This study explores the function of two highly conserved isoforms of the Arabidopsis thaliana 26S proteasome subunit RPT2 that participate in proteasome-mediated protein turnover. The data point to a specific role for RPT2 in stem fasciation, male and female gametogenesis, and leaf, root, trichome, and pollen development, along with a common role for RP subunits in histone dynamics and nucleosome assembly.
Abstract
The regulatory particle (RP) of the 26S proteasome contains a heterohexameric ring of AAA-ATPases (RPT1-6) that unfolds and inserts substrates into the core protease (CP) for degradation. Through genetic analysis of the Arabidopsis thaliana gene pair encoding RPT2, we show that this subunit plays a critical role in 26S proteasome assembly, histone dynamics, and plant development. rpt2a rpt2b double null mutants are blocked in both male and female gamete transmission, demonstrating that the subunit is essential. Whereas rpt2b mutants are phenotypically normal, rpt2a mutants display a range of defects, including impaired leaf, root, trichome, and pollen development, delayed flowering, stem fasciation, hypersensitivity to mitomycin C and amino acid analogs, hyposensitivity to the proteasome inhibitor MG132, and decreased 26S complex stability. The rpt2a phenotype can be rescued by both RPT2a and RPT2b, indicative of functional redundancy, but not by RPT2a mutants altered in ATP binding/hydrolysis or missing the C-terminal hydrophobic sequence that docks the RPT ring onto the CP. Many rpt2a phenotypes are shared with mutants lacking the chromatin assembly factor complex CAF1. Like caf1 mutants, plants missing RPT2a or reduced in other RP subunits contain less histones, thus implicating RPT2 specifically, and the 26S proteasome generally, in plant nucleosome assembly.
INTRODUCTION
Plants, like other eukaryotes, rely extensively on the ubiquitin (Ub)-26S proteasome system (UPS) to selectively control the abundance of key regulatory proteins and to remove aberrant polypeptides and normal proteins deemed no longer necessary (Smalle and Vierstra, 2004; Finley, 2009; Vierstra, 2009). Within the UPS, chains of Ubs are assembled onto appropriate substrates by a highly polymorphic, ATP-dependent conjugation cascade that connects Ub via an isopeptide bond to one or more accessible lysines within the substrate or to previously appended Ubs. The resulting polyubiquitylated proteins are then recognized and degraded by the 26S proteasome with the concomitant release of the Ub moieties for reuse.
The 26S proteasome is a ~2.5-MD, ATP-dependent protease composed of two functionally distinct complexes, the 20S core protease (CP) and the 19S regulatory particle (RP) (Voges et al., 1999; Finley, 2009; Bohn et al., 2010). The CP is a broad-spectrum, Ub- and ATP-independent protease assembled from four stacked heptameric rings, each containing seven α-subunits or seven β-subunits (Arabidopsis thaliana PAA-PAG and PBA-PBG, respectively) in a C2 symmetric α1-7β1-7β1-7α1-7 configuration. Upon assembly, a central chamber is created that houses six peptidase catalytic sites provided by the β1 (PBA), β2, (PBB), and β5 (PBE) subunits. Access to this chamber is restricted by two narrow axial pores, which are generated by the peripheral α-subunit rings to allow only unfolded substrates to enter (Groll et al., 2000). Passage through the pore is further gated by flexible N-terminal extensions appended to several α-subunits (α2-4 or PAB-PAD) that permit substrate entry and peptide exit (Köhler et al., 2001; Smith et al., 2007). Through this self-compartmentalized design, CP proteolysis is restricted to only those polypeptides that are deliberately unfolded and imported.
The 20 or more subunit RP docks to each end of the CP through contacts with the α-subunit rings (Finley, 2009; Gallastegui and Groll, 2010). It imparts both ATP dependence and substrate specificity to the CP, especially with regard to proteins modified with Ub polymers. Following substrate identification via several Ub receptors intrinsic to the RP (van Nocker et al., 1996; Husnjak et al., 2008; Schreiner et al., 2008) and a collection of extraproteasomal Ub binding proteins that shuttle ubiquitylated cargo to the RP (Finley, 2009; Farmer et al., 2010; Fu et al., 2010), the CP α-ring gate is opened, the Ub moieties are removed, and the substrates are unfolded and directed into the CP lumen.
The RP can be dissected further into base and lid subcomplexes (Glickman et al., 1998). The base contains a hexameric ring of AAA-ATPases designated RPT1-6 and three non-ATPase subunits, RPN1, RPN2, and RPN10. The lid is composed of RPN3, 5-13, and 15, plus an assortment of accessory proteins present at substoichiometric levels that aid in 26S particle assembly, substrate selection, ubiquitylation, and Ub recycling (Finley, 2009; Book et al., 2010). The RPN10 and RPN13 subunits in particular are the main receptors that identify substrates tagged with poly-Ub chains (van Nocker et al., 1996; Fu et al., 1998; Elsasser et al., 2004; Husnjak et al., 2008), whereas the RPN11 subunit has a deubiquitylation activity that helps remove Ubs bound to targets (Verma et al., 2002). The function(s) of most of the remaining RPN proteins is not yet clear.
Within the RP, the RPT ring plays a prominent role in RP-CP docking and substrate unfolding and import into the CP lumen (Finley, 2009; Tomko et al., 2010; Inobe et al., 2011). It sits on the CP α-ring and controls CP gating by moving the α-ring N-terminal extensions (Köhler et al., 2001; Smith et al., 2007; Rabl et al., 2008). Several RPT subunits (RPT1, 2, 3, and 5 in Arabidopsis; Fu et al., 1999) have a C-terminal HbYX extension (hydrophobic residue-Tyr-any amino acid) that helps template the RP onto the CP by plugging into intersubunit pockets formed upon assembly of the CP α-ring (Smith et al., 2007; Park et al., 2009, 2011; Roelofs et al., 2009). Like other members of the AAA superfamily, the RPT subunits contain an AAA cassette with the signature Walker A and Walker B P-loop motifs necessary for ATP binding and ATP hydrolysis, respectively. ATP binding promotes RP-CP assembly, while ATP hydrolysis is proposed to drive substrate unfolding and subsequent import (Smith et al., 2007, 2011; Inobe et al., 2011).
In addition to its functions within the RP, the RPT ring may also have nonproteasomal activities (Kodadek, 2010). For example, a free form of the yeast RP base containing the RPT1-6, RPN1, and RPN2 subunits has been detected bound to transcriptionally active regions of DNA and in association with the nucleotide excision DNA repair machinery (Gillette et al., 2001; Gonzalez et al., 2002). In these capacities, the minimal RP complex is hypothesized to exploit the AAA-ATPase unfoldase activities of the RPT ring as a chaperone to strip transcriptional or DNA repair regulators bound to chromatin.
Despite the expected sixfold pseudosymmetry of the RPT ring, studies on the RPT2 subunit in yeast (Saccharomyces cerevisiae) have shown that it has a special role in RP assembly and activity. Its HbYX motif in particular is key to stabilizing the CP-RP interface (Smith et al., 2007; Park et al., 2009, 2011) by binding in a fixed manner to a specific cavity between the α3 and α4 subunits of the CP α-ring (Tian et al., 2011). Additionally, whereas the ATPase activities of other RPT subunits are not essential for enhancing the peptidase activity of the CP or its ability to degrade ubiquitylated proteins, that from the RPT2 subunit is required (Rubin et al., 1998). Further analyses revealed that the ATPase activity of RPT2 is specifically needed for opening the CP gate, presumably by moving the α-ring extensions (Köhler et al., 2001). Moreover, while the other five RPT subunits from Arabidopsis or from the protist Trypanosoma brucei can fully complement the corresponding yeast rptΔ mutants, both RPT2 orthologs cannot, despite higher sequence identity, implying that a strict structural requirement exists for RPT2 in the assembled RPT ring (Fu et al., 1999; Li et al., 2002).
The RPT2 subunit, which is encoded by two paralogous genes in both Arabidopsis and rice (Oryza sativa; Fu et al., 1999; Shibahara et al., 2004; Book et al., 2010), also appears to have a unique function(s) in the plant 26S proteasome. For example, forward and reverse genetic screens have connected the RPT2a isoform to shoot apical meristem (SAM) and root apical meristem (RAM) development, cell elongation, organ size, trichome branching, DNA endoreduplication, oxidative stress protection, and responses to zinc deficiency (Ueda et al., 2004, 2011; Kurepa et al., 2008, 2009; Sonoda et al., 2009; Sako et al., 2010; Sakamoto et al., 2011). Some of these phenotypes are distinct from those induced by compromising RPN subunits and other subunits in the RPT ring (Smalle et al., 2002, 2003; Brukhin et al., 2005; Huang et al., 2006; Book et al., 2009; Gallois et al., 2009), suggesting that RPT2 has a unique role in the plant 26S proteasome or that it also participates in other nonproteasomal activities. Here, we further explore these possibilities with an in-depth genetic analysis of the RPT2a and b isoforms in Arabidopsis. Our data demonstrate that RPT2 is essential for assembly of the plant 26S proteasome and that the RPT2a and b isoforms are functionally equivalent. Strikingly, many of the rpt2a phenotypes, including a substantial reduction in histone levels, are shared with mutants defective in the chromatin assembly factor 1 (CAF1) complex required for nucleosome assembly (Kaya et al., 2001; Exner et al., 2006). Similar attenuation of histones was observed for other RP subunit mutants, strongly suggesting that RPT2 and other components of the RP are important for robust nucleosome assembly.
RESULTS
The RPT2 Subunit Is Synthesized from Two Active Loci in Arabidopsis
Prior studies established that the RPT2 subunit is encoded by two nearly identical genes, RPT2a and RPT2b, in the Arabidopsis ecotype Columbia-0 (Col-0) (Fu et al., 1999; Ueda et al., 2004; Kurepa et al., 2008; Sonoda et al., 2009). Each locus encodes a 443–amino acid polypeptide with obvious Walker A and Walker B P-loop motifs that are characteristic of the AAA superfamily (see Supplemental Figure 1 online). Also of potential importance are a possible myristoylation site at Gly-2 immediately distal to the N terminus, which may help link the 26S proteasome to membranes upon modification (Shibahara et al., 2002; Boisson et al., 2003), and a consensus HbYX motif at the C terminus, which likely promotes docking of the RP to the CP (Smith et al., 2007; Park et al., 2009, 2011). RPT2a and RPT2b are located on syntenic blocks in chromosomes 4 and 2, respectively, with the encoded proteins differing by only four amino acids (not three as previously described; Sonoda et al., 2009), suggesting that they arose from the recent whole scale duplication of the Arabidopsis genome (Ermolaeva et al., 2003). This extraordinary conservation extends to orthologs from other plant species, with amino acid sequence identities ranging from 90 to 96% when comparing Arabidopsis RPT2a and b to relatives from poplar (Populus trichocarpa), rice, maize (Zea mays), Physcomitrella patens, and Selaginella moellendorffii (see Supplemental Figure 1 online).
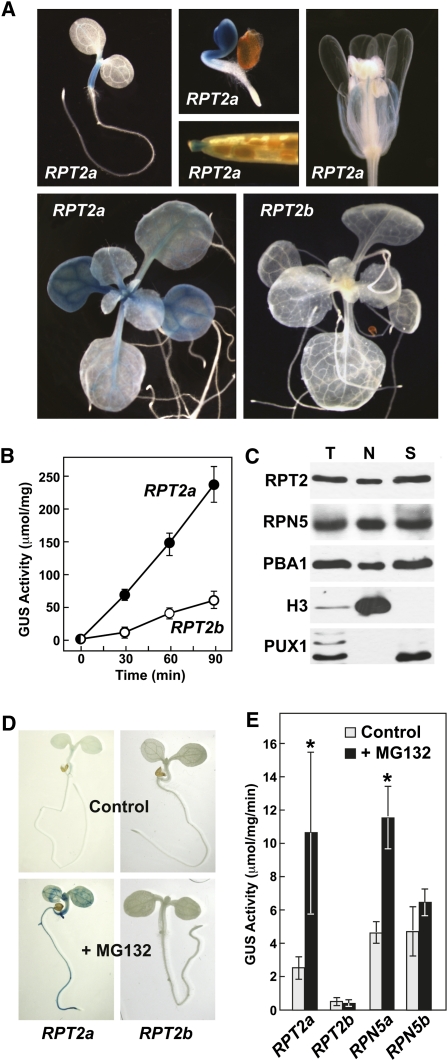
Expression patterns derived from RT-PCR, ESTs, and the Genevestigator DNA microarray database (Zimmermann et al., 2004) revealed that both loci are widely expressed in many tissues (see Supplemental Figure 2 online; Kurepa et al., 2008; Sonoda et al., 2009), with RPT2a expression estimated to be ~2 times higher than RPT2b based on EST counts (132 versus 70 ESTs, respectively). To confirm these expression patterns, we generated multiple transgenic Arabidopsis lines stably expressing the β-glucuronidase (GUS) reporter under the direction of the RPT2a or RPT2b promoters. As shown in Figure 1A, an ~2-kb fragment upstream of the RPT2a initiation ATG codon drove readily detectable GUS expression in a variety of tissues, whereas a comparable upstream fragment from RPT2b was much less active. Quantitative 4-methylumbelliferyl-β-d-glucuronide (MUG) assays, collectively involving 30 independent lines each, determined that the RPT2a promoter is approximately fourfold more active than the RPT2b promoter (Figure 1B).
Figure 1.
Expression Patterns and Subcellular Localization of Arabidopsis RPT2a and RPT2b Genes and Proteins.
(A) Expression patterns of RPT2apro:GUS and RPT2bpro:GUS fusions in various tissues. Examples include 1-week-old and 3-d-old seedlings for RPT2a, 2-week-old seedlings for RPT2a and b, and a developing silique and flower from RPT2a.
(B) Quantitative expression of RPT2apro:GUS and RPT2bpro:GUS. GUS activities were determined by MUG assays of crude extracts obtained from 10-d-old seedlings. Shown is a time course of the MUG reaction generated with each transgenic line. Average activity (±se) was determined from the analysis of 30 independent lines each assayed in triplicate.
(C) Localization of RPT2 protein in the cytosol and nucleus. Fourteen-day-old wild-type seedlings were homogenized and the total extract (T) was fractionated into the nuclear (N) and soluble fractions (S) by Percoll gradient centrifugation. Fractions were subjected to immunoblot analysis with antibodies against 26S proteasome subunits (RPT2, RPN5, and PBA1) and the nuclear and cytosolic markers histone H3 and PUX1, respectively.
(D) Effect of the proteasome inhibitor MG132 on RPT2a and RPT2b expression. Four-day-old RPT2apro:GUS and RPT2bpro:GUS seedlings were treated for 36 h with 100 μM MG132 or an equivalent amount of DMSO (control) and then stained overnight with X-Gluc.
