This work reveals that rice KNOX genes OSH1 and OSH15 are required for shoot apical meristem (SAM) formation and maintenance and that KNOX gene expression is positively and directly regulated by OSH1 through evolutionarily conserved cis-elements. This is thought to be a novel mechanism that is indispensable for self-maintenance of the SAM.
Abstract
Self-maintenance of the shoot apical meristem (SAM), from which aerial organs are formed throughout the life cycle, is crucial in plant development. Class I Knotted1-like homeobox (KNOX) genes restrict cell differentiation and play an indispensable role in maintaining the SAM. However, the mechanism that positively regulates their expression is unknown. Here, we show that expression of a rice (Oryza sativa) KNOX gene, Oryza sativa homeobox1 (OSH1), is positively regulated by direct autoregulation. Interestingly, loss-of-function mutants of OSH1 lose the SAM just after germination but can be rescued to grow until reproductive development when they are regenerated from callus. Double mutants of osh1 and d6, a loss-of-function mutant of OSH15, fail to establish the SAM both in embryogenesis and regeneration. Expression analyses in these mutants reveal that KNOX gene expression is positively regulated by the phytohormone cytokinin and by KNOX genes themselves. We demonstrate that OSH1 directly binds to five KNOX loci, including OSH1 and OSH15, through evolutionarily conserved cis-elements and that the positive autoregulation of OSH1 is indispensable for its own expression and SAM maintenance. Thus, the maintenance of the indeterminate state mediated by positive autoregulation of a KNOX gene is an indispensable mechanism of self-maintenance of the SAM.
INTRODUCTION
One of the more remarkable differences between animals and plants is the body plan. While animals complete their organogenesis during embryogenesis, plants form a simple structure consisting of two distinctive regions at opposite poles. One is the shoot apical meristem (SAM) and the other is the root apical meristem. Both meristems are organized groups of undifferentiated cells responsible for postembryogenic organ formation (Steeves and Sussex, 1989). The SAM is a dome-shaped tissue located at the growing tip of a shoot. After embryogenesis, lateral organs, such as leaves and flowers, are continuously formed from the flank of the SAM. Branches and stems are also derived from the SAM. Thus, the SAM is an ultimate source of all aboveground organs and therefore must be maintained throughout the life cycle. The elucidation of the mechanism involved in SAM maintenance is a fundamental issue in developmental biology.
Class I Knotted1-like homeobox (KNOX) genes, which encode homeodomain (HD) transcription factors, play indispensable roles in the SAM. Loss-of-function mutants of KNOX genes, such as knotted1 (kn1) in maize (Zea mays) and SHOOT MERISTEMLESS in Arabidopsis thaliana, lack the SAM, indicating that KNOX genes are essential for the formation and/or maintenance of the SAM (Long et al., 1996; Vollbrecht et al., 2000). KNOX genes negatively regulate biosynthesis of the phytohormone gibberellin (GA), which acts in differentiated organs to promote cell elongation and morphogenesis. In maize, KN1 directly activates GA2 oxidase1 (GA2ox1), the gene coding for a GA inactivation enzyme (Bolduc and Hake, 2009). A tobacco (Nicotiana tabacum) KNOX protein Nicotiana tabacum homeobox 15 also negatively regulates GA production in the SAM by direct repression of a GA biosynthetic enzyme gene, GA20ox1 (Sakamoto et al., 2001). Another hormonal pathway regulated by KNOX is cytokinin (CK). CK functions to activate cell division and shoot formation. In Arabidopsis and rice (Oryza sativa) , KNOX proteins positively regulate CK production through induction of the CK biosynthesis enzyme, adenosine phosphate isopentenyltransferase (IPT) (Jasinski et al., 2005; Yanai et al., 2005; Sakamoto et al., 2006). Thus, KNOX genes are considered to inhibit cell differentiation in the SAM by decreasing and increasing the amount of GA and CK, respectively.
KNOX genes are expressed in the shoot apex specifically and are excluded from differentiated organs (Jackson et al., 1994). Several genes required for repression of KNOX gene expression in leaves have been identified. Among them, the MYB transcription factor ROUGH SHEATH2 (RS2) in maize, its Arabidopsis putative ortholog ASYMMETRIC LEAVES1 (AS1), and the plant-specific LOB family protein AS2 have been well studied. In their loss-of-function mutants, KNOX genes are ectopically expressed in leaves (Timmermans et al., 1999; Tsiantis et al., 1999; Byrne et al., 2000; Ori et al., 2000; Semiarti et al., 2001; Iwakawa et al., 2002). AS1 and AS2 are thought to form a complex that directly represses transcription of the KNOTTED-like genes from Arabidopsis thaliana 1 (KNAT1)/BREVIPEDICELLUS (BP) and KNAT2 (Guo et al., 2008). RS2 and AS1 were shown to interact with a chromatin remodeling protein, HIRA (Phelps-Durr et al., 2005). Therefore, these proteins may establish and/or maintain the repressed state of KNOX loci via chromatin remodeling. In addition to this, proper activities of the phytohormone auxin, YABBY genes, and Polycomb genes are also required to repress KNOX gene expression in leaves (Kumaran et al., 2002; Katz et al., 2004; Hay et al., 2006).
Despite the critical function of KNOX genes in SAM maintenance, little is known of the mechanisms that activate KNOX gene expression. In this study, we isolated a loss-of-function mutant of one of the rice KNOX genes, Oryza sativa homeobox1 (OSH1). Genetic studies using osh1 in combination with d6, a loss-of-function mutant of another rice KNOX gene, OSH15, revealed the indispensable role of KNOX genes in SAM formation and maintenance in rice. Expression levels of KNOX genes were found to depend not only on CK but also on KNOX genes themselves. Furthermore, we demonstrated that OSH1 positively regulates the expression of all five KNOX genes tested in a direct manner through evolutionarily conserved cis-elements and that the positive autoregulation of OSH1 is essential for the expression of itself and SAM maintenance. Taken together, we propose that the positive autoregulation of a KNOX gene is one of the indispensable mechanisms of self-maintenance of the SAM.
RESULTS
Isolation of a Loss-of-Function Mutant of OSH1
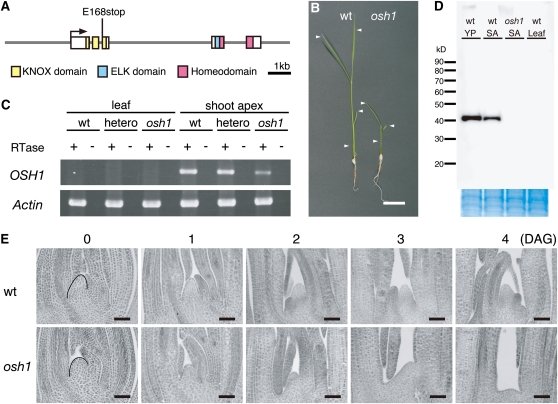
In the rice genome, there are five functional class I KNOX genes, OSH1, OSH6, OSH15, OSH43, and OSH71 (Sentoku et al., 1999; Sato et al., 2001). Among them, OSH15 has been studied by loss-of-function analysis and was revealed to be essential for internode elongation during the reproductive phase, but not for SAM maintenance (Sato et al., 1999). To provide further insight into the function of KNOX genes, we isolated a loss-of-function mutant of OSH1, an ortholog of maize kn1, using the Target-Induced Local Lesion In Genomes (TILLING) system of rice that we developed previously (Suzuki et al., 2008). From screening 1456 N-methyl-N-nitrosourea (MNU)-mutagenized M2 lines, we found one line that had a nucleotide substitution from G to T at the first nucleotide of the third exon, resulting in a premature stop codon (Figure 1A). This mutant osh1 allele (osh1-1) is predicted to encode a protein that lacks the HD essential for DNA binding. Homozygous osh1 mutant plants terminated growth soon after germination (Figure 1B). We performed a complementation test of osh1 and confirmed that normal morphology was recovered by introducing a copy of the wild-type OSH1 (see Supplemental Figure 1 online). Therefore, the osh1-1 allele is likely to be a loss-of-function mutant of OSH1 and was used in further analyses.
Figure 1.
Loss-of-Function Mutant of OSH1 Does Not Maintain the SAM after Germination.
(A) OSH1 structure and the mutation in osh1. Gray lines indicate introns and 5′ and 3′ regions. Boxes indicate exons. Conserved domains of OSH1 are labeled in color as shown below the gene structure.
(B) Phenotypes of wild-type (wt) (left) and osh1 (right) seedlings at 10 DAG. Arrowheads indicate the position of each leaf.
(C) RT-PCR analysis for OSH1 and Actin (loading control) in leaves and shoot apices of wild-type and osh1 plants at 10 DAG. This experiment was performed two times independently.
