Abstract
Simian varicella virus (SVV) is a primate herpesvirus that is closely related to varicella-zoster virus (VZV), the causative agent of varicella (chickenpox) and herpes zoster (shingles). Epizootics of simian varicella occur sporadically in facilities housing Old World monkeys. This review summarizes the molecular properties of SVV. The SVV and VZV genomes are similar in size, structure, and gene arrangement. The 124.5 kilobase pair (kbp) SVV genome includes a 104.7 kbp long (L) component covalently linked to a short (S) component which includes a 4.9 kbp unique short (US) segment flanked by 7.5 kbp inverted repeat sequences. SVV DNA encodes 69 distinct open reading frames (ORFs), three of which are duplicated within the viral inverted repeats. The viral genome is coordinately expressed and immediate early (IE), early, and late genes have been characterized. Genetic approaches have been developed to create SVV mutants, which will be used to study the role of SVV genes in viral pathogenesis, latency, and reactivation. In addition, SVV expressing foreign genes are being investigated as potential recombinant varicella vaccines.
Introduction
Simian varicella virus (SVV) produces a natural varicella-like disease in non-human primates. Simian varicella was initially reported in 1967 as an erythematous disease occurring in a colony of vervet monkeys (Cercopithecus aethiops) at the Liverpool School of Medicine (Clarkson et al. 1967). Since then, simian varicella outbreaks have occurred sporadically in primate facilities housing Old World monkeys, including vervet and patas (Erythrocebus patas) monkeys, and pig-tailed, Japanese, cynomolgous, and rhesus macaque monkeys (Macaca sp.) (Soike 1992; Gray 2004, 2008). In some of these outbreaks, the monkeys exhibited relatively mild clinical symptoms including fever and vesicular skin rash, similar to typical chickenpox in children. Other epizootics have been associated with a more severe disease characterized by a hemorrhagic rash, visceral dissemination, and high morbidity and mortality rates. After the acute disease is resolved, SVV establishes a latent infection within host neural ganglia and may reactivate later in life to cause a secondary disease, analogous to VZV-mediated herpes zoster (Mahalingam et al. 1991, 1992). SVV outbreaks are often initiated by viral reactivation from a latently infected monkey.
The etiologic agent is a primate herpesvirus that is genetically related to varicella-zoster virus (VZV). SVV is classified as Cercopithecus herpesvirus 9, a member of the Alphaherpesvirus family and the Varcellovirus genus along with VZV, equine herpesvirus types 1 and 4 (EHV-1, EHV-4), pseudorabies virus (PRV), Marek’s disease virus (MDV), and bovine herpesvirus type 1 (BHV-1). The virus has previously been classified as Delta herpesvirus (DHV), Liverpool vervet virus (LVV), Medical Lake macaque virus (MLMV), and patas herpesvirus (PHV), based upon geographic location of the epizootic or the monkey species involved, but molecular analysis has confirmed that these are isolates of the same virus, designated as SVV (Gray and Gusick 1996).
In addition to its importance as a veterinary disease, the genetic relatedness of SVV and VZV and the similarities between simian and human varicella pathogenesis and clinical symptoms make SVV infection of nonhuman primates a useful experimental model to study the molecular basis of varicella pathogenesis and to evaluate potential antiviral agents and vaccines (Gray 2004). Recent reviews have described simian varicella epidemiology, pathogenesis, latency, and clinical aspects of disease (White et al. 2001; Dueland 1998; Gray, 2003, 2004, 2008). This review will focus on the molecular properties of SVV including SVV morphology, DNA structure, genetic content, gene expression, and mutagenesis of the SVV genome.
Cell-Associated Nature of SVV
A discussion of the molecular properties of SVV should include an appreciation of the cell-associated nature of the virus. SVV is propagated in vitro in African green monkey kidney cells (Vero, BSC-1, or CV-1). Viral induced cytopathic effect (cpe) including rounded, swollen, and fused, multinucleated cells is evident during lytic infection of cell monolayers (Figure 1A). Cell death occurs within 24 hours and infected cells detach from the monolayer. Electron microscopic analysis of infected cells reveals viral nucleocapsids within the cell nucleus (Figure 1B). However, as virions progress into the cell cytoplasm a majority accumulate within vacuoles and are degraded by lysosomal enzymes. A mixture of intact and aberrant degraded virions are released into the extracellular media which has a relatively low titer of infectious virus (102–104 plaque forming units [pfu]/ml). Likewise, VZV infection of cultured cells is characterized by virion degradation and low infectious virus titers. This phenomenon is an unusual feature of SVV and VZV replication in cell culture, as herpes simplex virus type 1 (HSV-1) virions are not degraded in this manner in the same cell types.
Figure 1.
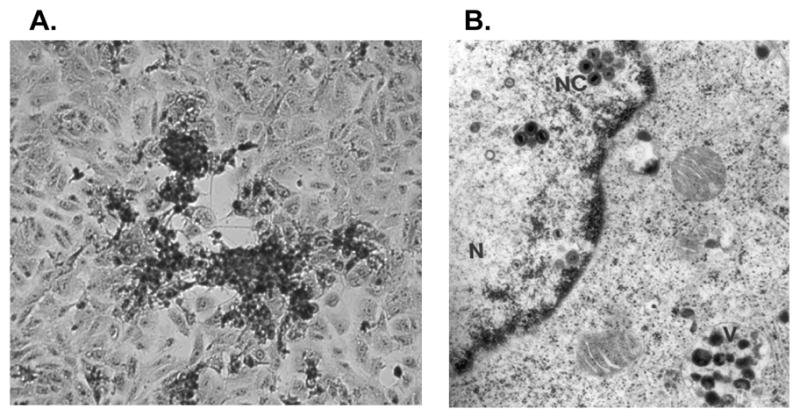
SVV-infected Vero cells. (A) Cytopathic effect in a SVV-infected Vero cell monolayer (20X). (B) Electron microscopy of a SVV-infected Vero cell. SVV nucleocapsids (NC) within the cell nucleus (N) and degraded virions within cell vacuoles (V) are shown. Reprinted with permission by Springer Science and Business Media from Oakes and d’Offay in Virus Diseases in Laboratory and Captive Animals 1988; 163–174. Copyright Kluwer Academic/Plenum Publishers.
Studies of SVV growth in cell culture have determined optimal conditions for generating maximal viral titers (Schmidt 1982). Yields of cell-free SVV generated from infected Vero or BSC-1 cells are generally less than 105 pfu/ml. Similarly, high titer VZV cell-free preparations are not feasible. By comparison, HSV-1 infected Vero cell preparations may routinely yield cell-free virus yields of >108 pfu/ml. Therefore, SVV is serially passaged in culture by transfer of infected cells rather than with cell-free virus and virus stocks are cryopreserved at −70° C or in liquid nitrogen as infected cells. Studies on the molecular properties of SVV are limited by this inability to generate high titer virus stocks of infectious cell-free SVV and the inability to produce synchronous, high multiplicity per cell infection in cell culture.
