This study identifies cinnamyl alcohol dehydrogenase as the predominant enzyme that catalyzes the final step in S lignin biosynthesis in woody angiosperm xylem and challenges the widely held view that sinapyl alcohol dehydrogenase regulates S lignin biosynthesis in angiosperms.
Abstract
The manipulation of lignin could, in principle, facilitate efficient biofuel production from plant biomass. Despite intensive study of the lignin pathway, uncertainty exists about the enzyme catalyzing the last step in syringyl (S) monolignol biosynthesis, the reduction of sinapaldehyde to sinapyl alcohol. Traditional schemes of the pathway suggested that both guaiacyl (G) and S monolignols are produced by a single substrate-versatile enzyme, cinnamyl alcohol dehydrogenase (CAD). This was challenged by the discovery of a novel sinapyl alcohol dehydrogenase (SAD) that preferentially uses sinapaldehyde as a substrate and that was claimed to regulate S lignin biosynthesis in angiosperms. Consequently, most pathway schemes now show SAD (or SAD and CAD) at the sinapaldehyde reduction step, although functional evidence is lacking. We cloned SAD from tobacco (Nicotiana tabacum) and suppressed it in transgenic plants using RNA interference–inducing vectors. Characterization of lignin in the woody stems shows no change to content, composition, or structure, and S lignin is normal. By contrast, plants additionally suppressed in CAD have changes to lignin structure and S:G ratio and have increased sinapaldehyde in lignin, similar to plants suppressed in CAD alone. These data demonstrate that CAD, not SAD, is the enzyme responsible for S lignin biosynthesis in woody angiosperm xylem.
INTRODUCTION
The biotechnological manipulation of lignin biosynthesis for efficient biofuel production from plant lignocellulosic biomass offers significant commercial and societal benefits. However, despite more than 20 years of intensive study originally aimed at improving plant raw materials for paper pulping and as animal forage, critical aspects of the lignin biosynthetic pathway are still uncertain. A major current issue is to fully understand how syringyl (S) lignin is made. Early versions of the lignin biosynthetic pathway indicated that the hydroxylation and methylation reactions converting guaiacyl (G) lignin precursors into S lignin precursors occurred at the level of the cinnamic acids by conversion of ferulate into sinapate (Higuchi, 1985). A major advance has been the more recent realization that, in fact, these reactions preferentially occur at a later position in the pathway, most likely converting coniferaldehyde into sinapaldehyde, the direct precursor of the S lignin monomer sinapyl alcohol (Chen et al., 1999; Humphreys et al., 1999; Osakabe et al., 1999; Li et al., 2000). Although most of the data prompting this revision came from enzyme functional studies performed in vitro, it is completely consistent with the results of in planta manipulation of the expression of genes catalyzing these reactions; ferulate 5-hydroxylase (F5H; also named coniferaldehyde 5-hydroxylase or Cald5H) and caffeate O-methyltransferase (COMT, alternatively called 5-hydroxyconiferaldehyde O-methyltransferase or AldOMT) (Baucher et al., 2003). Thus, coniferaldehyde is now accepted as being a key entry point into S lignin biosynthesis in angiosperms, providing the precursor for sinapaldehyde production by F5H and COMT. Attention has subsequently been focused on the identity of the enzyme catalyzing the last step in S monolignol biosynthesis, the reduction of sinapaldehyde to sinapyl alcohol.
For a long time, it was believed that both G and S monolignols (coniferyl and sinapyl alcohol), were produced by reduction of coniferaldehyde and sinapaldehyde via the action of a single enzyme, cinnamyl alcohol dehydrogenase (CAD). The discovery in aspen (Populus tremuloides) of a novel sinapyl alcohol dehydrogenase (SAD), related to but distinct from CAD, that is expressed during S lignin synthesis and that preferentially uses sinapaldehyde as a substrate, called this belief into question (Li et al., 2001). The claim that the last step of syringyl monolignol biosynthesis in angiosperms is regulated by this novel SAD gene was persuasive and has since proliferated in the literature such that most pathway diagrams now indicate SAD to be active in the conversion of sinapaldehyde to sinapyl alcohol (Boerjan et al., 2003; Ko and Han, 2004; Jørgensen et al., 2005; Shi et al., 2010; Umezawa, 2010).
Despite the attractiveness of this theory, several groups have challenged the existence of a specific SAD, particularly in Arabidopsis thaliana. It is notable that a comprehensive examination of the kinetic properties and substrate preferences of all CAD-like genes in Arabidopsis revealed that several had activity against sinapaldehyde but that none had a specific requirement for it (Kim et al., 2004). However, Arabidopsis has been criticized as a model for wood formation (Shi et al., 2010), because wood is not readily obtained from it, and its guaiacyl-rich lignin composition is not typical of that of woody angiosperms, such as aspen. Nevertheless, even when comparing across species, the subfamily of enzymes to which SAD is most closely related share certain key determinants but exhibit diverse substrate specificities, using a variety of hydroxycinnamaldehydes and benzaldehydes with varying efficiencies, and not all are competent to use sinapaldehyde (Bomati and Noel, 2005). Most importantly, independent studies, many predating the discovery of aspen SAD, indicate that both coniferaldehyde and sinapaldehyde accumulate in lignin in CAD-suppressed transgenic or mutant plants, suggesting that CAD and not SAD is responsible for sinapaldehyde reduction during lignification (Halpin et al., 1994; Ralph et al., 1998, 2001; Kim et al., 2002; Sibout et al., 2003, 2005).
Unfortunately, all of the evidence either supporting or contradicting the existence of an S lignin-specific SAD is circumstantial or indirect, based either on enzyme functional data obtained in vitro or on the effects of CAD-deficiency in vivo. Both types of data can be misleading. Enzyme assays in vitro cannot reproduce the complex and compartmentalized interactions that occur within a living cell and can only suggest the likely specificities of a given enzyme in vivo. Similarly, most descriptions of CAD-deficient plants include no analysis of the knock-on effects on the expression of other genes, such as SAD, and it is possible that some of the metabolic changes in these plants are secondary effects unrelated to the specific catalytic function of CAD.
Direct evidence on the role of SAD, obtained from plants where the gene has been knocked down or mutated, is needed to clarify the important issue of its potential involvement in lignin biosynthesis. This is important both to ensure accuracy in the literature and particularly to correctly inform efforts to exploit the potential for manipulating lignin to improve plant biomass for bioenergy applications. Several studies already illustrate that genetic manipulation of lignin can have beneficial effects on the release of fermentable sugars for biofuel production (Chen and Dixon, 2007; Gomez et al., 2010). Resolution of discrepancies in the literature and production of accurate schemes for the lignin biosynthetic pathway are critical to facilitate further useful lignin modification and to improve our understanding of complex metabolic systems in plants, of which the lignin biosynthetic grid is one of the best studied examples. In this article, we describe the existence, expression, and cloning of SAD in tobacco (Nicotiana tabacum), a true wood-producing angiosperm that closely parallels poplar (Populus spp) and aspen in the effects of genetic manipulations to lignin biosynthetic genes (Franke et al., 2000; O’Connell et al., 2002; Ralph et al., 2008a). We have used RNA interference to severely suppress SAD expression in tobacco and have used these plants to directly evaluate the proposed role of SAD in S lignin biosynthesis.
RESULTS
Tobacco Contains a SAD Similar to Aspen SAD
Tobacco is an ideal model woody plant for rapid determination of the roles of various genes in lignification using reverse genetics strategies. To ultimately use such a strategy to illuminate the involvement of SAD in lignin biosynthesis in angiosperms, we first set out to investigate the existence of a SAD in lignifying tissue in tobacco using an antiserum raised against aspen SAD. This antiserum was previously shown to recognize SAD in a wide range of angiosperm species (Li et al., 2001). Preliminary studies demonstrated that the aspen SAD antiserum also recognized a specific protein band in tobacco stem extracts (see Supplemental Figure 1 online). To determine whether the tobacco SAD protein had a localization pattern similar to that of aspen SAD and consistent with a role in lignification, SAD and CAD localization studies were performed on tobacco stem sections. To ensure that only native SAD was being detected by the SAD antiserum, SAD localization studies were also performed on CAD-deficient stems (CAD antisense plants) and yielded identical results to those reported here for the wild-type stems. Cross sections (70 to 100 μm) of stem were visualized by immunofluorescence microscopy after incubation with rabbit antiserum raised against either (a) tobacco CAD or (b) aspen SAD followed by Alexa Fluor 594-conjugated goat anti-rabbit IgG secondary antibody. Control sections incubated with nonimmunized rabbit serum gave no fluorescence signal. Sections incubated with anti-CAD or anti-SAD serum revealed that both proteins predominantly localize to the ray parenchyma cells in xylem, which can be identified by reference to a phloroglucinol-stained stem section (Figure 1A). In the cambial and phloem area of the sections, CAD (Figure 1B) is restricted to parenchyma cells extending outward from the rays and forming an outer border surrounding bundles of phloem cells that are not labeled. By contrast, SAD (Figure 1C) has a more uniform distribution in this area, suggesting that, at this stage of stem maturation, SAD is also expressed in phloem. The localization of tobacco CAD to parenchyma cells that provide lignin precursors to adjacent xylem vessels and xylem and phloem fibers corresponds with the previously determined expression pattern of the eucalyptus (Eucalyptus gunnii) CAD promoter (Feuillet et al., 1995) and aspen CAD protein (Li et al., 2001). The overlapping localization of tobacco SAD to the same cell types and additionally to phloem cells is consistent with a potential role in lignin biosynthesis and corresponds to the localization pattern reported for SAD in the sixth internode of aspen (Li et al., 2001).
Figure 1.
CAD and SAD Localization in Tobacco Stems.
Cross sections of tobacco stem (A) stained with phloroglucinol or incubated with antiserum raised against (B) tobacco CAD, (C) aspen SAD, or (D) nonimmunized rabbit serum and visualized by immunofluorescence microscopy. p, phloem; pf, phloem fiber; rp, ray parenchyma; vc, vascular cambium.
Bars = 100 μm.