(E) Quantitation of GUS activity generated upon treatment of 4-d-old RPT2apro:GUS and RPT2bpro:GUS seedlings with 100 μM MG132. Average levels of GUS were determined by MUG assay of three independent lines assayed in triplicate (±sd). The asterisks indicate a significant difference between control and MG132-treated samples (Student’s t test, P < 0.05). The RPT5apro:GUS and RPT5bpro:GUS lines used for comparison were as previously described (Book et al., 2009).
Many genes encoding 26S proteasome subunits are members of a proteasome stress regulon that is coordinately upregulated when the particle is impaired genetically or blocked by proteasomal inhibitors, such as MG132 (Yang et al., 2004; Book et al., 2009; Gallois et al., 2009; Kurepa et al., 2009). We confirmed that RPT2a, but not RPT2b, is part of this negative feedback regulon. Whereas GUS activity in RPT2apro:GUS seedlings was significantly elevated by treatment with 100 μM MG132, as seen by both histochemical and fluorescence-based activity assays, GUS activity in RPT2bpro:GUS seedlings was unaffected (Figures 1D and 1E). A similar but less exaggerated differential effect was also seen when comparing the MG132 response from the RPN5a and RPN5b promoters (Figure 1E).
Both RPT2a and RPT2b proteins were detected by mass spectrometry in purified Arabidopsis 26S proteasomes, demonstrating that each assembles into the complete particle (Yang et al., 2004; Book et al., 2010). In agreement with both nuclear and cytosolic locations for the complex (Book et al., 2009), RPT2 and other CP and RP subunits could be detected immunologically in fractions enriched for either compartment (Figure 1C). The anti-RPT2 antibodies used here were generated against RPT2a but recognize the RPT2b isoform equally well, as judged by immunoblot analysis of recombinant proteins (see Supplemental Figure 3 online). In an attempt to determine whether RPT2a and RPT2b have distinct intracellular locations, we examined the possibility of using green fluorescent protein (GFP) fusions as subcellular reporters. However, fusions of GFP to either the N or C terminus of RPT2a and RPT2b failed to phenotypically rescue the corresponding null mutants, thus negating their reliability (data not shown).
Mutational Analyses Identify Numerous Processes Affected by RPT2
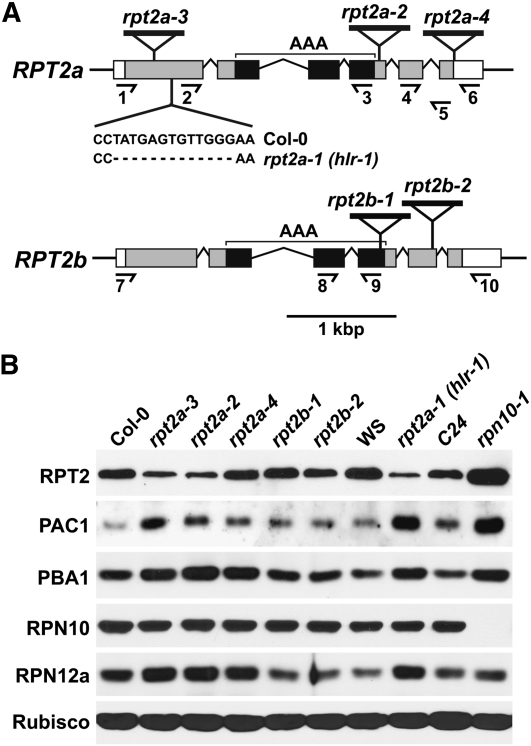
To define the full range of functions controlled by RPT2a and RPT2b, we expanded the collection of rpt2 mutant alleles described previously with two new lines (Figure 2A). To unify the nomenclature, the halted root1 (hlr1) deletion mutant identified by Ueda et al. (2004) in the Wassilewskija (Ws) background was designated as rpt2a-1, while the rpt2a-2, rpt2a-3, and rpt2b-1 T-DNA mutants in the Col-0 background used the names established by Kurepa et al. (2009). New alleles in the Col-0 background were rpt2a-4 and rpt2b-2 that each contain a T-DNA insertion near the 3′ end of the respective coding regions (Figure 2A). All five of the T-DNA insertions blocked accumulation of the full-length RPT2a or RPT2b transcripts as determined by RT-PCR (see Supplemental Figure 4A online). Based on their effects on transcript organization, most of the mutants appeared to represent null alleles. The only exception was rpt2a-4; it contains a T-DNA insert immediately downstream of the translation termination codon, indicating that a partial transcript encompassing the full RPT2a coding region is possible.
Figure 2.
Description of Mutants in the RPT2a and RPT2b Genes.
(A) Organization of the RPT2a and RPT2b genes highlighting the positions of the T-DNA insertion mutants and the rpt2a-1 (hlr-1) nucleotide deletion. Introns are indicated by lines. The coding region and untranslated regions are shown in black/gray and white boxes, respectively. The AAA-ATPase casette (AAA) is in black. The half arrows locate the positions of primers used for RT-PCR in Supplemental Figure 4A online. The nucleotide sequence deleted in the rpt2a-1 (hlr-1) mutant is shown compared with that for wild-type RPT2a.
(B) Immunoblot detection of RPT2 and other 26S proteasome subunits in homozygous rpt2a and rpt2b mutants compared with their corresponding wild types: Col-0 for rpt2a-2, rpt2a-3, rpt2a-4, rpt2b-1, and rpt2b-2; Ws for rpt2a-1 (hlr-1); and C24 for rpn10-1. Equal protein loading was confirmed by probing the blots with anti-Rubisco antibodies.
In agreement with the approximately fourfold higher expression of RPT2a versus RPT2b, homozygous rpt2a-1, rpt2a-2, and rpt2a-3 seedlings had approximately one-quarter the RPT2 protein level as the corresponding wild types, whereas the RPT2 levels in the rpt2b-1 and rpt2b-2 seedlings appeared unaffected (Figure 2B). The only exception to the rpt2a collection was the rpt2a-4 allele, which appeared to have near-normal RPT2 levels consistent with the likelihood that a full-length protein was translated. Reduced RPT2 levels in turn appeared to compromise 26S proteasome activity, as judged by the upregulation of other members of the proteasome stress regulon. As shown in Figure 2B, levels of the PAC1 (α3), PBA1 (β1), and RPN12a proteins were noticeably higher in the rpt2a-1, rpt2a-2, and rpt2a-3 lines compared with their wild-type parents, a response similar to that described previously for rpn10-1 plants (Smalle et al., 2003; Yang et al., 2004).
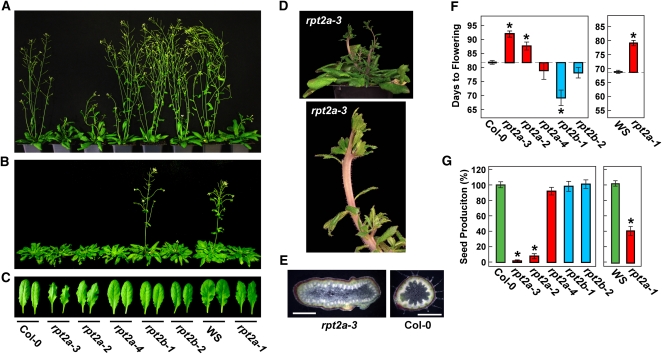
As in previous reports (Ueda et al., 2004; Huang et al., 2006; Kurepa et al., 2009; Sonoda et al., 2009), we observed a wide range of phenotypic defects generated by strong rpt2a mutants (rpt2a-1, rpt2a-2, and rpt2a-3) but not rpt2b mutants, including shorter roots, narrow serrated rosette leaves, increased trichome branching, and increased size for several organs, including cotyledons, leaves, flowers, and seeds (Figure 3; see Supplemental Figure 5 online). A slight but significant effect on root growth and trichome branching was seen for rpt2a-4 plants, in agreement with its likely weak affect on RPT2a protein accumulation (see Supplemental Figure 5 online). In contrast with previous studies (Kurepa et al., 2009; Sonoda et al., 2009), we found that the organ size increases of strong rpt2a mutants were highly variable and sometimes undetectable, suggesting that the growth conditions needed to elicit this defect are subtle. In addition, we observed several new rpt2 phenotypes. Most notable were strong fasciation of the inflorescence stem, especially when grown in short-day (SD) photoperiods, delayed flowering in long-day (LD) photoperiods, and substantially reduced fertility for rpt2a-1, rpt2a-2, and rpt2a-3 plants (Figure 3). By contrast, the rpt2b-1 and rpt2b-2 plants were relatively normal phenotypically, with the only obvious defect being an acceleration of flowering time in SDs (Figures 3B and 3F).
Figure 3.
Phenotypic Analysis of rpt2a and rpt2b Mutants.
(A) rpt2a mutants have delayed flowering in LDs; 42-d-old plants are shown.
(B) rpt2b mutants display accelerated flowering in SDs; 80-d-old plants are shown.
(C) Abnormal rosette leaf morphology of rpt2a seedlings grown under SDs. Newly expanded leaves from 75-d-old plants grown in SDs are shown.
(D) rpt2a-3 plants grown under SDs develop fasciated inflorescence stems.
(E) Cross section of a fasciated stem from rpt2a-3 and wild-type Col-0 plants. Bars = 1 mm.
(F) Effect of rpt2 mutations on flowering time under SDs. Each bar represents the average number of days before emergence of the inflorescence stem from 16 plants (±se).
(G) Effect of rpt2a mutations on seed production. Bars show the relative number of seeds from 20 plants (±se) compared with those from the corresponding wild types. In both (F) and (G), the asterisks represent a significant difference between the wild type and mutants (analysis of variance [ANOVA] followed by Tukey’s multiple comparisons post-test, P < 0.05).
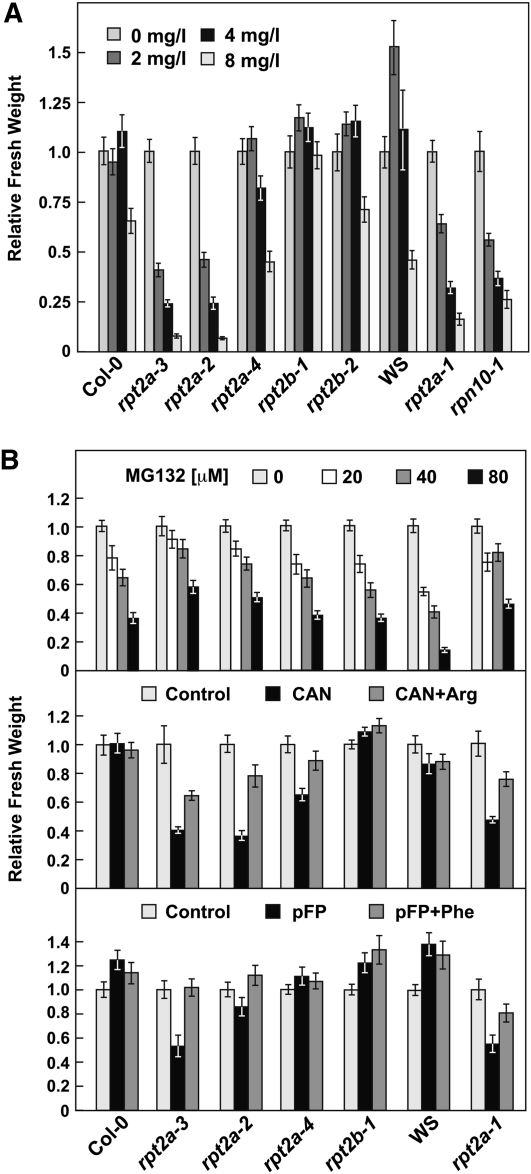
In agreement with the studies of Kurepa et al. (2008, 2009), plants homozygous for the three strong rpt2a alleles, but not rpt2b-1 or rpt2b-2, were hyposensitive to MG132 but hypersensitive to amino acid analogs, such as canavanine and p-fluorophenylalanine, whose incorporation generates abnormal proteins requiring the 26S proteasome for removal (Figure 4). Increased sensitivities to 5 mM canavanine or p-fluorophenylalanine were suppressed significantly if the rpt2a seedlings were simultaneously exposed to higher concentrations (25 mM) of the corresponding normal amino acid (Arg and Phe, respectively), demonstrating that the hypersensitivities were related to the analog’s effect on protein stability, rather than nonspecific toxicity. Like rpn10-1 mutants (Smalle et al., 2003), the strong rpt2a mutants, but not rpt2b mutants, were more sensitive to the DNA-damaging agent mitomycin C (Figure 4A). The hypersensitivity to amino acid analogs, coupled with activation of the proteasome stress regulon, raised the possibility that rpt2a mutants have higher levels of Ub-protein conjugates due to crippled 26S proteasome capacity. However, the levels of free Ub monomer, free Ub polymers, and Ub-protein conjugates appeared unchanged in rpt2a-2 versus wild-type plants, suggesting that the overall dynamics of ubiquitylation are not dramatically altered upon inactivation of RPT2a (see Supplemental Figure 7D online).
Figure 4.
Sensitivity of rpt2a Mutants to Mitomycin C, MG132, and Amino Acid Analogs.
(A) Sensitivity of rpt2 plants to mitomycin C. The relative fresh weight of at least 10 untreated versus treated plants (±se) was measured after 21 d.
(B) Sensitivity of rpt2a plants to various concentrations of MG132, 5 μM canavanine (CAN), or 5 μM p-fluorophenylalanine (pFP). For CAN and pFP, a set of plants was also exposed to the analog plus 25 μM of their respective normal amino acid, Arg and Phe. Relative fresh weight of at least 10 untreated versus treated plants (±se) was measured after 21 d.
