Several protocols and methods were developed to study the in vivo structure, ultrastructure, and physiology of sieve tubes. The ultrastructure of sieve tubes differs significantly from what has been shown so far. The impact on phloem function is discussed.
Abstract
Since the first ultrastructural investigations of sieve tubes in the early 1960s, their structure has been a matter of debate. Because sieve tube structure defines frictional interactions in the tube system, the presence of P protein obstructions shown in many transmission electron micrographs led to a discussion about the mode of phloem transport. At present, it is generally agreed that P protein agglomerations are preparation artifacts due to injury, the lumen of sieve tubes is free of obstructions, and phloem flow is driven by an osmotically generated pressure differential according to Münch’s classical hypothesis. Here, we show that the phloem contains a distinctive network of protein filaments. Stable transgenic lines expressing Arabidopsis thaliana Sieve-Element-Occlusion-Related1 (SEOR1)–yellow fluorescent protein fusions show that At SEOR1 meshworks at the margins and clots in the lumen are a general feature of living sieve tubes. Live imaging of phloem flow and flow velocity measurements in individual tubes indicate that At SEOR1 agglomerations do not markedly affect or alter flow. A transmission electron microscopy preparation protocol has been generated showing sieve tube ultrastructure of unprecedented quality. A reconstruction of sieve tube ultrastructure served as basis for tube resistance calculations. The impact of agglomerations on phloem flow is discussed.
INTRODUCTION
All organisms, in particular multicellular ones, need to maintain functional coherence. They must coordinate activities and processes that occur in their various parts and integrate a variety of stimuli from the outside to produce meaningful responses. In land plants, the phloem tissue is thought to play an essential role in organismal coordination.
The phloem tissue of angiosperms consists of phloem parenchyma cells, sieve elements, and companion cells. Sieve elements assemble into sieve tubes, which form a continuous microfluidics network throughout the plant body. The primary function of the phloem is the long-distance distribution of photoassimilates and signals. For rapid movement of large fluid volumes, tube systems are used in many natural and artificial systems. To support urban centers, we use pipelines for water, oil, sewage, etc. In animals, circulatory tube systems translocate nutrients and waste to be exchanged at dedicated locations. In basically all known cases, the driving force for flow is a pressure differential that may be positive (e.g., garden hose) or negative (e.g., xylem). Thus, it appears intuitive that the driving force to distribute photoassimilates in the phloem would follow similar mechanisms, and it is not surprising that an osmotically generated pressure differential is the central element of Münch’s pressure flow hypothesis (Münch, 1927, 1930).
However, on closer inspection, there are some striking differences between the phloem and other systems. To minimize resistance, the tube should be free of obstructions and the walls should be smooth. This is relatively easy to realize when flow occurs through the extracellular matrix. The phloem, however, is the only long-distance transport system where flow occurs intercellularly in the symplast. Thus, constituents required to maintain tube integrity, such as organelles, are located in the path of flow. Although the cellular infrastructure has been minimized by loss of the nucleus, the vacuole, ribosomes, Golgi, and the cytoskeleton, sieve elements are not empty tubes but contain smooth endoplasmic reticulum (ER), mitochondria, sieve element plastids, and phloem proteins (P proteins; Knoblauch and Peters, 2010).
Independent of the length of the tube, a single internal obstruction may increase the resistance of the tube to the point of complete flow stoppage. Obstructions can be used for flow control, for example, by a stopcock, but it bears some risks if a clot is formed unintentionally (e.g., stroke and heart attacks). Since the first descriptions of the phloem, clots in the lumen and often on the sieve plate were commonly observed. Initially, these clots were designated as slime (Hartig, 1854). Later, they were renamed P proteins due to their proteinaceous nature (Cronshaw, 1975). When transmission electron microscopy (TEM) became available, a surprising variety of P proteins were discovered. They were characterized as amorphous, crystalline, filamentous, tubular, and fibrillar (for an overview, see Evert, 1990). The higher resolution, however, did not change the fact that they were most commonly found in the lumen or inside the sieve plate pores, which led to one of the most controversial discussions in plant physiology of the last century. Some investigators believed that electron micrographs represented the in vivo state. Because bulk flow through occluded pores could not be driven by pressure gradients, alternative translocation hypotheses were developed, such as the electroosmotic theory (e.g., Fensom, 1957; Spanner, 1958, 1970; Siddiqui and Spanner, 1970). Other authors, however, believed that P proteins shown in many micrographs were dislocated during tissue preparation. Sometimes, plates had open pores after gentle preparation (e.g., Fellows and Geiger, 1974; Fisher, 1975; Russin and Evert, 1985). This led to the conclusion that sieve tubes form a continuous path and that phloem flow can be driven by an osmotically generated pressure differential (Thompson, 2006). However, convincing evidence has not been shown.
The major reason for this controversy is in the nature of phloem anatomy and the resulting difficulties with in vivo observations of sieve tubes. The phloem is generally embedded in layers of ground tissue preventing direct observation of cellular features. Therefore, some degree of invasive preparation is required. The sieve tube system builds a network in the plant body, and the exceptionally high turgor (Turgeon, 2010) causes an immediate effect over large distances when a tube is severed. This led to an overwhelming amount of ultrastructural data accounting for different degrees of injury, but it is not clear if uninjured sieve tubes have ever been observed in TEM micrographs.
Recently, we isolated three genes expressing phloem-specific P proteins involved in the formation of forisomes (Pélissier et al., 2008). Forisomes are contractile P protein bodies occurring in faboid legumes. They were suggested to reversibly block sieve tubes in case of injury (Knoblauch et al., 2001, 2003). We designated the gene family Sieve-Element-Occlusion (SEO; Pélissier et al., 2008). We found homologous genes of unknown function in other plant species, including Arabidopsis thaliana (At3g01680; Pélissier et al., 2008), and designated them Sieve-Element-Occlusion-Related (SEOR). Recently, Rüping et al. (2010) suggested calling genes involved in forisome formation SEO-F (for Sieve Element Occlusion by Forisomes). In our opinion, there is neither a reason nor a justification to rename the gene families. The SEO family implies forisome genes and SEOR signifies homologous genes in nonfabaceae families as originally described by Pélissier et al. (2008).
Without a clear understanding of the underlying construction of the sieve tube system, it will be impossible to properly understand its functional principles. Therefore, we intended to elucidate the ultrastructure of uninjured sieve tubes by TEM by comparing our findings to those obtained in in vivo studies by confocal microscopy.
RESULTS
Because electron microscopy samples are under high vacuum, samples have to be fixed and dehydrated. Since the structure of proteins, membranes, and other cellular components is often defined by their interaction with water molecules, dehydration may lead to artifacts. The degree of artifacts varies with cell and protein type. To draw appropriate conclusions, an in vivo reference is most helpful. Unfortunately, such a reference is lacking for sieve tubes. Sieve tube components are usually invisible in the light microscope because of their size and/or lack of contrast. In addition, it would be important that sieve tubes be observed without preparation, which is unfortunately usually impossible because of the anatomy of the plant. So far, not a single study has shown cellular features of the phloem without preparation of the tissue. Even the method that allowed us to investigate individual uninjured sieve elements in broad bean (Vicia faba) at high resolution requires removal of cortical cell layers (Knoblauch and van Bel, 1998). Our aim for this study, however, was to investigate sieve tubes without any mechanical intervention.
In aboveground organs, in addition to being embedded in a thick layer of ground tissue, the phloem is covered by more or less opaque, pigmented cells, making a direct observation impossible. The cells in roots of many plant species, however, are relatively transparent. To study the phloem, a thin cortical layer is beneficial, since cell wall–cytoplasm interfaces lead to reflection and refraction phenomena. In this regard, Arabidopsis thaliana appears ideal. The cortical layer in primary roots is just three cell layers thick, and the root cells do not contain significant amounts of polyphenolics and other compounds that would significantly affect optical properties. The small size of Arabidopsis sieve tubes is a drawback, but with high-end instrumentation, subcellular structures can be visualized.
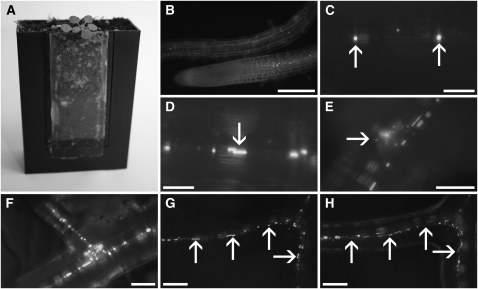
We expected that the forisome homolog gene SEOR1 in Arabidopsis encodes a specific P protein. We cloned the gene, including its endogenous promoter, fused yellow fluorescent protein (YFP) to its C terminus and generated transgenic Arabidopsis lines. To study roots in vivo, we used microscopy rhizosphere chambers (Micro-ROCs) and grew Arabidopsis plants expressing SEOR1-YFP for structural studies. The chambers consist of plant pots with a cover glass as one of the side walls, optimized for high resolution. Root growth is funneled along the cover glass by a porous mesh, while root hairs are in direct contact with soil. In contrast with glass-bottom Petri dishes, where plants are grown in an artificial medium under sterile conditions and at 100% humidity, Micro-ROCs allow direct visualization of the root system in a natural soil environment, which also includes symbionts. Maximum resolution without any preparation or manipulation of the tissue is possible (Figure 1A).
Figure 1.
Epifluorescence of SEOR1-YFP in Living Roots.
(A) An Arabidopsis plant grown in a Micro-ROC. The root hairs of the plant are in contact with the soil, while the roots are forced to grow along the cover slip.
(B) A root tip and a young part of a root as observed by epifluorescence in a Micro-ROC. Cells were stained with synapto-red to visualize cell outline. Bright spots along the root are SEOR1-YFP fusion proteins. The image is a single frame of Supplemental Movie 1 online.
(C) and (D) Higher magnification of SEOR1-YFP fusion proteins (C). In young vascular tissue, the proteins appear as round amorphous bodies (arrows), which increase in size and become elongated in consecutive slightly older areas ([D], arrow).
(E) Early indication of root branch formation is the abundance of SEOR1-YFP bodies beside the file (arrow).
(F) After the root tip broke through the cortical layer, a new vascular file formed.
(G) A root containing numerous amorphous bodies in a file (arrows).
(H) Ten hours later, the amorphous bodies have developed into more defined structures (arrows).
Bars = 150 μm in (B), 25 μm in (C) and (D), 50 μm in (E) and (F), and 100 μm in (G) and (H).
Development and Structure of SEOR1
YFP fluorescence was first detectable in the differentiation zone of young roots (Figure 1B). After elongation, spherical amorphous protein agglomerates were found inside the cells (Figure 1C). Time-lapse movies revealed quick movements of the bodies in actively growing root regions (see Supplemental Movie 1 online). In the course of further development, the protein bodies increased in size and became elongated (Figure 1D).