Cloning and Phylogenetic Analysis of Tobacco SAD
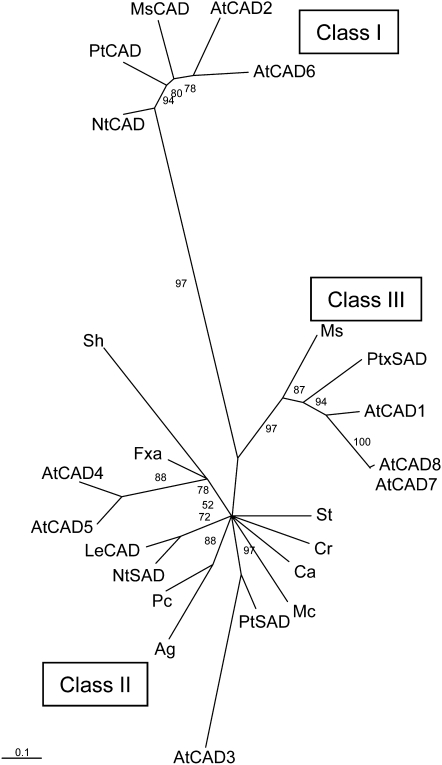
To enable us to design primers to facilitate cloning of the tobacco SAD, a BLAST search (Altschul et al., 1997) was performed using the aspen SAD protein sequence to identify related sequences in protein databases. Many SAD-like sequences were identified, including the protein products of three Elicitor-inducible (Eli3) genes (two from Arabidopsis and one from tomato [Solanum lycopersicum]), which showed high sequence identity (~70%) to aspen SAD. The SAD and SAD-like nucleotide sequences were aligned with four CAD sequences (from tobacco, tomato, eucalyptus, and alfalfa [Medicago sativa]), and regions of sequence conserved in the SADs but not in the CADs were chosen for degenerate primer design (see Methods). The products amplified from tobacco genomic DNA with these primers were used to design gene-specific primers for Rapid Amplification of cDNA Ends of cDNA prepared from 8-week-old stem xylem RNA. Finally, specific primers were designed to the ends of the obtained cDNA-derived fragments and were used to amplify entire cDNAs. Five closely related cDNAs were isolated. In amino acid sequence, these cDNAs showed 91 to 96% identity to each other, 71 to 75% identity to aspen SAD, and only 54% amino acid identity to tobacco CAD. Overall, the extremely similar Nicotiana tabacum (Nt) SAD2 and Nt SAD4 protein sequences (>96% identical) are most closely related to that of aspen SAD. It is common for tobacco to contain two near-identical copies of a gene, one homeolog inherited from each of the two ancestral progenitor species of the tobacco allotetraploid genome. Subsequent BLAST searches of the recently released tobacco genome sequence revealed no additional genes with a closer relationship to aspen SAD than the Nt SAD genes described here. Thus, if all angiosperms require a specific SAD for lignification, as proposed by Li et al. (2001), the Nt SAD genes are the only obvious candidate orthologs of aspen SAD in the tobacco genome.
Phylogenetic analysis using the Tree-Puzzle software (Schmidt et al., 2002) reveals that all five tobacco SAD proteins (for clarity, illustrated by one label, Nt SAD) and aspen SAD (Populus tremuloides [Pt] SAD) cluster together on a branch that is distinct from that containing the “true” CADs involved in developmental lignification, denoted class I CADs by Raes et al. (2003) (Figure 2; see Supplemental Data Set 1 online). Both the tobacco and aspen SADs cluster with the class II CADs, a group of enzymes with diverse substrate preferences that includes several proteins associated with defense responses, such as the elicitor-inducible Eli3 proteins from Arabidopsis, celery (Apium graveolens), and parsley (Petroselinum crispum). The phylogeny supports the evolutionary relationship between aspen and tobacco SAD proteins and confirms that the genes we have isolated from tobacco are the true orthologs of the aspen gene.
Figure 2.
Phylogenetic Tree of CAD and SAD Proteins.
CAD and SAD protein sequences were aligned using Clustal W (see Supplemental Data Set 1 online), and a phylogenetic tree was produced using a maximum likelihood method (Tree-Puzzle 5.0) with quartet puzzling support values. The amino acid substitution matrix used was JTT. Rate heterogeneity among sites was modeled using a gamma distribution with four categories. The tree was viewed in Tree-View v.1.6.6. The three major clades, denoted Class I, Class II, and Class III CADs by Raes et al. (2003),are indicated. Arabidopsis CAD genes are named according to Raes et al. (2003). Ag, Apium graveolens; At, Arabidopsis thaliana; Ca, Camptotheca acuminate; Cr, Catharanthus roseus; Fxa, Fragaria × ananassa; Le, Solanum lycopersicum; Mc, Mesembryanthemum crystallinum; Ms Medicago sativa; Nt, Nicotiana tabacum; Pc, Petroselinum crispum; Pt, Populus tremuloides; Ptx, Populus tremula × Populus tremuloides; Sh, Stylosanthes humilis; St, Solanum tuberosum.
Bar = 0.1 amino acid substitutions.
Homology Modeling of Nt SAD2 and Nt SAD5
Homology models of SAD2 and SAD5 were generated using the Phyre Web server (Kelley and Sternberg, 2009) using the high-resolution crystal structure of Pt SAD as the template (Bomati and Noel, 2005). The model of SAD2 suggests that the active site is, in essence, identical to that of the aspen SAD enzyme. Four essential residues, Cys-50, Ser-52, His-72, and Cys-166, identified by Bomati and Noel (2005) as necessary for tetrahedral coordination of the catalytic Zn2+ ion plus a coordinating water molecule (see Supplemental Figure 2 online), are strictly conserved in the SAD2 (and the homologous Nt SAD4) active site. The high degree of conservation also extends to residues involved in binding the cofactor and in forming a network of interactions in and around the active site. By contrast, the model of SAD5 reveals significant active site differences caused by the substitution of His-72 and Cys-166 (Cys-165 in SAD2) with Tyr-72 and Asn-165 of SAD5. The model of SAD5 predicts that the hydroxyl group of Tyr-72 would be positioned precisely where the catalytic Zn2+ binds in Pt SAD, and by implication also in SAD2 (see Supplemental Figure 2 online). These alterations therefore render it highly unlikely that SAD5 (and the similar Nt SAD1 and Nt SAD3 enzymes) can function in sinapaldehyde reduction. Thus, only SAD2 (and Nt SAD4) are likely to be functional orthologs of the aspen SAD enzyme, whereas Nt SAD1, Nt SAD3, and Nt SAD5 are merely “SAD-like” and were subsequently denoted as Nt SAD-L1, Nt SAD-L3, and Nt SAD-L5 when sequences were submitted to the Genbank database.
The homology models reveal active site residue substitutions in tobacco SADs similar to those that distinguish aspen SAD from classical CAD-like enzymes. According to the models of Bomati and Noel (2005), the two families of enzymes have complementary but opposite geometric substitution patterns for key active site aromatic residues. Aspen SAD has bulky aromatic residues (Trp-61 and Phe-289) on the left wall of the active site and small hydrophobic residues (Leu-122 and Gly-302) at the base and on the right wall of the active site. By contrast, aspen CAD has hydrophobic residues (Leu-61 and Pro-289) on the left wall and bulky aromatic residues (Trp-122 and Phe-302) on the right wall and at the base of the active site. These substitution patterns completely change the topology of the active site and may contribute to different substrate binding modes in classical CADs and SAD-like enzymes (Bomati and Noel, 2005). Tobacco SAD2 totally adheres to the configuration of residues expected in the active site of a SAD enzyme with bulky aromatic residues on the left wall (Trp-61 and Phe-288, exactly as in aspen SAD) and hydrophobic residues (Met-122 and Ala-301) at the base and right wall of the active site.
Substrate Preference of Tobacco CAD and SAD
The relative substrate preferences of tobacco CAD and SAD for coniferaldehyde and sinapaldehyde were evaluated by assaying the activity of the purified recombinant proteins after expression of CAD19 (encoding lignification-related tobacco CAD19), SAD2, and SAD-L5 coding sequences in Escherichia coli. Consistent with the predictions of the molecular modeling, SAD-L5 had no detectable activity toward coniferaldehyde or sinapaldehyde. By contrast, CAD19 and SAD2 exhibited activity toward both coniferaldehyde and sinapaldehyde. Although the Km values were considerably lower for CAD19 than they were for SAD2, neither enzyme showed a clear preference for either substrate, as indicated by similar Kcat/Km values (Table 1). Comparison of the tobacco SAD with the previously described aspen SAD (Li et al., 2001) revealed that the turnover number (Kcat) for sinapaldehyde substrate was virtually identical (4.3 s−1 for tobacco SAD and 200 min−1 or 3.3 s−1 for the aspen SAD). Thus, in addition to a close phylogenetic relationship to the aspen SAD gene and the high immunological and amino acid sequence identity between the SAD2 and aspen SAD proteins, the tobacco enzyme can also use both coniferaldehyde and sinapaldehyde as substrates, confirming that it is a true functional ortholog.
Table 1.
Kinetic Properties of Recombinant Tobacco CAD and SAD
| Enzyme | Substrate | Km (μM) | Vmax (μmol/min/mg) | Kcat (s−1) | Vmax/Km | Kcat/Km |
| CAD | Coniferaldehyde | 5.44 ± 1.32 | 25.3 ± 1.72 | 17.6 | 4.65 | 3.23 |
| CAD | Sinapaldehyde | 3.41 ± 1.11 | 18.1 ± 1.56 | 12.6 | 5.32 | 3.70 |
| SAD | Coniferaldehyde | 86.8 ± 19.8 | 3.12 ± 0.263 | 2.18 | 0.0359 | 0.0251 |
| SAD | Sinapaldehyde | 102 ± 19.8 | 6.19 ± 0.312 | 4.33 | 0.0607 | 0.0425 |
Data are means and se corresponding to triplicate analyses. CAD19 and SAD2 were expressed in E. coli, and the purified proteins were assayed using coniferaldehyde and sinapaldehyde as substrates. Values are means ± se for three independent reactions.
Production of Plants Suppressed in SAD Expression
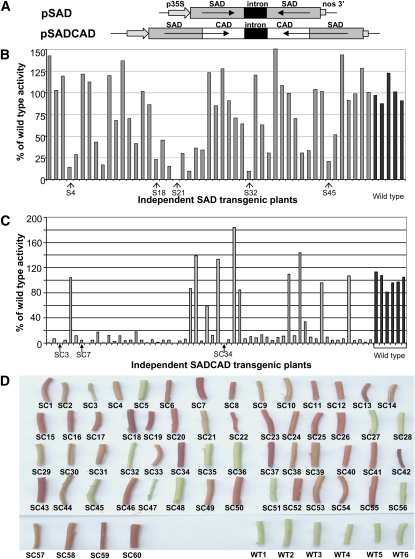
To directly assess the role of SAD in lignin biosynthesis via reverse genetics, two hairpin RNA-producing gene-silencing vectors (Wesley et al., 2001) were designed to suppress expression of (i) SAD and (ii) SAD and CAD simultaneously. Both constructs should target all five tobacco SAD genes for suppression because of the high degree of homology between them. For the coordinate suppression of SAD and CAD, we used a strategy previously tested by us in conventional nonhairpin producing vectors where fusing partial sense sequences for different genes within an expression cassette was sufficient to effect suppression of multiple targets (Abbott et al., 2002). The simultaneous suppression of both SAD and CAD was performed to assess whether SAD and CAD might act redundantly to convert sinapaldehyde to sinapyl alcohol in lignin biosynthesis. For the preparation of the vectors, a 619-bp partial SAD cDNA was introduced alone, or fused to a 586-bp partial CAD cDNA, into a donor vector (pDONR201) to make an entry clone before being recombined into the gene-silencing vector (pHellsgate8). The resultant constructs are shown in Figure 3A. These were introduced into tobacco via Agrobacterium-mediated transformation. More than 50 independent transgenic plants were regenerated for each construct, and those transformed with the pSAD construct were called “SAD” or “S” plants, whereas those transformed with the pSADCAD construct were denoted as “SAD CAD” or “SC” plants.