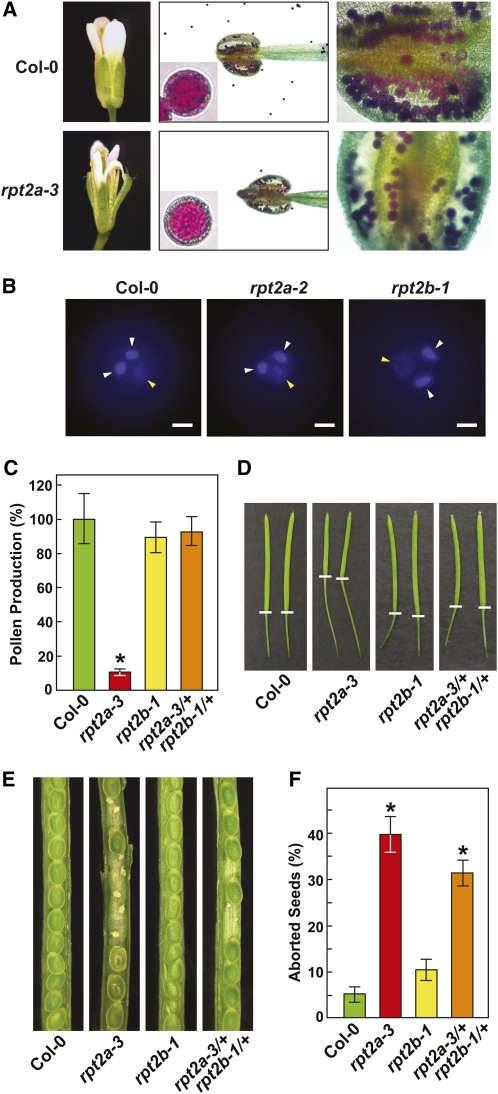
Reduced fertility for the rpt2a-1, rpt2a-2, and rpt2a-3 mutants was likely manifested at several levels. Detailed analysis of homozygous rpt2a-3 plants revealed subtle differences in flower morphology, which could disturb pollination (Figure 5A). Even though the anthers and maturing pollen appeared relatively normal, pollen production was dramatically reduced in rpt2a-3 flowers compared with those from wild-type, homozygous rpt2b-1, and double heterozygous rpt2a-3/+ rpt2b-1/+ flowers (Figure 5C). However, all mature pollen examined from rpt2a-2 anthers, like those from wild-type and rpt2b-1 anthers (at least 60 grains inspected for each), were fully expanded and contained the large tube nucleus together with the pair of smaller sperm nuclei (Figure 5B). Consequently, we conclude that the defect in pollen production in rpt2 plants occurs early in male gametogenesis, but once this block is passed, the development of the surviving pollen grains proceeds normally. Reduced fertilization of self-crossed homozygous rpt2a-3 flowers was observed by the appearance of shorter siliques that contained substantially more aborted seeds (40% aborted seeds versus just 5% for the wild type; Figures 5D to 5F). Often only the funiculus and small remnants of ovule tissue were found in the empty spaces adjacent to expanding seeds, suggesting that the lack of RPT2a impairs late female gametogenesis or early embryogenesis. We also noticed increased pedicel lengths on rpt2a-3 plants (Figure 5D), similar to that described for Arabidopsis mutants removing the RPN8b subunit of the RP lid (Huang et al., 2006).
Figure 5.
Characterization of Reproductive Organs from rpt2a-3 and rpt2b-1 Plants.
(A) Abnormal floral development and pollen production for rpt2a-3 plants. Left panels, mature flowers. Middle panels, Alexander dye staining of individual pollen grains and anthers from young flowers. Right panels, close-up pictures of the anthers stained with Alexander’s dye.
(B) The 4′,6-diamidino-2-phenylindole staining of mature pollen. The tube and sperm nuclei are indicated by the yellow and white arrowheads, respectively. Bar = 5 μm.
(C) Quantification of pollen production from homozygous rpt2a-3 and rpt2b-1 flowers and double heterozygous rpt2a-3/+ rpt2b-1/+ flowers. Each bar shows the average number of pollen grains from 10 plants assayed in triplicate (±se) compared with that from wild-type Col-0.
(D) Decreased silique elongation but increased pedicel elongation of rpt2a-3 plants. White bar marks the junction between the silique and pedicel.
(E) Homozygous mutants affecting RPT2a substantially increase seed abortion. Pictured are siliques from self-fertilized wild-type Col-0, homozygous rpt2a-3 and rpt2b-1, and heterozygous rpt2a-3/+ rpt2b-1/+ flowers.
(F) Quantification of seed abortion in self-fertilized homozygous rpt2a-3 flowers. Each bar (±se) represents the average percentage of aborted seeds from 20 siliques collected from 20 plants grown in LDs. In both (C) and (F), the asterisks indicate a significant difference between Col-0 wild type and the mutants (ANOVA followed by Tukey’s multiple comparisons post-test, P < 0.01 and P < 0.05, respectively).
RPT2 Is Essential for Male and Female Gamete Transmission
Interestingly, increased seed abortion was also seen in selfed rpt2a-3/+ rpt2b-1/+ flowers (Figures 5E and 5F), indicating that embryogenesis is impaired when total RPT2 is low or that male and/or female gametogenesis is blocked when both RPT2a and RPT2b are missing. To explore the latter scenario, we self-crossed rpt2a-3/+ rpt2b-1/+ double heterozygotes and examined progeny segregation by genomic PCR genotyping. Instead of finding the normal Mendelian distribution of wild-type and mutant alleles, we failed to find any progeny that were either homozygous for both rpt2a-3 and rpt2b-1 or heterozygous for one allele and homozygous for the other (Table 1). The absence of these allelic combinations is consistent with an inviability of double mutant gametes. To further support this possibility, we conducted reciprocal crosses using pollen from double heterozygous plants to fertilize wild-type ovules and vice versa. Genotyping of the resulting progeny in each cross failed to detect rpt2a-3/+ rpt2b-1/+ individuals, indicating that male and female gamete transmission both require at least one normal copy of RPT2 (Table 2). Thus, like several other subunits of the Arabidopsis 26S proteasome (Brukhin et al., 2005; Huang et al., 2006; Book et al., 2009, 2010), including the companion AAA-ATPase subunit RPT5 (Gallois et al., 2009), RPT2 can be defined as an essential component of the plant particle.
Table 1.
rpt2a rpt2b Double Mutants Are Inviable
| Genotypes of Progeny from Selfed AaBb Parenta | |||
| Genotype | No.b | % | Expected (%)c |
| AABB | 14 | 11 | 8 |
| AaBB | 23 | 17 | 17 |
| AABb | 35 | 26 | 17 |
| AaBb | 35 | 26 | 33 |
| aaBB | 12 | 9 | 8 |
| AAbb | 14 | 11 | 8 |
| Aabb | 0 | 0 | 17 |
| aaBb | 0 | 0 | 17 |
| aabb | 0 | 0 | 8 |
Each cross used the rpt2a-3 and rpt2b-1 alleles.
Total individuals genotyped = 133.
Expected genotypes if all combinations are viable.
Table 2.
rpt2a rpt2b Double Mutants Are Gametophytic Lethal
| Genotypes of Progeny from AaBb × Wild-Type Col-0 Crossa | ||
| Genotype | AaBb Egg × Wild-Type Pollen No. | Wild-Type Egg × AaBb Pollen No. |
| AABB | 32 | 24 |
| AaBB | 12 | 23 |
| AABb | 20 | 15 |
| AaBb | 0 | 0 |
| Total individuals genotyped | 64 | 62 |
Each cross used the rpt2a-3 and rpt2b-1 alleles.
RPT2a and RPT2b Proteins Are Functionally Equivalent
High sequence conservation (99% identity) strongly implied that RPT2a and b are functionally redundant. However, attempts by Sonoda et al. (2009) to phenotypically rescue the rpt2a-2 allele with an RPT2b cDNA expressed under the control of the RPT2a promoter failed, initially suggesting that the paralogs are not equivalent. To reexamine these results, we generated four complementation transgenes that express the full-length RPT2a or RPT2b coding regions either under the control of their own promoter or that from their paralog and introduced them into the rpt2a-2 background. The transgenes were designed to express RPT2a or RPT2b without modification of their N or C termini with epitope tags, given the potential importance of both ends to RPT2 function combined with our failure to rescue rpt2a plants with GFP-tagged versions (see above), and then were transformed into heterozygous rpt2a-2 plants to avoid the poor fertility of the homozygote. Multiple independent lines homozygous for both the rpt2a-2 allele and the transgenes were identified in selfed F2 populations by genomic PCR and confirmed to have near-wild-type levels of RPT2 protein as detected by immunoblot analysis (see Supplemental Figure 6A online).
In contrast with previous work (Sonoda et al., 2009), we found that all four of the transgenes tested (RPT2apro:RPT2a, RPT2apro:RPT2b, RPT2bpro:RPT2a, and RPT2bpro:RPT2b) rescued most, if not all, of the signature rpt2a-2 phenotypic abnormalities. The complemented rpt2a-2 plants had normal rosette leaf shape and flowering time in LDs, near-normal root length, and a wild-type distribution of trichome branch numbers regardless of whether the RPT2a or b cDNAs were connected to the RPT2a or b promoters (see Supplemental Figures 6B to 6E online). The complemented plants also had near-normal levels of other 26S proteasome subunits, indicating that the proteasome stress caused by inadequate RPT2 levels was relaxed (see Supplemental Figure 6A online). Taken together, our results imply that the RPT2a and RPT2b proteins have the same functions within the 26S proteasome and that the rpt2a phenotypes arise from insufficient expression of this subunit, which is likely caused by the weaker strength of the RPT2b promoter and its immunity to proteasome stress.
Loss of RPT2a Destabilizes the 26S Proteasome
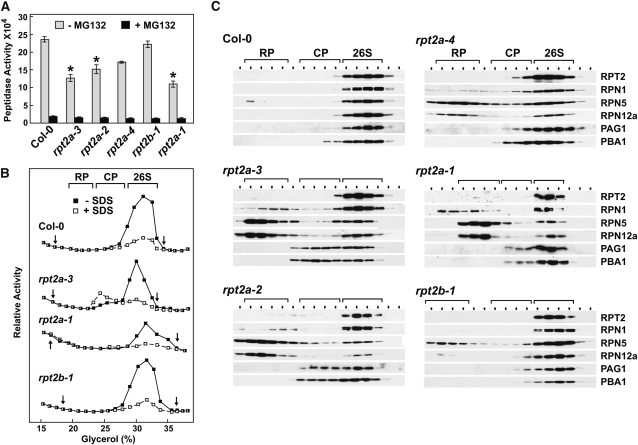
Based on the expected importance of RPT2 in completing the six-membered RPT ring and in promoting the connection between the RP base and the CP through its C-terminal HbYX motif (Rubin et al., 1998; Köhler et al., 2001; Smith et al., 2007; Park et al., 2009, 2011), we predicted that reduced RPT2 protein levels in the rpt2a backgrounds would compromise RP assembly and, thus, 26S proteasome accumulation. To test for an effect on overall proteasome activity, we assayed the collection of rpt2a/b mutants for total CP peptidase activity in the presence or absence of MG132, using the diagnostic substrate Suc-LLVY-AMC (Yang et al., 2004). In agreement with the strength of each rpt2a allele, homozygous rpt2a-1, rpt2a-2, and rpt2a-3 seedlings, but not rpt2a-4 and rpt2b-1 seedlings, had significantly less MG132-sensitive peptidase activity compared with wild-type seedlings (Figure 6A).
Figure 6.
rpt2a Mutants Destabilize the Arabidopsis 26S Proteasome.
26S proteasomes were enriched from 10-d-old liquid-grown seedlings of the indicated genotypes and subjected to glycerol gradient fractionation.
(A) Total 26S proteasome activity in crude extracts. Equal amounts of crude extracts (based on total protein) were assayed for CP peptidase activity (±MG132) using the substrate Suc-LLVY-AMC. Each bar represents the average of triplicate assays (±sd). The asterisks indicate a significant difference between peptidase activity in the Col-0 wild type and mutants in the absence of MG132 (ANOVA followed by Tukey’s multiple comparisons post-test, P < 0.01).
(B) Profile of CP peptidase activity across the glycerol gradient. Activity was measured in the absence or presence of 0.02% SDS using the substrate Suc-LLVY-AMC. The activity scale for each profile was adjusted to generate a near-equal height for the peak activity. The arrows delineate the range of fractions that were used for the immunoblot analyses in (C).
(C) Immunoblot analyses of glycerol gradient fractions with antibodies prepared against various RP (RPT2, RPN1, RPN5, and RPN12a) and CP (PAG1 and PBA1) subunits of the 26S proteasome. The locations of the 26S proteasome and the CP and RP subcomplexes, as determined by peptidase activity and immunoblot analysis, are indicated by the brackets.
To assay for defects in 26S complex assembly, we subjected proteasome-enriched fractions from mutant and wild-type seedlings to glycerol gradient centrifugation, which can resolve free CP and RP particles from the intact 26S proteasome (Glickman et al., 1998; Yang et al., 2004; Book et al., 2009). The sedimentation profiles of the complexes were then compared by peptidase activity and by immunoblot assays for various RP lid (RPN5 and RPN12a), RP base (RPN1 and RPT2), and CP (PAG1 and PBA1) subunits. The peptidase assays were also conducted with 0.02% SDS, which preferentially inhibits the 26S proteasome relative to the free CP (Yang et al., 2004).
A single proteasome peak was detected in wild-type Col-0 plants, which, based on its SDS-sensitive peptidase activity and subunit composition, represented the intact 26S particle (Figures 6B and 6C). rpt2b-1 plants had a similar profile with the exception of a smaller sized peak containing RPN5. This additional peak could reflect a modest accumulation of free RP or RPN5 integrated into other complexes related to the RP lid (e.g., the COP9 signalosome), as reported for its yeast counterpart (Yu et al., 2011). By contrast, all the rpt2a mutants had a dramatically altered profile, with a new peak evident that represents free RP and a shoulder appearing proximal to the 26S proteasome peak that contains free CP (Figures 6B and 6C). RPT2 protein was detected only in the 26S particle fractions and not in the free RP fractions. This segregation implied that the rpt2a plants assemble all available RPT2b polypeptides into the holocomplex and that the pool of remaining proteasome subunits are relegated to assembling free CP and a partial RP devoid of RPT2. The proportion of RP subunits in the free RP peak correlated with phenotypic severity of the rpt2a mutations (i.e., the rpt2a-1, rpt2a-2, and rpt2a-3 mutants had the strongest effects and the rpt2a-4 mutant had the weakest).
