This study reveals that the Arabidopsis thaliana microtubule-destabilizing protein MDP25 regulates hypocotyl elongation. The microtubule-destabilizing activity of MDP25 is regulated by calcium, which causes MDP25 to disassociate from the plasma membrane and move into the cytosol. We identified a novel microtubule mechanism that regulates hypocotyl cell elongation in Arabidopsis.
Abstract
The regulation of hypocotyl elongation is important for plant growth. Microtubules play a crucial role during hypocotyl cell elongation. However, the molecular mechanism underlying this process is not well understood. In this study, we describe a novel Arabidopsis thaliana microtubule-destabilizing protein 25 (MDP25) as a negative regulator of hypocotyl cell elongation. We found that MDP25 directly bound to and destabilized microtubules to enhance microtubule depolymerization in vitro. The seedlings of mdp25 mutant Arabidopsis lines had longer etiolated hypocotyls. In addition, MDP25 overexpression resulted in significant overall shortening of hypocotyl cells, which exhibited destabilized cortical microtubules and abnormal cortical microtubule orientation, suggesting that MDP25 plays a crucial role in the negative regulation of hypocotyl cell elongation. Although MDP25 localized to the plasma membrane under normal conditions, increased calcium levels in cells caused MDP25 to partially dissociate from the plasma membrane and move into the cytosol. Cellular MDP25 bound to and destabilized cortical microtubules, resulting in their reorientation, and subsequently inhibited hypocotyl cell elongation. Our results suggest that MDP25 exerts its function on cortical microtubules by responding to cytoplasmic calcium levels to mediate hypocotyl cell elongation.
INTRODUCTION
Etiolation occurs as buried seedlings fully elongate their hypocotyls upward in search of the soil surface. Light perception signifies soil emergence and significantly inhibits hypocotyl elongation. The cotyledons unfold and the photosynthetic growth process begins. Thus, the regulation of hypocotyl elongation is crucial for plant growth and development. Hypocotyl cells elongate quickly without division during postgermination and are widely used as a model to study the regulation of cell elongation (Gendreau et al., 1997; Wang et al., 2002; Tsuchida-Mayama et al., 2010). Hypocotyl cells begin their elongation in the basal region. They proceed in the acropetal direction and reach their maximum length when grown in the dark. The elongation of hypocotyl cells is strongly influenced by both external and internal cues. Numerous studies have detailed the mechanisms involved in hypocotyl cell elongation regulated by light, phytohormones, transcription factors, and the cytoskeleton (Wang et al., 2002; Ehrhardt, 2008; Niwa et al., 2009).
Microtubules are involved in cell elongation, expansion, division, and plant morphogenesis (Thitamadee et al., 2002; Smith and Oppenheimer, 2005; Ehrhardt and Shaw, 2006; Buschmann and Lloyd, 2008). Cortical microtubules regulate cell elongation by orientating cellulose fibrils and cellulose fibril arrays, thereby influencing the mechanical properties of the cell wall (Baskin, 2005; Paredez et al., 2006; Somerville, 2006; Lloyd and Chan, 2008). Long-term time-lapse imaging has revealed that clockwise and counterclockwise rotations are important dynamic features of cortical microtubules in growing hypocotyl cells (Chan et al., 2007). The parallel array of cortical microtubules is transversely oriented to the hypocotyl longitudinal growth axis in elongating hypocotyl cells and longitudinally oriented when elongation has stopped (Le et al., 2005). Thus, the regulation of the organization and dynamics of cortical microtubules is crucial for hypocotyl cell growth.
Microtubule regulatory proteins regulate the organization and dynamics of microtubules (Hamada, 2007; Kaloriti et al., 2007; Buschmann and Lloyd, 2008; Sedbrook and Kaloriti, 2008). Mutations in these regulatory proteins usually result in abnormal plant growth and cell morphogenesis by altering microtubule organization and dynamics. For example, a point mutation in Arabidopsis thaliana MICROTUBULE ORGANIZATION1 (MOR1)/GEMINI POLLEN1 induces organ twisting and isotropic cell expansion in the roots by altering the dynamic instability of microtubules (Whittington et al., 2001; Twell et al., 2002). In addition, Arabidopsis SPIRAL1 (SPR1) is involved in hypocotyl cell elongation. The expression pattern of SPR1 in etiolated hypocotyls correlates with hypocotyl cell growth status, and SPR1 overexpression increases hypocotyl length, suggesting a positive regulatory role of SPR1 in hypocotyl cell elongation (Nakajima et al., 2004).
Since the elongation of hypocotyl cells is strongly inhibited by light, a negative regulatory mechanism must exist during this process. Calcium is involved in multiple signaling pathways that regulate plant cell physiology and cellular responses to environmental cues (Dodd et al., 2010). Calcium influx is a crucial event in the growth inhibition pathway, particularly in light-mediated hypocotyl growth inhibition. For example, blue light induces Ca2+ influx across the plasma membrane and results in a transient increase in [Ca2+]cyt in Arabidopsis seedling hypocotyls (Baum et al., 1999). This transient increase in [Ca2+]cyt is required for the ability of blue light to inhibit hypocotyl growth (Shinkle and Jones, 1988; Folta et al., 2003). Despite accumulating evidence that indicates the involvement of calcium in light-mediated regulation of hypocotyl elongation, no downstream effector has been identified that responds to calcium elevation and directly inhibits cell elongation. In addition, no microtubule regulatory protein has been identified as a downstream negative regulator of hypocotyl elongation that directly alters microtubule organization and dynamics.
It is reported that two hydrophilic cation binding proteins, Arabidopsis plasma membrane–associated cation binding protein 1 (PCaP1) and PCaP2, stably bind to the plasma membrane and to calcium (Ide et al., 2007; Nagasaki et al., 2008; Kato et al., 2010a). PCaP2 is a microtubule-associated protein previously named MAP18, which binds and regulates microtubules (Wang et al., 2007). PCaP1 is homologous with MAP18 in Arabidopsis and exhibits similar biochemical properties as MAP18 with regard to cation binding in vitro (Nagasaki et al., 2008; Kato et al., 2010a). However, it is unknown whether PCaP1 is a novel microtubule regulatory protein and how it functions in plant cells.
In this study, we demonstrate that PCaP1 is a novel microtubule-destabilizing protein with a molecular mass of ~25 kD, which we renamed MDP25 to refer to its function on microtubules. MDP25 directly bound to and destabilized microtubules by depolymerization. The expression and protein levels of MDP25 revealed its positional pattern along etiolated hypocotyls, and MDP25 overexpression resulted in significant overall shortening of hypocotyl cells, suggesting that MDP25 plays a crucial role in the negative regulation of hypocotyl cell elongation. Calcium regulates the effect of MDP25 on microtubules in hypocotyl cells by causing it to dissociate from the plasma membrane and move into the cytosol. Our study suggests that the cellular levels of MDP25 and its function in the presence of calcium play important roles in modulating hypocotyl elongation by regulating microtubule organization.
RESULTS
MDP25 Directly Binds to Microtubules in Vitro
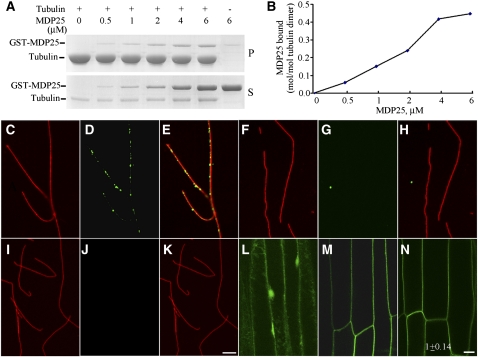
Although MDP25 is a protein in Arabidopsis that is homologous with MAP18 (Nagasaki et al., 2008; Kato et al., 2010a), there is no evidence that it directly affects microtubules. To determine whether this protein binds to microtubules, we expressed MDP25 in bacterial cells and purified the fusion protein. A cosedimentation experiment assessed the binding of MDP25 to paclitaxel-stabilized microtubules. The glutathione S-transferase (GST)-tagged MDP25 fusion protein was purified from Escherichia coli strain BL21 (DE3), and high-quality fusion proteins (see Supplemental Figure 1 online) were used in the following microtubule interaction assay. Increasing concentrations (0.5, 1, 2, 4, and 6 μMΜ) of GST-MDP25 were incubated with preformed 5 μMΜ paclitaxel-stabilized microtubules at room temperature for 20 min and then centrifuged. SDS-PAGE analysis showed that MDP25 bound to and cosedimented with the microtubules. Saturation occurred when the concentration of MDP25 reached ~4 μMΜ (Figure 1A). The amount of bound protein was quantified by densitometry. The molar binding ratio between MDP25 and the tubulin dimer was ~0.45:1 at saturation (Figure 1B). To further confirm the ability of this protein to bind to microtubules, in vitro immunofluorescence labeling was performed with an anti-GST antibody. The fusion protein was incubated with preformed rhodamine-labeled and paclitaxel-stabilized microtubules, stained with anti-GST antibody and secondary antibody, and detected by spinning disk microscopy. Our results revealed that the protein was distributed specifically along microtubules, exhibiting a dot-like structure (Figures 1C to 1E). No such structure was detected when denatured MDP25 (Figures 1F to 1H) or GST alone (Figures 1I to 1K) was incubated with microtubules. Therefore, we conclude that this protein is able to bind to microtubules in vitro. In addition, microtubules in the presence of MDP25 exhibited single filament patterns with no microtubule-bundling activity.
Figure 1.
MDP25 Directly Binds to Microtubules in Vitro.
(A) GST-MDP25 fusion protein was cosedimented with paclitaxel-stabilized microtubules. GST-MDP25 mostly appeared in the supernatant in the absence of microtubules (S) but cosedimented with microtubules into the pellets (P).
(B) The results of the quantitative analysis of the binding between GST-MDP25 and microtubules are shown. The amount of protein was determined by gel scanning.
(C) to (K) Immunofluorescence staining was performed to investigate the binding of MDP25 to microtubules in vitro. Red fluorescence signals indicate rhodamine-labeled microtubules, and green signals indicate GST-MDP25 probed with anti-GST and fluorescein isothiocyanate–conjugated secondary antibodies. Microtubules (C), MDP25 (D), and merged image (E). Microtubules (F), denatured MDP25 (G), and merged image (H). Microtubules (I), GST (J), and merged image (K). Notice that the microtubules exhibited a single-filament pattern, and no microtubule bundles were observed in the presence of MDP25 fusion protein.
(L) to (N) The plasma membrane localization of MDP25 occurred independently of cortical microtubules. Seedlings of PMDP25:MDP25:GFP were treated with 10 μM oryzalin for 60 min. P35S:GFP (L), PMDP25:MDP25:GFP (M), and seedlings of (M) treated with oryzalin (N). The GFP fluorescence intensity was averaged from the measurements of at least 14 cells after background subtraction, and the result in (N) is expressed as the ratio of fluorescence intensity of treated cells to untreated cells.
