This study demonstrates that Arabidopsis thaliana Lumen Thiol Oxidoreductase1 (LTO1) catalyzes disulfide bond formation in the thylakoid lumen of the chloroplast. Loss of LTO1 function yields an assembly defect in photosystem II, a photosynthetic complex that requires a disulfide bond in a structural subunit localized in the lumen.
Abstract
Here, we identify Arabidopsis thaliana Lumen Thiol Oxidoreductase1 (LTO1) as a disulfide bond–forming enzyme in the thylakoid lumen. Using topological reporters in bacteria, we deduced a lumenal location for the redox active domains of the protein. LTO1 can partially substitute for the proteins catalyzing disulfide bond formation in the bacterial periplasm, which is topologically equivalent to the plastid lumen. An insertional mutation within the LTO1 promoter is associated with a severe photoautotrophic growth defect. Measurements of the photosynthetic activity indicate that the lto1 mutant displays a limitation in the electron flow from photosystem II (PSII). In accordance with these measurements, we noted a severe depletion of the structural subunits of PSII but no change in the accumulation of the cytochrome b6f complex or photosystem I. In a yeast two-hybrid assay, the thioredoxin-like domain of LTO1 interacts with PsbO, a lumenal PSII subunit known to be disulfide bonded, and a recombinant form of the molecule can introduce a disulfide bond in PsbO in vitro. The documentation of a sulfhydryl-oxidizing activity in the thylakoid lumen further underscores the importance of catalyzed thiol-disulfide chemistry for the biogenesis of the thylakoid compartment.
INTRODUCTION
Thiol-disulfide chemistry is an essential process for the biogenesis of the bacterial periplasm, the mitochondrial intermembrane space (IMS), and the thylakoid lumen. Strikingly, each compartment appears to have unique redox enzymes that oxidize sulfhydryls (thio-oxidation) and reduce disulfide bonds (thio-reduction) in target proteins (Herrmann et al., 2009; Depuydt et al., 2011; Kadokura and Beckwith, 2010).
In the periplasmic spaces of most proteobacteria, the thio-oxidizing pathway consists of a disulfide bond–catalyzing system defined by soluble DsbA and membrane-bound DsbB (Dsb for disulfide bond) (Heras et al., 2009; Depuydt et al., 2011; Kadokura and Beckwith, 2010). DsbA catalyzes disulfide bridge formation on Cys-containing substrates that are translocated across the membrane into the periplasmic space. DsbB operates by recycling reduced DsbA to its oxidized form with transfer of the electrons to quinones, which are membrane-soluble redox carriers in the respiratory chain. A central component of the thio-reducing pathway is the thiol-disulfide transporter DsbD/CcdA (Ccd for cytochrome c deficiency). This protein conveys reducing power from the cytosol to several periplasmic protein targets whose activity requires reduced thiols. DsbD/CcdA maintains the reduction state of oxidoreductases that shuffle disulfide bonds that are incorrectly formed and protect proteins containing a single Cys from hyperoxidation (Depuydt et al., 2011; Kadokura and Beckwith, 2010). DsbD/CcdA is also needed to reduce the active site of a disulfide reductase involved in the assembly of cytochromes c, a class of metalloproteins with a heme covalently attached to a CXXCH motif. The accepted view is that the CXXCH motif is first oxidized by the Dsb machinery and then reduced by the disulfide reductase to provide free sulfhydryls for the heme attachment (Bonnard et al., 2010; Sanders et al., 2010).
While the presence of thiol-metabolizing pathways is well established in bacteria, there was little support for the operation of thiol-based chemistry in the mitochondrial IMS and the thylakoid lumen, which are topologically equivalent to the bacterial periplasm. Recent discoveries in both organelles have now changed this perception. In mitochondria, Mia40p/Erv1p (Mia for mitochondrial intermembrane space import and assembly; Erv for essential for respiration and viability) proteins were found to be key enzymes of a disulfide relay system driving the import of Cys-rich proteins into the IMS (Depuydt et al., 2011; Riemer et al., 2011; Sideris and Tokatlidis, 2010). Although unrelated in sequence, Mia40p/Erv1p are functionally equivalent to bacterial DsbA/DsbB. Mia40p introduces disulfide bonds into protein targets and is recycled back to its oxidized form by the flavoprotein Erv1p, which transfers the electrons to cytochrome c, a soluble redox shuttle in the IMS. By analogy to the bacterial pathways, the participation of thio-reducing factors in the IMS is expected. The flavoprotein Cyc2p (Cyc for cytochrome c) and CcmH (Ccm for cytochrome c maturation), an oxidoreductase implicated in cytochrome c maturation, were proposed to act as a disulfide reductase, but this still awaits experimental validation (Bernard et al., 2005; Meyer et al., 2005; Corvest et al., 2010).
In the thylakoid lumen, the involvement of a thio-reducing pathway was established through classical and reverse genetics approaches. Components of this pathway include a thiol/disulfide membrane transporter of the CcdA/DsbD family and CCS5/HCF164 (cytochrome c synthesis/high chlorophyll fluorescence), a membrane-anchored, lumen-facing, thioredoxin-like protein. These proteins define a trans-thylakoid pathway for the delivery of reductants from stroma to lumen (Lennartz et al., 2001; Page et al., 2004; Motohashi and Hisabori, 2006, 2010; Gabilly et al., 2010, 2011). Operation of the trans-thylakoid pathway is needed to reduce disulfides in target proteins, a process essential for photosynthesis (Lennartz et al., 2001; Page et al., 2004; Gabilly et al., 2010, 2011).
The identities of the thio-oxidizing catalysts in the lumen are currently unknown, and no DsbA- or DsbB-like enzymes can be detected in the genomes of cyanobacteria, which are the presumed ancestors of chloroplasts. However, disulfide bonded proteins are present in this compartment and include not only known structural components, such as PsbO, a subunit of photosystem II (PSII; Burnap et al., 1994; Betts et al., 1996; Wyman and Yocum, 2005), and Rieske, a subunit of the cytochrome b6f complex (Carrell et al., 1997), but also molecules participating in the assembly/regulation of the photosynthetic chain (Gupta et al., 2002; Gopalan et al., 2004). The operation of one or more disulfide bond–forming catalysts in the thylakoid lumen is supported by the finding that bacterial alkaline phosphatase (PhoA), an enzyme requiring two disulfide bonds for activity, is active when targeted to this compartment in tobacco (Nicotiana tabacum; Sone et al., 1997; Bally et al., 2008).
Recently, a novel class of disulfide bond–forming enzymes has been proposed to control disulfide bond formation. This class has similarity to VKOR (for vitamin K epoxide reductase) and was recognized in cyanobacteria (Singh et al., 2008; Li et al., 2010), some bacterial phyla lacking the typical DsbAB components (Dutton et al., 2008, 2010; Wang et al., 2011), and the green lineage (Grossman et al., 2010). The VKOR-like proteins were defined based on the presence of a redox domain containing two Cys pairs. This domain is related to the one present in VKOR, an integral membrane protein of the endoplasmic reticulum (ER) (Goodstadt and Ponting, 2004; Dutton et al., 2008; Furt et al., 2010). VKOR is well studied for its involvement in the reduction of vitamin K, a phylloquinone required as a cofactor for the γ-carboxylation of clotting factors in blood (Tie and Stafford, 2008). The recent identification of TMX (for Transmembrane oxidoreductase), a membrane-anchored thioredoxin-like protein, as a redox partner of VKOR suggests that the enzymatic activity of VKOR is also linked to oxidative folding of proteins in the ER lumen (Schulman et al., 2010). One current view is that TMX (and possibly other disulfide bond–forming proteins, such as PDI, the ER resident protein disulfide isomerase) catalyzes disulfide bond formation in protein targets and is recycled to its oxidized form via thiol-disulfide relay within the VKOR domain, the final electron acceptor being vitamin K epoxide in the membrane (Rishavy et al., 2011; Schulman et al., 2010).
In the plant Arabidopsis thaliana, a VKOR-like protein localizes to the plastid (Furt et al., 2010) and is detected at the thylakoid membrane by proteomic analysis (Zybailov et al., 2008). However, its function in plastid biogenesis has so far remained elusive (Furt et al., 2010). In this article, we investigated the function of plastid Lumen Thiol Oxidoreductase1 (LTO1), the Arabidopsis VKOR-like protein. Genetic and biochemical studies indicate that LTO1 is required for the assembly of PSII through the formation of a disulfide bond in PsbO, a subunit of the PSII oxygen-evolving complex (OEC) that resides in the lumen.
RESULTS
LTO1, a VKOR-Like Protein at the Thylakoid Membrane, Is Conserved in Photosynthetic Eukaryotes
A protein displaying a membrane domain with similarity to VKOR and fused to a thioredoxin-like domain was identified in all sequenced genomes of photosynthetic eukaryotes (Tie and Stafford, 2008). We name this protein LTO1. LTO1-like proteins are predicted to be polytopic membrane polypeptides with five to six transmembrane domains and contain seven strictly conserved Cys residues (with the exception of the Volvox and Chlorella proteins) (see Supplemental Figure 1 online). Four of the Cys residues are arranged in two motifs: CXXC and WCXXC. The WCXXC motif is part of a thioredoxin-like domain that is absent from VKOR and some bacterial VKOR-like proteins (Goodstadt and Ponting, 2004).
Topological Analysis of LTO1
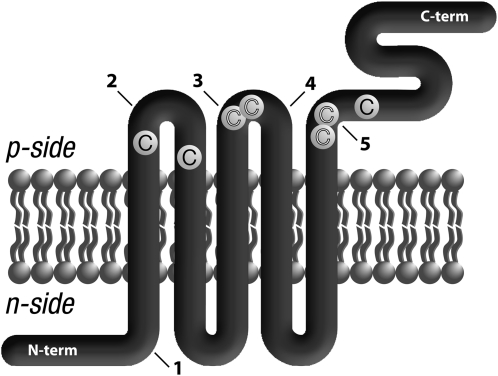
To generate a topological model of plastid LTO1, we employed PhoA and LacZ reporters in Escherichia coli (Manoil, 1991). We already demonstrated the reliability of this approach in establishing the topological arrangement of three proteins in the thylakoid membrane, which is analogous in a bioenergetic sense to the bacterial plasma membrane (Dreyfuss et al., 2003; Hamel et al., 2003; Page et al., 2004). We opted to engineer PhoA-LacZα sandwich fusions at positions predicted to be at extramembrane locations (see Supplemental Figure 1 online). High PhoA activities indicate a p-side location for the insertion site of the fusion, since PhoA is active only in the periplasm. Reciprocally, fusions with high LacZ activity confirm association of the α and ω fragments of β-galactosidase in the cytoplasm; therefore, an n-side location of the fusion site can be deduced. As shown in Table 1, PhoA activities for fusions 2, 3, 4, and 5 were 2.5- to 67-fold higher than the LacZ activities, and a periplasmic location was assigned for the corresponding positions in Arabidopsis LTO1. Note that it is likely that the stability of each sandwich fusion accounts for the difference in PhoA activities. Fusion 1 displayed a high LacZ activity but a low PhoA activity, an indication that the fusion point is located in the cytoplasm. Based on the n- and p-side topological analogy between the compartments of the bacteria (cytoplasm/periplasm) and those of the thylakoid (stroma/lumen), we deduced that the N and C termini of LTO1 face the stroma and the lumen, respectively, while domains containing redox motifs and conserved Cys residues are exposed to the lumen (Figure 1).