Figure 3.
Production and Characterization of SAD and SAD CAD Transgenics.
(A) Constructs introduced into tobacco to suppress expression of SAD (pSAD) or of SAD and CAD simultaneously (pSADCAD).
(B) Sinapaldehyde-reducing activity in primary transformants harboring the pSAD construct and wild-type plants. Plants selected for further study are indicated by arrows (S4, S18, S21, S32, S45).
(C) Sinapaldehyde-reducing activity in primary transformants harboring the pSADCAD construct and in wild-type plants. Plants selected for further study are indicated by arrows (SC3, SC7, SC34).
(D) Color of woody xylem of stem sections of SAD CAD and wild-type plants after removal of outer epidermal and cortical tissues. Plant number is shown underneath each section.
Plants were grown for 4 weeks in tissue culture followed by 2 weeks in the greenhouse before stem extracts were used for early evaluation of SAD and CAD suppression. We were aware that results of enzyme assays alone might be hard to interpret fully, because both SAD and CAD can use sinapaldehyde (see above), and there is evidence of substantial substrate-mediated kinetic inhibition of both enzymes (Li et al. 2001; Bomati and Noel, 2005). Nevertheless, we rationalized that effective gene suppression might still be evident in reduced enzyme activity, even if the degree of that reduction was masked by other activities, and so enzyme assays were performed on the entire SAD and SAD CAD populations as an initial screen.
Of 49 SAD plants initially assayed, 23 had clearly diminished sinapaldehyde-reducing activity, with seven having a more than 80% reduction. Five independent plants with residual sinapaldehyde-reducing activity ranging from 1 to 22% of the wild type were selected for further study (Figure 3B, plants S4, S18, S21, S32, and S45). Activity was even more suppressed in the SAD CAD plant population, in which both enzymes capable of reducing sinapaldehyde should be depleted. From a total of 60 plants assayed, 48 plants had more than 80% lower sinapaldehyde-reducing activity compared with the wild-type plants (Figure 3C). Coniferyl alcohol dehydrogenase activity (see Supplemental Figure 3 online) followed a similar pattern, being severely suppressed in the 48 plants that showed low sinapaldehyde reduction and remaining high in plants with high sinapaldehyde-reducing activity. The assay data were visually reinforced by the unusual red color in the woody xylem of these 48 plants (Figure 3D). This phenotype is well-documented as being characteristic of plants with severely suppressed CAD activity (Halpin et al., 1994; Pilate et al., 2002) and has been suggested to result from colored products of the CAD substrates, coniferaldehyde, and/or sinapaldehyde (Higuchi et al., 1994; Ralph et al., 2008a, 2008b). By comparison, none of the plants suppressed in SAD alone (i.e., transformed with the pSAD construct) showed any change to xylem color compared with the wild type. Three plants showing various degrees of color from pink (plant SC3) to red (SC7, SC34), and in which both SAD and CAD activity were reduced to below 10% of normal levels, were selected for further study. All subsequent analyses were performed on populations of progeny of each of the selected SAD and SAD CAD plants.
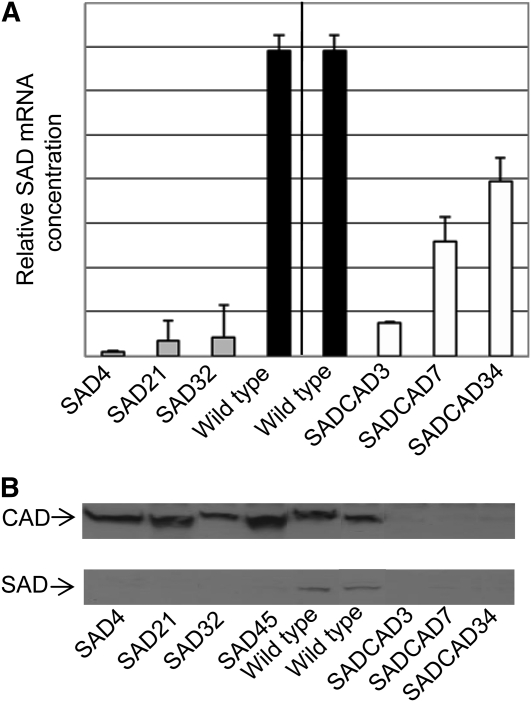
Given the potential complications inherent in enzyme assays of crude protein extracts, particularly the fact that assays performed in vitro may not truly differentiate between CAD and SAD, because both enzymes can use the same substrates, we performed two further tests. Both RT-PCRs and immunoblots were used to confirm the identity of the genes/proteins suppressed in a selection of the SAD and SAD CAD plants. Real-time RT-PCR performed with primers specific for the five SAD genes or for CAD confirmed that SAD transcripts were reduced in all SAD and SAD CAD lines evaluated (Figure 4A). In the SAD plant lines, SAD transcript levels were reduced by 93 to 98% (indicating, as expected, that all five closely related genes were suppressed), whereas in the SAD CAD lines, SAD transcripts were higher but nonetheless considerably reduced compared with the levels in the wild-type plants. CAD transcripts were also reduced in the SAD CAD lines compared with the wild type (see Supplemental Figure 4 online), confirming the enzyme assay data. Immunoblots (Figure 4B) confirmed that SAD and CAD protein levels were similarly altered. The SAD protein band evident in wild-type tobacco stems was hardly detectable in extracts from the SAD and SAD CAD plants, whereas levels of CAD protein were normal in SAD plants but greatly reduced in SAD CAD plants. These data confirm that we have successfully and severely suppressed expression of the gene that encodes the tobacco xylem protein that corresponds immunologically to aspen SAD.
Figure 4.
SAD and CAD Expression in SAD and SAD CAD Lines.
(A) Relative SAD mRNA concentration in stems of the wild-type, SAD, and SAD CAD plants evaluated by real time RT-PCR. Means and sd for three separate experiments on independent plants of three SAD and three SAD CAD lines were calculated using the standard curve method (User Bulletin no. 2, ABI PRISM 7700 Sequence Detection System software).
(B) Immunoblots of stem extracts from the wild-type, SAD, and SAD CAD lines probed with antiserum raised against tobacco CAD (Top) or aspen SAD (Bottom).
SAD-Suppressed Plants Have No Detectable Changes in Lignin
The phenotypic consequences of SAD suppression were monitored throughout development. Some SAD and SAD CAD lines initially grew more slowly than wild-type plants, but average heights after 10 weeks of growth were normal except for lines SAD CAD7 and SAD CAD34, which were 10 to 20% shorter than the wild type. Leaves of some lines showed slight loss of symmetry, whereas roots seemed longer and less colored than wild-type roots. The basis of these phenotypes is under further study.
The effect of SAD suppression on lignin biosynthesis was investigated by several complementary techniques. Because of the complex and heterogenous structure of lignin, no single technique yields complete, unbiased data, and several methods need to be used to get representative data on the content, structure, and composition of the polymer (Halpin, 2004).
Quantitative analysis of the lignin content of woody xylem tissues was determined by the gravimetric Klason method and by a complementary spectrophotometric procedure that relies on the solubilization of lignin, the acetyl bromide procedure. Both of these lignin determinations were performed on extractive-free xylem (EXR). Klason analysis revealed a lignin content of approximately 22% in both the wild-type and transgenic extractive-free stems, although line SAD CAD34 had a slight reduction in Klason lignin (Table 2). Small decreases in Klason lignin have previously been seen in some lines of tobacco suppressed in CAD alone (Halpin et al., 1994, see line 40). Acetyl bromide determinations on SAD and SAD CAD plants revealed no appreciable difference in lignin content between the wild type and the transgenic lines.
Table 2.
Lignin Content and Structure in the Wild Type and Transgenic Lines
| Line | Klason lignin KL (% DW) | AcBr Lignin (A280/gDW) | Alkali (A280/gDW) | Thioacidolysis Total Yield (μmol/g Klason Lignin) | S:G Thioacidolysis Molar Ratio |
| Wild type | 22.44 ± 0.40 | 91.61 ± 4.11 | 58.81 ± 2.02 | 1241 ± 119 | 1.03 ± 0.03 |
| SAD S4 | 22.90 ± 0.11 | 99.43 ± 1.84 | 66.31 ± 1.31 | 1222 ± 77 | 0.99 ± 0.02 |
| SAD S18 | 21.75 ± 0.46 | 88.94 ± 3.25 | 57.20 ± 2.36 | 1271 ± 56 | 1.09 ± 0.56 |
| SAD S21 | 21.92 ± 0.21 | ND | ND | 1287 ± 67 | 1.03 ± 0.02 |
| SAD S32 | 22.10 ± 0.25 | ND | ND | 1204 ± 58 | 1.02 ± 0.05 |
| SAD S45 | 22.23 ± 0.45 | ND | ND | 1315 ± 145 | 1.05 ± 0.04 |
| SAD CAD SC3 | 22.35 ± 0.18 | 100.93 ± 1.86 | 105.05 ± 4.44 | 975 ± 21 | 0.61 ± 0.03 |
| SAD CAD SC7 | 21.54 ± 0.45 | 108.99 ± 0.60 | 155.59 ± 13.36 | 641 ± 115 | 0.46 ± 0.08 |
| SAD CAD SC34 | 20.81 ± 0.52 | 85.04 ± 3.98 | 149.75 ± 17.03 | 875 ± 140 | 1.10 ± 0.09 |
Data are means and se corresponding to biological triplicates (three plants per line). Klason and acetyl bromide lignins were determined on EXR samples collected from the base of the stems of 10-week-old tobacco plants. Thioacidolyses are run from the same EXR samples, and the yields are expressed in μmoles per gram of Klason lignin. AcBR, acetyl bromide; DW, dry weight; gDW, g dry weight; ND, not determined.
Mild alkaline hydrolysis of the EXR can reveal information about the structure of the lignin polymer, because only a proportion of the linkages within lignin are susceptible to breakage under these conditions. The proportion of lignin structures that could be degraded by such alkali treatment was similar in the wild type and both SAD lines tested (S4 and S18), according to the absorbance values of the lignin-derived low molecular weight phenolics that were released (Table 2). By comparison, all three SAD CAD lines (SC3, SC7, and SC34) had significantly increased susceptibility of lignin to alkaline degradation, similar to the increased alkali solubility previously described in CAD antisense plants (Halpin et al., 1994; Baucher et al., 1996; Vailhé et al., 1998) (Table 2).