(D) Immunoblotting analysis of OSH1 protein in the wild type and osh1. Total nuclear protein extracts from 5-mm young panicles, shoot apices, and young leaf primordia at 10 DAG were loaded in each lane. Coomassie blue staining of membrane after immunoblotting shown at a bottom indicates that samples were loaded equally. SA, shoot apex; YP, young panicle.
(E) Shoot apex sections of the wild type and osh1 during germination. The SAM before germination is indicated with a black line.
Bars = 2 cm in (D) and 50 μm in (E).
To investigate the effect of this mutation on OSH1 mRNA accumulation, we examined transcript expression by RT-PCR. In the mutant shoot apex at 10 d after germination (DAG), a normal-sized transcript was detected, but the expression level was reduced compared with that of the wild type (Figure 1C). To examine OSH1 protein accumulation, we performed immunoblot analysis using an anti-OSH1 antibody that specifically recognizes the N terminus of OSH1. The premature stop codon in the third exon predicts a truncated form of OSH1 to be 17 kD. In the young panicle and shoot apex from wild-type plants, normal-sized OSH1 was detected at around 40 kD. On the other hand, neither a normal-sized nor a truncated form of OSH1 was detected in the mutant shoot apex (Figure 1D). OSH1 was not detected in wild-type leaves as previously determined (Jackson et al., 1994; Sentoku et al., 1999). Thus, these results indicate that the osh1-1 allele is a null.
OSH1 Is Essential for Shoot Meristem Maintenance during the Vegetative Phase
To study the function of OSH1 in SAM formation and maintenance, the phenotype of osh1 plants at various stages of development was observed. During embryogenesis, all embryos from heterozygous osh1 plants initiated a normal-appearing SAM and three embryonic leaves as in wild-type plants (n = 30). The defect was first observed in the mature embryo at 0 DAG (Figure 1E), in which osh1 had a slightly smaller SAM than the wild type. At 1 to 3 DAG, the dome shape of the osh1 mutant SAM gradually disappeared with the growth of the third leaf primordia. At 4 DAG, osh1 had a remnant bulge at the former SAM region, and after that, no additional leaves were formed. At 10 DAG, >90% of osh1 mutants terminated their growth with only three leaves produced during embryogenesis, compared with the wild type that continued to produce leaves (Figure 1B). However, a low frequency of osh1 plants produced four to six leaves (2.5%) or one to two leaves (3.9%) (see Supplemental Table 1 online). In very rare cases (<1%), osh1 mutants formed more than six leaves and continued to grow for about a month, although such plants also lost the SAM and terminated leaf formation eventually. Similar variability was documented with the maize kn1 loss-of-function mutants, although the penetrance of the shootless phenotype was much less (Vollbrecht et al., 2000). Thus, these results indicate that OSH1 is essential for SAM maintenance after germination but not for SAM formation during embryogenesis in rice.
OSH1 and OSH15 Are Redundantly Required for SAM Formation during Embryogenesis
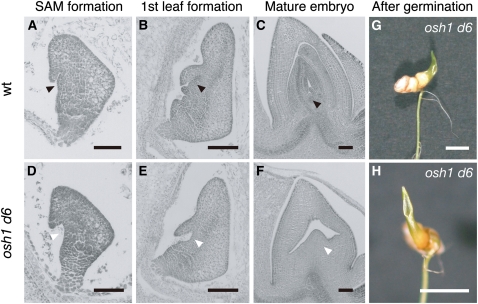
To examine the contribution of other KNOX genes to SAM formation, we observed embryogenesis of the osh1 d6 double mutants. OSH15 is expressed in the meristem, and d6, a loss-of-function mutant of OSH15, has a dwarf phenotype (Sato et al., 1999; Sentoku et al., 1999). In the wild type, the zygote continued cell division to form a globular embryo by 3 d after pollination (DAP). At 4 DAP, the SAM and coleoptile primordium appeared as a protrusion on the ventral side of the embryo (Figure 2A). The first leaf primordium developed from the flank of the SAM at 5 DAP (Figure 2B), and the second and third leaf primordia successively formed within another 3 to 4 d (Figure 2C). Subsequently, the embryo matured and entered dormancy (Itoh et al., 2005). Embryos from plants heterozygous for osh1 and homozygous for d6 had no visible abnormalities at 3 DAP (n = 30). However, in about one-fourth of examined embryos at 4 DAP, the coleoptile primordium was formed normally, but the region where SAM formation would occur was hollow and protrusion of the SAM was not observed (n = 7 out of 30 embryos; Figure 2D). Although the coleoptile developed as in normal embryos, no leaf primordia were initiated (Figure 2E). Even after embryo maturation, no organs were observed in osh1 d6 embryos except a coleoptile and a remnant bulge in the former SAM position (Figures 2F to 2H). Thus, osh1 d6 double mutant embryos failed to form the SAM, indicating that redundant functions of OSH1 and OSH15 were essential for the initiation of the SAM.
Figure 2.
OSH1 and OSH15 Are Redundantly Required for SAM Formation.
(A) to (C) Sections of wild-type (wt) embryos at 4 DAP (A), 5 DAP (B), and 10 DAP (C). Black arrowheads indicate the position of the SAM.
(D) to (F) Sections of osh1 d6 double mutant embryos at 4 DAP (D), 5 DAP (E), and 10 DAP (F). White arrowheads indicate the position where SAM formation should be observed in the wild type.
(G) An osh1 d6 double mutant at 10 DAG (cf. to Figure 1D).
(H) Top view of (G).
Bars = 100 μm in (A) to (F) and 5 mm in (G) and (H).
Shoot Regeneration from osh1 and osh1 d6 Mutant Callus
In Arabidopsis, it has been reported that key transcription factors for the SAM formation and maintenance, including KNOX genes, are coordinately expressed during shoot regeneration (Gordon et al., 2007). To examine the importance of KNOX gene function during de novo formation of the SAM, we observed the phenotype of shoots regenerated from the wild type, osh1, d6, and their double mutants.
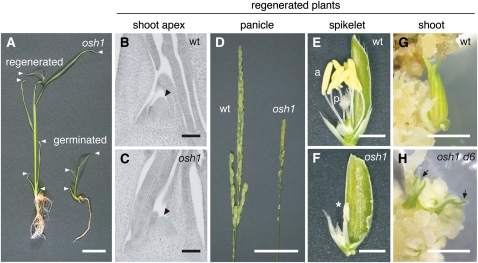
In the wild type and d6 single mutant, normal shoots were regenerated, and no abnormality was observed at later stages, except for the dwarf phenotype of d6 (Sato et al., 1999). In the case of osh1 single mutants, normal shoots were regenerated from callus. These plants could maintain the SAM and formed more than three leaves, unlike germinated osh1 mutants (Figures 3A to 3C). The phenotype of regenerated osh1 varied depending on their growth conditions. When they were grown in field conditions, they lost the SAM and terminated their growth prior to flowering (n = 56). However, almost all regenerated osh1 mutants could grow until reproductive phase in light- and temperature-controlled growth chambers (n = 43 out of 49 regenerated osh1 mutants). Regenerated osh1 mutant plants had a reduced number of spikelets with abnormally fused anthers (Figures 3D to 3F) and were completely sterile. Thus, regenerated osh1 mutants showed moderate phenotypes compared with germinated osh1 mutants. When we tried to regenerate plants from osh1 d6 double mutant callus, only leaf-like structures were formed and no shoot formation was observed at even 3 months after the induction of regeneration (Figure 3H). Therefore, as is the case for SAM formation during embryogenesis, OSH1 and OSH15 are redundantly required for de novo shoot formation in callus. Previously, it has been reported that Arabidopsis stm mutants can be rescued to grow until reproductive development when they were treated with CK (Yanai et al., 2005). Because the medium for shoot regeneration contains a high concentration of CK, it is possible that CK enabled the regenerated osh1 mutant to maintain the SAM in the absence of OSH1 function. However, the failure of the osh1 d6 double mutant callus to form shoots on the regeneration medium indicates that, even with a high concentration of CK, the function of KNOX genes is essential for SAM formation and/or maintenance.
Figure 3.
OSH1 and OSH15 Are Redundantly Required for SAM Formation and Maintenance during de Novo Shoot Regeneration from Callus.
(A) Regenerated (left) and germinated (right) seedlings of osh1. White arrowheads indicate the position of each leaf.
(B) and (C) Sections of shoot apices of the regenerated wild-type (B) and osh1 plants (C). Arrowheads indicate the SAM.
(D) Panicles of regenerated plants of the wild type (wt) (left) and osh1 (right).