Bars = 10 μm in (K) and (N).
A previous study suggested that MDP25 locates to the plasma membrane via N-myristoylation (Nagasaki et al., 2008). Cortical microtubules are usually considered to predominantly bind and securely fasten to the plasma membrane by linker proteins (Hardham and Gunning, 1978), and the experimental results above demonstrated that MDP25 binds to microtubules. Therefore, we investigated whether this plasma membrane localization is related to cortical microtubules. The plasma membrane localization of MDP25 was examined using hypocotyl epidermal cells of PMDP25:MDP25:green fluorescent protein (GFP) transgenic lines (Figure 1M) or protoplasts isolated from MDP25-GFP transgenic lines stained with FM4-64 dye (see Supplemental Figure 2 online). A fluorescence signal was detected at the plasma membrane but not at other cell membranes, which is consistent with previous reports that used transient expression or anti-MDP25 antibody (Ide et al., 2007; Nagasaki et al., 2008). The cells were then treated with the microtubule-disrupting drug oryzalin (10 μM) for 60 min. No difference in the MDP25-GFP fluorescence signal on the plasma membrane was detected (Figure 1N). Additionally, the protoplasts isolated from MDP25-GFP transgenic Arabidopsis were treated with oryzalin, followed by double immunostaining with antitubulin and anti-GFP antibodies. We found that the MDP25-GFP fluorescent signal was still located on the plasma membrane when most cortical microtubules were disrupted (see Supplemental Figure 3 online), suggesting that the targeting of MDP25 to the plasma membrane was independent of the presence of cortical microtubules.
MDP25 Inhibits Tubulin Polymerization
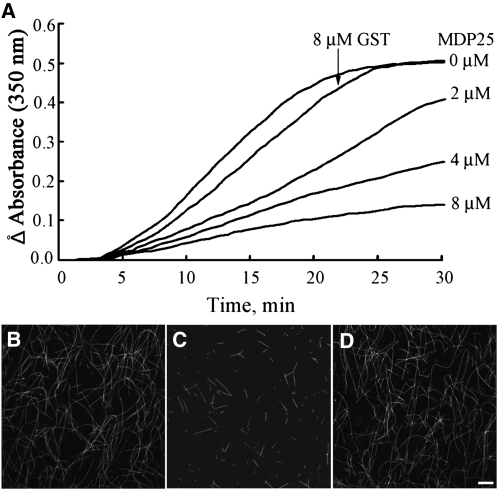
To determine the effect of MDP25 on microtubule polymerization, we performed turbidimetric analysis in vitro. Various concentrations (0, 2, 4, and 8 μMΜ) of GST-MDP25 fusion protein were added to a 30 μM tubulin solution, and changes in turbidity were monitored. The turbidity of the tubulin assembly decreased in the presence of MDP25 compared with the absence of MDP25 or the presence of 8 μMΜ GST (Figure 2A), indicating that the microtubule mass was reduced by MDP25. The assembly rate of tubulin, reflected by the slope of the assembly curves, also decreased in a concentration-dependent manner with the addition of MDP25.
Figure 2.
MDP25 Inhibits Tubulin Assembly in Vitro.
(A) Various concentrations of GST-MDP25 were added to a 30 μM tubulin solution, and turbidity was monitored during polymerization. The tubulin polymerization rate and mass of the microtubules at a steady state of tubulin polymerization decreased in a concentration-dependent manner when added to MDP25. The addition of 8 μM GST did not affect the mass of microtubules in a steady state of tubulin assembly.
(B) to (D) Microtubules were polymerized in a 20 μM rhodamine-labeled tubulin solution in the absence of GST-MDP25 (B), in the presence of 8 μM GST-MDP25 (C), and in the presence of 10 μM GST (D). Bar in (D) = 10 μm for (B) to (D).
To confirm this result, microtubules polymerized from rhodamine-labeled tubulin incubated with or without MDP25 were also observed by microscopy. Fewer microtubules were detected in the presence of 8 μMΜ GST-MDP25 fusion protein (Figure 2C) compared with microtubules in the absence of MDP25 (Figure 2B) or in the presence of 10 μMΜ GST protein (Figure 2D). This finding is consistent with the results of the turbidimetric assay and suggests that MDP25 functions as a microtubule destabilizer. Additionally, it also indicates that the tag (GST or GFP) does not significantly disturb the effect of MDP25 on microtubules (see Supplemental Figure 4 online).
MDP25 Induces Microtubule Depolymerization
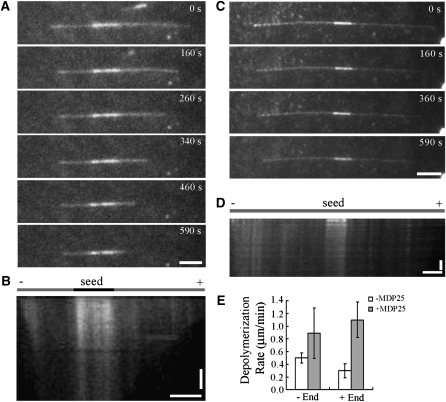
To further elucidate how MDP25 affects microtubules, rhodamine-labeled microtubules were imaged using total internal reflection fluorescence microscopy (TIRFM). Paclitaxel-stabilized and rhodamine-labeled microtubules were broken into small pieces that were used as microtubule seeds. After centrifuging to remove paclitaxel, rhodamine-labeled and unlabeled tubulins were added to the polymerization assay at a 3:1 ratio for 30 min in PIPES-KOH, EGTA, and MgCl2 (PEM) buffer plus 1 mM GTP. For the visual fluorescence analysis of microtubule depolymerization, purified GST-MDP25 was incubated with rhodamine-labeled microtubules at room temperature. The polarity of microtubules was judged by the length of the microtubules at the two ends of preformed microtubule seeds.
The microtubule depolymerization assays were monitored for 10 min (Figure 3). Microtubules disassembled at a rate of ~0.26 μm/min (plus end) and 0.46 μm/min (minus end) in the absence of MDP25 (Figures 3C to 3E; see Supplemental Movie 1 online). However, the rate increased to ~1.16 μm/min (plus end) and 0.89 μm/min (minus end) after the addition of 80 nM GST-MDP25 (Figures 3A, 3B, and 3E; see Supplemental Movie 2 online). This result demonstrated that MDP25 regulates microtubule dynamics by depolymerizing microtubules.
Figure 3.
MDP25-Dependent Microtubule Depolymerization in Vitro.
(A) and (C) MDP25 directly induced microtubule depolymerization in a TIRFM assay. Preformed rhodamine-labeled microtubules were incubated with 80 nM MDP25 (A) or 0 nM MDP25 (C), and the images of immobilized microtubules were recorded for 10 min at 10-s intervals. A few fluorescent images at different time points during the recording process are shown.
(B) and (D) Kymographs show the effect of preformed microtubules depolymerized by MDP25 or without MDP25, respectively.
(E) An analysis summary of at least 30 microtubules of each sample shows the depolymerization rate at both microtubule ends.
Horizontal bars = 5 μm; vertical bars = 2 min. Error bars indicate ± sd.
MDP25 Functions as a Negative Regulator of Hypocotyl Elongation
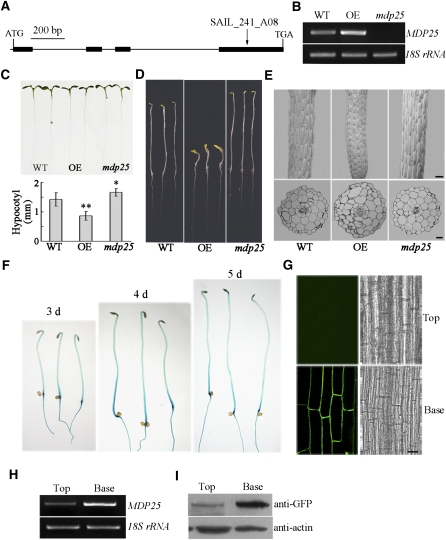
To analyze the function of MDP25 in Arabidopsis, a T-DNA insertion mutant mdp25 (SAIL_241_A08) and MDP25-GFP–overexpressing seedlings were obtained. The homozygous mdp25 mutant contained a T-DNA insertion in the exon, and no full-length transcript was detected by RT-PCR (Figures 4A and 4B). However, a partial transcript that is upstream of the T-DNA insertion site was identified (see Supplemental Figure 5 online). Nevertheless, the mdp25 phenotype indicated that the function of MDP25 was abolished or dramatically affected in the mutant (Figures 4C and 4D). In addition, we obtained another MDP25 T-DNA insertion allele (i.e., mdp25-1-SALK_022955) with a T-DNA insertion in the intron, which expressed a reduced level of MDP25 RNA. This mutant exhibited a similar phenotype as mdp25 (see Supplemental Figures 6A and 6B online).
Figure 4.
MDP25 Negatively Regulates Hypocotyl Cell Elongation.
(A) Physical structure of Arabidopsis MDP25. MDP25 contains four exons and three introns, which are represented by filled boxes and lines, respectively. The position of a T-DNA insertion mutant, designated mdp25 (T-DNA line SALK_241_A08), is noted by arrows above the diagram.
(B) RT-PCR analysis of full-length transcripts of MDP25 in seedlings of the wild-type Columbia ecotype (WT), MDP25-GFP transgenic Arabidopsis line (OE), and mdp25 mutant, with 18S rRNA used as a control.
(C) and (D) The mdp25 mutant has longer hypocotyls (t test, *P < 0.05), whereas the hypocotyls are shorter in MDP25-GFP transgenic Arabidopsis grown on half-strength Murashige and Skoog in continuous light for 7 d ([C]; t test, **P < 0.01) or the dark (D) for 6 d. The graph shows the average hypocotyl length measured from at least 22 seedlings under light growth. Error bars indicate ±se.
(E) Shapes of hypocotyl cells were disordered in MDP25-GFP transgenic Arabidopsis and mdp25 seedlings. The top panels show the scanning electron microscopy images, and the bottom panels show the light microscopy images of hypocotyl cross sections.
(F) to (I) MDP25 was mainly expressed in the nongrowing region of dark-grown hypocotyls of Arabidopsis.
(F) Histochemical GUS staining of PMDP25:GUS:TMDP25 transgenic seedlings grown in the dark for 3, 4, and 5 d.
(G) Confocal images of hypocotyls of 4-d-old dark-grown PMDP25:MDP25:GFP transgenic plants. The epidermal cells of a cell file of the hypocotyl were numbered from the base to the top. “Base” represents the region of 1 to 10 cells, and “Top” represents the region of 11 to 20 cells.