SVV Morphology and Relatedness of SVV and VZV
SVV virions are isolated from infected BSC-1 or Vero cells by zonal centifugation on glycerol gradients followed by differential centrifugation on glycerol/potassium tartrate gradients (Fletcher, III and Gray 1992). Purified SVV virions have a calculated buoyant density of 1.21 g/ml, identical to the density of VZV virions isolated by the same method. Electron microscopy of purified SVV virions reveals particles with typical herpesvirus morphology consisting of a ≈100 nm nucleocapsid with an electron dense core containing the viral DNA genome. The nucleocapsid is surrounded by a viral membrane envelope giving the virion an overall size of 170–200 nm, similar to the size of VZV. SVV virions consist of at least thirty polypeptide species ranging in size from 16 to >200 kilodaltons (kDa) as indicated by SDS-polyacrylamide gel electrophoresis (SDS-PAGE) analysis (Fletcher, III and Gray 1992). The polypeptide profiles of SVV and VZV purified virions are strikingly similar. At least four of the SVV virion polypeptides ranging in size from 113 K to 46 K are antigens immunoprecipitated with immune serum from SVV-infected monkeys. SVV includes at least six glycoproteins (46–115 kDa) as indicated by immunoprecipitation analysis of infected cell lysates.
The SVV virion and infected cell proteins and glycoproteins share cross-reacting epitopes with VZV as indicated by the extensive immunoprecipitation of SVV proteins with VZV immune serum derived from herpes zoster patients (Fletcher, III and Gray 1992). Other studies have confirmed the antigenic relatedness of SVV and VZV. Immune serum from SVV-infected monkeys neutralizes VZV and reacts with VZV antigens as indicated by immunoflourescence, complement fixation, and immunoblot assays (Felsenfeld and Schmidt 1977,1975; Blakely et al. 1973). In addition, while VZV inoculation of patas monkeys does not result in clinical disease, it induces cross-reacting antibodies to SVV antigens and immune protection against simian varicella disease following SVV challenge (Felsenfeld and Schmidt 1979).
A comparison of the restriction endonuclease profiles of SVV and VZV DNA confirmed that SVV and VZV are distinct herpesviruses (Gray et al. 1992). However, Southern blot DNA hybridizations performed under conditions of varied stringency demonstrated the genetic similarity of SVV and VZV and determined that the SVV and VZV genomes share 70–75% DNA homology (Gray and Oakes 1984). In addition, the SVV and VZV DNAs are co-linear with respect to genome organization as indicted by hybridizations conducted with labeled SVV and VZV restriction endonuclease DNA fragments (Pumphrey and Gray 1992). These experimental findings have been confirmed by comparison of the DNA sequence of the SVV and VZV genomes (Gray et al. 2001).
The SVV Genome
SVV DNA is purified from viral nucleocapsids isolated by centrifugation of infected Vero cell lysates through glycerol gradients (Gray et al. 1992). The molecular size of SVV DNA was initially estimated be ≈121 to 125 kilobases (kb) by electron microscopy, pulse field electrophoresis, and restriction endonuclease analyses (Gray et al. 1992; Clarke et al. 1992). Subsequent DNA sequence analysis has determined the SVV genome to be 124,785 bp in size (Gray et al. 2001), only slightly smaller than the 124,884 bp VZV DNA. The viral DNA has a buoyant density of 1.700 g/ml, corresponding to a guansine plus cytosine (G +C %) content of 40.8%, as determined by CsCl gradient isopycnic banding (Clarke et al. 1992). DNA sequence analysis has subsequently confirmed the SVV DNA content to be 40.4% G + C, compared to 46% G + C for VZV DNA (Table 1). Transfection of Vero or CV-1 cell monolayers with purified SVV DNA yields viral plaques within ten days indicating the infectious nature of SVV DNA (Clarke et al. 1992).
Table 1.
SVV-VZV Genome Comparison
| Size (bp) | G + C % | |||
|---|---|---|---|---|
| SVV | VZV1 | SVV | VZV1 | |
| TRS/IRS | 7557 | 7319.5 | 65.0 % | 59.0 % |
| US | 4,904 | 5,232 | 39.1 % | 42.8 % |
| Total S | 20,018 | 19,871 | 58.6 % | 54.7 % |
| TRL/IRL | 65 | 88.5 | 69.3% | 68.4 % |
| UL | 104,036 | 104,836 | 38.3% | 44.3% |
| TE2 | 666 | No | 53.8 % | - |
| Total L | 104,767 | 105,013 | 38.3 % | 44.3 % |
| Total Genome | 124,785 | 124,884 | 40.4% | 46.0 % |
Data derived from (Davison and Scott, 1986)
Terminal element- SVV genome left end.
The structure of SVV DNA was resolved using a variety of approaches including electron microscopy, and restriction endonuclease, lambda exonuclease, and Southern blot hybridization analyses (Gray et al. 1992; Clarke et al. 1992). The presence of inverted repeat sequences, typical for herpesvirus DNAs, was initially indicated by electron microscopy of denatured and re-annealed SVV DNA. SVV molecules consisted of a ≈7.2 kb double-stranded stem continuous with a ≈5.2 kb single-stranded loop and terminating with a long, ≈100 kb single-stranded sequence. Southern blot hybridization and restriction endonuclease analyses were used to confirm the inverted repeat sequences and to construct restriction endonuclease maps of the SVV genome. These studies indicated that the SVV genome consists of a ≈100 kb long (L) component covalently linked to a ≈20 kb short (S) component. The S component includes 7.5 kb terminal and internal inverted repeat sequences (IRS and TRS) which bracket the 4.9 kb unique short (US) component (Figure 2). The detection of 0.5 molar DNA bands in restriction endonuclease profiles, as well as PCR and hybridization analysis of SVV DNA, revealed that the L component can invert relative to the S component and that the SVV genome exists in two major isomeric forms (Gray et al. 1992; Clarke et al. 1995b). Subsequent DNA sequence analysis of the entire SVV genome more precisely defined the SVV DNA size and structure and confirmed that the SVV and VZV genomes are similar in size and DNA structure (Table 1) (Fletcher and Gray 1993; Gray et al. 1995, 2001).
Figure 2.
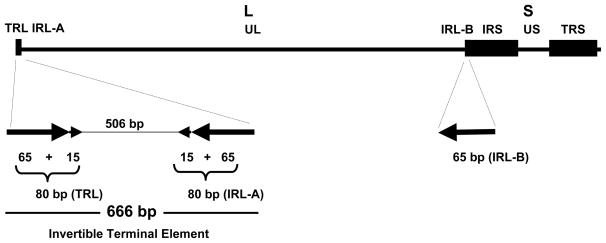
Structure of SVV genome. The 124.7 kb SVV genome consists of a long (L) component covalently linked to a short (S) component. The L component includes a 104.0 kb unique long (UL) segment bracketed by repeat sequences. The S component includes a 4.9 kb unique short (US) segment flanked by 7.5 kb terminal (TRS) and internal (IRS) repeat sequences. The SVV left end has a 666 bp terminal element which includes a 506 bp unique sequence flanked by 80 bp inverted repeats (TRL and IRL-A), of which 65 bp are also present at the right end of the UL segment (IRL-B) (data revised from Mahalingam and Gray, 2007).
The only significant difference between the structure of SVV and VZV DNA occurs at the left end of the viral genomes. The left terminus of the SVV genome possesses a 666 bp terminal element that includes a 506 bp unique sequence bracketed by 80 bp inverted repeat sequences (Figure 2) (Mahalingam and Gray 2007). A portion of this inverted repeat, 65 bp, is also present at the junction of the L and S components, such that the SVV L component is flanked by 65 bp inverted repeat sequences, similar to the 88 bp inverted repeat sequences that bracket the VZV L component. The 666 bp terminal element inverts relative to the SVV DNA UL component. Analysis of the genomes of various SVV isolates indicates that the terminal element is conserved, but may vary in size as the MLM and LVV isolates of SVV possess a 712 bp terminal element which includes an additional 46 bp repeat within the unique sequence. The terminal element was also confirmed to exist in SVV DNA isolated from tissues of acutely and latently infected monkeys. The EHV-1, EHV-4, and PRV varicellovirus genomes also include a analogous terminal element at the left end of their genomes (Mahalingam and Gray 2007). In contrast, the VZV genome does not include a similar element at the left terminus.
