zea mays L. endosperm. Crop Sci. 13: 491–493 [Google Scholar]
- Shen H., Zheng X., Luecke S., Green M.R. (2010). The U2AF35-related protein Urp contacts the 3′ splice site to promote U12-type intron splicing and the second step of U2-type intron splicing. Genes Dev. 24: 2389–2394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheridan W.F., Neuffer M.G. (1982). Maize developmental mutants: Embryos unable to form leaf primordia. J. Hered. 73: 318–329 [Google Scholar]
- Sørensen M.B., Mayer U., Lukowitz W., Robert H., Chambrier P., Jürgens G., Somerville C., Lepiniec L., Berger F. (2002). Cellularisation in the endosperm of Arabidopsis thaliana is coupled to mitosis and shares multiple components with cytokinesis. Development 129: 5567–5576 [DOI] [PubMed] [Google Scholar]
- Spielbauer G., Armstrong P., Baier J.W., Allen W.B., Richardson K., Shen B., Settles A.M. (2009). High-throughput near-infrared reflectance spectroscopy for predicting quantitative and qualitative composition phenotypes of individual maize kernels. Cereal Chem. 86: 556–564 [Google Scholar]
- Springer N.M. (2009). Small RNAs: How seeds remember to obey their mother. Curr. Biol. 19: R649–R651 [DOI] [PubMed] [Google Scholar]
- Tamura K., Dudley J., Nei M., Kumar S. (2007). MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol. Biol. Evol. 24: 1596–1599 [DOI] [PubMed] [Google Scholar]
- Tanabe N., Kimura A., Yoshimura K., Shigeoka S. (2009). Plant-specific SR-related protein atSR45a interacts with spliceosomal proteins in plant nucleus. Plant Mol. Biol. 70: 241–252 [DOI] [PubMed] [Google Scholar]
- Tanaka H., Onouchi H., Kondo M., Hara-Nishimura I., Nishimura M., Machida C., Machida Y. (2001). A subtilisin-like serine protease is required for epidermal surface formation in Arabidopsis embryos and juvenile plants. Development 128: 4681–4689 [DOI] [PubMed] [Google Scholar]
- Tillemans V., Dispa L., Remacle C., Collinge M., Motte P. (2005). Functional distribution and dynamics of Arabidopsis SR splicing factors in living plant cells. Plant J. 41: 567–582 [DOI] [PubMed] [Google Scholar]
- Tronchère H., Wang J., Fu X.D. (1997). A protein related to splicing factor U2AF35 that interacts with U2AF65 and SR proteins in splicing of pre-mRNA. Nature 388: 397–400 [DOI] [PubMed] [Google Scholar]
- Twine N.A., Janitz K., Wilkins M.R., Janitz M. (2011). Whole transcriptome sequencing reveals gene expression and splicing differences in brain regions affected by Alzheimer’s disease. PLoS ONE 6: e16266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ule J., et al. (2005). Nova regulates brain-specific splicing to shape the synapse. Nat. Genet. 37: 844–852 [DOI] [PubMed] [Google Scholar]
- Volk T., Israeli D., Nir R., Toledano-Katchalski H. (2008). Tissue development and RNA control: “HOW” is it coordinated? Trends Genet. 24: 94–101 [DOI] [PubMed] [Google Scholar]
- Wang B.B., Brendel V. (2006). Genomewide comparative analysis of alternative splicing in plants. Proc. Natl. Acad. Sci. USA 103: 7175–7180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S., Okamoto T. (2009). Involvement of polypyrimidine tract-binding protein (PTB)-related proteins in pollen germination in Arabidopsis. Plant Cell Physiol. 50: 179–190 [DOI] [PubMed] [Google Scholar]
- Weijers D., Van Hamburg J.P., Van Rijn E., Hooykaas P.J.J., Offringa R. (2003). Diphtheria toxin-mediated cell ablation reveals interregional communication during Arabidopsis seed development. Plant Physiol. 133: 1882–1892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wise A.A., Liu Z., Binns A.N. (2006). Three methods for the introduction of foreign DNA into Agrobacterium. Methods Mol. Biol. 343: 43–53 [DOI] [PubMed] [Google Scholar]
- Yang S., Johnston N., Talideh E., Mitchell S., Jeffree C., Goodrich J., Ingram G. (2008). The endosperm-specific ZHOUPI gene of Arabidopsis thaliana regulates endosperm breakdown and embryonic epidermal development. Development 135: 3501–3509 [DOI] [PubMed] [Google Scholar]
- Yoo S.D., Cho Y.H., Sheen J. (2007). Arabidopsis mesophyll protoplasts: a versatile cell system for transient gene expression analysis. Nat. Protoc. 2: 1565–1572 [DOI] [PubMed] [Google Scholar]