Abstract
Purpose
RAF inhibitors are effective against melanomas with BRAF V600E mutations but may induce keratoacanthomas (KAs) and cutaneous squamous cell carcinomas (cSCCs). The potential of these agents to promote secondary malignancies is concerning. We analyzed cSCC and KA lesions for genetic mutations in an attempt to identify an underlying mechanism for their formation.
Methods
Four international centers contributed 237 KA or cSCC tumor samples from patients receiving an RAF inhibitor (either vemurafenib or sorafenib; n = 19) or immunosuppression therapy (n = 53) or tumors that developed spontaneously (n = 165). Each sample was profiled for 396 known somatic mutations across 33 cancer-related genes by using a mass spectrometric–based genotyping platform.
Results
Mutations were detected in 16% of tumors (38 of 237), with five tumors harboring two mutations. Mutations in TP53, CDKN2A, HRAS, KRAS, and PIK3CA were previously described in squamous cell tumors. Mutations in MYC, FGFR3, and VHL were identified for the first time. A higher frequency of activating RAS mutations was found in tumors from patients treated with an RAF inhibitor versus populations treated with a non–RAF inhibitor (21.1% v 3.2%; P < .01), although overall mutation rates between treatment groups were similar (RAF inhibitor, 21.1%; immunosuppression, 18.9%; and spontaneous, 17.6%; P = not significant). Tumor histology (KA v cSCC), tumor site (head and neck v other), patient age (≤ 70 v > 70 years), and sex had no significant impact on mutation rate or type.
Conclusion
Squamous cell tumors from patients treated with an RAF inhibitor have a distinct mutational profile that supports a mechanism of therapy-induced tumorigenesis in RAS-primed cells. Conceivably, cotargeting of MEK together with RAF may reduce or prevent formation of these tumors.
INTRODUCTION
At least 50% of all melanomas carry an activating mutation in the BRAF oncogene.1 In the advanced setting, the treatment of these melanomas with the selective RAF inhibitors vemurafenib (formerly PLX4032 or RG7204) and GSK2118436 has yielded response rates of 50% to 80%2–4 and an improvement in overall survival when compared with conventional chemotherapy.5 Similar to patients treated with other small-molecule kinase inhibitors, patients treated with a selective RAF inhibitor frequently experience skin toxicities.6 However, a striking distinction of these agents has been the development of skin tumors in the form of keratoacanthomas (KAs) or cutaneous squamous cell carcinomas (cSCCs) in up to approximately 25% of patients.2,4,5 These lesions most frequently develop within 8 to 12 weeks of beginning therapy. Similar treatment-related skin neoplasms have been described with the structurally unrelated multikinase inhibitor sorafenib.7,8 Sorafenib has been reported to have pan-RAF inhibitory properties,9 although the overall cellular potency of this compound against RAF proteins is much less pronounced when compared with selective inhibitors.10 Perhaps not surprisingly, sorafenib-induced skin tumors occur much less frequently and are more delayed in onset.7,8 Together, these observations suggest that RAF inhibition may play a direct role in the development of skin tumors.
The concept that a targeted therapy that blocks an oncogenic pathway in one cell type may promote tumorigenesis in another is both novel and potentially concerning. Given that RAF inhibitors will likely gain widespread use in melanoma and perhaps other cancers, deciphering the molecular basis of inhibitor-induced cutaneous neoplasms is essential. One potential mechanism is suggested by recent preclinical experiments demonstrating that while RAF inhibitors inhibit mitogen-activated protein kinase (MAPK) signaling in BRAF-mutant cancer cells, they may also cause a paradoxical increase in MAPK signaling in the context of mutated or activated RAS. Toward this end, RAS mutations have previously been identified in actinic keratoses11–13—premalignant skin lesions with the potential to transform into cSCCs.14 We therefore hypothesized that RAS activation in certain cutaneous cell subpopulations might interact with RAF inhibitor therapy to promote cell proliferation, ultimately resulting in KAs and cSCCs.
To test this hypothesis, we used a mass spectrometric genotyping platform (OncoMap) to generate mutational profiles for KA and cSCC lesions that developed in patients treated with an RAF inhibitor. As a comparator, we evaluated similar tumors that developed spontaneously or in the setting of immunosuppressive therapy.
METHODS
Tumor Specimens
Archival formalin-fixed paraffin-embedded (FFPE) cSCC and KA tumor specimens were collected from four international centers: the University of Essen, Essen, Germany; the Peter MacCallum Cancer Centre, East Melbourne, Australia; the University Hospital Zurich, Zurich, Switzerland; and the Gustave Roussy Institute, Villejuif, France. These samples were enriched for tumors that developed in patients undergoing treatment with an RAF inhibitor (vemurafenib or sorafenib) or immunosuppressive therapy for solid organ or bone marrow transplantations. Relevant clinical data were obtained from patients' medical records, and all samples were de-identified before analysis. The study was conducted with the approval of local institutional review boards.
DNA Preparation
Each tumor specimen was independently reviewed by two dermatopathologists to confirm the diagnosis. Tumor-rich areas (> 70% purity) were dissected and scraped from consecutive unstained FFPE slides. Genomic DNA was extracted by using the Qiagen DNeasy extraction kit (Qiagen, Valencia, CA) per the manufacturer's instructions. DNA quality was assessed by quantification with Picogreen and polymerase chain reaction amplification of fragments 100 to 200 base pairs (bp) long.