Within infected cells, SVV DNA appears to exist as circular or concatemeric genomic molecules (Clarke et al. 1995b). Compared to SVV virion DNA, SVV genomes with connected termini are increased in infected BSC-1 cell DNA as indicated by PCR employing terminal primers from the left and rightward ends of the viral genome. DNA sequence analysis reveals an additional G + C base pair across the connected termini indicating that SVV genome termini possess an unpaired 3′ nucleotide which might enable SVV DNA circulation (Clarke et al. 1995b). Circulation may facilitate SVV DNA replication by a rolling circle mechanism that generates genomic DNA head-to-tail concatemers. While the structure of SVV DNA during latency is unknown, the termini of VZV genomes are connected in latently infected ganglia suggesting that latent VZV DNA exists in an extrachromosomal, circular configuration (Clarke et al. 1995a).
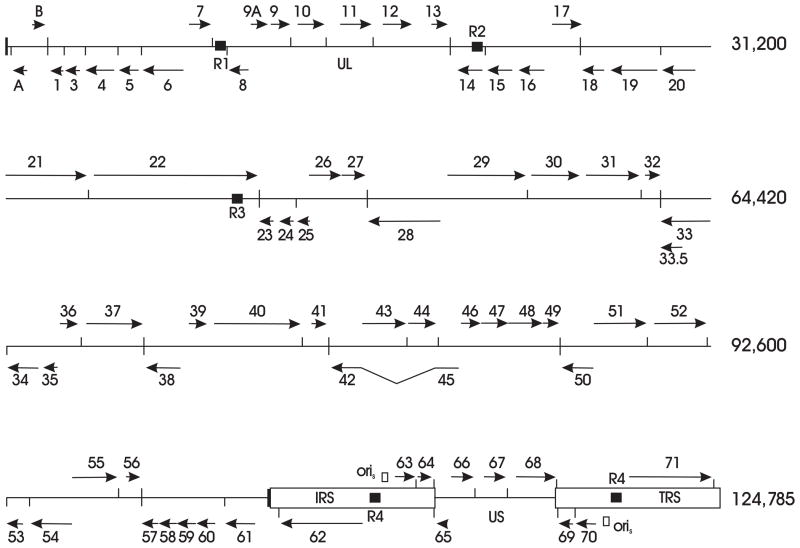
Tandem direct repeat elements, which are typical for herpesvirus genomes, are present in SVV DNA as they are in VZV DNA. In the SVV genome, these repeat elements are designated as R1, R2, R3, and R4 (Figure 3)(Gray et al. 2001). Located between ORFs 7 and 8, SVV R1 is an A + T rich 37 bp element that is repeated three times. SVV R2, a G + C rich 83 bp element that is repeated twice, is present within the viral glycoprotein C (gC) gene (ORF 14). The most complex and largest the SVV tandem repeats is SVV R3, which includes eight 12 bp repeats and seventeen 9 bp repeats found within SVV ORF 22. SVV R4 is a G + C rich 16 bp sequence that is repeated seven times and is present in a noncoding region between ORFs 62 and 63. The VZV genome has reiterations corresponding in location to the SVV R2, R3, and R4 repeat elements, although the DNA sequences of the SVV and VZV tandem repeats are different (Davison and Scott 1986). VZV DNA does not have a reiteration corresponding in location to the SVV R1, but instead the VZV R1 is located within ORF 11. VZV DNA also includes R5, an 88 bp element located between ORF 60 and 61, that is not present in SVV DNA. The function of these conserved repeat elements in SVV and other herpesvirus genomes is not understood.
Figure 3.
The SVV gene map. SVV ORFs are designated as horizontal arrows indicating gene location on each DNA strand. Potential polyadenylation sites are shown as vertical lines. The putative origins of replication (oriS) are indicated as open boxes. The R1, R2, R3, and R4 repeat elements are shown as black boxes.
The SVV and VZV genomes are co-linear with respect to gene organization. The SVV genome encodes 69 distinct ORFs, three of which are duplicated within the S component inverted repeat sequences (Table 2, Figure 3) (Gray et al. 2001). Corresponding SVV and VZV genes share extensive homology with predicted proteins having from 27% to 75% amino acid identity. The only significant difference in SVV and VZV gene content occurs in the left terminus of the viral DNAs (Mahalingam et al. 2000). SVV DNA lacks a homolog to the VZV ORF 2, a gene that is non-essential for in vitro replication and dispensible for the virus to establish latent infection of neural ganglia (Sato et al. 2002). The SVV DNA left terminus includes an 882 bp ORF A that is not present in the VZV genome. The SVV ORF A encodes a truncated homolog of the SVV ORF 4 and the VZV ORF 4, a viral transactivator of viral gene expression (Perera et al. 1994), although the SVV ORF A has not been demonstrated to transactivate SVV promoters (Gray, W.L., unpublished data). The SVV ORF B corresponds to the VZV ORF S/L, located at the VZV DNA left terminus, which encodes a cytoplasmic viral protein of unknown function (Kemble et al. 2000).
Table 2.
