The identification and characterization of a glutamyl-tRNA reductase binding protein (GluTRBP) results in a new model for spatial separation of 5-aminolevulinic acid (ALA) formation. GluTRBP-bound GluTR is attached to the thylakoid membrane and synthesizes ALA that is likely directed into the heme-synthesizing pathway.
Abstract
5-Aminolevulinic acid (ALA) is the universal precursor for tetrapyrrole biosynthesis and is synthesized in plants in three enzymatic steps: ligation of glutamate (Glu) to tRNAGlu by glutamyl-tRNA synthetase, reduction of activated Glu to Glu-1-semialdehyde by glutamyl-tRNA reductase (GluTR), and transamination to ALA by Glu 1-semialdehyde aminotransferase. ALA formation controls the metabolic flow into the tetrapyrrole biosynthetic pathway. GluTR is proposed to be the key regulatory enzyme that is tightly controlled at transcriptional and posttranslational levels. We identified a GluTR binding protein (GluTRBP; previously called PROTON GRADIENT REGULATION7) that is localized in chloroplasts and part of a 300-kD protein complex in the thylakoid membrane. Although the protein does not modulate activity of ALA synthesis, the knockout of GluTRBP is lethal in Arabidopsis thaliana, whereas mutants expressing reduced levels of GluTRBP contain less heme. GluTRBP expression correlates with a function in heme biosynthesis. It is postulated that GluTRBP contributes to subcompartmentalized ALA biosynthesis by maintaining a portion of GluTR at the plastid membrane that funnels ALA into the heme biosynthetic pathway. These results regarding GluTRBP support a model of plant ALA synthesis that is organized in two separate ALA pools in the chloroplast to provide appropriate substrate amounts for balanced synthesis of heme and chlorophyll.
INTRODUCTION
The tetrapyrrole biosynthetic pathway provides the vital cofactors and pigments for photoautotrophic growth (chlorophyll), for several essential redox reactions in electron transport chains (heme), for N- and S-assimilation (siroheme), and for photomorphogenic processes (phytochromobilin). The first committed molecule of the pathway, 5-aminolevulinic acid (ALA), is synthesized via the so-called C4 (Shemin) pathway involving ALA synthase that catalyzes condensation of succinyl-CoA and Gly and releases carbon dioxide and ALA (Schiffmann and Shemin, 1957). This pathway is found in mitochondria of eukaryotes lacking plastids (e.g., animals and fungi) as well as some groups of bacteria (e.g., α-proteobacteria; Beale, 1999). A second and evolutionary older pathway of ALA synthesis has been found in most bacteria, cyanobacteria, and plastids. This C5-pathway of ALA synthesis includes conversion of Glu by three enzymes, glutamyl-tRNA synthetase (GluRS), glutamyl-tRNA reductase (GluTR), and Glu-1-semialdehyde aminotransferase (GSAT). Eight molecules of ALA are ultimately combined to a cyclic tetrapyrrole. The oxidation of protoporphyrinogen by protoporphyrinogen oxidase (PPO) is unequivocally the first enzymatic step that takes place at the membrane (Joyard et al., 2009). Metal chelation reactions of protoporphyrin divide the pathway, leading to the formation of the main end products chlorophyll and heme (Tanaka and Tanaka, 2007).
A challenge in tetrapyrrole biosynthesis is the need for fine control of the distribution of metabolic intermediates to allow synthesis of different amounts of end products and avoid accumulation of photoreactive intermediates. This pathway responds quickly to diurnal and environmental changes (Papenbrock et al., 1999), but the regulated distribution of metabolites to the chlorophyll and heme-synthesizing branches is not well understood. ALA formation is the rate-limiting step for influx into the entire pathway, and GluTR activity is tightly regulated at both the transcriptional and posttranslational levels. Expression of genes encoding GluTR (e.g., Arabidopsis thaliana HEMA1, HEMA2, and HEMA3) is influenced by hormones, circadian rhythm, light signals, sugars, and also by feedback mechanisms originating from the Mg branch of the tetrapyrrole biosynthetic pathway (Papenbrock et al., 2000a, 2000b; Tanaka and Tanaka, 2007). At the posttranslational level, two mechanisms are known to regulate GluTR activity in planta. Heme was shown to inhibit GluTR activity in vitro (Pontoppidan and Kannangara, 1994; Vothknecht et al., 1998; Srivastava et al., 2005), indicating a regulatory feedback mechanism originating from the heme branch of tetrapyrrole biosynthesis. Downregulation of ALA synthesis is mediated by interaction between GluTR and the negative regulator FLUORESCENT (FLU) (Meskauskiene et al., 2001; Goslings et al., 2004). It is proposed that the enzymatic inactivation of the ternary complex formed by NADPH, NADPH-dependent protochlorophyllide oxidoreductase (POR) and protochlorophyllide, is communicated to ALA synthesis via FLU in darkness (Richter et al., 2010). The regulatory interrelation of the two branches for heme and chlorophyll biosynthesis with ALA formation is still extensively discussed (Cornah et al., 2003; Mochizuki et al., 2010).
Synthesis of tetrapyrrole end products takes place where they are needed most. The plant tetrapyrrole biosynthetic pathway is predominantly located in plastids (Tanaka and Tanaka, 2007; Masuda and Fujita, 2008; Grimm, 2010). GluTR, GSAT, and enzymes up to PPO have been found in the stromal fraction, while the subsequent enzymes of the heme and chlorophyll-synthesizing branches are mainly located in the organellar envelope and thylakoid membranes (Rolland et al., 2003; Joyard et al., 2009). Whereas almost all genes encoding enzymes of tetrapyrrole biosynthesis have been identified and the operation of tetrapyrrole metabolism has been unequivocally demonstrated in plants (Tanaka and Tanaka, 2007; Grimm, 2010; Mochizuki et al., 2010), the organization of the pathway remains to be clarified. Considering the high concentration of proteins in living cells, an organization that promotes associations in subcompartmental protein complexes to enhance the efficiency and flexibility of metabolic complexes can be postulated (Lunn, 2007).
Elucidation of the subcellular localization and the structural and regulatory framework of the pathway would provide a major advance in understanding tetrapyrrole biosynthesis in planta. In a search for proteins that function in tetrapyrrole metabolism, we identified a protein with tight association to GluTR. We propose a function for the GluTR binding protein (GluTRBP; previously called PROTON GRADIENT REGULATION7 [PGR7]) in the organization of ALA synthesis to regulate the distribution of ALA for heme and chlorophyll biosynthesis.
RESULTS AND DISCUSSION
Identification of a GluTRBP
GluTR is the target enzyme for regulating ALA and tetrapyrrole biosynthesis in response to environmental and endogenous signals (Beale, 1999; Tanaka and Tanaka, 2007). Currently two proteins are known to interact with GluTR. GSAT binding to GluTR has been shown (Lüer et al., 2005; Nogaj and Beale, 2005) and interpreted in the framework of metabolic channeling within ALA synthesis (Moser et al., 2001). FLU, a negative regulator of ALA synthesis, also interacts with GluTR (Meskauskiene et al., 2001; Goslings et al., 2004). In a search for additional regulatory mechanisms, we screened an Arabidopsis cDNA library for proteins interacting with GluTR by means of the yeast two-hybrid method. Following generation of 3.5 million primary yeast transformants, we identified 17 clones that interacted with the bait. Sequencing the prey cDNAs revealed a single gene (At3g21200) encoding a GluTRBP. The encoded protein was previously described as PGR7 (Jung et al., 2010). The protein sequence displays a putative N-terminal plastid transit sequence of 41 amino acids as predicted by the TargetP algorithm (Emanuelsson et al., 2007). Since the interaction with GluTR was initially demonstrated with the precursor of GluTRBP, we confirmed the interaction of GluTR with the mature GluTRBP by multiple yeast two-hybrid bait and prey clone combinations (Table 1). GluTRBP interacted with GluTR encoded by both HEMA1 and HEMA2. This is in contrast with FLU, which interacts only with HEMA1-encoded GluTR (Goslings et al., 2004). GluTRBP did not bind to the other two enzymes involved in ALA formation, GluRS and GSAT (Table 1). Interaction of GluTR and GluTRBP was confirmed in vitro by pull-down experiments (see Supplemental Figure 1 online) as well as in planta by bimolecular fluorescence complementation (BiFC; Figure 1). The latter approach not only confirms the translocation of At-GluTRBP and At-GluTR into tobacco (Nicotiana tabacum) chloroplasts and the interaction of both proteins within the organelle but also indicates the potential of GluTRBP as well as GluTR (Moser et al., 2001) to form homodimers (Table 1).
Table 1.
Protein Interactions between At-GluTR and At-GluTRBP Shown by Yeast Two-Hybrid Assays
| Gal4-AD |
|||||||
| LexA-BD | Empty Plasmid | At-GluTRBP | At-GluTRBP without TP | At-GluTR without TP (AtHEMA1) | At-GluTR without TP, without HBD (At-HEMA1) | At-GSAT | At-GluRS |
| Empty plasmid | − | − | − | − | − | − | − |
| At-GluTRBP | − | + | + | + | + | − | − |
| At-GluTRBP without TP | − | + | + | + | + | − | − |
| At-GluTR without TP (HEMA1) | − | + | + | − | − | − | − |
| At-GluTR without TP, without HBD (At-HEMA1) | − | + | + | − | − | − | − |
| At-GluTR (At-HEMA2) | − | + | + | − | − | − | − |
| At-GSAT | − | − | − | − | − | − | − |
| At-GluRS | − | − | − | − | − | − | − |
Protein interactions between At-GluTRBP and At-GluTR (encoded by either At-HEMA1 or At-HEMA2) were tested by different bait (columns) and prey (rows) protein combinations. Both proteins were cloned as full-length proteins as well as mature proteins without transit peptide (TP). Additionally, At-GluTR lacking its putative HBD also shows binding to At-GluTRBP. No protein interactions were detected using At-GSAT and At-GluRS as bait or prey proteins. Protein interactions are indicated by + and lack of interaction by −. Some of the data summarized here are shown in Supplemental Figures 2B and 6B online.
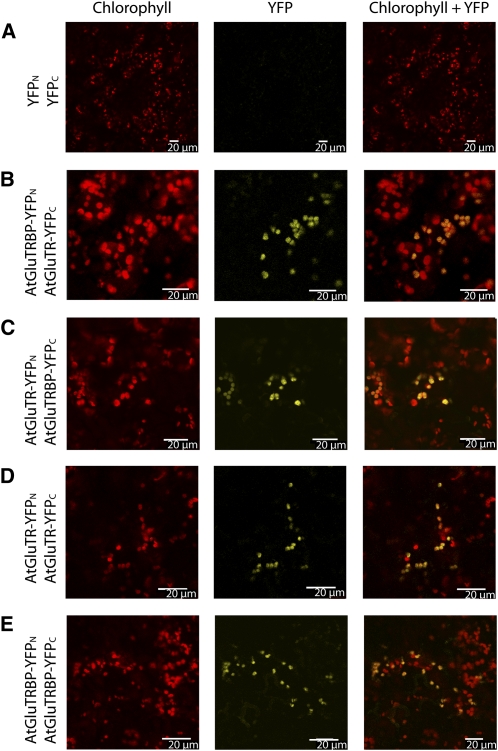
Figure 1.
BiFC Assay for At-GluTR and At-GluTRBP Confirms the Interaction of Both Proteins in Planta.
(A) Transient coexpression of the N- and C-terminal part of YFP does not restore YFP fluorescence in tobacco cells.
(B) to (E) YFP fluorescence is restored when At-GluTRBP is fused to the N-terminal part of YFP and coexpressed with an AtGluTR-YFPC fusion protein (B), or At-GluTR is fused to the N-terminal part of YFP and coexpressed with an AtGluTRBP-YFPC fusion protein (C), or At-GluTR is fused to both YFPN and YFPC (D), and At-GluTRBP is fused to both YFPN and YFPC (E). YFP fluorescence colocalizes with chlorophyll fluorescence. Magnification ×700 to 800.
