Abstract
Chemo-enzymatic methods for covalently crosslinking carrier proteins with partner enzymes within modular synthases holds promise for elucidating and engineering metabolic pathways. Our efforts to crystallize the ACP-KS complexes of fatty acid synthases have been complicated by difficulties in the purification of the crosslinked complex from the other proteins in the reaction. Here we present a solution that employs an orthogonal purification strategy to achieve the quantity and level of purity necessary for further studies of this complex.
In fatty acid and polyketide synthases (FASs and PKSs), the growing fatty acid or natural product is tethered to the acyl carrier protein (ACP) as it is modified by several enzymes responsible for carbon unit loading, condensation, and reductions of the resulting ketone.1 In our studies of these modular synthases, we have investigated the specific protein-protein interactions that allow ACP to associate with each of its partner domains. Few of the reported crystal structures of fatty acid and polyketide synthases have been able to resolve these interactions because of the highly disordered structure of ACP.2,3 While several ketosynthase structures have been reported,4,5,6 interactions with ACP have only been investigated through modeling studies.7 A greater understanding of these binding events will be crucial to engineering FAS and PKS systems to synthesize tailor-made natural products. Toward this end, we have sought to covalently cross-link ACP to its various partner domains to facilitate crystallization efforts.
Previously, we reported the one-pot chemo-enzymatic synthesis of carrier proteins, in which E. coli ACP may be covalently labeled with prosthetic groups using synthetic analogs of pantetheine.8 This in vitro reaction scheme utilizes three of the CoA biosynthetic enzymes from E. coli to modify analogs of CoA biosynthetic precursors. Specifically, the reaction includes pantothenate kinase (PanK or CoaA), phosphopanthetheine adenynyl transferase (PPAT or CoaD) and dephospho-CoA kinase (DPCK or CoaE). The promiscuous phosphopantetheinyl transferase Sfp from B. subtilis then transfers the modified phosphopantetheine group from the CoA derivative to a conserved serine residue on ACP. These enzymes have been shown to accept a wide variety of synthetic pantetheine analogs.9 Of the diverse applications for this system, modification of ACP with analogs that irreversibly bind partner proteins in the modular synthases, thereby covalently crosslinking ACP to partner proteins and locking them in a constrained conformation, offer a window into protein-protein interactions (Fig. 1).10
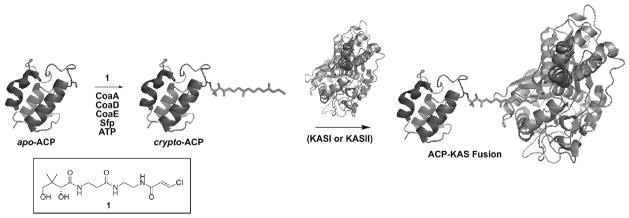
Figure 1.
The carrier protein–ketosynthase crosslinking reaction. ACP is loaded with a prosthetic phosphopantetheinyl group through the actions of the ATP-dependent CoA biosynthetic enzymes and Sfp in the one-pot chemo-enzymatic synthesis. The structure of the chloroacrylate pantetheine analog (1) used in these crosslinking studies is also shown. Protein-protein affinity allows the active site cysteine of the KS domain (KASI or KASII) to attack the trans-chloroacrylate functionality, thereby forming a covalent bond to crosslink ACP to KS.