(H) and (I) RT-PCR (H) and immunoblot (I) against anti-GFP antibody show that MDP25 was highly expressed in the basal region and minimally expressed in the top region of etiolated hypocotyls. 18S rRNA or actin was used as a loading control. Three biological replicates were performed and led to similar results.
Bars = 100 μm in (E) and (G).
[See online article for color version of this figure.]
Line 6 of MDP25-GFP–overexpressing Arabidopsis seedlings (OE6) was selected for cell and microtubule analysis, and the transcription level of MDP25 was considerably enhanced in the overexpressing line (Figure 4B). Immunoblot assay further showed that a band at ~64 kD, the molecular mass of the MDP25-GFP fusion protein, was specifically identified by an anti-GFP antibody in total extracts of MDP25-GFP–overexpressing Arabidopsis plants, suggesting that the MDP25-GFP was properly expressed in vivo (see Supplemental Figure 7 online).
Observations of 1-week-old seedlings of MDP25-overexpresssing lines showed that the hypocotyl length was dramatically reduced. Hypocotyls were longer in mdp25 mutant seedlings than in wild-type seedlings when grown in continuous light and even more so when the seedlings were grown in the dark. Statistical analysis using a paired Student’s t test indicated that this difference was significant (Figures 4C and 4D; see Supplemental Figure 8 online). To confirm that the hypocotyl phenotype of mdp25 was linked to the T-DNA insertion, mdp25 mutants were transformed with a construct expressing MDP25-GFP driven by its native promoter. The expression level of MDP25-GFP in the transgenic line (C-mdp25) was similar to MDP25 in the wild type, and the longer hypocotyl phenotype of mdp25 was suppressed upon the expression of this fusion (see Supplemental Figures 9A and 9B online). This observation also indicated that the MDP25-GFP fusion protein was functional. In addition, mdp25-1 showed slightly longer hypocotyls than the wild type (see Supplemental Figure 6 online), demonstrating that the phenotype of the mdp25 mutant is related to MDP25 expression level.
Scanning electron microscopy inspection of the hypocotyls revealed that the epidermal cells that overexpressed MDP25 were shorter and had a spindle shape. The cortex cells of MDP25-overexpressing Arabidopsis hypocotyls were shorter and slightly wider, and the cells of mdp25 mutants were longer compared with the wild type (Table 1). Further observations by cross-section microscopy revealed an overall swelling pattern, and the cell layers were obviously disordered in MDP25-overexpressing Arabidopsis. However, the epidermal cell shape and cortex cells of mdp25 mutant hypocotyls showed weak abnormalities compared with the wild type (Figure 4E). Thus, MDP25 plays a negative regulatory role in hypocotyl cell elongation.
Table 1.
Size of Hypocotyl Cells of MDP25 Transgenic Arabidopsis Grown in the Light for 7 d
| Parameter of Hypocotyl Cells | Wild Type | MDP25 Overexpressing | mdp25 |
| Length of hypocotyl cortex cells (μm) | 124.68 ± 34.55 (n = 100) | 60.80 ± 10.01 (n = 110) | 145.61 ± 29.14 (n = 100) |
| Width of hypocotyl cortex cells (μm) | 28.55 ± 5.88 (n = 100) | 33.28 ± 5.50 (n = 110) | 28.32 ± 4.82 (n = 100) |
Hypocotyl cells elongate quickly without cell division, and hypocotyls that are grown in the dark are excellent models for examining cell elongation. The precise growing region during hypocotyl development has been well defined. For example, the cells at the top of the hypocotyl region begin to expand 3 to 4 d postgermination, while the five basal cells stop growing (Gendreau et al., 1997). If a protein plays a positive role in hypocotyl growth, then it is expected to be highly expressed at the top region but not at the basal region of etiolated hypocotyls. To analyze the expression pattern of MDP25 at the tissue level, a construct was made in which the β-glucuronidase (GUS) reporter gene was placed under the ~1.0-kb MDP25 promoter. This construct (PMDP25:GUS) was introduced into wild-type plants using Agrobacterium tumefaciens–mediated transformation. Twenty-five independent transgenic lines were stained for GUS activity. GUS staining and confocal microscopy revealed that MDP25 was mostly expressed in the basal region of hypocotyls after 3 to 5 d of growth in the dark (GUS or PMDP25:MDP25:GFP transgenic Arabidopsis seedlings; Figures 4F and 4G; see Supplemental Figure 10 online). This region is considered to be a nongrowing region after 4 d of growth in the dark (Gendreau et al., 1997). To verify this result, RNA and total protein were purified from the basal and top regions of hypocotyls from seedlings grown in the dark for 4 d (wild-type and PMDP25:MDP25:GFP transgenic lines, respectively). 18S rRNA and actin were used as loading controls. The RT-PCR and immunoblot results showed that MDP25 was more highly expressed in the basal region than in the top region of etiolated hypocotyls (Figures 4H and 4I), further demonstrating that MDP25 functions as a negative regulator of hypocotyl elongation.
MDP25 Alters Cortical Microtubule Orientation by Destabilizing Microtubules
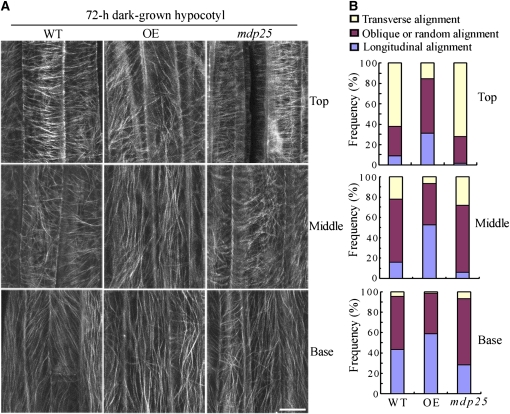
Since MDP25 had a destabilizing effect on microtubules, it is possible that the cortical microtubule array in hypocotyl cells is altered in response to the level of MDP25. Transverse and longitudinal cortical microtubule patterns are related to the rate of elongation in etiolated hypocotyl epidermal cells, and dark-growth hypocotyls can be used to examine the correlation between cell elongation and cortical microtubule organization (Le et al., 2005). Thus, we observed cortical microtubules in hypocotyl epidermal cells of MDP25 transgenic Arabidopsis and mdp25 mutants with a yellow fluorescence protein–tubulin background after 72 h of growth in the dark. Parallel arrays of cortical microtubules were mostly transversely oriented to the longitudinal hypocotyl growth axis in the upper and middle regions of hypocotyls in the wild type and mdp25 mutants. By contrast, most of the cells in MDP25-overexpressing Arabidopsis hypocotyls had random, oblique, or longitudinal arrays (Figures 5A and 5B), consistent with significant inhibition of hypocotyl cell elongation.
Figure 5.
The Cortical Microtubule Array Is Greatly Altered in Etiolated Epidermal Hypocotyl Cells of MDP25-Overexpressing Seedlings.
(A) Cortical microtubules in etiolated hypocotyl epidermal cells of MDP25-overexpressing (OE) and mdp25 seedlings with a yellow fluorescent protein–tubulin background in different regions (upper hypocotyl region, middle hypocotyl region, and basal cells) were observed by confocal microscopy after growth in the dark for 72 h. WT, wild type. Bar = 10 μm.
(B) Frequency of microtubule orientation patterns in different regions of etiolated hypocotyl epidermal cells of the wild type, OE, and the mdp25 mutant (n > 90 cells).
[See online article for color version of this figure.]
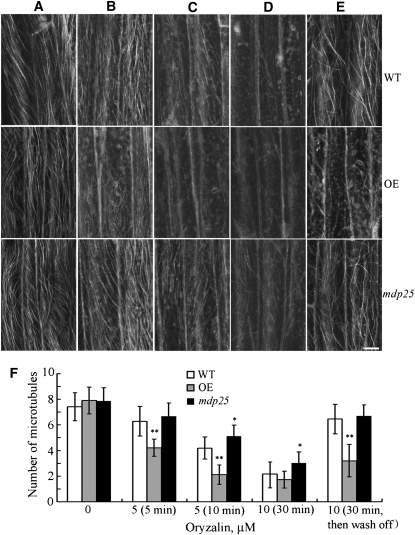
To determine the effect of MDP25 on cortical microtubule-mediated hypocotyl cell elongation, the microtubule-disrupting drug oryzalin was applied to epidermal hypocotyl cells of wild-type, MDP25-overexpressing, and mdp25 mutant Arabidopsis. Since MDP25 was strongly expressed in the basal region of etiolated hypocotyls, we used epidermal cells in the basal region to compare the stability of cortical microtubules. To quantify the effect of oryzalin on the stability of cortical microtubules in wild-type, MDP25-overexpressing, and mdp25 mutant Arabidopsis, we counted the number of cortical microtubules in hypocotyl epidermal cells. A paired Student’s t test was used to identify any significant differences. Our results revealed that cortical microtubules were similar in density before drug treatment in the epidermal cells of wild-type, MDP25-overexpressing, and mdp25 mutant plants (Figures 6A and 6F). However, the number of cortical microtubules in epidermal cells of wild-type, MDP25-overexpressing, and mdp25 mutant plants was significantly different after drug treatment (Figure 6F). Specifically, microtubules were disrupted in MDP25-overexpressing epidermal cells treated with 5 μM oryzalin for 5 min (Figures 6B and 6F), whereas microtubules in wild-type and mdp25 mutant cells were largely unaffected. Increased oryzalin concentration and treatment time resulted in the disruption of most cortical microtubules in both wild-type and MDP25-overexpressing cells. However, cortical microtubules remained relatively unaffected in mdp25 mutant cells (Figures 6C, 6D, and 6F). Cortical microtubules recovered in the cells of wild-type and mdp25 hypocotyls when oryzalin was washed off after the treatment, but many cortical microtubules remained disrupted in the cells of MDP25-overexpressing hypocotyls (Figures 6E and 6F). Thus, microtubules were more sensitive to oryzalin treatment in MDP25-overexpressing cells and less sensitive when the MDP25 expression level was reduced. These results confirm that MDP25 functions as a microtubule destabilizer.
Figure 6.
Cortical Microtubules Are Hypersensitive in MDP25-Overexpressing Arabidopsis Cells but More Resistant in mdp25 Mutant Cells to Oryzalin Treatment.
(A) to (D) Cortical microtubules were observed in epidermal cells in the basal region of etiolated hypocotyls in wild-type (WT), MDP25-overexpressing (OE), and mdp25 mutant seedlings after treatment with 0 μM oryzalin (A), 5 μM oryzalin for 5 min (B), 5 μM oryzalin for 10 min (C), and 10 μM oryzalin for 30 min (D).