ATP Binding/Hydrolysis Sites and the HbYX Motif Are Important for RPT2 Activity
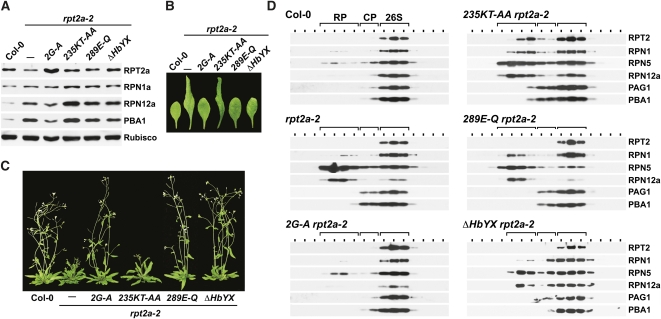
RPT2 has a number of functions/activities within the yeast 26S proteasome, including RPT ring enclosure, CP-RP docking, CP channel opening, substrate unfolding, and potential myristoylation-dependent localization of the particle (Rubin et al., 1998; Köhler et al., 2001; Boisson et al., 2003; Smith et al., 2007; Park et al., 2009). To examine how each might impact the Arabidopsis complex, we attempted to rescue rpt2a-2 seedlings with RPT2a mutant proteins potentially compromised in one or more of these activities. The 2G-A mutation removed the potential myristoylation site at Gly-2, the 235KT-AA and 289E-Q mutations disrupted the Walker A and Walker B P-loop motifs in the AAA cassette, respectively, and the ΔHbYX mutant removed the three C-terminal amino acids that help yeast RPT2 dock with the CP α-subunit ring (see Supplemental Figure 1 online). Plants homozygous for the rpt2a-2 allele and the mutant transgenes expressed under the control of the RPT2a promoter were identified in selfed F2 populations by genomic PCR (see Supplemental Figure 4C online). Multiple independent lines that increased the pool of total RPT2 protein near to that of the wild type were identified for each variant (Figure 7A; data not shown). For the 289E-Q and ΔHbYX variants, we also confirmed the accumulation of mutant protein by their slightly altered SDS-PAGE mobility compared with wild-type RPT2a (see Supplemental Figure 7A online). All four mutants expressed in the rpt2a-2 background had a normal nuclear versus cytoplasmic distribution of RPT2 and other proteasome subunits, indicating that the nuclear import of most, if not all, 26S proteasome subunits proceeds normally in these lines (see Supplemental Figure 8 online).
Figure 7.
Effects of Various Site-Directed Mutants on RPT2a Activity.
Heterozygous rpt2a-2 mutants were transformed with the mutant RPT2a transgenes expressed under the control of the RPT2a promoter and allowed to self, and then plants homozygous for the rpt2a-2 and transgene loci were identified by PCR (see Supplemental Figure 4C online).
(A) Immunoblot analysis of transgenic rpt2a-2 plants stably expressing the 2G-A, 235KT-AA, 289E-Q, and ΔHbYX variants of RPT2a. Crude protein extracts were prepared from 10-d-old plants and subjected to immunoblot analysis with antibodies against RPT2a, RPN1a, RPN12a, and PBA1. Equal protein loading was confirmed by probing the blots with anti-Rubisco antibodies.
(B) Phenotypic rescue of abnormal leaf shape. Newly expanded leaves from 7-week-old plants grown in LDs are shown.
(C) Phenotypic rescue of inflorescence development. Seven-week-old plants homozygous for the rpt2a-2 mutation without or with the rescue transgenes are shown.
(D) Integrity of the 26S proteasome in rpt2a-2 plants expressing various site-directed mutants of RPT2a. Crude extracts were separated by glycerol gradient centrifugation. The fractions were then subjected to immunoblot analysis with antibodies prepared against various RP (RPT2, RPN1, RPN5, and RPN12) and CP (PAG1 and PBA1) subunits of the 26S proteasome. The locations of the 26S proteasome and the CP and RP subcomplexes are indicated by the brackets.
Phenotypic and biochemical analyses of these mutant lines showed that only the 2G-A variant could adequately replace wild-type RPT2a. The 2G-A rpt2a-2 plants were fully rescued for rosette leaf morphology, root growth, and flowering time in LDs, were partially rescued for trichome branch numbers and root growth, showed relaxed proteasome stress as assessed by near-wild-type levels of other 26S proteasome subunits, and restored efficient assembly of the 26S particle (Figure 7; see Supplemental Figure 7 online). The collective results implied that myristoylation at the proposed Gly-2 site is not essential to the Arabidopsis 26S proteasome, but more subtle roles based on the incomplete rescue of some phenotypes cannot be ruled out.
By contrast, analysis of the 235KT-AA rpt2a-2 and 289E-Q rpt2a-2 lines revealed that most activities of RPT2 require the Walker A ATP binding motif and to a lesser extent the Walker B P-loop ATP-hydrolysis motif. The 235KT-AA transgene failed to, or poorly, rescued rpt2a-2 plants with respect to leaf morphology, flowering time, root growth, and trichome branch numbers, despite increasing the overall level of RPT2 protein (Figures 7A to 7C; see Supplemental Figures 7B and 7C online). Accumulation of the 235KT-AA protein also exacerbated proteasome stress, as indicated by an additional increase of other subunits from the proteasome stress regulon (RPN1a, RPN12a, and PBA1) compared with rpt2a-2 plants. Interestingly for 235KT-AA rpt2a-2 plants, a large amount of RPT2 protein was detected in the region containing free RP in addition to that associated with the 26S proteasome (Figure 7D). Assuming that the former pool represents the 235KT-AA variant, we conclude that it can assemble a complete RPT ring but that the resulting RP cannot bind the CP without the functional ATP binding motif from RPT2a. Conversely, expression of the 289E-Q variant rescued most of the rpt2a-2 phenotypic defects, either completely or partially (Figure 7; see Supplemental Figure 7 online). Despite the phenotypic rescue, the 289E-Q rpt2a-2 plants still displayed proteasome stress associated with inadequate RPT2, and the pool of resulting proteasomes readily dissociated into free CP and RP, indicating that assembly of the 26S complex with the 289E-Q protein remained compromised (Figures 7A and 7D).
Similar to the situation in yeast (Park et al., 2009), analysis of the ΔHbYX rpt2a-2 lines showed that the C-terminal HbYX motif of RPT2 is also important for Arabidopsis 26S proteasome assembly. Although the ΔHbYX mutant protein could fully or partially rescue many of the rpt2a-2 phenotypes (with the exception of trichome branching and proteasome stress), it did not fully stabilize the 26S particle (Figure 7; see Supplemental Figure 7 online). Substantial amounts of free RP and CP could be identified in extracts from ΔHbYX rpt2a-2 plants. The differential phenotypic effects of the 235KT-AA versus the 289E-Q and ΔHbYX mutants imply that the ATP binding activity of RPT2 has a particularly important role in RP assembly and function in planta, thus resembling yeast RPT2 in which this activity is essential (Rubin et al., 1998; Köhler et al., 2001; Smith et al., 2007).
We imagined that defective 26S proteasomes generated by incorporation of the various RPT2a mutants might globally compromise Ub conjugate turnover and thus elevate their levels, along with a corresponding reduction in the level of the free Ub monomer. However, immunoblot analysis of crude extracts from rpt2a-2 plants expressing these variants revealed that the pools of monomeric Ub, free Ub polymers, and Ub-protein conjugates were unaltered by the lack of RPT2a or the incorporation of these variants (see Supplemental Figure 7D online). Thus, the rpt2a phenotypes likely reflect more subtle effects on proteasome activity or role(s) of RPT2 independent of Ub or outside of the proteasome.
rpt2a Mutants Resemble Those Disrupting the CAF1 Complex
A survey of numerous Arabidopsis developmental mutants found a surprising phenotypic similarity between young rpt2a mutants and those eliminating components of the CAF1 complex. The heterotrimeric CAF1 complex consists of the FASCIATA1 (FAS1), FAS2, and MULTICOPY SUPPRESSOR OF IRA1 polypeptides and plays a critical role in heterochromatin structure through its activity as a histone chaperone during de novo nucleosome assembly (Ramirez-Parra and Gutierrez, 2007; Corpet and Almouzni, 2009). Whereas metazoan caf1 null mutants are lethal (Höek and Stillman, 2003; Song et al., 2007; Takami et al., 2007), comparable Arabidopsis caf1 mutants are viable but show dramatic defects related to SAM and RAM organization (Leyser and Furner, 1992; Kaya et al., 2001), perturbed cell differentiation and cell cycle progression (Exner et al., 2006; Chen et al., 2008), and misregulated genome transcription and stability (Endo et al., 2006; Kirik et al., 2006; Ono et al., 2006; Mozgová et al., 2010).
Similar to the rpt2a mutants described here and elsewhere (Ueda et al., 2004; Kurepa et al., 2009; Sonoda et al., 2009), the fas1-4 T-DNA knockdown and fas2-4 T-DNA null mutants (confirmed here by RT-PCR; see Supplemental Figure 4D online) have elongated, serrated leaves and display fasciated stems (Exner et al., 2006; Mozgová et al., 2010). They also develop trichomes with increased branch number, though this phenotype is not as strong as in rpt2a mutants (see Supplemental Figures 5A and 5B online; Exner et al., 2008), and like rpt2a mutants have stunted root growth (see Supplemental Figure 5C online). The increased trichome branching for both fas1/2 and rpt2a mutants appears to be causally related to increased ploidy driven by extra rounds of endoreduplication and in both cases is sensitive to exogenous Suc (Exner et al., 2008; Kurepa et al., 2009; Sonoda et al., 2009). As a further confirmation, we found that null fas1-4 and fas2-4 seedlings are hypersensitive to mitomycin C and canavanine but less sensitive to MG132, like the strong alleles of rpt2a (see Supplemental Figure 9 online). As expected for subunits in a singular complex, fas1 fas2 double mutants have the same phenotype as the single mutants (Exner et al., 2006). However, we note that the inflorescence morphology of adult fas1-4 and fas2-4 plants is distinct from rpt2a plants, indicating that the developmental defects induced by inactivation of these two complexes are not completely overlapping (see Supplemental Figure 10 online).
To examine whether the CAF1 complex and RPT2 work in parallel or are genetically additive, we attempted to generate double homozygous lines containing fas1-4 or fas2-4 with rpt2a-3 or rpt2b-1 by self-crosses of double heterozygous plants. As shown in Figure 8A, all four double homozygous mutants are viable. Homozygous fas1-4 rpt2b-1 and fas2-4 rpt2b-1 seedlings were detected in the selfed progeny near the expected 1/16 segregation frequency (eight out of 115 for fas1-4 rpt2b-1 and six out of 98 for fas2-4 rpt2b-1) and were phenotypically indistinguishable to single homozygous fas1-4 or fas2-4 seedlings. However, homozygous fas1-4 rpt2a-3 and fas2-4 rpt2a-3 combinations displayed dramatically compromised development and were rare based on this phenotype in segregating populations from selfed double heterozygous parents (four out of 216 and three out of 214, respectively). The double mutant seedlings that did germinate arrested cotyledon expansion and root development soon after emergence from the seed and accumulated high concentrations of anthocyanins in the cotyledons, which is indicative of severe stress (Figure 8A). The fas1-4 rpt2a-3 and fas2-4 rpt2a-3 seedlings failed to generate true leaves or reproductive structures beyond the cotyledons even after months of growth. Collectively, the phenotypes implied that the CAF1 complex and RPT2 play synergistic roles in Arabidopsis seedling development.
Figure 8.
rpt2a-3 and fas Mutants Have a Synergistic Effect on Arabidopsis Development and Have Reduced Histone Levels.
(A) Phenotype of 2-week-old fas1-4 rpt2a-3 and fas2-4 rpt2a-3 double mutant seedlings compared with wild-type Col-0, rpt2a-3, fas1-4, and fas2-4 single and rpt2b-1 fas1-4 and rpt2b-1 fas2-4 double mutant seedlings grown on agar in continuous light.
(B) Immunoblot analysis of the various fas and rpt2 mutants for histones (H1, H2B, and H3) and various subunits of the 26S proteasome. Crude protein extracts were prepared from 10-d-old plants and subjected to immunoblot analysis for the indicated proteins. Equal protein loading was confirmed by probing the blots with anti-PUX1 and anti-Rubisco antibodies.
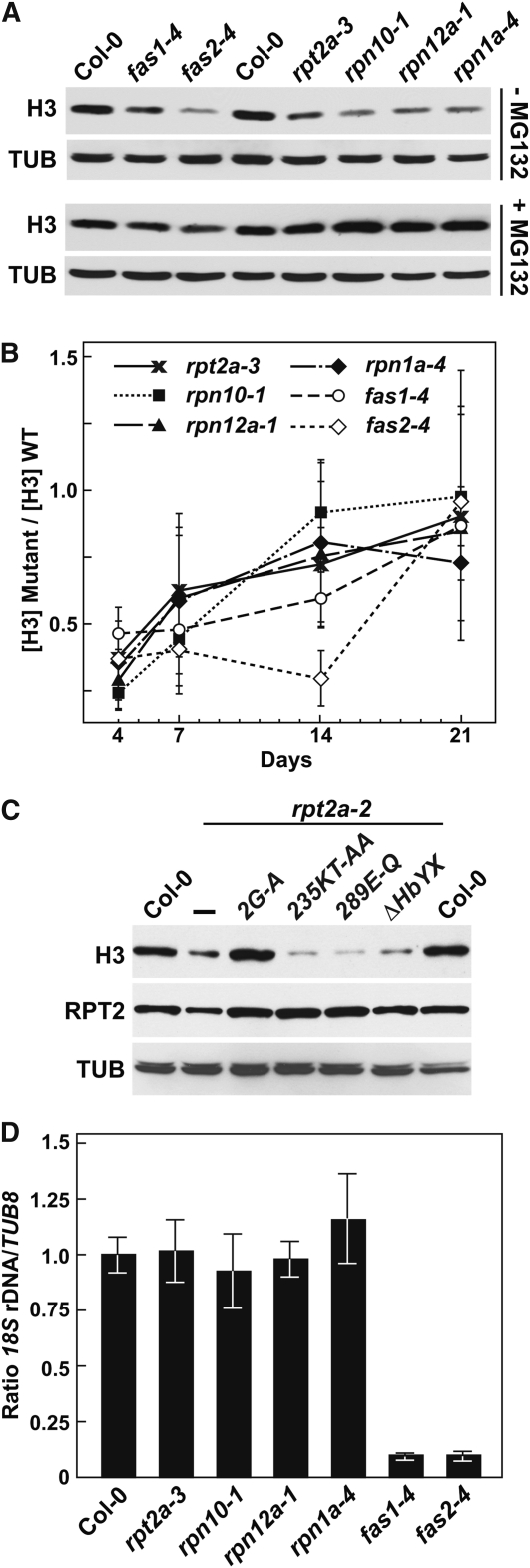
The primary function of the CAF1 complex is to help chaperone histones H3 and H4 from the cytoplasm into the nucleus and then promote their integration into the octameric nucleosome core (Corpet and Almouzni, 2009). As predicted, we found that fas1-4 and fas2-4 seedlings have reduced histone levels (Figure 8C). Similar analysis of the rpt2a-3 and rpt2b-1 mutants revealed that insufficient amounts of RPT2 also compromise histone accumulation. Immunoblot analysis of 10-d-old seedlings showed that the levels of the nucleosome core histones H2B and H3 and the linker histone H1 were reduced approximately threefold in rpt2a-3 seedlings compared with wild-type and rpt2b-1 seedlings (Figure 8C). Decreased histone levels in fas1-4, fas2-4, and rpt2a-3 plants could have arisen by reduced expression of the corresponding genes. However, both RT-PCR and quantitative real-time PCR demonstrated that none of the three mutations affected histone H3 transcript abundance, thus implying that posttranscriptional events were responsible (see Supplemental Figures 4E and 4F online). The similar effects on histone abundance in the rpt2a and fas1/2 mutants were contrasted by their differential effects on the proteasome stress regulon. Unlike the rpt2a-3 mutant, the fas1-4 and fas2-4 mutants did not upregulate the levels of various proteasome subunits nor that of the CP regulator PA200 (Figure 8C).
