Abstract
Several recent studies have found that selection acts on synonymous mutations at the beginning of genes to reduce mRNA secondary-structure stability, presumably to aid in translation initiation. This observation suggests that a metric of relative mRNA secondary-structure stability, ZΔG, could be used to test whether putative genes are likely to be functionally important. Using the Escherichia coli genome, we compared the mean ZΔG of genes with known functions, genes with known orthologs, genes where function and orthology are unknown, and pseudogenes. Genes in the first two categories demonstrated similar levels of selection for reduced stability (increased ZΔG), whereas for pseudogenes stability did not differ from our null expectation. Surprisingly, genes where function and orthology were unknown were also not different from the null expectation, suggesting that many of these open reading frames are not functionally important. We extended our analysis by constructing a Bayesian phylogenetic mixed model based on data from 145 prokaryotic genomes. As in E. coli, genes with no known function had consistently lower ZΔG, even though we expect that many of the currently unannotated genes will ultimately have their functional utility discovered. Our findings suggest that functional genes tend to evolve increased ZΔG, whereas nonfunctional ones do not. Therefore, ZΔG may be a useful metric for identifying genes of potentially important function and could be used to target genes for further functional study.
Keywords: synonymous mutations, protein function, translation
Introduction
Synonymous mutations, which do not cause changes to the protein encoded by a gene, are often referred to as silent mutations. Evidence has accumulated, however, that these mutations can have an important effect on phenotype (Chamary et al. 2006; Kimchi-Sarfaty et al. 2007; Zhang et al. 2010, see Plotkin and Kudla 2011; Sauna and Kimchi-Sarfaty 2011 for recent reviews). One recently discovered selective force on synonymous mutations arises from the mRNA secondary structure of transcribed genes. Using variants of green fluorescent protein that differed only by synonymous mutations, Kudla et al. (2009) found that variants with high levels of mRNA secondary structure produced lower amounts of protein. Indeed, mRNA secondary structure was the main source of variation in protein expression for that study. A second experimental study constructed ribosomal protein mutants containing different nonsynonymous and synomymous mutations and, subsequently, measured fitness via growth and competition assays (Lind et al. 2010). The distribution of fitness effects for both types of mutations were surprisingly similar, with most mutations being mildly deleterious. The conclusion from that study was that the similarity in the fitness distribution was due to the same underlying cause previously suggested by Kudla et al. (2009): changes in mRNA secondary structure. Although these studies suggest that there is a strong link between mRNA secondary structure and protein abundance, a third experimental study (Welch et al. 2009) found a much weaker effect and argued for codon usage bias as the primary determinant of protein abundance. Computational evidence for the importance of mRNA secondary structure includes a study by Gu et al. (2010), who analyzed the mRNA stability at the beginning of genes in a wide variety of cellular organisms spanning the tree of life; they found a general pattern of reduced mRNA stability at the beginning of coding sequences relative to null expectations. Tuller et al. (2010) found similar results in Escherichia coli and Saccharomyces cerevisiae, whereas Zhou and Wilke (2011) found a similar pattern in dsDNA viruses. Collectively, these studies suggest that mRNA stability at the 5′ coding region of genes is an important trait under selection in a wide variety of organisms.
Since the completion of the E. coli genome, biologists have amassed an incredible amount of functional knowledge about what its approximately 4,300 open reading frames (ORFs) are used for (Blattner et al. 1997). Of these ORFs, 86% are annotated with their known or believed function. Most of the remaining ORFs are believed to code for functional proteins because orthologous genes have been found in other organisms. There is only a small number of ORFs—about 100—for which it is not known whether a protein is made and, if so, whether it has a function. In other genomes, functional knowledge is much less complete. A survey of GenBank files for 310 prokaryotic genomes finds that 28% of genes are purely hypothetical, meaning that they have start and stop codons but nothing else is known about them. Thus, it would be useful to have a metric to identify which putative genes are likely to be functionally important, so they can be studied further.
We investigated whether genes of various categories displayed different levels of selection for reduced mRNA stability near the start codon. As expected, we found for E. coli that genes with known function generally had reduced levels of mRNA stability, whereas known pseudogenes displayed no evidence of selection. Genes with orthologs but no identified function displayed selection similar to genes with known function. By contrast, the remaining ORFs lacking functional knowledge and orthology had mRNA secondary-structure stability similar to the ORFs of known pseudogenes. We validated our finding of reduced selection in genes of unknown function using data from 145 prokaryotic genomes; in general, ORFs with a known or predicted function had less stability compared with genes of unknown function.
Materials and Methods
Data Sources
We obtained 126 bacterial and 19 archaeal annotated genomes from the NCBI FTP server (ftp://ftp.ncbi.nih.gov/). We selected genomes that had previously been analyzed by Gu et al. (2010) and for which 16S ribosomal RNA was available in the Comparative RNA website (Cannone et al. 2002). A list of the 145 genomes analyzed is provided as supplementary table 1, Supplementary Material online. As in Gu et al. (2010), we focused on coding sequences longer than 50 bases.