(E) After the treatment in (D), oryzalin was washed off, and cortical microtubules were imaged 2 h later. Bar = 10 μm.
(F) Quantification of cortical microtubules in hypocotyl epidermal cells of the wild type, OE, and mdp25 mutant by ImageJ software (n > 50 cells of each sample). Vertical scale represents the number of cortical microtubules across a fixed line (~10 μm) vertical to the orientation of most cortical microtubules in the cell. t tests were performed to compare the number of cortical microtubules in hypocotyl epidermal cells of OE and mdp25 with that of the wild type at the same conditions. **P < 0.01 and * P < 0.05 by t test. Error bars represent ±sd.
Twenty-Three Residues of the N Terminus Are Essential for Targeting MDP25 to Microtubules
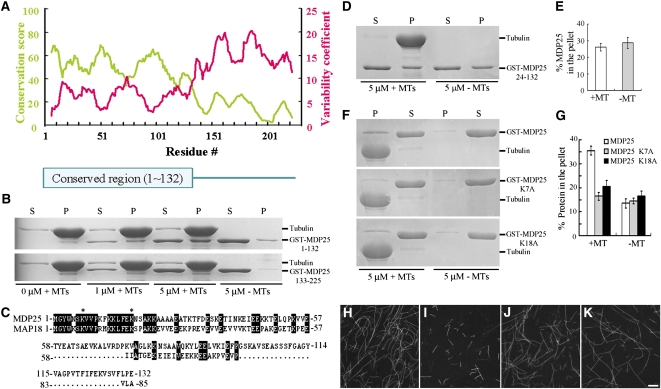
Arabidopsis MDP25 and MAP18 contain the developmentally regulated plasma membrane protein (DREPP) domain and do not exhibit obvious similarities to other known microtubule regulatory proteins. Proteins with the DREPP domain have been identified in several plant species, and these proteins are not related to prototypical microtubule regulatory proteins from animals and fungi (see Supplemental Figure 11, Supplemental Data Set 1, and Supplemental References 1 online). To determine whether the DREPP domain confers microtubule binding activity, the residue conservation and variability of the MDP25 DREPP domain was divided into fragment 1 (amino acids 1 to 132, MDP25 1–132) and fragment 2 (amino acids 133 to 225, MDP25 133–225) using a bioinformatics assay (Figure 7A). Fragment 1 is the most evolutionarily conserved section of MDP25 across various plant species. A GST fusion protein of MDP25 1–132 and MDP25 133–225 was expressed in and purified from bacterial cells, and a cosedimentation assay was performed to assess the microtubule binding activity of the two fragments. SDS-PAGE revealed that MDP25 1–132, but not MDP25 133–225, could bind to microtubules (Figure 7B), suggesting that a protein that contains the DREPP domain may have microtubule binding activity.
Figure 7.
Residues 1 to 23 of MDP25 Are Essential for Targeting MDP25 to Microtubules.
(A) Residue conservation and variability of the MDP25 DREPP domain. The conservation score and variability coefficient were averaged over a sliding window of 11 residues, which are shown in the panel as a green curve relating to the left y axis and red curve relating to the right y axis. The position of the conserved region (i.e., residues with higher conservation scores and lower variability coefficients) is labeled.
(B) The conserved domain of MDP25 across plant species is responsible for microtubule (MT) binding. GST-MDP25 1–132 but not GST-MDP25 133–225 cosedimented with paclitaxel-stabilized microtubules. P, pellet; S, supernatant.
(C) Alignment of amino acid sequences 1 to 132 of MDP25 with MAP18. The asterisks show the sites in the MDP25 amino acid sequence that were mutated (Lys to Ala).
(D) GST-MDP25 24–132 did not cosediment with paclitaxel-stabilized microtubules.
(E) The amount of MDP25 24–132 was estimated following gel density scanning in the absence or presence of 5 μM microtubules from three independent experiments. The amount of MDP25 24–132 in the pellet is expressed as a percentage of total MDP25 24–132.
(F) Compared with wild-type MDP25, less mutated MDP25 fusion protein cosedimented with paclitaxel-stabilized microtubules.
(G) The results of the quantitative analysis of the amount of GST-MDP25 fusion protein in the pellets are shown. The amount of protein was determined by gel scanning from three independent experiments (means ± sd, n = 3).
(H) to (K) Microtubules were polymerized in a 20 μM rhodamine-labeled tubulin solution in the absence of GST-MDP25 (H), in the presence of 8 μM GST-MDP25 (I), in the presence of 10 μM MDP25 K7A (J), or in the presence of 10 μM MDP25 K18A (K). Bar in (K) =10 μm for (H) to (K).
[See online article for color version of this figure.]
The alignment of the amino acid sequences of MDP25 and MAP18 revealed that the N-terminal region was the most conserved (Figure 7C). It has been suggested that residues 1 to 23 of the N terminus of MDP25 are responsible for its targeting to the membrane (Kato et al., 2010b). To investigate the potential role of these residues in regulating microtubules, deletion analyses were performed. Cosedimentation and gel density scanning analyses showed that the truncated protein MDP25 24–132 did not bind to microtubules in vitro (Figures 7D and 7E). We further mutated the two Lys residues of the MDP25 1–23 residues into Ala (Figure 7C, asterisks). The two mutated MDP25 proteins exhibited similarly decreased microtubule binding activity compared with wild-type MDP25, with a significant amount of mutated MDP25 in the supernatant but very little in the pellets with the microtubules (Figures 7F and 7G). To confirm this result, microtubules polymerized from rhodamine-labeled tubulins incubated with wild-type or mutated MDP25 proteins were also observed by microscopy. Fewer microtubules were detected in the presence of 8 μMΜ wild-type MDP25 fusion protein (Figure 7I), but no obvious difference in microtubule density was found between the presence of 10 μMΜ mutated MDP25 proteins (Figures 7J and 7K) and the absence of MDP25 (Figure 7H). Altogether, these results demonstrate that these residues play a crucial role in the binding of MDP25 to microtubules.
Increased Cellular Calcium Disassociates MDP25 from the Plasma Membrane to Destabilize Cortical Microtubules
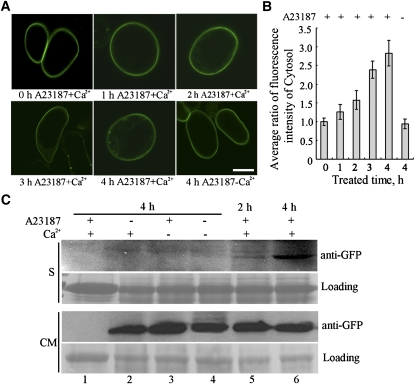
While MDP25 predominantly localizes to the plasma membrane, it also physically regulates cortical microtubules in the cell. A previous report showed that Ca2+ can dissociate MDP25 from the plasma membrane in vitro (Ide et al., 2007). Therefore, calcium may be a key factor in regulating the interaction of MDP25 and cortical microtubules in cells. We tested whether calcium regulates the translocation of MDP25 from the plasma membrane to the cytosol in cells. MDP25-GFP transgenic Arabidopsis suspension cells were treated with the calcium-ionophore A23187. Calcium levels were measured in cells using protoplasts isolated from seedlings that expressed the calcium indicator yellow Cameleon 3.6. The calcium level in the cells increased and reached ~1.63 × 10−7 M after treatment with A23187 plus Ca2+ (see Supplemental Figure 12 and Supplemental References 2 online). The fluorescence intensity ratio of MDP25-GFP in the cytosol between untreated and treated cells 1, 2, 3, and 4 h after treatment was measured. The average intensity of the GFP signal in the cytosol increased after treatment (Figures 8A and 8B), although a strong MDP25-GFP signal was still detected on the plasma membrane, suggesting that MDP25-GFP proteins partially disassociated from the plasma membrane into the cytosol.
Figure 8.
Calcium Partially Dissociates MDP25 from the Plasma Membrane to the Cytosol.
(A) MDP25-GFP transgenic suspension cells were treated with A23187 with or without free Ca2+ for 1, 2, 3, or 4 h. Bar = 20 μm.
(B) GFP signals of MDP25 protein were quantified and are expressed as the ratio of the average fluorescence intensity of the cytosol in each cell. The data represent the means ± sd of three independent experiments; 22 suspension cells were analyzed per sample.
(C) The blot assay showed that the amount of MDP25 protein increased in the soluble fraction when the cells were treated with A23187 plus Ca2+ for 2 and 4 h. Lane 1, wild-type cells; lanes 2 to 6, MDP25-GFP transgenic cells.
[See online article for color version of this figure.]
The soluble and membrane-enriched fractions were then isolated from the cells in the presence of A23187 plus Ca2+. A protein gel blot assay showed that the concentration of MDP25-GFP protein increased in the soluble fraction after 2 and 4 h of treatment (Figure 8C, lanes 5 and 6), which was detected with anti-GFP antibody. A low MDP25-GFP signal was detected in the soluble fraction when the cells were treated with Ca2+ alone or with or without A23187 in the absence of Ca2+ for 4 h (Figure 8C, lanes 2 to 4). MDP25 was not detected by anti-GFP in the control samples from wild-type cells (Figure 8C, lane 1). These results demonstrate that elevated [Ca2+]cyt can partially disassociate MDP25 from the plasma membrane to the cytosol.
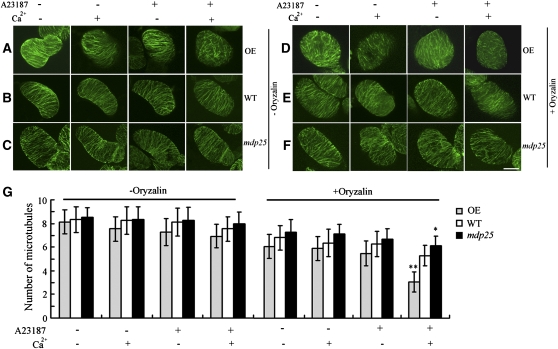
Oryzalin was used to investigate the effect of increasing cytosolic MDP25 concentrations on cortical microtubule stabilization. To quantify the effect of oryzalin on the stability of cortical microtubules, the number of cortical microtubules was counted in suspension cells. The number of cortical microtubules did not significantly change in wild-type, MDP25-overexpressing, or mdp25 suspension cells in the presence or absence of A23187 or Ca2+ (Figures 9A to 9C and 9G). In addition, cortical microtubules exhibited parallel arrays in the presence or absence of A23187 or Ca2+ alone in MDP25-overexpressing, wild-type, and mdp25 suspension cells. However, in the presence of A23187 plus Ca2+, most cortical microtubules were disordered in MDP25-overexpressing but not in wild-type or mdp25 cells (Figures 9A to 9C). Microtubule stabilization was tested by treatment with a lower concentration of oryzalin (1 μM). After treatment with A23187 plus Ca2+, most cortical microtubules were disrupted in MDP25-overexpressing cells (Figures 9D and 9G) but only mildly disrupted in wild-type suspension cells (Figures 9E and 9G) and unchanged in mdp25 mutant cells (Figures 9F and 9G). These observations suggest that more cortical microtubules were destabilized by MDP25 protein in the presence of increased [Ca2+]cyt.
