ABSTRACT
Nucleotide-binding domain, leucine-rich repeat containing proteins (NLRs) activate caspase-1 in response to a variety of bacterium-derived signals in macrophages. NLR-mediated activation of caspase-1 by Legionella pneumophila occurs through both an NLRC4/NAIP5-dependent pathway and a pathway requiring the adapter protein Asc. Both pathways are needed for maximal activation of caspase-1 and for the release of the cytokines interleukin-1β (IL-1β) and IL-18. Asc is not required for caspase-1-dependent pore formation and cell death induced upon infection of macrophages by L. pneumophila. Here, temporal and spatial localization of caspase-1-dependent processes was examined to better define the roles of Asc and NLRC4 during infection. Imaging studies revealed that caspase-1 localized to a single punctate structure in infected cells containing Asc but not in cells lacking this adapter. Both endogenous Asc and ectopically produced NLRC4 tagged with green fluorescent protein (GFP) were found to localize to caspase-1 puncta following L. pneumophila infection, suggesting that NLRC4 and Asc coordinate signaling through this complex during caspase-1 activation. Formation of caspase-1-containing puncta correlated with caspase-1 processing, suggesting a role for the Asc/NLRC4/caspase-1 complex in caspase-1 cleavage. In cells deficient for Asc, NLRC4 did not assemble into discrete puncta, and pyroptosis occurred at an accelerated rate. These data indicate that Asc mediates integration of NLR components into caspase-1 processing platforms and that recruitment of NLR components into an Asc complex can dampen pyroptotic responses. Thus, a negative feedback role of complexes containing Asc may be important for regulating caspase-1-mediated responses during microbial infection.
IMPORTANCE
Caspase-1 is a protease activated during infection that is central to the regulation of several innate immune pathways. Studies examining the macromolecular complexes containing this protein, known as inflammasomes, have provided insight into the regulation of this protease. This work demonstrates that the intracellular bacterium Legionella pneumophila induces formation of complexes containing caspase-1 by multiple mechanisms and illustrates that an adapter molecule called Asc integrates signals from multiple independent upstream caspase-1 activators in order to assemble a spatially distinct complex in the macrophage. There were caspase-1-associated activities such as cytokine processing and secretion that were controlled by Asc. Importantly, this work uncovered a new role for Asc in dampening a caspase-1-dependent cell death pathway called pyroptosis. These findings suggest that Asc plays a central role in controlling a distinct subset of caspase-1-dependent activities by both assembling complexes that are important for cytokine processing and suppressing processes that mediate pyroptosis.
Introduction
Activation of the cysteine protease caspase-1 is an important function of the innate immune system during the response to microbial pathogens and toxins. Upon activation of caspase-1, this protease is able to act on a large variety of downstream substrates, including the proinflammatory cytokines interleukin-1β (IL-1β) and IL-18 (1). Cleavage of these cytokines promotes their secretion from host cells, where they can signal to neighboring cells (1). In addition to cleavage of cytokines, active caspase-1 is able to induce pore formation in host cell membranes, leading to disruption of ion fluxes and osmotic lysis of the cell, or pyroptosis (2).
The ability of caspase-1 to cleave its target substrates is directly influenced by a repertoire of upstream sensor proteins comprised of the nucleotide-binding domain, leucine-rich repeat containing proteins (NLRs) and absent in melanoma 2 (Aim2) (3). These proteins are thought to initiate or be intermediates in signaling to caspase-1 following the detection of cytosolic factors produced by microbes or that indicate cellular dysfunction. The protein NLRC4 is thought to interact with caspase-1 directly through homotypic caspase recruitment domain (CARD) interactions following detection of microbial products in the cytosol, which include bacterial flagellin and the type III secretion system rod protein (4–6). In contrast, NLRP3, which lacks a CARD, interacts with an adapter protein called Asc following stimulation. Asc is a bipartite protein containing both a pyrin domain (PYD) and a CARD that is able to bridge the PYD of NLRP3 and the CARD of caspase-1 in order to form an activation complex (7). NLRP3 is thought to induce caspase-1 activation in response to a large variety of stimuli from both endogenous and microbial origins (8). In addition to NLRP3, the mammalian genome encodes many other NLRP proteins, which may function in caspase-1 activation or activation of other innate immune signaling pathways. In addition to the NLRP family of proteins, Aim2 has also been shown to activate caspase-1 through the adapter protein Asc following detection of DNA in the host cell cytosol (9, 10).
Bacterial pathogens encode a variety of molecules that might function as agonists for NLR proteins when present in the host cell cytosol. Thus, it is not surprising that bacteria induce caspase-1 activation through pathways involving multiple NLRs and Aim2. One such example is the intracellular pathogen Legionella pneumophila. This Gram-negative bacterium is normally found in freshwater environments, where it parasitizes amoebas (11). L. pneumophila is able to invade and replicate in alveolar macrophages of mammalian hosts upon aerosolization of water droplets containing these bacteria (12). Human infection can lead to a severe pneumonia known as Legionnaires’ disease (13, 14). L. pneumophila pathogenesis is dependent on a functional type IV secretion apparatus called Dot/Icm (15). This protein secretion system is utilized by the bacterium to translocate effector proteins into the host cell cytosol in order to manipulate host vesicular transport proteins and create a vacuole suitable for replication (16). Although the Dot/Icm secretion apparatus is critical for pathogenesis, microbial products delivered into the host cytosol by the secretion system are detected by host receptors and trigger innate immune responses during infection (17–22). One such response is the activation of caspase-1.
The L. pneumophila Dot/Icm system is required for activation of caspase-1 by two distinct mechanisms that can be separated genetically. One pathway requires the host protein NLRC4 and is dependent on bacterial flagellin (20, 23–25). This pathway is activated in response to L. pneumophila and also in response to Salmonella enterica serovar Typhimurium and Listeria monocytogenes (5, 6). NLRC4-mediated activation of caspase-1 during L. pneumophila infection requires an additional NLR protein known as NAIP5 (20, 26). This protein is thought to function cooperatively with NLRC4 in the response to L. pneumophila flagellin in murine macrophages (26, 27). In addition to the NLRC4-dependent activation pathway, there is an Asc-dependent pathway for caspase-1 activation in response to L. pneumophila infection that is independent of flagellin and NLRC4 (17). Protein sensors upstream of Asc remain unknown, and elimination of NLRP3 is not sufficient to block the Asc-dependent pathway during L. pneumophila infection (17, 28). Although both NLRC4 and Asc are required for maximal production of IL-1β and IL-18 in response to L. pneumophila (17), NLRC4-mediated pyroptosis by L. pneumophila does not require Asc (17). Similarly, activation of NLRC4 by L. pneumophila restricts bacterial replication, and Asc is not required for the caspase-1-dependent restriction of L. pneumophila replication (17, 20). These data suggest that NLRC4 and Asc regulate distinct caspase-1-dependent processes during L. pneumophila infection, with Asc being important for the processing and release of proinflammatory cytokines and NLRC4 being required to direct events that lead to pore formation and pyroptosis.