SVV open reading frames
| ORF | Start | Stop | Size (aa) | Size(kDa)1 | VZV % homol.2 | VZV Size (aa)3 | HSV-1 homolog | Putative Function4/Notes |
|---|---|---|---|---|---|---|---|---|
| A | 1807 | 926 | 293 | 33.5 | ---- | ---- | UL54 | Truncated homolog of ORF 4 |
| B | 2327 | 2671 | 114 | 12.7 | 29.9 | 157–224 | None | Homolog of VZV ORF S/L |
| 1 | 3036 | 2730 | 101 | 11.7 | 27.3 | 108 | None | Membrane protein |
| 2 | ---- | ---- | ---- | ---- | ---- | 238 | None | SVV DNA does not include a homolog of VZV ORF 2 |
| 3 | 3852 | 3301 | 183 | 19.7 | 63.7 | 179 | UL55 | Virion assembly |
| 4 | 5533 | 4121 | 470 | 54.3 | 43.2 | 452 | UL54 | Transcriptional activator, immediate early protein 2 |
| 5 | 6656 | 5643 | 337 | 42.0 | 59.3 | 340 | UL53 | Glycoprotein K |
| 6 | 9906 | 6661 | 1081 | 123.3 | 37.7 | 1083 | UL52 | Component of DNA helicase-primase complex |
| 7 | 9917 | 10612 | 231 | 25.3 | 72.9 | 259 | UL51 | Virion phosphoprotein |
| 8 | 12068 | 10881 | 395 | 44.9 | 38.2 | 396 | UL50 | DeoxyUTPase |
| 9A | 12037 | 12300 | 87 | 9.7 | 68.8 | 87 | UL49A | Glycoprotein N |
| 9 | 12405 | 13310 | 301 | 33.2 | 59.4 | 302 | UL49 | Tegument protein |
| 10 | 13537 | 14757 | 406 | 46.6 | 62.5 | 410 | UL48 | Transcriptional activator, tegument protein |
| 11 | 15076 | 17004 | 642 | 71.7 | 50.8 | 819 | UL47 | Tegument protein |
| 12 | 17150 | 19117 | 655 | 73.4 | 60.1 | 661 | UL46 | Tegument protein |
| 13 | 19243 | 20130 | 295 | 33.9 | 71.1 | 301 | None | Thymidylate synthetase |
| 14 | 21841 | 20219 | 540 | 60.6 | 43.8 | 560 | UL44 | Glycoprotein C |
| 15 | 23259 | 21994 | 421 | 47.3 | 36.2 | 406 | UL43 | Membrane protein |
| 16 | 24493 | 23336 | 385 | 42.8 | 46.8 | 408 | UL42 | Associated with DNA polymerase |
| 17 | 24719 | 26134 | 471 | 53.6 | 55.0 | 455 | UL41 | Host shutoff virion protein |
| 18 | 27117 | 26182 | 311 | 36.2 | 72.6 | 306 | UL40 | Ribonucleotide reductase, small subunit |
| 19 | 29471 | 27120 | 783 | 88.2 | 68.7 | 775 | UL39 | Ribonucleotide reductase, large subunit |
| 20 | 31068 | 29665 | 467 | 52.9 | 60.5 | 483 | UL38 | Capsid protein |
| 21 | 31243 | 34368 | 1041 | 116.8 | 52.0 | 1038 | UL37 | Tegument protein |
| 22 | 34559 | 42520 | 2653 | 294.8 | 48.9 | 2763 | UL36 | Tegument protein |
| 23 | 43870 | 43184 | 228 | 23.8 | 47.3 | 235 | UL35 | Capsid protein |
| 24 | 44596 | 43952 | 214 | 23.9 | 61.5 | 269 | UL34 | Membrane phosphoprotein |
| 25 | 45337 | 44876 | 153 | 17.4 | 45.2 | 156 | UL33 | Viral DNA cleavage/packaging |
| 26 | 45249 | 46967 | 572 | 65.1 | 61.3 | 585 | UL32 | DNA cleavage/packaging |
| 27 | 46894 | 47826 | 310 | 35.4 | 74.4 | 333 | UL31 | Nuclear phosphoprotein |
| 28 | 51268 | 47750 | 1172 | 113.2 | 65.7 | 1194 | UL30 | DNA polymerase |
| 29 | 51454 | 55038 | 1194 | 132.0 | 71.9 | 1204 | UL29 | Single-stranded DNA binding protein |
| 30 | 55125 | 57407 | 760 | 86.5 | 61.3 | 770 | UL28 | Viral DNA cleavage/packaging |
| 31 | 57266 | 60016 | 916 | 104.0 | 75.4 | 868 | UL27 | Glycoprotein B |
| 32 | 60152 | 60559 | 135 | 15.0 | 49.6 | 143 | None | Phosphoprotein |
| 33 | 62412 | 60646 | 588 | 65.1 | 64.4 | 605 | UL26 | Protease, capsid assembly protein |
| 34 | 64182 | 62443 | 579 | 65.7 | 61.1 | 579 | UL25 | Viral DNA cleavage/packaging |
| 35 | 64992 | 64246 | 248 | 28.7 | 49.7 | 258 | UL24 | Membrane protein |
| 36 | 65018 | 66031 | 337 | 37.9 | 52.3 | 341 | UL23 | Thymidine kinase |
| 37 | 66204 | 68762 | 852 | 96.8 | 55.5 | 841 | UL22 | Glycoprotein H |
| 38 | 70414 | 68813 | 533 | 59.8 | 60.3 | 541 | UL21 | Virion protein |
| 39 | 70693 | 71364 | 223 | 25.4 | 53.1 | 240 | UL20 | Envelope protein, viral egress |
| 40 | 71553 | 75731 | 1392 | 155.9 | 73.3 | 1396 | UL19 | Major capsid protein |
| 41 | 75826 | 76773 | 315 | 34.3 | 70.5 | 316 | UL18 | Capsid protein |
| 42/45 | 78005 82424 |
76821 81372 |
744 | 84.1 | 67.7 | 747 | UL15 | Spliced product5 Viral terminase |
| 43 | 78037 | 80073 | 678 | 75.9 | 47.3 | 676 | UL17 | Viral DNA cleavage/packaging |
| 44 | 80217 | 81299 | 360 | 39.8 | 69.3 | 363 | UL16 | Virion protein |
| 46 | 82536 | 83135 | 199 | 22.5 | 58.2 | 199 | UL14 | Tegument protein |
| 47 | 82988 | 84511 | 507 | 57.7 | 64.8 | 510 | UL13 | Protein Kinase |
| 48 | 84475 | 86004 | 509 | 57.8 | 56.2 | 551 | UL12 | Deoxyribonuclease |
| 49 | 86004 | 86252 | 82 | 9.2 | 50.0 | 81 | UL11 | Myristylated virion protein |
| 50 | 87656 | 86337 | 439 | 49.5 | 57.5 | 435 | UL10 | Glycoprotein M |
| 51 | 87665 | 90115 | 816 | 92.8 | 53.7 | 835 | UL9 | Origin binding protein |
| 52 | 90232 | 92529 | 765 | 86.0 | 50.4 | 771 | UL8 | Component of DNA helicase-primase complex |
| 53 | 93512 | 92598 | 304 | 34.5 | 56.6 | 331 | UL7 | Gamma-1 protein |
| 54 | 95610 | 93403 | 735 | 83.5 | 59.5 | 769 | UL6 | Viral DNA cleavage/packaging |
| 55 | 95645 | 98254 | 869 | 97.8 | 75.2 | 881 | UL5 | Component of DNA helicase-primase complex |
| 56 | 98326 | 98889 | 187 | 21.3 | 37.7 | 244 | UL4 | Gamma-2 protein |
| 57 | 99137 | 98919 | 72 | 8.2 | 39.5 | 71 | None | Nonessential VZV protein |
| 58 | 99745 | 99134 | 203 | 22.9 | 42.0 | 221 | UL3 | Phosphoprotein |
| 59 | 100661 | 99759 | 300 | 34.6 | 55.4 | 305 | UL2 | Uracil DNA glycosylase |
| 60 | 101092 | 100565 | 175 | 20.2 | 43.5 | 159 | UL1 | Glycoprotein L |
| 61 | 103781 | 102270 | 503 | 54.1 | 42.8 | 467 | RL2 | Transcriptional activator, repressor, immediate early protein 1 |
| 62 | 108458 | 104619 | 1279 | 136.8 | 58 | 1310 | RS1 | Transcriptional activator, immediate early protein 3 |
| 63 | 109844 | 110629 | 261 | 29.3 | 52 | 278 | US1 | Transcriptional activator, Immediate early protein 4 |
| 64 | 110851 | 111414 | 187 | 21.1 | 56 | 180 | US10 | Tegument phosphoprotein |
| 65 | 111948 | 111715 | 77 | 9.0 | 49 | 102 | US9 | Tegument phosphoprotein |
| 66 | 112388 | 113425 | 345 | 38.9 | 66 | 393 | US3 | Protein Kinase |
| 67 | 113656 | 114717 | 353 | 40.5 | 37 | 354 | US7 | Glycoprotein I |
| 68 | 114937 | 116751 | 604 | 67.6 | 47 | 623 | US8 | Glycoprotein E |
| 69 | 117408 | 116845 | 187 | 21.1 | 56 | 180 | US10 | Duplicate of orf 64 |
| 70 | 118415 | 117629 | 261 | 29.3 | 52 | 278 | US1 | Duplicate of orf 63 |
| 71 | 119801 | 123640 | 1279 | 136.8 | 58 | 1310 | ICP4 | Duplicate of orf 62 |
Predicted size based on amino acid (AA) sequence
Based on % amino acid identity with the homologous VZV protein
Derived from (Davison and Scott, 1986).