(E) and (F) Spikelets formed on regenerated plants of the wild type (E) and osh1 (F). a, anther; p, pistil; asterisk, abnormal short anther of osh1.
(G) and (H) Regenerating shoot from the wild type (G) and osh1 d6 (H). Arrows indicate leaf-like structures formed from osh1 d6 callus.
Bars = 2 cm in (A), 50 μm in (B) and (C), 5 cm in (D), and 2 mm in (E) to (H).
Reduced Expression of KNOX Genes in osh1 and osh1 d6
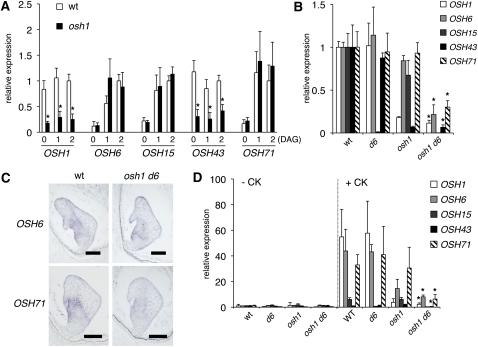
The phenotypic observations of osh1 mutants suggest that the contributions of individual rice KNOX genes to SAM maintenance varies depending on timing and environmental conditions. To investigate this possibility, we performed expression analyses of KNOX genes. During germination of wild-type plants, OSH1 and OSH43 were expressed constantly in all stages examined, whereas expression of OSH6, OSH15, and OSH71 was low at the mature embryo stage and increased just after germination (Figure 4A). In the osh1 mutant, OSH1 and OSH43 were significantly reduced at all stages, while expression of OSH6, OSH15, and OSH71 was induced after germination, as normal. Therefore, expression of all five KNOX genes is low in the mature embryo of osh1. This implies that the mutant shoot meristem temporally undergoes a state in which KNOX gene transcripts accumulate to very low levels.
Figure 4.
Expression Levels of KNOX Genes Were Reduced in osh Mutants.
(A) Expression of KNOX genes in germinating embryos. Relative expression level was calculated as the ratio to the expression level at 2 DAG in the wild type (wt) for each gene.
(B) Expression of KNOX genes in the SAM formation stage of the embryo at 5 DAP. Relative expression level was calculated as the ratio to the expression level in the wild type for each gene.
(C) In situ hybridization of OSH6 and OSH71 in wild-type and osh1 d6 embryos at around the stage of SAM formation. Bars = 100 μm.
(D) Expression of KNOX genes in callus before and 1 week after CK treatment. Relative expression level was calculated as the ratio to the expression level before CK treatment in the wild type for each gene.
In (A) to (C), the expression level of ubiquitin (ubq) was used as an internal control for normalization. The data represent the average of three independent biological replicates. Error bars represent the sd. Asterisks indicate significant reduction at P < 0.05 compared with the expression level in the wild type.
Next, we performed expression analysis during embryogenesis (Figure 4B). At the SAM formation stage, four KNOX genes except OSH15 were expressed at a normal level in d6 mutants. In osh1, as we observed during germination, the expression level of OSH43 was severely reduced, and OSH6 and OSH15 showed only slightly reduced expression, whereas OSH71 expression was comparable to that of the wild type. On the other hand, in osh1 d6 double mutant embryos, the expression of all five KNOX genes was reduced to one-third or less of wild-type values. In situ hybridization also confirmed the reduction of OSH6 and OSH71 expression in the osh1 d6 embryo (Figure 4C). Thus, KNOX gene transcripts barely accumulated in osh1 d6 embryos, which could lead to the failure of the SAM establishment.
We further performed expression analysis during de novo shoot formation from callus. From our previous studies, we found that KNOX gene expression was induced in callus by treatment with CK (Ito et al., 2001). Consistent with this, CK treatment induced the expression of OSH1, OSH6, OSH15, and OSH71 in wild-type callus, and OSH1, OSH6, and OSH71 were also induced to wild-type levels in d6 callus. In osh1 mutant callus, OSH6, OSH15, and OSH71 were also induced by CK, although the expression level of OSH6 was lower than in the wild type (Figure 4D). These data suggest that CK treatment bypassed the state of low KNOX gene expression in osh1 shoot apices during de novo shoot formation and could explain the phenotype difference between germination and regeneration.
In the osh1 d6 double mutant callus, not only was expression of OSH1, OSH6, and OSH43 reduced but also that of OSH15 and OSH71, even after CK treatment (Figure 4D). Thus, induction of KNOX genes by CK was impaired, which may be responsible for the failure of de novo shoot formation from callus in the osh1 d6 double mutant.
OSH1 Directly Binds to Five KNOX Loci through Evolutionarily Conserved cis-Elements
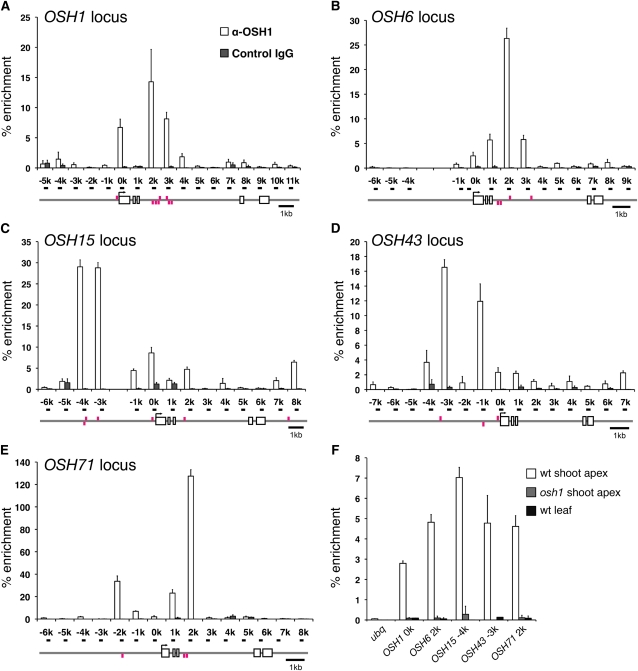
Our observation that expression of the other KNOX genes, OSH6, OSH43, and OSH71, was dependent on functional OSH1 and OSH15 in addition to CK suggested that KNOX genes might be positively autoregulated. Consistent with this hypothesis, we observed increased expression of KNOX genes in leaves of the 35S:OSH1 transgenic plants (see Supplemental Figure 2 online). To examine whether or not this regulation is mediated by KNOX proteins in a direct manner, we performed chromatin immunoprecipitation (ChIP) assays using an anti-OSH1 antibody that specifically recognizes OSH1. Using young panicles in which OSH1 expression is broad and strong, we tested the enrichment of OSH1-bound DNA in ChIP samples using primer pairs designed at 1-kb intervals along the OSH1 locus as indicated in Figure 5A. A ChIP assay followed by quantitative PCR (qPCR) revealed that sequences immediately upstream of the transcription start site and in the 5′ half of the third intron were strongly enriched in the OSH1-bound chromatin fragments. In addition, strong enrichment was obtained in several regions of four other KNOX loci (Figures 5B to 5E). Enrichment in these loci was also found in their 5′ upstream regions and the third introns. These results indicate that OSH1 binds to all five KNOX loci in vivo in the inflorescence meristem.
Figure 5.
OSH1 Protein Binds to Five KNOX Loci in Vivo.
ChIP assay using anti-OSH1 antibody followed by qPCR analysis of the OSH1 (A), OSH6 (B), OSH15 (C), OSH43 (D), and OSH71 (E) locus. Gene structures are indicated below the graphs. Primers used in qPCR were named after their position relative to the transcription start site of each gene and are shown as black bars on the gene structures. Magenta bars above and below the gene structure represent conserved TGAC sequences in the plus and minus strands, respectively. The percentage of enrichment represents the amount of immunoprecipitated DNA relative to input DNA. Each data point is the average of three independent biological replicates. Error bars represent the sd.
(A) to (E) ChIP assay from 5-mm young panicles of the wild type. For a negative control, control rabbit IgG was used.
(F) ChIP assay from vegetative tissues. For a negative control, osh1 shoot apex and wild-type (wt) young leaf primordia were used.
To examine if OSH1 directly binds to KNOX loci in vegetative shoot meristems, we tested the enrichment in ChIP samples of vegetative tissues at 1 DAG. In all highly enriched regions observed in young panicles, a modest enrichment was also detected when we used wild-type shoot apex as the sample but not in the case of osh1 shoot apex and young leaves (Figure 5F). These results confirmed the specificity of our ChIP assay and that OSH1 binds to KNOX loci in vivo in all OSH1-expressing tissues examined.