Table 1.
Topological Analysis of Arabidopsis LTO1 from PhoA and LacZ Fusion Analysis
| Fusion | Position | LacZ | PhoA | Topology |
| 1 | 74 | 73 (±22) | 4 (±3) | n-side |
| 2 | 113 | 19 (±4) | 78 (±7) | p-side |
| 3 | 191 | 16 (±7) | 40 (±5) | p-side |
| 4 | 195 | 14 (±8) | 80 (±5) | p-side |
| 5 | 573 | 19 (±6) | 1269 (±28) | p-side |
Measurements of alkaline phosphatase (PhoA) and β-galactosidase (LacZ) activities of sandwich fusions at the indicated positions within LTO1. Measurements were taken on at least two separate bacterial clones and correspond to an average of three representative measurements. The ±sd is indicated for those measurements. Fusion numbers correspond to those indicated in Figure 1. Values are expressed in arbitrary units for LacZ and Miller units for PhoA. The p-side corresponds to the bacterial periplasm and the lumen of the chloroplast, while the n-side corresponds to the bacterial cytoplasm and the stroma of the chloroplast.
Figure 1.
Proposed Topological Arrangement of Plastid Arabidopsis LTO1.
The proposed topological arrangement of Arabidopsis LTO1 in the thylakoid membrane was deduced from the analysis of PhoA/LacZα sandwich fusions expressed in bacteria (Table 1). The p-side corresponds to the lumen of the thylakoid, and the n-side corresponds to the stroma of the chloroplast. LTO1 is represented as a thick thread with its transmembrane domains in the lipid bilayer and extramembranous regions. The LTO1 N-terminal end faces the stroma, while the C-terminal domain, corresponding to the thioredoxin-like domain, is exposed to the thylakoid lumen. The arrows indicate the positions of in-frame sandwich fusions with the Pho-Lac minireporter, which corresponds to alkaline phosphatase fused to the α fragment of β-galactosidase. Fusions at positions 74 (1), 113 (2), 191 (3), 195 (4), and 573 (5) are indicated on the drawing by arrows. Strictly conserved Cys residues are marked (C). Conserved Cys residues within CXXC motifs are highlighted in white.
LTO1 Partially Complements for Loss of DsbAB in Bacteria
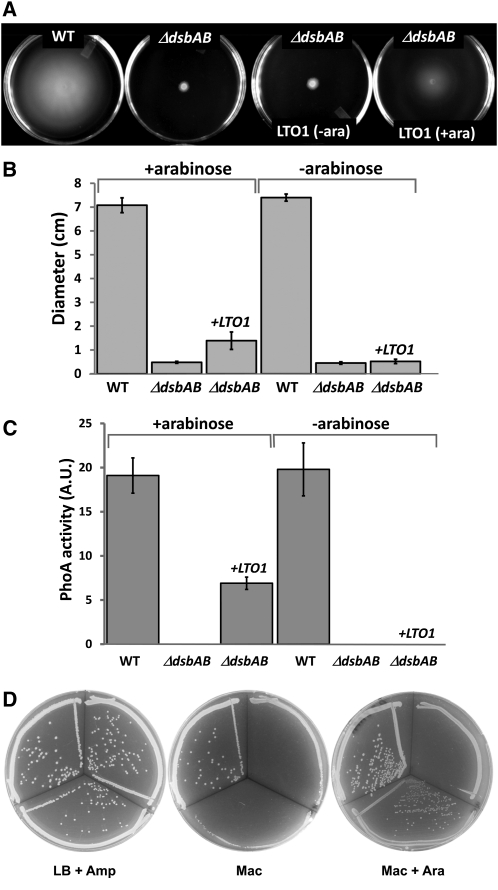
To provide further support for the proposed topology and function, we chose a heterologous complementation assay. In this assay, an E. coli dsbAB mutant was used to test the ability of LTO1 to restore disulfide bond formation in the periplasmic space. For this experiment, we chose an Ara-inducible promoter to drive the expression of full-length Arabidopsis LTO1 cDNA and monitored bacterial motility, PhoA activity, and growth on MacConkey agar, which are dependent upon DsbAB. When FlgI, a flagellar motor protein in the periplasmic space, is not disulfide bonded, the bacteria are nonmotile (Dailey and Berg, 1993). This loss of motility can be readily visualized on low concentration agar. As shown in Figures 2A and 2B, LTO1 partially restores the motility phenotype of a dsbAB mutant, demonstrating that the plant protein can promote disulfide bond formation. This restoration is Ara dependent, confirming that LTO1 is indeed responsible for the observed restored motility. The activity of periplasmic PhoA, which requires two intramolecular disulfide bonds (Sone et al., 1997), was also partially restored upon expression of the plant protein (Figure 2C). In addition, while E. coli strains lacking DsbA and/or DsbB grow poorly on MacConkey agar, the dsbAB mutant expressing LTO1 grows well on MacConkey agar supplemented with the inducer Ara (Table 2, Figure 2D).
Figure 2.
Arabidopsis LTO1 Partially Complements the E. coli dsbAB Mutant for Disulfide Bond Formation.
Strains are wild-type HK295 (pBAD24), ΔdsbAB HK329 (pBAD24), and ΔdsbAB HK329 (pLTO200).
(A) Complementation of the motility phenotype in dsbAB mutants by Arabidopsis LTO1. Representative motility phenotype of wild-type (WT), ΔdsbAB, and ΔdsbAB bacterial strains expressing LTO1 under an Ara-inducible promoter (+/−ara) on M9 solid media.
(B) Quantification of motility phenotype represented in (A). Each bar represents the sd from 10 independent plate measurements.
(C) Complementation of the phoA-deficient phenotype in dsbAB mutants by LTO1. Each bar represents the sd from three separate measurements of PhoA activity. A.U., arbitrary units.
(D) Complementation of the growth phenotype on MacConkey agar in dsbAB mutants by LTO1. In all plates in (D), the top left sector was inoculated with HK295 (pBAD24), the top right sector was inoculated with HK329 (pBAD24), and the bottom sector was inoculated with HK329 (pLTO200).
Table 2.
Efficiency of Plating on MacConkey Agar of dsbAB Strains Expressing Arabidopsis LTO1
| EOPa |
||||
| Strain | Relevant Genotype | LB + Amp | Mac | Mac + Ara |
| HK295(pBAD24) | Wild type | 1.0 | 1 | 0.1 |
| HK329(pBAD24) | ΔdsbAB | 1.0 | 10−5 | 10−4 |
| HK329 (pLTO200) | ΔdsbAB lto1+ | 0.1 | 10−5 | 0.1 |
Amp, ampicillin; Mac, MacConkey.
EOP (efficiency of plating) values were calculated with respect to the growth of the wild-type strain HK295 (pBAD24) on LB agar containing ampicillin.
We conclude that LTO1 partially compensates for the loss of bacterial DsbAB and displays sulfhydryl oxidizing activity. Our findings solidify the proposed lumenal location of the redox motifs and conserved Cys residues (Figure 1) and suggest that the relevant targets of action of LTO1 are also lumen localized.
Knockdown of Arabidopsis LTO1 Impairs Plant Growth
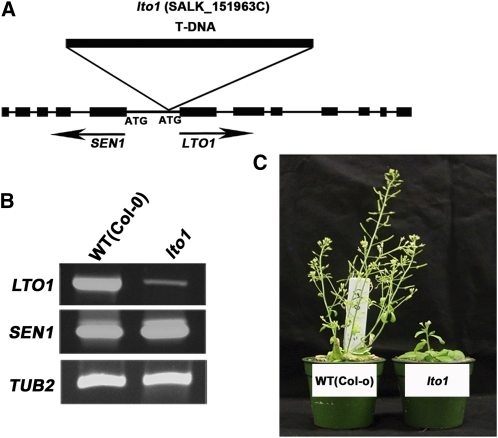
To address the question of LTO1 function at the thylakoid membrane, we chose a reverse genetics approach and identified a T-DNA insertion in the promoter region of Arabidopsis LTO1 (Figure 3A). Through PCR analysis and sequencing, we confirmed that the inserted T-DNA is located 45 bp from the predicted initiation codon of the LTO1 open reading frame (ORF). RT-PCR analysis showed that the LTO1 transcript accumulation is strongly reduced in the homozygous insertion line, suggesting that insertion of the T-DNA altered the transcriptional activity of LTO1 (Figure 3B). This promoter region is also shared by SEN1, which was shown to be involved in senescence (Schenk et al., 2005). However, we show that the transcription of SEN1 is not affected by the T-DNA insertion (Figure 3B). Homozygous lto1 mutants display a severe growth phenotype compared with the wild-type Columbia (Col) ecotype when grown on soil. The phenotype is not as drastic when grown on Suc-supplemented agar, indicating that the slow growth may be due to a defect in photosynthesis.
Figure 3.
Knockdown of LTO1 Results in a Growth Defect.
(A) Structure of LTO1 and SEN1. Black boxes represent exons and thin lines correspond to introns in SEN1 and LTO1. The initiation codons (ATG) of SEN1 and LTO1 are indicated. The position of the T-DNA insertion in the SALK_15193C line is indicated.
(B) Expression of LTO1. Transcripts corresponding to LTO1 and SEN1 were analyzed by RT-PCR in homozygous lto1 and wild-type (WT [Col-0]) plants. TUB2 was used as a control for constitutive expression. To ensure that amplification was in the linear range, the optimal number of cycles was determined for each couple of primers separately. PCR amplification products were separated by electrophoresis in agarose gel and ethidium bromide stained. The gel was imaged using an imaging system. Four independent biological replicates were performed for this experiment, and one representative is shown in the figure.
(C) The lto1 mutant displays a growth defect. Wild-type (Col-0) and lto1 homozygous plants are shown 6 weeks after seed germination on soil.
[See online article for color version of this figure.]
Loss of LTO1 Produces a Photosynthetic Defect
To test the hypothesis that the reduced growth seen in the mutant is a result of a photosynthetic defect, we conducted several spectroscopy tests on fresh leaves of lto1 plants. Illumination with far-red light preferentially excites photosystem I (PSI) over PSII, resulting in net oxidation of the plastoquinone (PQ) pool, cytochrome f, plastocyanin (PC), and P700 of PSI (Joliot and Joliot, 2005). The status of P700 was monitored by 10-μs flashes from a 705-nm LED. We found that the leaves of lto1 plants exhibited P700 oxidation by far-red light and rereduction immediately upon termination of illumination (see Supplemental Figure 2 online), indicating that they possessed functioning PSI. The absolute level of the P700 photobleaching signal in lto1 leaves was smaller than in wild-type leaves, but this was likely due to the fact that lto1 leaves tended to be thinner than wild-type leaves. We concluded that the function of PSI is not affected by the mutation. We did, however, notice that the net oxidation of P700 proceeded more quickly in the lto1 leaves. Moreover, they lacked a kinetic inflection point early in the time course (see Supplemental Figure 2A online), which is likely due to an influx of electrons into the electron transfer chain from a light-dependent source. The source is most likely PSII, as the rereduction of P700+ occurred with a higher rate in the lto1 mutant in the dark (see Supplemental Figure 2B and Supplemental Table 1 online). This would seem to rule out any defect in PC or cytochrome b6f levels. The higher rate of P700+ rereduction upon termination of illumination might be taken as an indication that thylakoids of lto1 plants are engaged in cyclic electron transfer to a much higher extent than in wild-type plants.