Figure 9.
Microtubule-Destabilizing Activity of MDP25 Is Enhanced in Cells When Cytoplasmic Calcium Levels Are Elevated.
Cortical microtubules are supersensitive to oryzalin after treatment with A23187 plus Ca2+ in MDP25-overexpressing suspension cells. MDP25-overexpressing cells (OE) ([A] and [D]), wild-type cells (WT) ([B] and [E]), mdp25 cells ([C] and [F]) in the absence of oryzalin ([A] to [C]; −Oryzalin) for 15 min and presence of oryzalin ([D] to [F]; +Oryzalin).
(A) to (C) The fluorescence images showed a parallel array of cortical microtubules in the presence or absence of A23187 or Ca2+ in MDP25-overexpressing, wild-type, and mdp25 suspension cells, but most cortical microtubules were disordered in the presence of A23187 plus Ca2+ in MDP25-overexpressing cells.
(D) Most cortical microtubules were disrupted in MDP25-overepxressing cells pretreated with A23187 plus Ca2+, but most were relatively normal in the presence of A23187 or Ca2+ alone with 1 μM oryzalin.
(E) and (F) However, in wild-type and mdp25 suspension cells, cortical microtubules showed little difference in sensitivity to oryzalin treatment when pretreated with A23187, Ca2+, or A23187 plus Ca2+. Compared with cortical microtubules in wild-type cells, cortical microtubules in mdp25 cells were less sensitive to oryzalin treatment when pretreated with A23187 plus Ca2+. Bar = 20 μm in (F).
(G) Quantification of cortical microtubules in Arabidopsis suspension cells of wild-type, MDP25-overexpressing, and mdp25 mutant cells by ImageJ software (n > 50 cells for each sample). Vertical scale represents the number of cortical microtubules across a fixed line (~10 μm) that is vertical to the orientation of most cortical microtubules in the cell. t tests were performed to compare the number of cortical microtubules in suspension cells of OE and mdp25 with that of the wild type at the same conditions. Error bars represent ±sd; **P < 0.01 and *P < 0.05 by t test.
[See online article for color version of this figure.]
To detect whether MDP25 interacts with cortical microtubules when it is disassociated from the plasma membrane, we performed double immunofluorescence microscopy using antitubulin and anti-GFP antibodies. MDP25 partially colocalized with cortical microtubules in MDP25-overexpressing suspension cells pretreated with A23187 plus Ca2+ (see Supplemental Figures 13A to 13C online). The distribution of MDP25 was the same in cells labeled with anti-GFP antibody without microtubule staining (see Supplemental Figures 13D to 13F online). In addition, according to Folta et al. (2003), [Ca2+]cyt levels significantly and transiently increase when etiolated hypocotyls are treated with light. Therefore, we performed immunostaining on the hypocotyls of dark-grown MDP25 transgenic Arabidopsis plants after they had been treated with light. Confocal microscopy showed that MDP25 partially colocalized with cortical microtubules in MDP25-overexpressing epidermal cells pretreated with light for 30 min (see Supplemental Figures 13G to 13I online). These results suggest that elevated levels of cytoplasmic calcium increase the cytoplasmic level of MDP25, which leads to the direct destabilization of microtubules.
DISCUSSION
Hypocotyl elongation is a major feature of plant growth. In this study, we demonstrated that Arabidopsis MDP25 is a novel microtubule-destabilizing factor directly involved in the negative regulation of hypocotyl cell elongation. MDP25 destabilizes cortical microtubules, and its microtubule-destabilizing activity in cells is regulated by the level of cellular calcium.
MDP25 Is a Novel Microtubule Destabilizing Protein
Microtubule regulatory proteins play important roles in the regulation of microtubule organization and dynamics (Sedbrook, 2004; Buschmann and Lloyd, 2008). Those proteins are considered to be microtubule stabilizers or destabilizers depending on their effect on microtubule stability (Heald and Nogales, 2002). For example, MOR1 and MAP65 proteins from Arabidopsis stabilize cortical microtubules (Whittington et al., 2001; Mao et al., 2005). A few microtubule destabilizers, such as katanin (Stoppin-Mellet et al., 2002, 2006), have been identified in plant cells, and they regulate microtubule stability to remodel different microtubule arrays in plant cells. Arabidopsis MAP18 also destabilizes cortical microtubules (Wang et al., 2007). Our results revealed that MDP25 exhibits the typical properties of a microtubule destabilizer both in vitro and in cells. We found that microtubules were more stable in mdp25 mutant cells and more unstable in MDP25-overexpressing cells, compared with wild-type cells. Interestingly, although the organization of cortical microtubules was altered, there was no obvious decrease in cortical microtubule density in MDP25-overexpressing cells. A similar phenomenon was found in Arabidopsis cells overexpressing MAP18 and in lefty mutant cells, in which cortical microtubules are destabilized (Thitamadee et al., 2002; Wang et al., 2007). These observations are consistent with the idea that MDP25 functions as a microtubule destabilizer.
Unlike katanin, which severs microtubules (Stoppin-Mellet et al., 2002), or Xenopus kinesin catastrophe modulator-1 and Xenopus kinesin superfamily protein-2 from frog eggs, which destabilize microtubules by targeting their ends (Desai et al., 1999), MDP25 induces depolymerization of preformed microtubules unevenly along their length. We found that the depolymerization rate of microtubules at the plus ends was similar to the minus end rate, suggesting that binding of MDP25 to microtubules might weaken the association between tubulins or protofilaments in the microtubules. However, the exact mechanism remains to be determined.
The Positional Expression of MDP25 Is Important for the Regulation of Hypocotyl Cell Elongation
MDP25 exhibits a positional expression pattern along etiolated hypocotyls and is mostly expressed at the nongrowing region. MDP25 overexpression inhibits the elongation of hypocotyls. Arabidopsis SPR1 protein and a key transcriptional factor of the brassinosteroid (BR) signal transduction pathway, BRASSINAZOLE RESISTANT1 (BZR1), also exhibit positional expression patterns along etiolated hypocotyls. However, they are mostly expressed at the fast-growing region, and the overexpression of these proteins results in longer hypocotyls (Wang et al., 2002; Nakajima et al., 2004). Thus, two opposite positional expression patterns exist along etiolated hypocotyls that regulate hypocotyl cell elongation in response to developmental and environmental cues.
MDP25 is expressed all along hypocotyl cells grown in the light, with no positional diversity (Ide et al., 2007). We detected MDP25 RNA levels in hypocotyls grown in continuous light or in the dark. Our RT-PCR analysis showed that MDP25 was more highly expressed in the light than in the dark (see Supplemental Figure 14 online), which is consistent with the role of MDP25 in the inhibition of hypocotyl elongation. These observations suggest that the light signaling pathway may be involved in the regulation of MDP25 expression to modulate hypocotyl growth. Additionally, BZR1 binds to the MDP25 promoter (Sun et al., 2010) and participates in the light signaling pathway by repressing the positive regulator GATA2 for Arabidopsis photomorphogenesis (Luo et al., 2010). Recently, a microarray assay showed that MDP25 expression levels were regulated by both BR and light, two crucial hypocotyl elongation regulation factors (Tang et al., 2008; Sun et al., 2010; Luo et al., 2010). Therefore, it is possible that MDP25 is involved in the crosstalk between the BR and light signaling pathways in the regulation of hypocotyl cell elongation. Whether and how BZR1 regulates MDP25 expression patterns along hypocotyls to mediate cell elongation is the subject of future studies.
The Destabilizing Activity of MDP25 Plays a Role in the Orientation of Cortical Microtubules and Regulation of Hypocotyl Elongation
The orientation of cortical microtubules is widely accepted to be interrelated with hypocotyl cell elongation (Le et al., 2005). However, how the orientation of cortical microtubules is regulated during hypocotyl cell elongation remains unclear. Microtubule regulatory proteins play important roles in the regulation of microtubule organization and dynamics (Sedbrook, 2004; Buschmann and Lloyd, 2008). These proteins regulate microtubule stability to remodel different microtubule arrays in plant cells.
However, it is unclear how microtubule stabilizers and destabilizers differentially contribute to the status of hypocotyl growth. Seedlings grown in a medium that contains the microtubule-disrupting drug propyzamide exhibit stunted and radially expanded hypocotyls (Le et al., 2005), indicating that the destabilization of cortical microtubules may result in the inhibition of hypocotyl cell elongation. Thus, microtubule regulatory proteins may play a role in the regulation of hypocotyl cell elongation by regulating the stability of cortical microtubules, which has been tested using mutants and transgenic lines. For example, overexpression of MAP18 induces a disordered cortical microtubule orientation in hypocotyl epidermal cells and inhibits hypocotyl elongation (Wang et al., 2007). By contrast, loss of function of multiple SPR1/SP1L genes, which encode microtubule stabilizer Arabidopsis SPR1, leads to highly disorganized and randomly oriented cortical microtubules in hypocotyl epidermal cells and decreased hypocotyl length (Nakajima et al., 2004, 2006). Our study also supports the possibility that the destabilization of cortical microtubules results in disordered microtubule orientation and consequently inhibits hypocotyl cell elongation.
While Arabidopsis MAP18 and MDP25 both contain the DREPP domain, their level of amino acid identity is low (sequence identity = 28.89%). Double mutants of MAP18 and MDP25 did not exhibit any additional phenotype on hypocotyl cell growth (see Supplemental Figures 15A and 15B online), suggesting that they have diverse physiological functions. Public microarray data (http://jsp.weigelworld.org/expviz/expviz.jsp; accessed July 5, 2011) and real-time PCR analysis revealed that MAP18 is strongly expressed in roots but weakly expressed in hypocotyls, suggesting that MDP25 and MAP18 affect different tissues and developmental/physiological processes (Kato et al., 2010a). Some microtubule regulatory protein mutants and transgenic Arabidopsis lines show broad phenotypes in most organ and cell types, whereas others exhibit phenotypes at specific tissues and certain cell types (Buschmann and Lloyd, 2008). Our results showed that seedlings with increased or decreased MDP25 expression levels had defects in hypocotyl tissues but not other tissues, suggesting that microtubule regulation may depend upon different proteins in different cells or tissue types.