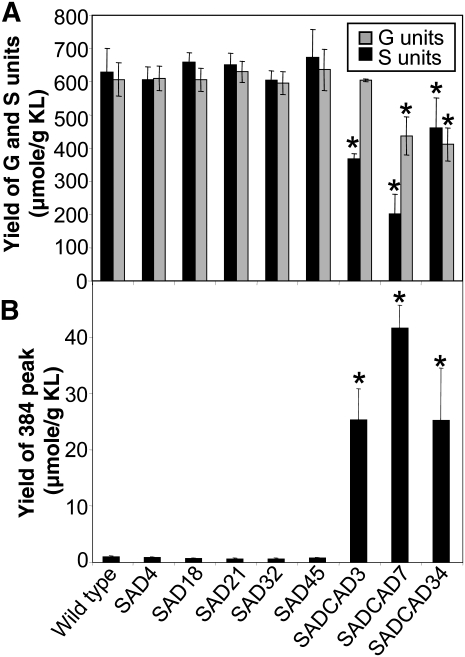
Lignin composition was investigated by thioacidolysis, a procedure that cleaves β-O-4 bonds in lignin. A similar yield of thioacidolysis monomers was released from extractive-free stems of SAD lines and wild-type plants, indicating a similar frequency of lignin β-O-4 bonds in both (Table 2). The amounts of S and G units recovered were unchanged in SAD lines compared with the wild-type plants (Figure 5), and the S:G ratio was close to 1 in both cases (Table 2). Indeed, no differences in either major or minor thioacidolysis products could be detected between SAD-suppressed lines and wild-type plants by this analysis. By contrast, SAD CAD lines differed from the wild type in several ways. Thioacidolysis yields were reduced in SAD CAD lines, indicating a change in the structure of the lignin and fewer β-O-4 bonds (Table 2). The amounts of thioacidolysis S monomers recovered from lignins were significantly reduced in all three SAD CAD lines, whereas the amounts of G monomers were reduced in two of the lines (Figure 5A). The greater proportional decrease in S unit yield compared with G unit yield means that the S:G thioacidolysis ratio is greatly reduced for the SAD CAD3 and SAD CAD7 lines (Table 2). Both reductions in thioacidolysis yield and a halving of S:G thioacidolysis ratio have been previously documented in plants suppressed in CAD alone (Chabannes et al., 2001). Indeed, the SAD CAD lines presented the typical lignin profile of plants deficient in CAD activity. Thus, according to a range of standard analyses, no changes to lignin were detected in severely SAD-suppressed plants, whereas plants suppressed in both SAD and CAD displayed only the changes to lignin structure and composition that have been previously described for plants suppressed in CAD alone. Our data therefore did not detect any evidence for mutually redundant roles of CAD and SAD in lignin biosynthesis.
Figure 5.
Thioacidolysis S and G Monomers and 384 Sinapaldehyde-Derived Compound in Lignin from SAD, SAD CAD, and the Wild-Type Plants.
Yields of thioacidolysis products were determined on EXR samples collected from the stem base of 10-week-old plants. The data are means and se of three plants per line.
(A) Yields of G and S lignin monomers.
(B) Yields of m/z 384 peak derived from sinapaldehyde incorporated into lignin. * significantly different to wild-type plants (P < 0.05).
Sinapaldehyde Accumulates in Lignin in SAD CAD but Not SAD Lines
The substrate for SAD’s proposed role in generating sinapyl alcohol for S lignin biosynthesis is sinapaldehyde, and one might therefore expect sinapaldehyde to accumulate when SAD activity is suppressed. Accumulating indene derivatives (peaks at mass-to-charge ratio [m/z] 384 and m/z 354) originating from sinapaldehyde and coniferaldehyde have previously been detected incorporated into lignin in the thioacidolysis products of CAD-antisense plants but are below the detection level in wild-type plants (Kim et al., 2002). Our thioacidolysis analysis of lignin in SAD and SAD CAD plants indicates that significant amounts of the sinapaldehyde indene derivative were present in the thioacidolysis products of all SAD CAD lines but that this was almost entirely absent in all of the SAD lines and in wild-type plants (Figure 5B). Similarly, the coniferaldehyde indene derivative was present in significant amounts only in lignin of the SAD CAD lines (see Supplemental Table 1 online). These data demonstrate that only plants suppressed in CAD activity accumulate sinapaldehyde-derived (and coniferaldehyde-derived) products in lignin. We must conclude that, although SAD is capable of reducing sinapaldehyde to sinapyl alcohol when fractionated crude plant extracts are assayed in vitro, within the complex and largely unknown spatial and metabolic organization of the lignifying cell, it is predominantly CAD that uses sinapaldehyde as a substrate for lignin biosynthesis in vivo.
Phenolic Profiling of SAD-Suppressed Tobacco Plants
Targeted metabolite profiling was performed on SAD plants to determine (a) whether intracellular sinapaldehyde pools were increased and (b) whether altered phenolic profiles would indicate which pathways were blocked by the deficiency in SAD. A comparative ultraperformance liquid chromatography-mass spectrometry (UPLC-MS) profiling of soluble phenolics was performed on methanol extracts from S4, S45, and wild-type plants. Analysis of extracts from stem xylem, cortex (i.e., soft tissues outside the woody xylem), leaves, and roots quantified 497, 867, 948, and 1471 m/z peaks, respectively. Data were subjected to rigorous statistical analysis using both principle component analysis and Student’s t tests. Only metabolites identified as differentially expressed by both statistical tests in all transgenic plants (i.e., five replicate plants of each of lines S4 and S45) compared with wild-type extracts were considered further. This joint statistical approach to evaluate the comparative profiling did not reveal any accumulation of sinapaldehyde or sinapic acid or of any other obvious aromatic that might be expected to accumulate if the lignin pathway is perturbed. No robust changes were pinpointed for xylem, bark, or root extracts, but one UPLC-MS peak (peak X) in leaf extracts was reduced by an average of 40% in all SAD plant samples compared with the wild type. Tandem mass spectrometry (MS/MS) fragmentation indicated that peak X is hexosylated dihydrodehydrodiconiferyl alcohol (DDDC) or isodihydrodehydrodiconiferyl alcohol (IDDDC), which may have arisen via reduction of the phenylcoumaran ring or the side-chain double bond of (hexosylated) DDDC (see Supplemental Figure 5 online). Because true differences may have been masked by the stringent significance threshold we initially used, we subsequently pursued a targeted search for hexosylated dehydrodiconiferyl alcohol (DDC) in the leaf profiles, from which hexosylated DDDC or IDDDC is derived. Manual integration of the corresponding chromatogram peak followed by a Student’s t test (α = 0.05) showed a decrease for hexosylated DDC in the transgenic lines, down to 55% of wild-type levels (see Supplemental Figure 5 online). None of these hexosylated phenylcoumarans were observed in xylem, bark, and root tissues, and no peaks associated with the aglycones, DDDC or IDDDC, and DDC, were observed in the leaf profiles. We conclude that SAD could therefore possibly play a role in the production or modification of DDC phenylcoumarans in tobacco.
DISCUSSION
We have cloned two tobacco orthologs of the aspen SAD gene first isolated by Li et al. (2001). No closer relatives of the aspen SAD could be found in the tobacco genome. The tobacco cDNAs encode proteins that share 72% amino acid identity with aspen SAD and are more closely related to it than to tobacco CAD, with which they share only 56% amino acid identity. Like aspen SAD, the tobacco SAD cDNAs were cloned from lignifying xylem, and immunolocalization indicates that tobacco SAD is localized to the same cell types as the aspen protein, most obviously the ray parenchyma cells. Expression of recombinant tobacco SAD shows that, like aspen SAD, it is capable of reducing both sinapaldehyde and coniferaldehyde. Our data therefore confirm the data of Li et al. (2001), demonstrating that a second enzyme distinct from CAD, but with overlapping ability to reduce sinapaldehyde, is expressed in the lignifying xylem of at least some woody dicots.
Although the existence of SAD in lignifying tissues is clear, its function is not. Crude extracts from plants suppressed in expression of the tobacco SADs have decreased capacity to reduce sinapaldehyde when assayed in vitro. Despite this, the biosynthesis of S lignin units proceeds normally in tobacco stem xylem even though SAD transcripts are reduced to 2 to 7% of normal levels and SAD protein is almost undetectable. In addition, metabolite profiling did not reveal accumulation of soluble sinapaldehydes in the SAD plants. Only when CAD is additionally suppressed are significant changes to lignin effected, making the polymer more susceptible to alkali extraction caused by an altered structure with increased incorporation of sinapaldehyde and coniferaldehyde units, reduced proportion of β-O-4 linked units, and reduced S:G ratio. These are well-documented characteristics of CAD suppression (Halpin et al., 1994; Baucher et al., 1996; Vailhé et al., 1998). No additional changes, or changes in the amplitude of these effects that could be assigned to concomitant SAD suppression, could be detected, despite the effectiveness with which both genes were suppressed by the chimeric pSADCAD construct.
To our knowledge, our results, from plants severely suppressed in SAD activity, provide the first direct and conclusive data to confirm other circumstantial evidence that SAD is not needed for the biosynthesis of S lignin units in dicots. Data from many different groups show that the accumulation in lignin of derivatives of sinapaldehyde, the precursor of S monomers, is a characteristic of severe CAD suppression (Halpin et al., 1994; Ralph et al., 1998, 2001; Kim et al., 2002; Sibout et al., 2003); indeed, the level of the indene derivative shows a close inverse correlation with CAD activity (Lapierre et al., 2004). Our data (see Supplemental Figure 1 online) demonstrate that SAD protein levels are not reduced in CAD-antisense tobacco lines that have previously been shown to accumulate sinapaldehyde (Halpin et al., 1994); therefore, reduction in expression of CAD alone is responsible for the accumulation of sinapaldehyde in lignin in these plants. A study of Arabidopsis cad mutants draws a similar conclusion. In double mutants in the lignification-related CAD genes showing large increases in sinapaldehyde in lignin, transcript levels of the genes most closely related to aspen SAD are not significantly altered (Sibout et al., 2005). Indeed, expression of a Populus tremula × tremuloides SAD (99.2% amino acid identity to aspen SAD) in these plants can restore only 18% of the S monomer yield (Sibout et al., 2005). Moreover, some plants may be able to bypass the requirement for a sinapaldehyde reduction step in lignin biosynthesis and alternatively produce sinapyl alcohol from coniferyl alcohol. Evidence for such a pathway comes from radiolabeled precursor feeding studies in several tree species (Chen et al., 1999, Matsui et al., 2000; Tsuji et al., 2004) and from investigation of the substrate specificities of Arabidopsis F5H and COMT (Humphreys et al., 1999). If such a pathway functions in vivo, even if only as a supplementary alternative route, it obviates the original arguments for the necessity of the existence of a specific SAD. Most significantly, a role for SAD in S lignin synthesis is not consistent with a recent attempt (Shi et al., 2010) to provide a comprehensive description of lignin biosynthesis in Populus trichocarpa, a close relative of aspen for which a genome sequence is available (Tuskan et al., 2006). Quantitative criteria were applied to identify, from the 45,555 gene model set of P. trichocarpa genome annotation, the genes most likely to be monolignol biosynthetic genes based on transcript abundance and specificity for differentiating xylem. P. trichocarpa genes involved in S lignin synthesis, CAld5H1 and COMT2 (encoding F5H and COMT, respectively), met these criteria, as did CAD1 (corresponding to aspen CAD), but CAD2 (the ortholog of aspen SAD) apparently did not, because it had low expression in all tissues tested. It was included in further analysis in any case, merely because of the belief from prior literature (Li et al., 2001) that it is important for the formation of sinapyl alcohol. However, collectively, the accumulating data from Arabidopsis, poplar, and tobacco argue forcefully that SAD does not play a major role in sinapaldehyde reduction during lignin biosynthesis in vivo; that role is reserved for CAD, and SAD cannot fully compensate if CAD is deficient.