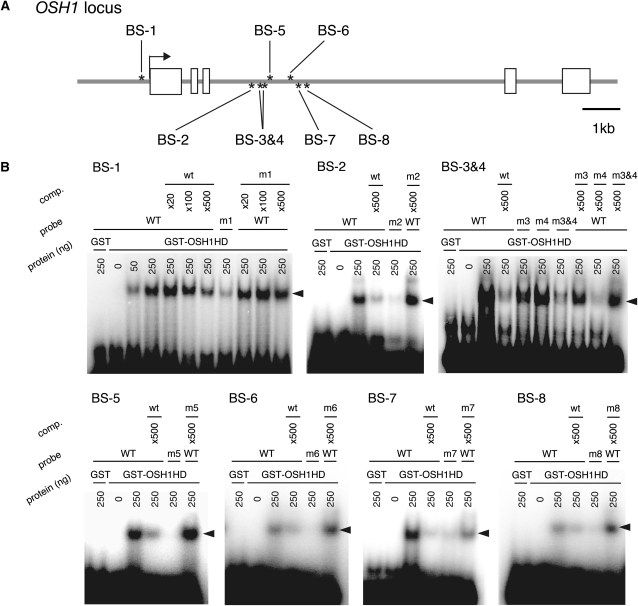
OSH1 Binding Sites Are Evolutionarily Conserved
Previously, it has been shown that KNOX proteins directly activate the expression of GA2ox1 through evolutionarily conserved TGAC core sequences (Bolduc and Hake, 2009). Therefore, we considered that if direct binding of OSH1 is important for transcriptional regulation of KNOX genes, the TGAC sequences should be conserved. To this end, we performed sequence comparison analysis between rice KNOX genes and their putative orthologs in Poaceae and searched for TGAC sequences in conserved regions. We found conserved TGAC sequences in each KNOX gene locus (magenta bars in Figure 5; see Supplemental Figure 3 online). The OSH1 locus had a single conserved sequence in the promoter region and seven in the 5′ half of the third intron. To test whether OSH1 directly binds to these cis-elements, we performed electrophoretic mobility shift assays (EMSAs) using a recombinant protein of the OSH1 HD fused to glutathione S-transferase (GST; Figure 6). In the case of binding site1 (BS-1) in the promoter region of OSH1, GST-OSH1HD bound to wild-type probes but hardly bound to mutant probes. In the competition assays, wild-type BS-1 cold DNA competed with radiolabeled BS-1 probe for GST-OSH1HD binding, but mutant BS-1 fragments did not, confirming that GST-OSH1HD specifically binds to BS-1. Similarly, GST-OSH1HD also specifically bound to the other seven BS sequences in the third intron of OSH1. Thus, these results indicate that OSH1 directly binds to these eight sequences in vitro.
Figure 6.
OSH1 Directly Binds to Conserved cis-Elements on the OSH1 Locus in Vitro.
(A) OSH1 binding sites (BS) on the OSH1 locus. Gray lines indicate introns and the 5′ and 3′ regions. Boxes indicate exons. Asterisks above and below the gene structure represent each BS, which contains a conserved TGAC sequence, in the plus and minus strands, respectively.
(B) EMSA using GST-OSH1HD. For mutant probes (m1-m8), evolutionarily conserved TGAC sequences were substituted with TCTC. Arrowheads represent the positions of shifted probes. GST alone was used as negative control, and “comp.” represents the competition assay using wild-type (WT) cold probes.
Several conserved TGAC sequences were also found in the other KNOX loci (see Supplemental Figure 3 online). As in OSH1, they were preferentially located in 5′ upstream regions and the third intron. Importantly, the distribution of these cis-elements coincided with that of highly enriched regions in ChIP assays. These results indicate that OSH1 directly binds to the five KNOX genes we examined, including OSH1 itself, via evolutionarily conserved cis-elements and suggest that transcriptional regulation of KNOX genes through these cis-elements has an important function.
Positive Autoregulation of OSH1 Is Essential for Its Expression and SAM Maintenance
Our results presented so far reveal the presence of positive autoregulation of OSH1. To investigate the importance of the autoregulation for transcriptional regulation of OSH1, we performed a reporter assay. Among 28 transgenic plants carrying a reporter gene, GFP-OSH1, in which green fluorescent protein was inserted into the N terminus of an OSH1 genomic DNA fragment, 22 lines showed GFP fluorescence in the shoot meristem, a finding that is in agreement with the endogenous OSH1 expression pattern previously reported (Figures 7A and 7B; Sentoku et al., 1999). Whereas no clear difference was observed when an OSH1 binding site located in the promoter region alone was mutated (see Supplemental Figure 4 online), none of 26 independent transgenic lines showed GFP fluorescence, and the reporter gene expression was almost completely abolished when all OSH1 binding sites were mutated (GFP-mOSH1; Figures 7A and 7C). These results indicate that the positive autoregulation of OSH1 through evolutionarily conserved OSH1 binding sites is essential for its proper expression.
Figure 7.
Positive Autoregulation of OSH1 Is Essential for Its Expression and SAM Maintenance.
(A) Reporter gene structures. Gray lines indicate introns and 5′ and 3′ regions. White and green boxes indicate exons and GFP, respectively. In the case of GFP-mOSH1, OSH1 binding sequences were substituted from TGAC to TCTC.
(B) GFP-OSH1 expression in the shoot apex.
(C) GFP-mOSH1 expression in the shoot apex.
(D) Regenerating shoots from osh1 d6 callus carrying GFP-OSH1 on regeneration medium.
(E) osh1 d6 callus carrying GFP-mOSH1 on regeneration medium. For regeneration, transgenic osh1 d6 calli were cultured on regeneration medium for 4 to 5 weeks.
(F) A rescued osh1 d6 plant carrying GFP-OSH1 grew until the reproductive phase.
(G) d6 and wild-type (wt) plants during the reproductive phase.
Bars = 100 μm in (B) and (C), 5 mm in (D) and (E), and 10 cm in (F) and (G).
Finally, we examined the biological importance of this autoregulation by rescue experiments using transgenic osh1 d6 double mutant callus. We observed the recovery of de novo shoot formation from osh1 d6 double mutant callus by the wild-type GFP-OSH1 gene (seven out of 58 independent transgenic osh1 d6 calli; Figure 7D) but not by GFP-mOSH1 (none of 53 independent transgenic osh1 d6 calli; Figure 7E). The osh1 d6 plants rescued by wild-type GFP-OSH1 were able to grow until the reproductive phase (Figure 7F). Thus, it was revealed that direct positive autoregulation of OSH1 was essential for SAM formation and/or maintenance.
DISCUSSION
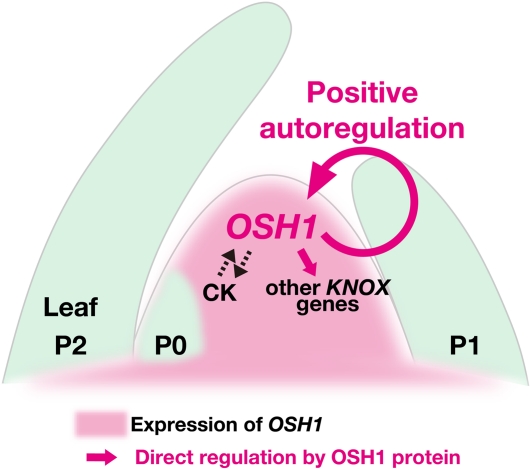
Here, we show that OSH1 plays an indispensable role in cooperation with its paralog, OSH15, in the establishment and maintenance of the SAM. Expression of KNOX genes is positively regulated by the phytohormone CK and directly regulated by OSH1 through evolutionarily conserved cis-elements. Furthermore, we demonstrate that the positive autoregulation of OSH1 is essential for its own expression and also for SAM maintenance. We propose that maintenance of the indeterminate state mediated by positive autoregulation of a KNOX gene is a mechanism for self-maintenance of the SAM that ensures the unique body plan of flowering plants (Figure 8).
Figure 8.
The Model of a KNOX Autoregulatory Loop for SAM Maintenance.
In this model, OSH1 expression is positively regulated via autoregulation. OSH1 protein directly and positively regulates its own expression and other KNOX genes in the SAM. KNOX genes and CK mutually activate each other, creating another positive autoregulatory loop.
Regulation of cell population size and maintenance of an indeterminate state are fundamental aspects of SAM function. Continuous cell division of stem cells at the center of the SAM and their daughter cells in surrounding regions replenishes the cell population. On the other hand, lateral organ initiation from the flank of the SAM consumes this cell population. Studies in Arabidopsis have shown that a feedback loop between a member of another class of homeobox genes, WUSCHEL, and the CLAVATA genes, which encode components of a signaling pathway, is essential for homeostasis of the stem cell population (Schoof et al., 2000). This regulatory feedback loop maintains the size of the stem cell population and has until now been the only described mechanism for self-regulation in the SAM.