Other RP Mutants Affect Histone Accumulation
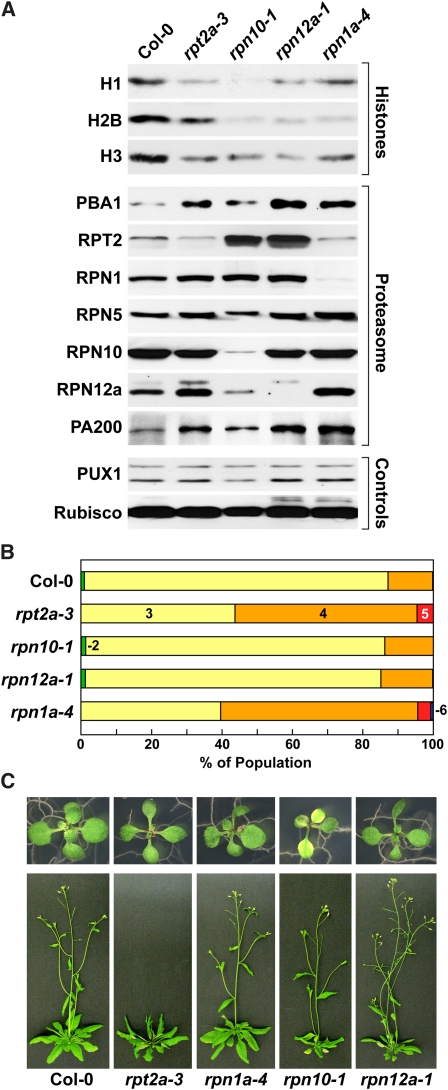
Some of the developmental abnormalities associated with rpt2a and fas1/2 mutations are shared with those affecting other RP subunits of the Arabidopsis 26S proteasome, notably leaf shape, trichome branching, root length, and MG132 hyposensitivity (Smalle et al., 2002, 2003; Huang et al., 2006; Book et al., 2009; Gallois et al., 2009; Kurepa et al., 2009). Consequently, it was possible that other RP subunit mutants, and not just those affecting RPT2 specifically, would dampen histone accumulation. To test this scenario, we compared the histone H1, H2B, and H3 levels in homozygous rpn10-1, rpn12a-1, and rpn1a-4 seedlings to those in rpt2a-3 seedlings. In each case, the homozygous line was confirmed immunologically by the absence or reduced amounts of the corresponding RPN/RPT protein (Figure 9A). Surprisingly, all the mutants also contained less of the three histones. As such, the entire RP complex, not just RPT2, likely contributes to histone dynamics.
Figure 9.
RP Mutants Have Differential Effects on Histone Levels and Arabidopsis Development.
(A) Immunoblot analysis of the various RP mutants for histones (H1, H2B, and H3) and various subunits of the 26S proteasome. Crude protein extracts were prepared from 10-d-old plants and subjected to immunoblot analysis for the indicated proteins. Equal protein loading was confirmed by probing the blots with anti-PUX1 and anti-Rubisco antibodies.
(B) Quantification of trichome branching. Each data set from 2-week-old seedlings represents the percentages of the total trichome population with specific branch numbers: 2 (green), 3 (yellow), 4 (orange), 5 (red), and 6 (blue) branches.
(C) Growth phenotypes of various RP mutants. Two- and five-week-old plants grown in LDs are shown.
Despite this common contribution to histone accumulation, the phenotypes of the RP mutant collection were not completely coincident. For example, whereas the rpt2a-3 and rpn1a-4 plants had increased trichome branch numbers, branching of the rpn10-1 and rpn12a-1 plants resembled the wild type (Figure 9B). Only rpt2a-3 plants had altered flowering time in LDs, while the rpn10-1 plants were the only RP mutants that display accelerated senescence in LDs (Figure 9C; Smalle et al., 2003).
In the absence of changes to histone H3 mRNA levels, the collection of RP mutants and/or fas1-4 and fas2-4 could dampen histone protein accumulation by accelerating their turnover, especially for the free forms. At least for the RP subunits, one possibility was increased breakdown by alternative proteasome complexes (such as the CP alone or with its PA200 regulator) that accumulate upon RP inactivation (Kurepa et al., 2008; Book et al., 2010). To test this possibility, we examined histone H3 levels in 7-d-old seedlings from the various rpn/rpt mutants with or without a 36-h pretreatment with 100 μM MG132. Whereas MG132 mildly increased H3 levels in the fas1-4 and fas2-4 lines, it increased H3 levels substantially in the rpt2a-3, rpn1a-4, rpn10-1, and rpn12a-1 lines (Figure 10A). In fact, MG132 elevated histone H3 content in the collection of RP mutants above that seen in the wild type, suggesting that the histones are degraded by a proteasomal route that is upregulated upon RP inactivation. We also noticed that the effect of the various fas1/2 and rpn/rpt mutants on histone H3 content was seedling age dependent, with the greatest effect seen in young seedlings. Whereas 4-d-old fas1/2 and rpn/rpt mutant seedlings had approximately threefold less histone H3 compared with the wild type, histone H3 levels were near equal to the wild type for all the mutants at 21 d (Figure 10B).
Figure 10.
RP and fas Mutants Have Differential Effects on Histone and 45S rDNA Levels.
(A) Restoration of histone H3 levels by exposing rpn/rpt mutant seedlings to MG132. Plants were grown on GM liquid medium for 5 d before incubation with 100 μM MG132 for 36 h. Crude extracts were immunoblotted for histone H3. Equal protein loading was confirmed by probing the blots with an anti-α-tubulin (αTUB) antibody.
(B) Recovery of histone H3 during seedling growth. Whole seedlings grown for 4, 7, 14, and 21 d were analyzed for the ratio of histone H3/α-tubulin (αTUB) as measured by immunoblot analysis of crude extracts. Each bar represents the average of triplicate assays (±sd). WT, wild type.
(C) Ability of various site-directed mutants of RPT2a to restore histone H3 levels. Crude extracts from 7-d-old wild-type Col-0, rpt2a-2, and rpt2a-2 seedlings expressing the various RPT2a mutations were immunoblotted with antibodies against histone H3 and RPT2a and the α-tubulin (αTUB) control. Mutant lines are described in Figure 7.
(D) Relative quantity of 45S rDNA evaluated by quantitative PCR in RP and fas mutants. Genomic DNAs were isolated from 4-d-old seedlings from 30 independent plants and subjected to quantitative PCR using 18S rDNA gene-specific primers (Mozgová et al., 2010). The signals were normalized by quantitative PCR with the TUB8 gene. Each bar indicates the average of triplicate assays (±sd).
Reduced histone levels in the collection of RP mutants could have been induced by inadequate levels of the assembled 26S proteasome particle or increased amounts of free RP acting in a CP-independent manner. To test these possibilities, we examined histone H3 accumulation in rpt2a-2 seedlings expressing the collection of RPT2a site-directed mutants that should compromise RPT2a activity (235KT-AA and 289E-Q) and/or its docking with the CP (ΔHbYX) (as shown in Figure 7 and Supplemental Figure 7 online). Whereas introduction of the RPT2a 2G-A mutant restored histone H3 levels back to that observed in wild-type plants, concomitant with a restoration of 26S particle assembly, none of the other three RPT2a mutants were comparably effective (Figure 10C). In fact, the least effective mutants were 289E-Q and ΔHbYX that interfere with 26S proteasome assembly, but in the case of the ΔHbYX mutant, should be benign with respect to its ATPase-directed activities. Taken together, it appears that the defects in histone accumulation for rpt2a plants are driven by impaired 26S proteasome assembly and not increased amounts of free RP lid and/or base working in another capacity.
CAF1 mutants also generate other chromosomal abnormalities that contribute to genome instability, including telomere shortening and loss of 45S rDNA repeats (Mozgová et al., 2010). Whereas we confirmed by quantitative PCR that the fas1-4 and fas2-4 mutants have dramatically reduced levels of 45S rDNA repeats, this problem was not detected with any of the RP mutants tested (Figure 10D). Thus, the RP mutants do not share all the genome abnormalities elicited by disruption of the CAF1 complex.
DISCUSSION
Despite the importance of the 26S proteasome to protein turnover, the organization of the RP subparticle and the function(s) of most of its subunits remain poorly understood. The best characterized is the heterohexameric AAA-ATPase RPT ring comprising part of the RP base. In yeast, this ring tethers the CP and RP subcomplexes, opens the axial CP channels, and then helps translocate unfolded substrates into the CP proteolytic chamber for breakdown (Finley, 2009). Whereas all RPT subunits are essential in yeast (Rubin et al., 1998), their necessity and functions in plants are largely unresolved, mainly because most subunits are expressed from duplicated genes, thus complicating genetic analyses (Fu et al., 1999; Shibahara et al., 2004; Book et al., 2010). To provide insights into the role(s) of each RPT subunit and test for possible nonredundant activities for the paralogous genes, we investigated the RPT2 subunit, which is encoded by two Arabidopsis loci. This subunit was expected have a special role in the RPT ring based on (1) its strong sequence conservation among eukaryotes (Fu et al., 1999), (2) its failure to allow cross-complementation between species (Fu et al., 1999; Li et al., 2002), and (3) the specific requirement of its ATPase activity for robust proteolysis by the 26S particle (Rubin et al., 1998; Köhler et al., 2001). From an in-depth genetic analysis of the RPT2a and RPT2b loci, we show here that the RPT2 subunit also has an essential role in Arabidopsis development and 26S proteasome assembly, along with an unexpected connection between RPT2 (and other RP subunits) and histone dynamics.
Prior work showed that the RPT2a locus has a particularly important role in Arabidopsis, with null mutants affected in root elongation, leaf/organ size, trichome branching, endoreduplication, and displaying a hyposensitivity to MG132 and zinc deficiency but a hypersensitivity to amino acid analogs (Ueda et al., 2004, 2011; Kurepa et al., 2008, 2009; Sonoda et al., 2009; Sako et al., 2010; Sakamoto et al., 2011). We extended this phenotypic description to also include roles for RPT2a in inflorescence stem fasciation, flowering time, fertility, and the sensitivity to the DNA damaging agent mitomycin C. Low fertility for homozygous null rpt2a lines arises from defects in flower morphology, reduced pollen production, and likely increased seed abortion. Malformed pollen grains or normal grains with less than the three expected nuclei were not observed in dehiscent anthers, indicating that the defect in male gametogenesis occurs early. From the analysis of rpt2a rpt2b double mutants, we further showed that the RPT2 subunit is essential to gamete function. Whereas rpt2a null mutants could still reproduce and rpt2b null mutants were without strong phenotypic consequences, double homozygous mutants could not be generated. Both the segregation analysis of progeny from selfed double heterozygous (rpt2a-3/+ rpt2b-1/+) plants and reciprocal crosses of heterozygous plants with the wild type failed to detect the simultaneous transmission of both mutations via the male or female gametes. Recent detailed microscopy analyses of male and female gametes from rpt2a-1/+ rpt2b-2/+ plants by Ueda et al. (2011) corroborated our findings, with additional data showing that problems in gametogenesis likely underpin the transmission defects. Thus, similar to the situation in yeast (Rubin et al., 1998), the Arabidopsis RPT2 subunit cannot be substituted in the RPT ring by any of the five remaining RPT members. Based on the defects in both male and female gametogenesis, cell size, and endoreduplication (Kurepa et al., 2009; Sonoda et al., 2009; this report), RPT2, like other RP subunits (RPN1, RPN5, RPN10, and RPN12a), plays a critical role during mitosis and meiosis in plants via its action within the RP.
In contrast with a previous study by Sonoda et al. (2009), we, like Ueda et al. (2011), demonstrate that the RPT2a and RPT2b proteins are functionally equivalent, in agreement with their strong sequence conservation (99% identity). Expression of the RPT2b cDNA under the control of its own promoter or that from RPT2a fully rescued the rpt2a-2 mutant phenotype. The question then arises as to why only the rpt2a single mutant plants have such strong developmental phenotypes. One explanation relates to expression strength of the two loci. Because the RPT2a gene is expressed approximately fourfold higher than RPT2b, we propose that only rpt2a plants fail to accumulate total RPT2 protein above a critical threshold needed to avoid phenotypic problems. A second possibility relates to the nonresponsiveness of the RPT2b locus to proteasome insufficiency (Figures 1D and 1E). Whereas rpt2b plants can upregulate RPT2a expression if needed via the proteasome stress regulon to overcome low RPT2 levels, rpt2a plants cannot upregulate RPT2b expression similarly. An identical situation likely exists for the RPN1a/RPN1b, RPN5a/RPN5b, and RPT5a/RPT5b gene pairs, where inactivation of the more highly expressed locus generates strong phenotypic defects even though the encoded proteins are, for the most part, functionally equivalent (Brukhin et al., 2005; Book et al., 2009; Gallois et al., 2009). Taken together, the strong phenotypic consequences of the rpt2a, rpt5a, rpn1a, and rpn5a mutants highlight the pivotal roles of expression strength and the proteasome stress regulon in coordinating the appropriate accumulation of each 26S proteasome subunit and the importance of sufficient 26S proteasome levels to normal plant physiology and development.
Complementation analyses of mutants disrupted in key motifs revealed that Arabidopsis RPT2 has a similar importance to the plant 26S proteasome as does its yeast counterpart. Substitutions in the Walker A ATP binding (235KT-AA) and the Walker B P-loop ATP-hydrolysis (289E-Q) sites and deletion of the CP docking HbYX motif (ΔHbYX) individually generated proteasomes with impaired assembly and/or stability, at least in vitro. Whereas 289E-Q and ΔHbYX mutants rescued most rpt2a-2 phenotypes, the 235KT-AA mutant did not, indicating that the ATP binding activity in RPT2 is especially critical for full function of the RPT ring, even though the five other RPT subunits could theoretically provide this activity. This selective importance likely reflects, at least in part, the key role for RPT2 in RP-CP docking and CP pore opening (Rubin et al., 1998; Köhler et al., 2001). Indeed, recent studies on the yeast 26S proteasome revealed the selective importance of RPT2 in stabilizing the CP/RP contact (Tian et al., 2011). Its HbYX extension binds to a pocket between α3 and α4 subunits of the CP, thereby fixing one-half of the asymmetric RPT-ring/α-ring interface. This contact, along with ATP hydrolysis, is proposed to allow RPT2 to displace the N-terminal tail of the α3 subunit, which is a key feature of the α-ring gate that controls substrate entry into the CP lumen. Interestingly, the proteasome stress regulon appears to be hyperactivated in 235KT-AA rpt2a-2 plants, suggesting that the 235KT-AA protein inhibits 26S proteasome activity dominantly when incorporated into the RPT ring. Despite the phenotypic abnormalities, it is worth noting that the pools of free Ub, free Ub polymers, and Ub-protein conjugates remained unaltered in the rpt2a-2 background with or without complementation with any of the site-directed RPT2a mutants. This result implies that the effects of rpt2a mutants on Ub dynamics might be compensated by other CP particles, such as the free CP or the CP-PA200 complex (Kurepa et al., 2008; Book et al., 2010), or the myriad of deubiquitylating enzymes in Arabidopsis.