Mass Spectrometric Genotyping
High-throughput mutation profiling was performed on each sample by using the OncoMap platform. As previously described,15 this approach interrogated 396 mutations across 33 known oncogenes and tumor suppressor genes. Genomic DNA obtained from tumor samples was initially screened by using iPLEX (Sequenom, San Diego, CA) genotyping, and candidate mutations were validated by using homogeneous mass extension chemistry on unamplified DNA. The cancer gene mutations interrogated by OncoMap were selected on the basis of a combination of historically documented mutation frequencies and their potential as therapeutic targets.15,16 Primers and probes were designed by using Sequenom MassARRAY Assay Design 3.0 software (Sequenom), as described previously.15,16
Statistical Methods
For categorical comparisons, we performed either Fisher's exact test or the χ2 test by using GraphPad Prism version 5.0 (GraphPad Software, La Jolla, CA). All tests were two-sided, and a threshold of P < .05 was used to define statistical significance.
RESULTS
Characteristics of Clinical Tumor Samples
A total of 237 FFPE clinical tumor specimens were evaluated for this study, consisting of 191 cSCCs and 46 KAs (Table 1). One hundred sixty-five samples were classified as “spontaneous” (ie, they originated in patients who did not receive either significant immunosuppressive therapy or small-molecule RAF inhibitor therapy); included were samples from four patients receiving cytotoxic chemotherapy, two patients taking thalidomide, one patient receiving interferon, and one patient who was HIV-positive. Patients undergoing significant immunosuppressive therapy contributed 53 samples (47 samples from solid organ transplantations, three from bone marrow transplantations, and one sample each from patients with severe psoriasis, aplastic anemia, and unknown), and 19 samples were derived from patients receiving small-molecule RAF inhibitors. Seven patients enrolled in phase I and II studies of vemurafenib contributed 10 lesions (four lesions from one patient), with lesions being excised between 48 and 107 days (mean, 70 days) after commencing vemurafenib. Nine samples were from patients receiving sorafenib for 3 to 9 months. Not unexpectedly, surrogate markers of higher ultraviolet radiation exposure (increased age, head and neck site) were statistically favored in the spontaneous versus nonspontaneous cohorts (P < .01), likely reflecting the lack of an additional precipitant when compared with the treatment groups.
Table 1.
Clinicopathologic Characteristics of Squamous Cell Tumor Samples
| Characteristic | RAF-Inhibitor Treated(n = 19) |
Immunosuppressed(n = 53) |
Spontaneous(n = 165) |
Total(N = 237) |
||||
|---|---|---|---|---|---|---|---|---|
| No. | % | No. | % | No. | % | No. | % | |
| Sex | ||||||||
| Male | 10 | 52.6 | 41 | 77.4 | 113 | 68.5 | 164 | 69.2 |
| Female | 9 | 47.4 | 12 | 22.6 | 52 | 31.5 | 73 | 30.8 |
| Age, years | ||||||||
| Range | 44-89 | 42-83 | 26-104 | 26-104 | ||||
| Mean | 64.6 | 64.2 | 76.9 | 73.1 | ||||
| SEM | 2.6 | 1.3 | 0.9 | 0.8 | ||||
| Site | ||||||||
| Head and neck | 5 | 26.3 | 24 | 45.3 | 102 | 61.8 | 131 | 55.3 |
| Trunk | 2 | 10.5 | 3 | 5.7 | 5 | 3.0 | 10 | 4.2 |
| Upper limb | 7 | 36.8 | 13 | 24.5 | 24 | 14.5 | 44 | 18.6 |
| Lower limb | 5 | 26.3 | 10 | 18.9 | 25 | 15.2 | 40 | 16.9 |
| Genital | — | — | 5 | 3.0 | 5 | 2.1 | ||
| Unknown | — | 3 | 5.7 | 4 | 2.4 | 7 | 3.0 | |
| Histology | ||||||||
| cSCC | 9 | 47.4 | 48 | 90.6 | 134 | 81.2 | 191 | 80.6 |
| KA | 10 | 52.6 | 5 | 9.4 | 31 | 18.8 | 46 | 19.4 |
Abbreviations: cSCC, cutaneous squamous cell carcinoma; KA, keratoacanthoma; SEM, standard error of the mean.
Mutations in cSCC and KA Detected by Mass Spectrometric Genotyping
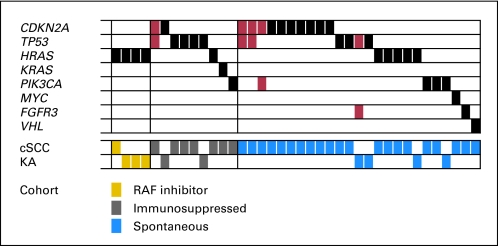
The OncoMap platform identified mutations in 38 of the 237 samples tested. Five samples exhibited co-occurring mutations; thus, a total of 43 mutations were detected (Fig 1; Table 2). The overall frequency of mutations was not significantly different between samples in the RAF inhibitor therapy, immunosuppression therapy, or spontaneous groups (21.1%, 18.9%, and 17.6%, respectively) nor did the frequency vary with patient age, sex, or tumor site. Furthermore, there was a similar rate and range of mutations between the cSCCs and KAs.
Fig 1.
Mutually exclusive and co-occurring mutations in human squamous cell tumors. Each column describes an individual sample with a detected mutation. Affected genes are listed in rows, with single mutations indicated by black bars and co-occurring mutations indicated by red bars. Histology (cutaneous squamous cell carcinoma [cSCC] and keratoacanthoma [KA]) and treatment cohort (RAF-inhibitor, gold; immunosuppressed, gray; spontaneous, blue) are detailed on the bottom two rows.