Calcium Regulates MDP25 Localization to Mediate Microtubule Stabilization
Although low levels of MDP25 are also detected in the cytosol using an anti-MDP25 antibody, MDP25 is predominantly located at the plasma membrane (Nagasaki et al., 2008). The N-terminal region of MDP25 (25 amino acid residues) binds to phosphatidylinositol phosphates when MDP25 is located on the plasma membrane (Kato et al., 2010b). Our truncation and point mutation analyses demonstrated that this region is also essential for targeting MDP25 to microtubules, suggesting that MDP25 cannot bind to microtubules when it is on the plasma membrane.
MDP25 binds to the plasma membrane via N-myristoylation (Nagasaki et al., 2008), and no transmembrane domain was predicted (transmembrane domain prediction was performed using the TMHMM server, http://www.cbs.dtu.dk/services/TMHMM/; accessed October 10, 2011), suggesting the possibility of a physical disassociation of MDP25 from the plasma membrane to the cytosol. A previous report showed that 100 mM Ca2+, but not 100 mM NaCl, completely disassociated MDP25 from the plasma membrane in vitro (Ide et al., 2007). Our data show that MDP25 partially dissociated from the plasma membrane and moved into the cytosol when [Ca2+]cyt levels increased, and an increase in cellular MDP25 levels resulted in microtubule destabilization, which consequently inhibited hypocotyl cell elongation. Thus, calcium plays a role in the regulation of MDP25 subcellular localization. The spatial and temporal regulation of cellular Ca2+ concentration is key in the protection of microtubules in the cytosol from the microtubule-destabilizing activity of MDP25.
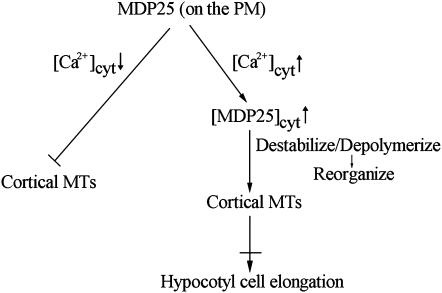
Calcium is widely known to play crucial roles in the regulation of plant cell physiology and cellular responses to environmental cues (Dodd et al., 2010). MAP18, the homolog of MDP25, has been suggested to be involved in the Ca2+-dependent signaling mechanism that induces cortical microtubule disassembly during leaf senescence (Keech et al., 2010). Many factors can increase [Ca2+]cyt in hypocotyl cells, such as light and gravistimulation. Under light/dark cycles, cytosolic Ca2+ displays sinusoidal variations and can reach peak concentrations of 300 to 700 nM (Johnson et al., 1995; Xu et al., 2007). Early and transient Ca2+ influx is a crucial event in light-induced hypocotyl growth inhibition (Folta et al., 2003). When etiolated wild-type, MDP25-overexpressing, and mdp25 mutant seedlings were treated with light and transferred to medium containing the Ca2+ chelator 1,2-bis(2-aminophenoxy)-ethane-N,N,N′,M′-tetraacetic acid (BAPTA), the hypocotyl length of MDP25-overexpressing seedlings was partially restored. However, hypocotyl length in the wild type and mdp25 mutants did not significantly change in the presence or absence of BAPTA (see Supplemental Figure 16A online). A similar analysis was also performed on map18 mdp25 double mutants, and they did not show any additional hypocotyl growth defect after treatment with BAPTA (see Supplemental Figure 16B online). Thus, MDP25 may exert its effects on microtubules via the transient elevation of cytosolic Ca2+ in response to multiple environmental cues. In addition, reducing the MAP18 expression level did not have much of an effect on the defective hypocotyl growth phenotype of the mdp25 mutant, indicating that MAP18 and MDP25 affect different physiological processes of Arabidopsis plant growth. We propose the following model for the function of MDP25 on cortical microtubules in cells (Figure 10). The plasma membrane represents a pool that sequesters MDP25 from microtubules to maintain microtubule stabilization. MDP25 is recruited to the cytosol when [Ca2+]cyt levels are elevated in response to multiple signals. Consequently, increased cytosolic levels of MDP25 induce microtubule destabilization by depolymerizing microtubules. This would result in microtubule reorganization with concomitant effects on cell elongation. Exactly how MDP25 physically binds to and regulates cortical microtubules is the subject of future study.
Figure 10.
Model of MDP25 Function on Cortical Microtubules in the Mediation of Cell Elongation.
During the resting state cytosolic calcium level, MDP25 dominantly binds to the plasma membrane, and its microtubule binding domain is covered in phosphatidylinositol phosphates. Increased cytosolic calcium levels induce partial MDP25 disassociation from the plasma membrane. Increased cytosolic MDP25 levels destabilize cortical microtubules (MTs) by depolymerizing microtubules, result in the disorganization of cortical microtubules, and further inhibit cell elongation.
METHODS
Isolation of MDP25 cDNA Clones from Arabidopsis thaliana
The full-length cDNA sequence of MDP25 was amplified by RT-PCR. GST-tagged fusion proteins were expressed and purified according to the manufacturer’s protocols. The protein concentration was determined using a Bio-Rad protein assay kit. Protein samples were analyzed by SDS-PAGE.
Microtubule Binding and Polymerization Assays
Porcine brain tubulin was purified and labeled with 5- and 6-carboxytetramethylrhodamine succinimidyl ester (NHS)-rhodamine as previously reported (Hyman, 1991; Castoldi and Popov, 2003). Microtubules were prepared, and an analysis of the cosedimentation of the fusion proteins with microtubules was performed according to previously published protocols (Mao et al., 2005). The purified proteins were centrifuged at 200,000g at 4°C for 20 min before use. Prepolymerized and paclitaxel-stabilized microtubules (5 μM) were incubated in PEM buffer plus paclitaxel (PEMT buffer; 1 mM MgCl2, 1 mM EGTA, 100 mM PIPES-KOH, and 20 μM paclitaxel, pH 6.9) with various concentrations of MDP25 fusion proteins at room temperature for 20 min. After centrifugation at 100,000g for 20 min, the supernatant and pellets were subjected to SDS-PAGE. The amount of fusion protein bound to the microtubules was determined by gel scanning, and the binding ratio was analyzed by the Alpha Image 2200 documentation and analysis system (Alpha Innotech Corporation).
Immunofluorescence staining experiments were performed to investigate the binding of MDP25 to microtubules. Rhodamine-labeled and paclitaxel-stabilized microtubules were incubated with GST-MDP25 at a molar ratio of 4:1 for 30 min at room temperature in PEMT buffer. The samples were then labeled with anti-GST antibody at a 1:500 dilution for 15 min. A secondary antibody of fluorescein isothiocyanate–conjugated goat anti-mouse immunoglobulin G (IgG; 1:500 dilution) was then added to the incubation for another 15 min, and the reaction was terminated by the addition of 1% (v/v) glutaraldehyde. The samples were transferred onto slides coated with 0.1% (v/v) poly-l-Lys. Fluorescence images of microtubules and MDP25 were visualized using a spinning disk microscope (Andor Technology) with a ×63 1.42–numerical aperture Olympus objective. Images were captured using Andor ImageQ software, version 1.1 (Andor Technology). MDP25 denatured in boiling water for 2 min and GST protein alone were used as controls.
For the microtubule polymerization assay, GST-MDP25 protein (0, 2, 4, and 8 μM) was added to a 30 μM tubulin solution in PEM buffer plus 1 mM GTP. GST (8 μM) was used as a control. Turbidimetric analysis was used to monitor microtubule polymerization by measuring absorbance at 350 nm at multiple time points. To further examine the effect of MDP25 on tubulin polymerization, 8 μM GST-MDP25 was incubated with 20 μM NHS-rhodamine–labeled tubulin at 35°C for 30 min. Glutaraldehyde (1% [v/v]) was added to terminate the reaction before observation with confocal microscopy, and 10 μM GST was used as a control.
Analysis of MDP25 Promoter:GUS Activity in Etiolated Seedlings
The MDP25 promoter fragment that contained 1011 bp upstream of the translation start site was amplified. After sequencing, the sequence was reconstructed into the pCAMBIA1391 vector. The construct was then transformed into Arabidopsis plants using Agrobacterium tumefaciens (strain GV3101). The homozygous etiolated seedlings were used for the histochemical localization of GUS activity in hypocotyl cells. The GUS staining procedure was performed according to Wang et al. (2007).
PMDP25:MDP25:GFP Construction and Overexpression in Arabidopsis
To visualized MDP25 in cells, a fragment 1011 bp upstream of the initiation codon (ATG) of MDP25 to the stop codon (TGA) was amplified and reconstructed into a pCAMBIA1390 vector. GFP was amplified and ligated at the C terminus of MDP25. To stably overexpress the GFP fusion protein in living cells, full-length MDP25 cDNA was amplified by PCR and subcloned into the pBI221 vector. Additionally, the cDNAs for MDP25 and GFP were amplified and reconstructed into the expression vector pCAMBIA1300, under the control of the 35S promoter and nopaline synthase terminator. The constructs were transformed into Arabidopsis plants by Agrobacterium (strain GV3101). The homozygous lines were used for the subsequent analyses.
RT-PCR and Immunoblot Analysis
To assess MDP25 transcript levels in mdp25 and mdp25-1 mutants, RT-PCR was performed. Total RNA was isolated using TRIzol reagent (Invitrogen). Three independent pairs of primers were used to determine the levels of full-length (5′-GCTGCTGAAGCTACCAAGACC-3′ and 5′-TCAAGGCTTTGGTGGTTCAG-3′) and partial MDP25 transcripts (5′-CTGAAGCTACCAAGACCTTTGAT-3′ and 5′-CCGAAGCTAGACGAAGCC-3′; 5′-GTGAAAGCTGAAGAACCTG-3′ and 5′-AAGGCTTTGGTGGTTCAGC-3′). The primer (5′-TCGTGGAGGAGGTCGTCGTGA-3′ and 5′-TTCAGCCACCGGAACAGCGG-3′) was used to assess MAP18 transcript levels in map18 (Salk_021652) and map18 mdp25 double mutants. 18S rRNA was used as a loading control (5′-CGGCTACCACATCCAAGGAA-3′ and 5′-GCTGGAATTACCGCGCGGCT-3′). The amplification products were visualized by ethidium bromide.
The hypocotyls of Columbia ecotype seedlings or PMDP25:MDP25:GFP transgenic homozygous seedlings grown in the dark for 4 d were used for the analysis. The hypocotyls from the basal region (approximately eight cells) were divided into two parts under a microscope. The primers used for the subsequent detection of MDP25 expression were 5′-CGCAGGACCGGTCACGTTCA-3′ and 5′-TTCAGCCACTGGCGCTGTCG-3′.