GluTRBP Is Bound to the Thylakoid Membrane
The At-GluTRBP precursor consisting of 317 amino acid residues did not exhibit sequence or structure similarities to known proteins of tetrapyrrole biosynthesis or other cellular functions. Since GluTRBP shows a 22% overall similarity to a putative pyridoxamine-5-phosphate oxidase (PPOX) encoded at locus At3g03890 (see Supplemental Figure 2A online), this sequence was cloned into the bait and prey plasmids and subjected to yeast two-hybrid experiments. PPOX was not able to interact with GluTR (see Supplemental Figure 2B online). The annotation of GluTRBP as a flavin mononucleotide (FMN) binding protein (FMN binding split barrel, InterPRO IPR009002, The Arabidopsis Information Resource) is likely based on the described weak similarity to PPOX. However, expression of At-GluTRBP in Escherichia coli did not result in a yellow-colored bacterial pellet as typically reported for flavoproteins (Kitamura et al., 1994). Moreover, biochemical and spectroscopy analyses using heterologously expressed and purified His- or glutathione S-transferase–tagged At-GluTRBP did not display FMN binding capacity (see Supplemental Figure 3 online).
Localization studies confirmed the N-terminal transit peptide and plastid translocation (Figure 2). Immunoblot analyses using a specific anti-AtGluTRBP antibody revealed that a minor amount of the protein was located in the stroma, while the majority of GluTRBP accumulated in the membrane fraction of Arabidopsis chloroplasts (Figure 2A). In vitro–translated, [35S]-labeled, full-length At-GluTRBP (apparent molecular mass 35.5 kD) was imported into intact chloroplasts isolated from 7-d-old pea (Pisum sativum) seedlings and was processed to its mature form (30.7 kD). Following chloroplast fractionation, At-GluTRBP was found in the stroma fraction as well as attached to thylakoids. At-GluTRBP was degraded in thylakoid membranes by thermolysin treatment, indicating that it is not an integral membrane protein or further translocated in the thylakoid lumen (Figure 2B). Immunogold labeling of GluTRBP in Arabidopsis chloroplasts revealed its preferential association with grana stacks (Figure 2C; see Supplemental Data Set 1 online). Although the primary sequence of At-GluTRBP did not contain membrane-spanning domains, the protein could not be removed from thylakoid membranes by washing with redox-active agents or salt-containing buffers (Figure 2D). Since ALA-forming activity and GluTR protein were originally detected and isolated from stromal extracts (Gough and Kannangara, 1977; Pontoppidan and Kannangara, 1994), GluTR has been reported to be a stroma protein. Nevertheless, ALA-forming enzymes were also described to be loosely bound to plastid membranes (Harel and Ne’eman, 1983). Immunoblot localization resulted in a minor amount of At-GluTR associated with the thylakoid membrane fraction, which could be removed by washing with oxidizing agents and salt-containing buffers (Figure 2D). Thus, we propose that GluTRBP functions as an anchor for a portion of GluTR to the thylakoid membrane.
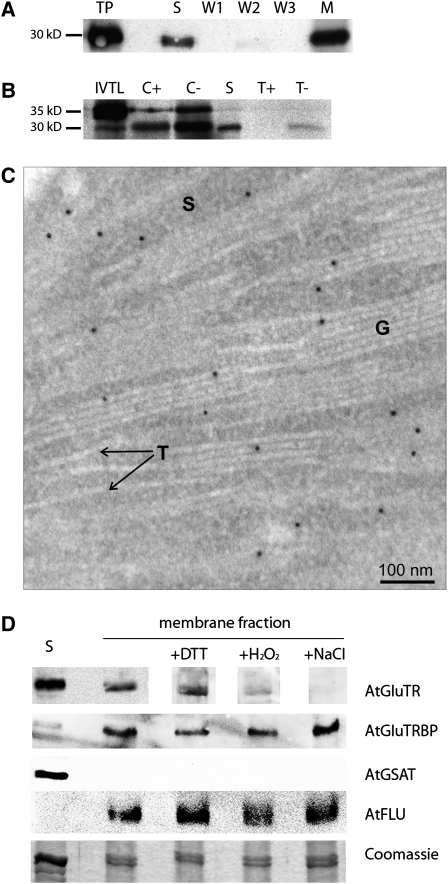
Figure 2.
At-GluTRBP Is Localized in the Chloroplast and Attached to the Thylakoid Membrane.
(A) In immunoblot analysis the 30-kD mature At-GluTRBP was detected in the stroma fraction (S) as well as in the membrane fraction (M) of isolated chloroplasts (W1 to W3, serial membrane washing fractions). An Arabidopsis total protein extract (TP) was used as control.
(B) In vitro full-length [35S]-labeled At-GluTRBP (IVTL) was incubated with isolated pea chloroplasts, resulting in two proteins with apparent molecular masses of 35.5 and 30.7 kD (C−). After thermolysin treatment of the chloroplasts (C+), only the mature 30.7-kD protein was protected against proteolytic digestion and found in the stroma fraction (S) as well as bound to membranes (T−). Thermolysin treatment of the membrane fraction (T+) digested At-GluTRBP.
(C) Immunogold localization of GluTRBP in chloroplasts of Arabidopsis leaves. GluTRBP is detected exclusively in the stroma (S) and the grana stacks (G) but not in the envelope or stroma thylakoids (T). See Supplemental Data Set 1 online for more pictures (e.g., for confirmation that GluTRBP is missing on envelope membranes and for additional controls). Magnification ×89,000.
(D) Localization of proteins within Arabidopsis chloroplasts. Chloroplasts from 4-week-old Arabidopsis plants were separated into stroma (S) and membrane fractions. Membranes were washed additionally with 10 mM DTT, 10 mM H2O2, or 4 M NaCl. GluTR, GluTRBP, GSAT, and FLU were detected by immunoblot analysis using specific antibodies. Parts of a Coomassie blue–stained 12% SDS-polyacrylamide gel were used as a loading control.
Further investigations were performed to identify chloroplast (membrane) protein complexes containing GluTRBP. Using intact chloroplasts isolated from mustard (Sinapis alba) seedlings, a protein complex with an approximate molecular mass of 300 kD was identified by Blue Native SDS-PAGE and immunoblot analysis using a specific antibody for GluTRBP (see Supplemental Figure 4 online). Unfortunately, the quality of all tested antibodies raised against GluTR did not allow an irrevocable identification of protein complexes containing GluTR. Moreover, identification of further constituents of the 300-kD protein complex was hindered by its low abundance in comparison to highly abundant photosystem II monomer and cytochrome b6f complexes, which showed similar mobility in the Blue Native PAGE (see Supplemental Table 1 and Supplemental Data Set 2 online). Proteomic analyses generally confirm the presence of tetrapyrrole biosynthetic enzymes in the aqueous and membranous phases of plastids (Rolland et al., 2003; Joyard et al., 2009). Regarding a putative ALA-forming protein complex, previous analyses of the Arabidopsis proteome failed to detect GluTRBP, GluTR, and GluRS, whereas GSAT was identified (Rolland et al., 2003; Friso et al., 2004; Peltier et al., 2006; Ytterberg et al., 2006; Joyard et al., 2009), indicating the instability or the low abundance of the former enzymes.
GluTRBP Is Found in All Chloroplast-Containing Organisms
In silico studies demonstrated a wide distribution of GluTRBP within chloroplast-containing organisms. An alignment of homologous proteins from Arabidopsis, tobacco, maize (Zea mays), rice (Oryza sativa), and grape (Vitis vinifera) showed homologies of 50 to 60% (see Supplemental Figure 5 online). Protein sequences similar to At-GluTRBP were found in the genomes of the moss Physcomitrella patens (PHYPADRAFT_143777 and 151198), green algae Ostreococcus lucimarinus and Ostreococcus tauri (OSTLU_41998, Ot11g00660), and Chlamydomonas reinhardtii (HQ456532). The C. reinhardtii homolog was cloned, and functional homology to At-GluTRBP was proven by interaction of Cr-GluTRBP with Cr-GluTR in the yeast two-hybrid system (see Supplemental Figures 6A and 6B online). Moreover, using heterologously expressed and purified Cr-GluTR and Cr-GluTRBP proteins, we determined binding constants of the CrGluTR-CrGluTRBP complex by a surface plasmon resonance technique (see Supplemental Table 2 and Supplemental Figure 7 online). The interaction of both proteins is characterized by a Kd of 195 nM that is within the typical range of native protein complexes (Archakov et al., 2003). It is noteworthy that the genomic sequence encoding the initially cloned Cr-GluTRBP was only annotated in the C. reinhardtii genome database version 2.0 (Joint Genome Institute). An update in 2007 (version 3.0; Merchant et al., 2007) omitted the corresponding genomic locus for Cr-GluTRBP and contained a novel locus coding for a protein showing 23% homology to At-GluTRBP and 22% to Cr-GluTRBP (CHLREDRAFT_76641; see Supplemental Figures 6C and 6D online). However, expression of both C. reinhardtii proteins is confirmed by the presence of several EST entries (see Supplemental Figure 6 online).
GluTRBP-like proteins are likely not present in photosynthetic bacteria since bacterial coding sequences display similarities of <7% (e.g., Synechococcus sp protein ID ACA99562) or 22% (Solibacter usitatus protein ID ABJ86901) to At-GluTRBP. These bacterial proteins are more similar to the putative PPOX (At3g03890). We conclude that eukaryotes or bacteria that synthesize ALA via the Shemin pathway do not express GluTRBP-like proteins. The occurrence of GluTRBP is associated with the C5 pathway of ALA synthesis in chloroplasts of algae, mosses, and plants (Beale, 1999, 2006). As GluTRBP acts as a membrane anchor for GluTR, it is suggested that GluTRBP contributes to the organization of ALA synthesis in a subcompartment located at the thylakoid membrane.
Expression Patterns of GluTRBP Resemble Those of Enzymes of Heme Metabolism
Greening of etiolated seedlings is one of the developmental processes that challenges regulation of tetrapyrrole biosynthesis on the level of expression and especially drive chlorophyll synthesis to its maximum. Analysis of expression profiles in the first hours of deetiolation of Arabidopsis seedlings revealed an abundance of HEMA1/GluTR that gradually increased with chlorophyll biosynthesis, while the level of GluTRBP/GluTRBP remained nearly constant (Figures 3A and 3B). In contrast with the diurnal oscillation of HEMA1/GluTR, expression of GluTRBP/GluTRBP did not oscillate in the light–dark cycle (Figures 3C and 3D). Expression analyses of GSA1 and GSA2/GSAT, LHCB1/LHCB1, LHCA1, and FLU (Figures 3A to 3D) confirm the light-induced and diurnal control of RNA and protein contents of chlorophyll biosynthetic enzymes. Analysis of GluTRBP expression in different tissues and developmental stages revealed its coexpression with HEMA1 but also its absence in Arabidopsis roots (Figure 3E).
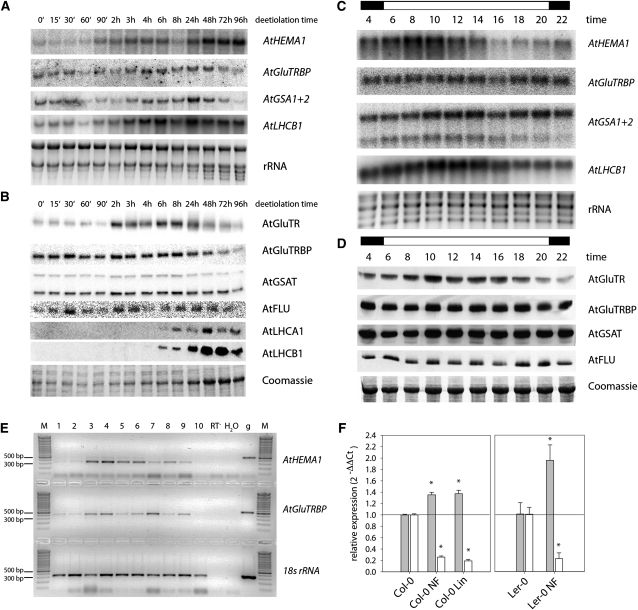
Figure 3.