One possible mechanism for regulation of caspase-1 is through spatial organization of complexes containing caspase-1, which would provide restricted access to substrates cleaved by this protease. Consistent with this hypothesis, Asc has been shown to mediate the localization of caspase-1 into distinct foci during Salmonella infection in macrophages (29). These foci are thought to be caspase-1 activation platforms known as inflammasomes (30). To better understand the roles of NLRC4 and Asc in controlling distinct caspase-1-dependent events that occur during infection of macrophages by L. pneumophila, the distribution and spatial regulation of inflammasome components were investigated.
RESULTS
Caspase-1 is recruited to punctate structures after macrophage infection by L. pneumophila.
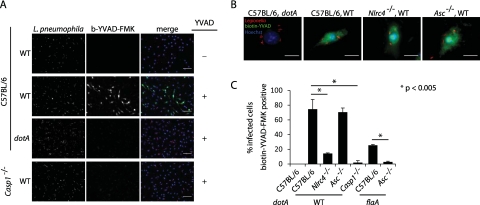
It had been previously shown that L. pneumophila activates caspase-1 through both Asc- and NLRC4-dependent mechanisms (17). Furthermore, given the differential roles of Asc and NLRC4 for pore formation and restriction of intracellular L. pneumophila replication, we hypothesized that Asc and NLRC4 may regulate caspase-1 activation in a spatial manner in order to elicit distinct downstream processes. To address this possibility, the localization of active caspase-1 in the context of Asc- or NLRC4-dependent signaling events was examined. Localization of active caspase-1 was monitored using a biotinylated caspase-1 substrate (biotin-YVAD-FMK) that binds irreversibly to the caspase-1 active site cysteine following activation. Bone marrow macrophages (BMMs) derived from either wild-type (C57BL/6) mice or mice lacking NLRC4, Asc, or caspase-1 were infected with L. pneumophila for 2 h and analyzed by fluorescence microscopy. Active caspase-1 was detected in wild-type macrophages infected with wild-type L. pneumophila but not the type IV secretion-deficient dotA mutant (Fig. 1A). Detection of active caspase-1 using biotin-YVAD-FMK was specific, as Casp1−/− macrophages did not exhibit detectable levels of staining following infection (Fig. 1A). In agreement with previous studies, L. pneumophila infection resulted in the activation of caspase-1 in the absence of NLRC4 or Asc alone, confirming that biotin-YVAD-FMK staining can be used to analyze the two genetically distinct pathways for caspase-1 activation (Fig. 1B and C). Interestingly, in the absence of Asc, the number of macrophages found to contain active caspase-1 was similar to that of wild-type macrophages (Fig. 1C). In contrast, significantly fewer NLRC4-deficient macrophages contained active caspase-1, suggesting that NLRC4 is the major determinant for caspase-1 activation during L. pneumophila infection (Fig. 1C). Furthermore, caspase-1 activation was not detectable in Asc−/− macrophages infected with flagellin-deficient L. pneumophila (Fig. 1C). These single-cell assays corroborate previous genetic studies and further demonstrate that L. pneumophila activates both NLRC4-dependent and NLRC4-independent Asc-dependent pathways for caspase-1 activation.
FIG 1 .
Detection of active caspase-1 during NLRC4- or Asc-dependent activation using biotin-YVAD-FMK. BMMs were infected with L. pneumophila at an MOI of 10 in the presence of biotin-YVAD-FMK for 2 hours. Bound biotin-YVAD-FMK was detected with streptavidin conjugated to Alexa Fluor 488. L. pneumophila were detected using a rabbit polyclonal antibody. (A) Wild-type or caspase-1-deficient (Casp1−/−) BMMs were infected with the wild type (WT) or L. pneumophila dotA. Images were obtained using a 20× lens. The scale bar equals 50 µm. (B) BMMs from wild-type, Asc-deficient (Asc−/−), or NLRC4-deficient (Nlrc4−/−) BMMs were infected with the wild type or L. pneumophila dotA. The scale bar equals 10 µm. (C) Quantification of biotin-YVAD-FMK-positive cells following infection of wild-type, Asc−/−, Nlrc4−/−, or Casp1−/− BMMs with wild-type L. pneumophila or L. pneumophila dotA or flaA. Data are represented as averages ± standard deviations for 3 coverslips. One hundred infected cells were scored per coverslip. Data are representative of four experiments.
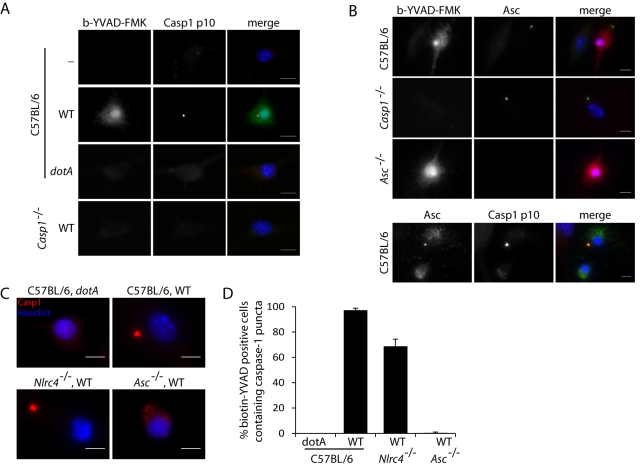
Areas of the cell where active caspase-1 localized were compared in wild-type, NLRC4-deficient, and Asc-deficient macrophages to elucidate potential differences in activation sites. Active caspase-1 appeared localized throughout the cytosol and nuclei of infected macrophages of all cell types tested (Fig. 1B). In addition, a portion of staining was concentrated to punctate structures in the cytosol of infected cells (Fig. 2A). These structures resembled the caspase-1 foci previously reported during infection of macrophages by S. enterica serovar Typhimurium (29, 31). These caspase-1 foci were found to be enriched in both active and inactive caspase-1, suggesting that they might comprise a platform that is used for caspase-1 activation (32). Macrophages were infected with L. pneumophila and costained both for active caspase-1 using biotin-YVAD-FMK and for total caspase-1 using a polyclonal antibody specific for caspase-1. The cellular pool of caspase-1 was found to localize primarily to these distinct foci during L. pneumophila infection (Fig. 2A).
FIG 2 .
Caspase-1 puncta induced by L. pneumophila require Asc but not NLRC4. BMMs were infected with the wild type or L. pneumophila dotA at an MOI of 10 in the presence of biotin-YVAD-FMK for 4 hours. Biotin-YVAD-FMK bound to active caspase-1 was detected with streptavidin conjugated to Alexa Fluor 488. (A) Wild-type or Casp1−/− BMMs were infected with L. pneumophila strains. Caspase-1 protein was detected using a rabbit polyclonal antibody to the p10 fragment of caspase-1. (B, top) BMMs from wild-type, Asc−/−, or Casp1−/− BMMs were infected with L. pneumophila and stained for Asc protein using a rabbit polyclonal antibody. (Bottom) Wild-type macrophages infected with L. pneumophila were stained for caspase-1 using a rabbit polyclonal antibody specific to the p10 fragment and for Asc using a goat polyclonal antibody. (C) BMMs from wild-type, Asc−/−, or Nlrc4−/− BMMs were infected with L. pneumophila strains and probed for caspase-1 using a rabbit polyclonal antibody to the p10 fragment of caspase-1. (D) Quantification of biotin-YVAD-FMK-positive cells containing caspase-1 puncta following infection of wild-type, Asc−/−, or Nlrc4−/− BMMs with L. pneumophila strains. Caspase-1 protein was detected using a rabbit polyclonal antibody to the p10 fragment. Data are represented as averages ± standard deviations for 3 coverslips. One hundred biotin-YVAD-FMK positive cells were scored per coverslip. Data are representative of three experiments. The scale bars equal 10 µm.