In our model, the stem cell population provides a source of cells for lateral organ initiation. All cells in the SAM accumulate KNOX proteins to maintain their indeterminate state (Jackson et al., 1994; Sentoku et al., 1999), and KNOX proteins bind directly to evolutionarily conserved cis-elements on KNOX loci to activate and/or maintain their expression. This autoregulation may provide robustness to maintain indeterminacy of the SAM, in which a complex network of transcription factors and phytohormones balances stem cell activity and organogenesis (Zhao et al., 2010). At a defined location in the meristem, this autoregulation is diminished and KNOX genes are downregulated.
In our observations, mutations in both OSH15 and OSH1 caused a severe reduction in the expression of other KNOX genes and resulted in the failure of SAM establishment, which was not the case for each single mutant. Members of KNOX genes are expressed in overlapping regions in the shoot meristems (Jackson et al., 1994; Sentoku et al., 1999), and overexpressors of each member show similar phenotypes to those of OSH1 and OSH15 (Ito et al., 2001). In addition, it has been reported that not only KN1, but also other members of KNOX protein, such as Gnarley1 and Liguleless3, directly bind to cis-elements in the intron of GA2ox1 in maize (Bolduc and Hake, 2009). Considering these facts, it is likely that members of the KNOX gene family have redundant functions in SAM maintenance; hence, it is plausible that not only OSH1 but also other members of the KNOX protein family participate in this autoregulation.
In Arabidopsis, strong stm alleles completely fail to establish the SAM during embryogenesis. By contrast, weak stm alleles have a smaller SAM than the wild type but continue organogenesis until reproductive development (Clark et al., 1996; Endrizzi et al., 1996). Mutations in other members of the KNOX gene family, such as KNAT1/BP and KNAT6, enhance the weak phenotype of stm-2, although stm-2 knat6-1 shows additional abnormalities in cotyledon separation (Byrne et al., 2002; Belles-Boix et al., 2006). Thus, members of Arabidopsis class I KNOX genes also have redundant functions in the SAM. It is an intriguing question to ask whether KNOX genes are also autoregulated in dicots. Given the conserved functions and similar expression patterns of KNOX genes between monocots and dicots, it is possible that the autoregulation is also conserved.
Previously, rice class I KNOX genes have been categorized into two groups based on their expression pattern in the SAM (Sentoku et al., 1999). OSH1 and OSH43 are in the first group, and they are expressed in the entire SAM except the epidermal layer. On the other hand, expression of a second group genes, OSH6, OSH15, and OSH71, is localized to the base of the SAM and boundaries between the SAM and incipient leaf primordia. Interestingly, we found that their expression profiles during germination also split into these two groups (Figure 4A). Genes in the first group are expressed constantly before and after germination. By contrast, the second group genes are greatly upregulated after germination. Therefore, these two KNOX groups might also be under the control of different regulatory mechanisms, although both are positively regulated by OSH1.
Seven OSH1 binding sites identified in this study are located in the 5′ half of the third intron of OSH1. This region also contains a cluster of sequences that are highly conserved between rice and maize (Inada et al., 2003). These conserved noncoding sequences (CNSs) are considered to have an important function in the repression of KNOX gene expression because transposon insertions and a 305-bp tandem duplication in these CNSs cause ectopic expression of KNOX genes in maize and barley (Hordeum vulgare), respectively (Greene et al., 1994; Müller et al., 1995). Our data reveal that these CNSs also contain important cis-elements for positive autoregulation of OSH1. Whether or not there are causal connections between the mutations and KNOX autoregulation, these facts indicate that this cluster of CNSs contains both enhancer and silencer elements. In addition, there might be many unidentified cis-elements in these CNSs; therefore, more detailed cis-element analyses are necessary to understand fully the mechanisms involved in transcriptional regulation of KNOX genes.
So far, it has been debated whether or not CK acts upstream of KNOX genes. In Arabidopsis, CK treatment enhances the expression of the ProKNAT2:β-glucuronidase reporter in the SAM (Hamant et al., 2002). By contrast, the induction of the CK biosynthetic gene IPT in Arabidopsis leaves or young seedlings does not increase the expression of KNOX genes (Craft et al., 2005). In this study, we observed a clear induction of KNOX genes by CK during shoot regeneration from callus. This result strongly suggests that KNOX genes are positively regulated by CK. Consistent with this notion, the expression of OSH1 is severely reduced in the floral meristem of the loss-of-function mutants of the LONELY GUY gene, which encodes an enzyme that catalyzes the final step of CK biosynthesis in rice (Kurakawa et al., 2007). The regulation of KNOX genes by CK could be limited only in the SAM context and in tissues competent to regenerate shoots, such as callus. On the other hand, it has been shown that KNOX genes activate the expression of IPT and promote the accumulation of CK in the SAM (Jasinski et al., 2005; Yanai et al., 2005; Sakamoto et al., 2006). Thus, these facts suggest that KNOX genes and CK facilitate each other, creating another positive autoregulatory loop (Figure 8).
In our observations, CK treatment did not enhance shoot regeneration from osh1 d6 callus carrying the GFP-mOSH1 construct (Figure 7). This result indicates that, at least during shoot regeneration from callus, the CK pathway is not enough to establish and/or maintain the SAM without the positive autoregulation of OSH1. It is possible that CK signaling activates GFP-mOSH1 expression once, but its stable expression could not be maintained because of lack of autoregulation. It is also an important question to ask what initially activates KNOX expression during embryogenesis. Given that CK positively regulates KNOX gene expression in callus and promotes shoot meristem function, this phytohormone is a likely candidate involved in the initial activation step. Another intriguing question is how KNOX gene expression is first downregulated in incipient differentiating organs. The phytohormone auxin has been reported to function in the negative regulation of KNOX genes. In lateral organ initials where KNOX genes are first downregulated, a local maximum of auxin accumulation is formed via the function of auxin transporters (Reinhardt et al., 2003). This local maximum of auxin concentration has also been shown to trigger lateral organ initiation. In addition, it has been reported that an impaired regulation of auxin signaling causes ectopic expression of KNOX genes in leaves (Hay et al., 2006). Thus, downregulation of KNOX genes and auxin is likely to be tightly linked in lateral organ initials. Recently, it has been reported that CK and auxin have antagonistic functions in the regulation of the stem cell niche (Zhao et al., 2010). Analogies between regulation of the stem cell niche and regulation of indeterminacy might exist. The elucidation of molecular links between these phytohormones and KNOX genes requires further investigation.
METHODS
Plant Material
Japonica rice (Oryza sativa) variety Taichung65 (T65) was used as the wild type. The osh1 mutant was derived from an MNU-mutagenized T65 population (Suzuki et al., 2008). Heterozygous plants for the osh1 mutation were backcrossed three times with wild-type plants and used in this study. The d6 mutant used in this study was the d6-tankanshirasasa allele described by Sato et al. (1999).
Screening for osh1 Using the TILLING System
Using the TILLING system described by Suzuki et al. (2008), 1456 lines of the M1 generation mutagenized by MNU were screened for mutations in the OSH1 open reading frame. Primer sets used to amplify OSH1 exon 2-3, exon 4, and exon 5 are listed in Supplemental Table 2 online. Target regions were amplified by PCR from the pooled genomic DNA library as described by Suzuki et al. (2008). The PCR products were denatured, reannealed, digested by Surveyor nuclease (Transgenomic), and analyzed as described by Suzuki et al. (2008). To identify the mutation in candidate lines, target regions were amplified by PCR as described above from genomic DNA of individual M2 plants and were sequenced.
Genotyping of osh1-1
A 198-bp PCR product was amplified from genomic DNA using 11S2-5 derived cleaved-amplified polymorphic sequence primers (see Supplemental Table 2 online). PCR was performed in the buffer conditions described by Suzuki et al. (2008) with 35 cycles of denaturation at 94°C for 20 s, annealing at 52°C for 20 s, and extension at 72°C for 5 s. PCR products were digested by 5 units of BslI (New England Biolabs) at 55°C for 1 h followed by electrophoresis on a 2% agarose gel.