We also examined directly the changes in the redox state of cytochrome b6f cofactors induced by far-red light. We found that both the oxidation of cytochrome f and the reduction of cytochrome b occurred to a greater extent in the lto1 plants during illumination, indicating a limitation in electron flow to the cytochrome b6f complex in the light (see Supplemental Figure 3 online). The kinetics of decay in the dark were roughly the same. In summary, there appears to be a limitation in light-driven electron flow to cytochrome b6f, which would imply a defect in PSII function.
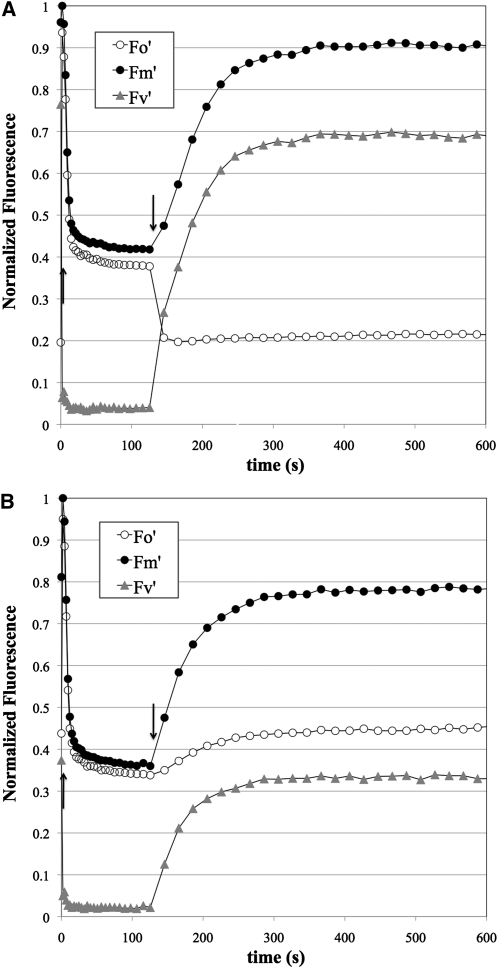
We tested this hypothesis by fluorescence induction analysis (Figure 4, Table 3). The rise in fluorescence emission from chlorophyll from the initial state in dark-adapted plants (F0) to the maximal level (FM) provoked by a 80-ms saturating pulse, otherwise known as the variable fluorescence (FV), is a function of the quantum efficiency of PSII (Rohacek and Bartak, 1999). The FV/FM ratio is taken as a measure of the maximal quantum yield of PSII. In wild-type leaves, FV/FM was 0.8, which is a typical value for healthy plants, and the variation was ~1 to 2% of this value. By contrast, the FV/FM ratio in lto1 leaves was ~0.5, with higher variability (~20%; Table 3). This indicates a rather severe defect in PSII. Upon continuous illumination, FV dropped precipitously due to a large decrease in FM (maximal fluorescence level in the light) in both wild-type and lto1 leaves. However, we found that the level of chlorophyll fluorescence before the saturating pulse (F0) was actually lower than F0 in lto1 plants, which is unusual (see Discussion). Nonphotochemical quenching in the lto1 leaves exhibited enormous variation and was not consistently higher or lower than wild-type leaves at the same light intensity (Table 3). While the lto1 leaves seemed to suffer from higher photoinhibition (loss of PSII activity due to illumination at high light fluxes) than wild-type leaves, the difference was not great (Table 3).
Figure 4.
The Photosynthetic Defect in the lto1 Mutant Is Due to a Limitation in the Electron Flow from PSII.
Fluorescence induction of representative leaves from wild-type (A) and lto1 (B) plants. Fluorescence was measured using a 10-μs pulse of 430-nm light and appropriate filters to transmit red light. Actinic illumination provided by 520-nm LEDs (1.4 mmol photons m−2 s−1) commenced immediately after taking F0 and FM values at time 0. At each time point, fluorescence immediately before (F0, open circles) and after (FM, closed circles) a 80-ms saturating pulse was measured during a brief window in which the actinic light was off (see Methods for details). After 2 min, the actinic light was extinguished and the recovery of fluorescence was followed in the same manner. The variable fluorescence at each point (FV = FM − F0) is also shown as gray triangles. All fluorescence values were normalized to FM; thus, what is shown is actually FM/FM (closed circles), F0/FM (open circles), and FV/FM (gray triangles). Upward and downward arrows indicate when the actinic light was on or off, respectively.
Table 3.
Quantum Efficiency, Nonphotochemical Quenching, and Photoinhibition in Wild-Type and lto1 Lines
| Wild Type | lto1 | |
| Actinic Light Intensity | Average ± sd, n = 6 | Average ± sd, n = 8 |
| 170 μE m−2 s−1 | ||
| FV/FM | 0.81 ± 0.01 | 0.49 ± 0.11 |
| NPQ | 1.17 ± 0.08 | 0.90 ± 0.79 |
| 1400 μE m−2 s−1 | ||
| FV/FM | 0.80 ± 0.01 | 0.48 ± 0.11 |
| NPQ | 1.370 ± 0.06 | 1.82 ± 1.15 |
| Photoinhibition (%) | 6.0 ± 1.3 | 9.9 ± 1.9 |
Fluorescence parameters FV/FM (maximum quantum yield of PSII), nonphotochemical quenching (NPQ), and photoinhibitions were calculated based on fluorescence measurements like those shown in Figure 4 using formulae described by Rohacek and Bartak (1999). FV/FM = (FM − F0)/FM, where F0 is the initial fluorescence emission after dark adaptation and immediately before a saturating pulse, FM is the maximal fluorescence emission immediately after the saturating pulse, and the variable fluorescence parameter, FV = FM − F0. NPQ = (FM − FM)/FM, where FM is the maximal fluorescence emission after adaptation to the actinic light (after 2 min). Photoinhibition = 1 – (FV/FM)/(FV/FM), where FV and FM are the variable and maximal fluorescence, respectively, after 2 min of exposure to 1400 μE m−2 s−1 actinic light followed by 8 min of dark recovery (i.e., irreversible loss of quantum efficiency).
Finally, we examined the function of ATP synthase using the decay of the carotene bandshift signal at 520 nm as a spectroscopic marker for the transmembrane electric field of the thylakoid membrane (Sacksteder and Kramer, 2000). The rate of decay is a measure of how fast the proton motive force is expended, which is primarily due to ATP synthase activity. Although we saw slight differences between the wild-type and mutant leaves, in terms of the multiphasic decay of the carotenoid bandshift signal, the overall decay was similar (see Supplemental Figure 4 online). Thus, we see no evidence for major defects in the function of ATP synthase. In conclusion, our spectroscopy analysis of lto1 leaves pinpoints PSII as the site of the photosynthetic defect caused by the lto1 mutation. We saw no problems with the function of the other three major thylakoid membrane complexes: PSI, cytochrome b6f, or ATP synthase.
Loss of LTO1 Impacts PSII Accumulation
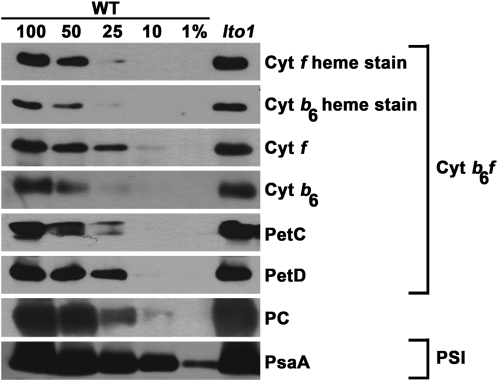
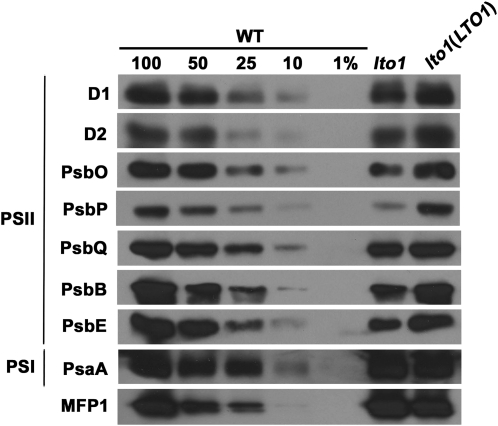
To further detail the impact on photosynthesis due to loss of LTO1, we performed immunoblot analyses using antibodies against the subunits of PSI, PSII, and the cytochrome b6f complex. We found that loss of LTO1 has no impact on the accumulation of the cytochrome b6f subunits, including the cytochromes b6 and f that contain a heme cofactor (Figure 5). The steady state level of PC, the electron carrier between the cytochrome b6f and PSI, was unaffected by the lto1 mutation (Figure 5). We also excluded a defect in PSI based on the fact that the abundance of PsaA, a core subunit whose accumulation is diagnostic of PSI assembly, is unchanged (Figure 5). By contrast, we noted a depletion of the PSII subunits, including D1, D2, PsbB (CP47), and PsbE (cytochrome b559), which are the core proteins of the reaction center (Figure 6). Interestingly, there is also a severe reduction in the levels of PsbO, PsbP, and PsbQ, three extrinsic proteins bound to the PSII core subunits on the lumenal side of the thylakoid membrane (Bricker and Frankel, 2011; Popelkova and Yocum, 2011). Together, these three proteins define a functional module called the OEC, which mediates the light-dependent oxidation of water.
Figure 5.
Accumulation of Cytochrome b6f and PSI Is Not Affected by Loss of LTO1.
Wild-type (WT) and lto1 homozygous plants were analyzed for the accumulation of cytochrome b6f subunits (cytochrome b6, cytochrome f, PetC, and PetD), PC, and PSI subunit PsaA via immunoblotting. Heme staining was also performed to analyze the accumulation of holoforms of cytochrome b6 and f. Total protein samples corresponding to 16 μg of chlorophyll were separated on a 12% SDS-acrylamide gel. For an estimation of the protein abundance in the lto1 mutant, dilutions of the wild-type sample were loaded on the gel. Gels were transferred to membranes before heme staining by chemiluminescence or immunodetection.
Figure 6.
Accumulation of PSII Subunits Is Impacted by Loss of LTO1.