Table 2.
Characteristics of Samples With Detected Mutations
| Patient | Sex | Age (years) | Histology | Site | Mutation Type | Patient Cohort | Treatment |
|---|---|---|---|---|---|---|---|
| 1 | Male | 89 | cSCC | Arm | HRAS Q61L | RAF kinase inhibitor | Vemurafanib |
| 2 | Female | 54 | KA | Back | HRAS Q61L | RAF kinase inhibitor | Vemurafanib |
| 3 | Male | 71 | KA | Chest | HRAS G12D; BRAF V600E | RAF kinase inhibitor | Vemurafanib |
| 4 | Male | 53 | KA | Thigh | HRAS Q61L | RAF kinase inhibitor | Sorafenib |
| 5 | Male | 59 | cSCC | Ear | CDKN2A P114L; TP53 R248W | Immunosuppression | Methotrexate, cyclosporin, efalizumab, adalimumab |
| 6 | Male | 82 | KA | Lower arm | CDKN2A R80* | Immunosuppression | Cyclosporin |
| 7 | Male | 67 | cSCC | Lower leg | TP53 R273H | Immunosuppression | Cyclosporin |
| 8 | Male | 68 | cSCC | Face | TP53 R248Q | Immunosuppression | Drugs not specified |
| 9 | Male | 74 | cSCC | Leg | TP53 R248W | Immunosuppression | Cyclosporin |
| 10 | Female | 61 | KA | N/A | TP53 R248Q | Immunosuppression | Tacrolimus, mycophenylate mofitil, azathioprine |
| 11 | Male | 60 | cSCC | Lower arm | HRAS Q61K | Immunosuppression | Cyclosporin |
| 12 | Male | 76 | cSCC | Hand | KRAS G12D | Immunosuppression | Cyclosporin |
| 13 | Female | 42 | cSCC | Chest | PIK3CA E545K | Immunosuppression | Rapamycin, sirolimus |
| 14 | Male | 80 | cSCC | Hand | CDKN2A H83Y; TP53 R248Q | Spontaneous | No treatment |
| 15 | Male | 69 | cSCC | Head | CDKN2A R58*; TP53 R248W | Spontaneous | No treatment |
| 16 | Male | 60 | cSCC | Leg | CDKN2A R80*; PIK3CA E545K | Spontaneous | Methotrexate, cisplatin, fluorouracil, bleomycin |
| 17 | Male | 81 | cSCC | Face | CDKN2A P114L | Spontaneous | No treatment |
| 18 | Male | 79 | cSCC | Face | CDKN2A P114L | Spontaneous | No treatment |
| 19 | Male | 72 | cSCC | Scalp | CDKN2A R58* | Spontaneous | No treatment |
| 20 | Male | 104 | cSCC | Head | CDKN2A R58* | Spontaneous | No treatment |
| 21 | Male | 71 | cSCC | Head | CDKN2A H83Y | Spontaneous | No treatment |
| 22 | Male | 77 | cSCC | Ear | CDKN2A P114L | Spontaneous | No treatment |
| 23 | Male | 65 | cSCC | Hand | CDKN2A E88* | Spontaneous | No treatment |
| 24 | Male | 81 | cSCC | Lower arm | TP53 R248W | Spontaneous | No treatment |
| 25 | Male | 83 | cSCC | Ear | TP53 R248W | Spontaneous | No treatment |
| 26 | Female | 88 | KA | Cheek | TP53 R248W; FGFR3 G370C | Spontaneous | No treatment |
| 27 | Female | 77 | KA | Cheek | TP53 R273C | Spontaneous | No treatment |
| 28 | Male | 81 | cSCC | Cheek | HRAS Q61L | Spontaneous | No treatment |
| 29 | Male | 75 | cSCC | Ear | HRAS Q61L | Spontaneous | No treatment |
| 30 | Female | 77 | cSCC | Genital | HRAS Q61L | Spontaneous | No treatment |
| 31 | Male | 75 | cSCC | Auricle | HRAS G12V | Spontaneous | No treatment |
| 32 | Female | 26 | KA | Lip | HRAS Q61L | Spontaneous | No treatment |
| 33 | Male | 77 | cSCC | Arm | PIK3CA E542K | Spontaneous | No treatment |
| 34 | Female | 81 | cSCC | Forehead | PIK3CA E545K | Spontaneous | No treatment |
| 35 | Female | 62 | KA | Lower Leg | PIK3CA H1047R | Spontaneous | No treatment |
| 36 | Female | 78 | cSCC | Upper arm | MYC A59V | Spontaneous | No treatment |
| 37 | Male | 57 | cSCC | Face | FGFR3 G370C | Spontaneous | No treatment |
| 38 | Male | 52 | cSCC | Lower leg | VHL P81S | Spontaneous | No treatment |
| 39 | Male | 61 | cSCC | Arm | BRAF V600E | RAF kinase inhibitor | Vemurafanib |
Abbreviations: cSCC, cutaneous squamous cell carcinoma; KA, keratoacanthoma; NA, not available.
Denotes stop codon.
Mutations were detected across eight different cancer-related genes. In keeping with previous studies,17) the most frequently involved genes were TP53 (n = 11), CDKN2A (n = 12), and the RAS isoforms HRAS (n = 10) and KRAS (n = 1). Not previously identified in cSCCs and KAs were mutations in PIK3CA (n = 5), FGFR3 (n = 2), MYC (n = 1), and VHL (n = 1).