Expression of Genes Related to ALA Biosynthesis and Photosynthesis during Deetiolation within a 24-h Light/Dark Cycle and in the Presence of Norflurazon or Lincomycin.
(A) and (B) Three-day-old etiolated Arabidopsis seedlings were transferred to light, and transcript and protein abundance of different tetrapyrrole biosynthesis genes was monitored during the first 96 h by RNA gel blots and immunoblots.
(C) and (D) Arabidopsis plants were grown under long-day conditions (14 h light/10 h dark) for 4 weeks, and changes in abundance of different transcripts and proteins were analyzed. The day–night cycle is indicated by white/black bars above of each panel.
(A) and (C) RNA gel blot using gene-specific probes for HEMA1, GluTRBP, GSA1+2, and LHCB1. Ethidium bromide–stained rRNA served as loading control.
(B) and (D) Immunoblots using specific antibodies against At-GluTR, At-GluTRBP, At-GSAT, At-FLU, At-LHCA1, and At-LHCB1. A detail of a Coomassie blue–stained 12% SDS-polyacrylamide gel served as loading control.
(E) Analysis of HEMA1 and GluTRBP expression in different tissues and developmental stages of Arabidopsis plants. Amounts of template used for the PCR reaction were adjusted using 18s rRNA. PCR products were separated electrophoretically in an ethidium bromide–stained agarose gel. Lane 1, 3-d-old etiolated seedlings; lane 2, 3-d-old etiolated seedlings illuminated for 30 min; lane 3, 5-d-old cotyledons; lane 4, primary leaves from 10-d-old seedlings; lane 5, second and third leaf of 18-d-old plants; lane 6, young rosette leaves of 27-d-old plants; lane 7, old rosette leaves of 27-d-old plants; lane 8, senescent leaves of 40-d-old plants; lane 9, inflorescences of 40-d-old plants; lane 10, roots of 24-d-old plants. g, genomic DNA as PCR control; H20, water as template was used as negative control; M, DNA standard; RT-, RNA not reverse transcribed as template was used as negative control.
(F) Relative quantification of GluTRBP (gray) and LHCB1.2 (white) mRNA levels in Arabidopsis wild-type Col-0 or Ler-0 treated with or without norflurazon (NF) or lincomycin (Lin) analyzed by RT-PCR. The mRNA levels were normalized by those of TUB2 mRNA and are given as 2−ΔΔCt ± sd of three biological replications. Asterisk marks a significant difference to wild-type Col-0 or Ler-0 (P < 0.05).
Expression of tetrapyrrole biosynthetic genes during the onset of greening was previously classified in different categories (Matsumoto et al., 2004). HEMA1 belongs to a cluster of Arabidopsis genes strongly induced upon greening. Genes encoding GSAT (GSA1 and GSA2) belong to a second cluster containing genes with a moderate induction of expression during deetiolation. Prominent members of a third gene cluster representing noninduced genes encode proteins of the heme metabolic pathway (HO1, HO3, HO4, and HY2 encoding isoforms of heme oxygenase), FC1 (encoding an isoform of ferrochelatase), and HEMA2. HEMA2 encodes the second GluTR isoform that purportedly plays a role in heme synthesis (Tanaka et al., 1996; Ujwal et al., 2002). Clustering genes by their 24-h expression pattern resulted in three classes, grouping genes with either circadian, diurnal, or nonrhythmic expression (Matsumoto et al., 2004). Expression of GluTRBP during greening of etiolated seedlings and of light/dark-grown green seedlings resembles expression of genes involved in heme metabolism (Figure 3). Inhibition of chloroplast biogenesis (e.g., by norflurazon), which is an inhibitor of carotenoid biosynthesis, was used to distinguish between genes related to heme and chlorophyll biosynthesis (Moulin et al., 2008). Expression of HEMA2, PPO2 (encoding an isoform of PPO), FC1, and HO1, HO2, and HY2 was either induced or not affected when Arabidopsis seedlings were treated with norflurazon. Hence, it was concluded that in contrast with chlorophyll, heme biosynthesis is stimulated when chloroplast development is inhibited by photooxidative stress that is scavenged by heme-containing enzymes (Moulin et al., 2008). We tested expression of GluTRBP in young seedlings treated with norflurazon as well as with lincomycin, an inhibitor of plastidial protein translation. GluTRBP expression was enhanced in Arabidopsis ecotypes Columbia-0 (Col-0) and Landsberg erecta (Ler-0) upon norflurazon as well as lincomycin treatment, which is in contrast with many photosynthetic genes (e.g., LHCB1.2; Figure 3F). Summing up, At-GluTRBP expression patterns support the hypothesis that the function of GluTRBP is related to heme rather than chlorophyll biosynthesis.
The 30 N-terminal amino acids of the mature barley (Hordeum vulgare) GluTR have been proposed to be a heme binding domain (HBD) and essential for feedback inhibition of GluTR activity by heme (Vothknecht et al., 1998). Inhibition of C. reinhardtii GluTR activity by heme required an additional factor (Srivastava et al., 2005). We speculated that GluTRBP was the postulated protein that enables heme-mediated feedback inhibition of ALA synthesis. However, At-GluTRBP readily interacts with truncated At-GluTR lacking the putative HBD (Table 1). Moreover, when ALA synthesis activity was assayed using purified recombinant Cr-GluTR and Cr-GSAT, Cr-GluTRBP neither positively or negatively affected Cr-GluTR activity nor mediated feedback inhibition of ALA synthesis by heme (see Supplemental Figure 8 online). Thus, we hypothesize that GluTRBP does not affect the enzymatic reactions but is rather involved in the spatial organization of tetrapyrrole biosynthesis by recruiting a membrane-bound GluTR to ensure ALA synthesis for the heme branch.
Reduction of GluTRBP Contents Causes Heme Deficiency
Several attempts to investigate the physiological function of GluTRBP by generating constitutive At-GluTRBP RNA interference (RNAi) knockdown mutants using the pAGRIKOLA system (Hilson et al., 2004) failed. We interpreted these results as an indication that lack of GluTRBP is lethal in Arabidopsis. The use of an ethanol-inducible RNAi system demonstrated that GluTRBP plays an essential role during greening of Arabidopsis seedlings. Induced expression of an At-GluTRBP RNAi transgene in germinating Arabidopsis seedlings resulted in a drastic depletion of GluTRBP that reached only 5% of the wild-type level followed by instantaneous cell death of seedlings (see Supplemental Figure 9 online). The two T-DNA insertion lines available in public databases (SALK_071358 and GABI_099E12) contain T-DNA insertions 5′ and 3′ of the At-GluTRBP coding sequence. Immunoblot analysis of homozygous individuals revealed that the mutants are not affected in GluTRBP abundance. Arabidopsis mutants with reduced content of GluTRBP were generated by expressing the GluTRBP open reading frame in sense or antisense orientation under control of the cauliflower mosaic virus 35S promoter. Stable homozygous lines could not be established since expression of the transgene was silenced starting from the T4 generation. However, T2 and T3 progenies of line antisense_#22 expressing GluTRBP antisense mRNA and line sense_#34 expressing GluTRBP in sense orientation showed reduced GluTRBP contents. These two lines contain still ~30 to 60% of wild-type GluTRBP levels. This moderate reduction of GluTRBP amounts may explain why both lines are more viable than GluTRBP-RNAi knockdown mutants. It is likely that a certain threshold level of GluTRBP is required in young seedlings to ensure their viability.
Changes in protein levels were observed in different developmental stages of the mutants, namely, in etiolated seedlings, 48 h after deetiolation as well as in 12-d-old seedlings (Figure 4A). With the exception of barely detectable growth retardation, the two lines did not display a different macroscopic phenotype compared with that of corresponding Arabidopsis wild-type Col-0 (Figure 4B) under standard growth conditions. Analysis of protochlorophyllide contents in 3-d-old etiolated GluTRBP antisense_#22 and sense_#34 lines revealed that lack of GluTRBP causes neither a reduction nor an increase in ALA-forming capacity (Figure 4C). Moreover, determination of pigment contents in the mutants 48 h after deetiolation shows no negative impact of GluTRBP deficiency on chlorophyll biosynthesis. By contrasts, mutants with reduced content of GluTRBP tend to overaccumulate chlorophyll within the first hours of greening (Figure 4D). Nevertheless, analysis of pigment and heme content of 2-week-old progenies of the two lines revealed a >50% reduced heme content compared with wild-type seedlings (Table 2) and slightly reduced chlorophyll a and b and carotenoid contents.
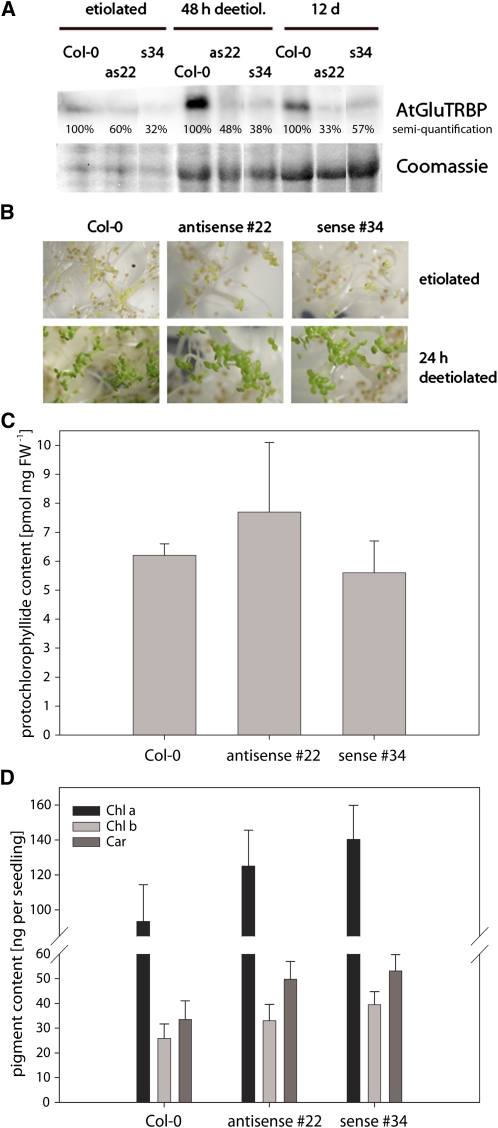
Figure 4.
Arabidopsis Mutants with Reduced GluTRBP Contents.
(A) Immunoblot analysis of Arabidopsis Col-0 and two mutant lines (GluTRBP_antisense_#22, as22 and GluTRBP_sense_#34, s34) expressing reduced amounts of GluTRBP. Reduction in GluTRBP contents can be followed in etiolated, 48 h deetiolated, and 12-d-old plants. A detail of a Coomassie blue–stained 12% SDS-polyacrylamide gel served as loading control.
(B) Both mutant lines expressing reduced amounts of GluTRBP are hardly distinguishable from the wild type grown under standard growth conditions (90 μmol photons m−2 s−1). Since GluTRBP knockout is lethal in the very early stages of cotyledon development (see Supplemental Figure 10 online), the phenotypes of 48 h deetiolated plantlets is shown as an example. Magnification ×2.
(C) Protochlorophyllide content of 3-d-old etiolated Arabidopsis Col-0 and the two mutant lines GluTRBP_antisense_#22 and GluTRBP_sense_#34.