Based on known VZV function or function of HSV-1 homolog (Arvin, 1996; Harper et al., 1998; and Subak-Sharpe and Dargan, 1998)
Predicted spliced gene including ORF 42 and 45 exons
Sensitive techniques have been developed for detection of SVV DNA. A PCR-based assay readily detects SVV DNA in skin rash specimens, peripheral blood lymphocytes, and other tissues of acutely infected monkeys (Gray et al. 1998). In addition, SVV DNA can be detected in neural ganglia of latently infected animals. The SVV PCR assay is specific, detecting DNA derived from various SVV isolates, but does not cross-react with DNA derived from VZV, HSV-1, or other primate herpesviruses. Real-time quantitative PCR assay may be used to determine SVV copy number in infected tissues (White et al. 2002a). These sensitive assays for SVV DNA detection are useful for rapid diagnosis of simian varicella in primate facilities and for investigating SVV pathogenesis.
SVV Gene Expression
Investigation of SVV gene expression is hampered by the cell-associated nature of SVV and the resulting inability to generate high titer infectious virus stocks for synchronous infection in cell culture. However, SVV gene expression, like that of other herpesviruses, is considered to be coordinately regulated into immediate early (IE), early, and late phases with the IE genes expressing regulatory proteins, the early genes encoding enzymes involved in viral DNA synthesis, and late genes expressing structural proteins and glycoproteins.
The SVV IE ORF 62, homolog of the HSV-1 ICP 4, encodes a regulatory protein that serves as a major transactivator of viral genes. The IE62 protein stimulated the SVV ORF 28 and ORF 29 early gene promoters by over 100-fold using a luciferase reporter gene assay (Ou and Gray 2006). A cellular transcription factor, the upstream stimulatory factor (USF), appears to play a critical role, as a USF DNA binding sequence (5′-CACGTG) within the ORF 28/29 bidirectional promoter is necessary for efficient IE62-mediated transactivation. The SVV IE62 also transactivates its own promoter, other SVV immediate early genes (ORFs 4 and 61), other early genes (ORF 21), and late genes (ORF 68) (Mahalingam et al. 2006, Gray, W.L., unpublished data).
The SVV ORFs 4, 61, and 63 are putative IE genes, based upon their VZV and HSV-1 homologs. The SVV ORF 61 protein, homolog to the HSV-1 ICP0, transactivates its own promoter, the ORF 62 IE promoter, early promoters (ORFs 28 and 29), and ORF 68 (gE) late gene promoter in transfected Vero cells (Gray et al. 2007). However, IE61-mediated transactivation is modest compared to IE62-mediated induction and ORF 61 is non-essential for replication in cell culture (Gray et al. 2007). The IE61 protein includes a RING finger motif at the amino-terminus and a nuclear localization signal sequence (NLS) at the carboxy-terminus both of which are required for transactivation. The SVV ORF 63 IE gene product did not transactivate the SVV ORF 21 early promoter upon co-transfection of Vero and Mewo cells (Mahalingam et al. 2006). The IE63 down-regulated IE62-mediated transactivation of the ORF 21 promoter in these cells, but augmented IE62-transactivation of this promoter in neuronal cell culture, indicating that the IE63 has a differential effect on the ORF 21 promoter depending on the cell type (Mahalingam et al. 2006). In limited studies to date, the SVV ORF 4 and its truncated homolog, ORF A, have not been demonstrated to transactivate SVV genes (Gray, W.L., unpublished data).
Expression of several SVV early and late genes has been characterized. The SVV thymidine kinase (ORF 36), uracil DNA glycosylase (ORF 59), and deoxyuridine nucleotidohydrolase (dUTPase, ORF 8) genes are demonstrated to express functional enzymes involved in viral DNA synthesis and repair (Pumphrey and Gray 1996; Ashburn and Gray 1999; Ward et al. 2009). SVV expresses several glycoproteins which are incorporated into the viral envelope and the surface and inner membranes of infected cells. The expression and molecular properties of the SVV gB, gC, gE, gH, and gL have been characterized, although the specific roles of these glycoproteins in viral attachment and penetration has not been studied (Pumphrey and Gray 1994, 1995; Gray and Byrne 2003; Gray et al. 2001; Ashburn and Gray 2001). Unlike HSV-1 and other alphaherpesviruses, SVV and VZV do not express a gD homolog.
All regions of the SVV genome are transcriptionally active during lytic infection of Vero cell monolayers. At least 53 distinct RNA species ranging in size from 9.2 to 0.8 kb were detected by Northern blot hybridization analysis using SVV restriction endonuclease probes representing the entire SVV genome (Gray et al. 1993). A SVV transcription map was constructed which revealed similarities between SVV and VZV gene expression. More recently, microarray assay was used to analyze viral gene transcription from 70 predicted SVV ORFs during lytic infection (Deitch et al. 2006). Using cloned DNA fragments generated from the 5′ and 3′ ends of each SVV ORF and polyadenylated RNA isolated from infected Vero cells at maximal cytopathic effect (3 days postinfection), viral transcripts mapping to each ORF were identified and their relative abundance was determined. The most abundant transcript mapped to SVV ORF 9. Interestingly, a similar array analysis of VZV transcription also identified the VZV ORF 9 as the most abundant transcript in infected BSC-1 cells (Cohrs et al. 2003). The VZV ORF 9 encodes a tegument protein that is essential for replication in cell culture and interacts with IE62 in the cytoplasm of infected cells (Cilloniz et al. 2007). Putative splice sites are predicted for SVV ORF 42/45 and ORF B (S/L) transcripts, but splicing of transcripts is unusual for processing of SVV and VZV RNA (Gray et al. 2001). Analysis of SVV promoter sequences have revealed conservation of the cis acting elements which control SVV and VZV gene expression (Fletcher, III and Gray 1994; Pumphrey and Gray 1994,1996).
SVV gene expression has been analyzed in tissues of African green monkeys with acute simian varicella disease (day 10–12 postinfection). SVV IE, early, and late transcripts and viral antigens were detected in the skin, lung, liver, spleen, and neural ganglia indicating expression of viral genes from across the viral genome and active viral replication in tissues of acutely infected monkeys (Gray et al. 2002). A persistent infection was described in African green monkeys experimentally infected by intratracheal inoculation with IE, early, and late transcripts detected for as long as 12 months postinfection (White et al. 2002b).
In contrast, viral gene expression was restricted in tissues of African green monkeys confirmed to be latently infected following intratracheal and subcutaneous SVV inoculation followed by a second SVV immunization (Ou et al. 2007). A latency associated transcript (LAT) oriented antisense to the SVV ORF 61 mRNA was consistently detected in neural ganglia, but not other tissues, derived from latently infected monkeys (Ou et al. 2007). Similarly, gene expression of HSV-1 and other neurotropic varicelloviruses, including EHV-1 and BHV-1, is limited to expression of a LAT antisense to an ICP0 homolog during latent infection. In contrast, several VZV transcripts (ORF 4, 21, 29, 62, 63, 66) may be detected in latently infected human ganglia (Cohen et al. 2006).