RAS Mutations Occur More Frequently in Tumors From Patients Treated With RAF Inhibitors
Tumors from the cohort of patients treated with an RAF inhibitor were enriched for HRAS mutations despite similar rates of total mutations between groups. Known activating mutations in HRAS (Q61L, G12D) were identified in 30% (95% CI, 10% to 61%) of samples from patients treated with vemurafenib and 11% (95% CI, < 0.01% to 46%) of samples from patients treated with sorafenib. Combined, HRAS mutations were found in four of the 19 samples from patients treated with an RAF inhibitor compared with six of 218 HRAS mutations (21.1% v 2.8%; P < .01) and a single KRAS mutant (all RAS mutations, 21.1% v 3.12%; P < .01) in samples treated with a non-RAF inhibitor. No NRAS mutations were identified in this study. Furthermore, in the cohort of patients treated with an RAF inhibitor, no activating mutations were identified in 11 receptor tyrosine kinases that are commonly mutated in human cancers and that function upstream of RAS (CSF1R, EGFR, ERBB2, FGFR1, FGFR2, FGFR3, FLT3, KIT, MET, PDGFRA, and RET).
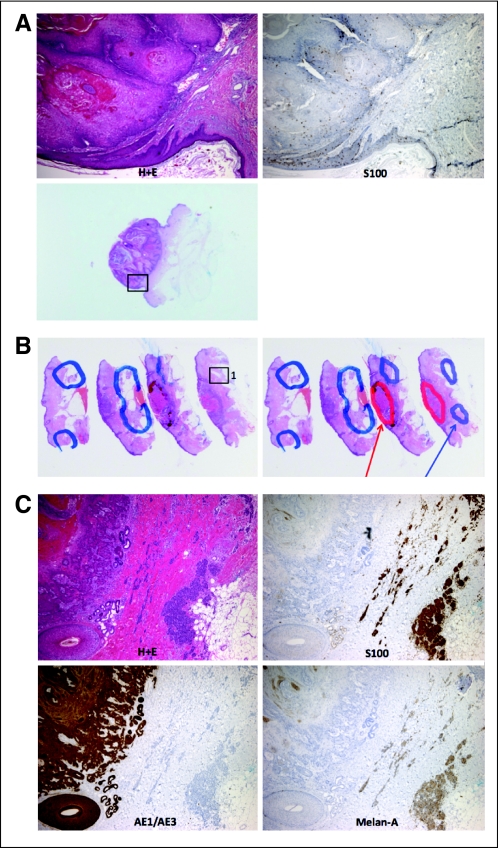
Surprisingly, we also identified BRAF V600E mutations in two samples from patients treated with vemurafenib for BRAF V600E-mutant metastatic melanoma (samples 3 and 39). Further immunohistochemical studies for AE1/AE3 (squamous cell carcinoma), S100, and melan-A (melanoma) identified clearly separate populations of malignant squamous cells and melanocytes in close proximity, with one sample showing evidence of metastatic melanoma cells within the lymphovascular space (Appendix Fig A1, online only). The BRAF mutations in these samples were therefore attributable to melanocytic contamination, and these mutations were not included in our statistical analysis.
DISCUSSION
Recent preclinical studies have identified the potential for selective RAF inhibitors to augment MAPK pathway activation in the context of activated RAS. In these systems, signaling occurred preferentially through C-RAF, with RAF inhibitors thought to induce a conformational change in C-RAF heterodimers or B-RAF homodimers that resulted in pathway hyperactivity.18–21 In our study, we found that squamous cell tumors arising in patients treated with an RAF inhibitor are enriched for RAS mutations when compared with those from untreated patients. This finding suggests that the formation of these tumors is not due to a direct mutagenic event of RAF inhibitor therapy but is caused, at least in part, by a pro-proliferative interaction between RAF inhibitors and latent RAS mutant keratinocytes.
Unlike in other common skin cancers such as melanoma or basal cell carcinoma, a predominant mutational driver has yet to be identified in cSCC. H-, K-, and NRAS mutations occur infrequently in cSCC from the general11,13,22,23 and immunosuppressed24 populations and are reported at similarly low frequencies (1% to 12%) in premalignant actinic keratoses.11–13,25 The presence of RAS mutations in these premalignant lesions (common in sun-damaged skin) suggests that ultraviolet radiation can induce RAS mutations in keratinocytes but that these mutations are not sufficient to induce malignant transformation. When RAS-activated keratinocytes are exposed to RAF inhibition, additional checks and balances may be exceeded, resulting in abnormal cell growth and, ultimately, progression to either KA or cSCC. This model would fit the clinical observations that the KA and cSCC lesions most commonly develop in the first 8 to 12 weeks after initiating RAF inhibitor therapy and are more frequent in patients with increased sun exposure, supporting the existing presence of a predisposing lesion. In some cases, these tumors have spontaneously regressed on treatment discontinuation. The mechanisms for this regression are not clear but could involve altered signaling, activation of a senescence-like program, or induction of an immune response to the KA and cSCC lesions. Regarding the latter, some studies have noted the presence of cytotoxic T lymphocytes, Langerhans cells, and CD30+ cells within KA lesions.26–29
So far, the cutaneous squamous cell tumors that have developed in patients receiving vemurafenib and GSK2118436 have all been well differentiated and have not metastasized or recurred after complete excision.4,30 Sorafenib is a multikinase inhibitor whose effects against RAF isoforms are considerably less pronounced in vivo than those of vemurafenib and GSK2118436, which may explain the lower incidence of skin tumors seen with this drug.7,9 Notably, sorafenib is ineffective against BRAF-mutant melanoma; this reduced in vivo potency may relate in part to its higher affinity for the inactive conformation of this kinase than the active form acquired with a BRAF V600E mutation.31 In the context of patients with advanced melanoma and its associated poor prognosis, development of KA or cSCC as a treatment-related adverse effect has been considered acceptable to both patients and clinicians. However, the confirmation of a clinically significant interaction between selective RAF inhibitors and RAS-activated cells raises several additional considerations.