This work highlights the caution that should be applied when interpreting the biological significance of enzyme assays performed in vitro. We have shown that whole stem extracts of our transgenic plants can have significant reductions in sinapaldehyde-reducing activity, but this does not necessarily translate (in the case of the SAD plants) into deficiencies in sinapyl alcohol supply for lignin biosynthesis in woody xylem tissues. There are several possible explanations for this. Whole stems at the age sampled here contain more epidermal, cortical, and pith tissue than developing xylem, and low levels of SAD expression can be detected in these tissues by immunoblot. Consequently, the enzyme assays are not reporting directly on sinapaldehyde-reducing activity in lignifying xylem alone. Indeed, neither sinapaldehyde nor coniferaldehyde can normally be detected in soluble phenolic extracts from xylem (Dauwe et al., 2007), suggesting that, in vivo, either (a) very low levels of substrate are sufficient to maintain sinapyl alcohol production and the slow accumulation of lignin over the lifetime of a plant or (b) a substrate-channeling mechanism exists to guide newly synthesized sinapaldehyde directly into lignin synthesis. Enzyme assays performed in vitro in an excess of exogenously added substrate cannot inform on either of these possibilities. Furthermore, the apparent disappearance of most sinapaldehyde-reducing activity in the SAD plants despite normal CAD levels may simply be misleading. Both CAD and SAD have been reported to exhibit substantial substrate inhibition kinetics (Lauvergeat et al., 1995; Li et al., 2001; Bomati and Noel, 2005). Indeed, Lauvergeat et al. (1995), working with eucalyptus CAD, noted that lack of detection of CAD activity may merely reflect particularly strong substrate inhibition promoted by the high substrate concentrations used for assays in vitro. Bomati and Noel (2005) likewise urged caution in interpreting SAD activity data based on their discovery of unusual kinetic properties and substantial substrate inhibition that cause significant loss in turnover at higher substrate concentrations. These phenomena may help explain how our assays of the primary transformant plants could predominantly report only on the activity of SAD, if it was the case that CAD was inhibited to a higher degree than SAD by the high sinapaldehyde concentration used. All of these issues highlight the absolute necessity of focusing on measurable effects on the lignin polymer itself before assigning a role in its biosynthesis to specific genes or enzymes.
Similarly, assays performed in vitro on recombinant proteins do not necessarily identify an enzyme’s true substrate, they merely suggest its likely relative preference in vivo toward the few substrates tested in the assay. Consequently, predicting the function of an enzyme from its activity in vitro, even when associated with localization data (as in the case of aspen SAD) is fraught with potential error and likely to be misleading. Accumulating data suggest that the SAD gene family is not highly conserved across species in the way that the CAD family is and that many duplication and deletion events occurred during its evolutionary history (Guo et al., 2010). Genes within the SAD family can have different expression patterns and substrate specificities (Kim et al., 2004, 2007; Barakat et al., 2009), implying some divergence of function. Indeed, Bomati and Noel (2005) demonstrated that just two changes in the active site of aspen SAD were sufficient to completely alter specificity. A mutant aspen SAD where Leu-122 was replaced by Trp and Gly-302 was replaced by Phe was no longer highly specific for sinapaldehyde but was more specific for coniferaldehyde (Bomati and Noel, 2005). Overall, the SAD family does not seem to have the conservation of sequence, function, or substrate specificity expected of genes essential to lignin biosynthesis, and instead several family members have been suggested to be involved in stress or defense responses (Somssich et al., 1996; Brill et al., 1999; Montesano et al., 2003). It has been hypothesized that SAD might be a dehydrogenase that directs monolignol precursors toward plant defense and that experimentation in planta would be critical to the experimentally challenging objective of defining its true physiological function (Bomati and Noel, 2005). We present just such in planta data here and demonstrate that, although tobacco SAD maintains a functional relationship to aspen SAD, being able to use both coniferaldehyde and sinapaldehyde as substrates, it plays no detectable role in lignin biosynthesis.
If tobacco SAD is not needed for biosynthesis of S lignin units, what alternative function might it serve in lignifying tissues? Phenolic profiling of wild-type and SAD-suppressed plants by UPLC-MS suggest that SAD could possibly play a role in the production or modification of DDC phenylcoumarans, or neolignans. Although a wide spectrum of different lignans exist in plants, their functions are unclear, although roles in defense are indicated by the biological activity of many lignans, which may have antimicrobial, antifungal, and antifeedant properties, and some have antitumor and antimitotic activity toward mammalian cells. In particular, DDC glucosides have been shown to have cytokinin-like activity in tobacco (Orr and Lynn, 1992). In this study, reductions in DDC derivatives were detected in leaf extracts in SAD-suppressed plants. Consistent with this observation, subsequent work using plants expressing a β-glucuronidase gene fused to the SAD promoter demonstrated that SAD is expressed in both leaf veins and lamina.
Our results have relevance to contemporary theories regarding the evolution of S lignin biosynthesis in angiosperms. It has been proposed that SAD and S lignin units may have emerged together during angiosperm evolution (Li et al., 2001; Peter and Neale, 2004). This idea is consistent with the traditional supposition that the absence of S lignin units in conifers is caused by the low affinity of gymnosperm CADs for sinapaldehyde. However, several commentators have challenged such a generalization and have pointed out that some exceptional gymnosperms can make S lignin units and/or have CADs that display activity toward sinapaldehyde (Campbell and Sederoff, 1996; Whetten et al., 1998; Sibout et al., 2005; Uzal et al., 2009). Moreover, a couple of recent reports demonstrate the presence of low levels of S lignin in the red alga Calliarthron cheilosporioides (Martone et al., 2009) and in the lycophyte Selaginella moellendorffii (Weng et al., 2008), suggesting possible independent evolution of S lignin biosynthesis in different plant lineages (Weng and Chapple, 2010). A recent study of the evolutionary history and functional differentiation of the CAD/SAD gene family (Guo et al., 2010) confirmed that the clade containing aspen SAD was angiosperm-specific but suggested that the relaxed evolutionary selection within the clade, plus the high substrate versatility, pointed to relatively specialized functions for individual genes in processes such as stress resistance. These data and our evidence presented here indicate that SAD was not critical to the evolution of S lignin unit biosynthesis in angiosperms.
METHODS
Plant Growth Conditions
Tobacco (Nicotiana tabacum cv Samsun) was grown in tissue culture on solid Murashige and Skoog medium with 3% Suc at 25°C with a 16-h photoperiod (100 μmol m−2 s−2). At 4 weeks, plants were transferred to compost in the greenhouse for further growth and analysis.
SAD Homology Modeling
Homology models of Nt SAD2 and Nt SAD5 were generated using the Phyre Web server (Kelley and Sternberg, 2009). The most suitable template was the high-resolution crystal structure of Pt SAD, Protein Data Bank code 1YQD (Bomati and Noel, 2005). The high degree of amino acid sequence conservation (>70%) provides confidence in the resulting models. Structures were examined using the graphics visualization software Crystallographic Object-Oriented Toolkit (Emsley and Cowtan, 2004).
Activity of Recombinant Tobacco CAD and SAD
Coding sequences of TCAD19, TSAD2, and TSAD5 were amplified by PCR using primers to incorporate convenient restriction sites (see Supplemental Table 2 online). After cloning and sequencing, the products were transferred into pET52b+ vector (Novagen) to fuse a His tag at the C terminus of the cloned sequence. The obtained clones, pET52b-TCAD19 and pET52b-TSAD2, were introduced into Escherichia coli strain Bl21(DE3) (Novagen). After isopropyl-β-d-thiogalactopyranoside induction, cells were lysed, and the soluble recombinant proteins were purified using Ni-NTA columns (Novagen). The eluted fractions were concentrated, and the buffer was exchanged to 100 mM Tris-HCl (pH 8.8) using Amicon Ultra-15 filters (Millipore) before further purification of recombinant proteins by anion exchange chromatography on Q HyperD 20 resin prepacked into an optima 5/10 column (BioSepra). Elution was achieved using a linear gradient to 0.5 M KCl in 100 mM Tris (pH 8.8) followed by a step gradient to 1 M KCl. Fractions containing active recombinant protein were pooled and concentrated, and the buffer was exchanged to 100 mM Tris-HCl (pH 7.5) and 5 mM DTT.
The kinetic parameters of the purified proteins were determined using triplicate reaction mixtures at 30°C containing 33 mM sodium/potassium-phosphate buffer (pH 6.5), 0.5 mM NADPH, and increasing amounts of coniferaldehyde or sinapaldehyde and monitoring at A400. Initial reaction rates were fitted to the Michaelis-Menten equation to determine steady state kinetic parameters using the software Enzyme Kinetics module of SigmaPlot (SYSTAT Software).
Immunoblots
Protein extracts (30 to 50 μg) were separated by 12% SDS-PAGE along with prestained protein markers (New England Biolabs). Proteins were transferred to nitrocellulose membranes using a Trans-Blot SD semidry blotter (Bio-Rad), treated with blocking buffer (2% dried milk in 200 mM Tris base, 75 mM NaCl, 0.1% Tween-20, pH 7.6), incubated with primary antibody (anti-CAD 1:10000 or anti-SAD 1:3000), washed three times, and then incubated with 1:10000 dilution of horseradish peroxidase-conjugated anti-rabbit IgG (New England Biolabs). Detection was performed using the Phototope-HRP Western Blot Detection Kit (New England Biolabs).
Fluorescence Microscopy
Cross sections (70 to 100 μm) of stem from 12-week-old plants cut on a Vibraslice machine were placed on Superfrost Polylysine slides and blocked in 1% BSA (in PBS, pH 7.2, 0.5% Tween-20) for 45 min. Sections were incubated for 1 h at room temperature in a 1:50 dilution of rabbit antiserum raised against (a) tobacco CAD or (b) aspen SAD. After washing, the sections were incubated with a 1:250 dilution Alexa Fluor 594-conjugated goat anti-rabbit IgG for 45 min, then washed again before mounting in Hydromount and viewing with a Leica SP2 confocal laser scanning system equipped with an inverted microscope. Normal rabbit serum was used as the primary antibody for the controls.