An early indication of branch root development is the appearance of additional small protein bodies beneath the sieve tube (Figure 1E). The bodies increase in size (Figure 1F) until the branch root breaks through the cortical layer and branch root sieve tubes are formed (Figure 1G). At certain locations, a sudden and significant alteration in the shape of the protein bodies can be observed. The oval, amorphous bodies condense and transform into defined filamentous structures (Figure 1H). Both amorphous bodies and filamentous structures are usually aligned in files.
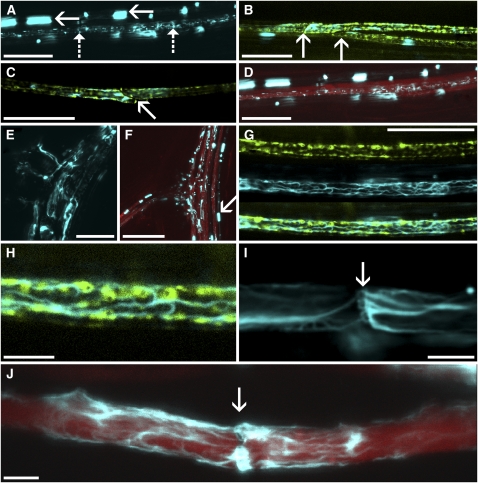
To study subcellular localization, we investigated living root sieve tubes by confocal microscopy. Besides the prominent amorphous bodies, fine strands became visible (Figure 2A). To identify the cell type in which the fine strands occur, we generated a double transgenic line expressing SEOR1-YFP and green fluorescent protein (GFP) tagged to the ER under control of the Medicago truncatula SEO2 promoter. This promoter is known to be sieve element specific, which allows the unequivocal distinction of sieve elements and companion cells (Knoblauch and Peters, 2010). Filaments are restricted to sieve elements (Figures 2B and 2C), while amorphous protein may occur outside a file, probably in young developing tubes. To determine the location of actively translocating sieve tubes, we loaded leaves of transgenic Arabidopsis plants grown in Micro-ROCs with carboxyfluorescein diacetate (CFDA; Wright and Oparka, 1997) and observed transport in uninjured roots. Translocation occurred in cells containing filamentous proteins (Figure 2D).
Figure 2.
In Vivo Observation of Sieve Tube Structure.
(A) SEOR1-YFP fusion protein distribution within vascular bundles shows files containing amorphous bodies (solid arrows) and files containing fine strands (dashed arrows).
(B) and (C) GFP specifically tagged to the sieve tube ER (green) reveals that SEOR1-YFP (cyan) is located in sieve elements. Arrows point toward sieve plates.
(D) Loading of phloem with CFDA (red) shows that fine SEOR1-YFP filaments (cyan) are located within mature, translocating sieve tubes. Amorphous bodies are located outside of translocating files.
(E) and (F) In older root tissue, a large amount of SEOR1-YFP is abundant in sieve tubes. Consecutive files lead into branch roots (E). Before dispersion, amorphous SEOR1-YFP bodies (arrow) are indicative of young developing sieve tubes and do not translocate CFDA (red).
(G) and (H) At highest resolution, the ER (green) is surrounded by a fine SEOR1-YFP filament meshwork (cyan).
(I) SEOR1-YFP filaments cover and/or traverse a sieve plate (arrow), outlining the sieve plate pores.
(J) Despite the presence of filaments (cyan) in the pores, sieve tubes are fully functional, as indicated by translocation of CFDA (red).
Bars = 25 μm in (A) to (E) and (G), 75 μm in (F), and 5 μm in (H) to (J).
In older roots, SEOR1-YFP filaments became more prominent (Figures 2E and 2F). Amorphous protein bodies are located in neighboring and nontranslocating cell files, supporting the notion that these are young developing sieve elements (Figure 2F) and that sieve tubes become active after the proteins transform into filaments.
At highest resolution, a meshwork becomes visible that usually extends throughout the sieve element (Figure 2G). The meshwork and ER cover a significant fraction of the sieve tube membrane (Figure 2H) and are closely associated. At the sieve plate, the meshwork traverses the sieve plate pores, outlining their location (Figures 2I and 2J).
We reinvestigated the literature and our own vast collection of sieve tube micrographs but failed to find any structures in electron micrographs of Arabidopsis and other species that resembled the meshworks found in our confocal images of mature translocating tubes. We therefore decided to reinvestigate sieve tube ultrastructure.
TEM of Sieve Tubes
The formation of artifacts in sieve tubes due to preparation and fixation for electron microscopy has been discussed in numerous publications (e.g., Spanner, 1978; Evert, 1982). A large number of fixation protocols, including chemical and freeze fixation of plant and callus sieve tubes has been tested (e.g., Cronshaw and Esau, 1967; Wooding, 1969; Sjolund and Shih, 1983). A crucial step, however, is the preparation before fixation. Since electron microscopy generally requires small samples, the tissue is usually sectioned. This procedure induces artifacts before fixation is even initiated. Therefore, we decided to fix entire plants to prevent prefixation artifacts.
Chemical fixation may be suboptimal because of the slow diffusion of the fixative through multiple tissue layers. Ultrafast freezing procedures require vitrification of the tissue; otherwise, water-crystal formation leads to complete distortion of cellular features. Although high-pressure freezing provides superior vitrification to a depth of up to 500 μm and represents the optimum procedure for tissue cryofixation, the maximum sample size is an area of 1 × 2 mm, too small for any plant (Bozzola and Russell, 1999). Other freezing techniques such as jet freezing or plunge freezing usually vitrify the outer 5 to 40 μm of the tissue at best. Even in very small plants such as Arabidopsis, the phloem is never closer than 50 μm to the surface. However, the phloem has one major advantage over other tissues. It carries a high concentration of an intrinsic cryoprotectant: Suc.
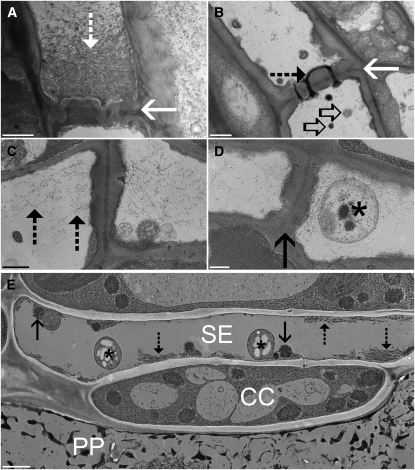
Standard fixation of Arabidopsis leaf and stem segments after excision leads to the typical precipitation of P proteins on the sieve plate (Figure 3A), which has been seen before in many other plant species. Often the pores are filled with protein filaments and lined with a thick layer of callose (e.g., Wooding, 1969; Figure 3B). We compared chemical fixation of excised tissue with chemical fixation of whole young plants. P proteins in uncut sieve tubes were more evenly distributed throughout the lumen, and the organelles were usually intact (Figures 3C and 3D). The appearance resembled tubes after gentle preparation (Ehlers et al., 2000). However, no structure could be found that matched the strands observed by confocal microscopy in living tubes.
Figure 3.
TEM of Sieve Tubes in Arabidopsis.
(A) and (B) Standard chemical fixation of tissue sections of Arabidopsis shows the typical abundance of P protein filaments (dashed arrow) in front of the sieve plate ([A], solid arrow) or in the sieve plate pores (B). Remnants of sieve element plastids ([B], open arrows) can be found around the sieve plate.
(C) and (D) Standard fixation of whole Arabidopsis plants resembles images after gentle preparation. Protein filaments (dashed arrows) are located in the lumen of the sieve element, but a sieve element plastid (asterisk) in front of the sieve plate (solid arrow) is intact.
(E) Arabidopsis phloem tissue after plunge freezing of entire plants. Phloem parenchyma cells (PP) are completely destroyed by the freezing procedure, but sieve elements (SE) and companion cells (CC) show unprecedented preservation. Sieve element plastids (asterisk) and mitochondria (solid arrows) are well preserved. Most importantly, protein filaments (dashed arrows) are not randomly located in the lumen but consist of longitudinally aligned filaments at the margins of the cells.
Bars = 1000 nm in (A), (B), and (E) and 500 nm in (C) and (D).
We then took young Arabidopsis plants in the four to eight leaf state and plunge froze them in slush nitrogen (~63K). Subsequently, the tissue was freeze-substituted in aldehyde fixative containing acetone and postfixed in osmium tetroxide (see Methods for a detailed protocol). Initially, preservation was poor and it turned out that plants had to be grown at 100% humidity either in soil or on Petri dishes to achieve appropriate preservation. Such growth conditions prevented the formation of a thick cuticle that obviously represents a significant freezing barrier. In some cases, it is beneficial to add 0.1 to 0.5% water to the glutaraldehyde-containing acetone fixative. The water supports preservation and easier sectioning of the tissue.
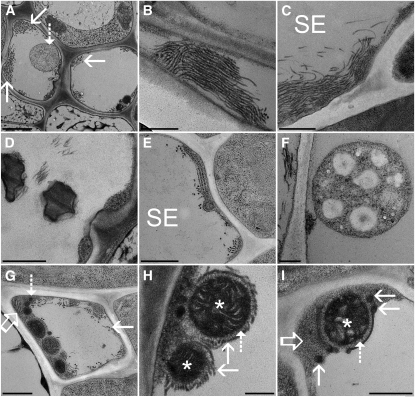
In plunge-frozen and freeze-substituted tissue, parenchyma cells surrounding the sieve elements and companion cells are severely damaged and no subcellular structures are preserved (Figure 3E). However, unprecedented preservation is achieved in sieve elements and companion cells of source leaves. Sieve element plastids and mitochondria are intact. Most importantly, protein filaments, 20 nm (± 1.7 nm) in diameter and often forming bundles, are located at the margin of the cells (Figure 3E), while the fine filaments in the lumen, usually found after chemical fixation (cf. Figures 3A and 3C), are absent. In accordance with confocal images (Figures 2G and 2I), filament bundles are preferentially oriented longitudinally to the sieve tube axis (Figures 4A to 4C). Bundles may consist of <10 to >100 individual filaments (Figure 4A). Tangential and longitudinal sections suggest that the filaments are relatively flexible, may bend backward, and often are not strictly aligned in parallel (Figures 4B and 4C).
Figure 4.
Fine Structure of Arabidopsis Sieve Tubes.
(A) Cross section of an Arabidopsis vascular bundle showing two sieve elements. Large bundles of filaments (solid arrows) are located at the margins of the cells. Filaments and sieve element plastids (dashed arrow) fill a significant portion of the tube lumen.
(B) and (C) Tangential section through the marginal layer of a sieve element showing aligned filaments in a bundle (B). While the filaments are usually aligned in parallel to the sieve elements’ (SE) long axis, they appear flexible and may bend backward (C).
(D) Sieve plate pores are unobstructed and do not contain any detectable callose.
(E) Cross section of a sieve element (SE) showing stacked ER cisternae. The ER is usually not as well preserved as in standard fixed tissue. It appears to descend into a less defined amorphous ground matrix.
(F) A sieve element plastid with a smooth surface in direct contact with sieve tube sap.