Asc is required for the recruitment of caspase-1 to punctate structures after L. pneumophila infection of macrophages.
To determine if other inflammasome components were located in the caspase-1-containing foci, L. pneumophila-infected macrophages were stained using a polyclonal antibody specific for Asc, and the localization of Asc was visualized by fluorescence microscopy. Similar to the localization pattern observed for caspase-1, fluorescence microscopy indicated that Asc relocalized into a punctate structure following L. pneumophila infection (Fig. 2B). Importantly, Asc puncta were formed upon infection of Casp1−/− macrophages (Fig. 2B), indicating that the formation of this Asc-containing structure was independent of caspase-1. Lastly, costaining revealed that caspase-1 and Asc colocalized in these punctate structures (Fig. 2B). These data suggest that infection by L. pneumophila triggers formation of Asc puncta, which then recruit caspase-1.
In order to examine the individual contributions of NLRC4 and Asc to caspase-1-dependent punctum formation, macrophages deficient in NLRC4 or Asc were infected with L. pneumophila and stained for caspase-1. These data show that Asc was required for efficient formation of caspase-1 puncta during L. pneumophila infection (Fig. 2C and D). In contrast, NLRC4 was dispensable for the formation of caspase-1 puncta, albeit formation of caspase-1 puncta was less frequent in the absence of NLRC4 (Fig. 2C and D), further illustrating that L. pneumophila infection stimulates multiple pathways that lead to caspase-1 activation. In summary, these data demonstrate spatially distinct patterns of caspase-1 activation in the cell that correlate with distinct platforms regulated by NLRC4 and Asc.
Asc is important for caspase-1 cleavage but not for caspase-1 activation following L. pneumophila infection of macrophages.
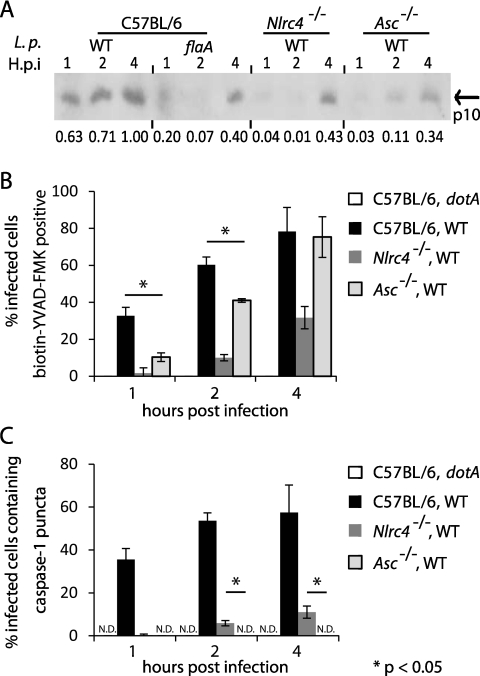
Although the biotin-YVAD-FMK staining indicates that activation of caspase-1 by NLRC4 does not appear to require Asc, it has been shown that Asc is critical for cleavage of caspase-1 after NLR activation (33–35). Thus, the role of Asc in the activation and cleavage of caspase-1 was assessed during L. pneumophila infection. Macrophages derived from Asc-deficient and NLRC4-deficient mice were infected for different lengths of time with L. pneumophila. Immunoblot analysis measuring the appearance of the p10 subunit of caspase-1 was used to detect cleavage, and biotin-YVAD-FMK staining was used to assess activation of caspase-1. In C57BL/6 macrophages infected with L. pneumophila, active and cleaved caspase-1 enzymes were detected at 1 h postinfection, and the levels of each increased gradually over the 4 h of infection (Fig. 3A and B). Caspase-1 cleavage was not detectable in NLRC4-deficient macrophages until 4 h postinfection (Fig. 3A), which correlated with the appearance of active caspase-1 in these macrophages (Fig. 3B). In contrast, in Asc-deficient macrophages, active caspase-1 was observed at 2 h postinfection; however, relatively low levels of cleaved caspase-1 were detectable at these early times (Fig. 3A and B). When Asc-deficient macrophages were compared to control C57BL/6 macrophages at 4 h postinfection, the percentages of macrophages that stained positive for active caspase-1 were similar (Fig. 3B). Consistent with Asc being important for caspase-1 cleavage, the levels of cleaved caspase-1 produced by infected Asc-deficient macrophages were greatly diminished compared to those produced by infected macrophages derived from control C57BL/6 mice (Fig. 3A). NLRC4-deficient macrophages showed diminished levels of active caspase-1 by biotin-YVAD-FMK staining at 4 h postinfection; however, the levels of cleaved caspase-1 from the NLRC4-deficient macrophages were much higher than those observed for the Asc-deficient cells at this time (Fig. 3A and B). Additionally, the detection of active caspase-1 in Asc-deficient macrophages occurred independently of the detectable puncta containing caspase-1 that was observed in the cells containing Asc (Fig. 3C). In contrast, cleavage of caspase-1 was difficult to detect in the Asc-deficient cells, which showed a strong defect in the formation of caspase-1-containing puncta (Fig. 3A and C). Lastly, the temporal delay in caspase-1 activation and cleavage observed in NLRC4-deficient macrophages correlated with the kinetics of punctum formation (Fig. 3A and C). These data suggest that Asc is not required for the activation of caspase-1 upon NLRC4 stimulation by L. pneumophila; however, Asc does play an important role in the recruitment and cleavage of caspase-1 in the puncta formed upon stimulation of the NLRC4 activation pathway and the pathway that is independent of NLRC4.
FIG 3 .
Asc is important for caspase-1 processing but not caspase-1 activation. Wild-type, Asc−/−, or Nlrc4−/− BMMs were infected with the L. pneumophila wild-type, dotA, or flaA strain at an MOI of 10 for various times. (A) Immunoblot analysis of combined culture supernatants and lysates to detect the p10 fragment of cleaved caspase-1. Numbers below the lanes indicate the relative densities of p10 bands for each sample compared to those of the wild type at 4 hours postinfection after normalizing to a loading control band for each sample on the same gel. (B and C) Macrophages were incubated with biotin-YVAD-FMK for 15 minutes prior to infection. Biotin-YVAD-FMK bound to active caspase-1 was detected using streptavidin conjugated to Alexa Fluor 488. (B) Infected cells positive for biotin-YVAD-FMK were scored. (C) Infected cells containing caspase-1 puncta were scored. Caspase-1 protein was detected using a rabbit polyclonal antibody to the p10 fragment. Data are represented as averages ± standard deviatiosn for 3 coverslips in each experiment. One hundred infected cells were scored per coverslip in each experiment. ND, not detected. Data are representative of at least two experiments.