We have reported the synthesis of phosphopantetheine analogs containing epoxide and chloroacrylate (1) functionalities in place of the natural thiol group.10 These analogs, when loaded onto ACP, have been shown to specifically and irreversibly bind to the active site cysteine in ACP-dependent ketosynthase domains from FAS and type II PKS (Fig. 1).10 These crosslinking reactions are specifically driven by the protein-protein interactions between ACP and the KS, as evidenced by the result that similarly modified carrier proteins from other biosynthetic systems, such as non-ribosomal peptide synthases, do not crosslink with the KS domain. Elucidating a crystal structure of the two domains constrained in their associative conformation will allow us to visualize these binding interactions. However, producing a large amount of sufficiently pure ACP-KS complex has posed significant obstacles. Isolating quantities of a chemically-modified protein complex from a mixture containing seven recombinant protein species offered a unique challenge. All of the enzymes used in our previous work were recombinantly labeled with His6 tags, and many potential partner proteins are similarly labeled. Previous purification attempts included isolating ACP-KS from the crude reaction mixture with a combination of ion exchange and size exclusion chromatography. In these cases, the purified fractions showed residual amounts of the reaction enzymes, indicating that additional purification would be necessary (Fig. 2A). To solve this problem, we chose a purification strategy that involved the subcloning and expression of the necessary CoA biosynthetic enzymes with orthogonal affinity purification tags. Gateway technology (Invitrogen) offered a rapid sub-cloning system to assess multiple fusion tags from vectors that express the gene product with varying N- and C-terminal affinity labels.11 This strategy should have applications for any reaction where several proteins from an enzyme pathway are utilized in vitro and subsequent protein separation is needed.
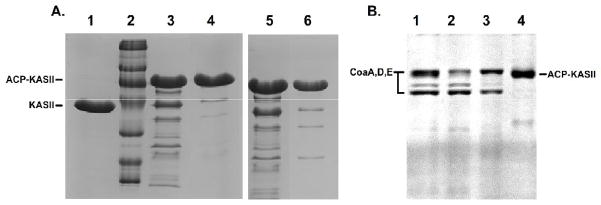
Figure 2.
(a) Size exclusion (left) and ion-exchange chromatography (right) were used to purify the ACP-KS complex from the other enzymes in the reaction. The unreacted KS domain is shown in lane 1 and a size standard is in lane 2. The crude reaction mixture, with all His6-tagged enzymes is shown in lanes 3 and 5. The fractions containing the ACP-KS fusion purified by traditional chromatography are shown in lanes 4 and 6. The purified fractions still show residual amounts of enzymes from the reaction. (b) The purification of ACP-KS from the reaction using orthogonal enzymes. The crude reaction mixture is seen in lane 1, where CoaA, CoaD, and CoaE appear as bands. Native Sfp is sufficiently dilute to not appear as a band. Nickel resin flowthrough is shown in lane 2. 50mM and 200mM elutions of the nickel resin are shown in lanes 3 and 4. In lane 4, only ACP-KS elutes from the resin.
The open reading frame (ORF) for E. coli CoaA was PCR amplified from a pCA24N construct using primers containing a recognition sequence for the Gateway recombinase at the 5′ end and 21–27 bases complementary to the CoaA gene (Fig. S1).12 The gene for CoaD was PCR amplified from a pET24b construct, 13 and the gene for CoaE was PCR amplified from of a pCA24N construct in a similar manner.12 A pUC8 construct of Sfp was used as the template in its PCR amplification.14 The PCR products were excised from 1% agarose gels, filtered and purified by ethanol precipitation. The purified PCR products were then recombined into the pDONR207 entry vector using BP Clonase (Invitrogen). Electrocompetent DH5α cells were transformed with these entry vectors, and positive clones were selected with LB + gentamycin. The entry plasmids coding for CoaA, CoaD, CoaE, and Sfp were purified with a QiaPrep kit (Qiagen).