(G) A cross section through a sieve element showing a variety of sieve tube components, such as mitochondria, P protein filaments (solid arrow), ER (open arrow), and electron-dense vesicles (dashed arrow) embedded in an amorphous ground matrix.
(H) Two mitochondria (asterisks) covered by a halo of proteins (dashed arrow) that attach them to protein filaments (solid arrow).
(I) In other cases, mitochondria (asterisk) are surrounded by membranes from which electron-dense vesicles (solid arrows) may bud off. Again, membranes are not in direct contact with the mitochondria but are attached by small proteins (dashed arrows). The electron-dense vesicles and mitochondria are usually embedded in the amorphous ground matrix (open arrow), while P protein filaments and sieve element plastids are always in contact with sieve tube sap.
Bars = 1000 nm in (A), 500 nm in (B) to (E), (G), and (I), and 250 nm in (F) and (H).
Sieve plate pores are often unobstructed (but see below), and there is no indication of callose deposition around the pores (Figure 4D). In accordance with investigations after chemical fixation, the ER is organized in stacks (Figure 4E). Sieve element plastids have a smooth surface and are not in close contact with other structures or organelles (Figures 3E, 4A, and 4F). By contrast, mitochondria are always embedded in a parietal layer (Figure 4G), and they are always surrounded by a “halo” of 34.5 nm (± 8 nm; Figures 4H and 4I) to which other structures, such as protein filaments (Figure 4H) or membranes (Figure 4I), are attached. In addition, it appears that there is an amorphous ground matrix of the parietal layer embedding all other structures (Figures 4G to 4I). The nature of this matrix is obscure. In some cases, it looks as though ER membranes disintegrate or transform into this amorphous structure (Figure 4E). The layer, however, could also consist of parietal proteins found in other plant species (e.g., Knoblauch and van Bel, 1998). In addition to mitochondria, smaller, electron-dense vesicles can frequently be found in the parietal ground matrix (Figures 4G to 4I), which seem to bud off of membrane structures. The nature of the membranes is yet unclear. They may be constituents of the ER, but they often appear more electron dense and significantly better preserved than the ER, suggesting a different molecular composition.
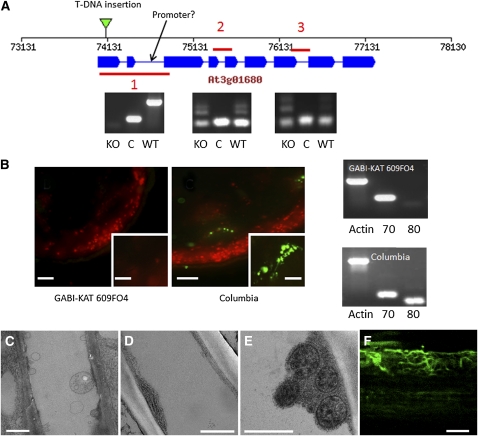
To verify that the filaments and bundles are formed by SEOR1, we investigated the Arabidopsis T-DNA insertion mutant GABI-KAT 609F04. The T-DNA insertion is located in the first exon (Figure 5A). PCR experiments verified that the protein is effectively knocked out, but truncated mRNAs are formed. PlantpromoterDB 2.0 predicted a possible weak promoter in the second intron, which might lead to the formation of the observed truncated mRNAs. However, the mutant did not show antigenicity to the P protein–specific antibody RS21 (Toth and Sjolund, 1994; Toth et al., 1994), while the phloem in wild-type plants was well labeled (Figure 5B). The neighboring gene, At3g01670, which shows high homology to At3g01680, was not affected by the T-DNA insertion (Figure 5B). TEM images confirmed the absence of SEOR filaments, while all other structures found in wild-type plants were present (Figures 5C to 5E). Complementation by transformation of the mutant with SEOR1-GFP led to recovery of filament generation in the mutant. The complemented line showed reduced fluorescence compared with the SEOR1-YFP line; however, bundles of filaments resembling those of SEOR1-YFP plants could clearly be visualized (Figure 5F).
Figure 5.
SEOR1 Mutant-DNA Insertion Line.
(A) A representation of the Arabidopsis gene At3g01680 indicating the location of the T-DNA insertion in the GABI-KAT 609F04 line and the location of a possible weak promoter indicated by analysis using PlantpromoterDB 2.0 (http://ppdb.agr.gifu-u.ac.jp/ppdb/cgi-bin/index.cgi). Also shown are three sections amplified by RT-PCR showing that a truncated mRNA product containing sections 2 and 3 is produced in the T-DNA insertion mutant. C, amplification control; KO, GABI-KAT 609f04; WT, wild-type Arabidopsis line Columbia.
(B) Immunolocalization using a P protein–specific antibody indicates P proteins are absent in GABI-KAT 609F04 (insets are higher magnification images of single vascular bundles), and RT-PCR analysis shows the expression of the adjacent gene At3g01670 (70) is unaffected in the At3G01680 (80) T-DNA insertion mutant (Actin serves as an amplification control).
(C) TEM micrograph of SEOR1 T-DNA insertion mutant after standard chemical fixation. Filaments filling the lumen of the sieve tube as shown in Figure 3 are absent.
(D) and (E) TEM micrographs of At SEOR1 T-DNA insertion mutant after freeze substitution of whole plants. At SEOR1 filaments are absent, but all other structures, such as ER, mitochondria, and clamps proteins surrounding the mitochondria, are present.
(F) Transformation of KO:GABI-KAT 609f04 with At SEOR1-GFP leads to filament formation.
Bars = 100 μm in (B) (inset = 20 μm), 1 μm in (C), 500 nm in (D) and (E), and 3 μm in (F).
Obstructions in Sieve Tubes
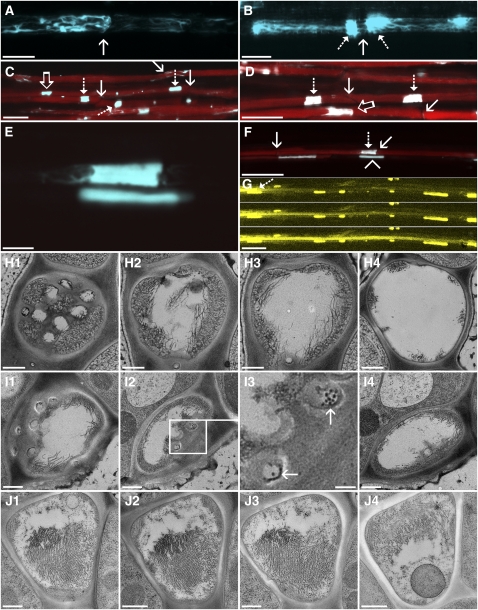
Frequently, we noticed in confocal images that SEOR1-YFP forms agglomerates filling significant portions of the tube diameter at, or close to, the sieve plate. The appearance of these agglomerates is extremely variable. Some sieve tubes may contain large bundles (Figure 6A), while others have agglomerates on both sides of the sieve plate and filaments spanning through the pores (Figure 6B). In many cases, however, multiple large agglomerations fill the entire lumen of the tube (Figures 6C and 6D). We loaded the phloem with CFDA and, surprisingly, independent of the amount of protein in the tube lumen, all sieve tubes were fully functional (Figures 6C and 6D). The structural state of the protein filling the lumen is different in mature and young sieve tubes. Developing sieve tubes contain amorphous protein bodies (Figure 6E, lower body), while the agglomerations in mature sieve tubes consist mainly of filaments and bundles, indicated by their extensions (Figure 6E, upper body; see also Figures 6H to 6J). Despite the large amount of protein within the flow path, the tube contains CFDA (Figure 6F). The lower tube is still in development with isolated sieve elements and a sieve element in the transition phase (Figure 6F, lower file, left).
Figure 6.
Obstructions in Arabidopsis Sieve Tubes.
(A) to (D) Protein agglomerations (cyan) in the lumen of sieve tubes are variable. In many sieve elements, filaments are located at the margins of the cells.
(A) The presence of filaments on the sieve plate (arrow) outlines their location.
(B) A larger agglomeration of P protein (dashed arrows) on both sides of a sieve plate (solid arrow). The P protein agglomerations fray out into filaments. Some of the filaments connect through the sieve plate.
(C) and (D) Overview images of CFDA (red) translocating sieve tubes containing massive P protein agglomerations. Sieve plates (solid arrows) are often not directly covered with P protein agglomerations. Some agglomerations appear to completely fill the lumen of the tube (dashed arrows), while others only cover part of it (open arrows).
(E) Two P protein agglomerates. The upper agglomeration frays out into filaments. Some darker spots indicate the location of organelles, in this case most likely mitochondria. The lower agglomerate is completely amorphous.
(F) The same sieve tubes as shown in (E). The upper file is fully mature and translocates CFDA (red) despite the presence of the large P protein agglomeration (dashed arrow) in front of the sieve plate (solid arrow). The lower tube is not fully mature. The amorphous P protein body (arrowhead) has not transformed into strands and is not translocating CFDA, while the next sieve element on the left in the same file is in the transition phase.
(G) Three consecutive images of a FRAP experiment. The dashed arrow indicates the location of the P protein agglomeration shown in (E). The tube has been bleached by the laser and quickly refills after decrease of the laser energy indicating transport.
(H) Four TEM images of a serial section of a sieve tube in the area of the sieve plate.
(H1) to (H4) A cross section through the plate shows several open pores in the center, while significant portions at the margin of the plate are covered with filaments (H1). In consecutive sections (~1 μm apart from each other), filaments fill >50% of the lumen (H2) and move toward the membrane (H3) until they form discrete bundles (H4).
(I) Serial section through an Arabidopsis sieve plate, oriented in a slight angle in relation to the sieve tube.
(I1) and (I2) While most pores are open (I1), filaments are present on the plate (I2).
(I3) Higher magnification of sieve pores in the sieve plate shown in (I2) (box). SEOR1 filaments can be seen in some pores ([I3], arrows).
(I4) A few micrometers behind the plate, filaments move toward the margins.
(J) Four images of a serial section through the lumen of a sieve tube containing an agglomeration. Major parts of the lumen are filled with P protein filaments, but a channel is unobstructed. The filaments are mostly oriented in parallel and have a pseudocrystalline appearance. A sieve element plastid is abundant in J4. The distance from (J1) to (J4) is 7 μm.
Bars = 10 μm in (A) and (B), 25 μm in (C), (D), (F), and (G), 5 μm in (E), and 500 nm in (H) to (J).
Since CFDA is loaded in leaves and diffuses into source tissue until it reaches the phloem, no distinct front but a gradual increase in fluorescence results in the transport phloem. The problem becomes especially obvious when neighboring companion and parenchyma cells light up almost as fast as the sieve tubes. Since the quality and speed of loading is dependent on multiple factors, it has not yet been possible to standardize the procedure to always obtain the same loading. Therefore, we were not yet able to exclude the possibility that sieve tubes containing large agglomerations did not actually translocate but that the fluorescence diffused from neighboring tubes.