(D) Pigment content (Chl a, chlorophyll a; Chl b, chlorophyll b; and Car, carotenoid) of 48 h deetiolated Arabidopsis Col-0 and lines GluTRBP_antisense_#22 and GluTRBP_sense_#34. Seedlings were grown in darkness for 3 d before they were transferred to light.
(C) and (D) Given are means ± sd. t tests were performed, but differences between wild-type Col-0 and mutant lines were not significant (P > 0.05).
Table 2.
Heme and Pigment Contents and Chlorophyll a/b Ratio of 14-d-Old Arabidopsis Mutants with Reduced Content of GluTRBP
| Arabidopsis Line | Heme Content (% Col-0) | Pigment Content Chlorophyll a (% Col-0) Chlorophyll b (% Col-0) Carotenoid (% Col-0) | Chlorophyll a/b Ratio |
| Wild-type Col-0 | 100 ± 7 | 100 ± 9 | 3.8 ± 0.2 |
| 100 ± 10 | |||
| 100 ± 9 | |||
| Antisense #22 | 47 ± 8* | 74 ± 4* | 4.0 ± 0.1 |
| 71 ± 5* | |||
| 74 ± 4* | |||
| Sense #34 | 51 ± 5* | 80 ± 2* | 4.1 ± 0.0* |
| 75 ± 1* | |||
| 81 ± 3* |
AtGluTRBP_antisense #22, AtGluTRBP_sense #34, and the corresponding wild-type Col-0 were grown under standard conditions (long day, 90 μmol photons m−2 s−1, 23°C) for 2 weeks. Tetrapyrrole contents are given in percentage of Col-0 ± sd. Asterisk marks a significant difference to wild-type Col-0 (P < 0.05).
When these mutants were transferred to high light (~300 μmol photons m−2 s−1), a drastic, stress-related growth retardation was observed (Figure 5). Phenotypes of the germinating T3 progenies of the selected transgenic lines were subdivided into three groups: (1) segregated wild-type seedlings, (2) seedlings with an intermediate phenotype characterized by a strong growth arrest and reduced pigmentation, and (3) nonviable seedlings with drastically bleached leaves (Figure 5A). Seedlings of the second group entirely arrested in growth and were scarcely able to develop the true leaves. However, when these seedlings were subjected to standard growth conditions following the high-light treatment, they returned to normal plant development (Figure 5B). We assign these high-light-associated phenotypic aberrations to heme deficiency (Table 2) coupled with high-light-induced oxidative stress (Rossel et al., 2002; Krieger-Liszkay et al., 2008). In line with these observations, it is speculated that the slightly elevated chlorophyll accumulation within the first 48 h of greening in transgenic lines with reduced expression of GluTRBP (Figure 4D) is attributed to an increased availability of GluTR for chlorophyll biosynthesis.
Figure 5.
High-Light Stress Phenotype of Arabidopsis Mutants with Reduced GluTRBP Contents.
(A) Phenotype of line AtGluTRBP_antisense_#22 grown under high-light (300 μmol photons m−2 s−1) conditions (line AtGluTRBP_sense_#34 shows a similar phenotype). Besides a segregating wild-type (left in the left panel), two additional phenotypes were distinguishable among the T3 progenies of the transgenic lines. A strong phenotype results in plant death (right plant), and an intermediate phenotype causes growth retardation (medium in the left panel and left in the right panel). Both transgenic phenotypes are connected to the expression of the GluTRBP transgene, as shown by PCR of genomic DNA from three individual seedlings of each of the three different phenotypes (wild-type-like, intermediate or strong) upon stress (indicated by arrows). +, The diluted binary plasmid applied to Arabidopsis transformation was used as positive control PCR template; H20, water as template was used as negative control. Control PCRs using primers to amplify GUN4 were performed to demonstrate the quality of the template DNA.
(B) Phenotype of the lines AtGluTRBP_antisense_#22 and AtGluTRBP_sense_#34 5, 14, 30, and 45 d after germination under high-light conditions. Individual seedlings among the mutant progenies that grew phenotypically like the wild type were removed. When light intensity was lowered to 90 μmol photons m−2 s−1 (NL), 30 d after germination recovery of the growth retardation phenotype of the mutant was observed. The remaining seedlings showed the mutant phenotype, and the presence of the transgene was confirmed by PCR.
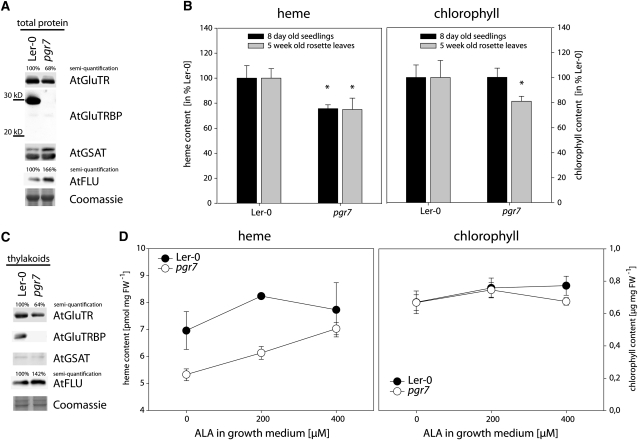
The recently described Arabidopsis mutant pgr7 reveals reduced nonphotochemical quenching (Jung et al., 2010). Due to a point mutation, pgr7 expresses a truncated GluTRBP lacking the 84 C-terminal amino acids, resulting in a protein that is either unstable or not recognized by the available anti-AtGluTRBP antibodies (Figure 6A). A direct function of PGR7 was not indicated in this previous report, but pgr7 was affected in photosynthetic electron transport between QA of photosystem II and P700 of photosystem I, most likely by an impaired cytochrome b6f complex (Jung et al., 2010). We analyzed the heme contents of pgr7 and found a reduction by 25% compared with its corresponding wild-type Ler-0 in 8-d-old seedlings as well as in 5-week-old rosette leaves (Figure 6B). As a cytochrome b6f monomer requires four essential heme molecules, it is proposed that reduced efficiency of photosynthetic electron transport in pgr7 is a consequence of heme deficiency. Moreover, slightly reduced chlorophyll contents in pgr7 may be a secondary effect caused by inefficient electron transport and adjustments of the photosynthetic apparatus. The phenotype of pgr7 resembled that of lines #22 and #34 with modified GluTRBP expression. They were indistinguishable from the corresponding wild type under standard growth conditions (Figure 4B) except for barely detectable growth retardation (Jung et al., 2010). It is remarkable that 8-d-old pgr7 mutants show reduced GluTR (68%) and increased FLU (166%) contents compared with the Arabidopsis wild-type Ler-0 (Figure 6A). These changes may be attributed to fine-tuning of ALA-forming capacity in response to perturbed heme biosynthesis. However, similar to the transgenic Arabidopsis lines with silenced GluTRBP expression, the chlorophyll contents of pgr7 mutants are unaffected in 8-d-old seedlings and only slightly reduced in 5-week-old rosette leaves (Figure 6B). It was reported that the dysfunctional mutant PGR7 did not influence the abundance of photosynthetic protein complexes (Jung et al., 2010). This is consistent with our analysis of chloroplast ultrastructure. Chloroplasts were analyzed from both transgenic Arabidopsis lines, which are characterized by expression of reduced GluTRBP contents and high-light-dependent growth retardation (Figure 5), but we could not observe a significant alteration of their ultrastructure or abundance (see Supplemental Figure 10 online). These findings are consistent with previously published reports, which showed normal chloroplast ultrastructure and no thylakoid disintegration, although a severe chlorophyll reduction of up to 30% content was determined (Murray and Kohorn, 1991; Andersson et al., 2003).
Figure 6.
Arabidopsis pgr7 Mutants Lack Functional GluTRBP, Show Reduced Heme Contents, and Have Less Thylakoid-Bound GluTR.
(A) Immunoblot analysis of total protein extracts of 8-d-old seedlings of Arabidopsis Ler-0 and pgr7. A point mutation in GluTRBP of pgr7 terminates the coding sequence 84 triplets before the endogenous stop codon, resulting in a theoretical size of a truncated protein of around 21 kD. Neither the 30-kD wild type nor the truncated GluTRBP is immunologically detectable in pgr7. A detail of a Coomassie blue–stained 12% SDS-polyacrylamide gel served as loading control.
(B) Heme and chlorophyll (a and b) contents of 8-d-old seedlings and 5-week-old rosette leaves of Arabidopsis wild-type Ler-0 and pgr7 mutants. Asterisk marks a significant difference to wild-type Ler-0 (P < 0.05).
(C) Immunoblot analysis of thylakoid membranes of 8-d-old seedlings of Arabidopsis Ler-0 and pgr7. Purity of thylakoid preparations is shown by absence of GSAT and presence of FLU. The amount of thylakoid-bound GluTR is reduced in pgr7.
(D) Arabidopsis wild-type Ler-0 and pgr7 were grown for 8 d on MS containing 0, 200, or 400 μM ALA before heme and chlorophyll contents were determined.
(A) and (C) The protein amounts of GluTR and FLU were quantified and given in the figure.
(B) and (D) Given are means ± sd.
However, analysis of the thylakoid-bound portion of GluTR revealed that the expression of a truncated GluTRBP in pgr7 went along with a reduced amount of thylakoid-bound GluTR that reached only 64% of the amount in the corresponding wild-type Ler-0 (Figure 6C). Since pgr7 contains more FLU protein than the wild type (Figures 6A and 6C), more GluTR can likely be bound to the thylakoid membrane via FLU. Moreover, feeding of pgr7 with up to 400 μM exogenous ALA resulted in an increase of noncovalently bound heme equal to the wild-type level (Figure 6D). These moderate amounts of ALA supplied with the medium are not sufficient to increase greatly the heme contents in the Arabidopsis wild-type Ler-0 and do not significantly change chlorophyll accumulation in Ler-0 and pgr7. Since exogenous ALA can bypass heme deficiency caused by lack of functional GluTRBP, it is concluded that GluTRBP does not have a role in heme biosynthesis downstream of ALA formation nor in assembly or stability of heme-containing proteins but rather acts specifically on the level of ALA formation.
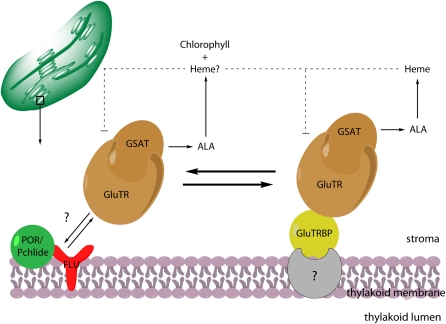
GluTRBP-Mediated Spatial Separation of ALA Synthesis: A Model
Though technical limitations (e.g., quality of available GluTR antibodies) did not allow unambiguously identification of native complexes of GluTR and GluTRBP proteins on the thylakoid membrane of Arabidopsis chloroplasts, we propose based on the presented data a model in which GluTRBP forms a thylakoid-associated anchor for GluTR and contributes to the organization of ALA synthesis to separate synthesis of ALA dedicated either to heme or to chlorophyll biosynthesis (Figure 7). GluTRBP binds a minor portion of GluTR. We suggest that both proteins belong to a complex that facilitates channeling of ALA into the Fe branch. This mechanism would still ensure heme biosynthesis when other GluTR molecules are posttranslationally inactivated by feedback inhibition of the chlorophyll branch (e.g., by the action of the negative regulator FLU). However, it cannot be entirely excluded that GluTRBP additionally functions for other processes in chloroplasts or contributes to heme biosynthesis without an interaction with GluTR.
Figure 7.
A Model for the Role of GluTRBP in the Spatial Separation of GluTR and ALA Synthesis for Tetrapyrrole Biosynthesis in Chloroplasts.