Asc recruits NLRC4 to punctate structures following L. pneumophila infection of macrophages.
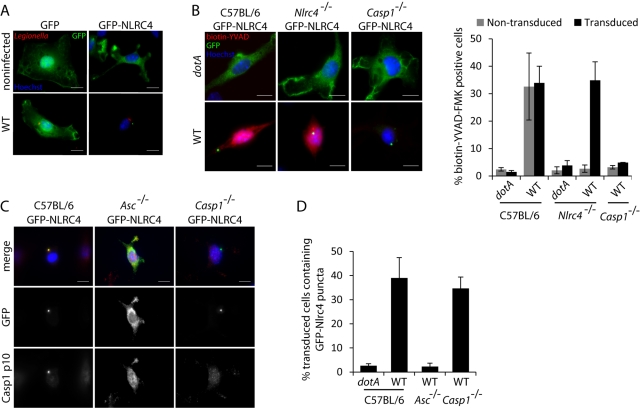
Although Asc was not necessary for activation of caspase-1 following stimulation of NLRC4, the observation that activation of NLRC4 resulted in the recruitment of caspase-1 to Asc-containing puncta raised the question of whether NLRC4 was also recruited to these structures. Bone marrow-derived cells were transduced with a retroviral vector encoding the green fluorescent protein (GFP)-NLRC4 protein so that NLRC4 localization could be visualized in macrophages. In unstimulated macrophages, the GFP-NLRC4 protein was dispersed throughout the cytosol, similar to cells expressing GFP alone (Fig. 4A). The GFP-NLRC4 protein localized to a single punctate structure following infection of macrophages by L. pneumophila (Fig. 4A), and this GFP-NLRC4 structure resembled the puncta observed previously that contained Asc and caspase-1. In control macrophages producing GFP alone, there was no localization of GFP to punctate structures following infection (Fig. 4A). Complementation assays showed that production of GFP-NLRC4 in Nlrc4−/− macrophages restored the ability of these cells to activate caspase-1 in response to wild-type L. pneumophila and that localization of GFP-NLRC4 to a punctate structure correlated with stimulation of the pathway leading to caspase-1 activation (Fig. 4B).
FIG 4 .
NLRC4 is recruited to Asc-dependent punctate structures during L. pneumophila infection. Mouse bone marrow cells from wild-type, Asc−/−, Nlrc4−/−, or Casp1−/− mice were transduced with a retrovirus encoding either GFP or GFP-NLRC4. Transduced macrophages were infected with L. pneumophila strains at an MOI of 10 for 1 hour. (A) Wild-type macrophages expressing GFP or GFP-NLRC4 were infected with L. pneumophila and stained for GFP using a mouse polyclonal antibody and for L. pneumophila using a rabbit polyclonal antibody. (B) Macrophages expressing GFP-NLRC4 were infected with L. pneumophila in the presence of biotin-YVAD-FMK. Biotin-YVAD-FMK bound to active caspase-1 was detected using streptavidin conjugated to Alexa Fluor 594. (Right) Quantification of data obtained. Data are represented as averages ± standard deviations for 3 coverslips in each experiment. One hundred cells were scored per coverslip in each experiment. (C) Macrophages expressing GFP-NLRC4 were infected with L. pneumophila and probed for GFP using a mouse polyclonal antibody and for caspase-1 using a rabbit polyclonal antibody to the p10 fragment. Data are representative of at least two experiments. The scale bars equal 10 µm. (D) Quantification of GFP-NLRC4 puncta from the data obtained from experiments shown in panel C. Data are represented as averages ± standard deviations for 3 coverslips in each experiment. One hundred transduced cells were scored per coverslip in each experiment.
Because the GFP-NLRC4 complexes were morphologically similar to the punctate structures containing Asc that appeared in cells after stimulation of caspase-1 activation pathways, studies were conducted to determine if Asc was required for recruitment of GFP-NLRC4 to these punctate structures. GFP-NLRC4 was produced in Asc−/− macrophages. Localization of GFP-NLRC4 to punctate structures was not observed in the Asc-deficient macrophages after infection by L. pneumophila (Fig. 4C and D). Importantly, the localization of GFP-NLRC4 to punctate structures after stimulation of macrophages by L. pneumophila did not require caspase-1 (Fig. 4C and D), indicating that recruitment of GFP-NLRC4 to these structures by Asc did not require interactions with caspase-1. Thus, Asc could play a role in modulating NLRC4 function through recruitment of this protein into the punctate structures formed upon stimulation of a caspase-1 activation pathway.
NLRC4 recruitment into Asc-dependent punctate structures is not required for pyroptosis.
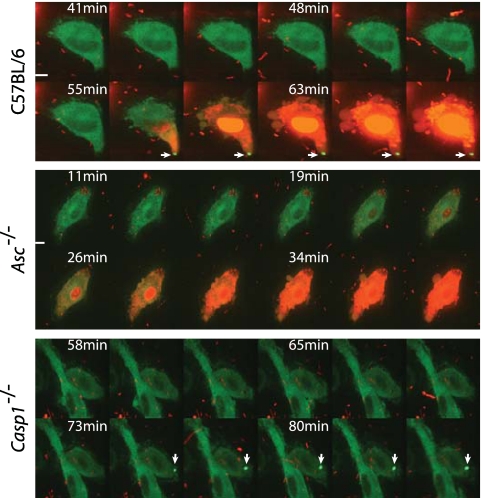
Although NLRC4 is recruited to an Asc-containing complex upon stimulation of this pathway by L. pneumophila, previous studies indicated that Asc is dispensable for pore formation and pyroptosis (17). To determine whether pyroptosis involves a major relocalization of NLRC4, the dynamics of GFP-NLRC4 localization was analyzed during induction of pyroptosis by L. pneumophila. Pyroptosis was assessed using two assays. The first assay measured the formation of pores in the plasma membrane of macrophages that resulted in the staining of macrophages by the membrane-impermeable dye propidium iodide (PI). The second assay measured the release of lactate dehydrogenase (LDH) from macrophages that results from cell lysis. Production of GFP-NLRC4 restored pore formation and LDH release in NLRC4-deficient macrophages but not in caspase-1-deficient macrophages following L. pneumophila infection (see Fig. S1 in the supplemental material). Thus, GFP-NLRC4 complements the defect in pyroptosis observed for the NLRC4-deficient macrophages. The dynamics of GFP-NLRC4 localization and the kinetics of pyroptosis were analyzed upon infection of macrophages with L. pneumophila using live-cell imaging. The dynamics of GFP-NLRC4 changed upon stimulation of macrophages with L. pneumophila. The diffuse cytosolic pool of GFP-NLRC4 observed in unstimulated macrophages relocalized into a single punctate structure following infection with L. pneumophila (Fig. 5; see also Movie S1 in the supplemental material). Macrophages became positive for propidium iodide (PI) and appeared to undergo pyroptosis shortly after recruitment of GFP-NLRC4 into these punctate structures. Studies using Asc-deficient macrophages showed that this redistribution of GFP-NLRC4 into punctate structures was not necessary for pyroptosis. Pyroptosis as measured by PI staining was not adversely affected in Asc-deficient macrophages upon infection by L. pneumophila; however, no redistribution of GFP-NLRC4 was apparent (Fig. 5; Movie S2), indicating that the formation of NLRC4 puncta is not required for pyroptosis. Importantly, the formation of GFP-NLRC4 puncta could be observed in control caspase-1-deficient macrophages following infection by L. pneumophila, but these cells remained PI impermeable, indicating a defect in pyroptosis (Fig. 5; Movie S3). These data demonstrate that macrophage pyroptosis induced by L. pneumophila requires NLRC4 and caspase-1 but does not require Asc or the redistribution of NLRC4 into a morphologically distinct activation platform resembling the Asc-containing punctate structure.