The most concerning of these is the potential for secondary malignancies other than those of the skin. RAS mutations are estimated to occur in 30% of all human cancers, with a significant further proportion having either activating mutations or overexpression of upstream receptor tyrosine kinases (such as HER2, c-KIT, and EGFR32). Furthermore, apart from occurring in actinic keratoses, RAS mutations also occur as early genetic events in a range of premalignant lesions. For instance, KRAS mutations represent early genetic events in colon carcinogenesis: they are present in up to 50% of colonic adenomas.33,34 They are also found at increasing frequency with progressively higher grades of pancreatic intraepithelial neoplasia, correlating with increasing risk of progressing to carcinoma35 and are more commonly detected in the bronchial washings of smokers compared with those of nonsmokers.36 In principle, exposure of these cell subpopulations to RAF inhibitors could promote clonal expansion and propel them toward permanent malignant transformation.
Why have no such extracutaneous tumors been detected thus far? One possible reason might be that the median treatment duration of vemurafenib and GSK2118436 (6 to 9 months) has not been long enough for these lesions to manifest. Alternatively, some such events may have been misinterpreted as melanoma progression. It is also possible that RAS-mutant cell populations in other organs may not undergo the sustained proliferation typical of fully malignant neoplasms. Concerns of possible tumorigenic complications may become heightened by the inevitable transition of these agents into the adjuvant setting, in which treatment duration could last 1 to 2 years, and the clinical impact of any secondary malignancies might be increased. Thus, for patients participating in the initial adjuvant studies of these agents, careful surveillance will be essential. Aggressive histologic characterization (which may include tumor mutational profiling) may be needed for new lesions that arise during adjuvant treatment using selective RAF inhibitors.
Our findings also illustrate how proposed strategies to overcome resistance and potential strategies to prevent secondary tumor development may converge going forward. To date, several mechanisms of secondary RAF inhibitor resistance have been postulated. These include acquired mutations in NRAS and overexpression of PDGFR-β and IGFR, both of which can operate upstream of RAS.37,38 These models may suggest a switch to C-RAF–driven MAPK signaling39 that is operant in some cases. Conceivably, such resistance mechanisms might be circumvented through the development of more potent RAF inhibitors that abrogate the mechanism of RAF activation observed with existing compounds, or alternatively by blocking signaling downstream of both B-RAF and C-RAF by targeting MEK or ERK. Clinical studies of concomitant RAF and MEK inhibitors have commenced in an attempt to prolong the effectiveness of (or overcome resistance to) RAF blockade in BRAF V600E-mutant melanomas. Preliminary results suggest that this combination may reduce the incidence of RAF inhibitor–induced KA and cSCC lesions.40 We speculate that such combinations may also suppress proliferation of RAS-activated nonmelanoma cell populations elsewhere in the body.
More generally, our study provides one of the largest mutational studies of cSCCs and KAs reported to date. Consistent with the previous literature, the most frequently mutated genes were TP53 and CDKN2A.41,42 However, we detected substantially lower rates of mutations in these genes (4.6% and 5.1%, respectively) compared with those previously reported (44.4% and 24.5%, respectively), most likely reflecting the known limitation of genotyping-based platforms in detecting mutations in tumor suppressor genes. Inactivating mutations are more diverse and therefore harder to cover with multiplexed, mutation-specific assays when compared with the limited number of functional activating mutations in oncogenes. For instance, our assays covered only 11% of TP53 and 24% of CDKN2A mutations previously identified in cSCC, whereas 100% of previously described H-, K-, and N-RAS mutations were assessed. Of note, PTCH1 was reportedly mutated in more than 5% of cSCC samples; however, this finding was limited to only one study,43 in which all three nonsilent mutations occurred in patients with a history of multiple basal cell carcinomas (PTCH1 was not assessed in our study). We identified novel mutations in four genes: PIK3CA, FGFR3, MYC, and VHL, but these occurred in no more than 2% of samples. No difference was identified in the rate or types of mutations between cSCCs and KAs. The histologic and biologic distinction between these entities remains an area of controversy for dermatopathologists.44,45
Finally, although we have identified mutations in RAS in roughly 20% of squamous cell tumors that developed during therapy with an RAF inhibitor, tumorigenic mechanisms operant in RAS-negative KA and cSCC lesions remain unclear. It is possible that the frequency of RAS mutations in treatment-induced KAs and cSCCs will rise as larger patient cohorts treated at maximal RAF inhibitor doses are analyzed. Additional mechanisms may involve activation of upstream effectors (eg, receptor tyrosine kinases) by gene amplification or overexpression, which were not examined here. The application of more comprehensive mutation profiling technologies such as targeted, massively parallel sequencing may shed additional light on the full spectrum of genomic alterations that drive the biology of these squamous cell tumors. Several other mechanisms have been proposed by which tumors may develop in the presence of sorafenib, with most being related to its multikinase activity.46,47 How such mechanisms might relate to those linked to selective RAF inhibitors remains unclear.