It is suggested that GluTRBP contributes to the spatial separation of ALA synthesis for heme and chlorophyll synthesis. GluTRBP anchors a small part of the total amount of GluTR at the thylakoid membrane. ALA formed in this subcompartment is dedicated to heme biosynthesis, when in parallel the majority of GluTR is located in the stroma of plastids to provide ALA for chlorophyll biosynthesis. A direct physical interaction of POR and FLU has not been demonstrated yet. However, in response to accumulating protochlorophyllide, the dominant part of ALA biosynthesis is inactivated when GluTR interacts with FLU. The FLU-dependent inactivation is proposed not to interfere with the GluTR-GluTRBP portion belonging to the separated ALA synthesis. Thus, heme synthesis can independently continue. Consequently, heme may act as feedback inhibitor on ALA biosynthesis through transcriptional or posttranslational control.
This model integrates the previous ideas on specific ALA pools (Huang and Castelfranco, 1990) and provides a regulatory mechanism for the synthesis of varying amounts of heme and chlorophyll in green tissue during light–dark cycles and plant development. Moreover, regulatory feedback cues will address different branches of the parallel tetrapyrrole pathway from ALA biosynthesis to heme or chlorophyll synthesis. Our model is in agreement with the concept that the majority of GluTR activity contributes to chlorophyll biosynthesis and is posttranslationally regulated by FLU (Meskauskiene et al., 2001; Richter et al., 2010). Additionally, heme acts as feedback regulator for both fractions of GluTR either localized in the stroma or in a specific subcompartment that is closely attached to the thylakoid membrane and organizes ALA biosynthesis dedicated to heme synthesis. At present, our model enables answers about the spatial and temporal organization of tetrapyrrole biosynthesis. A minor portion of GluTR is attached to the thylakoid membrane by GluTRBP and potentially continues formation of ALA even when the majority of GluTR is posttranslationally inactivated. Thereby, GluTRBP is not involved in the inactivation or inhibition of enzymes of the Mg branch of tetrapyrrole biosynthesis (e.g., Mg chelatase or POR) in darkness. In fact, the more continuously expressed GluTRBP is suggested to function in the spatial separation of a small portion of GluTR in the chloroplasts. Subsequently, ALA that is more constantly synthesized in this subcompartment is further metabolized and channeled into the heme biosynthetic pathway, while the chlorophyll biosynthesis is inhibited.
Joyard et al. (2009) proposed a model of spatial organization of tetrapyrrole biosynthesis within the chloroplast. Following their ideas, ALA synthesis and the successive enzymatic steps from ALA to protoporphyrinogen IX are located in the stroma of the chloroplasts. Starting from protoporphyrinogen IX oxidation, the reactions were relocated to thylakoid and envelope membranes. Interestingly, localization of GluTR was not part of this model, and the authors discuss that GluTR was never found in proteomic studies. Our idea of a spatial separation of ALA synthesis in chloroplasts forms an extension of the existing model. The placement of the GluTRBP into a model of tetrapyrrole metabolism stands at the beginning of our understanding of compartmentalized metabolic pathways allowing a more specific and dynamic control of metabolic activities. At present, additional metabolic subcompartments within the chloroplasts not only for GluTR but also for other enzymes of the tetrapyrrole biosynthetic pathway can be hypothesized but lack experimental evidence. A spatial separation of defined sets of enzymes or multienzyme complexes of the metabolic pathway is proposed to provide an additional regulatory mechanism to separate a fraction of a given enzyme activity while the residual activity is differently controlled. This mechanism can fine-tune metabolic activities and contribute to a balanced allocation of metabolites for the synthesis of different end products of a branched pathway. The organization in subcompartments necessitates more scaffold or anchoring proteins. The presented work directs the attention to additional organizing principles of the tetrapyrrole biosynthesis metabolic pathway and opens new insights into intraorganellar structures enabling defined localizations of enzymes dedicated to either chlorophyll or heme synthesis. Future studies will address the organization of tetrapyrrole biosynthesis in further protein complexes.
METHODS
Yeast Two-Hybrid System
Protein coding sequences were cloned into the multiple cloning sites of pBTM117c (Bartel et al., 1993; Han and Colicelli, 1995; Wanker et al., 1997) or pGAD10 (BD Biosciences). An Arabidopsis thaliana cDNA library was cloned into the plasmids pGAD10-GW-A, pGAD10-GW-B, and pGAD10-GW-C1 as described by Bürkle et al. (2005). Yeast strain L40ccua (Goehler et al., 2004) was used for protein interaction assays. Compositions of media used for yeast clone cultivation and selection are given in Supplemental Table 3 online. Autoactivations of all bait containing yeast clones were tested according to protocols given by MacDonald (2001).
Small-Scale Yeast Transformation
Three milliliters of medium (yeast extract peptone dextrose supplemented with 20 mg/liter adenine hemisulfate, SD-Trp, or SD-Leu; see Supplemental Table 3 online) was inoculated with one yeast colony (L40ccua, L40ccua_bait, or L40ccua_prey) and incubated overnight at 30°C and 300 rpm. Yeast cells were harvested from 1 mL culture by short centrifugation (5 s) and suspended in 506 μL PLATE mix (450 μL 45% polyethylene glycol 4000, 50 μL 1 M lithium acetate, 5 μL 1 M Tris-HCl, pH 7.5, and 1 μL 0.5 M EDTA, pH 8.0). One microgram of plasmid DNA, 2 μL denatured salmon sperm DNA (10 mg mL−1, boiled for 5 min, then cooled on ice), and 20 μL 1 M DTT were added. Suspension was mixed and incubated for at least 6 h at room temperature before cells were heat shocked at 42°C for 10 min. Cells were harvested by short centrifugation, plated on transformation selection medium (SD-Trp, SD-Leu, SD-Leu-Trp; see Supplemental Table 3 online), and incubated at 30°C until yeast colonies were grown.
Large-Scale Yeast Transformation
To screen a cDNA library for protein interaction partners of At-GluTR, the yeast clone L40ccua_ AtGluTR_w/o_TP_pBTM117c was transformed in a large-scale experiment according to Woods and Gietz (2001). One hundred milliliters of SD-Trp was inoculated with L40ccua_ AtGluTR_w/o_TP_pBTM117c and incubated at 30°C and 300 rpm for 2 d. OD545 was determined and a volume corresponding to 1.5·109 cells (1 OD545 = 107 cells mL−1) was harvested by centrifugation (3000g, 10 min, 10°C) and suspended in 300 mL 2× YPAD. Cultures were incubated at 30°C and 300 rpm until OD545 reached 2. Subsequently, cells were harvested by centrifugation (3000g, 10 min, 10°C), washed with water three times, and finally suspended in 10 mL 100 mM lithium acetate. After 15 min incubation at 30°C, cells were harvested again (3000g, 10 min, 10°C), and suspended in 14.4 mL 50% polyethylene glycol 3350, 2.16 mL 1 M lithium acetate, 0.6 mL denatured salmon sperm DNA, and 100 μg prey-plasmid DNA (pGAD10-GW_cDNA_library) in 4.44 mL water. After incubation at 30°C for 30 min, cells were heat shocked at 42°C for 45 min. Yeast cells were harvested by centrifugation (3000g, 10 min, 10°C), suspended in 10 mL water, and plated on SD-Trp-Leu-His-Ura. Plates were incubated at 30°C until colonies were grown.
Identification of Prey Sequences in Yeast Clones
Growing of yeast colonies on SD-Trp-Leu-His-Ura indicates protein–protein interaction of the known bait (At-GluTR) with an unknown prey. To identify the coding prey sequence, yeast plasmid isolation was performed according to protocols given by MacDonald (2001). Highly electro-competent Escherichia coli (ElectroSHOX; Bioline) was transformed with the plasmid mix originating from one a single yeast clone and plated on Luria-Bertani medium containing 100 μg mL−1 ampicillin. At least 24 E. coli plasmid preparations per transformation were performed according to Birnboim and Doly (1979). Yeast clone L40ccua_AtGluTR_w/o_TP_pBTM117c was retransformed with plasmids isolated from E. coli to verify interaction with At-GluTR. Prey plasmids causing an interaction with At-GluTR were sequenced with pGAD10-specific primers (5′-GCGCTTTCACAACCAATTGC-3′ and 5′-TGATTGGAGACTTGACCAAACC-3′).
Plasmids Used for Expression of Proteins in Yeast, E. coli, Tobacco, and Arabidopsis
Subcloning into expression plasmids was initiated by PCR amplification of gene-specific sequences from Arabidopsis or Chlamydomonas reinhardtii cDNA using primers given in Supplemental Table 4 online and a Phusion High-Fidelity DNA polymerase according to the manufacturer’s protocols (Finnzymes). Recognition sequences from the multiple cloning sites of target plasmids were chosen, and sequences were fused to the 5′ end of the primers. When necessary, artificial start codons (ATG) were introduced. Amplified sequences were subcloned into pGEM-T (Promega) or pJET1.2 (Fermentas) according to the protocols of the manufacturers. Subcloned cDNA sequences were finally cloned in the destination plasmids using the chosen restriction enzymes.
The multiple cloning sites of plasmids pBTM117c (Wanker et al., 1997), pGAD10 (BD Biosciences Clontech), and pGEX-2T (GE Healthcare) were modified by introduction of additional restriction enzyme recognition sequences to ease cloning (Czarnecki, 2010; Hackenberg, 2010). BiFC plasmids pSPYNE and pSPYCE are derived from Walter et al. (2004). Enhanced efficiency of in vitro transcription/translation of Arabidopsis proteins in rabbit reticulocyte system was achieved by the use of a pBAT22 derivate (pBAT22rev; Annweiler et al., 1991) encoding a 5′-rabbit-globin-leader sequence for increased translation efficiency. Moreover, a typical Kozak consensus sequence of vertebrates (C ACC atg GCG; Joshi et al., 1997) was introduced to force translation start from the primary ATG site. Expression plasmids pGEX-2T and pQE80L were obtained from GE Healthcare and Qiagen, respectively. Plant binary expression plasmid pGLI is a derivate of pGBTV (Becker et al., 1992) lacking the β-glucuronidase sequence (B. Hedtke, unpublished data). Recombinant proteins used in this work are listed in Supplemental Table 5 online. Expression of recombinant proteins in E. coli and purification are described in Supplemental Methods 1 online.
BiFC
BiFC assays were performed according to Walter et al. (2004). Agrobacterium tumefaciens (strain GV2260; Hellens et al., 2000) was transformed with pSPYNE, pSPYCE, AtGluTR_pSPYNE, AtGluTR_pSPYCE, AtGluTRBP_pSPYNE, and AtGluTRBP_pSPYCE and selected on YEB medium (0.5% [w/v] beef extract, 0.5% [w/v] peptone, 0.5% [w/v] sucrose, 0.1% [w/v] yeast extract, 2 mM MgSO4, pH 7.0, and, if necessary, 1.5% [w/v] agar) containing 25 μg mL−1 rifampicin and 50 μg mL−1 kanamycin. A 3-mL preculture of YEB medium with 25 μg mL−1 rifampicin and 50 μg mL−1 kanamycin was inoculated with each A. tumefaciens clone and incubated at 28°C and 250 rpm overnight. A fresh 50-mL culture containing additional 20 μM acetosyringone (3′,5′-dimethoxy-4'-hydroxyacetophenon) was inoculated with each A. tumefaciens clone incubated at 28°C and 250 rpm overnight. Cells were pelleted by centrifugation (5500g, 20 min) washed twice with water to remove any remaining antibiotics, suspended in infiltration medium (10 mM MgCl2, 10 mM MES, pH 5.7, and 100 μM acetosyringone) until OD600 was 1.0, and incubated at room temperature for 4 h. Equal volumes of the A. tumefaciens suspensions containing pSPYNE or pSPYCE plasmids were mixed and infiltrated into Nicotiana benthamiana Domin leaves (Bendahmane et al., 2000). Infiltrated plants were dark incubated for 72 h before leaf segments from infiltrated areas were analyzed for reconstitution of yellow fluorescent protein (YFP) fluorescence by a confocal laser scanning microscope (excitation at 514 nm, YFP emission 530 to 555 nm, chlorophyll emission 600 to 700 nm).