The protein extracts were prepared from two parts of hypocotyls of PMDP25:MDP25:GFP transgenic seedlings as described above. The blots were probed with anti-GFP antibody (Roche) at a dilution of 1:5000 with TBST (50 mM Tris, 150 mM NaCl, and 0.05% Tween 20, pH 7.5) and alkaline phosphatase–conjugated goat anti-mouse IgG secondary antibody (Sigma-Aldrich) at a dilution of 1:10,000. Actin was used as a loading control and was detected by antiactin antibody (Sigma-Aldrich).
Microtubule Depolymerization Activity of MDP25 Assessed by TIRFM Assay
To determine the polarity of preformed microtubules, rhodamine-labeled tubulins without unlabeled tubulins were polymerized into rhodamine-labeled microtubules and used as seeds. A rhodamine-labeled tubulin dimer (90 μM) was incubated in 100 mM PEM buffer plus 1 mM GTP for 30 min at 35°C. After centrifugation and resuspension in the PEM buffer, the microtubules were broken into small seeds. The microtubule seeds were incubated with a mixture of rhodamine-labeled and -unlabeled tubulin dimer at a molar ratio of 1:3 plus 1 mM GTP for 30 min at 35°C and then centrifuged and resuspended in PEM buffer with 8 mM vitamin C to a tubulin-dimer concentration of 5 μM.
For the visual fluorescence analysis of microtubule depolymerization, 80 nM purified GST-MDP25 was incubated at room temperature with 5 μM of the rhodamine-labeled polarity-marked microtubules described above. One microliter of the mixture was gently pipetted onto 18 × 18-mm cover slips coated with antitubulin antibody, which bound the microtubules to the cover slip surface without obvious release. The depolymerization assays were monitored using an Olympus IX81 microscope and a total internal reflection fluorescence microscope (Olympus) equipped with an Andor iXon charge-coupled device camera (Andor Technology) and a ×100 1.4–numerical aperture Olympus objective. Total internal reflection fluorescence illumination was performed at λ = 568 nm. The exposure time was 100 ms. Images were captured using Andor ImageQ software, version 1.1 (Andor Technology) and analyzed with Image J 1.38 software (http://rsb.info.nih.gov/ij; accessed October 12, 2011).
Preparation of Suspension Cells
Arabidopsis seedlings were grown on medium (4.4 g/L Murashige and Skoog salt, 30 g/L Suc, 1 mg/L 2,4-D, 0.1 mg/L 6-benzylaminopurine, and 1% agar) for 2 weeks. The leaves were then cut and stored at 21°C on the medium in the dark until calli formed after 1 to 2 months. Calli were pressed into small-cell aggregates and diluted in liquid culture medium (4.4 g/L Murashige and Skoog salt [Sigma-Aldrich], 30 g/L Suc, 2 mg/L 2,4-D, and 0.1 mg/L 6-benzylaminopurine). The suspension was kept in the dark at 23°C on a rotor set to 130 rpm and transferred into new liquid medium every week to obtain a stable suspension culture.
MDP25-GFP Translocation and Oryzalin Treatment in Suspension Cell Assay
Three days after subculturing, the suspension cells were washed with 3% Suc three times to remove the medium and incubated with 20 μM A23187 plus 1 mM CaCl2 at room temperature for 1, 2, 3, and 4 h. The cells treated with 20 μM A23187 without CaCl2 for 4 h were used as a control. The samples were observed by confocal microscopy. The images were analyzed by ImageJ (http://rsb.info.nih.gov/ij/). To calculate the ratio of the cytoplasmic signal of MDP25-GFP for the untreated to treated cells, a small area of fixed size (3 μm × 2 μm) was selected, and the integrated densities within the cytoplasm were measured, with the values of central vacuoles as a background. Three repeated measures were performed for each cell, and the average of the background values was subtracted from the average values for the cytoplasmic signals. The cytoplasmic signal ratio of treated to untreated cells was then calculated from at least 22 cells from each treatment.
For oryzalin treatment, the cells were incubated with 20 μM A23187 plus 1 mM CaCl2 at room temperature for 2 h and then treated with 1 μM oryzalin for 15 min. After fixation with 4% paraformaldehyde, microtubule immunostaining was performed according to Li et al. (2006).
Quantification of Cortical Microtubules in the Cell
To quantify the numbers of cortical microtubules, ImageJ software was employed. To count the number of cortical microtubules in cells, a vertical line in the orientation of the most cortical microtubules of fixed length (10 μm) was drawn, and the number of cortical microtubules across this line was counted. Five repeated measures were performed for each cell, and at least 50 cells from each treatment were used. The values were recorded and the significance of difference was analyzed using a paired Student’s t test.
Immunostaining of MDP25 in Hypocotyl Epidermal Cells
Three-day-old etiolated MDP25-GFP–overexpressing seedlings were treated with light for 30 min and immediately fixed in 4% (v/v) paraformaldehyde and 0.1% (v/v) glutaraldehyde in PEM buffer (50 mM PIPES, 2 mM EGTA, and 2 mM MgCl2, pH 6.9, containing 0.05% [v/v] Triton X-100) for 50 min at room temperature. The samples were washed three times in this buffer and subsequently digested with 1% (w/v) pectinase (Sigma-Aldrich), 1% (w/v) cellulose (Sigma-Aldrich) and 0.4 M mannitol dissolved in PEM buffer for 1 h at 37°C. After rinsing in PEM buffer three times, the cotyledon and part of the root were removed. Autofluorescence caused by free aldehydes from glutaraldehyde fixation was reduced by treatment with 1 mg/mL NaBH4 in PBS for 20 min followed by treatment with 50 mM Gly in PBS (incubation buffer) for 30 min. The samples were incubated with rabbit GFP antibody diluted 1:200 in PBS containing 3% (w/v) BSA and 0.1% (w/v) Tween 20 at 4°C overnight. The samples were washed three times with PBS buffer before adding mouse anti-β-tubulin monoclonal antibody (Sigma-Aldrich) diluted 1:800 in PBS containing 3% (w/v) BSA and 0.1% (w/v) Tween 20 at 4°C overnight. Secondary antibodies were Alexa-488 conjugated goat anti-rabbit IgG (Molecular Probes) and tetramethylrhodamine-5(and 6-)-isothiocyanate–conjugated goat anti-mouse IgG (Sigma-Aldrich), both diluted 1:500 in PBS containing 3% (w/v) BSA and 0.1% (w/v) Tween 20, and the samples were incubated at 35°C for 2 h. After rinsing four times in PBS, the samples were mounted in 0.1% (w/v) paraphenylene diamine (Sigma-Aldrich P-5412) in 1:1 PBS-glycerol, pH 9. Fluorescent images were collected with a Zeiss 510 META confocal microscope.
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative under the following accession numbers: MDP25, At4g20260; and MAP18, At5g44610.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. GST-Tagged Fusion Protein of MDP25: Expression and Purification.
Supplemental Figure 2. MDP25 Is Detected at the Plasma Membrane but Not at Other Cell Membranes in MDP25-GFP–Overexpressing Lines.
Supplemental Figure 3. Plasma Membrane Localization of MDP25 Is Independent of the Presence of Cortical Microtubules.
Supplemental Figure 4. GFP or GST Tag Does Not Affect MDP25-Induced Inhibition of Tubulin Polymerization in Vitro.
Supplemental Figure 5. Identification of Partial Transcripts of MDP25 in the mdp25 Mutant.
Supplemental Figure 6. Abnormal Etiolated Hypocotyl Elongation in Another MDP25 T-DNA Insertion Line (SALK_022955).
Supplemental Figure 7. MDP25-GFP Is Properly Expressed in Vivo.
Supplemental Figure 8. Etiolated Hypocotyls of mdp25 Are Significantly Longer Than Those of the Wild Type.
Supplemental Figure 9. Longer Hypocotyl Phenotype of mdp25 Is Completely Suppressed by Expression of MDP25-GFP Driven by Its Native Promoter.
Supplemental Figure 10. Fluorescence Image of MDP25-GFP Distribution in the 4-d-old Dark-Grown Hypocotyl of PMDP25:MDP25:GFP Transgenic Plants.
Supplemental Figure 11. Phylogenetic Tree of the Plant DREPP Protein Family.
Supplemental Figure 12. Elevated Cytosolic Ca2+ Levels Induced by A23187 Plus Ca2+ Treatment.
Supplemental Figure 13. MDP25 Partially Colocalized with Cortical Microtubules in Cells.
Supplemental Figure 14. MDP25 Is Highly Expressed in Light-Grown Hypocotyls but Minimally Expressed in Dark-Grown Hypocotyls.
Supplemental Figure 15. The map18 mdp25 Double Mutant Did Not Exhibit an Additional Hypocotyl Growth Defect Phenotype.
Supplemental Figure 16. Growth of Light-Treated Hypocotyls from MDP25 Transgenic Arabidopsis Is Partially Restored When Grown on Ca2+ Chelator Medium.
Supplemental Data Set 1. The Clustal Alignment of the DREPP Proteins and the Outgroup Protein ZP01619125.
Supplemental Movie 1. Depolymerization of Preformed Microtubules.
Supplemental Movie 2. MDP25 Induces Microtubule Depolymerization.
Supplemental Movie Legends 1. Legends for Supplemental Movies 1 and 2.
Supplemental References 1. Supplemental References for Supplemental Figure 11.
Supplemental References 2. Supplemental References for Supplemental Figure 12.
Acknowledgments
We thank Christopher J. Staiger (Purdue University) and Patrick J. Hussey (Durham University, UK) for critically commenting on the manuscript. We thank the ABRC (The Ohio State University) for providing the T-DNA insertion lines. This research was supported by grants from the National Basic Research Program of China (2012CB114200 to M.Y.), the Natural Science Foundation of China (31070258 to T.M.; 30830058 and 30721062 to M.Y.), and 111 Project (B06003), Chinese Universities Scientific Fund (2011JS108 to T.M.).
AUTHOR CONTRIBUTIONS
T.M. designed the project. J.L., X.W., T.Q., Y.Z., X.L., J. S., Y.Z., L.Z., and Z.Z. performed specific experiments and analyzed data. T.M. wrote the article. M.Y. and T.M. revised and edited the article.