In summary, exposure to selective RAF inhibitors may lead to pro-proliferative effects on RAS-primed cells. This has already manifested clinically in the form of squamous cell tumors, but the potential may also exist to promote growth of other extracutaneous neoplasms (and to promote resistance in melanoma) by the same mechanism. Cotargeting of MEK (or, in the future, ERK) together with RAF may block this effect. Thus, compound MAP kinase pathway inhibition may simultaneously enhance antitumor efficacy and restrict pro-neoplastic adverse effects of single-agent RAF inhibition.
Appendix
Fig A1.
Tumor samples with BRAF V600E mutations demonstrating colocalized cutaneous squamous cell tumors and melanoma. (A) Sample 3: hematoxylin and eosin (H+E) stain demonstrating classical keratoacanthoma histology, with an interspersed S100-positive melanocytic population. (B) Low-power and (C) high-power images of H+E stains from sample 39. AE1/AE3-positive cells (squamous cell carcinoma) are demonstrated immediately adjacent to S100-positive and melan-A–positive cells (melanoma).
Footnotes
See accompanying article on page 329
Supported by Grants No. DP2OD002750 and R33CA126674 from the National Institutes of Health, National Cancer Institute; by the Starr Cancer Consortium; by the Melanoma Research Alliance (L.A.G.); and by Grant No. PASMP3-127676/1 from the Swiss National Science Foundation (P.A.O. and L.A.G.).
Authors' disclosures of potential conflicts of interest and author contributions are found at the end of this article.
AUTHORS' DISCLOSURES OF POTENTIAL CONFLICTS OF INTEREST
Although all authors completed the disclosure declaration, the following author(s) indicated a financial or other interest that is relevant to the subject matter under consideration in this article. Certain relationships marked with a “U” are those for which no compensation was received; those relationships marked with a “C” were compensated. For a detailed description of the disclosure categories, or for more information about ASCO's conflict of interest policy, please refer to the Author Disclosure Declaration and the Disclosures of Potential Conflicts of Interest section in Information for Contributors.
Employment or Leadership Position: None Consultant or Advisory Role: Caroline Robert, GlaxoSmithKline (C), Roche (C); Grant A. McArthur, GlaxoSmithKline (U), Roche (U); Levi A. Garraway, Foundation Medicine (C), Novartis (C), Daiichi Sankyo (C) Stock Ownership: Levi A. Garraway, Foundation Medicine Honoraria: Caroline Robert, GlaxoSmithKline, Roche; Levi A. Garraway, Boehringer Ingelheim, Constellation Pharmaceuticals, Merck, Novartis Research Funding: Levi A. Garraway, Novartis Expert Testimony: None Other Remuneration: None
AUTHOR CONTRIBUTIONS
Conception and design: Patrick A. Oberholzer, Damien Kee, Caroline Robert, Günther F.L. Hofbauer, Grant A. McArthur, Dirk Schadendorf, Levi A. Garraway
Financial support: Levi A. Garraway
Provision of study materials or patients: Damien Kee, Caroline Robert, Günther F.L. Hofbauer, Grant A. McArthur, Dirk Schadendorf
Collection and assembly of data: Patrick A. Oberholzer, Damien Kee, Piotr Dziunycz, Antje Sucker, Nyam Kamsukom, Robert Jones, Christine Roden, Clinton J. Chalk, Kristin Ardlie, Emanuele Palescandolo, Adriano Piris, Laura E. MacConaill, Caroline Robert, Günther F.L. Hofbauer, Grant A. McArthur
Data analysis and interpretation: Patrick A. Oberholzer, Damien Kee, Grant A. McArthur
Manuscript writing: All authors
Final approval of manuscript: All authors
REFERENCES
- 1.Platz A, Egyhazi S, Ringborg U, et al. Human cutaneous melanoma: A review of NRAS and BRAF mutation frequencies in relation to histogenetic subclass and body site. Mol Oncol. 2008;1:395–405. doi: 10.1016/j.molonc.2007.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Flaherty KT, Puzanov I, Kim KB, et al. Inhibition of mutated, activated BRAF in metastatic melanoma. N Engl J Med. 2010;363:809–819. doi: 10.1056/NEJMoa1002011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kefford R, Arkenau H, Brown MP, et al. Phase I/II study of GSK2118436, a selective inhibitor of oncogenic mutant BRAF kinase, in patients with metastatic melanoma and other solid tumors. J Clin Oncol. 2010;28(suppl; abst 8503):611s. [Google Scholar]
- 4.Ribas A, Kim KB, Schuchter LM, et al. BRIM-2: An open-label, multicenter phase II study of vemurafenib in previously treated patients with BRAF V600E mutation-positive metastatic melanoma. J Clin Oncol. 2011;29(suppl; abst 8509):528s. [Google Scholar]
- 5.Chapman PB, Hauschild A, Robert C, et al. Improved survival with vemurafenib in melanoma with BRAF V600E mutation. N Engl J Med. 2011;364:2507–2516. doi: 10.1056/NEJMoa1103782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Robert C, Soria JC, Spatz A, et al. Cutaneous side-effects of kinase inhibitors and blocking antibodies. Lancet Oncol. 2005;6:491–500. doi: 10.1016/S1470-2045(05)70243-6. [DOI] [PubMed] [Google Scholar]