Isolation and Fractionation of Arabidopsis Chloroplasts
Isolation of intact chloroplasts from Arabidopsis followed the protocols from Kunst (1998). Fifty grams of Arabidopsis leaf material were harvested 1 h after onset of light and incubated in ice-cold water for 30 min. After drying, leaf tissues were disrupted in a modified Warring-Blender (Kannangara et al., 1977) in the presence of 500 mL homogenization buffer (0.45 M sorbitol, 20 mM Tricine, pH 8.4, 10 mM EDTA, 10 mM NaHCO3, and 0.1% [w/v] BSA) with three pulses of 5 s each. The homogenate was filtered through Miracloth (Calbiochem) and centrifuged for 2 min at 2000g. The supernatant was decanted, and the chloroplast-containing pellet was carefully suspended in 1 mL buffer RB (0.3 M sorbitol, 20 mM Tricine, pH 7.6, 5 mM MgCl2, and 2.5 mM EDTA). Intact chloroplasts were purified by a Percoll gradient (50% [v/v] Percoll in RB, centrifuged at 43,400g for 30 min) by overlaying the gradient with the chloroplast suspension and centrifugation at 13,300g for 6 min. Intact chloroplasts were removed, washed two times in buffer RB, and sedimented by centrifugation (3300g, 2 min). Subsequently, chloroplasts were suspended in 200 μL PBS (20 mM sodium phosphate buffer and 150 mM NaCl, pH 7.4) and lysed on ice for 10 min. Stroma and thylakoid fractions were separated by centrifugation at 16,000g for 30 min. Thylakoids were suspended in 2 mL PBS and aliquoted into 4 volumes. Aliquots were washed three times with 1 mL of PBS or PBS containing 10 mM DTT or 10 mM H2O2 or 4 M NaCl, respectively, and finally suspended in 200 μL PBS. Protein concentrations were determined according to Bradford (1976).
In Vitro Transcription and Translation
In vitro transcription/translation of At-GluTR and At-GluTRBP was performed using plasmids AtGluTR_pBAT22rev, AtGluTRBP_w/o_TP_pBAT22rev, and AtGluTRBP _pBAT22rev and the TNT Quick Coupled Transcription/Translation System (Promega) in presence of [35S]-Met (1000 Ci mmol−1; Hartmann Analytic) according to protocols provided by the manufacturer.
Chloroplast Import Assay
Seeds of pea (Pisum sativum) were watered for 24 h and germinated on wet vermiculite for 7 d at 23°C and 90 μmol photons m−2 s−1 under long-day conditions (14 h light/10 h dark). Isolation of intact chloroplasts and subsequently performed import assay of [35S]-marked At-GluTRBP followed the protocols given by Brock et al. (1993) and Clausmeyer et al. (1993). Chloroplasts fraction proteins were separated electrophoretically on a 12% SDS-polyacrylamide gel. Detection of [35S]-Met was performed using a phosphor imager.
RNA Gel Blot Analysis
Arabidopsis total RNA was extracted using the Invisorb Spin Plant Mini Kit (Invitek). Ten grams of RNA was separated on 1.5% agarose gels containing formaldehyde and transferred to nylon membrane (Hybond N+; GE Healthcare) by capillary blot according to Sambrook and Russell (2001). Membranes were hybridized with [32P]-dCTP–labeled (DecaLabel DNA labeling kit; Fermentas) gene-specific probes amplified from Arabidopsis cDNA using primers given in Supplemental Table 6 online. Hybridizations were performed using church buffer at 65°C (Church and Gilbert, 1984; Sambrook and Russell, 2001), and a phosphor imager system was used for detection of radioactive signals. Unless only single experiments were shown in the figures, these experiments were repeated at least two times.
Quantitative Real-Time PCR
For quantitative RT-PCR, cDNA was synthesized from 2 μg total RNA using oligo(dT18) primers and amplified with the SensiMix SYBR No-ROX kit (Bioline) according to the manufacturer’s instructions using primers listed Supplemental Table 7 online. The following PCR conditions were used: 5 min 95°C, followed by 40 cycles with 5 s at 95°C, 10 s at 55°C, and 20 s at 72°C. The relative levels of expression were calculated in relation to TUB2 (At5g62690) as the housekeeping gene and given as 2−ΔΔCt (cycle threshold) (Livak and Schmittgen, 2001; Schmittgen and Livak, 2008).
Immunoblot Analysis
Arabidopsis proteins were extracted from 100 mg plant tissue ground in liquid nitrogen and suspended in 500 μL buffer containing 56 mM Na2CO3, 56 mM DTT, 2% (w/v) SDS, 12% (w/v) sucrose, and 2 mM EDTA. Protein extracts were denatured at 70°C for 20 min and centrifuged for 10 min at 16,000g. Protein concentrations of the supernatant were determined according to Bradford (1976) after trichloracetic acid precipitation. Therefore, 25 μL protein samples were mixed with 500 μL ice-cold 10% (w/v) trichloracetic acid, incubated on ice for 15 min, and centrifuged at 4°C at 16,000g for 10 min. Pellets were dissolved in 50 μL 0.1 n NaOH. Ten micrograms of total protein was electrophoretically separated using 12% SDS containing polyacrylamide gels in a discontinuous Tris-Gly buffer system (Laemmli, 1970) according to standard protocols (Harlow and Lane, 1988; Sambrook and Russell, 2001). After electrophoresis, gels were stained with Coomassie Brilliant Blue R 250, or proteins were transferred to nitrocellulose membranes (Hybond-C; GE Healthcare) according to protocols supplied by the manufacturer. Membrane-bound proteins were detected and immunologically quantified using standard protocols (Harlow and Lane, 1988; Sambrook and Russell, 2001), with antibodies given in Supplemental Table 8 online, and ECL detection. Signal intensities were quantified by two-dimensional densitometry using AIDA image analyzer software (Raytest). Unless only single experiments were shown in the figures, these experiments were repeated at least two times.
Ultrastructural Analysis and Immunogold Localization Experiments
Ultrastructural analysis and immunogold labeling were performed as described by Tognetti et al. (2006), except that an FEI Tecnai G2 Sphera transmission electron microscope (FEI Company) was used for ultrastructural analysis at 120 kV. Controls are provided in Supplemental Data Set 1 online.
Determination of Pigment and Heme Contents
Extraction and determination of pigments and heme from 4-week-old Arabidopsis plants followed the protocols from Lichtenthaler (1987). One hundred milligrams of leaf material was homogenized in 80% (v/v) acetone containing 10 μM KOH and centrifuged (16,000g, 10 min). Extraction of the pellets was repeated until they became white and supernatants were combined. Quantification of chlorophylls a and b as well as total carotenoids was performed spectrophotometrically at the wavelengths 470, 645, 663, and 720 nm, and concentrations were calculated according to Lichtenthaler (1987).
Extraction of protochlorophyllide was performed according to the protocol of Koski and Smith (1948). Briefly, 100 to 300 mg of etiolated Arabidopsis wild-type and mutant seedlings were treated with steam for 2 min, ground in liquid nitrogen, and extracted three times in alkaline acetone (9 volumes 100% acetone: 1 volume 0.1 n NH4OH). Protochlorophyllide contents were determined by fluorescence spectroscopy (excitation at 433 nm and fluorescence emission at 632 to 633 nm). For calibration, a protochlorophyllide standard was extracted from 7-d-old etiolated barley (Hordeum vulgare) leaves (Koski and Smith, 1948) and quantified using extinction coefficient in diethyl ether at 623 nm of 35,600 M−1 cm−1 (Dawson et al., 1986).
For extraction of heme, 50 mg leaf material was ground in 5 mL ice-cold 90% (v/v) acetone and 10% (v/v) 0.1 M NH4OH to remove pigments. After centrifugation (10,000g, 10 min), noncovalently bound heme was extracted from the pellets using 1 mL 80% (v/v) acetone, 16% (v/v) DMSO, and 4% (v/v) concentrated HCl. After centrifugation (10,000g, 10 min), heme extraction of the pellet was repeated and both supernatants were combined. Quantification of the heme content in the extracts followed the protocols of Masuda and Takahashi (2006) and Takahashi and Masuda (2009). Briefly, extracts were diluted in 10 μM KOH up to 4000-fold. Twenty microliters of diluted extracts were mixed with 80 μL 125 mM Tris-HCl, pH 8.4, containing 3.125 nM horseradish peroxidase apoenzyme (Sigma-Aldrich) and incubated for 30 min at room temperature. One hundred microliters of very sensitive Western blot detection reagent (Applichem) was added and incubated according to the manufacturer’s instructions. Luminescence generated by reconstituted horseradish peroxidase was detected and quantified using a charge-coupled device camera.
Generation of Transgenic Arabidopsis with Reduced Contents of AtGluTRBP
A. tumefaciens (strain GV2260) were transformed with plasmids AtGluTRBP_sense_pGL1 and AtGluTRBP_antisense_pGL1, respectively, and positive clones were selected on YEB medium containing 25 μg mL−1 rifampicin and 50 μg mL−1 kanamycin. Stable transformation of Arabidopsis Col-0 was performed by floral dip (Clough and Bent, 1998). Seeds were sawn on soil, and transgenic plants were selected by repeated application (days 8, 11, 14, and 18 after germination) of 0.1% (w/v) BASTA solution (Bayer CropScience). DNA of transgenic plants was extracted according to Weigel and Glazebrook (2002), and presence of the transgene was tested by PCR using primers specific for the cauliflower mosaic virus 35S promoter (5′-ATCCTTCGCAAGACCCTTCC-3′) and GluTRBP in sense and antisense orientation, respectively (5′-TGGAGACCATACCCGAA-3′, sense orientation; 5′-GACCCACAAGCCCTTT-3′, antisense orientation). Control PCRs were performed using primers for specific amplification of GUN4 (5′-ATGGCGACCACAAACTCTCTC-3′; 5′-TCAGAAGCTGTAATTTGTTTTAAACACT-3′).
Generation of Transgenic Arabidopsis with Ethanol-Inducible AtGluTRBP-RNAi
Ethanol-induced expression of an AtGluTRBP-RNAi construct in Arabidopsis was based on the system reported by Chen et al. (2003). Plasmid pBinΔalcR_AtGluTRBP_RNAi was generated by cloning of a 242-bp fragment of the GluTRBP exon 5′ amplified with the specific primers 5′-AGATCTAACAAAGATGATATGGATGG-3′ and 5′-CTCGAGCTCCTCCCTTGTGAGAATGA-3′ (primers include recognition sites of the restriction enzymes BglII and XhoI). A. tumefaciens (strain GV2260) was transformed with pBinΔalcR_AtGluTRBP_RNAi, and positive clones were selected on YEB medium containing 25 μg mL−1 rifampicin and 50 μg mL−1 kanamycin. Stable transformation of Arabidopsis Col-0 was performed by floral dip (Clough and Bent, 1998). Surface-sterilized seeds were germinated on solid Murashige and Skoog (MS) medium (Murashige and Skoog, 1962) containing 100 μg mL−1 kanamycin. Selected transformants were analyzed for transgene insertion by PCR strategies. Induction of transgene expression was performed 3 d after germination by adding 1 mL 2% (v/v) ethanol into an applicable hole in the medium.