The accumulation of PSII subunits (D1, D2, PsbO, PsbP, PsbQ, PsbB, and PsbE) in the lto1 mutant, the lto1 mutant complemented by the LTO1-C-MYC cDNA (under the control of the 35S promoter), and wild-type (WT) plants was analyzed via immunoblot analyses. PSI subunit PsaA and MFP1 (for MAR binding filament-like protein 1), a thylakoid-associated nucleoid binding protein, were used as control (Jeong et al., 2003). Thylakoid proteins corresponding to 7 μg of chlorophyll were separated on a 12% SDS-acrylamide gel. For an estimation of the protein abundance in the lto1 mutant, dilutions of the wild-type sample were loaded on the gel. Gels were transferred to membranes before immunodetection with antisera against PSII subunits PsaA and MFP1.
A construct expressing the LTO1 cDNA engineered with a C-terminal C-myc tag complements both the photoautotrophic growth and PSII defects when introduced in the lto1 mutant (Figures 6 and 7, Table 4). This demonstrates that the molecular lesion in LTO1 is responsible for the PSII-deficient phenotype. Using an anti-C-myc antibody to probe thylakoid membrane extracts, we were able to detect a 40-kD band corresponding to LTO1 in the lines complemented with the cDNA expressing construct (see Supplemental Figure 5 online).
Figure 7.
Complementation of the lto1 Photosynthetic Defect by LTO1 cDNA.
Phenotypes of the wild type (WT), two independent lto1 lines expressing the LTO1-C-MYC cDNA under the control of the 35S promoter (A and B), and lto1 homozygous plants after 3 weeks of growth on soil.
[See online article for color version of this figure.]
Table 4.
Quantum Efficiency of Wild-Type, lto1, and LTO1-Complemented lto1 Lines
| Strain | Medium | FV/FM (±sd), n = 5 |
| Wild type | MS | 0.828 ± 0.007 |
| lto1 | MS | 0.581 ± 0.068 |
| lto1 + LTO1 | MS | 0.831 ± 0.005 |
| Wild type | MS + Suc | 0.815 ± 0.008 |
| lto1 | MS + Suc | 0.522 ± 0.113 |
| lto1 + LTO1 | MS + Suc | 0.828 ± 0.003 |
Maximum quantum yield of PSII (FV/FM) was measured as described in Table 3 and Methods. Each type of plant was grown from seeds on Murashige and Skoog (MS) or Murashige and Skoog + 2% Suc medium under moderate light (~80 μmol PAR photons m−2 s−1). Leaves from five separate plants were removed and immediately assayed.
The Thioredoxin-Like Domain of LTO1 Interacts with PsbO1 and PsbO2
The finding that LTO1 exhibits sulfhydryl oxidizing activity and is required for PSII accumulation suggests that the formation of one or more disulfide bonds is needed for the biogenesis of this photosynthetic complex. Based on our heterologous complementation experiments, it is likely this step occurs on the lumenal side of the thylakoid membrane. Interestingly, PsbO, a lumenal subunit of the OEC, carries a single disulfide bond that is critical for PSII assembly and activity (Burnap et al., 1994; Betts et al., 1996; Wyman and Yocum, 2005). To test the hypothesis that PsbO is a relevant target of LTO1, we decided to see if the thioredoxin-like soluble domain of LTO1 (LTO1sol), postulated to carry the sulfhydryl oxidizing activity, could interact with PsbO in a yeast two-hybrid assay. LTO1sol was used as bait and PsbO1 and PsbO2, the two isoforms of Arabidopsis PsbO (Peltier et al., 2002; Schubert et al., 2002), were used as prey in a GAL4-based two-hybrid system. In one study, the second Cys in the WCXXC motif was shown to be critical in detecting the interaction between thioredoxins and their targets using a yeast two-hybrid assay (Vignols et al., 2005). However, we found that both wild-type (WCSHC) and mutant (WCSHS) forms of LTO1sol could interact with either PsbO1 or PsbO2, based on the recovery of GAL4-dependent adenine/His prototrophies and aureobasidin A resistance in the yeast reporter strain (Figure 8). As expected, none of these polypeptides alone elicit such a response (Figure 8). This established both PsbO1 and PsbO2 as relevant targets of LTO1 action in vivo.
Figure 8.
PsbO1 and PsbO2 Interact with the Thioredoxin-Like Domain of LTO1 in a Yeast Two-Hybrid Assay.
The thioredoxin-like domain of wild-type (WCSHC) or mutant (WCSHS) LTO1 constitutes the bait and is expressed as a fusion with the GAL4 DNA binding domain from the TRP1-based pGBKT7 vector. Arabidopsis PsbO1 or PsbO2 constitute the prey and are expressed as a fusion with the GAL4 activation domain from the LEU2-based pGADT7 vector. The yeast Y2HGold reporter strain was cotransformed with various combinations of two-hybrid plasmids as indicated on the top right side of the figure (“Empty” refers to the pGBKT7 or pGADT7 vectors). The yeast transformants were tested for adenine/His prototrophies and resistance to aureobasidin A, which depend on reconstitution of an active GAL4. One representative transformant for each combination was plated on solid medium lacking Leu and Trp (SD-L-W), incubated for 3 d at 28°C, and replicated on medium lacking Leu, Trp, adenine, and His (SD-L-W-A-H) or lacking Leu, Trp, adenine, and His with aureobasidin A (SD-L-W-A-H+AbA) and incubated at 28°C for 5 d. GAL4-dependent adenine and His prototrophies and resistance to aureobasidin A indicate interaction between PsbO2 and wild-type LTO1 and also between PsbO1 and mutant LTO1.
The Soluble Domain of LTO1 Can Catalyze Disulfide Bond Formation in PsbO
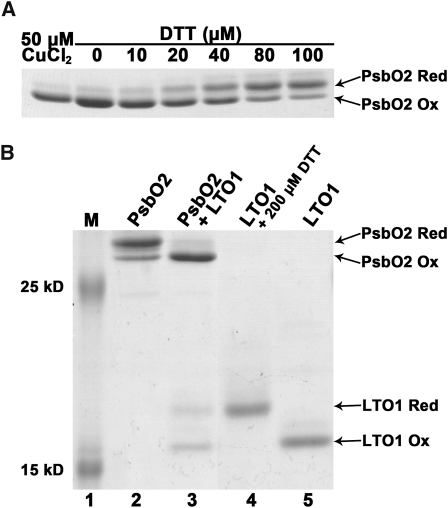
To test the possibility that the single Cys pair present in PsbO1 and PsbO2 could be oxidized by LTO1, we purified recombinant forms of LTO1sol and PsbOs and performed in vitro redox assays. We attempted to purify PsbO1 as a recombinant protein, but the resulting polypeptide was proteolytically processed when expressed in bacteria. Recombinant PsbO2, on the other hand, was purified in its oxidized form and the disulfide bond present in the molecule could be chemically reduced in a dose-dependent fashion by the action of DTT (Figure 9A). To test the ability of oxidized LTO1sol to introduce a disulfide bond in PsbO2, we incubated air-oxidized LTO1sol with reduced PsbO2. This resulted in the oxidation of PsbO2 (Figure 9B, lane 3). Concomitant with the oxidation of PsbO, oxidized LTO1sol was converted to its reduced form (Figure 9B, lane 3), an expected finding if LTO1sol catalyzes disulfide bond formation. Quantification of the oxidized and reduced species indicates that one molecule of reduced PsbO2 was acted upon by one molecule of oxidized LTO1. It should be noted that, even though DTT treatment fully reduced PsbO2, a small fraction of reduced PsbO2 can become reoxidized, presumably because of traces of oxygen present in solution (Figure 9B, lane 2). Based on our redox assays, we conclude that the thioredoxin-like domain of LTO1 can catalyze disulfide bond formation in the PsbO2 target.
Figure 9.
The Thioredoxin-Like Domain of LTO1 Catalyzes Disulfide Bond Formation in PsbO2.
Recombinant proteins (PsbO2 and the thioredoxin-like domain of LTO1) were treated with AMS, separated by nonreducing SDS-PAGE (15%), and visualized by Coomassie blue staining. The positions of reduced (PsbO2 Red) and oxidized (PsbO2 Ox) forms of PsbO2 and reduced (LTO1 Red) and oxidized (LTO1 Ox) forms of LTO1 are indicated by arrows. AMS is an alkylating reagent, and AMS treatment of exposed thiols in PsbO2 and LTO1 results in an increased molecular mass of the alkylated molecules. Only reduced PsbO2 or LTO1 reacts with AMS.
(A) DTT-dependent reduction of the disulfide in PsbO2. Lane 1, recombinant PsbO2 was fully oxidized by incubation with 50 μM CuCl2. Lanes 2 to 7, air-oxidized PsbO2 was reduced by 1 h incubation with increasing concentrations of DTT.
(B) LTO1-dependent oxidation of PsbO2 sulfhydryls. Prestained protein ladder (Fermentas) was used in lane 1. Lane 2, reduced PsbO2 after removal of DTT; lane 3, reduced PsbO2 (10 μM) mixed with oxidized LTO1 thioredoxin-like domain (16 μM); lane 4, DTT-reduced LTO1 thioredoxin-like domain; and lane 5, air-oxidized LTO1. Quantification using ImageJ software indicates that 72% of reduced PsbO2 (7.2 μM) becomes oxidized, whereas 44% of oxidized LTO1 (7.04 μM) is converted to its reduced form.
DISCUSSION
The Plastid VKOR-Like Protein Defines a Trans-Thylakoid Thio-Oxidation Pathway
In this article, we explored the function of LTO1, a plastid VKOR-like protein in Arabidopsis. In plastids, VKOR-like proteins are present at the thylakoid membrane and carry a C-terminal, thioredoxin-like domain typical of oxidoreductases belonging to the PDI family (Ellgaard and Ruddock, 2005). By analogy to VKOR in the ER, a plastid VKOR-like protein is presumed to participate in a transmembrane thio-oxidation pathway. One key question in terms of deducing the function of a plastid VKOR-like protein is to define its topological arrangement within the thylakoid membrane, particularly with respect to the domains predicted to catalyze redox chemistry. Using bacterial topological reporters, a lumenal localization was assigned for the two Cys pairs present in the VKOR-like domain and the C-terminal thioredoxin-like domain of LTO1 (Figure 1). This result is in accordance with the topological model deduced for a mycobacterial VKOR-like protein and the crystal structure of a cyanobacterial VKOR-like protein. Both of these methods place the functional domains in the periplasm, which is analogous to the plastid lumen (Dutton et al., 2008; Li et al., 2010; Wang et al., 2011).
In proteobacteria, DsbA introduces disulfide bonds in Cys-containing targets, while DsbB recycles DsbA to its oxidized form with transfer of electrons to a quinone (Depuydt et al., 2011; Kadokura and Beckwith, 2010). A role in disulfide bond formation for LTO1 is inferred from the fact that the plant protein is able to substitute for the function of DsbAB in bacteria (Figure 2). Further support for the involvement of LTO1 in sulfhydryl oxidation in the thylakoid lumen comes from our evidence that lumen resident PsbO can be oxidized by the thioredoxin-like domain of the protein in our in vitro assay (Figure 9). Previous studies with mycobacterial and cyanobacterial VKOR-like proteins have demonstrated that the thioredoxin-like domain carries DsbA-like activity, while the VKOR-like central domain is functionally equivalent to DsbB (Singh et al., 2008; Dutton et al., 2010; Wang et al., 2011). In vitro assays show that the Arabidopsis LTO1 thioredoxin-like domain can transfer electrons to its VKOR-like central domain (Furt et al., 2010). This supports the view that the plastid protein operates in a manner similar to bacterial VKOR-like proteins in regard to thiol-disulfide chemistry.