- 7.Arnault JP, Wechsler J, Escudier B, et al. Keratoacanthomas and squamous cell carcinomas in patients receiving sorafenib. J Clin Oncol. 2009;27:e59–e61. doi: 10.1200/JCO.2009.23.4823. [DOI] [PubMed] [Google Scholar]
- 8.Dubauskas Z, Kunishige J, Prieto VG, et al. Cutaneous squamous cell carcinoma and inflammation of actinic keratoses associated with sorafenib. Clin Genitourin Cancer. 2009;7:20–23. doi: 10.3816/CGC.2009.n.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wilhelm SM, Carter C, Tang L, et al. BAY 43-9006 exhibits broad spectrum oral antitumor activity and targets the RAF/MEK/ERK pathway and receptor tyrosine kinases involved in tumor progression and angiogenesis. Cancer Res. 2004;64:7099–7109. doi: 10.1158/0008-5472.CAN-04-1443. [DOI] [PubMed] [Google Scholar]
- 10.Whittaker S, Kirk R, Hayward R, et al. Gatekeeper mutations mediate resistance to BRAF-targeted therapies. Sci Transl Med. 2010;2:35ra41. doi: 10.1126/scitranslmed.3000758. [DOI] [PubMed] [Google Scholar]
- 11.Zaravinos A, Kanellou P, Spandidos DA. Viral DNA detection and RAS mutations in actinic keratosis and nonmelanoma skin cancers. Br J Dermatol. 2010;162:325–331. doi: 10.1111/j.1365-2133.2009.09480.x. [DOI] [PubMed] [Google Scholar]
- 12.Nindl I, Gottschling M, Krawtchenko N, et al. Low prevalence of p53, p16(INK4a) and Ha-ras tumour-specific mutations in low-graded actinic keratosis. Br J Dermatol. 2007;156(suppl 3):34–39. doi: 10.1111/j.1365-2133.2007.07857.x. [DOI] [PubMed] [Google Scholar]
- 13.Spencer JM, Kahn SM, Jiang W, et al. Activated ras genes occur in human actinic keratoses, premalignant precursors to squamous cell carcinomas. Arch Dermatol. 1995;131:796–800. [PubMed] [Google Scholar]
- 14.Ko CJ. Actinic keratosis: Facts and controversies. Clin Dermatol. 2010;28:249–253. doi: 10.1016/j.clindermatol.2009.06.009. [DOI] [PubMed] [Google Scholar]
- 15.MacConaill LE, Campbell CD, Kehoe SM, et al. Profiling critical cancer gene mutations in clinical tumor samples. PLoS One. 2009;4:e7887. doi: 10.1371/journal.pone.0007887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Thomas RK, Baker AC, Debiasi RM, et al. High-throughput oncogene mutation profiling in human cancer. Nat Genet. 2007;39:347–351. doi: 10.1038/ng1975. [DOI] [PubMed] [Google Scholar]
- 17.Wellcome Trust Sanger Institute: COSMIC database. http://www.sanger.ac.uk/genetics/CGP/cosmic/
- 18.Hatzivassiliou G, Song K, Yen I, et al. RAF inhibitors prime wild-type RAF to activate the MAPK pathway and enhance growth. Nature. 2010;464:431–435. doi: 10.1038/nature08833. [DOI] [PubMed] [Google Scholar]
- 19.Heidorn SJ, Milagre C, Whittaker S, et al. Kinase-dead BRAF and oncogenic RAS cooperate to drive tumor progression through CRAF. Cell. 2010;140:209–221. doi: 10.1016/j.cell.2009.12.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Poulikakos PI, Zhang C, Bollag G, et al. RAF inhibitors transactivate RAF dimers and ERK signalling in cells with wild-type BRAF. Nature. 2010;464:427–430. doi: 10.1038/nature08902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Halaban R, Zhang W, Bacchiocchi A, et al. PLX4032, a selective BRAF(V600E) kinase inhibitor, activates the ERK pathway and enhances cell migration and proliferation of BRAF melanoma cells. Pigment Cell Melanoma Res. 2010;23:190–200. doi: 10.1111/j.1755-148X.2010.00685.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Campbell C, Quinn AG, Rees JL. Codon 12 Harvey-ras mutations are rare events in non-melanoma human skin cancer. Br J Dermatol. 1993;128:111–114. doi: 10.1111/j.1365-2133.1993.tb15137.x. [DOI] [PubMed] [Google Scholar]
- 23.Corominas M, Kamino H, Leon J, et al. Oncogene activation in human benign tumors of the skin (keratoacanthomas): Is HRAS involved in differentiation as well as proliferation? Proc Natl Acad Sci U S A. 1989;86:6372–6376. doi: 10.1073/pnas.86.16.6372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pélisson I, Chardonnet Y, Euvrard S, et al. Low incidence of c-Ha-ras gene mutations in benign and malignant cutaneous lesions from transplant recipients. Int J Cancer. 1993;55:915–920. doi: 10.1002/ijc.2910550607. [DOI] [PubMed] [Google Scholar]
- 25.Taguchi M, Tsuchida T, Ikeda S, et al. Alterations of p53 gene and Ha-ras gene are independent events in solar keratosis and squamous cell carcinoma. Pathol Int. 1998;48:689–694. doi: 10.1111/j.1440-1827.1998.tb03969.x. [DOI] [PubMed] [Google Scholar]
- 26.Batinac T, Zamolo G, Hadzisejdic I, et al. A comparative study of granzyme B expression in keratoacanthoma and squamous cell carcinoma. J Dermatol Sci. 2006;44:109–112. doi: 10.1016/j.jdermsci.2006.07.003. [DOI] [PubMed] [Google Scholar]