The entry clones were subcloned into several expression vectors for fusion protein construction with the Gateway system (data not shown), but only the MBP constructs provided uniform soluble, active protein across the three CoA biosynthetic enzymes. To create these MBP fusion proteins, the entry vectors were recombined with the destination vector coding for an N-terminal maltose binding protein (MBP) fusion using LR Clonase (Invitrogen).11 Electrocompetent BL21 cells were transformed with the products and selected on ampicillin. One liter cultures were grown containing glucose + LB media, and protein expression was induced with the addition of 1 mM IPTG. After 2–4 hours of expression, the cells were harvested and lysed using a French pressure cell. The crude lysates were centrifuged, and the MBP fusion proteins were isolated by batch binding with 1 mL of amylose resin (NEB) for 1 hour. The resin was loaded onto a column and washed with 20 mL of amylose resin wash buffer (20 mM Tris, 200 mM NaCl, 1 mM EDTA). The MBP-CoaA, D, and E, and MBP-Sfp were eluted from the resin using 10 mL amylose wash buffer containing 10 mM maltose in 1 mL fractions. A large majority of the protein was eluted in the first 5 mL (Fig. S2). As only catalytic amounts of the enzymes are required, a one liter culture yielded sufficient enzyme to modify several hundred milligrams of carrier protein.
To assay for activity of the CoA enzymes, we performed a one-pot chemoenzymatic modification of ACP with a fluorescent dansyl pantetheine analog.8 In order for the fluorescent dansyl group to load onto ACP, all three of the ATP-dependent CoA enzymes are needed to convert the dansyl-pantetheine into dansyl-CoA, which is then loaded onto ACP by Sfp. The His-tagged CoA enzymes used in previous studies were used as positive control to test each enzyme individually. His-tagged Sfp was used in all CoA biosynthetic enzyme activity assays. The contents of the reaction were then run on a 10% acrylamide gel and visualized under UV light. The activities of the three CoA enzymes were confirmed by the fluorescence of the band correlating to ACP (Fig. S3). The kinetic efficiencies of His6-CoaA and MBP-CoaA were also compared using an assay described previously,9 in which the consumption of ATP is coupled to the consumption of NADH through the actions of pyruvate kinase and lactate dehydrogenase, and the reaction rate is monitored by absorbance at 340 nm. The two enzymes showed very similar Michaelis-Menten plots (Fig. S4). Our results demonstrate that the MBP fusion domain does not significantly diminish enzyme efficiency.
The PPTase activity of the MBP-Sfp fusion was assayed through the reaction of ACP with fluorescent dimethylaminocoumarin-labeled-CoA.9 A fluorescent band correlating to ACP would confirm the PPTase activity of the MBP-Sfp. However, no fluorescent ACP was seen in this reaction (Fig. S5). We presume that the large MBP tag might interfere with the ability of ACP to associate with Sfp. We therefore sub-cloned Sfp with a smaller glutathione-S-transferase (GST) tag at the N-terminus. This construct was inactive and insoluble (Fig. S6). We further created a biotinylation fusion (Avidity), which is a smaller 17-residue peptide sequence that becomes biotinylated when expressed in AVB101 cells.15 However, the N-terminal biotinylated Sfp was also inactive (Fig. S7). We conclud that Sfp is not amenable to N-terminal fusions, so we next attempted C-terminal fusions of Sfp. Specifically, the stop codon in the reverse primer was removed, and a ribosomal binding sequence was inserted just upstream of the start codon on the forward primer. A C-terminal Sfp-GST fusion expressed as an insoluble protein (Fig. S8). Given the recalcitrance of Sfp toward orthogonal labeling, we finally chose to express and purify Sfp in its native, unlabeled form, as only minimal amounts of this enzyme are needed in the catalytic crosslinking reactions.
Using a pUC8-Sfp construct,14 native Sfp was expressed in BL21 cells. The crude lysate from this expression was evaluated for PPTase activity using the assay described above, and native Sfp was found to be active (Fig. S9). This cell lysate was subjected to ion exchange chromatography using DE52 resin (Millipore), and the fractions containing Sfp were precipitated with 80% ammonium sulfate (Fig. S10). The resuspended protein was then subjected to size exclusion chromatography using S-100 resin, yielding pure Sfp (Fig. S11). With the MBP-tagged CoA biosynthetic enzymes and native Sfp in hand, purification of the ACP-KS complex was next performed.