Cloning of Tobacco SAD
RNA was purified from dissected xylem taken from the base of an 8-week-old tobacco stem using the SV Total RNA Isolation System (Promega). cDNA ends were amplified using the GeneRacer Kit (Invitrogen). The SAD and SAD-like nucleotide sequences were aligned with four CAD sequences (from tobacco, tomato [Solanum lycopersicum], eucalyptus [Eucalyptus gunnii], and alfalfa [Medicago sativa]). To increase the chances of ultimately amplifying all potential SAD-like genes from tobacco xylem, regions of sequences that were conserved in SAD and SAD-like sequences but not in CAD sequences were chosen for degenerate primer design. Amplification of tobacco genomic DNA with these primers resulted in a single product of 354 bp that had high sequence identity with aspen SAD (70% identity at the amino acid level). Using the sequence of the tobacco fragment, two gene-specific primers and two nested primers were designed with which to amplify the ends of the gene from tobacco cDNA prepared from the RNA of 8-week-old stem xylem. Fragments of approximately 600 bp for the SAD 5′ end and approximately 900 bp for the SAD 3′ end were isolated, cloned into Zero Blunt TOPO vector (Invitrogen), sequenced, and aligned against aspen SAD. New specific primers were then designed to the ends of the tobacco SAD sequence, and the entire cDNA was amplified from total stem cDNA.
Phylogenetic Analysis
Protein sequences with homology to tobacco CAD and SAD genes were identified by BLAST searches (Altschul et al., 1997), and the top hits were used to construct a phylogenetic tree. The sequences were aligned using Clustal W (Thompson et al., 1994). The output file was run in Tree-Puzzle 5.0 using quartet puzzling (Schmidt et al., 2002) to estimate the phylogenetic tree. This is a maximum likelihood method, which provides good support values for the clusters within the tree. The amino acid substitution matrix used was JTT (Jones et al., 1992). Rate heterogeneity among sites was modeled using a gamma distribution with four categories. The tree was viewed in Tree-View v.1.6.6 (Page, 1996). Alignments are provided as Supplemental Data Set 1 online.
Preparation of pSAD and pSADCAD Constructs
Partial sequences of SAD cDNA and CAD cDNA were amplified by PCR and independently introduced into the donor vector (pDONR201) using Gateway technology (Invitrogen). Both entry clones were digested with Hpa1 and Msc1 endonucleases and then religated to fuse together SAD and CAD partial sequences in one pDONR201 vector. These two entry clones were then recombined with pHellsgate8, a modified version of pHellsgate (Wesley et al., 2001), to make pSAD and pSADCAD, respectively.
Production of Transgenic Plants
Transgenic tobacco plants were produced by Agrobacterium-mediated transformation as described in O’Connell et al. (2002). After regenerated plants had gone through two rounds of rooting on selective medium, they were transferred to nonselective medium for 4 weeks and then transferred to compost and grown in the greenhouse for 2 weeks before being screened for activity of SAD and CAD. Selected plants were clonally propagated to yield small populations of identical individuals for further analysis.
Determination of SAD Activity in Crude Plant Extracts
Stem samples were ground in liquid nitrogen, extracted with 100 mM Tris-HCl, pH 7.5, 20 mM β-mercaptoethanol, then centrifuged at 20,000g, 4°C for 15 min. The total protein concentration of the supernatant was determined using the Bradford assay. SAD activity was determined by monitoring the reduction of sinapaldehyde to sinapyl alcohol. Reactions (1 mL) contained 20 to 50 μL plant extract, 100 mM KNa phosphate buffer, pH 6.25, 0.01 mM sinapaldehyde, and 0.01 mM NADPH, and the decrease in absorbance at 340 nm was measured for 3 to 5 min at 30°C.
Real-Time PCR Analysis of SAD and SAD CAD Plants
Total RNA was extracted from 100 mg of stem tissue using the RiboPure RNA Kit (Ambion). The RNA was treated with DNaseI and then reverse transcribed into first-strand cDNA using Superscript III reverse transcriptase (Invitrogen). Reactions (25 μL) were set up in triplicate with 1 μL cDNA (diluted 1:10), 12.5 μL 2× SYBR Green reporter dye, and 300 nM of SAD, CAD, or ubiquitin (UBI) UBI primers. Control reactions for each primer set contained no cDNA. The reactions were run on the ABI Prism 7700 Sequence Detector at 95°C for 15 min, followed by 40 cycles at 95°C for 15 s, 60°C for 30 s, and 72°C for 30 s. Primers used: NtUbi-F = GGA CCA GCA GAG GTT GAT CT; NtUbi-R = TCA GCC AAG GTC CTT CCA T; NtCAD-F = CAT TTT GGT TTT AAT CAG AGT GGA; NtCAD-R = CCC ATA TGT CCA ACT CCT CCT; NtSAD-F = GAC CCT GAC CAA ATG CAG; NtSAD-R = GTG CGC CAA C(C/T)A TTA CAA GC.
Alkali Extraction of EXR
Xylem samples (1 g) from the base of 10-week-old tobacco stems were dried at room temperature, weighed, milled in a SPEX Certiprep freezer mill, and reweighed. Samples (300 mg) were extracted in a Soxhlet extractor with toluene: ethanol (1:1 v/v, 25 cycles), ethanol (25 cycles), then water overnight, before air-drying and weighing of the EXR. For alkali extraction, 10 mg EXR was shaken in 0.7 mL 1 M NaOH in the dark for 2 h at room temperature. A total of 70 μL of concentrated HCl was added to reduce the pH to below 3. The mixture was extracted twice with 0.7 mL ethyl acetate. Upper organic phases were combined and dried and then resuspended in 1 mL methanol for the determination of UV absorbance at 280 nm.
Acetyl Bromide Lignin Determination
The method of Iiyama and Wallis (1990) was used. EXR (5 mg) was extracted in 2.4 mL 25% (v/v) acetyl bromide in acetic acid with 100 μL perchloric acid at 70°C for 30 min, mixing every 10 min. The samples were cooled on ice, the contents were transferred to a 50-mL volumetric flask containing 10 mL 2 M NaOH and 12 mL glacial acetic acid, and the flask was made up to volume with glacial acetic acid. The UV absorption of the solution was determined at 280 nm.
Klason Lignin Analysis
The base of 10-week-old tobacco stems was collected (three plants per line) and ground to pass a 0.5-mm sieve, and solvent was extracted in a Soxhlet extractor (toluene-ethanol 2:1, ethanol, and H2O). The Klason lignin determination of the EXR was run in duplicate for each sample by the standard procedure (Dence, 1992). The se between analytical duplicates was always less than 1% of the mean value.
Thioacidolysis
Lignin structure was investigated using thioacidolysis as previously described (Lapierre et al., 1999). All the experiments were run in duplicate, and 20 mg from the extract-free samples was prepared for Klason lignin analysis. The lignin-derived compounds were identified by gas chromatography–mass spectrometry of their trimethylsilyl derivatives. The quantitative evaluation of indenes originating from sinapaldehyde and coniferaldehyde that have undergone 8-O-4 coupling during lignification was performed from ion chromatograms reconstructed at m/z 384 and 354, respectively. The determination of the conventional G and S monomers was done from their prominent benzylic ions at m/z 269 or 299, respectively. All the calculations were done with the response factors of the main G and S monomers, relative to the C22 internal standard, and the yields were expressed on the basis of the extract-free lignin content of the samples.
UPLC-MS Profiling
The wild type and transgenic tobacco lines SAD4 and SAD45 were grown for 10 weeks in the greenhouse (16-h photoperiod at 22 to 25°C) and harvested just before flowering. After homogenization of leaf, xylem, bark, and root tissues in liquid nitrogen, liquid-liquid extraction was performed as previously described (Meyermans et al., 2000; Morreel et al., 2006).
Phenolics present in the aqueous phase were analyzed on a reversed phase Acquity BEH C18 (2.1 × 150 mm, 1.7 μm; Waters) column, heated at 40°C, using an Acquity Ultrahigh Performance Liquid Chromatography (Waters) instrument coupled with an Acquity 2996 photodiode array detector (Waters) and a LCQ Classic ion trap mass spectrometer (ThermoQuest). A gradient from 0.1% triethylammonium acetate (pH 5, buffer A) to acetonitrile/methanol (75/25, v/v, to which 0.1% triethylammonium acetate was added, buffer B) was used for separation. At a flow rate of 0.2 mL/min, buffer A decreased from 95% to 50% in 25 min, and to 0% in 5 min. Ionization occurred in the negative mode using an Atmospheric Pressure Chemical Ionization source using the following conditions: capillary temperature 150°C, vaporizer temperature 350°C, sheath gas 25 (arb), aux gas 3 (arb), source current 5 μA. Quantification was based on full mass spectrometry (MS) (maximum ion time, 50 ms; number of microscans, 1) data acquired in the m/z range from 100 to 1000 D. For structure elucidation, full MS scans were interrupted with data-dependent MS/MS scans (maximum ion time, 50 ms; number of microscans, 2; collision energy, 35%).
Full MS data were integrated with the XCMS package (R version 2.4.1) using only the xcmsSet (fwhm 7, max 20, snthresh 5, step 0.1, steps 3, mzdiff 0.5) and group (bw 10, mzwid 0.5) functions. Statistical analysis was performed with Student’s t tests (α = 0.001), using a Welch correction in the case of heteroscedasticity, and principal component analysis with the prcomp function available in R version 2.4.1. All peaks were centered and scaled to unit variance. Peaks with loading factors that had an absolute value more than 2 sd away from the mean loading value for a particular principal component were considered as major contributors to that principal component.
MS Structure Elucidations
Peak X, eluting at 12.8 min, showed a MS peak at m/z 581. MS/MS analysis yielded a base peak at m/z 521, resulting from the loss of the acetate adduct and three other major daughter ions at m/z 359, probably caused by the further loss of a hexose moiety, and at m/z 341 and 329, both latter ions likely derived from the m/z 359 ion by the detachment of H2O and CH3OH, respectively. The similarity of the MS/MS fragmentation behavior of the aglycone moiety to that of (8–5)-DDC (parent ion at m/z 357 and major daughter ions at m/z 339 and 327) (Morreel et al., 2004a, 2004b) suggests that peak X is hexosylated DDDC or IDDDC. These structures might arise from the reduction of the phenylcoumaran ring or the side chain double bond of hexosylated DDC.
A targeted search for hexosylated DDC (m/z 579) in the UPLC-MS chromatograms revealed a peak at 12.6 min. Because MS/MS fragmentation afforded the same neutral losses as observed for peak X, and because the m/z 357, 339, and 327 daughter ions referred to the presence of a DDC moiety (Morreel et al., 2004a, 2004b), this compound was identified as hexosylated DDC.