Arabidopsis Mutants and Plant Growth Conditions
Arabidopsis wild-type Col-0 or Ler-0 and mutant plants were grown in soil in grow banks (CLF Plant Climatics) at 23°C and ~90 or 300 μmol photons m−2 s−1. Surface-sterilized seedlings of Arabidopsis wild-type Col-0 or Ler-0 were plated on MS medium with 0.8% agar with or without 1 μM norflurazon or 500 μM lincomycin. After incubation at 4°C for 3 d, plates were transferred to darkness at 23°C for another 3 d. Etiolated plants were transferred to light (90 μmol photons m−2 s−1) for the time period indicated. Arabidopsis mutant pgr7 was initially described by Jung et al. (2010). Surface-sterilized seedlings of Arabidopsis wild-type Ler-0 and pgr7 were plated on MS medium with 0.8% agar with 0, 200, and 400 μM ALA. After incubation at 4°C for 3 d, plates were transferred to light (90 μmol photons m−2 s−1) at 23°C for 8 d.
Accession Numbers
The GluTRBP homologous protein mRNA and amino acid sequence data from this article can be found in the GenBank/EMBL data libraries under the following accession numbers: C. reinhardtii, HQ456532; and Nicotiana tabacum, HQ456533 and HQ456534.
Supplemental Data
The following materials are available in the online version of this article:
Supplemental Figure 1. Verification of Protein-Protein Interaction between At-GluTR and At-GluTRBP by Protein Pull-Down.
Supplemental Figure 2. Comparison of At-GluTRBP and At3g03890.
Supplemental Figure 3. Spectroscopy Analyses of Recombinant At-GluTRBP.
Supplemental Figure 4. Identification of a Protein Complex Containing GluTRBP in the Thylakoids of Mustard (Sinapis alba).
Supplemental Figure 5. Sequence Alignment of Homologous GluTRBP Proteins.
Supplemental Figure 6. Identification of an At-GluTRBP Homolog in Chlamydomonas reinhardtii.
Supplemental Figure 7. Analysis of Protein Interaction between Cr-GluTR and Cr-GluTRBP Using the Surface Plasmon Resonance Technique (Biacore).
Supplemental Figure 8. Application of Cr-GluTRBP in a Cr-GluTR Activity Assay.
Supplemental Figure 9. Lethal Phenotype of AtGluTRBP RNAi Knockdown.
Supplemental Figure 10. Chloroplast Ultrastructure of Stressed At-GluTRBP Mutants.
Supplemental Table 1. Identification of Proteins by Mass Spectrometry.
Supplemental Table 2. Binding Constants of the Chlamydomonas GluTR-GluTRBP Interaction.
Supplemental Table 3. Composition of Media Used for the Yeast Two-Hybrid System.
Supplemental Table 4. List of Generated Expression Plasmids.
Supplemental Table 5. List of Recombinant Proteins Used in This Study.
Supplemental Table 6. List of Probes for RNA Gel Blots.
Supplemental Table 7. List of Primers for Real-Time PCR.
Supplemental Table 8. List of Antibodies Used in This Study.
Supplemental Methods 1. Supplemental Methods Used in the Supplemental Data.
Supplemental Data Set 1. Controls for GluTRBP Immunogold Labeling (10-nm Gold) of the Arabidopsis Wild Type.
Supplemental Data Set 2. Mass Spectrometry Data.
Acknowledgments
We thank Lukas Bürkle for providing the Arabidopsis cDNA library, Krishna K. Niyogi for pgr7 seeds, Stefan Frielingsdorf, Yvonne Pörs, and Lars Dietzel for technical and experimental assistance, and Thomas J. Buckhout for critically reading the manuscript. Parts of this work were supported by grants of the Deutsche Forschungsgemeinschaft (SFB 429 and FOR 804 to B.G.).
AUTHOR CONTRIBUTIONS
O.C. and B.G. designed the research. O.C., B.H., M.M., M.R., A.R., and Y.S. performed research. M.M. and T.P. contributed new analytical tools. O.C., B.H., T.P., and B.G. analyzed data. O.C. and B.G. wrote the article.
References
- Andersson J., Wentworth M., Walters R.G., Howard C.A., Ruban A.V., Horton P., Jansson S. (2003). Absence of the Lhcb1 and Lhcb2 proteins of the light-harvesting complex of photosystem II - effects on photosynthesis, grana stacking and fitness. Plant J. 35: 350–361 [DOI] [PubMed] [Google Scholar]
- Annweiler A., Hipskind R.A., Wirth T. (1991). A strategy for efficient in vitro translation of cDNAs using the rabbit beta-globin leader sequence. Nucleic Acids Res. 19: 3750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Archakov A.I., Govorun V.M., Dubanov A.V., Ivanov Y.D., Veselovsky A.V., Lewi P., Janssen P. (2003). Protein-protein interactions as a target for drugs in proteomics. Proteomics 3: 380–391 [DOI] [PubMed] [Google Scholar]
- Bartel P.L., Chien C.T., Sternglanz R., Fields S. (1993). Using the two-hybrid system to detect protein protein interactions. In Cellular Interactions in Development: A Practical Approach., D.A. Hartley, ed (Oxford, UK: Oxford University Press), pp. 153–179 [PubMed] [Google Scholar]
- Beale S.I. (1999). Enzymes of chlorophyll biosynthesis. Photosynth. Res. 60: 43–73 [Google Scholar]
- Beale S.I. (2006). Biosynthesis of 5-aminolevulinic acid. In Chlorophylls and Bacteriochlorophylls: Biochemistry, Biophysics, Functions and Applications, B. Grimm, R.J. Porra, W. Rudiger, and H. Scheer, eds (Dordrecht, The Netherlands: Springer), pp. 147–158 [Google Scholar]
- Becker D., Kemper E., Schell J., Masterson R. (1992). New plant binary vectors with selectable markers located proximal to the left T-DNA border. Plant Mol. Biol. 20: 1195–1197 [DOI] [PubMed] [Google Scholar]
- Bendahmane A., Querci M., Kanyuka K., Baulcombe D.C. (2000). Agrobacterium transient expression system as a tool for the isolation of disease resistance genes: application to the Rx2 locus in potato. Plant J. 21: 73–81 [DOI] [PubMed] [Google Scholar]
- Birnboim H.C., Doly J. (1979). A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res. 7: 1513–1523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradford M.M. (1976). A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72: 248–254 [DOI] [PubMed] [Google Scholar]
- Brock I.W., Hazell L., Michl D., Nielsen V.S., Møller B.L., Herrmann R.G., Klösgen R.B., Robinson C. (1993). Precursors of one integral and five lumenal thylakoid proteins are imported by isolated pea and barley thylakoids: Optimisation of in vitro assays. Plant Mol. Biol. 23: 717–725 [DOI] [PubMed] [Google Scholar]
- Bürkle L., Meyer S., Dortay H., Lehrach H., Heyl A. (2005). In vitro recombination cloning of entire cDNA libraries in Arabidopsis thaliana and its application to the yeast two-hybrid system. Funct. Integr. Genomics 5: 175–183 [DOI] [PubMed] [Google Scholar]
- Chen S., Hofius D., Sonnewald U., Börnke F. (2003). Temporal and spatial control of gene silencing in transgenic plants by inducible expression of double-stranded RNA. Plant J. 36: 731–740 [DOI] [PubMed] [Google Scholar]
- Church G.M., Gilbert W. (1984). Genomic sequencing. Proc. Natl. Acad. Sci. USA 81: 1991–1995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clausmeyer S., Klösgen R.B., Herrmann R.G. (1993). Protein import into chloroplasts. The hydrophilic lumenal proteins exhibit unexpected import and sorting specificities in spite of structurally conserved transit peptides. J. Biol. Chem. 268: 13869–13876 [PubMed] [Google Scholar]
- Clough S.J., Bent A.F. (1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Cornah J.E., Terry M.J., Smith A.G. (2003). Green or red: What stops the traffic in the tetrapyrrole pathway? Trends Plant Sci. 8: 224–230 [DOI] [PubMed] [Google Scholar]
- Czarnecki O. (2010). Die Synthese der 5-Aminolävulinsäure in Pflanzen: Charakterisierung eines neuen GluTR-bindenden Proteins und dessen Rolle bei der Mikrokompartimentierung der Tetrapyrrolbiosynthese. (Berlin: Mensch & Buch Verlag) [Google Scholar]
- Dawson R.C.M., Elliott D.C., Elliot W.H., Jones K.M. (1986). Data for Biochemical Research. (Oxford, UK: Oxford University Press) [Google Scholar]
- Emanuelsson O., Brunak S., von Heijne G., Nielsen H. (2007). Locating proteins in the cell using TargetP, SignalP and related tools. Nat. Protoc. 2: 953–971 [DOI] [PubMed] [Google Scholar]
- Friso G., Giacomelli L., Ytterberg A.J., Peltier J.B., Rudella A., Sun Q., Wijk K.J. (2004). In-depth analysis of the thylakoid membrane proteome of Arabidopsis thaliana chloroplasts: New proteins, new functions, and a plastid proteome database. Plant Cell 16: 478–499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goehler H., et al. (2004). A protein interaction network links GIT1, an enhancer of huntingtin aggregation, to Huntington’s disease. Mol. Cell 15: 853–865 Erratum. Mol. Cell 22: 287 [DOI] [PubMed] [Google Scholar]
- Goslings D., Meskauskiene R., Kim C., Lee K.P., Nater M., Apel K. (2004). Concurrent interactions of heme and FLU with Glu tRNA reductase (HEMA1), the target of metabolic feedback inhibition of tetrapyrrole biosynthesis, in dark- and light-grown Arabidopsis plants. Plant J. 40: 957–967 [DOI] [PubMed] [Google Scholar]
- Gough S.P., Kannangara C.G. (1977). Synthesis of delta-aminolevulinate by a chloroplast stroma preparation from greening barley leaves. Carlsberg Res. Commun. 42: 459–464 [Google Scholar]
- Grimm B. (2010). Control of the metabolic flow in tetrapyrrole.biosynthesis: Regulation of expression and activity of enzymes in the Mg branch of tetrapyrrole biosynthesis. In The Chloroplast, Basics and Applications., C.A. Rebeiz, C. Benning, H.J. Bohnert, H. Daniell, J.K. Hoober, H.K. Lichtenthaler, A.R. Portis, and B.C. Tripathy, eds (Dordrecht, The Netherlands: Springer), pp. 39–53 [Google Scholar]
- Hackenberg D. (2010). Untersuchung der Funktion von Mitgliedern der NF-Y-Genfamilie während der Antwort auf abiotischen Stress in Arabidopsis thaliana. (Berlin: Mensch & Buch Verlag) [Google Scholar]
- Han L., Colicelli J. (1995). A human protein selected for interference with Ras function interacts directly with Ras and competes with Raf1. Mol. Cell. Biol. 15: 1318–1323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harel E., Ne’eman E. (1983). Alternative routes for the synthesis of 5-aminolevulinic acid in maize leaves. II. Formation from glutamate. Plant Physiol. 72: 1062–1067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harlow E., Lane D. (1988). Antibodies: A Laboratory Manual. (Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press) [Google Scholar]
- Hellens R., Mullineaux P., Klee H. (2000). Technical Focus: A guide to Agrobacterium binary Ti vectors. Trends Plant Sci. 5: 446–451 [DOI] [PubMed] [Google Scholar]
- Hilson P., et al. (2004). Versatile gene-specific sequence tags for Arabidopsis functional genomics: Transcript profiling and reverse genetics applications. Genome Res. 14: 2176–2189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang L., Castelfranco P.A. (1990). Regulation of 5-aminolevulinic acid (ALA) synthesis in developing chloroplasts: III. Evidence for functional heterogeneity of the ALA pool. Plant Physiol. 92: 172–178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joshi C.P., Zhou H., Huang X., Chiang V.L. (1997). Context sequences of translation initiation codon in plants. Plant Mol. Biol. 35: 993–1001 [DOI] [PubMed] [Google Scholar]