Genetic Manipulation of the SVV Genome- SVV Mutants and Recombinant Viruses
Development of genetic approaches to insert site-specific mutations into the SVV genome is important to elucidate the roles of viral genes in replication and pathogenesis. The inability to generate high titer infectious virus due the cell-associated nature of SVV creates challenges for genetic manipulation of the SVV genome. The initial approach for generating SVV mutants was based upon insertion of a reporter gene encoding the green fluorescent protein (GFP), into the SVV genome by homologous recombination. A SVV gC deletion mutant was constructed by insertional mutagenesis using the GFP gene (Gray and Byrne 2003). A cassette consisting of the GFP gene (under control of the human cytomegalovirus [HCMV] IE promoter) flanked by SVV gC sequences was transfected into SVV-infected BSC-1 cells and the recombinant SVV gC−/GFP mutant was selected by plaque-purification of fluorescent cells. The SVV gC−/GFP mutant replicated efficiently in cell culture indicating that the SVV gC is nonessential for in vitro replication. Similarly, a SVV mutant was constructed by homologous recombination insertion of the GFP reporter gene into the SVV genome between SVV ORFs 66 and 67 (Mahalingam et al. 1998). This recombinant SVV replicated efficiently in Vero cells and in lung tissues of an infected African green monkey. While successful, genetic manipulation of the SVV genome by homologous recombination and insertion of a reporter gene is laborious, requiring extensive plaque purification. In addition, the presence of the reporter gene may complicate evaluation of the SVV mutants in studies of viral pathogenesis, so additional steps are required to remove the foreign gene from the viral genome.
The development of a cosmid-based recombination system was an advance in the ability to genetically manipulate the SVV genome (Gray and Mahalingam 2005). Following a genetic approach used to alter VZV DNA (Cohen and Seidel 1993), the entire SVV genome in four overlapping DNA fragments, each 32–38 kilobase pairs (kbp) in size, was cloned into cosmid vectors. Co-transfection of all four of the SVV cosmids (Cos A, Cos B, Cos C, and Cos D) into Vero or CV-1 cells results in homologous recombination of the overlapping DNA ends and generation of infectious virus plaques within 9 – 12 days post-transfection (Figure 4). A SVV gC− mutant was generated by insertion of a 30 bp oligonucleotide containing translational stop codons into a unique KpnI site within ORF 14 of Cos A, followed by co-transfection of a Vero cell monolayer with Cos A/gC−, Cos B, Cos C, and CosD. Analysis of the infectious SVV gC− virus that was generated verified the mutation within ORF 14 and confirmed that the SVV gC is nonessential for replication in cell culture. A more versatile approach for inserting gene specific mutations within SVV cosmids employs rec A-assisted restriction endonuclease (RARE) cleavage (Ferrin and Camerini-Otero 1991). RARE-mediated deletion of 1.3 kb from the SVV gene 61 within Cos D, followed by co-transfection of Vero cells with the other three cosmids generated a SVV ORF 61 deletion mutant. This SVV ORF61del multiplied only slightly less efficiently than wild-type SVV in CV-1 cells, indicating that SVV 61 transactivation is not critical for in vitro replication (Gray et al. 2007). SVV mutants defective in dUTPase (ORF 8) and UDG (ORF 59) were also generated using RARE mutagenesis and the SVV cosmid recombination system (Ward et al. 2009). While the SVV dUTPase and UDG are non-essential for replication in cell culture, these enzymes may be important for viral DNA synthesis and repair in neuronal cells and play a role in SVV latency and/or reactivation.
Figure 4.
Creation of SVV mutants and recombinant viruses using the SVV cosmid-based recombination system. The relative genomic location of each 32–38 kb SVV fragment cloned into cosmid vectors is indicated. As an example, the arrow indicates the location for insertion of translational stop codons or a foreign gene into the unique KpnI site of ORF 14 (gC gene) within CosA. Following co-transfection of Vero cells, genetic recombination occurs resulting in the generation of infectious recombinant virus harboring the specified mutation or foreign gene.
The SVV cosmid recombination system has also been used to insert foreign genes into the SVV genome. A recombinant SVV expressing simian immunodeficiency virus (SIV) antigens was constructed by insertion of the SIV env and gag genes along with the HCMV IE promoter into the SVV genome at a unique KpnI site within ORF 14 (gC) of Cos A followed by co-transfection of CV-1 cells with the other three cosmids. Immunization of African green monkeys with the rSVV/SIVenv, gag recombinant virus induced antibody and cellular immune responses to SIV gag and env antigens as well as to SVV antigens (Ou et al. 2007). An additional study generated a recombinant SVV expressing the respiratory syncytial virus (RSV) G and M antigens and the rSVV/RSVG, M recombinant virus induced antibody responses to the RSV antigens in immunized monkeys (Ward et al. 2008). These studies provide support for the potential use of recombinant VZV vaccines expressing antigens of other pathogens.
Most recently, the entire SVV genome has been cloned into a bacterial artificial chromosome (BAC) permitting stable maintenance of SVV DNA in E. coli (Gray, W.L., unpublished data). The SVV BAC was constructed by insertion of the BAC vector into Cos A within the intergenic sequence between SVV ORFs 12 and 13, followed by co-transfection of CV-1 cells with CosA-BAC along with Cos B, Cos C, and Cos D. Generation of the SVV-BAC represents an advance over the SVV cosmid system as it avoids the necessity of transfecting four large independent cosmids into the same cell for the required recombination. The BAC sequence is flanked by loxP sites, so that the BAC vector can be excised from the SVV genome by Cre recombinase. The SVV BAC system permits genetic manipulation (site-specific point mutations, insertions, and deletions) of SVV DNA using efficient techniques including Red-mediated recombination (Tischer et al., 2006). This approach was recently used to generate a SVV mutant with a deletion in ORF 10, which encodes a homolog of the HSV-1 VP16 transactivator protein (Gray, W.L., unpublished data). These studies employing SVV mutants will provide a foundation to elucidate the molecular basis of SVV pathogenesis, latency, and reactivation.
Acknowledgments
Research conducted by the author was supported by Public Health Service Grant RO1 AI052373 of the National Institutes of Health.