FIG 5 .
NLRC4 relocalization to Asc-dependent punctate structures is not required for pyroptosis. Mouse bone marrow cells from wild-type, Asc−/−, or Casp1−/− mice were transduced with retroviruses encoding GFP or GFP-NLRC4. Transduced macrophages were infected with broth culture-grown L. pneumophila expressing DsRed at an MOI of 5. GFP-NLRC4 relocalization was monitored using live-cell microscopy with a spinning-disk confocal microscope. Pore formation and pyroptosis were monitored by using propidium iodide uptake and loss of cell membrane integrity as readouts. Cells were incubated in the presence of propidium iodide at 6 µg/ml. Images were acquired every minute. Galleries contain images of the projected confocal stacks at the indicated time points for each experiment. Arrows indicate GFP-NLRC4 punctate structures. The scale bars equal 5 µm. Images are representative of at least 3 experiments.
Asc dampens the pyroptotic response to L. pneumophila.
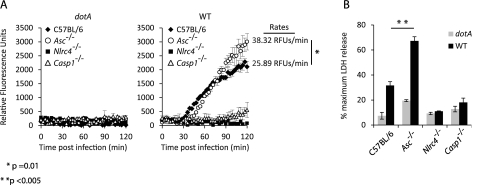
Infection of NLRC4-deficient macrophages by L. pneumophila resulted in the formation of a caspase-1 activation platform containing Asc; however, caspase-1-dependent pore formation was not observed in the NLRC4-deficient macrophages, suggesting that formation of the Asc platform used to activate caspase-1 was not sufficient to promote pyroptosis. The inability of the Asc platform to promote pyroptosis suggests that this complex may promote other caspase-1-dependent processes, such as cytokine processing and secretion, and compete with the Asc-independent processes, such as pyroptosis. This suggests that recruitment of NLRC4 and caspase-1 into the large punctate structure formed by Asc could modulate the pyroptotic potential of NLRC4. To test this possibility, the kinetics of pore formation in macrophages were compared to determine whether the presence of Asc affects the rates of pyroptosis in response to L. pneumophila infection. Pyroptosis was measured in real time using fluorescence spectroscopy to measure PI uptake by macrophages during L. pneumophila infection. These data revealed that Asc-deficient macrophages became permeable to propidium iodide at a significantly higher rate than wild-type macrophages (Fig. 6A). Interestingly, a slight delay in the initial propidium iodide uptake was observed in Asc-deficient cells compared to that observed in control macrophages (Fig. 6A), which was consistent with the delay in caspase-1 activation observed in Asc-deficient cells (Fig. 3B). Both the rate and the maximum intensity of PI staining, however, were higher in the Asc-deficient macrophages. Consistent with the real-time assays measuring pore formation, LDH release assays showed higher levels of cell lysis for the Asc-deficient macrophages than for control macrophages following infection with L. pneumophila (Fig. 6B). These data suggest that recruitment of NLRC4 into Asc-containing puncta dampens the pyroptotic functions associated with this NLR.
FIG 6 .
Asc dampens NLRC4-dependent pore formation and pyroptosis. BMMs from wild-type, Asc−/−, Nlrc4−/−, or Casp1−/− mice were infected with broth-grown L. pneumophila strains at an MOI of 2. (A) BMMs infected with the wild type or L. pneumophila dotA were incubated in the presence of propidium iodide (6 µg/ml) to monitor pore formation. PI fluorescence was measured every 3 minutes using a 96-well microplate reader. Data are represented as averages ± standard deviations of 3 wells after subtraction of noninfected samples in each experiment. Rates of pore formation for Asc−/− or wild-type cells in response to wild-type L. pneumophila were calculated from the initial 60 minutes following detection of PI fluorescence above background levels. (B) Cell death of BMMs infected with the wild type or L. pneumophila dotA was monitored by quantifying LDH release at 2 hours postinfection. Values represent the percentages of LDH released compared to the cells lysed with Triton X-100. Data points are averages ± standard deviations. Data are representative of at least two experiments.
DISCUSSION
Caspase-1 activation in response to microbe-derived signals can stimulate a diverse number of cellular processes. The ranges in signaling pathways and host responses triggered by caspase-1 activation likely reflect the large number of substrates in the cell that are susceptible to cleavage by the active protease (36). Although a large number of proteins are cleaved by caspase-1 in vitro, demonstration of cleavage of these cellular factors by caspase-1 in vivo has not been straightforward. One possible reason for this could be that the activation of caspase-1 and the access the active protein has to substrates could be regulated both spatially and temporally in vivo. This hypothesis is strengthened by studies examining caspase-1-dependent responses to microbial pathogens, which can differentially activate these diverse signaling pathways. Studies on L. pneumophila have demonstrated two different caspase-1 activation pathways that utilize different adapter proteins (17). NLRC4-mediated activation of caspase-1 by L. pneumophila is required for pyroptotic cell death and restriction of bacterial intracellular replication, whereas NLRC4-independent activation of caspase-1 is mediated by an Asc-dependent pathway and is not associated with pyroptosis or restriction of L. pneumophila intracellular replication (17, 20). Thus, L. pneumophila provides a genetically tractable system in which distinct caspase-1-dependent processes can be analyzed.
Staining L. pneumophila-infected macrophages with the reagent biotin-YVAD-FMK was used to address the possibility that caspase-1-mediated activities can be differentially regulated through targeting the active protease to discrete regions of the cell containing different substrates or to complexes that can recruit specific substrates. These studies revealed diffuse cytosolic distribution of the active protease following NLRC4-dependent or Asc-dependent signaling alone (Fig. 1B). These data do not disprove the hypothesis that spatial regulation of caspase-1 activation could influence substrate selection, as it remains possible that initial targeting of the protein could be important for cleavage of select substrates. However, diffusion may then disperse that active protease after activation, making it difficult to identify the regions of initial activation.
Although active caspase-1 was diffusely localized in macrophages, previous studies and data provided here indicate that much of the total caspase-1 is recruited into a discrete punctate structure in the cell upon stimulation of caspase-1 activation pathways (29, 30). Both caspase-1 and Asc localized to this structure following L. pneumophila infection (Fig. 2A and B). The association of caspase-1 into punctate structures required Asc but not NLRC4, consistent with assembly of this complex being a general consequence of caspase-1 activation and not being associated with activation by a single NLR protein member. The rate at which these punctate structures formed and the number of cells that stained positive for these punctate structures following L. pneumophila infection were reduced in NLRC4-deficient cells. Thus, the stimulation of NLRC4 by L. pneumophila provided a signal that resulted in rapid assembly of these structures, and this indicates that NLRC4-dependent signaling integrates into the formation of Asc complexes containing caspase-1 upon infection of macrophages by L. pneumophila.