RPT2 has a predicted N-myristoylation site within an MGXXX(S/T) consensus sequence (Sorek et al., 2009) and some (Shibahara et al., 2002; Boisson et al., 2003), but not other (Book et al., 2010), mass spectrometry studies have detected the RPT2-myristic acid adduct in the plant 26S particle. Complementation studies with the RPT2a 2G-A mutant implied that myristoylation, if it occurs, is not essential for most phenotypic functions of RPT2, assembly of the RP with the CP, or for nuclear/cytoplasmic partitioning of the 26S particle. However, we note that the 2G-A rpt2a-2 rescued plants were not completely wild-type for trichome branch numbers and root length despite accumulating high levels of the RPT2a 2G-A protein, suggesting that this modification and its potential effect on 26S proteasome localization might still be relevant for a subset of 26S particles.
A surprising conclusion to our genetic analyses of RPT2a was the coherence of many rpt2a null mutant phenotypes with those eliminating the FAS1 and FAS2 subunits of the CAF1 complex involved in assembling histones H3 and H4 into nucleosomes. These included effects observed here and elsewhere on root growth, stem fasciation, leaf serration, trichome branching, endoreduplication, hyposensitivity to MG132, and a hypersensitivity to amino acid analogs and DNA-damaging agents (Leyser and Furner, 1992; Kaya et al., 2001; Ueda et al., 2004; Exner et al., 2006, 2008; Kurepa et al., 2008, 2009; Sonoda et al., 2009; Mozgová et al., 2010; Sako et al., 2010). As expected for a role of the CAF1 complex in de novo nucleosome assembly, we found that fas1-4 and fas2-4 plants have severely reduced levels of core (H2B and H3) and linker (H1) histones. Consistent with the phenotypic similarities, we discovered that rpt2a-3 but not rpt2b-1 seedlings also have reduced H1, H2B, and H3 levels. We imagine that the increased ploidy observed for rpt2a plants, especially in trichome cells (Kurepa et al., 2009; Sonoda et al., 2009; Sako et al., 2010), would further exacerbate the effect of low histone levels on the proper packaging of DNA into nucleosome-condensed chromatin.
Presumably, a number of chromatin-associated functions would then be compromised by altered histone dynamics, including cell division, cell/tissue differentiation, and SAM and RAM maintenance, which might underlie many of the observed rpt2a phenotypes. For example, several studies have shown that altered histone activities, generated by either histone H3 trimethylation, Ub-mediated H2B turnover, or insertion of the histone H2A variant H2A.Z, profoundly affect flowering time in Arabidopsis via modification of key loci such as FLOWERING LOCUS C (Deal et al., 2007; Cao et al., 2008; Tamada et al., 2009). Whereas the repressive trimethylation and ubiquitylation histone modifications can accelerate flowering time, presence of the H2A.Z variant or the reduced histone levels in the caf1 and rpn/rpt mutants observed here can be connected to delayed flowering. The intriguing possibility that caf1 and rpn/rpt mutants specifically alter the dynamics of ubiquitylated histone H2B is currently under investigation. However, we emphasize that not all fas1/2 phenotypes are shared with rpt2a mutants (e.g., inflorescence morphology and 45S rDNA replication [this report; Mozgová et al., 2010]), indicating that the CAF1 complex also works in processes outside of those involving RPT2, the RP, or the 26S proteasome, and vice versa.
Additional analysis with other RP mutants showed that impaired histone accumulation is not specific for RPT2 but might reflect a general defect in RP function. Taken together, the data raise the possibility that many of the phenotypes common among the various RP mutants, like those induced by CAF1 complex mutations, are related to altered histone dynamics, leading to reduced nucleosome levels. Such altered dynamics might be expected to have numerous consequences, including increased chromatin instability, altered accessibility of the transcriptional machinery to genes, and impaired DNA replication and repair. However, it should be emphasized that we could at least partially uncouple some of the rpt2a phenotypes from histone levels. While leaf and inflorescence development was largely rescued by introduction of the 289E-Q and ΔHbYX variants into the rpt2a-2 background, the effects on histone H3 accumulation were not.
Whereas low histone levels in CAF1 mutants can be easily explained based on the role of the complex in nucleosome assembly, why RP mutants have less histones appears counterintuitive, given that they should increase protein levels by attenuating 26S proteasome–mediated turnover. Our observations that the fas1-4 rpt2a-3 and fas2-4 rpt2a-3 double mutants are much more strongly compromised than the single mutants suggest that the CAF1 and RP regulate a common outcome despite diametrically opposite activities. The most parsimonious explanation is that the RP as part of the 26S proteasome together with the CAF1 complex are required for robust nucleosome recycling. In the absence of the 26S proteasome, other less-regulated proteasomes accumulate to degrade histones more indiscriminately. These alternative proteasomes could include the free CP and the CP-PA200 complex that work independently of Ub and whose levels rise when the 26S particle is limiting (Yang et al., 2004; Kurepa et al., 2008; Gallois et al., 2009; Book et al., 2010; this report). Blocking their action by MG132 might explain how histone H3 levels rise back to wild-type levels when rpt2a-2 seedlings are exposed to the inhibitor. An alternative scenario is that increased levels of free RP particles driven by CP-RP instability enhance histone turnover directly by accelerating their extraction from chromatin (Gillette et al., 2001; Gonzalez et al., 2002). However, our failure to increase histone H3 levels in rpt2a-2 plants by expressing the ΔHbYX variant that should promote the accumulate of free RP subparticles but not abrogate the assembly or AAA ATPase unfoldase functions of the RPT ring argue against this possibility. Whatever the mechanism, it is notable that histone H3 levels rise back to near that of the wild type in older rpt2a, fas1, and fas2 plants, suggesting that the dynamics of nucleosome assembly/disassembly slow as plants mature, possibly in line with slower cell division rates.
More generally, a role for the entire 26S proteasome in histone recycling is bolstered by studies with yeast and metazoan histones, which showed that most, including H3 and H4, are polyubiquitylated in vivo and degraded via a Ub-dependent process and that the levels of ubiquitylated histones are strictly controlled to block negative effects on chromatin stability (Collins et al., 2004; Singh et al., 2009; Lopes da Rosa et al., 2011). We and others have confirmed that Arabidopsis histones H1 and H2B are ubiquitylated, thus implying that a similar mode of regulation exists in plants (Fleury et al., 2007; Liu et al., 2007; Sridhar et al., 2007; Saracco et al., 2009). Such a role for the 26S proteasome in nucleosome cycling may explain the pleiotropic phenotypes generated by a number of RP subunit mutants and further highlights the importance of the UPS in chromatin dynamics.
METHODS
Plant Material and Growth Conditions
The three T-DNA insertion alleles of RPT2a (rpt2a-2, SALK_005596; rpt2a-3, SALK_130019; rpt2a-4, SALK_135391) and the two T-DNA insertion alleles of RPT2b (rpt2b-1, SALK_043450; rpt2b-2, SALK_052875) in the Arabidopsis thaliana ecotype Col-0 were generated by T-DNA insertional mutagenesis (Alonso et al., 2003) and obtained from the ABRC. Homozygous lines were derived from self-fertilized heterozygous plants following at least two backcrosses to the Col-0 wild type using genomic PCR with gene- and T-DNA–specific primers to track the mutants. Primers used for genotyping were P1 (T-DNA left border), P2 and P3 (rpt2a-2), P4 and P5 (rpt2a-3), P6 and P7 (rpt2a-4), P8 and P9 (rpt2b-1), and P10 and P11 (rpt2b-2). (Oligonucleotide primers used throughout this study are listed in Supplemental Table 1 online.) The hlr-1 deletion allele of RPT2a in the Ws background (Ueda et al., 2004) was provided by Kiyotaka Okada (Kyoto University, Japan). The rpn10-1 and rpn12a-1 exon-trap lines in the C24 background were as previously described (Smalle et al., 2002, 2003), as were the rpn1a-4, fas1-4, and fas2-4 lines in the Col-0 background (Exner et al., 2006; Wang et al., 2009). Introgression of the various mutants was accomplished by crossing; homozygous lines were then identified by PCR genotyping of individuals from segregating F2 populations.
For most assays, seeds were surface sterilized, exposed to 4°C in the dark for 3 d, and then germinated on 1% agar containing half-strength Murashige and Skoog medium (Sigma-Aldrich), 1% Suc, and 0.05% MES, pH 5.7. Seedlings were transferred to soil 2 to 3 weeks after germination and grown to maturity at 22°C under continuous white light or either LD (16 h light/8 h dark) or SD (8 h light/16 h dark) photoperiods. Sensitivity to mitomycin C, canavanine, p-fluorophenylalanine (Sigma-Aldrich), and MG132 (benozylcarbonyl-Leu-Leu-Leu-al; Enzo Life Sciences) was determined by measuring the fresh weight of 21-d-old seedlings grown on Murashige and Skoog medium containing various concentrations of the inhibitors.
Trichome branching was examined on the middle region of the first and second rosette leaves from at least 20 14-d-old plants grown on solid Gamborg’s B5 medium (GM; Sigma-Aldrich) in LDs. Images of representative trichomes were collected with an FEI Quanta 200 environmental scanning electron microscope. Fresh tissue was placed on a rotating stage set to cooling (4°C), and samples were scanned at 25 kV under 3.11 Torr pressure. Pollen development was examined by Alexander staining of anthers just prior to dehiscence (Gallois et al., 2009). The relative number of pollen grains per plant was determined by vortexing 10 anthers from individual plants in 200 μL of Alexander staining solution (10 mL 95% ethanol, 1 mL 1% malachite green [in 95% ethanol], 5 mL 1% fuchsin acid [in water], 0.5 mL 1% orange G [in water], 5 g phenol, 5 g chloral hydrate, 2 mL glacial acetic acid, 25 mL glycerol, and 50 mL water [Alexander, 1969]). The number of pollen in 10 μL of this solution was then counted microscopically. Anthers from 10 different plants were assayed for each genotype. Seed abortion was assessed from 20 individual siliques harvested from 20 different plants per genotype. Pollen nuclei staining with 4′,6-diamidino-2-phenylindole was performed as previously described (Doelling et al., 2007).
Analysis of RPT2a/b, FAS1/2, and Histone H3 Expression
For RT-PCR, total RNA was extracted from leaves of 4-week-old plants using Trizol reagent (Gibco BRL) according to the manufacturer's protocol, treated with RQ1 DNase (Promega), and then isolated by phenol/chloroform/isoamyl alcohol precipitation. RNA was reverse transcribed using Superscript II reverse transcriptase (Invitrogen) with an oligo(dT) primer. The RT products were then subjected to PCR using pairs of gene-specific primers for RPT2a (P12 and P14, P13 and P16, P13 and P17, and P15 and P17), RPT2b (P18 and P20, P19 and P21, and P22 and P23), PAE2 (P24 and P25), FAS1 (P26 and P27), FAS2 (P28 and P29), and UBC21 (P30 and P31). Quantitative real-time PCR amplification of the histone H3 gene (At5g10400) was performed with the MyiQ single-color real-time PCR detection system using iQ SYBR Green Supermix (Bio-Rad) and the primer pair P32 and P33. Relative expression was calculated by the comparative threshold cycle method using reactions with the TUB8 transcript (At5g23860) as an internal control (primers P34 and P35).
Fusions between the promoter regions of RPT2a/b and the coding region of GUS were generated by PCR amplification of promoter fragments from genomic Col-0 DNA using primers that were designed to end immediately upstream of the translation start sites for RPT2a (P36 and P12) and RPT2b (P37 and P18). The products (2131 bp for RPT2a and 2004 bp for RPT2b) were cloned in-frame upstream of the GUS coding region present in the pCAMBIA3301 vector (Curtis and Grossniklaus, 2003), which was then introduced into wild-type Col-0 plants by the floral dip method using the Agrobacterium tumefaciens strain GV3101 (Smalle et al., 2002). Basta-resistant seedlings were screened for GUS expression by histochemical staining with the substrate 5-bromo-4-chloro-3-indolyl-β-d-glucuronic acid (X-Gluc; Sigma-Aldrich). At least 30 independent insertion lines were examined for each construction to avoid artifacts generated by the site of transgene insertion. Stained microscopic images were collected after an overnight exposure to X-Gluc. To compare expression strength of the RPT2a and RPT2b promoters, GUS activity was quantified in crude extracts prepared from 10-d-old seedlings by a fluorescence-based MUG assay (Walker and Vierstra, 2007). The average GUS expression was determined from the analysis of 30 independent transgenic lines for each promoter. For each line, T1 progeny seedlings, obtained from independently transformed T0 plants, were grown for 10 d in solid GM medium under LDs. Twenty T1 seedlings were homogenized together for each assay, with the assay then performed in triplicate. The effects of 100 μM MG132 on RPT2a/b and RPN5a/b expression was determined by MUG assay using three independent lines for each promoter:GUS fusion generated above or from Book et al. (2009), respectively.
Immunoblot Analyses
Antibodies against RPT2a, RPN1a, RPN5a, RPN10, RPN12a, PAC1, PAG1, PBA1, PA200, Ub, ribulose-1,5-bis-phosphate carboxylase/oxygenase (Rubisco), and PUX1 were as described (van Nocker et al., 1996; Smalle et al., 2002; Rancour et al., 2004; Yang et al., 2004; Book et al., 2010). Antigenicity of the anti-RPT2a antibodies against RPT2a and RPT2b was tested with recombinant N-terminal 6His-tagged proteins expressed from full-length cDNAs. The cDNAs were amplified from RT-PCR reactions using primers P38 to P41 and introduced into the pRSET A plasmid (Invitrogen). Immunoblot analysis with anti-5His antibodies (Qiagen) was used to confirm equal loading. Antibodies against histones H2B and H3 were purchased from Abcam (AB1790 and AB1791, respectively), whereas antibodies against histone H1 were obtained from Millipore (05-457, clone AE-4). Immunoblot analyses were performed as according to Book et al. (2010). Primary antibodies were visualized with horseradish peroxidase–conjugated goat anti-rabbit antibodies, and radiographic signals were detected with Classic Autoradiography Film (MidSci), with exposure times kept within the linear range of the film. Immunoblot quantifications of histone H3 levels were determined by densitometric scans of the blots, using the signals generated with an anti-αTUB mouse monoclonal antibody (T9026; Sigma-Aldrich) as the loading control.