To unequivocally prove that the tubes are actively translocating, we conducted studies on real-time movement of fluorescent dyes within individual sieve tubes. We grew plants in Micro-ROCs, loaded them with CFDA, and photobleached CFDA in the tube to produce a distinct front of fluorescence and imaged refilling at 0.3-s intervals (fluorescence recovery after photobleaching [FRAP]). Since the laser of a confocal microscope can be directed with pixel size accuracy, precise areas can be targeted. Figure 6G shows three frames of a FRAP experiment of the tube shown in Figures 6E and 6F. Refilling occurs at a velocity of ~60 μm s−1 downstream of the obstruction (see Supplemental Movie 2 online). There is no other sieve tube or lateral sieve plate that would allow bypassing the agglomeration. We therefore conclude that transport occurs through agglomerations.
Currently, phloem translocation is thought to be driven by an osmotically generated pressure differential. Sieve tubes supposedly provide a channel of adequately low hydraulic resistance permitting pressure differential driven flow. The presence of agglomerations in the flow path necessitated a reevaluation of the feasibility of a pressure flow. To calculate the increase of resistance by obstructions, the resolution of confocal microscopy is insufficient. Only TEM permits precise measurements. TEM sections, on the other hand, are only in the range of 80-nm thick and the agglomerations are rare in comparison to unobstructed areas. To section through a single sieve element of 120-μm length, 1500 individual cross sections are required, and on average, only every tenth sieve element contains an agglomeration. By conducting serial cross sectioning, we were able to find two sieve plates and one area of obstruction. At the sieve plate, numerous filaments are located that traverse the pores (Figure 6H1). Right behind the sieve plate, ~55% of the lumen is obstructed by filaments (Figure 6H2). With increasing distance, the filaments are located further toward the margins (Figure 6H3) until they form distinct bundles (Figure 6H4). Sieve plates in Arabidopsis are often not strictly perpendicular to the tube axis (Figure 6I1). The sieve plate in Figure 6I also contains filaments on the plate. While some pores are open, others contain several filaments that obstruct a certain percentage of the pore’s lumen (Figure 6I3). The only agglomeration we found so far is shown in Figure 6J. The distance between image Figure 6J1 and 6J4 is ~7 μm. Major parts of the lumen are filled with filaments with the exception of an area of ~0.5 × 1 μm. Average filament diameter is 21.8 ± 2.5 nm (n = 100) with 6.1 ± 1.3 nm (n = 43) spacing between filaments within the agglomeration.
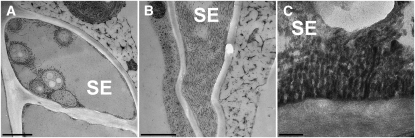
To see if the abundance of filaments and bundles is a general feature of dicot sieve tubes, we investigated two nonrelated plant species. While Arabidopsis has developed into an important model plant, ultrastructural studies are very limited. We chose tobacco (Nicotiana tabacum), since extensive ultrastructural data are available, and black cottonwood (Populus trichocarpa) as model tree species. Preservation of some ultrastructural features was not as good as in Arabidopsis, and modification of the fixation protocol will be required in the future. However, filaments and bundles can be seen in the periphery of sieve elements (Figures 7A to 7C). In contrast with Arabidopsis, sieve element plastids of tobacco are decorated with filaments (Figure 7A). The size of the filaments differed slightly from that of Arabidopsis with an average of 18.68 ± 2.1 nm in tobacco and 23.88 ± 2.1 nm in black cottonwood.
Figure 7.
SEOR1-Like Filaments in Tobacco and Black Cottonwood.
(A) A cross section through a tobacco sieve element (SE) shows several sieve element (SE) plastids covered with Arabidopsis SEOR1 filaments and bundles.
(B) A tangential section through an Arabidopsis sieve element along the organelle containing layer close to the plasma membrane. A large SEOR1 bundle of multiple filaments covers the membrane.
(C) Longitudinal section through a black cottonwood sieve tube. The preservation is not as good as in Arabidopsis and tobacco, but Arabidopsis SEOR1-like filaments are visible.
Bars = 1000 nm (A) and (B) and 150 nm in (C).
Sieve Tube Structure and Its Impact on Phloem Translocation
The structures found in whole-plant freeze substitutions differ significantly from what has been described using other preparation and fixation protocols. The large amount of Arabidopsis SEOR1 filaments in the translocation path inevitably leads to the question of its impact on tube resistance. The question is: Can a pressure flow, as discussed by Münch (1930), drive phloem translocation? Figure 8 shows a schematic summary of our findings in Arabidopsis sieve tubes. In recent phloem flow calculation models, sieve tubes were expected to be empty tubes and the space occupied by organelles and other structures is considered to be below the error of sieve tube geometry measurements (Thompson and Holbrook, 2003; Mullendore et al., 2010). In reality, however, even in areas without significant SEOR1 accumulation, a major fraction of the tube lumen turned out to be unavailable for translocation. Usually, up to 30% (Figure 4G) are occupied by sieve tube constituents. Even in the wider tubes of tobacco, up to 35% (Figure 7A) of the lumen is filled with sieve element plastids and other organelles. On top of this, SEOR1 agglomerations need to be included in the calculations.
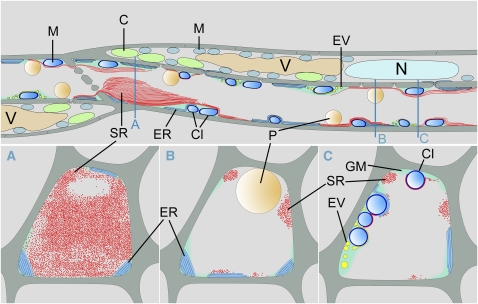
Figure 8.
Schematic Reconstruction of an Arabidopsis Sieve Tube.
Reconstruction of the structure of a sieve element-companion cell complex as found in in vivo confocal studies and after freeze substitution of whole plants. Sieve elements contain ER, mitochondria covered with clamp proteins, and electron-dense vesicles. While those structures are usually embedded in an amorphous ground matrix, SEOR1 filaments and sieve element plastids are always in direct contact with the sieve tube sap. A SEOR1 agglomeration is shown in front of a plate that does not fill the entire lumen of the sieve element. Companion cells contain all organelles typical for a plant cell, but only nucleus, vacuoles, chloroplasts, and mitochondria are shown. Blue lines indicate the location of a cross section for (A) to (C). C, chloroplast; Cl, clamp proteins; EV, electron-dense vesicles; GM, ground matrix; M, mitochondria; N, nucleus; P, plastid; SR, SEOR1 filaments; V, vacuole.
To quantify the effects of the organelles and SEOR1 filaments on the flow, we calculate the influence of these on the hydrostatic pressure difference between source and sink tissues required to drive the observed flow. For simplicity, we consider a single sieve tube as a proxy for the phloem and model the translocation pathway as consisting of a collection of approximately cylindrical sieve tube elements lying end to end separated by sieve plates. This approach has been widely used in previous studies of phloem transport (for example, see Thompson and Holbrook, 2003, and references therein). The relation between the hydrostatic pressure drop Δp between source and sink and the volumetric flow rate Q through the sieve tube is the hydraulic equivalent of Ohm’s law (Bruus, 2008)
Here, the volumetric flow rate Q = UA is the product of the flow velocity U and cross-section area A of the sieve element, and R is the hydraulic resistance of the phloem translocation pathway. Due to the abundance of sieve tube constituents at the margins, we estimate that between 65 and 100% of the area is open to flow, such that the cross-section area A lies in the range A ≃ (4.5 – 7.1) μm2. Typical flow speeds observed are of the order U ≃ 100 μm/s, which yields Q ≃ (450 – 710) μm3/s. When calculating the resistance R in Equation 1, we take into account three major components: (1) the tube lumen including organelles, (2) the sieve plate, and (3) the SEOR1 agglomerations. Assuming that the translocation pathway consists of N ≃ 1250 identical sieve tube elements, 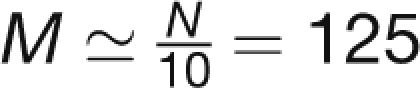 of which contain a SEOR1 agglomeration, we write the resistance of the phloem translocation pathway R as
of which contain a SEOR1 agglomeration, we write the resistance of the phloem translocation pathway R as
Here, Rlumen is the resistance of a single sieve tube element lumen, Rplate is the resistance of a single sieve plate separating adjacent sieve elements, and Rplug is the resistance of a single SEOR1 agglomeration. Please refer to the Supplemental Appendix online for a detailed discussion of how these resistance values are determined and Table 1 for a list of characteristic values of the parameters used. As shown in Table 2, we find typical values of the terms in Equation 2: 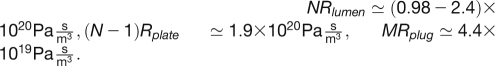 While the contribution from the lumen and plate resistances are of comparable magnitude, the contribution from the SEOR1 agglomerations is somewhat smaller, reflecting the fact that these are only found in every ~10 sieve tube elements. We finally have for the total resistance in Equation 2 that
While the contribution from the lumen and plate resistances are of comparable magnitude, the contribution from the SEOR1 agglomerations is somewhat smaller, reflecting the fact that these are only found in every ~10 sieve tube elements. We finally have for the total resistance in Equation 2 that 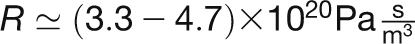 and find from Equation 1 that the pressure drop required to drive the flow over a distance of 15 cm lies in the range Δp ≃ (0.21 – 0.23) MPa.
and find from Equation 1 that the pressure drop required to drive the flow over a distance of 15 cm lies in the range Δp ≃ (0.21 – 0.23) MPa.
Table 1.
List of Parameters for Flow Calculations
| Parameter | Symbol/Expression | Value, Unit, Reference |
| Sieve tube cross-section area | 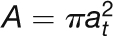 |
m2 |
| Effective sieve tube cross-section area | 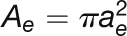 |
m2 |
| At SEOR1 agglomeration cross-section area | 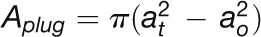 |
m2 |
| Effective sieve tube radius | ae | 1.2 μm |
| At SEOR1 filament radius | af | 10 nm |
| At SEOR1 agglomeration opening radius | ao | 0.5 μm |
| Average sieve pore radius | ap | 156 nm |
| Sieve tube radius | at | 1.5 μm |
| Effective sieve tube diameter | de | 2.4 μm |
| At SEOR1 filament diameter | df | 20 nm |
| At SEOR1 agglomeration opening diameter | do | 1 μm |
| Sieve tube diameter | dt | 3.0 μm |
| Observed flow speed | U | 100 μm s−1 |
| At SEOR1 filament separation distance | b | 6 nm |
| Permeability of At SEOR1 agglomeration | K | m2 |
| Length of plant | L | 15 cm |
| At SEOR1 agglomeration length | Lp | 6 μm |
| Sieve element length | Lt | 120 μm |
| Sieve plate thickness | l | 450 nm |
| Number of sieve elements | N | 1250 |
| Average number of sieve pores | Np | 15 |
| Number of At SEOR1 agglomerations | M | 125 |
| Volume flux | Q | m3 s−1 |
| Hydraulic resistance of the phloem translocation pathway | R | Pa s m−3 |
| Viscosity | η | 1.3 mPa s (Deeken et al., 2002; Hunt et al., 2009) |
| Nondimensional permeability of At SEOR1 agglomeration | 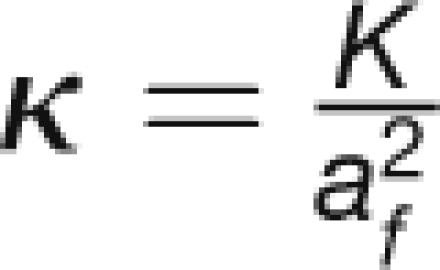 |
|
| Volume fraction occupied by filaments inside agglomeration | ϕ | 0.45 |
Reference is given next to parameter value when not measured by the authors.