References
- Ashburn CV, Gray WL. Identification and characterization of the simian varicella virus uracil DNA glycosylase. Arch Virol. 1999;144:2161–2172. doi: 10.1007/s007050050630. [DOI] [PubMed] [Google Scholar]
- Ashburn CV, Gray WL. Expression of the simian varicella virus glycoprotein L and H. Arch Virol. 2001;147:335–348. doi: 10.1007/s705-002-8323-6. [DOI] [PubMed] [Google Scholar]
- Blakely GA, Lourie B, Morton WG, Evans HH, Kaufmann AF. A varicella-like disease in macaque monkeys. J Infect Dis. 1973;127:617–625. doi: 10.1093/infdis/127.6.617. [DOI] [PubMed] [Google Scholar]
- Cilloniz C, Jackson W, Grose C, Czechowski D, Hay J, Ruyechan WT. The varicella-zoster virus (VZV) ORF 9 protein interacts with the IE62 major VZV transactivator. J Virol. 2007;81:761–774. doi: 10.1128/JVI.01274-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke P, Beer T, Cohrs R, Gilden DH. Configuration of latent varicella-zoster virus DNA. J Virol. 1995a;69:8151–8154. doi: 10.1128/jvi.69.12.8151-8154.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke P, Beer T, Gilden DH. Configuration and terminal sequences of the simian varicella virus genome. Virology. 1995b;207:154–159. doi: 10.1006/viro.1995.1061. [DOI] [PubMed] [Google Scholar]
- Clarke P, Rabkin SD, Inman MV, Mahalingam R, Cohrs R, Wellish M, Gilden DH. Molecular analysis of simian varicella virus DNA. Virology. 1992;190:597–605. doi: 10.1016/0042-6822(92)90897-x. [DOI] [PubMed] [Google Scholar]
- Clarkson MJ, Thorpe E, McCarthy K. A virus disease of captive vervet monkeys (Cercopithicus aethiops) caused by a new herpes virus. Arch Gesamte Virusforsch. 1967;22:219–234. doi: 10.1007/BF01240517. [DOI] [PubMed] [Google Scholar]
- Cohen JI, Seidel KE. Generation of varicella-zoster virus (VZV) and viral mutants from cosmid DNAs: VZV thymidylate synthetase is not essential for replication in vitro. Proc Natl Acad Sci USA. 1993;90:7376–7380. doi: 10.1073/pnas.90.15.7376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen JI, Straus SE, Arvin AM. Varicella-zoster virus replication, pathogenesis, and management. In: Knipe DM, et al., editors. Field’s Virology. Lippincott Williams & Wilkins; Philadelphia: 2006. pp. 2774–2840. [Google Scholar]
- Cohrs RJ, Hurley MP, Gilden DH. Array analysis of viral gene transcription during lytic infection of cells in tissue culture with varicella-zoster virus. J Virol. 2003;77:11718–11732. doi: 10.1128/JVI.77.21.11718-11732.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davison AJ, Scott JE. The complete DNA sequence of varicella-zoster virus. J Gen Virol. 1986;67:1759–1816. doi: 10.1099/0022-1317-67-9-1759. [DOI] [PubMed] [Google Scholar]
- Deitch SB, Gilden DH, Wellish M, Smith J, Cohrs R, Mahalingam R. Array analysis of simian varicella virus gene transcription in productively infected cell in tissue culture. J Virol. 2006;79:5315–5325. doi: 10.1128/JVI.79.9.5315-5325.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dueland AN. Simian varicella virus infection in primates as a model for human varicella zoster virus infection. Herpes. 1998;5:76–78. [Google Scholar]
- Felsenfeld AD, Schmidt NJ. Immunological relationship between delta herpesvirus of patas monkeys and varicella-zoster virus of humans. Infect Immun. 1975;12:261–266. doi: 10.1128/iai.12.2.261-266.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felsenfeld AD, Schmidt NJ. Antigenic relationships among several simian varicella-like viruses and varicella-zoster virus. Infect Immun. 1977;15:807–812. doi: 10.1128/iai.15.3.807-812.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felsenfeld AD, Schmidt NJ. Varicella-zoster virus immunizes patas monkeys against simian varicella-like disease. J Gen Virol. 1979;42:171–178. doi: 10.1099/0022-1317-42-1-171. [DOI] [PubMed] [Google Scholar]
- Ferrin LJ, Camerini-Otero RD. Selective cleavage of human DNA: RecA-assisted restriction endonuclease (RARE) cleavage. Science. 1991;254:1494–1497. doi: 10.1126/science.1962209. [DOI] [PubMed] [Google Scholar]
- Fletcher TM, III, Gray WL. Simian varicella virus: Characterization of virion and infected cell polypeptides and the antigenic cross-reactivity with varicella-zoster virus. J Gen Virol. 1992;73:1209–1215. doi: 10.1099/0022-1317-73-5-1209. [DOI] [PubMed] [Google Scholar]
- Fletcher TM, Gray WL. DNA sequence and genetic organization of the unique short (Us) region of the simian varicella virus genome. Virology. 1993;193:762–773. doi: 10.1006/viro.1993.1185. [DOI] [PubMed] [Google Scholar]
- Fletcher TM, III, Gray WL. Transcriptional analysis of two simian varicella virus glycoprotein genes which are homologous to varicella-zoster virus gpl (gE) and gpIV (gl) Virology. 1994;205:352–359. doi: 10.1006/viro.1994.1652. [DOI] [PubMed] [Google Scholar]
- Gray WL. Pathogenesis of simian varicella virus. J Med Virol. 2003;70:S4–S8. doi: 10.1002/jmv.10312. [DOI] [PubMed] [Google Scholar]
- Gray WL. Simian varicella: a model for human varicella-zoster virus infections. Rev Med Virol. 2004;14:363–381. doi: 10.1002/rmv.437. [DOI] [PubMed] [Google Scholar]
- Gray WL. Simian varicella in Old World monkeys. Comp Med. 2008;58:22–30. [PMC free article] [PubMed] [Google Scholar]
- Gray WL, Byrne BH. Characterization of the simian varicella virus glycoprotein C, which is nonessential for in vitro replication. Arch Virol. 2003;148:537–545. doi: 10.1007/s00705-002-0945-9. [DOI] [PubMed] [Google Scholar]
- Gray WL, Davis K, Ou Y, Ashburn C, Ward TM. Simian varicella virus gene 61 encodes a viral transactivator but is non-essential for in vitro replication. Arch Virol. 2007;152:553–563. doi: 10.1007/s00705-006-0866-0. [DOI] [PubMed] [Google Scholar]
- Gray WL, Gusick N, Fletcher TM, Pumphrey CY. Characterization and mapping of simian varicella virus transcripts. J Gen Virol. 1993;74:1639–1643. doi: 10.1099/0022-1317-74-8-1639. [DOI] [PubMed] [Google Scholar]
- Gray WL, Gusick NJ. Viral isolates derived from simian varicella epizootics are genetically related but distinct from other primate herpesviruses. Virology. 1996;224:161–166. doi: 10.1006/viro.1996.0517. [DOI] [PubMed] [Google Scholar]
- Gray WL, Gusick NJ, Ek-kommonen C, Kempson SE, Fletcher TM. The inverted repeat regions of the simian varicella virus and varicella-zoster virus genomes have a similar genetic organization. Virus Res. 1995;39:181–193. doi: 10.1016/0168-1702(95)00091-7. [DOI] [PubMed] [Google Scholar]
- Gray WL, Mahalingam R. A cosmid-based system for inserting mutations and foreign genes into the simian varicella virus genome. J Virol Meth. 2005;130:89–94. doi: 10.1016/j.jviromet.2005.06.013. [DOI] [PubMed] [Google Scholar]
- Gray WL, Mullis LB, Soike KF. Expression of the simian varicella virus glycoprotein E. Virus Res. 2001;79:27–37. doi: 10.1016/s0168-1702(01)00281-7. [DOI] [PubMed] [Google Scholar]
- Gray WL, Mullis LB, Soike KF. Viral gene expression during acute simian varicella virus infection. J Gen Virol. 2002;83:841–846. doi: 10.1099/0022-1317-83-4-841. [DOI] [PubMed] [Google Scholar]
- Gray WL, Oakes JE. Simian varicella virus DNA shares homology with human varicella-zoster virus DNA. Virology. 1984;136:241–246. doi: 10.1016/0042-6822(84)90263-0. [DOI] [PubMed] [Google Scholar]