Accession Numbers
Sequence data from this article and for the phylogenetic analysis shown in Figure 2 can be found in the EMBL/GenBank data libraries under the following accession numbers: Ag (Apium graveolens), Q38707; At (Arabidopsis thaliana) CAD1, P42734, At4g39330; At CAD2, P48523, At3g19450; At CAD3, O65621, At4g37970; At CAD4, Q02971, At4g37980; At CAD5, Q02972, At4g37990; At CAD6, O49482, At4g34230; At CAD7, Q9SJ25, At2g21730; At CAD8, Q9SJ10, At2g21890; Ca (Camptotheca acuminate), Q7XAB2; Cr (Catharanthus roseus), Q6V4H0; Fxa (Fragaria × ananassa), AAD10327; Le CAD, X92855; Mc (Mesembryanthemum crystallinum), P93257; Ms (Medicago sativa), O82515; Ms CAD, CAA79625; Nt CAD, CAA44217; Nt SAD, ABD73280 (SAD-L1), ADD81205 (SAD-2), ADD81206 (SAD-L3), ADD81207 (SAD-4), ADD81208 (SAD-L5); Pc, P42754; Pt CAD, AAF43140; Pt SAD, AAK58693; Ptx SAD, Q8L7U8; Sh (Stylosanthes humilis), Q43138; St (Solanum tuberosum), Q8H0L8.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Immunoblots of SAD and CAD in Tobacco Stem Extracts from the Wild Type and CAD-Antisense Plants.
Supplemental Figure 2. Active Sites of Pt SAD and Nt SAD5.
Supplemental Figure 3. CAD Activity in SAD CAD Lines.
Supplemental Figure 4. CAD Expression in SAD CAD Lines.
Supplemental Figure 5. Selected Ion Current (SIC) Chromatograms of a Methanol Extract from the Wild-Type Leaves.
Supplemental Table 1. Indene Derivatives of Coniferaldehyde and Sinapaldehyde in Lignin from SAD CAD Lines.
Supplemental Table 2. Primers Used for E. coli Expression of Recombinant CAD and SAD Proteins.
Supplemental Data Set 1. CLUSTAL 2.0.12 Multiple Sequence Alignment of CAD and SAD Protein Sequences.
Acknowledgments
We thank Vincent Chiang for the gift of the SAD antiserum, Peter M. Waterhouse for the gift of the pHellsgate8 vector, Frédéric Legée for running the wood extractions and the Klason determinations, Laurent Cezar for running the thioacidolysis experiments, Brigitte Pollet for supervising these analyses, Frank Wright for help with phylogenetic analysis, and Ingo Hein for help with real-time RT-PCR. This work was funded by the Biotechnology and Biological Sciences Research Council of the United Kingdom (P18182).
AUTHOR CONTRIBUTIONS
A.G. performed protein localization studies, W.N.H. performed the structural modeling, D.M. analyzed bioinformatic data, R.D.H. performed enzyme kinetics, C.L. performed lignin analysis, K.M. and W.B. performed phenolic profiling. All other research work and data analysis was performed equally by A.B and J.S., and they are joint first authors of the article. The research was designed and the article was written by C.H.
References
- Abbott J.C., Barakate A., Pinçon G., Legrand M., Lapierre C., Mila I., Schuch W., Halpin C. (2002). Simultaneous suppression of multiple genes by single transgenes. Down-regulation of three unrelated lignin biosynthetic genes in tobacco. Plant Physiol. 128: 844–853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul S.F., Madden T.L., Schäffer A.A., Zhang J., Zhang Z., Miller W., Lipman D.J. (1997). Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 25: 3389–3402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barakat A., Bagniewska-Zadworna A., Choi A., Plakkat U., DiLoreto D.S., Yellanki P., Carlson J.E. (2009). The cinnamyl alcohol dehydrogenase gene family in Populus: Phylogeny, organization, and expression. BMC Plant Biol. 9: 26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baucher M., Halpin C., Petit-Conil M., Boerjan W. (2003). Lignin: genetic engineering and impact on pulping. Crit. Rev. Biochem. Mol. Biol. 38: 305–350 [DOI] [PubMed] [Google Scholar]
- Baucher M., Chabbert B., Pilate G., Van Doorsselaere J., Tollier M.-T., Petit-Conil M., Cornu D., Monties B., Van Montagu M., Inze D., Jouanin L., Boerjan W. (1996). Red xylem and higher lignin extractability by down-regulating a cinnamyl alcohol dehydrogenase in Poplar. Plant Physiol. 112: 1479–1490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boerjan W., Ralph J., Baucher M. (2003). Lignin biosynthesis. Annu. Rev. Plant Biol. 54: 519–546 [DOI] [PubMed] [Google Scholar]
- Bomati E.K., Noel J.P. (2005). Structural and kinetic basis for substrate selectivity in Populus tremuloides sinapyl alcohol dehydrogenase. Plant Cell 17: 1598–1611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brill E.M., Abrahams S., Hayes C.M., Jenkins C.L.D., Watson J.M. (1999). Molecular characterisation and expression of a wound-inducible cDNA encoding a novel cinnamyl-alcohol dehydrogenase enzyme in lucerne (Medicago sativa L.). Plant Mol. Biol. 41: 279–291 [DOI] [PubMed] [Google Scholar]
- Campbell M.M., Sederoff R.R. (1996). Variation in lignin content and composition. Mechanisms of control and implications for the genetic improvement of plants. Plant Physiol. 110: 3–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chabannes M., Barakate A., Lapierre C., Marita J.M., Ralph J., Pean M., Danoun S., Halpin C., Grima-Pettenati J., Boudet A.M. (2001). Strong decrease in lignin content without significant alteration of plant development is induced by simultaneous down-regulation of cinnamoyl CoA reductase (CCR) and cinnamyl alcohol dehydrogenase (CAD) in tobacco plants. Plant J. 28: 257–270 [DOI] [PubMed] [Google Scholar]
- Chen F., Dixon R.A. (2007). Lignin modification improves fermentable sugar yields for biofuel production. Nat. Biotechnol. 25: 759–761 [DOI] [PubMed] [Google Scholar]
- Chen F., Yasuda S., Fukushima K. (1999). Evidence for a novel biosynthetic pathway that regulates the ratio of syringyl to guaiacyl residues in lignin in the differentiating xylem of Magnolia kobus DC. Planta 207: 597–603 [Google Scholar]
- Dauwe R., et al. (2007). Molecular phenotyping of lignin-modified tobacco reveals associated changes in cell-wall metabolism, primary metabolism, stress metabolism and photorespiration. Plant J. 52: 263–285 [DOI] [PubMed] [Google Scholar]
- Dence C. (1992). Lignin determination. In Methods in Lignin Chemistry, C. Dence and S. Lin, eds (Berlin: Springer-Verlag), pp. 29–34 [Google Scholar]
- Emsley P., Cowtan K. (2004). Coot: Model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60: 2126–2132 [DOI] [PubMed] [Google Scholar]
- Feuillet C., Lauvergeat V., Deswarte C., Pilate G., Boudet A., Grima-Pettenati J. (1995). Tissue- and cell-specific expression of a cinnamyl alcohol dehydrogenase promoter in transgenic poplar plants. Plant Mol. Biol. 27: 651–667 [DOI] [PubMed] [Google Scholar]
- Franke R., McMichael C.M., Meyer K., Shirley A.M., Cusumano J.C., Chapple C. (2000). Modified lignin in tobacco and poplar plants over-expressing the Arabidopsis gene encoding ferulate 5-hydroxylase. Plant J. 22: 223–234 [DOI] [PubMed] [Google Scholar]
- Gomez L.D., Whitehead C., Barakate A., Halpin C., McQueen-Mason S.J. (2010). Automated saccharification assay for determination of digestibility in plant materials. Biotechnol Biofuels 3: 23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo D.M., Ran J.-H., Wang X.-Q. (2010). Evolution of the Cinnamyl/Sinapyl Alcohol Dehydrogenase (CAD/SAD) gene family: The emergence of real lignin is associated with the origin of bona fide CAD. J. Mol. Evol. 71: 202–218 [DOI] [PubMed] [Google Scholar]
- Halpin C. (2004). Investigating and manipulating lignin biosynthesis in the post-genomic era. Adv. Bot. Res. 41: 63–106 [Google Scholar]
- Halpin C., Knight M.E., Foxon G.A., Campbell M.M., Boudet A.M., Boon J.J., Chabbert B., Tollier M.-T., Schuch W. (1994). Manipulation of lignin quality by down-regulation of cinnamyl alcohol dehydrogenase. Plant J. 6: 339–350 [Google Scholar]
- Higuchi T. (1985). Biosynthesis of lignin. In Biosynthesis and Biodegradation of Wood Components T. Higuchi, ed. (Orlando, FL: Academic Press), pp. 141–160 [Google Scholar]
- Higuchi T., Ito T., Umezawa T., Hibino T., Shibata D. (1994). Red-brown color of lignified tissues of transgenic plants with antisense CAD gene: Wine-red lignin from coniferyl aldehyde. J. Biotechnol. 37: 151–158 [Google Scholar]
- Humphreys J.M., Hemm M.R., Chapple C. (1999). New routes for lignin biosynthesis defined by biochemical characterization of recombinant ferulate 5-hydroxylase, a multifunctional cytochrome P450-dependent monooxygenase. Proc. Natl. Acad. Sci. USA 96: 10045–10050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iiyama K., Wallis A.F.A. (1990). Determination of lignin in herbaceous plants by an improved acetyl bromide procedure. J. Sci. Food Agric. 51: 145–161 [Google Scholar]
- Jones D.T., Taylor W.R., Thornton J.M. (1992). The rapid generation of mutation data matrices from protein sequences. Comput. Appl. Biosci. 8: 275–282 [DOI] [PubMed] [Google Scholar]
- Jørgensen K., Rasmussen A.V., Morant M., Nielsen A.H., Bjarnholt N., Zagrobelny M., Bak S., Møller B.L. (2005). Metabolon formation and metabolic channeling in the biosynthesis of plant natural products. Curr. Opin. Plant Biol. 8: 280–291 [DOI] [PubMed] [Google Scholar]
- Kelley L.A., Sternberg M.J.E. (2009). Protein structure prediction on the Web: A case study using the Phyre server. Nat. Protoc. 4: 363–371 [DOI] [PubMed] [Google Scholar]
- Kim S.J., Kim K.W., Cho M.H., Franceschi V.R., Davin L.B., Lewis N.G. (2007). Expression of cinnamyl alcohol dehydrogenases and their putative homologues during Arabidopsis thaliana growth and development: Lessons for database annotations? Phytochemistry 68: 1957–1974 [DOI] [PubMed] [Google Scholar]
- Kim H., Ralph J., Lu F.-C., Pilate G., Leplé J.-C., Pollet B., Lapierre C. (2002). Identification of the structure and origin of thioacidolysis marker compounds for cinnamyl alcohol dehydrogenase deficiency in angiosperms. J. Biol. Chem. 277: 47412–47419 [DOI] [PubMed] [Google Scholar]