RNA Extraction, RT-PCR, and qRT-PCR
In the case of shoot apex and callus, 10 shoot apices and 500 mg of callus were used for total RNA extraction, respectively. Total RNA was extracted using Concert Plant RNA reagent (Invitrogen), and mRNA was purified from 1 μg of total RNA using Dynabeads oligo(dT) 25 (Invitrogen) as described in the manufacturer’s protocol. Half of the purified mRNA was reverse transcribed by Superscript III reverse transcriptase (Invitrogen) in a 20-μL reaction mixture, and the other half was treated similarly without reverse transcriptase. In the case of expression analysis in embryos, individual embryos and endosperms were collected separately and genotyped as described above. cDNA was synthesized from 100 ng of total RNA that was extracted from 30 embryos of each genotype.
For RT-PCR analysis, 1 μL of cDNA was used in a 20-μL PCR reaction mixture using the primer set listed in Supplemental Table 2 online, and 5 μL of PCR products was electrophoresed on a 1% agarose gel.
For qRT-PCR, 1 μL of cDNA was used in a 25-μL PCR reaction mixture with SYBR Premix Ex Taq (Takara), and data were collected and analyzed using the TAKARA Thermal Cycler Dice Real Time System TP850. Primer sets used are listed in Supplemental Table 2 online. Data presented were the average of three biological replicates normalized using ubiquitin as the internal control.
Polyclonal Antibody Production and Affinity Purification
An N terminus–coding region of OSH1 (amino acids 1 to 238) was amplified from OSH1 cDNA and was cloned into pENTR/D-TOPO (Invitrogen) and sequenced. The insert was transferred into pDEST17 (Invitrogen) by an LR recombination reaction, and a hexahistidine-tagged protein was expressed in the BL21-AI Escherichia coli strain (Invitrogen) according to the manufacturer’s procedure. The hexahistidine-tagged OSH1 recombinant protein was purified using the HisTrap FF crude Kit (GE Healthcare) and used for immunizing rabbits (Medical and Biological Laboratories). For affinity purification, the 103–amino acid coding region from the first Met was amplified from OSH1 cDNA, and recombinant proteins were prepared as described above. The immune serum was affinity purified using this recombinant protein (Medical and Biological Laboratories).
Immunoblotting
Nuclear extracts prepared for ChIP assay (see ChIP assay section) were de-cross-linked at 75°C for 1 h, boiled at 100°C for 5 min, and electrophoresed on a 10% SDS-polyacrylamide gel. Proteins were transferred to an Immobilon-P membrane (Millipore) by electroblotting and immunodetected using anti-OSH1 antibody as described previously (Harlow and Lane, 1988).
Histological Analysis
Samples were fixed in FAA fixative solution (formalin:acetic acid:50% ethanol = 5:5:90) for 24 h at 4°C and dehydrated through a graded ethanol series from 70 to 100%. Ethanol was replaced with xylene, and samples were embedded in Paraplast Plus (McCormick Scientific). Ten-micrometer sections were applied to Matsunami adhesive microscope slides (MAS; Matsunami Glass) and were stained with hematoxylin (Wako).
In Situ Hybridization
Digoxigenin-labeled antisense RNA probes for OSH6 and OSH71 were prepared as described by Sentoku et al. (1999). Rice embryos were fixed in 4% paraformaldehyde, dehydrated, and embedded as described above. Probe hybridization, posthybridization washing, and immunological detection of digoxigenin were performed as previously described (Kouchi and Hata, 1993).
CK Treatment of Callus
Callus induced on N6D medium was transferred to Murashige and Skoog medium containing 10 μM synthetic CK 6-benzylaminopurine and cultured for 1 week. Callus was harvested before and after CK treatment and stored at −80°C until used.
ChIP Assay
Ten young panicles at the 5-mm stage, 200 shoot apices at 1 DAG, or 200 young leaf primordia at 1 DAG were used as samples for independent replicates. Shoot apices include the SAM, P1 leaf primordium, and developing stem below the SAM and P1 leaf primordium. Young leaf primordia include P2 and P3 leaves. A ChIP assay was performed as previously described (Saze et al., 2008) with minor modifications. Plant materials were cross-linked in fixation buffer (400 mM Suc, 10 mM Tris-HCl, pH8.0, 1 mM EDTA, and 1% formaldehyde) for 10 min under vacuum, and cross-linking was stopped by adding Gly to a final concentration of 100 mM and drawing vacuum for 5 min. Plant materials were washed with distilled water and stored at −80°C. Cross-linked materials were frozen, crushed to fine powder, suspended in extraction buffer 1 (400 mM Suc, 10 mM Tris-HCl, pH 8.0, 5 mM β-mercaptoethanol, and 1× Complete Proteinase Inhibitor Cocktail [Roche]), and filtered through two layers of mesh (50-μm pore size). Isolated nuclei were washed using extraction buffer 2 (250 mM Suc, 10 mM Tris-HCl, pH 8.0, 10 mM MgCl2, 1% Triton X-100, 5 mM β-mercaptoethanol, and 1× Complete Proteinase Inhibitor Cocktail) and extraction buffer 3 (1.7 M Suc, 10 mM Tris-HCl, pH 8.0, 0.15% Triton X-100, 2 mM MgCl2, 5 mM β-mercaptoethanol, and 1× Complete Proteinase Inhibitor Cocktail) and sonicated in 300 μL of nuclei lysis buffer (50 mM Tris-HCl, pH 8.0, 1% SDS, 10 mM EDTA, and 1× Complete Proteinase Inhibitor Cocktail) until the average genomic DNA fragment size became 500 bp using a sonicator (Branson Sonifier 250). Fragmented chromatin was split into two batches, and each batch was diluted with 1350 μL of ChIP dilution buffer (50 mM Tris-HCl, pH 8.0, 167 mM NaCl, 1.1% Triton X-100, 0.11% sodium deoxycholate, and 1× Complete Proteinase Inhibitor Cocktail), and 20 μL of diluted chromatin was stored for input control. One microgram of OSH1-specific polyclonal antibody or rabbit control IgG (Santa Cruz Biotechnology) was prebound to 40 μL of Dynabeads protein A (Invitrogen) according to the instructions of the manufacturer, and immunoprecipitation reactions were performed for 30 min at room temperature. The beads-chromatin complex was washed with low-salt radio-immunoprecipitation assay buffer (50 mM Tris-HCl, pH 8.0, 150 mM NaCl, 1 mM EDTA, 0.1% SDS, 0.1% sodium deoxycholate, 1% Triton X-100, and 1× Complete Proteinase Inhibitor Cocktail), high-salt radio-immunoprecipitation assay buffer (50 mM Tris-HCl, pH 8.0, 500 mM NaCl, 1 mM EDTA, 0.1% SDS, 0.1% sodium deoxycholate, 1% Triton X-100, and 1× Complete Proteinase Inhibitor Cocktail), lithium chloride buffer (250 mM LiCl, 1% Nonidet P-40, 1% sodium deoxycholate, 1 mM EDTA, and 10 mM Tris-HCl, pH 8.0), and two times with Tris EDTA buffer. The beads and input control were incubated at 65°C for more than 5 h in 200 μL of elution buffer (10 mM Tris-HCl, pH 8.0, 300 mM NaCl, 5 mM EDTA, and 0.5% SDS). The eluted chromatin and input control were treated with 20 mg of Proteinase K (Sigma-Aldrich) at 55°C for 1 h, and DNA was isolated by phenol-chloroform extraction and ethanol precipitation with 40 μg of glycogen (Roche). Finally, DNA was dissolved in 100 μL of Tris EDTA buffer. For quantification of the ChIP assay, 1 μL of DNA was used in a 25-μL PCR reaction mixture with SYBR Premix Ex Taq (Takara), and data were collected and analyzed using the Takara TP850 Thermal Cycler Dice Real Time System. Primer sets used are listed in Supplemental Table 2 online. Each experiment was repeated at least three times independently.
EMSA
The HD of the OSH1 (amino acids 264 to 326) coding region was amplified from OSH1 cDNA and cloned into pENTR/D-TOPO and sequenced. The insert was transferred into pDEST15 (Invitrogen) by an LR recombination reaction, and GST-OSH1HD was expressed in the BL21-AI and purified using Glutathione Sepharose 4B resin (GE Healthcare). To prepare double-strand DNA probes, two complementary oligonucleotides were mixed, heat-denatured at 95°C for 10 min, and reannealed by gradually cooling to room temperature. Double-strand DNA probes were labeled with [32P]ATP using T4 polynucleotide kinase and purified with G-25 sepharose (GE Healthcare). The EMSA binding reaction was performed as described previously (Bolduc and Hake, 2009) and separated on 6% nondenaturing PAGE gels in 0.5× Tris-borate-EDTA buffer.