Data presented here and in previous studies show that Asc is not required for caspase-1 activation in response to NLRC4-dependent signaling (17, 20); however, it is clear that cleavage of caspase-1 is significantly impaired in the absence of Asc (33–35). These data indicate that uncleaved and cleaved caspase-1 represent distinct forms of the active protease and may have distinct functions. Caspase-1 cleavage correlated with formation of the punctate structure containing Asc that recruited caspase-1. In the absence of Asc, both caspase-1 cleavage and punctum formation were significantly impaired, indicating that this structure is important for the caspase-1 cleavage process. The finding that puncta containing Asc were present in macrophages deficient for caspase-1 indicate that the assembly of this complex likely precedes caspase-1 recruitment and that assembly of this Asc complex is a consequence of stimulation of sensor proteins that regulate caspase-1 activation and is not directly mediated by caspase-1. These data suggest that the Asc-containing puncta are involved in the processing of caspase-1, but in the NLRC4 activation pathway, the Asc complex is not required for caspase-1 activation.
These results agree with recently published data indicating that Asc is important for caspase-1 processing in response to NLR activation (32). It was proposed that recruitment and cleavage of caspase-1 by Asc puncta is important for the processing and secretion of the proinflammatory cytokine IL-1β (32). Most pathways that trigger caspase-1 activation lead to IL-1β and IL-18 processing by macrophages, which is consistent with the Asc platform being shared by different caspase-1 activation pathways. Thus, the Asc platform may allow agonists sensed by different proteins that activate caspase-1 to be integrated into a central hub dedicated to the cleavage and secretion of these cytokines. This hypothesis is consistent with data presented here and published previously that indicate Asc is important for the cytokine response induced during L. pneumophila infection but not for NLRC4-mediated responses that restrict replication of L. pneumophila (17, 20).
The finding that GFP-NLRC4 localized to puncta containing Asc and caspase-1 following L. pneumophila infection was of importance because it provided direct evidence that an NLR protein can integrate into the Asc platform. Time-lapse microscopy showed clearly that in macrophages deficient for Asc, the localization of GFP-NLRC4 in punctate structures was not observed, even though these macrophages were able to sense infection by L. pneumophila and activate caspase-1-dependent processes that resulted in the formation of pores in the plasma membrane and induction of pyroptosis. Thus, NLRC4 appears to integrate both Asc-independent pyroptotic responses requiring caspase-1 and the processing of cytokines mediated by cleaved caspase-1 that requires the Asc platform. These data are in agreement with recent observations showing that a variant of caspase-1 that is unable to undergo proteolytic processing was activated and induced pyroptosis in response to Salmonella or Legionella infection of macrophages (32).
A role for Asc in dampening pyroptotic responses was revealed when the kinetics of pore formation induced during infection of macrophages by L. pneumophila in control and Asc-deficient cells were compared. Infection of the Asc-deficient macrophages augmented the rate and magnitude of pore formation and resulted in macrophages that were more susceptible to the NLRC4-dependent pyroptotic cell death induced by L. pneumophila. These data suggest that the ability of the Asc complex to recruit NLRC4 may interfere with the role of NLRC4 in promoting pyroptosis and facilitate cytokine processing by focusing activation of caspase-1 to the Asc platform. Thus, the scaffold assembled by Asc plays a role in cellular homeostasis by balancing caspase-1-dependent pyroptotic responses and caspase-1-dependent processing of cytokines.
MATERIALS AND METHODS
Bacterial strains and bone marrow-derived macrophage infections.
C57BL/6 mice were purchased from Jackson Laboratories. Nlrc4−/−, Asc−/−, and Casp1−/− mice have been described previously (34, 37–39). Bone marrow was collected from the femurs and tibiae of mice and cultured as described previously (17). Following differentiation, macrophages were replated in RPMI 1640 containing 10% fetal bovine serum (FBS) and 5% macrophage colony-stimulating factor (M-CSF), unless indicated otherwise. For infection studies, L. pneumophila thyA (Lp02) (40), a thymidine auxotroph derived from the L. pneumophila serogroup 1 strain Lp01, was used, along with the dotA (40) and flaA (23) isogenic mutants. Bacteria were cultured for 48 h on charcoal-yeast extract agar plates or overnight in liquid culture in ACES [N-(2-acetamido)-2-aminoethanesulfonic acid]-buffered yeast extract media supplemented with thymidine (100 µg/ml). For live imaging experiments, Lp02 strains expressing the fluorogenic protein DsRed-Express were used (41). Infections were performed by centrifuging bacteria onto macrophages for 5 min at 400 × g. More detailed protocols of macrophage culturing and bacterial infections can be found in Text S1 in the supplemental material.
Microscopy.
Macrophages were plated on glass coverslips (2 × 105 per coverslip) in 24-well dishes. Cells were infected with L. pneumophila at a multiplicity of infection (MOI) of 10 for the indicated times. In experiments involving biotin-YVAD staining of caspase-1, biotin-YVAD-FMK (ENZO Life Sciences) was added to macrophage media 15 minutes prior to infection at a final concentration of 25 µM. Primary antibodies used included rabbit anti-caspase-1 p10 (1:100 dilution; Santa Cruz Biotechnology), rabbit anti-Asc (1:1,000 dilution; AdipoGen), goat anti-Asc (1:100 dilution; Santa Cruz Biotechnology), and mouse anti-GFP (1:500 dilution; Roche). L. pneumophila was detected using either rabbit polyclonal or mouse polyclonal antibodies developed in this lab. Secondary antibodies used were Alexa Fluor conjugates from Invitrogen (1:500 to 1:2,000 dilutions). For detection of biotin-YVAD-FMK, streptavidin conjugated to Alexa Fluor 488 or 594 was used (1:1,000 dilution; Invitrogen). Detailed methods for immunofluorescent staining and live cell microscopy can be found in Text S1 in the supplemental material.
Immunoblot analysis.
Macrophages were added to 12-well plates (5 × 105 per well) and infected with L. pneumophila at an MOI of 10 for the indicated times. Proteins from culture supernatants and lysates were prepared as described previously (17), combined, subjected to SDS-PAGE analysis, and transferred to Immobilon-P membranes. Rabbit polyclonal antibodies raised against the p10 subunit of caspase-1 (Santa Cruz Biotechnology) and goat anti-rabbit secondary antibodies conjugated to horseradish peroxidase (Zymed) were used. ImageJ (NIH) was used for quantitation of protein levels in immunoblots. Scanned TIFF images of immunoblots were analyzed, and the integrated signal density of individual bands was calculated.
Pore formation and cell death assays.
Macrophages were added to 96-well plates (1 × 105 per well) and infected with broth-grown L. pneumophila at an MOI of 2 for 2 h. Infections were carried out in RPMI 1640 lacking phenol red. For cell death assays, supernatants were harvested for analysis of LDH released by dying cells. LDH levels in supernatants were quantified using the Cytotox 96 kit (Promega). For pore formation assays, cultures contained 20 mM HEPES and propidium iodide (6 µg/ml) and were incubated at 37°C in a Tecan Infinite M1000 fluorescent plate reader. Propidium iodide fluorescence was measured every 3 minutes.