As seen with bacterial VKOR-like proteins, it is expected that the sulfhydryl oxidizing activity of LTO1 is linked to the reduction of a quinone in the thylakoid membrane (Li et al., 2010). In vitro enzymatic assays indicate that the Arabidopsis VKOR-like protein is active in reducing phylloquinone but not PQ, two quinones found in plastids (Furt et al., 2010). The role of phylloquinone as a structural cofactor tightly bound to PSI is well documented (Brettel, 1997). However, it is unclear if phylloquinone participates in redox processes in addition to the known electron transfer reactions through PSI (Gross et al., 2006; Lohmann et al., 2006).
The Redox Activity of LTO1 Is Required for the Assembly of PSII
Detailed phenotypic analysis of the Arabidopsis lto1 mutant revealed that the function of the plastid VKOR-like protein is required for accumulation of PSII, a photosynthetic complex involved in the light-dependent reactions of photosynthesis (Figures 3 to 7). Moreover, analysis of the decay of variable fluorescence after a 1-ms flash in the presence of the QB site inhibitor DCMU was consistent with the idea that there was a higher amount of PSII reaction centers with damaged OECs in the lto1 mutant (see Supplemental Figure 6 online). We reasoned that catalysis of a disulfide bond in a lumenal target is a required step for the biogenesis of PSII. Interestingly, PsbO, a lumenal subunit of PSII required for stable assembly of the OEC, was shown to carry a single intramolecular disulfide that is strictly conserved in cyanobacteria and photosynthetic eukaryotes. In vitro studies established that this disulfide bond is critical to maintain the tertiary structure of PsbO (Tanaka et al., 1989; Betts et al., 1996; Wyman and Yocum, 2005). In chloroplast lumenal extracts, reduction of the disulfide in PsbO1 and PsbO2 targets the proteins for degradation. This indicates that the oxidation state of the Cys residues is a key determinant for the subunit stability in vivo (Hall et al., 2010). Further underscoring the importance of this disulfide, a mutation of one of the conserved Cys residues in cyanobacterial PsbO results in complete loss of the subunit and yields a PSII defect (Burnap et al., 1994). In Arabidopsis, loss of PsbO1 and PsbO2 impacts the stability of both extrinsic components of the OEC and core components of the reaction center (Yi et al., 2005). The resulting phenotype is similar to the one displayed by the lto1 mutant lines (Figure 6). Considering that PsbO is a relevant target of LTO1 activity (Figure 9) and that the redox state of the sulfhydryls appears to be determinant for the stability of this subunit (Burnap et al., 1994; Hall et al., 2010), it is conceivable that the PSII assembly deficiency in the lto1 mutant is caused by a sole defect in the oxidation of PsbO sulfhydryls. However, there are proteins required for PSII biogenesis that also have a disulfide bond, such as FKBP20-2, a lumen resident molecule (Lima et al., 2006). Therefore, we cannot rule out the possibility that additional factors might also contribute to the PSII-deficient phenotype in the lto1 mutant.
It is interesting that the fluorescence induction phenotype of the lto1 mutant strongly resembles that of Arabidopsis mutants with low amounts of functional PsbO protein. The quantum yield of PSII fluorescence is decreased significantly in mutants of PSBO1 (Murakami et al., 2002) and in RNA interference mutants largely lacking both PsbO1 and PsbO2 (Yi et al., 2005). However, the PSII maximal quantum yield in the lto1 mutant and in the psbO mutants is not as low as in mutants completely lacking PSII, where it effectively becomes zero (Bennoun et al., 1986). Moreover, in the psbO-1 mutant, Murakami et al. (2002) found that the fluorescence level transiently dropped below the F0 level during illumination, similar to what we saw in the lto1 mutant (Figure 4). This phenomenon is not limited to psbO mutants, as it was also seen recently with mutants in the cytochrome b559 subunit of PSII (Bondarava et al., 2010). One possibility is that the PQ pool is partially reduced in the dark in the lto1 mutant and in other mutants with low PSII activity, which would result in an increased F0 level. Upon illumination, PSI activity overwhelms PSII activity (due to the PSII deficiency), and the PQ pool becomes more oxidized, resulting in a fluorescence level lower than the level in the dark. We suggest that in such mutants, the cyclic electron flow pathway has been activated. This would allow the mutants to use PSI and cytochrome b6f to pump protons and thereby synthesize ATP in the absence of significant levels of linear electron flow due to the deficiency in PSII. This would explain both the elevated F0 level and the fact that P700+ is rereduced much faster in the lto1 mutants compared with the wild type (see Supplemental Figure 2 online), an expected finding if cyclic electron transfer is elevated (Finazzi and Forti, 2004; Iwai et al., 2010; Peeva et al., 2010).
Are There Other Relevant Targets of LTO1 Sulfhydryl Oxidizing Activity?
The lto1 mutant characterized during this study is a knockdown, and additional defects could be revealed in conditions of complete loss of LTO1 function (Figure 3). Conceivably, LTO1 regulates the redox state of additional Cys-containing proteins residing in the thylakoid lumen. An indication that further targets exist comes from the observation that a cyanobacterial VKOR-like null mutant displays a pleiotropic growth defect incompatible with a sole defect in PSII (Singh et al., 2008). However, it is not known if this phenotype can be attributed to impaired disulfide bond formation in targets residing in the thylakoid lumen and/or in the periplasm. Other possible targets of LTO1 are proteins containing lumen-facing Cys residues, which are active in the oxidized form. Only a few lumenal proteins with known thiol-dependent enzymatic activities have been identified. Examples of such are violaxanthin deepoxidase, an enzyme involved in dissipating excess light (Yamamoto and Kamite, 1972; Sokolove and Marsho, 1976), STT7/STN7, a kinase involved in adaptation to changes in light intensity (Depège et al., 2003; Lemeille et al., 2009), and FKBP13, a prolyl isomerase postulated to act as a foldase for the Rieske protein (Gupta et al., 2002; Gopalan et al., 2004). It has now become apparent that thiol-disulfide chemistry is a catalyzed process not only restricted to bacterial energy-transducing membranes but operating also on the lumenal side of the thylakoid membrane. Further genetic and biochemical dissection is needed to elaborate the thiol-metabolizing pathways and understand how they regulate the biogenesis of the thylakoid compartment.
METHODS
Bacterial Strains and Growth Media
Strains HK295 [F- Δara-714 galU galK Δ(lac)X74 rpsL thi] (Kadokura and Beckwith, 2002) and HK329 (HK295 ΔdsbA ΔdsbB) (Eser et al., 2009) were a kind gift from Jonathan Beckwith. Luria-Bertani (LB) broth, LB agar, and MacConkey agar were prepared as described previously (Silhavy et al., 1984). When indicated, ampicillin (50 μg/mL) and l-Ara were used.
Heterologous Complementation of the dsbAB Mutant
The LTO1 ORF was cloned, using the In-Fusion system (Clontech), into the Ara-inducible vector pBAD24 (Guzman et al., 1995) at NcoI and SphI sites to create plasmid pLTO200. Primers used for In-Fusion cloning and sequencing of the resulting plasmids are listed in a table of primers (see Supplemental Methods 1 and Supplemental References 1 online). The pBAD24 and pLTO200 plasmids were introduced into the ΔdsbAB HK329 strain and transformants assessed for complementation of motility and alkaline phosphatase (PhoA)–deficient phenotypes. For motility tests, 10 μL of an overnight culture grown in M9 medium was inoculated into the center of an M9 medium plate (0.25% agar), with or without 0.002% Ara. Plates were incubated overnight at 37°C, and complementation of the motility phenotype was quantified by measuring the diameter of the swarm in centimeters using a metric ruler. Ten plates were measured for both the control pBAD24 and pLTO200 in both plus and minus Ara conditions. The PhoA activities were measured as described by Manoil (1991) using overnight cultures in M9 medium, with or without Ara.
Efficiency of Plating Assay
Strains HK295 (pBAD24), HK329 (pBAD24), and HK329 (pLTO200) were grown at 37°C overnight in LB broth supplemented with ampicillin. Cultures were serially diluted 1:10 in LB broth, and ~2 μL of these dilutions was spotted onto LB agar and MacConkey agar either lacking or containing l-Ara (0.2%). Plates were incubated overnight at 37°C. Efficiency of plating values were calculated with respect to the growth of the wild-type strain HK295 (pBAD24) on LB agar.
Topological Analysis of LTO1 via PhoA/LacZα Sandwich Fusions
Four PhoA/LacZα sandwich fusion constructs (pLTO212-pLTO215) were generated using In-Fusion recombination with three PCR products as described by Zhu et al. (2007). PCR products were generated using the LTO1 cDNA or the complete PhoA/LacZα fusion cassette in pMA650 as templates (Alexeyev and Winkler, 1999). The PCR products were then cloned into the isopropyl 1-thio-β-d-galactopyranoside (IPTG)–inducible expression vector pMA657 (Alexeyev and Winkler, 1999) cut by BamHI and XbaI. The pLTO211 plasmid was also generated using In-Fusion recombination at SpeI in pLTO210, which was constructed by In-Fusion recombination of the LTO1 ORF into pMA657. PhoA activity measurements were performed as described by Manoil (1991). On the same cultures, LacZ activity measurements were performed as described by Brickman and Beckwith (1975). The leaky nature of the lac promoter allowed measurements to be performed without IPTG induction.
Yeast Two-Hybrid Experiments
The soluble domain of LTO1 (Gln-247 to Gln-376) was used as bait, and the corresponding sequence was cloned via the In-Fusion technique (Clontech) as a PCR fragment at the NdeI-SalI sites of the pGBKT7 vector. The sequence corresponding to the soluble domain of wild-type LTO1 was PCR amplified using LTO1-BD-F (5′-GATCTCAGAGGAGGACCTGCAACAACCAATCCCTTCACGC-3′) and LTO1-BD-R (5′-TGCGGCCGCTGCAGGTCGACGCTGAAGTTGATTGGTCTC-3′) as primers and the Arabidopsis thaliana LTO1 cDNA from the ABRC (The Ohio State University; #U25043) as template. Plasmids expressing the mutant form of LTO1 (WCSHS) were constructed via the QuickChange II site-directed mutagenesis kit (Stratagene). Arabidopsis PsbO1 and PsbO2 (Ser-30 to Glu-332 or Ser-29 to Glu-331, respectively) were used as prey, and the corresponding sequences were cloned via In-Fusion as a PCR fragment at the NdeI-XhoI of the pGADT7 vector. The sequence corresponding to PsbO1 was PCR amplified using PsbO1-AD-F (5′-CGACGTACCAGATTACGCTCAATCGACTCAAGCCGTCGGC-3′) and PsbO1-AD-R (5′-CGATTCATCTGCAGCTCGAGCCTCAAGTTGACCATACCACAC-3′), and the sequence corresponding to PsbO2 was PCR amplified using PsbO2-AD-F (5′-CGACGTACCAGATTACGCTCAATCAAACCAAACCGTGGGCAAG-3′) and PsbO2-AD-R (5′-CGATTCATCTGCAGCTCGAGCCTCAATCTGACCGTACCAAAC-3′) as primers and Arabidopsis wild-type cDNA as template. The yeast strain Y2HGold (Clontech) was used as a reporter.