The ACP-KS crosslinking reaction was initiated using the three MBP-CoA enzymes and native Sfp. Large excesses of apo-ACP and pantetheine analog 1 were used to ensure that all of the KS reacted to completion. ACP loading was performed in 50 mM sodium phosphate buffer (pH=7.2), 8 mM ATP, 12.5 mM MgCl2, 167 μM chloroacrylate-pantetheine (1), with 5 mg ACP, 4.5 μg Sfp, 135 μg MBP-CoA, 150 μg MBP-CoaD, and 165 μg MBP-CoaE in a total volume of 14 mL. This mixture was incubated for 1 hour at 37°C, then 400 μg KS in a volume of 1 mL was added to the reaction. After an additional hour of incubation at 37°C, 1.0 mL of Ni-NTA His-bind resin suspension (Qiagen) was added to the solution with agitation. After 1 hour of batch binding on ice, the resin was loaded onto a column and washed with 10 mL of 25 mM sodium phosphate (pH=7.2), 100 mM NaCl. The resin was subsequently washed with 10 mL of 50 mM imidazole in wash buffer to elute non-specifically bound proteins, and then the ACP-KS complex was eluted with 10 mL of 200 mM imidazole. The 50 mM wash and 200 mM elution were collected in 1 mL fractions. The second 1 mL fractions from each imidazole concentration showed the highest protein content by qualitative Bradford analysis and were analyzed by SDS-PAGE (Fig. 2B). The 200 mM imidazole fractions 2–6 contained essentially pure ACP-KS (Fig. S12). After pooling all of the fractions containing purified protein, absorbance at 280 nm was used to calculate a total yield of 98% (final total moles of ACP-KS per initial total moles of KS).
In summary, we have used recombinant methods to clone and express the CoA biosynthetic enzymes with orthogonal affinity tags. Sfp was not amenable to recombinant affinity tag fusions. The cloning of these enzymes using Gateway technology allowed for facile sub-cloning and expression to analyze various fusion systems. Using several parallel preparations like the one described here, we have the ability to purify milligram quantities of the ACP-KS complex for crystallographic studies. This strategy should have applications in any chemo-enzymatic reaction in which the isolation of a single modified protein out of a complex mixture is required. Using the strategy described here, we are currently purifying modified type II PKS carrier proteins to study binding events through structural studies with cognate partner proteins.
Supplementary Material
Acknowledgments
We thank Dr. Mike Austin and Prof. Joe Noel (Salk Institute) for initial studies with crosslinked enzymes. We thank Prof. Andrei Osterman (Burnham Institute) for supplying constructs of E. coli genes used in this study, and to Prof. Eli Chapman (The Scripps Research Institute) for chromatography assistance. Funding was provided by the American Cancer Society RSG-06-011-01-CDD.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Keating TA, Walsh CT. Initiation, Elongation, and Termination Strategies in Polyketide and Polypeptide Antibiotic Biosynthesis. Curr Op in Chem Bio. 1999;3(5):598–606. doi: 10.1016/s1367-5931(99)00015-0. [DOI] [PubMed] [Google Scholar]
- 2.Jenni S, Leibundgut M, Maier T, Ban N. Architecture of Fungal Fatty Acid Synthase at 5Å Resolution. Science. 2006;311:1263–1267. doi: 10.1126/science.1123251. [DOI] [PubMed] [Google Scholar]