Plasmid Construction and Transformation of Rice
For a genomic DNA construct containing the entire OSH1 gene and the 4-kb upstream region, the BAC clone AC145380 was digested using BamHI, and a 14-kb fragment containing a 4-kb 5′ upstream region and the entire OSH1 gene was cloned into the BamHI site of pUC19 (pUC19BmgOSH1). For the GFP-OSH1 reporter gene, the sGFP(S65T) (Chiu et al., 1996) coding region without a stop codon was inserted in front of the start codon of OSH1. To prepare inserts for the plasmids, we separately amplified each OSH1 genomic region or GFP using overlapping primers listed in Supplemental Table 2 online and fused neighboring PCR products one by one using their overlapping ends and outermost primers by additional PCR. To generate GFP-OSH1, a region from an ApaI site in the OSH1 promoter region to the OSH1 start codon, GFP, and a region from the OSH1 start codon to an MluI site in exon1 were amplified from pUC19BmgOSH1 and the cDNA clone of GFP using primer pairs listed in Supplemental Table 2 online. These PCR products were fused by additional PCR reactions using outermost primers, and the final PCR product was cloned into pCR-BluntII-TOPO (Invitrogen) and sequenced. This construct was digested with ApaI and MluI, and a 2.4-kb fragment containing GFP fused to the OSH1 N-terminal region was cloned into the ApaI-MluI site of pUC19BmgOSH1 in place of a 1.7-kb ApaI-MluI fragment of OSH1. This plasmid was designated as pUC19BmgGFP-OSH1. The entire OSH1 gene or GFP-OSH1 was digested by HindIII and KpnI and cloned into the HindIII-KpnI site of a binary vector, pBCH1 (Ito et al., 2001). Mutations in each OSH1 binding site were introduced by PCR. For the mBS-1 GFP-OSH1, the ApaI-BS-1 and BS-1-MluI region were amplified from pUC19BmgGFP-OSH1 and fused using primers listed in Supplemental Table 2 online. The final PCR product containing a mutation in BS-1 was cloned, sequenced, and transferred to pUC19BmgOSH1 as described above. This plasmid was designated as pUCmBS-1GFP-OSH1. This reporter gene was transferred to pBCH1 as described above. For GFP-mOSH1, a region between an MluI site in exon1 and an AccIII site in intron3 were separated into seven regions by OSH1 binding sites. These seven regions were amplified from pUC19BmgOSH1 and fused by additional PCR reactions. The resultant PCR products were cloned into pCR-BluntII-TOPO and sequenced. This construct was digested by MluI and AccIII, and a 3.4-kb fragment was cloned into an MluI-AccIII site of pUC10mBS-1GFP-OSH1. This reporter gene was transferred to pBCH1 as described above. These binary vectors were introduced into Agrobacterium tumefaciens strain EHA101 by electroporation. Using the Agrobacterium-mediated method (Hiei et al., 1994), rice calli were transformed, and transgenic plants regenerated from hygromycin-resistant calli were used in further studies.
Observation of GFP Fluorescence under Microscopy
Shoot apices of transgenic plants were embedded in 6% low melting temperature agarose (BMA) and cut into 30-μm sections using a microslicer DTK-1000 (DOSAKA). Sections were mounted in sterilized water, and GFP fluorescence was observed using a Biozero BZ-8000 fluorescence microscope (Keyence) with 470-nm excitation and 535-nm emission filters.
Accession Numbers
Genomic DNA sequence data of putative OSH1 orthologs of wheat (Triticum aestivum) and barley (Hordeum vulgare) can be found in the DNA Data Bank of Japan (http://www.ddbj.nig.ac.jp/) data library under the following accession numbers: wheat, AB182943; barley, X83518. Genomic DNA sequences of rice KNOX genes are collected from RAP-DB (http://rapdb.dna.affrc.go.jp/). Genomic DNA sequences of maize (Zea mays), sorghum (Sorghum bicolor), and Brachypodium distachyon were found in Phytozome v7.0 (http://www.phytozome.net/). The genomic location of rice, maize, sorghum, and Brachypodium sequences are indicated in Supplemental Table 3 online.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Complementation Test of osh1 by the OSH1 Gene.
Supplemental Figure 2. Expression of Endogenous KNOX Genes Was Increased in OSH1 Overexpressor Leaves.
Supplemental Figure 3. Sequence Comparison of Genomic DNA of KNOX Loci among Poaceae.
Supplemental Figure 4. Expression of GFP-OSH1 with a Single Mutation at the OSH1 Binding Site in the 5′ Upstream Region.
Supplemental Table 1. The Number of Leaves Formed in osh1 Mutants before Arrest of the Growth.
Supplemental Table 2. Oligonucleotide Sequences Used in This Study.
Supplemental Table 3. Genomic Location Used in Sequence Comparison Analysis.
Acknowledgments
We thank S. Hake and N. Bolduc for critical reading and comments on the manuscript. We thank T. Suzuki for technical advice for TILLING, S. Fukudome for technical assistance, and M. Shenton and members of the Kurata lab for helpful discussions and comments. This work was supported by a Grant-in-Aid for Scientific Research on Priority Area (18075009) from the Ministry of Education, Culture, Sports, Science, and Technology of Japan and a Research Fellowship of the Japan Society for the Promotion of Science for Young Scientist (22-1871).
AUTHOR CONTRIBUTIONS
K.T. and N.K. designed the research. K.T. performed the research. Y.I. and Y.S. contributed antibody production and plasmid construction, respectively. K.T. and N.K. wrote the article with help from Y.I. and Y.S.
References
- Belles-Boix E., Hamant O., Witiak S.M., Morin H., Traas J., Pautot V. (2006). KNAT6: An Arabidopsis homeobox gene involved in meristem activity and organ separation. Plant Cell 18: 1900–1907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolduc N., Hake S. (2009). The maize transcription factor KNOTTED1 directly regulates the gibberellin catabolism gene ga2ox1. Plant Cell 21: 1647–1658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byrne M.E., Barley R., Curtis M., Arroyo J.M., Dunham M., Hudson A., Martienssen R.A. (2000). Asymmetric leaves1 mediates leaf patterning and stem cell function in Arabidopsis. Nature 408: 967–971 [DOI] [PubMed] [Google Scholar]
- Byrne M.E., Simorowski J., Martienssen R.A. (2002). ASYMMETRIC LEAVES1 reveals knox gene redundancy in Arabidopsis. Development 129: 1957–1965 [DOI] [PubMed] [Google Scholar]
- Chiu W., Niwa Y., Zeng W., Hirano T., Kobayashi H., Sheen J. (1996). Engineered GFP as a vital reporter in plants. Curr. Biol. 6: 325–330 [DOI] [PubMed] [Google Scholar]
- Clark S.E., Jacobsen S.E., Levin J.Z., Meyerowitz E.M. (1996). The CLAVATA and SHOOT MERISTEMLESS loci competitively regulate meristem activity in Arabidopsis. Development 122: 1567–1575 [DOI] [PubMed] [Google Scholar]
- Craft J., Samalova M., Baroux C., Townley H., Martinez A., Jepson I., Tsiantis M., Moore I. (2005). New pOp/LhG4 vectors for stringent glucocorticoid-dependent transgene expression in Arabidopsis. Plant J. 41: 899–918 [DOI] [PubMed] [Google Scholar]
- Endrizzi K., Moussian B., Haecker A., Levin J.Z., Laux T. (1996). The SHOOT MERISTEMLESS gene is required for maintenance of undifferentiated cells in Arabidopsis shoot and floral meristems and acts at a different regulatory level than the meristem genes WUSCHEL and ZWILLE. Plant J. 10: 967–979 [DOI] [PubMed] [Google Scholar]
- Gordon S.P., Heisler M.G., Reddy G.V., Ohno C., Das P., Meyerowitz E.M. (2007). Pattern formation during de novo assembly of the Arabidopsis shoot meristem. Development 134: 3539–3548 [DOI] [PubMed] [Google Scholar]
- Greene B., Walko R., Hake S. (1994). Mutator insertions in an intron of the maize knotted1 gene result in dominant suppressible mutations. Genetics 138: 1275–1285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo M., Thomas J., Collins G., Timmermans M.C. (2008). Direct repression of KNOX loci by the ASYMMETRIC LEAVES1 complex of Arabidopsis. Plant Cell 20: 48–58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamant O., Nogué F., Belles-Boix E., Jublot D., Grandjean O., Traas J., Pautot V. (2002). The KNAT2 homeodomain protein interacts with ethylene and cytokinin signaling. Plant Physiol. 130: 657–665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harlow E., Lane D. (1988). Antibodies: A Laboratory Manual. (Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press) [Google Scholar]