Fractionation of Nuclei and Cytosol
Fractions enriched for nuclei and cytosol were prepared from 10 g of 7-d-old wild-type or rpt2a mutant seedlings grown in liquid GM using the Percoll gradient method (Folta and Kaufman, 2006) with modifications as previously described (Farmer et al., 2010). ATP (10 mM) was included throughout the protocol. Approximately equal amounts of the crude and soluble fractions and an ~40-fold concentrated nuclear fraction (as determined by Bradford assay; Bio-Rad) were mixed with SDS-PAGE sample buffer and subjected to SDS-PAGE and immunoblot analysis as above. Antibodies against histone H3 and PUX1 were used as markers for the nucleus and cytosol, respectively (Farmer et al., 2010).
Glycerol Gradient Fractionation
The 26S proteasome was partially purified from 10-d-old seedlings grown in liquid GM as previously described (Book et al., 2009). Plants were frozen to liquid nitrogen temperatures, pulverized, and ground in 1.25 volumes of Buffer A (20 mM Tris-HCl, pH 7.5, 10% glycerol, 2 mM ATP, 5 mM MgCl2, 1 mM DTT, 10 mM phosphocreatine, and 1 mg/mL creatine phosphokinase). Crude protein extracts were filtered through two layers of Miracloth, clarified for 20 min at 30,000g, adjusted to 10% in polyethylene glycol 8000, and mixed for 30 min at 4°C. The precipitate was collected by centrifugation at 12,000g for 15 min and resuspended in 300 μL of Buffer A. Following clarification, the resuspended pellet was loaded on top of an 11-mL 10-40% glycerol density gradient in Buffer A and centrifuged at 100,000g for 18 h at 4°C. Fractions (0.5 mL) were collected and assayed for proteasome activity with the substrate succinyl-Leu-Leu-Val-Tyr-7-amino-4-methylcoumarin (Suc-LLVY-AMC; Sigma-Aldrich) or subjected to SDS-PAGE and immunoblot analysis with various 26S proteasome subunit antibodies (Yang et al., 2004; Book et al., 2010).
rpt2a Complementation
The RPT2a and RPT2b complementation transgenes were generated by overlapping PCR. The promoter and full coding sequences of each were individually isolated by PCR of genomic and cDNA templates, respectively. The products were mixed and used as templates in a second round of PCR to generate the final promoter/cDNA combinations. The primers used to complete each construction are as follows: P42, P43, P46, and P47 (RPT2apro:RPT2a); P42, P45, P48, and P49 (RPT2apro:RPT2b); P43, P44, P50, and P51 (RPT2bpro:RPT2a); and P44, P45, P52, and P53 (RPT2bpro:RPT2b). Site-directed mutagenesis of RPT2a was performed by overlapping PCR with the RPT2apro:RPT2a cDNA using the following mutagenic primer pairs: P54 and P55 (2G-A), P56 and P57 (235KT-AA), P58 and P59 (289E-Q), and P47 and P60 (ΔHbYX), coupled with the terminal primers P42 and P43 (2G-A, 235KT-AA, and 289E-Q) and P42 and P60 (ΔHbYX). These reaction products were then subjected to a second round of PCR with the terminal primers. The PCR products were recombined into the pDONR221 plasmid using Gateway technology (Invitrogen) and sequence verified, and the resulting entry clones were then recombined into the complementation vector pEarleyGate302 (Earley et al., 2006). All constructs were introduced into heterozygous rpt2a-2 plants by the A. tumefaciens–mediated floral dip method; transgenic T0 plants were then selected by Basta resistance. Plants homozygous for the rpt2a-2 allele and the transgenes expressing wild-type or mutant forms of RPT2a/b were identified by PCR genotyping and by Basta resistance in selfed T1 populations. Primers used for genotyping were P61 and P62 (RPT2a gene), P61 and P1 (T-DNA insert), P63 and P64 (RPT2b gene), and combinations of P65 to P68 for the various RPT2a and RPT2b transgenes.
Quantification of 45S rDNA Repeats
Chromosomal DNA was isolated from 4-d-old seedlings and quantitated by absorbance at 260 nm. Quantitative PCR was performed by the comparative threshold cycle method described above using primers specific for 18S rDNA (primers P69 and P70) as described by Mozgová et al. (2010). The reactions were normalized using the TUB8 gene as an internal control (primers P34 and P35).
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative under the following accession numbers: At4g29040 (RPT2a), At2g20140 (RPT2b), At1g65470 (FAS1), and At5g64630 (FAS2).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Amino Acid Sequence Alignment of RPT2 Proteins.
Supplemental Figure 2. Expression Patterns of Arabidopsis RPT2a and RPT2b Genes.
Supplemental Figure 3. RPT2a and RPT2b Are Equally Recognized by Anti-RPT2a Antibodies.
Supplemental Figure 4. RT-PCR, Genomic PCR, and qPCR Analyses of rpt2 and fas Mutants and Rescued Lines.
Supplemental Figure 5. Effects of rpt2 and fas Mutations on Trichome Branching and Root Growth.
Supplemental Figure 6. Both RPT2a and RPT2b Can Rescue the rpt2a Mutant Phenotype.
Supplemental Figure 7. Effects of Several Site-Directed Mutants on RPT2a Activity.
Supplemental Figure 8. Site-Directed RPT2a Mutations Do Not Affect the Nuclear/Cytoplasmic Partitioning of the RPT2 Protein.
Supplemental Figure 9. Sensitivity of rpt2a-3, fas1-4, and fas2-4 Mutants to Mitomycin C, Canavanine, and MG132.
Supplemental Figure 10. Phenotype of Mature rpt2a-2, fas1-4, and fas2-4 Plants.
Supplemental Table 1. Oligonucleotide Primers Used in This Study.
Acknowledgments
We thank ABRC for providing the EST clones and SALK T-DNA lines for RPT2a and RPT2b, Lars Hennig for the fas1-4 and fas2-4 mutants, and Kiyotaka Okada for the rpt2a-1/hrl-1 mutant. The anti-PUX1 and anti-PA200 antibodies were provided by Sebastian Bednarek and Nicholas Gladman, respectively. This work was supported by grants from the U.S. Department of Energy Basic Energy Sciences Program (DE-FG02-88ER13968), the National Science Foundation Arabidopsis 2010 Program (MCB-0115870), and the Research Division of the University of Wisconsin College of Agricultural and Life Sciences to R.D.V., by a National Institutes of Health predoctoral fellowship to A.J.B. (5F31NS054563), and by a sabbatical fellowship from the Tsuruoka National College of Technology to A.M.
AUTHOR CONTRIBUTIONS
K.-H.L., A.M., and R.D.V. designed the research. K.-H.L, A.M., R.S.M., A.J.B., and L.M.F. performed the research. J.M.W. provided technical expertise. K.-H.L., R.S.M., and R.D.V. analyzed the data and wrote the article.
References
- Alexander M.P. (1969). Differential staining of aborted and nonaborted pollen. Stain Technol. 44: 117–122 [DOI] [PubMed] [Google Scholar]
- Alonso J.M., et al. (2003). Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301: 653–657 [DOI] [PubMed] [Google Scholar]
- Bohn S., Beck F., Sakata E., Walzthoeni T., Beck M., Aebersold R., Förster F., Baumeister W., Nickell S. (2010). Structure of the 26S proteasome from Schizosaccharomyces pombe at subnanometer resolution. Proc. Natl. Acad. Sci. USA 107: 20992–20997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boisson B., Giglione C., Meinnel T. (2003). Unexpected protein families including cell defense components feature in the N-myristoylome of a higher eukaryote. J. Biol. Chem. 278: 43418–43429 [DOI] [PubMed] [Google Scholar]
- Book A.J., Gladman N.P., Lee S.S., Scalf M., Smith L.M., Vierstra R.D. (2010). Affinity purification of the Arabidopsis 26S proteasome reveals a diverse array of plant proteolytic complexes. J. Biol. Chem. 285: 25554–25569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Book A.J., Smalle J.A., Lee K.H., Yang P., Walker J.M., Casper S., Holmes J.H., Russo L.A., Buzzinotti Z.W., Jenik P.D., Vierstra R.D. (2009). The RPN5 subunit of the 26S proteasome is essential for gametogenesis, sporophyte development, and complex assembly in Arabidopsis. Plant Cell 21: 460–478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brukhin V., Gheyselinck J., Gagliardini V., Genschik P., Grossniklaus U. (2005). The RPN1 subunit of the 26S proteasome in Arabidopsis is essential for embryogenesis. Plant Cell 17: 2723–2737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao Y., Dai Y., Cui S., Ma L. (2008). Histone H2B monoubiquitination in the chromatin of FLOWERING LOCUS C regulates flowering time in Arabidopsis. Plant Cell 20: 2586–2602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z., Tan J.L., Ingouff M., Sundaresan V., Berger F. (2008). Chromatin assembly factor 1 regulates the cell cycle but not cell fate during male gametogenesis in Arabidopsis thaliana. Development 135: 65–73 [DOI] [PubMed] [Google Scholar]
- Collins K.A., Furuyama S., Biggins S. (2004). Proteolysis contributes to the exclusive centromere localization of the yeast Cse4/CENP-A histone H3 variant. Curr. Biol. 14: 1968–1972 [DOI] [PubMed] [Google Scholar]
- Corpet A., Almouzni G. (2009). Making copies of chromatin: The challenge of nucleosomal organization and epigenetic information. Trends Cell Biol. 19: 29–41 [DOI] [PubMed] [Google Scholar]
- Curtis M.D., Grossniklaus U. (2003). A Gateway cloning vector set for high-throughput functional analysis of genes in planta. Plant Physiol. 133: 462–469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deal R.B., Topp C.N., McKinney E.C., Meagher R.B. (2007). Repression of flowering in Arabidopsis requires activation of FLOWERING LOCUS C expression by the histone variant H2A.Z. Plant Cell 19: 74–83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doelling J.H., Phillips A.R., Soyler-Ogretim G., Wise J., Chandler J., Callis J., Otegui M.S., Vierstra R.D. (2007). The ubiquitin-specific protease subfamily UBP3/UBP4 is essential for pollen development and transmission in Arabidopsis. Plant Physiol. 145: 801–813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Earley K.W., Haag J.R., Pontes O., Opper K., Juehne T., Song K., Pikaard C.S. (2006). Gateway-compatible vectors for plant functional genomics and proteomics. Plant J. 45: 616–629 [DOI] [PubMed] [Google Scholar]
- Elsasser S., Chandler-Militello D., Müller B., Hanna J., Finley D. (2004). Rad23 and Rpn10 serve as alternative ubiquitin receptors for the proteasome. J. Biol. Chem. 279: 26817–26822 [DOI] [PubMed] [Google Scholar]
- Endo M., Ishikawa Y., Osakabe K., Nakayama S., Kaya H., Araki T., Shibahara K.I., Abe K., Ichikawa H., Valentine L., Hohn B., Toki S. (2006). Increased frequency of homologous recombination and T-DNA integration in Arabidopsis CAF-1 mutants. EMBO J. 25: 5579–5590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ermolaeva M.D., Wu M., Eisen J.A., Salzberg S.L. (2003). The age of the Arabidopsis thaliana genome duplication. Plant Mol. Biol. 51: 859–866 [DOI] [PubMed] [Google Scholar]
- Exner V., Gruissem W., Hennig L. (2008). Control of trichome branching by chromatin assembly factor-1. BMC Plant Biol. 8: 54–65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Exner V., Taranto P., Schönrock N., Gruissem W., Hennig L. (2006). Chromatin assembly factor CAF-1 is required for cellular differentiation during plant development. Development 133: 4163–4172 [DOI] [PubMed] [Google Scholar]
- Farmer L.M., Book A.J., Lee K.H., Lin Y.L., Fu H., Vierstra R.D. (2010). The RAD23 family provides an essential connection between the 26S proteasome and ubiquitylated proteins in Arabidopsis. Plant Cell 22: 124–142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finley D. (2009). Recognition and processing of ubiquitin-protein conjugates by the proteasome. Annu. Rev. Biochem. 78: 477–513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleury D., et al. (2007). The Arabidopsis thaliana homolog of yeast BRE1 has a function in cell cycle regulation during early leaf and root growth. Plant Cell 19: 417–432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Folta K.M., Kaufman L.S. (2006). Isolation of Arabidopsis nuclei and measurement of gene transcription rates using nuclear run-on assays. Nat. Protoc. 1: 3094–3100 [DOI] [PubMed] [Google Scholar]
- Fu H., Doelling J.H., Rubin D.M., Vierstra R.D. (1999). Structural and functional analysis of the six regulatory particle triple-A ATPase subunits from the Arabidopsis 26S proteasome. Plant J. 18: 529–539 [DOI] [PubMed] [Google Scholar]
- Fu H., Lin Y.L., Fatimababy A.S. (2010). Proteasomal recognition of ubiquitylated substrates. Trends Plant Sci. 15: 375–386 [DOI] [PubMed] [Google Scholar]
- Fu H., Sadis S., Rubin D.M., Glickman M.H., van Nocker S., Finley D., Vierstra R.D. (1998). Multiubiquitin chain binding and protein degradation are mediated by distinct domains within the 26 S proteasome subunit Mcb1. J. Biol. Chem. 273: 1970–1981 [DOI] [PubMed] [Google Scholar]
- Gallastegui N., Groll M. (2010). The 26S proteasome: Assembly and function of a destructive machine. Trends Biochem. Sci. 35: 634–642 [DOI] [PubMed] [Google Scholar]
- Gallois J.L., Guyon-Debast A., Lécureuil A., Vezon D., Carpentier V., Bonhomme S., Guerche P. (2009). The Arabidopsis proteasome RPT5 subunits are essential for gametophyte development and show accession-dependent redundancy. Plant Cell 21: 442–459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillette T.G., Huang W., Russell S.J., Reed S.H., Johnston S.A., Friedberg E.C. (2001). The 19S complex of the proteasome regulates nucleotide excision repair in yeast. Genes Dev. 15: 1528–1539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glickman M.H., Rubin D.M., Coux O., Wefes I., Pfeifer G., Cjeka Z., Baumeister W., Fried V.A., Finley D. (1998). A subcomplex of the proteasome regulatory particle required for ubiquitin-conjugate degradation and related to the COP9-signalosome and eIF3. Cell 94: 615–623 [DOI] [PubMed] [Google Scholar]