- Gray WL, Pumphrey CY, Ruyechan WT, Fletcher TM. The simian varicella virus and varicella zoster virus genomes are similar in size and structure. Virology. 1992;186:562–572. doi: 10.1016/0042-6822(92)90022-h. [DOI] [PubMed] [Google Scholar]
- Gray WL, Starnes B, White MW, Mahalingam R. The DNA sequence of the simian varicella virus genome. Virology. 2001;284:123–130. doi: 10.1006/viro.2001.0912. [DOI] [PubMed] [Google Scholar]
- Gray WL, Williams RJ, Soike KF. Rapid diagnosis of simian varicella using the polymerase chain reaction. Lab Anim Sci. 1998;48:45–49. [PubMed] [Google Scholar]
- Kemble GW, Annunziato PW, Lungu O, Winter RE, Cha T, Silverstein SJ, Spaete RR. Open reading frame S/L of varicella-zoster virus encodes a cytoplasmic protein expressed in infected cells. J Virol. 2000;74:11311–11321. doi: 10.1128/jvi.74.23.11311-11321.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahalingam R, Clarke P, Wellish M, Dueland AN, Soike KF, Gilden DH, Cohrs R. Prevalence and distribution of latent simian varicella virus DNA in monkey ganglia. Virology. 1992;188:193–197. doi: 10.1016/0042-6822(92)90749-f. [DOI] [PubMed] [Google Scholar]
- Mahalingam R, Gilden DH, Wellish M, Pugazhenthi S. Transactivation of the simian varicella virus (SVV) open reading fram (ORF) 21 promoter by SVV ORF 62 is upregulated in neuronal cells but downregulated in non-neuronal cells by SVV ORF 63 protein. Virology. 2006;345:244–250. doi: 10.1016/j.virol.2005.08.045. [DOI] [PubMed] [Google Scholar]
- Mahalingam R, Gray WL. The simian varicella virus genome contains an invertible 665 base pair terminal element that is absent in the varicella-zoster virus genome. Virology. 2007;366:387–393. doi: 10.1016/j.virol.2007.04.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahalingam R, Smith D, Wellish M, Wolf W, Dueland AN, Cohrs R, Soike K, Gilden D. Simian varicella virus DNA in dorsal root ganglia. Proc Natl Acad Sci USA. 1991;88:2750–2752. doi: 10.1073/pnas.88.7.2750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahalingam R, Wellish M, White T, Soike K, Kleinschmidt-DeMasters BK, Gilden DH. Infectious simian varicella virus expressing the green fluorescent protein. J Neurovirol. 1998;4:438–444. doi: 10.3109/13550289809114543. [DOI] [PubMed] [Google Scholar]
- Mahalingam R, White T, Wellish M, Gilden D, Gray WL. Sequence analysis of the leftward end of simian varicella virus (EcoRI- I fragment) reveals the presence of an 8-bp repeat flanking the unique long segment and an 881-bp open reading frame that is absent in the varicella-zoster virus genome. Virology. 2000;274:420–428. doi: 10.1006/viro.2000.0465. [DOI] [PubMed] [Google Scholar]
- Ou Y, Davis KA, Traina-Dorge V, Gray WL. Simian varicella virus expresses a latency associated transcript that is antisense to ORF 61 (ICP0) mRNA in neural ganglia of latency infected monkeys. J Virol. 2007;81:8149–8156. doi: 10.1128/JVI.00407-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ou Y, Gray WL. The simian varicella virus gene 28 and 29 promoters share a common USF binding site and are induced by IE62 transactivation. J Gen Virol. 2006;87:1501–1508. doi: 10.1099/vir.0.81645-0. [DOI] [PubMed] [Google Scholar]
- Ou Y, Traina-Dorge V, Davis KA, Gray WL. Recombinant simian varicella vaccines induce immune responses to simian immunodeficiency virus (SIV) antigens in imunized vervet monkeys. Virology. 2007;364:291–300. doi: 10.1016/j.virol.2007.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perera LP, Kaushal S, Kinchington PR, Mosca JD, Hayward GS, Straus SE. Varicella-zoster virus open reading frame 4 encodes a transcriptional activator that is functionally distinct from that of herpes simplex virus homolog ICP27. J Virol. 1994;68:2468–2477. doi: 10.1128/jvi.68.4.2468-2477.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pumphrey CY, Gray WL. The genomes of simian varicella virus and varicella zoster virus are colinear. Virus Res. 1992;26:255–266. doi: 10.1016/0168-1702(92)90017-4. [DOI] [PubMed] [Google Scholar]
- Pumphrey CY, Gray WL. DNA sequence and transcriptional analysis of the simian varicella virus glycoprotein B gene. J Gen Virol. 1994;75:3219–3227. doi: 10.1099/0022-1317-75-11-3219. [DOI] [PubMed] [Google Scholar]
- Pumphrey CY, Gray WL. DNA sequence of the simian varicella virus (SVV) glycoprotein H (gH) gene and analysis of the SVV and varicella-zoster virus (VZV) gH transcripts. Virus Res. 1995;38:55–70. doi: 10.1016/0168-1702(95)00049-v. [DOI] [PubMed] [Google Scholar]
- Pumphrey CY, Gray WL. Identification and analysis of the simian varicella virus thymidine kinase gene. Arch Virol. 1996;141:43–55. doi: 10.1007/BF01718587. [DOI] [PubMed] [Google Scholar]
- Sato H, Pesnicak L, Cohen JI. Varicella-zoster virus open reading frame 2 encodes a membrane phosphoprotein that is dispensible for viral replication and for establishment of latency. J Virol. 2002;76:3575–3578. doi: 10.1128/JVI.76.7.3575-3578.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt NJ. Improved yields and assay of simian varicella virus, and a comparison of certain biological properties of simian and human varicella viruses. J Virol Meth. 1982;5:229–241. doi: 10.1016/0166-0934(82)90013-1. [DOI] [PubMed] [Google Scholar]
- Soike KF. Simian varicella virus infection in African and Asian monkeys. The potential for development of antivirals for animal diseases. Ann NY Acad Sci. 1992;653:323–333. doi: 10.1111/j.1749-6632.1992.tb19659.x. [DOI] [PubMed] [Google Scholar]
- Tischer BK, von Einem J, Kaufer B, Osterrieder N. Two-step Red-mediated recombination for versatile high-efficiency markerless DNA manipulation in Escherichia coli. Biotechniques. 2006;40:191–196. doi: 10.2144/000112096. [DOI] [PubMed] [Google Scholar]
- Ward TM, Traina-Dorge V, Davis KA, Gray WL. Recombinant simian varicella viruses expressing respiratory syncytial virus antigens are immunogenic. J Gen Virol. 2008;89:741–740. doi: 10.1099/vir.0.83453-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward TM, Williams MV, Traina-Dorge V, Gray WL. The simian varicella virus uracil glycosylase and dUTPase genes are expressed in vivo, but are non-essential for replication in cell culture. Virus Res. 2009;142:78–84. doi: 10.1016/j.virusres.2009.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White TM, Gilden DH, Mahalingam R. An animal model of varicella virus infection. Brain Pathol. 2001;11:475–479. doi: 10.1111/j.1750-3639.2001.tb00416.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White TM, Mahalingam R, Traina-Dorge V, Gilden DH. Persistence of simian varicella virus DNA in CD4+ and CD8+ blood mononuclear cells for years after intratracheal inoculation of African green monkeys. Virology. 2002a;303:192–198. doi: 10.1006/viro.2002.1664. [DOI] [PubMed] [Google Scholar]
- White TM, Mahalingam R, Traina-Dorge V, Gilden DH. Simian varicella virus DNA is present and transcribed months after experimental infection of adult African green monkeys. J Neurovirol. 2002b;8:191–205. doi: 10.1080/13550280290049705. [DOI] [PubMed] [Google Scholar]