Table 2.
Parameters Relevant for the Calculation of the Pressure Drop Δp in Equation 1/(A1)
| de (μm) | do (μm) | Rlumen (Pa sm−3) | NRlumen (Pa sm−3) | Rplate (Pa s/m3) | (N−1) × Rplate (Pa sm−3) | Rplug (Pa s/m3) | MRplug (Pa s/m3) | R (Pa s/m3) | Q (m3/s) | Δp (MPa) |
| 3.0 | 1.0(*) | 7.8 × 1016 | 9.8 × 1019 | 1.5 × 1017 | 1.9 × 1020 | 3.5 × 1017 | 4.4 × 1019 | 3.3 × 1020 | 7.1 × 10−16 | 0.23 |
| 3.0 | 0.50 | 7.8 × 1016 | 9.8 × 1019 | 1.5 × 1017 | 1.9 × 1020 | 5.2 × 1018 | 6.5 × 1020 | 9.4 × 1020 | 7.1 × 10−16 | 0.66 |
| 3.0 | 0.23 | 7.8 × 1016 | 9.8 × 1019 | 1.5 × 1017 | 1.9 × 1020 | 7.5 × 1019 | 9.4 × 1021 | 9.7 × 1021 | 7.1 × 10−16 | 6.8 |
| 3.0 | (†) | 7.8 × 1016 | 9.8 × 1019 | 1.5 × 1017 | 1.9 × 1020 | (†) | (†) | 2.9 × 1020 | 7.1 × 10−16 | 0.20 |
| 2.4 | 1.0(*) | 1.9 × 1017 | 2.4 × 1020 | 1.5 × 1017 | 1.9 × 1020 | 3.5 × 1017 | 4.4 × 1019 | 4.7 × 1020 | 4.5 × 10−16 | 0.21 |
| 2.4 | 0.50 | 1.9 × 1017 | 2.4 × 1020 | 1.5 × 1017 | 1.9 × 1020 | 5.2 × 1018 | 6.5 × 1020 | 1.1 × 1021 | 4.5 × 10−16 | 0.49 |
| 2.4 | 0.23 | 1.9 × 1017 | 2.4 × 1020 | 1.5 × 1017 | 1.9 × 1020 | 7.5 × 1019 | 9.4 × 1021 | 9.8 × 1021 | 4.5 × 10−16 | 4.4 |
| 2.4 | (†) | 1.9 × 1017 | 2.4 × 1020 | 1.5 × 1017 | 1.9 × 1020 | (†) | (†) | 4.3 × 1020 | 4.5 × 10−16 | 0.19 |
Calculated values of the lumen resistance Rlumen, plate resistance Rplate, agglomeration resistance Rplug, and total resistance R determined from Equations (A3), (A4), (A5), and (A2) (see Supplemental Appendix 1 and Supplemental References 1 online). The results are given for two values of the effective sieve tube diameter de and three values of the agglomeration opening diameter do. de = 3.0 μm corresponds to a completely empty sieve tube, and de = 2.4 μm corresponds to a sieve tube with only 65% of the area open to flow. Results marked with an asterisk indicate the measured value of do = 1 μm. Results marked with (†) indicate the case where no At SEOR1 agglomerations are present.
SEOR1 Function
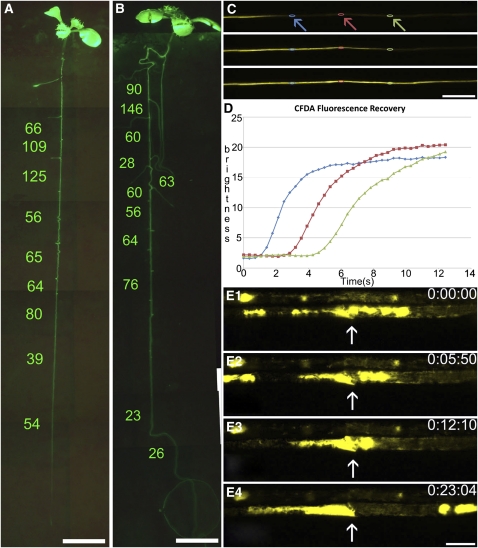
Since SEOR1 is located in the flow path of sieve tubes, we tested a potential influence on translocation. We used the homozygous SEOR1 T-DNA insertion mutant (GABI-KAT 609F04) and conducted flow velocity studies along intact roots. We studied the flow in eight independent plants of each wild type and T-DNA insertion mutant (Figures 9A to 9D). Velocities in the root system are variable in both lines. So far, no significant difference between insertion mutant and the wild type has been found (Figures 9A and 9B). We further measured the average sieve tube diameter of the two lines to see if fewer obstructions lead to a change in tube anatomy. Average sieve tube diameters did not differ significantly between the lines.
Figure 9.
In Vivo Flow and Injury Experiments.
(A) and (B) Comparison of phloem flow velocities along a main root of the Arabidopsis wild type (A) and SEOR1 T-DNA insertion mutant (B). The entire root system is visible in MicroROCs after loading with CFDA, permitting flow measurements in individual tubes by FRAP. No significant difference was found between mutant and wild-type plants.
(C) and (D) FRAP experiment on an individual tube. Three frames from Supplemental Movie 2 online (C). After bleaching of CFDA, the laser intensity was lowered and refilling of the tube was monitored at subsecond intervals. Regions of interest are marked along the tube (arrows and colors of arrows correspond to colors in graph), and fluorescence intensity is measured and graphed (D), giving a direct reading of flow velocity in the tube.
(E) and (E1) to (E4) Four frames of Supplemental Movie 3 online, showing the slow movement (flow is right to left) of SEOR1-YFP filaments through a sieve plate (arrow). Movement does not stop even after 23 min.
Bars = 5 mm in (A) and (B), 100 μm in (C), and 10 μm in (E).
Our study was initiated because the genomic SEOR1 sequence in Arabidopsis showed homology to genomic sequences of Medicago forisomes (Pélissier et al., 2008) and therefore was likely to be a yet unknown P protein. This relationship also suggested a similar function. Forisomes appear to form reversible agglomerations that temporarily stop sieve tube flow (Knoblauch et al., 2001, 2003; Peters et al., 2006). The transformation from the low volume to the high volume state of a forisome may be completed in 100 ms (Peters et al., 2008). Thus, we tested SEOR1 for a potential injury reaction. Initially, we observed intact sieve tubes for several hours by epifluorescence and confocal microscopy without any indication of dynamic behavior of SEOR1 bundles and agglomerations. Then, we tested different injury stimuli that are known to trigger the forisome reaction, such as local mechanical injury, distant burning of leaf tips, and local cold shocks (Furch et al., 2007; Thorpe et al., 2010). None of the treatments triggered any immediate reaction. Even direct application of 1 to 5 mM Ca2+ medium on ruptured sieve tubes or isolated SEOR1 bundles did not result in any structural changes.
Although we were not able to find reactions equivalent to that of forisomes, SEOR1 in Arabidopsis sieve tubes underwent an obvious structural alteration after tissue excision and standard fixation for TEM (Figures 3A and 3B). To understand the development of these structures, we conducted time-lapse movies of injured sieve tubes. A very slow movement of SEOR1 toward the sieve plate was observed in some cases. Surprisingly, the movement did not stop at the sieve plate but agglomerates continued to move through the plate for extended periods of time (Figure 9E; see Supplemental Movie 3 online).
DISCUSSION
Sieve Tube Ultrastructure
Fluorescent tagging of SEOR1 filaments permitted comparison of in vivo confocal micrographs of sieve tubes with TEM images collected from variably processed tissue. Freeze substitution of whole plants most accurately resembled the in vivo structure and location of components found in confocal images. Freeze substitution, however, has some limitations. For good preservation, a high sugar content that acts as antifreeze substance is necessary. In addition, a close location to the surface is required. So far, we were only able to preserve sieve tubes for TEM in source leaves. The coverage of root sieve tubes with rhizodermis, large cortical parenchyma cells, endodermis, and pericycle in combination with a lower sugar concentration in sink sieve elements has so far prevented us from studying root sieve tubes. By contrast, in vivo confocal investigations using Micro-ROCs are possible in roots only. Thus, we are currently not able to compare phloem structures in the same organs. On the other hand, removal of some cortical cell layers at the main vein of source leaves exposes uninjured sieve tubes, which show the same fine structure as found in root sieve tubes by confocal microscopy. We conclude that the different location probably has just a minor influence on sieve tube structure. Preservation of phloem tissue in larger plants may become increasingly difficult since sieve tubes are usually covered by a thicker tissue layer, which may increase problems with freezing artifacts. Specific treatment, such as localized chilling, which halts phloem translocation but not loading (Pickard and Minchin, 1992), might become necessary to increase sieve tube antifreeze concentrations. Specific protocols may have to be developed for different plant species. Usually, membrane structures were more difficult to preserve. This may be due to the acetone solvent. The addition of water or tannic acid helps to some extent, but in general, standard chemical fixation shows a more pronounced outline of ER stacks. Alterations of the fixation protocol might help solve this problem in the future. However, within those limitations, the method has proven most beneficial, since structures in TEM images match the location and distribution of structures found in translocating sieve tubes.
To understand the mechanism of, for example, long-distance transport, the interactions of sieve tubes with pathogens such as aphids or viruses and the interactions of sieve elements and companion cells, a good understanding of the cellular equipment available for those interactions is fundamental. Besides the well-known previously described sieve tube components, mitochondria, sieve element plastids, and ER, some new, frequently found components have to be added to our picture of sieve tube infrastructure. Mitochondria in Arabidopsis are always surrounded by a halo of small protein spikes that attach them to membranes and/or SEOR1 filaments (Figure 4). Similar clamps have been found in one earlier study in tomato (Solanum lycopersicum) and fava bean (Ehlers et al., 2000). Clamp proteins do not attach to all organelles in all species. In Arabidopsis, mitochondria are completely covered, while sieve element plastids lack clamps (Figure 4F). In Vicia and Solanum, clamps are present on all organelles, including the ER. In contrast with other organelles, mitochondria in Arabidopsis have a layer of clamp proteins each, doubling the distance between the organelles (Figure 4H).