References
- Baskin T.I. (2005). Anisotropic expansion of the plant cell wall. Annu. Rev. Cell Dev. Biol. 21: 203–222 [DOI] [PubMed] [Google Scholar]
- Baum G., Long J.C., Jenkins G.I., Trewavas A.J. (1999). Stimulation of the blue light phototropic receptor NPH1 causes a transient increase in cytosolic Ca2+. Proc. Natl. Acad. Sci. USA 96: 13554–13559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buschmann H., Lloyd C.W. (2008). Arabidopsis mutants and the network of microtubule-associated functions. Mol. Plant 1: 888–898 [DOI] [PubMed] [Google Scholar]
- Castoldi M., Popov A.V. (2003). Purification of brain tubulin through two cycles of polymerization-depolymerization in a high-molarity buffer. Protein Expr. Purif. 32: 83–88 [DOI] [PubMed] [Google Scholar]
- Chan J., Calder G., Fox S., Lloyd C. (2007). Cortical microtubule arrays undergo rotary movements in Arabidopsis hypocotyl epidermal cells. Nat. Cell Biol. 9: 171–175 [DOI] [PubMed] [Google Scholar]
- Desai A., Verma S., Mitchison T.J., Walczak C.E. (1999). Kin I kinesins are microtubule-destabilizing enzymes. Cell 96: 69–78 [DOI] [PubMed] [Google Scholar]
- Dodd A.N., Kudla J., Sanders D. (2010). The language of calcium signaling. Annu. Rev. Plant Biol. 61: 593–620 [DOI] [PubMed] [Google Scholar]
- Ehrhardt D.W. (2008). Straighten up and fly right: Microtubule dynamics and organization of non-centrosomal arrays in higher plants. Curr. Opin. Cell Biol. 20: 107–116 [DOI] [PubMed] [Google Scholar]
- Ehrhardt D.W., Shaw S.L. (2006). Microtubule dynamics and organization in the plant cortical array. Annu. Rev. Plant Biol. 57: 859–875 [DOI] [PubMed] [Google Scholar]
- Folta K.M., Lieg E.J., Durham T., Spalding E.P. (2003). Primary inhibition of hypocotyl growth and phototropism depend differently on phototropin-mediated increases in cytoplasmic calcium induced by blue light. Plant Physiol. 133: 1464–1470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gendreau E., Traas J., Desnos T., Grandjean O., Caboche M., Höfte H. (1997). Cellular basis of hypocotyl growth in Arabidopsis thaliana. Plant Physiol. 114: 295–305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamada T. (2007). Microtubule-associated proteins in higher plants. J. Plant Res. 120: 79–98 [DOI] [PubMed] [Google Scholar]
- Hardham A.R., Gunning B.E.S. (1978). Structure of cortical microtubule arrays in plant cells. J. Cell Biol. 77: 14–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heald R., Nogales E. (2002). Microtubule dynamics. J. Cell Sci. 115: 3–4 [DOI] [PubMed] [Google Scholar]
- Hyman A.A. (1991). Preparation of marked microtubules for the assay of the polarity of microtubule-based motors by fluorescence. J. Cell Sci. Suppl. 14(Suppl): 125–127 [DOI] [PubMed] [Google Scholar]
- Ide Y., Nagasaki N., Tomioka R., Suito M., Kamiya T., Maeshima M. (2007). Molecular properties of a novel, hydrophilic cation-binding protein associated with the plasma membrane. J. Exp. Bot. 58: 1173–1183 [DOI] [PubMed] [Google Scholar]
- Johnson C.H., Knight M.R., Kondo T., Masson P., Sedbrook J., Haley A., Trewavas A. (1995). Circadian oscillations of cytosolic and chloroplastic free calcium in plants. Science 269: 1863–1865 [DOI] [PubMed] [Google Scholar]
- Kaloriti D., Galva C., Parupalli C., Khalifa N., Galvin M., Sedbrook J.C. (2007). Microtubule associated proteins in plants and the processes they manage. J. Integr. Plant Biol. 49: 1164–1173 [Google Scholar]
- Kato M., Nagasaki-Takeuchi N., Ide Y., Maeshima M. (2010a). An Arabidopsis hydrophilic Ca2(+)-binding protein with a PEVK-rich domain, PCaP2, is associated with the plasma membrane and interacts with calmodulin and phosphatidylinositol phosphates. Plant Cell Physiol. 51: 366–379 [DOI] [PubMed] [Google Scholar]
- Kato M., Nagasaki-Takeuchi N., Ide Y., Tomioka R., Maeshima M. (2010b). PCaPs, possible regulators of PtdInsP signals on plasma membrane. Plant Signal. Behav. 5: 848–850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keech O., Pesquet E., Gutierrez L., Ahad A., Bellini C., Smith S.M., Gardeström P. (2010). Leaf senescence is accompanied by an early disruption of the microtubule network in Arabidopsis. Plant Physiol. 154: 1710–1720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le J., Vandenbussche F., De Cnodder T., Van Der Straeten D., Verbelen J.P. (2005). Cell elongation and microtubule behaviour in the Arabidopsis hypocotyl: Responses to ethylene and auxin. J. Plant Growth Regul. 24: 166–178 [Google Scholar]
- Li C.L., Chen Z.L., Yuan M. (2006). Actomyosin is involved in the organization of the microtubule preprophase band in Arabidopsis suspension cultured cells. J. Integr. Plant Biol. 48: 53–61 [Google Scholar]
- Lloyd C., Chan J. (2008). The parallel lives of microtubules and cellulose microfibrils. Curr. Opin. Plant Biol. 11: 641–646 [DOI] [PubMed] [Google Scholar]
- Luo X.M., et al. (2010). Integration of light- and brassinosteroid-signaling pathways by a GATA transcription factor in Arabidopsis. Dev. Cell 19: 872–883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao T.L., Jin L.F., Li H., Liu B., Yuan M. (2005). Two microtubule-associated proteins of the Arabidopsis MAP65 family function differently on microtubules. Plant Physiol. 138: 654–662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagasaki N., Tomioka R., Maeshima M. (2008). A hydrophilic cation-binding protein of Arabidopsis thaliana, AtPCaP1, is localized to plasma membrane via N-myristoylation and interacts with calmodulin and the phosphatidylinositol phosphates PtdIns(3,4,5)P(3) and PtdIns(3,5)P(2). FEBS J. 275: 2267–2282 [DOI] [PubMed] [Google Scholar]
- Nakajima K., Furutani I., Tachimoto H., Matsubara H., Hashimoto T. (2004). SPIRAL1 encodes a plant-specific microtubule-localized protein required for directional control of rapidly expanding Arabidopsis cells. Plant Cell 16: 1178–1190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakajima K., Kawamura T., Hashimoto T. (2006). Role of the SPIRAL1 gene family in anisotropic growth of Arabidopsis thaliana. Plant Cell Physiol. 47: 513–522 [DOI] [PubMed] [Google Scholar]
- Niwa Y., Yamashino T., Mizuno T. (2009). The circadian clock regulates the photoperiodic response of hypocotyl elongation through a coincidence mechanism in Arabidopsis thaliana. Plant Cell Physiol. 50: 838–854 [DOI] [PubMed] [Google Scholar]
- Paredez A.R., Somerville C.R., Ehrhardt D.W. (2006). Visualization of cellulose synthase demonstrates functional association with microtubules. Science 312: 1491–1495 [DOI] [PubMed] [Google Scholar]
- Sedbrook J.C. (2004). MAPs in plant cells: delineating microtubule growth dynamics and organization. Curr. Opin. Plant Biol. 7: 632–640 [DOI] [PubMed] [Google Scholar]
- Sedbrook J.C., Kaloriti D. (2008). Microtubules, MAPs and plant directional cell expansion. Trends Plant Sci. 13: 303–310 [DOI] [PubMed] [Google Scholar]
- Shinkle J.R., Jones R.L. (1988). Inhibition of stem elongation in cucumis seedlings by blue light requires calcium. Plant Physiol. 86: 960–966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith L.G., Oppenheimer D.G. (2005). Spatial control of cell expansion by the plant cytoskeleton. Annu. Rev. Cell Dev. Biol. 21: 271–295 [DOI] [PubMed] [Google Scholar]
- Somerville C. (2006). Cellulose synthesis in higher plants. Annu. Rev. Cell Dev. Biol. 22: 53–78 [DOI] [PubMed] [Google Scholar]
- Stoppin-Mellet V., Gaillard J., Vantard M. (2002). Functional evidence for in vitro microtubule severing by the plant katanin homologue. Biochem. J. 365: 337–342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoppin-Mellet V., Gaillard J., Vantard M. (2006). Katanin’s severing activity favors bundling of cortical microtubules in plants. Plant J. 46: 1009–1017 [DOI] [PubMed] [Google Scholar]
- Sun Y., et al. (2010). Integration of brassinosteroid signal transduction with the transcription network for plant growth regulation in Arabidopsis. Dev. Cell 19: 765–777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang W., Deng Z., Oses-Prieto J.A., Suzuki N., Zhu S., Zhang X., Burlingame A.L., Wang Z.Y. (2008). Proteomics studies of brassinosteroid signal transduction using prefractionation and two-dimensional DIGE. Mol. Cell. Proteomics 7: 728–738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thitamadee S., Tuchihara K., Hashimoto T. (2002). Microtubule basis for left-handed helical growth in Arabidopsis. Nature 417: 193–196 [DOI] [PubMed] [Google Scholar]
- Tsuchida-Mayama T., Sakai T., Hanada A., Uehara Y., Asami T., Yamaguchi S. (2010). Role of the phytochrome and cryptochrome signaling pathways in hypocotyl phototropism. Plant J. 62: 653–662 [DOI] [PubMed] [Google Scholar]
- Twell D., Park S.K., Hawkins T.J., Schubert D., Schmidt R., Smertenko A., Hussey P.J. (2002). MOR1/GEM1 has an essential role in the plant-specific cytokinetic phragmoplast. Nat. Cell Biol. 4: 711–714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., Zhu L., Liu B.Q., Wang C., Jin L.F., Zhao Q., Yuan M. (2007). Arabidopsis MICROTUBULE-ASSOCIATED PROTEIN18 functions in directional cell growth by destabilizing cortical microtubules. Plant Cell 19: 877–889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z.Y., Nakano T., Gendron J., He J., Chen M., Vafeados D., Yang Y., Fujioka S., Yoshida S., Asami T., Chory J. (2002). Nuclear-localized BZR1 mediates brassinosteroid-induced growth and feedback suppression of brassinosteroid biosynthesis. Dev. Cell 2: 505–513 [DOI] [PubMed] [Google Scholar]
- Whittington A.T., Vugrek O., Wei K.J., Hasenbein N.G., Sugimoto K., Rashbrooke M.C., Wasteneys G.O. (2001). MOR1 is essential for organizing cortical microtubules in plants. Nature 411: 610–613 [DOI] [PubMed] [Google Scholar]
- Xu X., Hotta C.T., Dodd A.N., Love J., Sharrock R., Lee Y.W., Xie Q., Johnson C.H., Webb A.A. (2007). Distinct light and clock modulation of cytosolic free Ca2+ oscillations and rhythmic CHLOROPHYLL A/B BINDING PROTEIN2 promoter activity in Arabidopsis. Plant Cell 19: 3474–3490 [DOI] [PMC free article] [PubMed] [Google Scholar]