- 3.Maier T, Jenni S, Ban N. Architecture of Mammalian Fatty Acid Synthase at 4.5Å Resolution. Science. 2006;311:1258–1262. doi: 10.1126/science.1123248. [DOI] [PubMed] [Google Scholar]
- 4.Olsen JG, Kadziola A, von Wettstein-Knowles P, Siggaard-Andersen M, Lindquist Y, Larsen S. The X-ray Crystal Structure of Beta-Ketoacyl [Acyl Carrier Protein] Synthase I. FEBS Letts. 1999;460:46–52. doi: 10.1016/s0014-5793(99)01303-4. [DOI] [PubMed] [Google Scholar]
- 5.Huang W, Jia J, Edwards P, Dehesh K, Schneider G, Lindquist Y. Crystal Structure of Beta-ketoacyl-acyl Carrier Protein Synthase II from E. coli Reveals the Molecular Architecture of Condensing Enzymes. EMBO J. 1998;17:1183–1191. doi: 10.1093/emboj/17.5.1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Qiu X, Janson CA, Kostantinidis AK, Nwagwu S, Silverman C, Smith WW, Khandekar S, Lonsdale J, Abdel-Meguid SS. Crystal Structure of Beta-ketoacyl-acyl Carrier Protein Synthase III. A Key Condensing Enzyme in Bacterial Fatty Acid Biosynthesis. J Biol Chem. 1999;274:36465–36471. doi: 10.1074/jbc.274.51.36465. [DOI] [PubMed] [Google Scholar]
- 7.Zhang YM, Rao MS, Heath RJ, Price AC, Olson AJ, Rock CO, White SW. Identification and Analysis of the Acyl Carrier Protein (ACP) Docking Site on Beta-ketoacyl-ACP Synthase III. J Biol Chem. 2001;274:8231–8238. doi: 10.1074/jbc.M008042200. [DOI] [PubMed] [Google Scholar]
- 8.Worthington AS, Burkart MD. One-pot Chemoenzymatic Synthesis of Reporter Modified Proteins. Org Biomol Chem. 2006;4:44–46. doi: 10.1039/b512735a. [DOI] [PubMed] [Google Scholar]
- 9.Meier JL, Mercer AC, Rivera H, Burkart MD. Synthesis and Evaluation of Bioorthogonal Pantetheine Analogs for In Vivo Protein Modification. J Am Chem Soc. 2006;128(37):12174–12184. doi: 10.1021/ja063217n. [DOI] [PubMed] [Google Scholar]
- 10.Worthington AS, Rivera H, Torpey JW, Alexander MD, Burkart MD. Mechanism-based Protein Cross-linking Probes to Investigate Carrier Protein-mediated Biosynthesis. ACS Chem Biol. 2006;1:687–691. doi: 10.1021/cb6003965. [DOI] [PubMed] [Google Scholar]
- 11.Braun P, Hu Y, Shen B, Halleck A, Koundinya M, Harlow E, LaBaer J. Proteome-scale purification of human proteins from bacteria. Proc Natl Acad Sci USA. 2002;99(5):2654–2659. doi: 10.1073/pnas.042684199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kitagawa M, Ara T, Arifuzzaman M, Ioka-Nakamichi T, Inamoto E, Toyonaga H, Mori H. Complete set of ORF clones of Escherichia coli ASKA library (a complete set of E. coli K-12 ORF archive): unique resources for biological research. DNA Res. 2005;12:291–299. doi: 10.1093/dnares/dsi012. [DOI] [PubMed] [Google Scholar]
- 13.Clarke KM, Mercer AC, La Clair JJ, Burkart MD. Vivo Reporter Labeling of Proteins via Metabolic Delivery of Coenzyme A Analogues. J Am Chem Soc. 2005;127:11234–5. doi: 10.1021/ja052911k. [DOI] [PubMed] [Google Scholar]
- 14.Nakano MM, Corbell N, Besson J, Zuber P. Isolation and characterization of sfp: a gene that functions in the production of the lipopeptide biosurfactant, surfactin, in Bacillus subtilis. Mol Gen Genet. 1992;232(2):313–321. doi: 10.1007/BF00280011. [DOI] [PubMed] [Google Scholar]
- 15.Tucker J, Grisshammer R. Purification of a Rat Neurotensin Receptor Expressed in E. coli. Biochem J. 1996;317(3):891–899. doi: 10.1042/bj3170891. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.