In addition, we found electron-dense vesicles of various sizes. The vesicles seem to bud off of membranes. The nature of the membranes and vesicles has yet to be established. Vesicles are always embedded in the ground matrix of the parietal layer.
The structures that clearly stand out are SEOR1 filaments. Sieve plate pores are mostly unobstructed, but large SEOR1 agglomerations exist in the lumen of some translocating sieve tubes. Agglomerations have frequently been observed previously. However, the ultrastructure and location of the agglomeration differs depending on the preparation used. After standard fixation of sectioned tissue, Arabidopsis sieve plate precipitates consist of fine 5- to 10-nm-thick filaments (Figures 3A and 3B). The precipitates are located on the plate or in the lumen, but no filaments are found at the margins that would match the location of in vivo confocal images. In tobacco, different forms of P proteins have been described to occur after standard preparation and fixation, including 23-nm tubules designated as P1 protein, 15-nm striated filaments designated as P2 protein, very fine filaments (Cronshaw and Esau, 1967; Gilder and Cronshaw, 1973), and crystalline filaments of ~100 nm diameter (Johnson, 1969). The diameters reported may vary depending on the study. All filaments are located in the lumen or the pores but do not form a distinct meshwork at the margins. In Cucurbita maxima, phloem protein1 (PP1) is a 96-kD protein that forms filaments, and PP2 is a 25-kD dimeric lectin that binds covalently to PP1 (Bostwick et al., 1992; Golecki et al., 1999). The structural component PP1 belongs to a gene family found only in cucurbits (Clark et al., 1997; Beneteau et al., 2010). By contrast, genes encoding SEOR proteins have been reported in many dicot families (Pélissier et al., 2008; Huang et al., 2009, Rüping et al., 2010).
In Arabidopsis and tobacco, freeze substitution of whole plants results in only one morphological form of P protein: filaments of ~20 nm diameter, which are absent in SEOR1 T-DNA insertion mutants. Our data suggest that many P protein structures described are alterations of SEOR proteins due to preparation and that P proteins usually exist in the form of SEOR filaments in active sieve tubes.
Phloem Translocation
The controversy about the mode of phloem translocation in the last century mainly revolved around the question of the abundance of P protein agglomerations inside the sieve tube lumen and inside sieve plate pores and has split phloem researchers into two groups. While investigators believing in occluded plates favored the electroosmotic theory (Fensom, 1957; Spanner, 1958, 1970), the pressure flow hypothesis was supported by the group believing in open pores and that occlusion is due to preparation artifacts (for an overview, see Knoblauch and Peters, 2010). Over the years, gentle preparation methods for TEM (e.g., Fisher, 1975; Turgeon et al., 1975; Lawton and Newman, 1979) and in vivo studies on translocating sieve tubes (Knoblauch and van Bel, 1998) supported an unobstructed sieve tube path. To date, an osmotically generated pressure flow is generally accepted as the mode of action of long-distance translocation in sieve tubes. In this context, our finding that massive SEOR1 agglomerates are a standard feature in the lumen of translocating sieve tubes in Arabidopsis is most surprising. In the end, both groups of investigators were right. Most of the pores are usually unobstructed, but massive agglomerates exist in the lumen. The resulting question is: Is a pressure differential driven flow possible?
We calculated the pressure differential required to drive flow through a 15-cm-long Arabidopsis sieve tube from a source leaf to the root at a velocity of 100 μm s−1 to be 0.2 MPa. The osmotic concentration of Arabidopsis sieve tube sap in source tissue can be taken from measurements of sap collected by stylectomy to be 0.7 M (Deeken et al., 2002; Hunt et al., 2009), which can generate a pressure of ~1.7 MPa or less, depending on the osmolarity of the apoplastic solution. Not all of this pressure is available for transport, since sink cells possess a turgor of ~0.7 MPa (Pritchard, 1996; Turgeon, 2010). This leaves a maximum of 1 MPa pressure differential for flow. For our calculations, we assumed all parameters to be at the lower end of observation and to favor pressure flow. All pores were assumed to be unobstructed, the average occupation of cross-sectional area with organelles was assumed to be only 20%, the surface of the parietal layer was assumed to be smooth, the channel in the SEOR1 agglomeration was assumed to be 1 μm in diameter, and the tubes were assumed to be circular. The calculated pressure differential required is ~0.2 MPa. Even if we assume less favorable conditions, the potential 1 MPa pressure differential leaves plenty of margin in comparison to the calculated 0.2 MPa required pressure, since the agglomeration opening would have to be smaller than 500 nm in diameter in every agglomeration to increase the required pressure to more than 1 MPa.
The situation, however, changes if we assume that SEOR1 agglomerations do not contain open channels. Unfortunately, serial sectioning for TEM is so labor intensive that we were only able to find a single SEOR1 agglomeration. This agglomeration had a channel of ~0.5 × 1 μm. In vivo confocal images, on the other hand, show a variety of agglomerations. Confocal resolution is ~230 nm and would have allowed us to identify the opening in the agglomeration shown in Figure 6J. In many confocal images, however, agglomerations appear to fill the entire lumen (Figures 6C to 6E), which would increase the required pressure significantly.
To calculate the impact of agglomerations on flow, the porosity of the material is critical. For our calculations, we assumed filaments in the agglomeration to be straight rods, with a smooth surface and without major interaction with the surrounding medium, similar to glass filaments. Proteins, especially if they contain a high percentage of charged amino acids, form a hydration shell and turn the surrounding water into a viscous layer, which increases the effective filament diameter significantly (Bánó and Marek, 2006). In this case, a single agglomeration without an opening would add considerable resistance to the flow and would most likely be sufficient to block the flow entirely. Assuming flow favoring conditions with filaments lacking hydration shells, ~10 agglomerations would be needed to increase the resistance to the point that the calculated pressure differential of 1 MPa would not be sufficient to drive flow at the measured velocities (Table 2).
In summary, despite the existence of large SEOR1 agglomerations in the lumen of sieve tubes, a pressure differential–driven flow appears feasible, given that the porosity of SEOR1 agglomerations is high. It is, however, surprising that there is no significant difference in transport velocity between the mutant and wild type. The existence of agglomerations necessarily has an influence on tube resistance and must result in a higher pressure differential in wild-type plants to maintain constant flow velocities.
The pressure flow hypothesis remains an issue of debate. While the tube anatomy does appear to scale with plant size (Jensen et al., 2011), pressure does not (Turgeon, 2010). Also, larger tubes with significantly lower resistance translocate at slower velocities than tubes with higher resistance (Mullendore et al., 2010). It appears necessary to conduct correlated determinations of translocation velocity, pressure differential, and sieve tube structure.
SEOR1 Function
It has been repeatedly suggested that P proteins are involved in sieve tube occlusion, and the discovery of forisome function supported this notion (Pélissier et al., 2008). On the other hand, it had also been questioned whether occlusion is the (only) function of P proteins (Sabnis and Sabnis, 1995).
The reaction of SEOR1 to injury is not comparable to the reaction found in forisomes. SEOR1 filaments do not show a detectable structural reaction to Ca2+ ions, nor do they react within milliseconds. TEM snapshots of injured sieve tubes gave the impression that SEOR1 occluded pores (e.g., Figure 3B). SEOR1 filaments were supposed to be compressed by callose formation to form a tight seal (Mullendore et al., 2010). In reality, there is a slow movement of SEOR1 through the pores, which can continue for at least 45 min. SEOR1 often moves out of the wound site and disappears in the surrounding medium. This explains why high concentrations of P protein filaments can be found in phloem exudates (Cronshaw et al., 1973) in different plant species, including tobacco. This argues against a targeted sealing mechanism and also suggests that callose occlusion is relatively inefficient, at least in Arabidopsis.
The structure of sieve tubes is probably the least understood of all major plant cell types. Although we have known of the existence of sieve element plastids for almost a century, and meanwhile have studied the function of all other plastid types in detail, we still have no indication of sieve element plastid function. The function of many other structures, including SEOR1, is also obscure. Because the phloem is a key player to maintain plant integrity, it will be crucial to obtain more detailed insights into the functions of its components. Sieve elements are far from being empty tubes. The existence of a protein filament meshwork that structurally resembles a cytoskeleton may lead to new insights in short- and long-distance signaling, plant–pathogen interaction, such as viral movement, and, among others, sieve element–companion cell interactions. Some of the sieve tube components might have a direct effect on translocation and/or flow control. The tools to investigate these open questions by in vivo studies on a cellular basis are now available.
METHODS
Plant Material for Freeze Substitution
Arabidopsis thaliana ecotype Columbia, SEOR1 T-DNA insertion mutant GABI-KAT 609F04, and tobacco (Nicotiana tabacum) were grown on 0.44% (w/v) Murashige and Skoog (MS) medium containing 87.6 mM Suc, 2.56 mM MES buffer, pH 5.8, and 0.8% (w/v) agar. Seeds were surface sterilized with 70% ethanol, plated, and cold treated overnight at 4°C before being placed into the growth chamber. The plants were grown at 25°C with a 16/8-h light/dark period. Black cottonwood (Populus trichocarpa) was grown in pots in a greenhouse at 23°C, with 60 to 70% relative humidity, and a 14/10-h light/dark period (daylight plus additional lamp light [model PL 90; PL Lighting Systems]) with a minimum irradiance of 150 μE m−2 s−1.
Micro-ROCs
Plants were grown in Micro-ROCs (Advanced Science Tools) in the greenhouse at a 14-h photoperiod, 300 to 400 μE m–2 s–1, at 20°C day and 15°C night. Plants were grown to the six- to eight-leaf stage for SEOR1-YFP imaging. FRAP plants were grown until the first true leaves matched the diameter of the cotyledons.
Plunge Freezing and Freeze Substitution
Liquid nitrogen was placed into a shallow, thick-sided polystyrene container and placed under vacuum for ~7 min until the nitrogen became slushy. Whole Arabidopsis plants, in the four-leaf state, were gently teased from the MS agar and rapidly plunged into the slush nitrogen. The frozen plants were then transferred to 2% glutaraldehyde in acetone with 0.1% water, 0.1% tannic acid, or 4% tannic acid in scintillation vials on dry ice. The plants in solution were transferred to −80°C for 24 h and then placed into a −20°C freezer while removing most of the dry ice. The solution was allowed to ramp up to −20°C over a period of at least 8 h. The plants were rinsed twice for 30 min with cooled (−20°C) acetone. Postfixation was achieved in cooled (−20°C) 2% OsO4 in acetone overnight. The material was ramped to 20°C over a period of 6 h. The OsO4 was rinsed with acetone two times for 30 min and exchanged for propylene oxide (PO). The plants were infiltrated with a soft recipe Spurr’s resin (SR; Bozzola and Russell, 1999) as follows: 3:1 PO:SR, 48 h; 2:1 PO:SR, 24 h; 1:1 PO:SR, 24 h; 1:2 PO:SR, 24 h; 1:3 PO:SR, 24 h; 100% SR, 24 h; 100% SR, 48 h; 100% SR, 24 h. Before each exchange, the samples were cycled three times in vacuum for 5 min each cycle. The samples were embedded in fresh SR and cured for 2 d at 60°C. Ultrathin sections (70 to 100 nm) were taken with an ultramicrotome (Reichert Ultracut R; Leica) and placed on formvar-coated slot grids. They were stained with a solution of 1% uranyl acetate and 0.01% potassium permanganate for 10 min and poststained for 6 min in Reynolds lead citrate (Reynolds, 1963). The sections were imaged on a FEI Tecnai G2 TEM (FEI Company) or a Philips CM200 UT Intermediate Voltage TEM (FEI Company).