- Kim S.J., Kim M.R., Bedgar D.L., Moinuddin S.G.A., Cardenas C.L., Davin L.B., Kang C.L., Lewis N.G. (2004). Functional reclassification of the putative cinnamyl alcohol dehydrogenase multigene family in Arabidopsis. Proc. Natl. Acad. Sci. USA 101: 1455–1460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ko J.H., Han K.H. (2004). Arabidopsis whole-transcriptome profiling defines the features of coordinated regulations that occur during secondary growth. Plant Mol. Biol. 55: 433–453 [DOI] [PubMed] [Google Scholar]
- Lapierre C., Pilate G., Pollet B., Mila I., Leplé J.C., Jouanin L., Kim H., Ralph J. (2004). Signatures of cinnamyl alcohol dehydrogenase deficiency in poplar lignins. Phytochemistry 65: 313–321 [DOI] [PubMed] [Google Scholar]
- Lapierre C., Pollet B., Petit-Conil M., Toval G., Romero J., Pilate G., Leple J.C., Boerjan W., Ferret V., De Nadai V., Jouanin L. (1999). Structural alterations of lignins in transgenic poplars with depressed cinnamyl alcohol dehydrogenase or caffeic acid O-methyltransferase activity have an opposite impact on the efficiency of industrial kraft pulping. Plant Physiol. 119: 153–164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauvergeat V., Kennedy K., Feuillet C., McKie J.H., Gorrichon L., Baltas M., Boudet A.M., Grima-Pettenati J., Douglas K.T. (1995). Site-directed mutagenesis of a serine residue in cinnamyl alcohol dehydrogenase, a plant NADPH-dependent dehydrogenase, affects the specificity for the coenzyme. Biochemistry 34: 12426–12434 [DOI] [PubMed] [Google Scholar]
- Li L., Popko J.L., Umezawa T., Chiang V.L. (2000). 5-hydroxyconiferyl aldehyde modulates enzymatic methylation for syringyl monolignol formation, a new view of monolignol biosynthesis in angiosperms. J. Biol. Chem. 275: 6537–6545 [DOI] [PubMed] [Google Scholar]
- Li L.G., Cheng X.F., Leshkevich J., Umezawa T., Harding S.A., Chiang V.L. (2001). The last step of syringyl monolignol biosynthesis in angiosperms is regulated by a novel gene encoding sinapyl alcohol dehydrogenase. Plant Cell 13: 1567–1586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martone P.T., Estevez J.M., Lu F.C., Ruel K., Denny M.W., Somerville C., Ralph J. (2009). Discovery of lignin in seaweed reveals convergent evolution of cell-wall architecture. Curr. Biol. 19: 169–175 [DOI] [PubMed] [Google Scholar]
- Matsui N., Chen F., Yasuda S., Fukushima K. (2000). Conversion of guaiacyl to syringyl moieties on the cinnamyl alcohol pathway during the biosynthesis of lignin in angiosperms. Planta 210: 831–835 [DOI] [PubMed] [Google Scholar]
- Meyermans H., et al. (2000). Modifications in lignin and accumulation of phenolic glucosides in poplar xylem upon down-regulation of caffeoyl-coenzyme A O-methyltransferase, an enzyme involved in lignin biosynthesis. J. Biol. Chem. 275: 36899–36909 [DOI] [PubMed] [Google Scholar]
- Montesano M., Hyytiäinen H., Wettstein R., Palva E.T. (2003). A novel potato defence-related alcohol: NADP+ oxidoreductase induced in response to Erwinia carotovora. Plant Mol. Biol. 52: 177–189 [DOI] [PubMed] [Google Scholar]
- Morreel K., Ralph J., Kim H., Lu F., Goeminne G., Ralph S., Messens E., Boerjan W. (2004a). Profiling of oligolignols reveals monolignol coupling conditions in lignifying poplar xylem. Plant Physiol. 136: 3537–3549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morreel K., Ralph J., Lu F., Goeminne G., Busson R., Herdewijn P., Goeman J.L., Van der Eycken J., Boerjan W., Messens E. (2004b). Phenolic profiling of caffeic acid O-methyltransferase-deficient poplar reveals novel benzodioxane oligolignols. Plant Physiol. 136: 4023–4036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morreel K., Goeminne G., Storme V., Sterck L., Ralph J., Coppieters W., Breyne P., Steenackers M., Georges M., Messens E., Boerjan W. (2006). Genetical metabolomics of flavonoid biosynthesis in Populus: A case study. Plant J. 47: 224–237 [DOI] [PubMed] [Google Scholar]
- O’Connell A., Holt K., Piquemal J., Grima-Pettenati J., Boudet A., Pollet B., Lapierre C., Petit-Conil M., Schuch W., Halpin C. (2002). Improved paper pulp from plants with suppressed cinnamoyl-CoA reductase or cinnamyl alcohol dehydrogenase. Transgenic Res. 11: 495–503 [DOI] [PubMed] [Google Scholar]
- Orr J.D., Lynn D.G. (1992). Biosynthesis of dehydrodiconiferyl alcohol glucosides: Implications for the control of tobacco cell growth. Plant Physiol. 98: 343–352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osakabe K., Tsao C.C., Li L.-G., Popko J.L., Umezawa T., Carraway D.T., Smeltzer R.H., Joshi C.P., Chiang V.L. (1999). Coniferyl aldehyde 5-hydroxylation and methylation direct syringyl lignin biosynthesis in angiosperms. Proc. Natl. Acad. Sci. USA 96: 8955–8960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Page R.D.M. (1996). TreeView: An application to display phylogenetic trees on personal computers. Comput. Appl. Biosci. 12: 357–358 [DOI] [PubMed] [Google Scholar]
- Peter G., Neale D. (2004). Molecular basis for the evolution of xylem lignification. Curr. Opin. Plant Biol. 7: 737–742 [DOI] [PubMed] [Google Scholar]
- Pilate G., et al. (2002). Field and pulping performances of transgenic trees with altered lignification. Nat. Biotechnol. 20: 607–612 [DOI] [PubMed] [Google Scholar]
- Raes J., Rohde A., Christensen J.H., Van de Peer Y., Boerjan W. (2003). Genome-wide characterization of the lignification toolbox in Arabidopsis. Plant Physiol. 133: 1051–1071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ralph J., Brunow G., Harris P.J., Dixon R.A., Schatz P.F., Boerjan W. (2008b). Lignification: Are lignins biosynthesized via simple combinatorial chemistry or via proteinaceous control and template replication? In Recent Advances in Polyphenol Research, Vol 1, F. Daayf, A. El Hadrami, L. Adam, and G.M. Ballance, eds (Oxford, United Kingdom: Wiley-Blackwell Publishing), pp. 36–66 [Google Scholar]
- Ralph J., Hatfield R.D., Piquemal J., Yahiaoui N., Pean M., Lapierre C., Boudet A.-M. (1998). NMR characterization of altered lignins extracted from tobacco plants down-regulated for lignification enzymes cinnamylalcohol dehydrogenase and cinnamoyl-CoA reductase. Proc. Natl. Acad. Sci. USA 95: 12803–12808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ralph J., Kim H., Lu F., Grabber J.H., Leplé J.-C., Berrio-Sierra J., Derikvand M.M., Jouanin L., Boerjan W., Lapierre C. (2008a). Identification of the structure and origin of a thioacidolysis marker compound for ferulic acid incorporation into angiosperm lignins (and an indicator for cinnamoyl CoA reductase deficiency). Plant J. 53: 368–379 [DOI] [PubMed] [Google Scholar]
- Ralph J., et al. (2001). Elucidation of new structures in lignins of CAD- and COMT-deficient plants by NMR. Phytochemistry 57: 993–1003 [DOI] [PubMed] [Google Scholar]
- Schmidt H.A., Strimmer K., Vingron M., von Haeseler A. (2002). TREE-PUZZLE: maximum likelihood phylogenetic analysis using quartets and parallel computing. Bioinformatics 18: 502–504 [DOI] [PubMed] [Google Scholar]
- Shi R., Sun Y.-H., Li Q., Heber S., Sederoff R., Chiang V.L. (2010). Towards a systems approach for lignin biosynthesis in Populus trichocarpa: Transcript abundance and specificity of the monolignol biosynthetic genes. Plant Cell Physiol. 51: 144–163 [DOI] [PubMed] [Google Scholar]
- Sibout R., Eudes A., Mouille G., Pollet B., Lapierre C., Jouanin L., Séguin A. (2005). CINNAMYL ALCOHOL DEHYDROGENASE-C and -D are the primary genes involved in lignin biosynthesis in the floral stem of Arabidopsis. Plant Cell 17: 2059–2076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sibout R., Eudes A., Pollet B., Goujon T., Mila I., Granier F., Séguin A., Lapierre C., Jouanin L. (2003). Expression pattern of two paralogs encoding cinnamyl alcohol dehydrogenases in Arabidopsis. Isolation and characterization of the corresponding mutants. Plant Physiol. 132: 848–860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Somssich I.E., Wernert P., Kiedrowski S., Hahlbrock K. (1996). Arabidopsis thaliana defense-related protein ELI3 is an aromatic alcohol:NADP+ oxidoreductase. Proc. Natl. Acad. Sci. USA 93: 14199–14203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson J.D., Higgins D.G., Gibson T.J. (1994). CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22: 4673–4680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuji Y., Chen F., Yasuda S., Fukushima K. (2004). The behavior of deuterium-labeled monolignol and monolignol glucosides in lignin biosynthesis in angiosperms. J. Agric. Food Chem. 52: 131–134 [DOI] [PubMed] [Google Scholar]
- Tuskan G.A., et al. (2006). The genome of black cottonwood, Populus trichocarpa (Torr. & Gray). Science 313: 1596–1604 [DOI] [PubMed] [Google Scholar]
- Umezawa T. (2010). The cinnamate/monolignol pathway. Phytochem. Rev. 9: 1–17 [Google Scholar]
- Uzal E.N., Gómez Ros L.V., Pomar F., Bernal M.A., Paradela A., Albar J.P., Ros Barceló A. (2009). The presence of sinapyl lignin in Ginkgo biloba cell cultures changes our views of the evolution of lignin biosynthesis. Physiol. Plant. 135: 196–213 [DOI] [PubMed] [Google Scholar]
- Vailhé M.A.B., Besle J.M., Maillot M.P., Cornu A., Halpin C., Knight M. (1998). Effect of down-regulation of cinnamyl alcohol dehydrogenase on cell wall composition and on degradability of tobacco stems. J. Sci. Food Agric. 76: 505–514 [Google Scholar]
- Weng J.K., Chapple C. (2010). The origin and evolution of lignin biosynthesis. New Phytol. 187: 273–285 [DOI] [PubMed] [Google Scholar]
- Weng J.K., Li X., Stout J., Chapple C. (2008). Independent origins of syringyl lignin in vascular plants. Proc. Natl. Acad. Sci. USA 105: 7887–7892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wesley S.V., et al. (2001). Construct design for efficient, effective and high-throughput gene silencing in plants. Plant J. 27: 581–590 [DOI] [PubMed] [Google Scholar]
- Whetten R.W., MacKay J.J., Sederoff R.R. (1998). Recent advances in understanding lignin biosynthesis. Annu. Rev. Plant Physiol. Plant Mol. Biol. 49: 585–609 [DOI] [PubMed] [Google Scholar]