In Vitro Redox Assay
The cloned and full-length AT3G50820 cDNA was used as template in a PCR to amplify the coding sequence corresponding to the PsbO2 protein with the following oligonucleotides as primers: pET-Psbo2-F (5′-CTTTAAGAAGGAGATATACATATGTCAAACCAAACCGTGGGCAAGTCCTT-3′) and pET-Psbo2-R (5′-CGGATCTCAGTGGTGGTGGTGGTGGTGCTCAATCTGACCGTACCAAACCC-3′). The DNA sequence encoding the soluble domain of Chlamydomonas reinhardtii LTO1 was codon optimized for expression in Escherichia coli using the DNA2.0 Gene Designer software (Villalobos et al., 2006). The optimized sequence was synthesized (Mr Gene). The resulting plasmid (pMA-LTO1opt) was used as template in PCR with NdeI- and XhoI-engineered oligonucleotides pET24b-LTO1opt-F (5′-CTTTAAGAAGGAGATATACATATGCCGGGCGCTGGTCAAGTGTACATCGA-3′) and pET24b-LTO1opt-R (5′-CGGATCTCAGTGGTGGTGGTGGTGGTGCTCGAGTGGGGCAACCGCAACTG-3′) as primers. The PCR products of both reactions were cloned at the NdeI-XhoI sites of the hexahistidinyl-tag vector pET24b (Novagen) using In-Fusion (Clontech), resulting in the pET24b-PsbO2 and pET24b-LTO1opt plasmids. For expression of the recombinant His6-tagged proteins, 1 liter of LB broth (with 30 μg/mL kanamycin) of E. coli BL21(DE3) strain (Novagen) carrying pET24b- PsbO2 or pET24b-LTO1opt was grown from a 20-mL LB broth overnight starter culture. To induce the recombinant proteins, IPTG was added to a final concentration of 0.5 mM at A600 = 0.4, and the cultures were further grown for 3 h at 37°C. Cells were then harvested by centrifugation at 4000g for 20 min at 4°C, and the pellet was stored at −20°C. Batch purification of the His6-tagged proteins was performed under denaturing conditions (6 M urea) using nickel-nitrilotriacetic acid resin (Qiagen). Recombinant PsbO2 and LTO1 were dialyzed in a refolding buffer (25 mM Tris, pH 7.5) and stored at −80°C.
PsbO2 in 25 mM Tris-HCl, pH 7.5, was reduced by 200 μM DTT during 1 h on ice. DTT was eliminated by buffer exchange using the Amicon Centriprep system (Ultracel-10 membrane; Millipore). Reduced PsbO2 (10 μM) in 25 mM Tris-HCl, pH 7.5, was incubated for 60 min at 25°C in the absence or presence of oxidized soluble LTO1 (16 μM) in 25 mM Tris-HCl, pH 7.5. After incubation, proteins were precipitated with trichloroacetic acid (final 5%), washed with ice-cold acetone and then dissolved in buffer containing 50 mM Tris-HCl, pH 6.8, 2% SDS, 10 mM 4-acetamido-4′-maleimidylstilbene-2,2′-disulfonic acid (AMS). After a 90-min incubation, reduced (AMS derivative) and oxidized forms of PsbO2 and LTO1 were separated by 15% nonreducing SDS-PAGE and visualized by Coomassie Brilliant Blue staining.
Growth of Arabidopsis
The Col-0 ecotype of Arabidopsis was used as the wild type. The T-DNA line SALK_151963C (lto1) was provided as a confirmed homozygote by ABRC. Seeds were surface sterilized and sown on Murashige and Skoog plates (with or without 2% Suc) or on soil and stratified at 4°C for 48 h in the dark before germination. Plants were grown in controlled-environment chambers at a room humidity of 50% and provided daily with 16 h of light (80 to 120 μE m−2 s−1) and 8 h of dark at 22°C.
Molecular Characterization of the lto1 Mutant Lines
The SALK_151963C (lto1) line was analyzed for the presence and orientation of the T-DNA via PCR using genomic DNA as a template and the following primer pairs: S-Exon1D (5′-ATGGAAACCACTGCTTTTAAC-3′) and Exon2R (5′-TCAGATGAAGAACATTTAATC-3′); RB1 (5′-AGTGTTTGACAGGATATATTG-3′) and Exon2R (5′-TCAGATGAAGAACATTTAATC-3′); RB1 (5′-AGTGTTTGACAGGATATATTG-3′) and Stop (5′-TTACTGAAGTTGATTGGTCTC-3′); and LB1 (5′-GCGTGGACCGCTTGCTGCAACT-3′) and S-Exon1D (5′- ATGGAAACCACTGCTTTTAAC-3′).
RNA Extraction and RT-PCR
The leaves of 21-d-old lto1 and wild-type plants propagated on soil were used for total RNA isolation (Iratni et al., 1997). One microgram of DNase I–treated RNA was reverse transcribed using 200 units of Superscript II reverse transcriptase (Invitrogen) according to the manufacturer’s protocol. The reaction was performed in the presence of 1 μL of 100 μM oligo(dT)18 in a total volume of 15 μL at 42°C for 50 min. Aliquots (1 μL) were used as template for the PCR reaction with LTO1-specific primers Exon1D (5′-ATGATGGCGAGGTTTGTTTCTG-3′) and Stop (5′-TTACTGAAGTTGATTGGTCTC-3′), SEN1-specific primers S-Exon1D (5′-ATGGAAACCACTGCTTTTAAC-3′) and S-Stop R (5′-TCACTCTTCTACCGGCAGCTC-3′), and TUB2-specific primers TUB2-F (5′-CTCAAGAGGTTCTCAGCAGTA-3′) and TUB2-R (5′-TCACCTTCTTCATCCGCAGTT-3′).
Plasmids for Plant Transformation
For the complementation experiments, the LTO1 full-length cDNA (#U25043) was obtained from the ABRC. The LTO1 coding sequence (excluding the stop codon) was amplified by PCR with platinum Pfx DNA polymerase (Invitrogen) using the primers pENT/LTO1-F (5′-CACCATGATGGCGAGGTTTGTTTC-3′) and pENTR/LTO1-R (5′-CTGAAGTTGATTGGTCTCATTTGCC-3′). The amplified product was cloned using Gateway Technology into the pENTR/D-TOPO vector (Invitrogen) and then transferred into the pGWB20 destination vector (T. Nakagawa, Shimane University, Matsue, Japan) previously linearized by XhoI. The resulting plasmid (p35S-LTO1-C-MYC) was transferred to Agrobacterium tumefaciens GV3101 by electroporation. The lto1 plants were transformed using the floral dip method (Clough and Bent, 1998), and selection of the T1 generation of transgenics was performed on Murashige and Skoog plates with kanamycin (30 μg·mL−1) and hygromycin B (30 μg·mL−1). Seedlings were transplanted onto soil 2 weeks after germination and grown in controlled-environment chambers.
Protein Preparation and Analysis
Total plant protein extracts were prepared following the method described by Hurkman and Tanaka (1986), and protein concentrations were determined using the Bio-Rad DC protein kit (with BSA as a standard). Total protein was extracted with an SDS-containing solubilization buffer, and chlorophyll concentration was then determined. When necessary, Coomassie Brilliant Blue–stained gels were used to assess equal loading. For immunoblot analyses, the proteins were separated by SDS-PAGE, blotted onto Immobilon-P membranes (Millipore), and immunodecorated with antibodies. Commercially available antibodies against PC, PsaA, D1, D2, PsbD, PsbO, PsbP, PsbQ (Agrisera), and C-myc (Sigma-Aldrich) were used. The anti-PetC and anti-PetD antibodies were provided by Barkan (University of Oregon). Antisera against cytochrome f, cytochrome b6, cytochrome b559, and MFP1 were gifts from Sabeeha Merchant (University of California, Los Angeles), Catherine deVitry (Centre National de la Recherche Scientifique), Karen Meierhoff (Heinrich-Heine-Universität), and Iris Meier (The Ohio State University), respectively.
Fluorescence Induction
A freshly cut leaf was placed in the leaf cuvette of a JTS-10 LED spectrometer (Bio-Logic). Fluorescence emission was measured using a 10-μs pulse of light from a 520-nm LED as the excitation source, and a 670-nm high-pass filter was placed in front of the sample detector. A BG39 filter was placed in front of the reference detector to measure the excitation pulse, allowing proper normalization of fluorescence emission. Actinic light and saturating pulses were provided by a 520-nm LED array. Before each run, leaves were allowed to dark adapt for 30 to 60 s followed by a 2-s treatment with far-red light (743-nm peak with full width at half maximum of 30 nm; photon flux = 25 mmol m−2 s−1) to drive complete oxidation of the PQ pool. At each time point, fluorescence was measured (F0 or F0), followed immediately by an 80-ms saturating pulse to fully oxidize the PQ pool, and then fluorescence was measured 100 μs after the pulse was over (FM or FM). The first pair of points should thus provide F0 and FM. The actinic light was turned on 100 ms later, fluorescence measurements were taken 2 s after that, and then 24 more pairs of measurements were taken in an exponentially spaced fashion for 2 min total. The actinic light was turned on 100 ms later, fluorescence measurements were taken 2 s after that, and then 24 more pairs of measurements were taken in an exponentially spaced fashion for 2 min total. The actinic light was then extinguished, and 24 more time points were taken every 20 s. Note that the actinic light was briefly turned off while fluorescence measurements were made: At each time point, the actinic light was turned off, the F0 measurement was taken 100 μs later, the 80-ms saturating pulse was given 20 μs later, and then 100 μs later the FM measurement was taken, 20 μs after which the actinic light was turned back on. Thus, all fluorescence measurements were taken in the absence of any light besides the excitation source, avoiding complications or the necessity of subtraction of additional signals induced by the actinic or saturating lights. We found that FM after 2 s of actinic light was slightly higher (~4%) than FM in the wild-type case, indicating that the 80-ms pulse is not completely saturating. The difference was greater in the case of the lto1 mutant, consistent with the idea that it has a smaller antenna size. Thus, for the purposes of normalization of the data shown in Figure 4, we used the FM value measured at 2 s, which was the highest fluorescence value measured in all cases.
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers ACN43307 and ACN43308 for C. reinhardtii LTO1 cDNAs and accession number AAM65737 for Arabidopsis LTO1 cDNA.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Alignment of LTO1-Like Proteins from Photosynthetic Eukaryotes.
Supplemental Figure 2. P700 Oxidation and Rereduction Kinetics.
Supplemental Figure 3. Oxidation and Reduction of Cytochrome b6 and Cytochrome f.