Epifluorescence Microscopy
Epifluorescence microscopy was performed with a Leica DM LFSA microscope or with a Leica MZ8 stereomicroscope. Images and movies were recorded with a Leica DFC 300FX-cooled charge-coupled device camera. To show the outline of root cells, 0.1 μg/mL synapto-red (EMD Chemicals) was added to agar plates. For synapto red and YFP double-labeled tissue, a Leica filter cube I3 was used, and for YFP or CFDA detection, a GFP filter cube was used.
Confocal Microscopy
All confocal laser scanning microscope images were obtained with a Leica TCS SP5. Respective excitation and emission for YFP, GFP, GFP5, and CFDA were 514 argon/520 to 550, 488 argon/500 to 600, 405 diode/475 to 530, and 488 argon/490 to 515. Subsequent processing used ImageJ for time series and Leica LAS AF Lite software for images. For flow velocity, measurements were conducted with plants in the four-leaf state grown in micro-ROCs.
FRAP
CFDA was loaded into the first true leaves and cotyledons by half clipping and applying 20 μL 1:5 (v/v) 50 mg mL−1 CFDA in acetone to water. Loaded sieve elements in the primary root were manually photobleached at 488 nm at maximum laser intensity, pinhole at Airy 3, and at ×8 zoom, starting apically and moving toward the hypocotyl. A 3-frames per second time series, to record refilling of the sieve element, immediately followed the reduction of the laser power to 15% and zoom to ×1. Region-of-interest intensities were generated using Leica LAS AF Lite software.
Cloning and Transformation
SEOR1-YFP
The Modular Binary Construct System (gift from Christopher G. Taylor) was used for all constructs. The K4 adapter made from 5′-TTCGGATCCACTAGTTCTGCTGCTGGTTCTGCTGCTGGTTCTGGGGGATCCCTT-3′ and 5′-AAGGGATCCCCCAGAACCAGCAGCAGAACCAGCAGCAGAACTAGTGGATCCGAA-3′, which contains a unique SpeI restriction site was cloned into the BamHI site of a modified AKK 1435 vector containing the YFP gene and sequenced for directionality. SEOR1, minus the stop codon, and its 1500-bp promoter region was amplified from BAC clone F4P13 with 5′-TCGGTACCGAACTAATACACAAGTAACACAAGT-3′ and 5′-TTCACTAGTGAAGTTGTAGTTCTCGTCTT-3′. This was ligated into the AKK 1435 shuttle vector at the KpnI and SpeI sites. The PacI promoter-gene fusion cassette was then ligated into the AKK 1426b binary vector containing in planta glufosinate resistance (Thompson et al., 1987). The construct was used to transform Arabidopsis ecotype Columbia via Agrobacterium rhizogenes 18r12v using the floral dip method (Clough and Bent, 1998), and the transformed seeds were screened with daily spraying of 0.003% glufosinate ammonium (Sigma-Aldrich) and 0.05% Silwet L-77. T2 generations were screened by epifluorescence microscopy to identify homozygous lines.
GFP5-ER
GFP5-ER was amplified from pBINmGFP5ER (Haseloff et al., 1997) with primers 5′-TTCAAGCTTAAGGAGATATAACAATGAAGACTA-3′ and 5′-TTCGGATCCGATCTAGTAACATAGATGACACC-3′ and subsequently cloned into AKK 1408 at the 3′ end of the 2047-bp Medicago truncatula SEO2 promoter (Pélissier et al., 2008). The Pro-Mt-SEO2-GFP5-ER cassette was then cloned into binary vector AKK 1426b via SdaI. Arabidopsis expressing At SEOR1-YFP was transformed with Pro-Mt-SEO2-GFP5-ER by A. rhizogenes 18r12v using the floral dip method (Clough and Bent, 1998) with seeds screened on MS plates containing 50 μg mL−1 kanamycin.
SEOR1-GFP
Approximately 1000 bp of promoter sequences extending 5′ from, but not including, the translation start codon of SEOR1 were PCR amplified from the bacterial artificial chromosome F4P13. The amplicons were initially cloned into the pGEM-T easy vector, and specific primers were designed to subclone the amplicons into SalI and XbaI restriction sites located 3 bp 5′ of the translation initiation codon of the GUS reporter gene (uidA) in the pGPTV-Kan binary vector (Becker et al., 1992). The enhanced GFP gene was PCR amplified and subcloned into the pGPTV-Kan binary vector in place of the uidA gene using the SmaI and KpnI restriction sites, and these primers also created a multiple cloning site at the 3′ end of the enhanced GFP gene. Subsequently, the SEOR1 open reading frame was PCR amplified and subcloned into this multiple cloning site (KpnI and ApaI). The binary vectors were transformed into Agrobacterium tumefaciens strain GV3101 and used to transform Arabidopsis by the floral dip method (Clough and Bent, 1998). Transgenic plants were then screened on kanamycin-supplemented media.
T-DNA Insertion Mutants
T-DNA insertions in SEOR1 were identified using T-DNA Express (http://signal.salk.edu/cgi-bin/tdnaexpress). Seeds for GABI-KAT 609F04 (SEOR1 knockout) were obtained from the Genomanalyse im Biologischen System Pflanze. Plants from the original seed stocks or one generation later were screened to identify individual homozygous plants using the PCR-based screening technique according to the method of Siebert et al. (1995). The GABI-KAT 609F04 mutant contained a second T-DNA insertion so plants were allowed to self-fertilize and plants homozygous for the SEOR1 insertion alone were identified.
Successful knockout of the gene was confirmed using RT-PCR. In brief, total RNA was extracted using the Trizol method, and total RNA was reverse transcribed using SuperScript II according to the manufacturer’s instructions. Partial, intron-spanning sections of the gene were amplified using gene-specific primers, including section 1 (1 to 351 bp) 5′-ATGGAGTCGCTGATCAAGTC-3′ and 5′-TATCTCGCAGGCAACACGAT-3′, section 2 (860 to 988) 5′-ACCATCTCGCTGAGACCTTGAGG-3′ and 5′-GGCCGTGAGAATCTTCATGTTATCA-3′, section 3 (1494 to 1659) 5′-GAGAGAGACCTTTTCCCTTAACCTCA-3′ and 5′-TTCACGTTGGAATCTTTGGCC-3′, and subsequently visualized on a 1.6% agarose gel containing ethidium bromide.
Immunolocalization
Cross sections of unfixed floral stems from Arabidopsis Columbia plants and GABI-KAT 609F04 were cut with a vibrating microtome (Vibratome) at 50 μm and collected in PBS. Sections were washed twice in 10 mM PBS and then incubated for 30 min in blocking buffer (PBS with 3% nonfat dry milk). Sections were washed twice more with PBS and incubated for 45 min with the RS21 primary monoclonal antibody in blocking buffer (1:100). After incubation with primary antibody, the sections were washed three times with PBS and then incubated in PBS with ALEXA 488-nm fluorescently tagged secondary goat anti-mouse antibody (Molecular Probes) (1:250). Finally, the labeled sections were washed twice with PBS and once with nanopure water and observed under a Nikon E600 epifluorescence microscope, with an excitation wavelength of 490 nm and an emission wavelength of 512 nm.
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under the following accession numbers: At3g01680 (SEOR1) and GK-609F04-021864 (GABI-KAT 609F04).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Movie 1. SEOR1 in Root Tip.
Supplemental Movie 2. Real-Time Imaging of Phloem Flow.
Supplemental Movie 3. SEOR1 Movement in Injured Sieve Tubes.
Supplemental Movie Legends. Legends for Supplemental Movies 1 to 3.
Supplemental Appendix 1. Mathematical Derivation of Expressions for the Three Resistances given in Equation (2).
Supplemental References 1. Supplemental References for Supplemental Appendix 1.
Acknowledgments
We thank Karl J. Oparka (University of Edinburgh) and Winfried S. Peters (Indiana University–Purdue University Fort Wayne) for helpful discussions and critical reading of the manuscript. We acknowledge technical support from Washington State University’s Franceschi Microscopy and Imaging Center and thank Valerie Lynch-Holm, Christine Davitt, and Chuck Cody (Washington State University) for technical assistance. We thank four anonymous reviewers for helpful comments. This work was supported by National Science Foundation Integrated Organismal Systems Grants 0818182 and 1022106.
AUTHOR CONTRIBUTIONS
D.R.F., D.L.M., and M.K. designed and conducted confocal and electron microscopy experiments. D.R.F., T.J.R.-E., and H.C.P. performed cloning and transformation. J.A.A. and G.A.T. analyzed the T-DNA insertion mutant GABI-KAT 609F04 and complementation plants. K.H.J. analyzed results and calculated pressures based on microscopy data. M.K. wrote the article with participation of all the authors.