- 27.Bayer-Garner IB, Ivan D, Schwartz MR, et al. The immunopathology of regression in benign lichenoid keratosis, keratoacanthoma and halo nevus. Clin Med Res. 2004;2:89–97. doi: 10.3121/cmr.2.2.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fernandez-Flores A. CD30+ cells in regressing keratoacanthoma and in non-keratoacanthomatous squamous cell carcinoma. Bratisl Lek Listy. 2008;109:508–512. [PubMed] [Google Scholar]
- 29.Korenberg R, Penneys NS, Kowalczyk A, et al. Quantitation of S100 protein-positive cells in inflamed and non-inflamed keratoacanthoma and squamous cell carcinoma. J Cutan Pathol. 1988;15:104–108. doi: 10.1111/j.1600-0560.1988.tb00528.x. [DOI] [PubMed] [Google Scholar]
- 30.Lacouture ME, McArthur GA, Chapman PB, et al. PLX4032 (RG7204), a selective mutant RAF inhibitor: Clinical and histologic characteristics of therapy-associated cutaneous neoplasms in a phase I trial. J Clin Oncol. 2010;28(suppl; abstr 8592):634s. [Google Scholar]
- 31.Hauschild A, Agarwala SS, Trefzer U, et al. Results of a phase III, randomized, placebo-controlled study of sorafenib in combination with carboplatin and paclitaxel as second-line treatment in patients with unresectable stage III or stage IV melanoma. J Clin Oncol. 2009;27:2823–2830. doi: 10.1200/JCO.2007.15.7636. [DOI] [PubMed] [Google Scholar]
- 32.Bos JL. ras oncogenes in human cancer: A review. Cancer Res. 1989;49:4682–4689. [PubMed] [Google Scholar]
- 33.Fearon ER, Vogelstein B. A genetic model for colorectal tumorigenesis. Cell. 1990;61:759–767. doi: 10.1016/0092-8674(90)90186-i. [DOI] [PubMed] [Google Scholar]
- 34.Vogelstein B, Fearon ER, Hamilton SR, et al. Genetic alterations during colorectal-tumor development. N Engl J Med. 1988;319:525–532. doi: 10.1056/NEJM198809013190901. [DOI] [PubMed] [Google Scholar]
- 35.Löhr M, Klöppel G, Maisonneuve P, et al. Frequency of K-ras mutations in pancreatic intraductal neoplasias associated with pancreatic ductal adenocarcinoma and chronic pancreatitis: A meta-analysis. Neoplasia. 2005;7:17–23. doi: 10.1593/neo.04445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yakubovskaya MS, Spiegelman V, Luo FC, et al. High frequency of K-ras mutations in normal appearing lung tissues and sputum of patients with lung cancer. Int J Cancer. 1995;63:810–814. doi: 10.1002/ijc.2910630611. [DOI] [PubMed] [Google Scholar]
- 37.Nazarian R, Shi H, Wang Q, et al. Melanomas acquire resistance to B-RAF(V600E) inhibition by RTK or N-RAS upregulation. Nature. 2010;468:973–977. doi: 10.1038/nature09626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Villanueva J, Vultur A, Lee JT, et al. Acquired resistance to BRAF inhibitors mediated by a RAF kinase switch in melanoma can be overcome by cotargeting MEK and IGF-1R/PI3K. Cancer Cell. 2010;18:683–695. doi: 10.1016/j.ccr.2010.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Johannessen CM, Boehm JS, Kim SY, et al. COT drives resistance to RAF inhibition through MAP kinase pathway reactivation. Nature. 2010;468:968–972. doi: 10.1038/nature09627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Infante JR, Falchook GS, Lawrence DP, et al. Phase I/II study to assess safety, pharmacokinetics, and efficacy of the oral MEK 1/2 inhibitor GSK1120212 (GSK212) dosed in combination with the oral BRAF inhibitor GSK2118436 (GSK436) J Clin Oncol. 2011;29(suppl; abst CRA8503):526s. [Google Scholar]
- 41.Boukamp P. Non-melanoma skin cancer: What drives tumor development and progression? Carcinogenesis. 2005;26:1657–1667. doi: 10.1093/carcin/bgi123. [DOI] [PubMed] [Google Scholar]
- 42.Pacifico A, Goldberg LH, Peris K, et al. Loss of CDKN2A and p14ARF expression occurs frequently in human nonmelanoma skin cancers. Br J Dermatol. 2008;158:291–297. doi: 10.1111/j.1365-2133.2007.08360.x. [DOI] [PubMed] [Google Scholar]
- 43.Ping XL, Ratner D, Zhang H, et al. PTCH mutations in squamous cell carcinoma of the skin. J Invest Dermatol. 2001;116:614–616. doi: 10.1046/j.1523-1747.2001.01301.x. [DOI] [PubMed] [Google Scholar]
- 44.Robert C, Arnault JP, Mateus C. RAF inhibition and induction of cutaneous squamous cell carcinoma. Curr Opin Oncol. 2011;23:177–182. doi: 10.1097/CCO.0b013e3283436e8c. [DOI] [PubMed] [Google Scholar]
- 45.Ko CJ. Keratoacanthoma: Facts and controversies. Clin Dermatol. 2010;28:254–261. doi: 10.1016/j.clindermatol.2009.06.010. [DOI] [PubMed] [Google Scholar]
- 46.Williams VL, Cohen PR, Stewart DJ. Sorafenib-induced premalignant and malignant skin lesions. Int J Dermatol. 2011;50:396–402. doi: 10.1111/j.1365-4632.2010.04822.x. [DOI] [PubMed] [Google Scholar]
- 47.Kwon EJ, Kish LS, Jaworsky C. The histologic spectrum of epithelial neoplasms induced by sorafenib. J Am Acad Dermatol. 2009;61:522–527. doi: 10.1016/j.jaad.2008.10.043. [DOI] [PubMed] [Google Scholar]