Supplemental Figure 4. Decay of Signal at 520 nm upon Termination of Actinic Light.
Supplemental Figure 5. Accumulation of LTO1-C-myc in the LTO1 Complemented lto1 Line.
Supplemental Figure 6. Decay of Flash-Induced Fluorescence in the Presence of DCMU.
Supplemental Table 1. Parameters of P700 Oxidation and Rereduction.
Supplemental Table 2. Parameters of Decay of Flash-Induced Fluorescence Rise in the Presence of DCMU.
Supplemental Methods 1. Spectroscopy Measurements and Table of Primers.
Supplemental References 1. Supplemental References for the Supplemental Methods 1.
Acknowledgments
This work is supported by a Muscular Dystrophy Association (4727) grant, a National Science Foundation grant (MCB-0920062) to P.P.H., an M. Russell technology grant to S.C., and Grant DE-FG02-98ER20314MOD012 to K.R. from the Division of Chemical Sciences, Geosciences, and Biosciences, Office of Basic Energy Sciences of the U.S. Department of Energy. We thank Terry M. Bricker for the gift of antibodies, Rebecca Lamb for critical reading of the manuscript, and Rebecca Davis for performing the experiments shown in Figure 2D. We also thank the ABRC for the Arabidopsis LTO1 cDNA.
AUTHOR CONTRIBUTIONS
P.P.H., K.R., and N.R. designed the experiments, analyzed the data, and wrote the article. M.K. and S.C. performed the bulk of the experiments with the exception of the in vivo spectroscopy measurements, which were performed by K.R.
References
- Alexeyev M.F., Winkler H.H. (1999). Membrane topology of the Rickettsia prowazekii ATP/ADP translocase revealed by novel dual pho-lac reporters. J. Mol. Biol. 285: 1503–1513 [DOI] [PubMed] [Google Scholar]
- Bally J., Paget E., Droux M., Job C., Job D., Dubald M. (2008). Both the stroma and thylakoid lumen of tobacco chloroplasts are competent for the formation of disulphide bonds in recombinant proteins. Plant Biotechnol. J. 6: 46–61 [DOI] [PubMed] [Google Scholar]
- Bennoun P., Spierer-Herz M., Erickson J., Girard-Bascou J., Pierre Y., Delosme M., Rochaix J.D. (1986). Characterization of photosystem II mutants of Chlamydomonas reinhardii lacking the psbA gene. Plant Mol. Biol. 6: 151–160 [DOI] [PubMed] [Google Scholar]
- Bernard D.G., Quevillon-Cheruel S., Merchant S., Guiard B., Hamel P.P. (2005). Cyc2p, a membrane-bound flavoprotein involved in the maturation of mitochondrial c-type cytochromes. J. Biol. Chem. 280: 39852–39859 [DOI] [PubMed] [Google Scholar]
- Betts S.D., Ross J.R., Hall K.U., Pichersky E., Yocum C.F. (1996). Functional reconstitution of photosystem II with recombinant manganese-stabilizing proteins containing mutations that remove the disulfide bridge. Biochim. Biophys. Acta 1274: 135–142 [DOI] [PubMed] [Google Scholar]
- Bondarava N., Gross C.M., Mubarakshina M., Golecki J.R., Johnson G.N., Krieger-Liszkay A. (2010). Putative function of cytochrome b559 as a plastoquinol oxidase. Physiol. Plant. 138: 463–473 [DOI] [PubMed] [Google Scholar]
- Bonnard G., Corvest V., Meyer E.H., Hamel P.P. (2010). Redox processes controlling the biogenesis of c-type cytochromes. Antioxid. Redox Signal. 13: 1385–1401 [DOI] [PubMed] [Google Scholar]
- Brettel K. (1997). Electron transfer and arrangement of the redox cofactors in photosystem I. Biochim. Biophys. Acta 1318: 322–373 [Google Scholar]
- Bricker T.M., Frankel L.K. (2011). Auxiliary functions of the PsbO, PsbP and PsbQ proteins of higher plant Photosystem II: a critical analysis. J. Photochem. Photobiol. B 104: 165–178 [DOI] [PubMed] [Google Scholar]
- Brickman E., Beckwith J. (1975). Analysis of the regulation of Escherichia coli alkaline phosphatase synthesis using deletions and phi80 transducing phages. J. Mol. Biol. 96: 307–316 [DOI] [PubMed] [Google Scholar]
- Burnap R.L., Qian M., Shen J.R., Inoue Y., Sherman L.A. (1994). Role of disulfide linkage and putative intermolecular binding residues in the stability and binding of the extrinsic manganese-stabilizing protein to the photosystem II reaction center. Biochemistry 33: 13712–13718 [DOI] [PubMed] [Google Scholar]
- Carrell C.J., Zhang H., Cramer W.A., Smith J.L. (1997). Biological identity and diversity in photosynthesis and respiration: Structure of the lumen-side domain of the chloroplast Rieske protein. Structure 5: 1613–1625 [DOI] [PubMed] [Google Scholar]
- Clough S.J., Bent A.F. (1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Corvest V., Murrey D.A., Bernard D.G., Knaff D.B., Guiard B., Hamel P.P. (2010). c-type cytochrome assembly in Saccharomyces cerevisiae: A key residue for apocytochrome c1/lyase interaction. Genetics 186: 561–571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dailey F.E., Berg H.C. (1993). Mutants in disulfide bond formation that disrupt flagellar assembly in Escherichia coli. Proc. Natl. Acad. Sci. USA 90: 1043–1047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Depège N., Bellafiore S., Rochaix J.D. (2003). Role of chloroplast protein kinase Stt7 in LHCII phosphorylation and state transition in Chlamydomonas. Science 299: 1572–1575 [DOI] [PubMed] [Google Scholar]
- Depuydt M., Messens J., Collet J.-F. (2011). How proteins form disulfide bonds. Antioxid. Redox Signal. 15: 49–66 [DOI] [PubMed] [Google Scholar]
- Dreyfuss B.W., Hamel P.P., Nakamoto S.S., Merchant S. (2003). Functional analysis of a divergent system II protein, Ccs1, involved in c-type cytochrome biogenesis. J. Biol. Chem. 278: 2604–2613 [DOI] [PubMed] [Google Scholar]
- Dutton R.J., Boyd D., Berkmen M., Beckwith J. (2008). Bacterial species exhibit diversity in their mechanisms and capacity for protein disulfide bond formation. Proc. Natl. Acad. Sci. USA 105: 11933–11938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutton R.J., Wayman A., Wei J.-R., Rubin E.J., Beckwith J., Boyd D. (2010). Inhibition of bacterial disulfide bond formation by the anticoagulant warfarin. Proc. Natl. Acad. Sci. USA 107: 297–301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellgaard L., Ruddock L.W. (2005). The human protein disulphide isomerase family: Substrate interactions and functional properties. EMBO Rep. 6: 28–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eser M., Masip L., Kadokura H., Georgiou G., Beckwith J. (2009). Disulfide bond formation by exported glutaredoxin indicates glutathione’s presence in the E. coli periplasm. Proc. Natl. Acad. Sci. USA 106: 1572–1577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finazzi G., Forti G. (2004). Metabolic flexibility of the green alga Chlamydomonas reinhardtii as revealed by the link between state transitions and cyclic electron flow. Photosynth. Res. 82: 327–338 [DOI] [PubMed] [Google Scholar]
- Furt F., Oostende C., Widhalm J.R., Dale M.A., Wertz J., Basset G.J. (2010). A bimodular oxidoreductase mediates the specific reduction of phylloquinone (vitamin K) in chloroplasts. Plant J. 64: 38–46 [DOI] [PubMed] [Google Scholar]
- Gabilly S.T., Dreyfuss B.W., Karamoko M., Corvest V., Kropat J., Page M.D., Merchant S.S., Hamel P.P. (2010). CCS5, a thioredoxin-like protein involved in the assembly of plastid c-type cytochromes. J. Biol. Chem. 285: 29738–29749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabilly S.T., Kropat J., Karamoko M., Page M.D., Nakamoto S.S., Merchant S.S., Hamel P.P. (2011). A novel component of the disulfide-reducing pathway required for cytochrome c assembly in plastids. Genetics 187: 793–802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodstadt L., Ponting C.P. (2004). Vitamin K epoxide reductase: Homology, active site and catalytic mechanism. Trends Biochem. Sci. 29: 289–292 [DOI] [PubMed] [Google Scholar]
- Gopalan G., He Z., Balmer Y., Romano P., Gupta R., Héroux A., Buchanan B.B., Swaminathan K., Luan S. (2004). Structural analysis uncovers a role for redox in regulating FKBP13, an immunophilin of the chloroplast thylakoid lumen. Proc. Natl. Acad. Sci. USA 101: 13945–13950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gross J., Cho W.K., Lezhneva L., Falk J., Krupinska K., Shinozaki K., Seki M., Herrmann R.G., Meurer J. (2006). A plant locus essential for phylloquinone (vitamin K1) biosynthesis originated from a fusion of four eubacterial genes. J. Biol. Chem. 281: 17189–17196 [DOI] [PubMed] [Google Scholar]
- Grossman A.R., Karpowicz S.J., Heinnickel M., Dewez D., Hamel B., Dent R., Niyogi K.K., Johnson X., Alric J., Wollman F.A., Li H., Merchant S.S. (2010). Phylogenomic analysis of the Chlamydomonas genome unmasks proteins potentially involved in photosynthetic function and regulation. Photosynth. Res. 106: 3–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta R., Mould R.M., He Z., Luan S. (2002). A chloroplast FKBP interacts with and affects the accumulation of Rieske subunit of cytochrome bf complex. Proc. Natl. Acad. Sci. USA 99: 15806–15811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guzman L.M., Belin D., Carson M.J., Beckwith J. (1995). Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J. Bacteriol. 177: 4121–4130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall M., Mata-Cabana A., Akerlund H.E., Florencio F.J., Schröder W.P., Lindahl M., Kieselbach T. (2010). Thioredoxin targets of the plant chloroplast lumen and their implications for plastid function. Proteomics 10: 987–1001 [DOI] [PubMed] [Google Scholar]
- Hamel P.P., Dreyfuss B.W., Xie Z., Gabilly S.T., Merchant S. (2003). Essential histidine and tryptophan residues in CcsA, a system II polytopic cytochrome c biogenesis protein. J. Biol. Chem. 278: 2593–2603 [DOI] [PubMed] [Google Scholar]
- Heras B., Shouldice S.R., Totsika M., Scanlon M.J., Schembri M.A., Martin J.L. (2009). DSB proteins and bacterial pathogenicity. Nat. Rev. Microbiol. 7: 215–225 [DOI] [PubMed] [Google Scholar]
- Herrmann J.M., Kauff F., Neuhaus H.E. (2009). Thiol oxidation in bacteria, mitochondria and chloroplasts: Common principles but three unrelated machineries? Biochim. Biophys. Acta 1793: 71–77 [DOI] [PubMed] [Google Scholar]
- Hurkman W.J., Tanaka C.K. (1986). Solubilization of plant membrane proteins for analysis by two-dimensional gel electrophoresis. Plant Physiol. 81: 802–806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iratni R., Diederich L., Harrak H., Bligny M., Lerbs-Mache S. (1997). Organ-specific transcription of the rrn operon in spinach plastids. J. Biol. Chem. 272: 13676–13682 [DOI] [PubMed] [Google Scholar]