We obtained mRNA abundance data for E. coli from Ragavan et al. (2011). This data set reported mRNA expression level per individual nucleotide. We converted these data into gene expression levels by calculating the mean mRNA expression level over all nucleotide positions in the coding sequence of each gene. We considered genes with expression level below the genome-wide median as lowly expressed and all others as highly expressed.
We calculated codon adaptation index (CAI) values for E. coli genes using the CodonW program (http://codonw.sourceforge.net/). We considered genes with CAI below the genome-wide median as low-CAI genes and all others as high-CAI genes.
We obtained the E. coli core genome from Touchon et al. (2009).
RNA Secondary Structure Stability
The folding free energy of RNA (ΔG), a measure of how much secondary structure is present, was estimated by RNAfold (Hofacker et al. 1994) using default parameters. We compared the observed secondary structure stability to 1,000 randomly permutated mRNA sequences to obtain a statistical deviation from a null sequence distribution, denoted as ZΔG. Within coding sequences, permutations were performed such that the protein sequence for a given gene was maintained, but synonymous codons within the gene were reshuffled (Gu et al. 2010). This method controls for codon usage, GC content, and the protein sequence. ZΔG is simply the difference between the observed mRNA secondary structure stability and the mean stability of 1,000 randomly reshuffled coding sequences, normalized to the standard deviation of the stability null distribution.
For E. coli, we generally considered mRNA stability for the first 30 bases of a coding sequence, as Gu et al. (2010) had previously shown that lowered stability occurred primarily in this region. For the remaining genomes, we collected the ZΔG values for the first 30 bases in each ORF from Gu et al. (2010), available at http://openwetware.org/wiki/Wilke:Data_Sets.
In certain analyses, we also examined ZΔG for regions upstream of a coding region, using a sliding window approach for 30 base windows starting 10, 20, and 30 bases upstream of the coding region. The reshuffling method used to generate the null stability distribution in noncoding regions was different from that used for ORFs: the 30 bases upstream of an ORF were reshuffled at random, whereas in ORFs synonymous codons were reshuffled throughout the ORF.
Programming was done using a combination of the Python and Cython (Behnel et al. 2011) programming languages.
Gene Function Categorization
We parsed the E. coli GenBank file using Biopython (Cock et al. 2010), assigning genes a functional category based on their annotation. Due to E. coli’s long use as a model organism, the E. coli gene annotation was fairly standardized. Information about the function of a gene was typically contained in the “product” annotation. Genes were designated as conserved if orthologs were known but no functional annotation was available. Putative genes were coding regions with no functional knowledge or evidence of homology. Finally, we identified known pseudogenes as a separate class.
The remaining genomes used in this study were less consistently annotated. Thus, for the remaining organisms, we considered only genes of known or predicted function, conserved genes, and genes of unknown function.
Our raw data for E. coli, including functionality annotation and ZΔG values, are provided as supplementary table 2, Supplementary Material online. The contents of the table columns is explained in the supplementary text, Supplementary Material online.
Comparative Phylogenetics
We obtained alignments for the highly conserved 16S ribosomal RNA from the Comparative RNA website (Cannone et al. 2002). If multiple sequences were available for a species, we generated a consensus sequence. We then built a maximum-likelihood tree in RAxML, using the combined bootstrap-treesearching method (Stamatakis 2006). We then constructed an ultrametric tree from the RAxML output by using a semiparametric penalized likelihood approach implemented in the R package “ape” (Sanderson 2002; Paradis et al. 2004; R Development Core Team 2010). This method uses a smoothing parameter, λ, to control how much evolutionary rates vary across a tree. As suggested in Sanderson (2002), we used a range of λ values; our final tree was generated with the λ that minimized a cross-validation statistic. The cross-validation statistic was calculated by eliminating each tip in succession and taking the sum of squared differences between the branch lengths in the reduced tree and the full tree (Paradis et al. 2004). The final phylogenetic tree is provided as supplementary file, Supplementary Material online.
We constructed Bayesian phylogenetic mixed models (BPMMs) based on Markov chain Monte Carlo estimates using our ultrametric tree and the R package “MCMCglmm” (Hadfield 2010; Hadfield and Nakagawa 2010; R Development Core Team 2010). Priors for all parameters were uninformative. MCMC chains were run for a total of 60,000 iterations, discarding the first 10,000 generations as the burn-in period. We then sampled every 25 iterations to generate a posterior distribution of 2,000 samples. We assessed convergence visually using the R “coda” package (Plummer et al. 2006), as well as formally diagnosing convergence with the Heidelberger–Welch and Geweke tests (Heidelberger and Welch 1983; Geweke 1992).
Predicting Gene Functionality
We constructed logistic regression models in R (R Development Core Team 2010) to assess the ability of various predictors to classify E. coli genes as putatively functional versus putatively nonfunctional. To test the predictive power of these models, we used the E. coli core genome plus pseudogenes as the test data set and all other genes as the training data set. We considered as the core genome the genes common to 28 distinct E. coli genomes (Touchon et al. 2009). We evaluated predictive power by calculating the area under the curve (AUC) of receiver operating characteristic (ROC) curves for a given model.