References
- Bánó M., Marek J. (2006). How thick is the layer of thermal volume surrounding the protein? Biophys. Chem. 120: 44–54 [DOI] [PubMed] [Google Scholar]
- Becker D., Kemper E., Schell J., Masterson R. (1992). New plant binary vectors with selectable markers located proximal to the left T-DNA border. Plant Mol. Biol. 20: 1195–1197 [DOI] [PubMed] [Google Scholar]
- Beneteau J., Renard D., Marché L., Douville E., Lavenant L., Rahbé Y., Dupont D., Vilaine F., Dinant S. (2010). Binding properties of the N-acetylglucosamine and high-mannose N-glycan PP2-A1 phloem lectin in Arabidopsis. Plant Physiol. 153: 1345–1361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bostwick D.E., Dannenhoffer J.M., Skaggs M.I., Lister R.M., Larkins B.A., Thompson G.A. (1992). Pumpkin phloem lectin genes are specifically expressed in companion cells. Plant Cell 4: 1539–1548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bozzola J.J., Russell L.D. (1999). Electron Microscopy: Principles and Techniques for Biologists. (Sudbury, MA: Jones & Bartlett Learning) [Google Scholar]
- Bruus H. (2008). Theoretical Microfluidics. (Cambridge, UK: Cambridge University Press) [Google Scholar]
- Clark A.M., Jacobsen K.R., Bostwick D.E., Dannenhoffer J.M., Skaggs M.I., Thompson G.A. (1997). Molecular characterization of a phloem-specific gene encoding the filament protein, phloem protein 1 (PP1), from Cucurbita maxima. Plant J. 12: 49–61 [DOI] [PubMed] [Google Scholar]
- Clough S.J., Bent A.F. (1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Cronshaw J. (1975). P-proteins. In Phloem Transport, J.D.S. Aronoff, P.R. Gorman, L.M. Srivastava, and C.M. Swanson, eds (New York, London: Plenum), pp. 79–115 [Google Scholar]
- Cronshaw J., Esau K. (1967). Tubular and fibrillar components of mature and differentiating sieve elements. J. Cell Biol. 34: 801–815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cronshaw J., Gilder J., Stone D. (1973). Fine structural studies of P-proteins in Cucurbita, Cucumis, and Nicotiana. J. Ultrastruct. Res. 45: 192–205 [DOI] [PubMed] [Google Scholar]
- Deeken R., Geiger D., Fromm J., Koroleva O., Ache P., Langenfeld-Heyser R., Sauer N., May S.T., Hedrich R. (2002). Loss of the AKT2/3 potassium channel affects sugar loading into the phloem of Arabidopsis. Planta 216: 334–344 [DOI] [PubMed] [Google Scholar]
- Ehlers K., Knoblauch M., van Bel A.J.E. (2000). Ultrastructural features of well-preserved and injured sieve elements: Minute clamps keep the phloem transport conduits free for mass flow. Protoplasma 214: 80–92 [Google Scholar]
- Evert R.F. (1982). Sieve-tube structure in relation to function. Bioscience 32: 789–795 [Google Scholar]
- Evert R.F. (1990). Dicotyledons. In Sieve Elements, H.D.B.R.D. Sjolund, ed (Berlin: Springer), pp. 103–137 [Google Scholar]
- Fellows R.J., Geiger D.R. (1974). Structural and physiological changes in sugar beet leaves during sink to source conversion. Plant Physiol. 54: 877–885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fensom D.S. (1957). The bioelectric potentials of plants and their functional significance. Can. J. Bot. 35: 573–582 [Google Scholar]
- Fisher D.B. (1975). Structure of functional soybean sieve elements. Plant Physiol. 56: 555–569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furch A.C.U., Hafke J.B., Schulz A., van Bel A.J.E. (2007). Ca2+-mediated remote control of reversible sieve tube occlusion in Vicia faba. J. Exp. Bot. 58: 2827–2838 [DOI] [PubMed] [Google Scholar]
- Gilder J., Cronshaw J. (1973). Adenosine triphosphatase in the phloem of Cucurbita. Planta 110: 189–204 [DOI] [PubMed] [Google Scholar]
- Golecki B., Schulz A., Thompson G.A. (1999). Translocation of structural P proteins in the phloem. Plant Cell 11: 127–140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartig T. (1854). Über die Querscheidewände zwischen den einzelnen Gliedern der Siebröhren in Cucurbita pepo. Bot. Z. 12: 51–54 [Google Scholar]
- Haseloff J., Siemering K.R., Prasher D.C., Hodge S. (1997). Removal of a cryptic intron and subcellular localization of green fluorescent protein are required to mark transgenic Arabidopsis plants brightly. Proc. Natl. Acad. Sci. USA 94: 2122–2127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang S., et al. (2009). The genome of the cucumber, Cucumis sativus L. Nat. Genet. 41: 1275–1281 [DOI] [PubMed] [Google Scholar]
- Hunt E., Gattolin S., Newbury H.J., Bale J.S., Tseng H.-M., Barrett D.A., Pritchard J. (2009). A mutation in amino acid permease AAP6 reduces the amino acid content of the Arabidopsis sieve elements but leaves aphid herbivores unaffected. J. Exp. Bot. 61: 55–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen K.H., Lee J., Bohr T., Bruus H., Holbrook N.M., Zwieniecki M.A. (2011). Optimality of the Münch mechanism for translocation of sugars in plants. J. R. Soc. Interface 8: 1155–1165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson R.P.C. (1969). Crystalline fibrils and complexes of membranes in the parietal layer in sieve elements. Planta 84: 68–80 [DOI] [PubMed] [Google Scholar]
- Knoblauch M., Noll G.A., Müller T., Prüfer D., Schneider-Hüther I., Scharner D., Van Bel A.J.E., Peters W.S. (2003). ATP-independent contractile proteins from plants. Nat. Mater. 2: 600–603 [DOI] [PubMed] [Google Scholar]
- Knoblauch M., Peters W.S. (2010). Münch, morphology, microfluidics - Our structural problem with the phloem. Plant Cell Environ. 33: 1439–1452 [DOI] [PubMed] [Google Scholar]
- Knoblauch M., Peters W.S., Ehlers K., van Bel A.J.E. (2001). Reversible calcium-regulated stopcocks in legume sieve tubes. Plant Cell 13: 1221–1230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knoblauch M., van Bel A.J.E. (1998). Sieve tubes in action. Plant Cell 10: 35–50 [Google Scholar]
- Lawton D.M., Newman Y.M. (1979). Ultrastructure of phloem in young runner-bean stem: Discovery, in old sieve elements on the brink of collapse, of parietal bundles of P-protein tubules linked to the plasmalemma. New Phytol. 82: 213–222 [Google Scholar]
- Mullendore D.L., Windt C.W., Van As H., Knoblauch M. (2010). Sieve tube geometry in relation to phloem flow. Plant Cell 22: 579–593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Münch E. (1927). Versuche über den Saftkreislauf. Ber. Deut. Bot. Ges. 45: 340–356 [Google Scholar]
- Münch E. (1930). Die Stoffbewegungen in der Pflanze. (Jena, Germany: Fischer) [Google Scholar]
- Pélissier H.C., Peters W.S., Collier R., van Bel A.J.E., Knoblauch M. (2008). GFP tagging of sieve element occlusion (SEO) proteins results in green fluorescent forisomes. Plant Cell Physiol. 49: 1699–1710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters W.S., Knoblauch M., Warmann S.A., Pickard W.F., Shen A.Q. (2008). Anisotropic contraction in forisomes: Simple models won’t fit. Cell Motil. Cytoskeleton 65: 368–378 [DOI] [PubMed] [Google Scholar]
- Peters W.S., van Bel A.J.E., Knoblauch M. (2006). The geometry of the forisome-sieve element-sieve plate complex in the phloem of Vicia faba L. leaflets. J. Exp. Bot. 57: 3091–3098 [DOI] [PubMed] [Google Scholar]
- Pickard W.F., Minchin P.E. (1992). The nature of the short-term inhibition of stem translocation produced by abrupt stimuli. Funct. Plant Biol. 19: 471–480 [Google Scholar]
- Pritchard J. (1996). Aphid stylectomy reveals an osmotic step between sieve tube and cortical cells in barley roots. J. Exp. Bot. 47: 1519–1524 [Google Scholar]
- Reynolds E.S. (1963). The use of lead citrate at high pH as an electron-opaque stain in electron microscopy. J. Cell Biol. 17: 208–212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rüping B., Ernst A.M., Jekat S.B., Nordzieke S., Reineke A.R., Müller B., Bornberg-Bauer E., Prüfer D., Noll G.A. (2010). Molecular and phylogenetic characterization of the sieve element occlusion gene family in Fabaceae and non-Fabaceae plants. BMC Plant Biol. 10: 219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russin W.A., Evert R.F. (1985). Studies on the leaf of Populus deltoides (Salicaceae): Ultrastructure, plasmodesmatal frequency, and solute concentrations. Am. J. Bot. 72: 1232–1247 [Google Scholar]
- Sabnis D.D., Sabnis H.M. (1995). Phloem proteins: Structure, biochemistry and function. In The Cambial Derivatives (Encyclopedia of Plant Anatomy, Vol. 9), M. Iqbal, ed (Berlin: Borntraeger), pp. 271–292 [Google Scholar]
- Siddiqui A.W., Spanner D.C. (1970). The state of the pores in functioning sieve plates. Planta 91: 181–189 [DOI] [PubMed] [Google Scholar]
- Siebert P.D., Chenchik A., Kellogg D.E., Lukyanov K.A., Lukyanov S.A. (1995). An improved PCR method for walking in uncloned genomic DNA. Nucleic Acids Res. 23: 1087–1088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sjolund R.D., Shih C.Y. (1983). Freeze-fracture analysis of phloem structure in plant tissue cultures. II. The sieve element plasma membrane. J. Ultrastruct. Res. 82: 189–197 [DOI] [PubMed] [Google Scholar]
- Spanner D.C. (1958). The translocation of sugar in sieve tubes. J. Exp. Bot. 9: 332–342 [Google Scholar]
- Spanner D.C. (1970). The electro-osmotic theory of phloem transport in the light of recent measurements on Heracleum phloem. J. Exp. Bot. 21: 325–334 [Google Scholar]
- Spanner D.C. (1978). Sieve-plate pores, open or occluded. A critical review. Plant Cell Environ. 1: 7–20 [Google Scholar]
- Thompson C.J., Movva N.R., Tizard R., Crameri R., Davies J.E., Lauwereys M., Botterman J. (1987). Characterization of the herbicide-resistance gene bar from Streptomyces hygroscopicus. EMBO J. 6: 2519–2523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson M.V. (2006). Phloem: the long and the short of it. Trends Plant Sci. 11: 26–32 [DOI] [PubMed] [Google Scholar]
- Thompson M.V., Holbrook N.M. (2003). Application of a single-solute non-steady-state phloem model to the study of long-distance assimilate transport. J. Theor. Biol. 220: 419–455 [DOI] [PubMed] [Google Scholar]
- Thorpe M.R., Furch A.C.U., Minchin P.E.H., Föller J., Van Bel A.J.E., Hafke J.B. (2010). Rapid cooling triggers forisome dispersion just before phloem transport stops. Plant Cell Environ. 33: 259–271 [DOI] [PubMed] [Google Scholar]
- Toth K.F., Sjolund R.D. (1994). Monoclonal antibodies against plant P-protein from plant tissue cultures. II. Taxonomic distribution of cross reactivity. Am. J. Bot. 81: 1378–1383 [Google Scholar]
- Toth K.F., Wang Q., Sjolund R.D. (1994). Monoclonal antibodies against plant P-protein from plant tissue cultures. I. Microscopy and biochemical analysis. Am. J. Bot. 81: 1370–1377 [Google Scholar]
- Turgeon R. (2010). The puzzle of phloem pressure. Plant Physiol. 154: 578–581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turgeon R., Webb J.A., Evert R.F. (1975). Ultrastructure of minor veins in Cucurbita pepo leaves. Protoplasma 83: 217–232 [Google Scholar]
- Wooding F.B.P. (1969). P protein and microtubular systems in Nicotiana callus phloem. Planta 85: 284–298 [DOI] [PubMed] [Google Scholar]
- Wright K.M., Oparka K.J. (1997). Metabolic inhibitors induce symplastic movement of solutes from the transport phloem of Arabidopsis roots. J. Exp. Bot. 48: 1807–1814 [Google Scholar]