- Iwai M., Takizawa K., Tokutsu R., Okamuro A., Takahashi Y., Minagawa J. (2010). Isolation of the elusive supercomplex that drives cyclic electron flow in photosynthesis. Nature 464: 1210–1213 [DOI] [PubMed] [Google Scholar]
- Jeong S.Y., Rose A., Meier I. (2003). MFP1 is a thylakoid-associated, nucleoid-binding protein with a coiled-coil structure. Nucleic Acids Res. 31: 5175–5185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joliot P., Joliot A. (2005). Quantification of cyclic and linear flows in plants. Proc. Natl. Acad. Sci. USA 102: 4913–4918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadokura H., Beckwith J. (2002). Four cysteines of the membrane protein DsbB act in concert to oxidize its substrate DsbA. EMBO J. 21: 2354–2363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadokura H., Beckwith J. (2010). Mechanisms of oxidative protein folding in the bacterial cell envelope. Antioxid. Redox Signal. 13: 1231–1246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemeille S., Willig A., Depège-Fargeix N., Delessert C., Bassi R., Rochaix J.-D. (2009). Analysis of the chloroplast protein kinase Stt7 during state transitions. PLoS Biol. 7: e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lennartz K., Plücken H., Seidler A., Westhoff P., Bechtold N., Meierhoff K. (2001). HCF164 encodes a thioredoxin-like protein involved in the biogenesis of the cytochrome b(6)f complex in Arabidopsis. Plant Cell 13: 2539–2551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W., Schulman S., Dutton R.J., Boyd D., Beckwith J., Rapoport T.A. (2010). Structure of a bacterial homologue of vitamin K epoxide reductase. Nature 463: 507–512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lima A., Lima S., Wong J.H., Phillips R.S., Buchanan B.B., Luan S. (2006). A redox-active FKBP-type immunophilin functions in accumulation of the photosystem II supercomplex in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 103: 12631–12636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohmann A., Schöttler M.A., Bréhélin C., Kessler F., Bock R., Cahoon E.B., Dörmann P. (2006). Deficiency in phylloquinone (vitamin K1) methylation affects prenyl quinone distribution, photosystem I abundance, and anthocyanin accumulation in the Arabidopsis AtmenG mutant. J. Biol. Chem. 281: 40461–40472 [DOI] [PubMed] [Google Scholar]
- Manoil C. (1991). Analysis of membrane protein topology using alkaline phosphatase and beta-galactosidase gene fusions. Methods Cell Biol. 34: 61–75 [DOI] [PubMed] [Google Scholar]
- Meyer E.H., Giegé P., Gelhaye E., Rayapuram N., Ahuja U., Thöny-Meyer L., Grienenberger J.M., Bonnard G. (2005). AtCCMH, an essential component of the c-type cytochrome maturation pathway in Arabidopsis mitochondria, interacts with apocytochrome c. Proc. Natl. Acad. Sci. USA 102: 16113–16118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motohashi K., Hisabori T. (2006). HCF164 receives reducing equivalents from stromal thioredoxin across the thylakoid membrane and mediates reduction of target proteins in the thylakoid lumen. J. Biol. Chem. 281: 35039–35047 [DOI] [PubMed] [Google Scholar]
- Motohashi K., Hisabori T. (2010). CcdA is a thylakoid membrane protein required for the transfer of reducing equivalents from stroma to thylakoid lumen in the higher plant chloroplast. Antioxid. Redox Signal. 13: 1169–1176 [DOI] [PubMed] [Google Scholar]
- Murakami R., Ifuku K., Takabayashi A., Shikanai T., Endo T., Sato F. (2002). Characterization of an Arabidopsis thaliana mutant with impaired psbO, one of two genes encoding extrinsic 33-kDa proteins in photosystem II. FEBS Lett. 523: 138–142 [DOI] [PubMed] [Google Scholar]
- Page M.L.D., Hamel P.P., Gabilly S.T., Zegzouti H., Perea J.V., Alonso J.M., Ecker J.R., Theg S.M., Christensen S.K., Merchant S. (2004). A homolog of prokaryotic thiol disulfide transporter CcdA is required for the assembly of the cytochrome b6f complex in Arabidopsis chloroplasts. J. Biol. Chem. 279: 32474–32482 [DOI] [PubMed] [Google Scholar]
- Peeva V.N., Tóth S.Z., Cornic G., Ducruet J.M. (2010). Thermoluminescence and P700 redox kinetics as complementary tools to investigate the cyclic/chlororespiratory electron pathways in stress conditions in barley leaves. Physiol. Plant 144: 83–97 [DOI] [PubMed] [Google Scholar]
- Peltier J.B., Emanuelsson O., Kalume D.E., Ytterberg J., Friso G., Rudella A., Liberles D.A., Söderberg L., Roepstorff P., von Heijne G., van Wijk K.J. (2002). Central functions of the lumenal and peripheral thylakoid proteome of Arabidopsis determined by experimentation and genome-wide prediction. Plant Cell 14: 211–236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popelkova H., Yocum C.F. (2011). PsbO, the manganese-stabilizing protein: analysis of the structure-function relations that provide insights into its role in photosystem II. J. Photochem. Photobiol. B 104: 179–190 [DOI] [PubMed] [Google Scholar]
- Riemer J., Fischer M., Herrmann J.M. (2011). Oxidation-driven protein import into mitochondria: Insights and blind spots. Biochim. Biophys. Acta 1808: 981–989 [DOI] [PubMed] [Google Scholar]
- Rishavy M.A., Usubalieva A., Hallgren K.W., Berkner K.L. (2011). Novel insight into the mechanism of the vitamin K oxidoreductase (VKOR): Electron relay through Cys43 and Cys51 reduces VKOR to allow vitamin K reduction and facilitation of vitamin K-dependent protein carboxylation. J. Biol. Chem. 286: 7267–7278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohacek K., Bartak M. (1999). Technique of the modulated chlorophyll fluorescence: basic concepts, useful parameters, and some applications. Photosynthetica 37: 339–363 [Google Scholar]
- Sacksteder C.A., Kramer D.M. (2000). Dark-interval relaxation kinetics (DIRK) of absorbance changes as a quantitative probe of steady-state electron transfer. Photosynth. Res. 66: 145–158 [DOI] [PubMed] [Google Scholar]
- Sanders C., Turkarslan S., Lee D.-W., Daldal F. (2010). Cytochrome c biogenesis: The Ccm system. Trends Microbiol. 18: 266–274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schenk P.M., Kazan K., Rusu A.G., Manners J.M., Maclean D.J. (2005). The SEN1 gene of Arabidopsis is regulated by signals that link plant defence responses and senescence. Plant Physiol. Biochem. 43: 997–1005 [DOI] [PubMed] [Google Scholar]
- Schubert M., Petersson U.A., Haas B.J., Funk C., Schröder W.P., Kieselbach T. (2002). Proteome map of the chloroplast lumen of Arabidopsis thaliana. J. Biol. Chem. 277: 8354–8365 [DOI] [PubMed] [Google Scholar]
- Schulman S., Wang B., Li W., Rapoport T.A. (2010). Vitamin K epoxide reductase prefers ER membrane-anchored thioredoxin-like redox partners. Proc. Natl. Acad. Sci. USA 107: 15027–15032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sideris D.P., Tokatlidis K. (2010). Oxidative protein folding in the mitochondrial intermembrane space. Antioxid. Redox Signal. 13: 1189–1204 [DOI] [PubMed] [Google Scholar]
- Silhavy T.J., Berman M.L., Enquist L.W. (1984). Experiments with Gene Fusions. (Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press) [Google Scholar]
- Singh A.K., Bhattacharyya-Pakrasi M., Pakrasi H.B. (2008). Identification of an atypical membrane protein involved in the formation of protein disulfide bonds in oxygenic photosynthetic organisms. J. Biol. Chem. 283: 15762–15770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sokolove P.M., Marsho T.V. (1976). Ascorbate-independent carotenoid de-epoxidation in intact spinach chloroplasts. Biochim. Biophys. Acta 430: 321–326 [DOI] [PubMed] [Google Scholar]
- Sone M., Kishigami S., Yoshihisa T., Ito K. (1997). Roles of disulfide bonds in bacterial alkaline phosphatase. J. Biol. Chem. 272: 6174–6178 [DOI] [PubMed] [Google Scholar]
- Tanaka S., Kawata Y., Wada K., Hamaguchi K. (1989). Extrinsic 33-kilodalton protein of spinach oxygen-evolving complexes: Kinetic studies of folding and disulfide reduction. Biochemistry 28: 7188–7193 [DOI] [PubMed] [Google Scholar]
- Tie J.K., Stafford D.W. (2008). Structure and function of vitamin K epoxide reductase. Vitam. Horm. 78: 103–130 [DOI] [PubMed] [Google Scholar]
- Vignols F., Bréhélin C., Surdin-Kerjan Y., Thomas D., Meyer Y. (2005). A yeast two-hybrid knockout strain to explore thioredoxin-interacting proteins in vivo. Proc. Natl. Acad. Sci. USA 102: 16729–16734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villalobos A., Ness J.E., Gustafsson C., Minshull J., Govindarajan S. (2006). Gene Designer: A synthetic biology tool for constructing artificial DNA segments. BMC Bioinformatics 7: 285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., Dutton R.J., Beckwith J., Boyd D. (2011). Membrane topology and mutational analysis of Mycobacterium tuberculosis VKOR, a protein involved in disulfide bond formation and a homologue of human vitamin K epoxide reductase. Antioxid. Redox Signal. 14: 1413–1420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyman A.J., Yocum C.F. (2005). Structure and activity of the photosystem II manganese-stabilizing protein: role of the conserved disulfide bond. Photosynth. Res. 85: 359–372 [DOI] [PubMed] [Google Scholar]
- Yamamoto H.Y., Kamite L. (1972). The effects of dithiothreitol on violaxanthin de-epoxidation and absorbance changes in the 500-nm region. Biochim. Biophys. Acta 267: 538–543 [DOI] [PubMed] [Google Scholar]
- Yi X., McChargue M., Laborde S., Frankel L.K., Bricker T.M. (2005). The manganese-stabilizing protein is required for photosystem II assembly/stability and photoautotrophy in higher plants. J. Biol. Chem. 280: 16170–16174 [DOI] [PubMed] [Google Scholar]
- Zhu B., Cai G., Hall E.O., Freeman G.J. (2007). In-fusion assembly: seamless engineering of multidomain fusion proteins, modular vectors, and mutations. Biotechniques 43: 354–359 [DOI] [PubMed] [Google Scholar]
- Zybailov B., Rutschow H., Friso G., Rudella A., Emanuelsson O., Sun Q., van Wijk K.J. (2008). Sorting signals, N-terminal modifications and abundance of the chloroplast proteome. PLoS ONE 3: e1994. [DOI] [PMC free article] [PubMed] [Google Scholar]