- Gonzalez F., Delahodde A., Kodadek T., Johnston S.A. (2002). Recruitment of a 19S proteasome subcomplex to an activated promoter. Science 296: 548–550 [DOI] [PubMed] [Google Scholar]
- Groll M., Bajorek M., Köhler A., Moroder L., Rubin D.M., Huber R., Glickman M.H., Finley D. (2000). A gated channel into the proteasome core particle. Nat. Struct. Biol. 7: 1062–1067 [DOI] [PubMed] [Google Scholar]
- Höek M., Stillman B. (2003). Chromatin assembly factor 1 is essential and couples chromatin assembly to DNA replication in vivo. Proc. Natl. Acad. Sci. USA 100: 12183–12188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang W., Pi L., Liang W., Xu B., Wang H., Cai R., Huang H. (2006). The proteolytic function of the Arabidopsis 26S proteasome is required for specifying leaf adaxial identity. Plant Cell 18: 2479–2492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Husnjak K., Elsasser S., Zhang N., Chen X., Randles L., Shi Y., Hofmann K., Walters K.J., Finley D., Dikic I. (2008). Proteasome subunit Rpn13 is a novel ubiquitin receptor. Nature 453: 481–488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inobe T., Fishbain S., Prakash S., Matouschek A. (2011). Defining the geometry of the two-component proteasome degron. Nat. Chem. Biol. 7: 161–167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaya H., Shibahara K.I., Taoka K.I., Iwabuchi M., Stillman B., Araki T. (2001). FASCIATA genes for chromatin assembly factor-1 in Arabidopsis maintain the cellular organization of apical meristems. Cell 104: 131–142 [DOI] [PubMed] [Google Scholar]
- Kirik A., Pecinka A., Wendeler E., Reiss B. (2006). The chromatin assembly factor subunit FASCIATA1 is involved in homologous recombination in plants. Plant Cell 18: 2431–2442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kodadek T. (2010). No splicing, no dicing: Non-proteolytic roles of the ubiquitin-proteasome system in transcription. J. Biol. Chem. 285: 2221–2226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Köhler A., Cascio P., Leggett D.S., Woo K.M., Goldberg A.L., Finley D. (2001). The axial channel of the proteasome core particle is gated by the Rpt2 ATPase and controls both substrate entry and product release. Mol. Cell 7: 1143–1152 [DOI] [PubMed] [Google Scholar]
- Kurepa J., Toh-E A., Smalle J.A. (2008). 26S proteasome regulatory particle mutants have increased oxidative stress tolerance. Plant J. 53: 102–114 [DOI] [PubMed] [Google Scholar]
- Kurepa J., Wang S., Li Y., Zaitlin D., Pierce A.J., Smalle J.A. (2009). Loss of 26S proteasome function leads to increased cell size and decreased cell number in Arabidopsis shoot organs. Plant Physiol. 150: 178–189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leyser H.M., Furner I.J. (1992). Characterization of three shoot apical meristem mutants of Arabidopsis thaliana. Development 116: 397–403 [Google Scholar]
- Li Z., Zou C.B., Yao Y., Hoyt M.A., McDonough S., Mackey Z.B., Coffino P., Wang C.C. (2002). An easily dissociated 26S proteasome catalyzes an essential ubiquitin-mediated protein degradation pathway in Trypanosoma brucei. J. Biol. Chem. 277: 15486–15498 [DOI] [PubMed] [Google Scholar]
- Liu Y., Koornneef M., Soppe W.J. (2007). The absence of histone H2B monoubiquitination in the Arabidopsis hub1 (rdo4) mutant reveals a role for chromatin remodeling in seed dormancy. Plant Cell 19: 433–444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopes da Rosa J., Holik J., Green E.M., Rando O.J., Kaufman P.D. (2011). Overlapping regulation of CenH3 localization and histone H3 turnover by CAF-1 and HIR proteins in Saccharomyces cerevisiae. Genetics 187: 9–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mozgová I., Mokroš P., Fajkus J. (2010). Dysfunction of chromatin assembly factor 1 induces shortening of telomeres and loss of 45S rDNA in Arabidopsis thaliana. Plant Cell 22: 2768–2780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ono T., Kaya H., Takeda S., Abe M., Ogawa Y., Kato M., Kakutani T., Mittelsten Scheid O., Araki T., Shibahara K.I. (2006). Chromatin assembly factor 1 ensures the stable maintenance of silent chromatin states in Arabidopsis. Genes Cells 11: 153–162 [DOI] [PubMed] [Google Scholar]
- Park S., Kim W., Tian G., Gygi S.P., Finley D. (2011). Structural defects in the regulatory particle-core particle interface of the proteasome induce a novel proteasome stress response. J. Biol. Chem. 286: 36652–36666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park S., Roelofs J., Kim W., Robert J., Schmidt M., Gygi S.P., Finley D. (2009). Hexameric assembly of the proteasomal ATPases is templated through their C termini. Nature 459: 866–870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabl J., Smith D.M., Yu Y., Chang S.C., Goldberg A.L., Cheng Y. (2008). Mechanism of gate opening in the 20S proteasome by the proteasomal ATPases. Mol. Cell 30: 360–368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramirez-Parra E., Gutierrez C. (2007). The many faces of chromatin assembly factor 1. Trends Plant Sci. 12: 570–576 [DOI] [PubMed] [Google Scholar]
- Rancour D.M., Park S., Knight S.D., Bednarek S.Y. (2004). Plant UBX domain-containing protein 1, PUX1, regulates the oligomeric structure and activity of Arabidopsis CDC48. J. Biol. Chem. 279: 54264–54274 [DOI] [PubMed] [Google Scholar]
- Roelofs J., Park S., Haas W., Tian G., McAllister F.E., Huo Y., Lee B.H., Zhang F., Shi Y., Gygi S.P., Finley D. (2009). Chaperone-mediated pathway of proteasome regulatory particle assembly. Nature 459: 861–865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubin D.M., Glickman M.H., Larsen C.N., Dhruvakumar S., Finley D. (1998). Active site mutants in the six regulatory particle ATPases reveal multiple roles for ATP in the proteasome. EMBO J. 17: 4909–4919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakamoto T., Kamiya T., Sako K., Yamaguchi J., Yamagami M., Fujiwara T. (2011). Arabidopsis thaliana 26S proteasome subunits RPT2a and RPT5a are crucial for zinc deficiency-tolerance. Biosci. Biotechnol. Biochem. 75: 561–567 [DOI] [PubMed] [Google Scholar]
- Sako K., Maki Y., Imai K.K., Aoyama T., Goto D.B., Yamaguchi J. (2010). Control of endoreduplication of trichomes by RPT2, a subunit of the 19S proteasome regulatory particle in Arabidopsis. J. Plant Res. 123: 701–706 [DOI] [PubMed] [Google Scholar]
- Saracco S.A., Hansson M., Scalf M., Walker J.M., Smith L.M., Vierstra R.D. (2009). Tandem affinity purification and mass spectrometric analysis of ubiquitylated proteins in Arabidopsis. Plant J. 59: 344–358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schreiner P., Chen X., Husnjak K., Randles L., Zhang N., Elsasser S., Finley D., Dikic I., Walters K.J., Groll M. (2008). Ubiquitin docking at the proteasome through a novel pleckstrin-homology domain interaction. Nature 453: 548–552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibahara T., Kawasaki H., Hirano H. (2002). Identification of the 19S regulatory particle subunits from the rice 26S proteasome. Eur. J. Biochem. 269: 1474–1483 [DOI] [PubMed] [Google Scholar]
- Shibahara T., Kawasaki H., Hirano H. (2004). Mass spectrometric analysis of expression of ATPase subunits encoded by duplicated genes in the 19S regulatory particle of rice 26S proteasome. Arch. Biochem. Biophys. 421: 34–41 [DOI] [PubMed] [Google Scholar]
- Singh R.K., Kabbaj M.H., Paik J., Gunjan A. (2009). Histone levels are regulated by phosphorylation and ubiquitylation-dependent proteolysis. Nat. Cell Biol. 11: 925–933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smalle J.A., Kurepa J., Yang P., Babiychuk E., Kushnir S., Durski A., Vierstra R.D. (2002). Cytokinin growth responses in Arabidopsis involve the 26S proteasome subunit RPN12. Plant Cell 14: 17–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smalle J.A., Kurepa J., Yang P., Emborg T.J., Babiychuk E., Kushnir S., Vierstra R.D. (2003). The pleiotropic role of the 26S proteasome subunit RPN10 in Arabidopsis growth and development supports a substrate-specific function in abscisic acid signaling. Plant Cell 15: 965–980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smalle J.A., Vierstra R.D. (2004). The ubiquitin 26S proteasome proteolytic pathway. Annu. Rev. Plant Biol. 55: 555–590 [DOI] [PubMed] [Google Scholar]
- Smith D.M., Chang S.C., Park S., Finley D., Cheng Y., Goldberg A.L. (2007). Docking of the carboxyl-termini of the proteasomal ATPases in the α-ring of the 20S proteasome opens the gate for substrate entry. Mol. Cell 27: 731–744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith D.M., Fraga H., Reis C., Kafri G., Goldberg A.L. (2011). ATP binds to proteasomal ATPases in pairs with distinct functional effects, implying an ordered reaction cycle. Cell 144: 526–538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song Y., He F., Xie G., Guo X., Xu Y., Chen Y., Liang X., Stagljar I., Egli D., Ma J., Jiao R. (2007). CAF-1 is essential for Drosophila development and involved in the maintenance of epigenetic memory. Dev. Biol. 311: 213–222 [DOI] [PubMed] [Google Scholar]
- Sonoda Y., Sako K., Maki Y., Yamazaki N., Yamamoto H., Ikeda A., Yamaguchi J. (2009). Regulation of leaf organ size by the Arabidopsis RPT2a 19S proteasome subunit. Plant J. 60: 68–78 [DOI] [PubMed] [Google Scholar]
- Sorek N., Bloch D., Yalovsky S. (2009). Protein lipid modifications in signaling and subcellular targeting. Curr. Opin. Plant Biol. 12: 714–720 [DOI] [PubMed] [Google Scholar]
- Sridhar V.V., Kapoor A., Zhang K., Zhu J., Zhou T., Hasegawa P.M., Bressan R.A., Zhu J.K. (2007). Control of DNA methylation and heterochromatic silencing by histone H2B deubiquitination. Nature 447: 735–738 [DOI] [PubMed] [Google Scholar]
- Takami Y., Ono T., Fukagawa T., Shibahara K.I., Nakayama T. (2007). Essential role of chromatin assembly factor-1-mediated rapid nucleosome assembly for DNA replication and cell division in vertebrate cells. Mol. Biol. Cell 18: 129–141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamada Y., Yun J.Y., Woo S.C., Amasino R.M. (2009). ARABIDOPSIS TRITHORAX-RELATED7 is required for methylation of lysine 4 of histone H3 and for transcriptional activation of FLOWERING LOCUS C. Plant Cell 21: 3257–3269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian G., Park S., Lee M.J., Huck B., McAllister F.E., Hill C.P., Gygi S.P., Finley D. (2011). An asymmetric interface between the regulatory and core particles of the proteasome. Nat. Struct. Mol. Biol. 18: 1259–1267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomko R.J., Funakoshi M., Schneider K., Wang J., Hochstrasser M. (2010). Heterohexameric ring arrangement of the eukaryotic proteasomal ATPases: Implications for proteasome structure and assembly. Mol. Cell 38: 393–403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueda M., Matsui K., Ishiguro S., Kato T., Tabata S., Kobayashi M., Seki M., Shinozaki K., Okada K. (2011). Arabidopsis RPT2a encoding the 26S proteasome subunit is required for various aspects of root meristem maintenance, and regulates gametogenesis redundantly with its homolog, RPT2b. Plant Cell Physiol. 52: 1628–1640 [DOI] [PubMed] [Google Scholar]
- Ueda M., Matsui K., Ishiguro S., Sano R., Wada T., Paponov I., Palme K., Okada K. (2004). The HALTED ROOT gene encoding the 26S proteasome subunit RPT2a is essential for the maintenance of Arabidopsis meristems. Development 131: 2101–2111 [DOI] [PubMed] [Google Scholar]
- van Nocker S., Deveraux Q., Rechsteiner M., Vierstra R.D. (1996). Arabidopsis MBP1 gene encodes a conserved ubiquitin recognition component of the 26S proteasome. Proc. Natl. Acad. Sci. USA 93: 856–860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verma R., Aravind L., Oania R., McDonald W.H., Yates J.R., III, Koonin E.V., Deshaies R.J. (2002). Role of Rpn11 metalloprotease in deubiquitination and degradation by the 26S proteasome. Science 298: 611–615 [DOI] [PubMed] [Google Scholar]
- Vierstra R.D. (2009). The ubiquitin-26S proteasome system at the nexus of plant biology. Nat. Rev. Mol. Cell Biol. 10: 385–397 [DOI] [PubMed] [Google Scholar]
- Voges D., Zwickl P., Baumeister W. (1999). The 26S proteasome: a molecular machine designed for controlled proteolysis. Annu. Rev. Biochem. 68: 1015–1068 [DOI] [PubMed] [Google Scholar]
- Walker J.M., Vierstra R.D. (2007). A ubiquitin-based vector for the co-ordinated synthesis of multiple proteins in plants. Plant Biotechnol. J. 5: 413–421 [DOI] [PubMed] [Google Scholar]
- Wang S., Kurepa J., Smalle J.A. (2009). The Arabidopsis 26S proteasome subunit RPN1a is required for optimal plant growth and stress responses. Plant Cell Physiol. 50: 1721–1725 [DOI] [PubMed] [Google Scholar]
- Yang P., Fu H., Walker J.M., Papa C.M., Smalle J.A., Ju Y.M., Vierstra R.D. (2004). Purification of the Arabidopsis 26S proteasome: biochemical and molecular analyses revealed the presence of multiple isoforms. J. Biol. Chem. 279: 6401–6413 [DOI] [PubMed] [Google Scholar]
- Yu Z., Kleifeld O., Lande-Atir A., Bsoul M., Kleiman M., Krutauz D., Book A.J., Vierstra R.D., Hofmann K., Reis N., Glickman M.H., Pick E. (2011). Dual function of Rpn5 in two PCI complexes, the 26S proteasome and COP9 signalosome. Mol. Biol. Cell 22: 911–920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmermann P., Hirsch-Hoffmann M., Hennig L., Gruissem W. (2004). GENEVESTIGATOR. Arabidopsis microarray database and analysis toolbox. Plant Physiol. 136: 2621–2632 [DOI] [PMC free article] [PubMed] [Google Scholar]