Retroviral transductions.
Murine Nlrc4 was cloned into the pEGFP(C2) vector (Clontech). GFP-NLRC4 and GFP alone were cloned into the pMSCV2.2 murine-specific retroviral vector (Clontech). 293T cells were used to package retroviruses as described previously (42). Bone marrow cells were transduced with virus-containing 293T supernatants on days 3 and 4 of differentiation.
Statistical analysis.
Statistical significances for immunofluorescence quantitation, pore formation, and LDH assays were calculated using the unpaired Student t test. Differences were considered statistically significant if the P value was <0.05.
SUPPLEMENTAL MATERIAL
Supplemental methods Download Text S1, DOCX file, 0.015 MB.
GFP-NLRC4 complements pore formation and pyroptosis in response to L. pneumophila infection. Mouse bone marrow cells from wild-type, Asc-deficient (Asc−/−), or caspase-1-deficient (Casp1−/−) mice were transduced with retroviruses encoding GFP or GFP-NLRC4. (a) BMMs infected at an MOI of 5 with the L. pneumophila wild-type, dotA, or flaA strain were incubated in the presence of propidium iodide (6 g/ml) to monitor pore formation. Fluorescence resulting from propidium iodide staining of macrophages was monitored using a 96-well microplate reader continuously during infection, with readings every 4 minutes. Data are represented as averages ± standard deviations for 3 wells after subtraction of noninfected samples in each experiment. (b) Cell death of BMMs infected with broth culture-grown the L. pneumophila wild-type, dotA, or flaA strain at an MOI of 5 was monitored by quantifying LDH release at 2 h postinfection. Values represent the percentages of LDH released compared to the cells lysed with Triton X-100. Data points are averages ± standard deviations. Data are representative of at least two experiments. Download Figure S1, PDF file, 0.46 MB.
Mouse bone marrow cells from wild-type (shown here), Asc-deficient (Asc−/−) (Movie S2), or caspase-1-deficient (Casp1−/−) (Movie S3) mice were transduced with retroviruses encoding GFP-NLRC4. Transduced macrophages were infected with broth culture-grown L. pneumophila expressing DsRed at an MOI of 5. GFP-NLRC4 relocalization was monitored using live-cell microscopy with a spinning-disk confocal microscope. Pore formation and pyroptosis were monitored by using propidium iodide uptake and loss of cell membrane integrity as readouts. Cells were incubated in the presence of propidium iodide at 6 g/ml. Images were acquired every minute, and movies are shown at 3 frames per second. Download Movie S1, MOV file, 5.199 MB.
Mouse bone marrow cells from Asc-deficient (Asc−/−) mice. Download Movie S2, MOV file, 8.327 MB.
Mouse bone marrow cells from caspase-1-deficient (Casp1−/−) mice. Download Movie S3, MOV file, 7.157 MB.
ACKNOWLEDGMENTS
We are grateful to Richard Flavell and Ruslan Medzhitov for providing the mice used in these studies.
This research was supported by an NSF predoctoral award (to C.L.C.) and by NIH grants R01-AI048770 and R01-AI041699 (to C.R.R.).
Footnotes
Citation Case CL, Roy CR. 2011. Asc modulates the function of NLRC4 in response to infection of macrophages by Legionella pneumophila. mBio 2(4):e00117-11. doi:10.1128/mBio.00117-11.
REFERENCES
- 1. Fantuzzi G, Dinarello CA. 1999. Interleukin-18 and interleukin-1 beta: two cytokine substrates for ICE (caspase-1). J. Clin. Immunol. 19:1–11 [DOI] [PubMed] [Google Scholar]
- 2. Fink SL, Cookson BT. 2006. Caspase-1-dependent pore formation during pyroptosis leads to osmotic lysis of infected host macrophages. Cell. Microbiol. 8:1812–1825 [DOI] [PubMed] [Google Scholar]
- 3. Davis BK, Wen H, Ting JP. 2010. The inflammasome NLRs in immunity, inflammation, and associated diseases. Annu. Rev. Immunol. 29:707–735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Miao EA, et al. 2010. Innate immune detection of the type III secretion apparatus through the NLRC4 inflammasome. Proc. Natl. Acad. Sci. U. S. A. 107:3076–3080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Franchi L, et al. 2006. Cytosolic flagellin requires Ipaf for activation of caspase-1 and interleukin 1beta in Salmonella-infected macrophages. Nat. Immunol. 7:576–582 [DOI] [PubMed] [Google Scholar]
- 6. Miao EA, et al. 2006. Cytoplasmic flagellin activates caspase-1 and secretion of interleukin 1beta via Ipaf. Nat. Immunol. 7:569–575 [DOI] [PubMed] [Google Scholar]
- 7. Lamkanfi M, Dixit VM. 2009. Inflammasomes: guardians of cytosolic sanctity. Immunol. Rev. 227:95–105 [DOI] [PubMed] [Google Scholar]
- 8. Schroder K, Tschopp J. 2010. The inflammasomes. Cell 140:821–832 [DOI] [PubMed] [Google Scholar]
- 9. Fernandes-Alnemri T, Yu JW, Datta P, Wu J, Alnemri ES. 2009. AIM2 activates the inflammasome and cell death in response to cytoplasmic DNA. Nature 458:509–513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Hornung V, et al. 2009. AIM2 recognizes cytosolic dsDNA and forms a caspase-1-activating inflammasome with ASC. Nature 458:514–518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Fields BS. 1996. The molecular ecology of legionellae. Trends Microbiol. 4:286–290 [DOI] [PubMed] [Google Scholar]
- 12. Horwitz MA, Silverstein SC. 1980. Legionnaires’ disease bacterium (Legionella pneumophila) multiples intracellularly in human monocytes. J. Clin. Invest. 66:441–450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Fraser DW, et al. 1977. Legionnaires’ disease: description of an epidemic of pneumonia. N. Engl. J. Med. 297:1189–1197 [DOI] [PubMed] [Google Scholar]
- 14. McDade JE, et al. 1977. Legionnaires’ disease: isolation of a bacterium and demonstration of its role in other respiratory disease. N. Engl. J. Med. 297:1197–1203 [DOI] [PubMed] [Google Scholar]
- 15. Nagai H, Roy CR. 2001. The DotA protein from Legionella pneumophila is secreted by a novel process that requires the Dot/Icm transporter. EMBO J. 20:5962–5970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Roy CR, Tilney LG. 2002. The road less traveled: transport of Legionella to the endoplasmic reticulum. J. Cell Biol. 158:415–419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Case CL, Shin S, Roy CR. 2009. Asc and Ipaf Inflammasomes direct distinct pathways for caspase-1 activation in response to Legionella pneumophila. Infect. Immun. 77:1981–1991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Nogueira CV, et al. 2009. Rapid pathogen-induced apoptosis: a mechanism used by dendritic cells to limit intracellular replication of Legionella pneumophila. PLoS Pathog. 5:e1000478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Shin S, et al. 2008. Type IV secretion-dependent activation of host MAP kinases induces an increased proinflammatory cytokine response to Legionella pneumophila. PLoS Pathog. 4:e1000220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Zamboni DS, et al. 2006. The Birc1e cytosolic pattern-recognition receptor contributes to the detection and control of Legionella pneumophila infection. Nat. Immunol. 7:318–325 [DOI] [PubMed] [Google Scholar]