- Joyard J., Ferro M., Masselon C., Seigneurin-Berny D., Salvi D., Garin J., Rolland N. (2009). Chloroplast proteomics and the compartmentation of plastidial isoprenoid biosynthetic pathways. Mol. Plant 2: 1154–1180 [DOI] [PubMed] [Google Scholar]
- Jung H.S., Okegawa Y., Shih P.M., Kellogg E., Abdel-Ghany S.E., Pilon M., Sjölander K., Shikanai T., Niyogi K.K. (2010). Arabidopsis thaliana PGR7 encodes a conserved chloroplast protein that is necessary for efficient photosynthetic electron transport. PLoS ONE 5: e11688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kannangara C.G., Gough S.P., Hansen B., Rasmussen J.N., Simpson D.J. (1977). Homogenizer with replaceable razor blades for bulk isolation of active barley plastids. Carlsberg Res. Commun. 42: 431–439 [Google Scholar]
- Kitamura M., Kojima S., Ogasawara K., Nakaya T., Sagara T., Niki K., Miura K., Akutsu H., Kumagai I. (1994). Novel FMN-binding protein from Desulfovibrio vulgaris (Miyazaki F). Cloning and expression of its gene in Escherichia coli. J. Biol. Chem. 269: 5566–5573 [PubMed] [Google Scholar]
- Koski V.M., Smith J.H. (1948). The isolation and spectral absorption properties of protochlorophyll from etiolated barley seedlings. J. Am. Chem. Soc. 70: 3558–3562 [DOI] [PubMed] [Google Scholar]
- Krieger-Liszkay A., Fufezan C., Trebst A. (2008). Singlet oxygen production in photosystem II and related protection mechanism. Photosynth. Res. 98: 551–564 [DOI] [PubMed] [Google Scholar]
- Kunst L. (1998). Preparation of physiologically active chloroplasts from Arabidopsis. In Arabidopsis Protocols. Methods in Molecular Biology, J. Martinez-Zapater and J. Salinas, eds (Totowa, NJ: Humana Press), pp. 43–48 [Google Scholar]
- Laemmli U.K. (1970). Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227: 680–685 [DOI] [PubMed] [Google Scholar]
- Lichtenthaler H.K. (1987). Chlorophylls and carotenoids: Pigments of photosynthetic membranes. Methods Enzymol. 148: 350–382 [Google Scholar]
- Livak K.J., Schmittgen T.D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods 25: 402–408 [DOI] [PubMed] [Google Scholar]
- Lüer C., Schauer S., Möbius K., Schulze J., Schubert W.D., Heinz D.W., Jahn D., Moser J. (2005). Complex formation between glutamyl-tRNA reductase and glutamate-1-semialdehyde 2,1-aminomutase in Escherichia coli during the initial reactions of porphyrin biosynthesis. J. Biol. Chem. 280: 18568–18572 [DOI] [PubMed] [Google Scholar]
- Lunn J.E. (2007). Compartmentation in plant metabolism. J. Exp. Bot. 58: 35–47 [DOI] [PubMed] [Google Scholar]
- MacDonald P.N. (2001). Two-Hybrid Systems. Methods and Protocols. (Totowa, NJ: Humana Press) [Google Scholar]
- Masuda T., Fujita Y. (2008). Regulation and evolution of chlorophyll metabolism. Photochem. Photobiol. Sci. 7: 1131–1149 [DOI] [PubMed] [Google Scholar]
- Masuda T., Takahashi S. (2006). Chemiluminescent-based method for heme determination by reconstitution with horseradish peroxidase apo-enzyme. Anal. Biochem. 355: 307–309 [DOI] [PubMed] [Google Scholar]
- Matsumoto F., Obayashi T., Sasaki-Sekimoto Y., Ohta H., Takamiya K., Masuda T. (2004). Gene expression profiling of the tetrapyrrole metabolic pathway in Arabidopsis with a mini-array system. Plant Physiol. 135: 2379–2391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merchant S.S., et al. (2007). The Chlamydomonas genome reveals the evolution of key animal and plant functions. Science 318: 245–250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meskauskiene R., Nater M., Goslings D., Kessler F., op den Camp R., Apel K. (2001). FLU: A negative regulator of chlorophyll biosynthesis in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 98: 12826–12831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mochizuki N., Tanaka R., Grimm B., Masuda T., Moulin M., Smith A.G., Tanaka A., Terry M.J. (2010). The cell biology of tetrapyrroles: A life and death struggle. Trends Plant Sci. 15: 488–498 [DOI] [PubMed] [Google Scholar]
- Moser J., Schubert W.D., Beier V., Bringemeier I., Jahn D., Heinz D.W. (2001). V-shaped structure of glutamyl-tRNA reductase, the first enzyme of tRNA-dependent tetrapyrrole biosynthesis. EMBO J. 20: 6583–6590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moulin M., McCormac A.C., Terry M.J., Smith A.G. (2008). Tetrapyrrole profiling in Arabidopsis seedlings reveals that retrograde plastid nuclear signaling is not due to Mg-protoporphyrin IX accumulation. Proc. Natl. Acad. Sci. USA 105: 15178–15183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murashige T., Skoog F. (1962). A revised medium for rapid growth and bio assays with tobacco tissue cultures. Physiol. Plant. 15: 473–497 [Google Scholar]
- Murray D.L., Kohorn B.D. (1991). Chloroplasts of Arabidopsis thaliana homozygous for the ch-1 locus lack chlorophyll b, lack stable LHCPII and have stacked thylakoids. Plant Mol. Biol. 16: 71–79 [DOI] [PubMed] [Google Scholar]
- Nogaj L.A., Beale S.I. (2005). Physical and kinetic interactions between glutamyl-tRNA reductase and glutamate-1-semialdehyde aminotransferase of Chlamydomonas reinhardtii. J. Biol. Chem. 280: 24301–24307 [DOI] [PubMed] [Google Scholar]
- Papenbrock J., Mock H.P., Kruse E., Grimm B. (1999). Expression studies in tetrapyrrole biosynthesis: Inverse maxima of magnesium chelatase and ferrochelatase activity during cyclic photoperiods. Planta 208: 264–273 [Google Scholar]
- Papenbrock J., Mock H.P., Tanaka R., Kruse E., Grimm B. (2000b). Role of magnesium chelatase activity in the early steps of the tetrapyrrole biosynthetic pathway. Plant Physiol. 122: 1161–1169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papenbrock J., Pfündel E., Mock H.P., Grimm B. (2000a). Decreased and increased expression of the subunit CHL I diminishes Mg chelatase activity and reduces chlorophyll synthesis in transgenic tobacco plants. Plant J. 22: 155–164 [DOI] [PubMed] [Google Scholar]
- Peltier J.B., Cai Y., Sun Q., Zabrouskov V., Giacomelli L., Rudella A., Ytterberg A.J., Rutschow H., van Wijk K.J. (2006). The oligomeric stromal proteome of Arabidopsis thaliana chloroplasts. Mol. Cell. Proteomics 5: 114–133 [DOI] [PubMed] [Google Scholar]
- Pontoppidan B., Kannangara C.G. (1994). Purification and partial characterisation of barley glutamyl-tRNA(Glu) reductase, the enzyme that directs glutamate to chlorophyll biosynthesis. Eur. J. Biochem. 225: 529–537 [DOI] [PubMed] [Google Scholar]
- Richter A., Peter E., Pörs Y., Lorenzen S., Grimm B., Czarnecki O. (2010). Rapid dark repression of 5-aminolevulinic acid synthesis in green barley leaves. Plant Cell Physiol. 51: 670–681 [DOI] [PubMed] [Google Scholar]
- Rolland N., Ferro M., Seigneurin-Berny D., Garin J., Douce R., Joyard J. (2003). Proteomics of chloroplast envelope membranes. Photosynth. Res. 78: 205–230 [DOI] [PubMed] [Google Scholar]
- Rossel J.B., Wilson I.W., Pogson B.J. (2002). Global changes in gene expression in response to high light in Arabidopsis. Plant Physiol. 130: 1109–1120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J., Russell D.W. (2001). Molecular Cloning: A Laboratory Manual. (Cold Spring Harbor, NY: Cold Spring Harbor Laboratory) [Google Scholar]
- Schiffmann E., Shemin D. (1957). Further studies on the utilization of aminolevulinic acid for porphyrin synthesis. J. Biol. Chem. 225: 623–628 [PubMed] [Google Scholar]
- Schmittgen T.D., Livak K.J. (2008). Analyzing real-time PCR data by the comparative C(T) method. Nat. Protoc. 3: 1101–1108 [DOI] [PubMed] [Google Scholar]
- Srivastava A., Lake V., Nogaj L.A., Mayer S.M., Willows R.D., Beale S.I. (2005). The Chlamydomonas reinhardtii gtr gene encoding the tetrapyrrole biosynthetic enzyme glutamyl-trna reductase: structure of the gene and properties of the expressed enzyme. Plant Mol. Biol. 58: 643–658 [DOI] [PubMed] [Google Scholar]
- Takahashi S., Masuda T. (2009). High throughput heme assay by detection of chemiluminescence of reconstituted horseradish peroxidase. Comb. Chem. High Throughput Screen. 12: 532–535 [DOI] [PubMed] [Google Scholar]
- Tanaka R., Tanaka A. (2007). Tetrapyrrole biosynthesis in higher plants. Annu. Rev. Plant Biol. 58: 321–346 [DOI] [PubMed] [Google Scholar]
- Tanaka R., Yoshida K., Nakayashiki T., Masuda T., Tsuji H., Inokuchi H., Tanaka A. (1996). Differential expression of two hemA mRNAs encoding glutamyl-tRNA reductase proteins in greening cucumber seedlings. Plant Physiol. 110: 1223–1230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tognetti V.B., Palatnik J.F., Fillat M.F., Melzer M., Hajirezaei M.R., Valle E.M., Carrillo N. (2006). Functional replacement of ferredoxin by a cyanobacterial flavodoxin in tobacco confers broad-range stress tolerance. Plant Cell 18: 2035–2050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ujwal M.L., McCormac A.C., Goulding A., Kumar A.M., Söll D., Terry M.J. (2002). Divergent regulation of the HEMA gene family encoding glutamyl-tRNA reductase in Arabidopsis thaliana: Expression of HEMA2 is regulated by sugars, but is independent of light and plastid signalling. Plant Mol. Biol. 50: 83–91 [DOI] [PubMed] [Google Scholar]
- Vothknecht U.C., Kannangara C.G., von Wettstein D. (1998). Barley glutamyl tRNAGlu reductase: mutations affecting haem inhibition and enzyme activity. Phytochemistry 47: 513–519 [DOI] [PubMed] [Google Scholar]
- Walter M., Chaban C., Schütze K., Batistic O., Weckermann K., Näke C., Blazevic D., Grefen C., Schumacher K., Oecking C., Harter K., Kudla J. (2004). Visualization of protein interactions in living plant cells using bimolecular fluorescence complementation. Plant J. 40: 428–438 [DOI] [PubMed] [Google Scholar]
- Wanker E.E., Rovira C., Scherzinger E., Hasenbank R., Wälter S., Tait D., Colicelli J., Lehrach H. (1997). HIP-I: A huntingtin interacting protein isolated by the yeast two-hybrid system. Hum. Mol. Genet. 6: 487–495 [DOI] [PubMed] [Google Scholar]
- Weigel D., Glazebrook J. (2002). Arabidopsis. A Laboratory Manual. (Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press) [Google Scholar]
- Woods R.A., Gietz R.D. (2001). High-efficiency transformation of plasmid DNA into yeast. Methods Mol. Biol. 177: 85–97 [DOI] [PubMed] [Google Scholar]
- Ytterberg A.J., Peltier J.B., van Wijk K.J. (2006). Protein profiling of plastoglobules in chloroplasts and chromoplasts. A surprising site for differential accumulation of metabolic enzymes. Plant Physiol. 140: 984–997 [DOI] [PMC free article] [PubMed] [Google Scholar]