- Hay A., Barkoulas M., Tsiantis M. (2006). ASYMMETRIC LEAVES1 and auxin activities converge to repress BREVIPEDICELLUS expression and promote leaf development in Arabidopsis. Development 133: 3955–3961 [DOI] [PubMed] [Google Scholar]
- Hiei Y., Ohta S., Komari T., Kumashiro T. (1994). Efficient transformation of rice (Oryza sativa L.) mediated by Agrobacterium and sequence analysis of the boundaries of the T-DNA. Plant J. 6: 271–282 [DOI] [PubMed] [Google Scholar]
- Inada D.C., Bashir A., Lee C., Thomas B.C., Ko C., Goff S.A., Freeling M. (2003). Conserved noncoding sequences in the grasses. Genome Res. 13: 2030–2041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito Y., Eiguchi M., Kurata N. (2001). KNOX homeobox genes are sufficient in maintaining cultured cells in an undifferentiated state in rice. Genesis 30: 231–238 [DOI] [PubMed] [Google Scholar]
- Itoh J., Nonomura K., Ikeda K., Yamaki S., Inukai Y., Yamagishi H., Kitano H., Nagato Y. (2005). Rice plant development: from zygote to spikelet. Plant Cell Physiol. 46: 23–47 [DOI] [PubMed] [Google Scholar]
- Iwakawa H., Ueno Y., Semiarti E., Onouchi H., Kojima S., Tsukaya H., Hasebe M., Soma T., Ikezaki M., Machida C., Machida Y. (2002). The ASYMMETRIC LEAVES2 gene of Arabidopsis thaliana, required for formation of a symmetric flat leaf lamina, encodes a member of a novel family of proteins characterized by cysteine repeats and a leucine zipper. Plant Cell Physiol. 43: 467–478 [DOI] [PubMed] [Google Scholar]
- Jackson D., Veit B., Hake S. (1994). Expression of maize KNOTTED1 related homeobox genes in the shoot apical meristem predicts patterns of morphogenesis in the vegetative shoot. Development 120: 405–413 [Google Scholar]
- Jasinski S., Piazza P., Craft J., Hay A., Woolley L., Rieu I., Phillips A., Hedden P., Tsiantis M. (2005). KNOX action in Arabidopsis is mediated by coordinate regulation of cytokinin and gibberellin activities. Curr. Biol. 15: 1560–1565 [DOI] [PubMed] [Google Scholar]
- Katz A., Oliva M., Mosquna A., Hakim O., Ohad N. (2004). FIE and CURLY LEAF polycomb proteins interact in the regulation of homeobox gene expression during sporophyte development. Plant J. 37: 707–719 [DOI] [PubMed] [Google Scholar]
- Kouchi H., Hata S. (1993). Isolation and characterization of novel nodulin cDNAs representing genes expressed at early stages of soybean nodule development. Mol. Gen. Genet. 238: 106–119 [DOI] [PubMed] [Google Scholar]
- Kumaran M.K., Bowman J.L., Sundaresan V. (2002). YABBY polarity genes mediate the repression of KNOX homeobox genes in Arabidopsis. Plant Cell 14: 2761–2770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurakawa T., Ueda N., Maekawa M., Kobayashi K., Kojima M., Nagato Y., Sakakibara H., Kyozuka J. (2007). Direct control of shoot meristem activity by a cytokinin-activating enzyme. Nature 445: 652–655 [DOI] [PubMed] [Google Scholar]
- Long J.A., Moan E.I., Medford J.I., Barton M.K. (1996). A member of the KNOTTED class of homeodomain proteins encoded by the STM gene of Arabidopsis. Nature 379: 66–69 [DOI] [PubMed] [Google Scholar]
- Müller K.J., Romano N., Gerstner O., Garcia-Maroto F., Pozzi C., Salamini F., Rohde W. (1995). The barley Hooded mutation caused by a duplication in a homeobox gene intron. Nature 374: 727–730 [DOI] [PubMed] [Google Scholar]
- Ori N., Eshed Y., Chuck G., Bowman J.L., Hake S. (2000). Mechanisms that control knox gene expression in the Arabidopsis shoot. Development 127: 5523–5532 [DOI] [PubMed] [Google Scholar]
- Phelps-Durr T.L., Thomas J., Vahab P., Timmermans M.C. (2005). Maize rough sheath2 and its Arabidopsis orthologue ASYMMETRIC LEAVES1 interact with HIRA, a predicted histone chaperone, to maintain knox gene silencing and determinacy during organogenesis. Plant Cell 17: 2886–2898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinhardt D., Pesce E.R., Stieger P., Mandel T., Baltensperger K., Bennett M., Traas J., Friml J., Kuhlemeier C. (2003). Regulation of phyllotaxis by polar auxin transport. Nature 426: 255–260 [DOI] [PubMed] [Google Scholar]
- Sakamoto T., Kamiya N., Ueguchi-Tanaka M., Iwahori S., Matsuoka M. (2001). KNOX homeodomain protein directly suppresses the expression of a gibberellin biosynthetic gene in the tobacco shoot apical meristem. Genes Dev. 15: 581–590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakamoto T., Sakakibara H., Kojima M., Yamamoto Y., Nagasaki H., Inukai Y., Sato Y., Matsuoka M. (2006). Ectopic expression of KNOTTED1-like homeobox protein induces expression of cytokinin biosynthesis genes in rice. Plant Physiol. 142: 54–62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato Y., Fukuda Y., Hirano H.Y. (2001). Mutations that cause amino acid substitutions at the invariant positions in homeodomain of OSH3 KNOX protein suggest artificial selection during rice domestication. Genes Genet. Syst. 76: 381–392 [DOI] [PubMed] [Google Scholar]
- Sato Y., Sentoku N., Miura Y., Hirochika H., Kitano H., Matsuoka M. (1999). Loss-of-function mutations in the rice homeobox gene OSH15 affect the architecture of internodes resulting in dwarf plants. EMBO J. 18: 992–1002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saze H., Shiraishi A., Miura A., Kakutani T. (2008). Control of genic DNA methylation by a jmjC domain-containing protein in Arabidopsis thaliana. Science 319: 462–465 [DOI] [PubMed] [Google Scholar]
- Schoof H., Lenhard M., Haecker A., Mayer K.F., Jürgens G., Laux T. (2000). The stem cell population of Arabidopsis shoot meristems in maintained by a regulatory loop between the CLAVATA and WUSCHEL genes. Cell 100: 635–644 [DOI] [PubMed] [Google Scholar]
- Semiarti E., Ueno Y., Tsukaya H., Iwakawa H., Machida C., Machida Y. (2001). The ASYMMETRIC LEAVES2 gene of Arabidopsis thaliana regulates formation of a symmetric lamina, establishment of venation and repression of meristem-related homeobox genes in leaves. Development 128: 1771–1783 [DOI] [PubMed] [Google Scholar]
- Sentoku N., Sato Y., Kurata N., Ito Y., Kitano H., Matsuoka M. (1999). Regional expression of the rice KN1-type homeobox gene family during embryo, shoot, and flower development. Plant Cell 11: 1651–1664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steeves T.A., Sussex I.M. (1989). Patterns in Plant Development, 2nd ed. (Cambridge, MA: Cambridge University Press) [Google Scholar]
- Suzuki T., Eiguchi M., Kumamaru T., Satoh H., Matsusaka H., Moriguchi K., Nagato Y., Kurata N. (2008). MNU-induced mutant pools and high performance TILLING enable finding of any gene mutation in rice. Mol. Genet. Genomics 279: 213–223 [DOI] [PubMed] [Google Scholar]
- Timmermans M.C., Hudson A., Becraft P.W., Nelson T. (1999). ROUGH SHEATH2: A Myb protein that represses knox homeobox genes in maize lateral organ primordia. Science 284: 151–153 [DOI] [PubMed] [Google Scholar]
- Tsiantis M., Schneeberger R., Golz J.F., Freeling M., Langdale J.A. (1999). The maize rough sheath2 gene and leaf development programs in monocot and dicot plants. Science 284: 154–156 [DOI] [PubMed] [Google Scholar]
- Vollbrecht E., Reiser L., Hake S. (2000). Shoot meristem size is dependent on inbred background and presence of the maize homeobox gene, knotted1. Development 127: 3161–3172 [DOI] [PubMed] [Google Scholar]
- Yanai O., Shani E., Dolezal K., Tarkowski P., Sablowski R., Sandberg G., Samach A., Ori N. (2005). Arabidopsis KNOXI proteins activate cytokinin biosynthesis. Curr. Biol. 15: 1566–1571 [DOI] [PubMed] [Google Scholar]
- Zhao Z., Andersen S.U., Ljung K., Dolezal K., Miotk A., Schultheiss S.J., Lohmann J.U. (2010). Hormonal control of the shoot stem-cell niche. Nature 465: 1089–1092 [DOI] [PubMed] [Google Scholar]