- 21. Ge J, et al. 2009. A Legionella type IV effector activates the NF-kappaB pathway by phosphorylating the IkappaB family of inhibitors. Proc. Natl. Acad. Sci. U. S. A. 106:13725–13730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Losick VP, Haenssler E, Moy MY, Isberg RR. 2010. LnaB: a Legionella pneumophila activator of NF-kappaB. Cell. Microbiol. 12:1083–1097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Ren T, Zamboni DS, Roy CR, Dietrich WF, Vance RE. 2006. Flagellin-deficient Legionella mutants evade caspase-1- and Naip5-mediated macrophage immunity. PLoS Pathog. 2:e18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Amer A, et al. 2006. Regulation of Legionella phagosome maturation and infection through flagellin and host Ipaf. J. Biol. Chem. 281:35217–35223 [DOI] [PubMed] [Google Scholar]
- 25. Molofsky AB, et al. 2006. Cytosolic recognition of flagellin by mouse macrophages restricts Legionella pneumophila infection. J. Exp. Med. 203:1093–1104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Lightfield KL, et al. 2008. Critical function for Naip5 in inflammasome activation by a conserved carboxy-terminal domain of flagellin. Nat. Immunol. 9:1171–1178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Lightfield KL, et al. 2011. Differential requirements for NAIP5 in activation of the NLRC4 inflammasome. Infect. Immun. 79:1606–1614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Sutterwala FS, Ogura Y, Zamboni DS, Roy CR, Flavell RA. 2006. NALP3: a key player in caspase-1 activation. J. Endotoxin Res. 12:251–256 [DOI] [PubMed] [Google Scholar]
- 29. Broz P, et al. 2010. Redundant roles for inflammasome receptors NLRP3 and NLRC4 in host defense against Salmonella. J. Exp. Med. 207:1745–1755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Martinon F, Burns K, Tschopp J. 2002. The inflammasome: a molecular platform triggering activation of inflammatory caspases and processing of proIL-beta. Mol. Cell 10:417–426 [DOI] [PubMed] [Google Scholar]
- 31. Fink SL, Bergsbaken T, Cookson BT. 2008. Anthrax lethal toxin and Salmonella elicit the common cell death pathway of caspase-1-dependent pyroptosis via distinct mechanisms. Proc. Natl. Acad. Sci. U. S. A. 105:4312–4317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Broz P, von Moltke J, Jones JW, Vance RE, Monack DM. 2010. Differential requirement for caspase-1 autoproteolysis in pathogen-induced cell death and cytokine processing. Cell Host Microbe 8:471–483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Franchi L, et al. 2007. Critical role for Ipaf in Pseudomonas aeruginosa-induced caspase-1 activation. Eur. J. Immunol. 37:3030–3039 [DOI] [PubMed] [Google Scholar]
- 34. Mariathasan S, et al. 2004. Differential activation of the inflammasome by caspase-1 adaptors ASC and Ipaf. Nature 430:213–218 [DOI] [PubMed] [Google Scholar]
- 35. Sutterwala FS, et al. 2007. Immune recognition of Pseudomonas aeruginosa mediated by the IPAF/NLRC4 inflammasome. J. Exp. Med. 204:3235–3245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Shao W, Yeretssian G, Doiron K, Hussain SN, Saleh M. 2007. The caspase-1 digestome identifies the glycolysis pathway as a target during infection and septic shock. J. Biol. Chem. 282:36321–36329 [DOI] [PubMed] [Google Scholar]
- 37. Kuida K, et al. 1995. Altered cytokine export and apoptosis in mice deficient in interleukin-1 beta converting enzyme. Science 267:2000–2003 [DOI] [PubMed] [Google Scholar]
- 38. Martinon F, Pétrilli V, Mayor A, Tardivel A, Tschopp J. 2006. Gout-associated uric acid crystals activate the NALP3 inflammasome. Nature 440:237–241 [DOI] [PubMed] [Google Scholar]
- 39. Sutterwala FS, et al. 2006. Critical role for NALP3/CIAS1/cryopyrin in innate and adaptive immunity through its regulation of caspase-1. Immunity 24:317–327 [DOI] [PubMed] [Google Scholar]
- 40. Berger KH, Isberg RR. 1993. Two distinct defects in intracellular growth complemented by a single genetic locus in Legionella pneumophila. Mol. Microbiol. 7:7–19 [DOI] [PubMed] [Google Scholar]
- 41. Ninio S, Celli J, Roy CR. 2009. A Legionella pneumophila effector protein encoded in a region of genomic plasticity binds to Dot/Icm-modified vacuoles. PLoS Pathog. 5:e1000278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Naviaux RK, Costanzi E, Haas M, Verma IM. 1996. The pCL vector system: rapid production of helper-free, high-titer, recombinant retroviruses. J. Virol. 70:5701–5705 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental methods Download Text S1, DOCX file, 0.015 MB.
GFP-NLRC4 complements pore formation and pyroptosis in response to L. pneumophila infection. Mouse bone marrow cells from wild-type, Asc-deficient (Asc−/−), or caspase-1-deficient (Casp1−/−) mice were transduced with retroviruses encoding GFP or GFP-NLRC4. (a) BMMs infected at an MOI of 5 with the L. pneumophila wild-type, dotA, or flaA strain were incubated in the presence of propidium iodide (6 g/ml) to monitor pore formation. Fluorescence resulting from propidium iodide staining of macrophages was monitored using a 96-well microplate reader continuously during infection, with readings every 4 minutes. Data are represented as averages ± standard deviations for 3 wells after subtraction of noninfected samples in each experiment. (b) Cell death of BMMs infected with broth culture-grown the L. pneumophila wild-type, dotA, or flaA strain at an MOI of 5 was monitored by quantifying LDH release at 2 h postinfection. Values represent the percentages of LDH released compared to the cells lysed with Triton X-100. Data points are averages ± standard deviations. Data are representative of at least two experiments. Download Figure S1, PDF file, 0.46 MB.
Mouse bone marrow cells from wild-type (shown here), Asc-deficient (Asc−/−) (Movie S2), or caspase-1-deficient (Casp1−/−) (Movie S3) mice were transduced with retroviruses encoding GFP-NLRC4. Transduced macrophages were infected with broth culture-grown L. pneumophila expressing DsRed at an MOI of 5. GFP-NLRC4 relocalization was monitored using live-cell microscopy with a spinning-disk confocal microscope. Pore formation and pyroptosis were monitored by using propidium iodide uptake and loss of cell membrane integrity as readouts. Cells were incubated in the presence of propidium iodide at 6 g/ml. Images were acquired every minute, and movies are shown at 3 frames per second. Download Movie S1, MOV file, 5.199 MB.
Mouse bone marrow cells from Asc-deficient (Asc−/−) mice. Download Movie S2, MOV file, 8.327 MB.
Mouse bone marrow cells from caspase-1-deficient (Casp1−/−) mice. Download Movie S3, MOV file, 7.157 MB.