Results
Reduced Structural Stability Generally Occurs in Genes of Known or Predicted Function
We began by assessing differences in mRNA stability between genes in the E. coli genome. For each gene, we calculated the change in free energy (ΔG) for the first 30 bases of the 5′ mRNA. We then compared this observed value with a null distribution of ΔG values calculated from random mRNA sequences that encoded the same protein, yielding a Z-score, ZΔG. Positive values indicate reduced mRNA secondary-structure stability relative to null expectations based on codon usage and GC content, whereas negative values indicate increased mRNA secondary-structure stability.
On average, we found that larger ZΔG values corresponded to less negative ΔG values (i.e., less stable secondary structure), as previously reported by Gu et al. (2010). For example, the average ΔG for a ZΔG window of width 1 centered around 0 was − 3.18. This value rose to − 1.51 as we centered the window around 1 and to − 1.09 as we centered the window around 2.
Our analysis included a total of 4,163 E. coli genes. The function of a large percentage of these genes is well understood. For other genes, their exact function is unknown but structural similarities to other proteins indicate some core functionality, such as being a repressor, transporter, or ligase. We classified all genes for which function was known or reasonably obvious to infer as genes of known or predicted function. There were 3,651 such genes. For other genes, no functional annotation is available but they have orthologs in other species. We refer to these genes as conserved genes and found 285 such genes. Finally, there were 127 genes of unknown function that lacked orthologs (we refer to these as genes of unknown function) and 100 known pseudogenes.
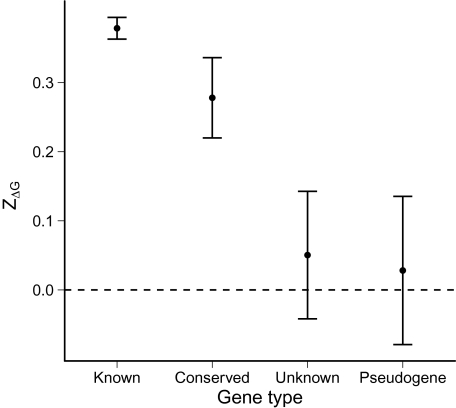
We calculated the mean ZΔG for all four of these subsets of genes (fig. 1). The mean ZΔG for genes of known or predicted function was 0.39, significantly higher than the null expectation (one-sample t test; t = 24.92, degrees of freedom [df] = 3,650, P < 10 − 15) and consistent with prior observations of reduced mRNA stability in the translation initiation region (Kudla et al. 2009; Gu et al. 2010). Likewise, the mean ZΔG in conserved genes was 0.28, significantly higher than zero (one-sample t test; t = 4.95, df = 284, P = 1.3×10 − 6) and not significantly different from genes of known function (two-sample t test; t = 1.77, df = 327.42, P = 0.079). This result was as expected, since evolutionarily conserved genes likely are expressed and have a function.
FIG. 1.—
Average ZΔG for different gene categories in Escherichia coli. Error bars are ± 1 standard error. The dashed line is the null expectation of ZΔG for coding sequences with randomly chosen codons. ZΔG is significantly different from 0 for known and conserved genes but not for unknown genes or pseudogenes.
If a positive mean ZΔG indicates selection for efficient translation, then we would expect that the mean ZΔG for pseudogenes should not differ from zero. Indeed, the mean ZΔG for the 100 known pseudogenes was not significantly different from zero (one-sample t test; mean ZΔG = 0.032, t = 0.30, df = 99, P = 0.76). Similarly, the 127 genes of unknown function, on average, had ZΔG values that were not significantly different from the null expectation (one-sample t test; mean ZΔG = 0.068, t = 0.73, df = 126, P = 0.47). Although it is possible that some of the genes in this category are functional, overall these results suggest that most are nonfunctional.
Collectively, we found that ZΔG was similar in genes with a known or predicted function and in genes with known orthologs but lacking a predicted function (fig. 1). Conversely, there was no evidence of selection for reduced mRNA stability in genes of completely unknown function or in pseudogenes.
Selection for Reduced mRNA Stability Extends Upstream of Genes
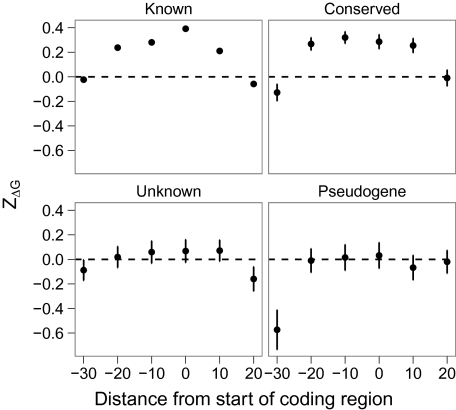
Certain noncoding regions before the beginning of a coding sequence are known to be important for translation initiation, most notably the Shine–Dalgarno sequence (Shine and Dalgarno 1975). We used a sliding window approach to determine whether the noncoding sequence upstream of ORFs contributed to mRNA destabilization. Sequences in these upstream regions were randomized by shuffling bases rather than codons. As in our earlier analysis, ORFs with a known or predicted function and conserved ORFs showed elevated ZΔG values, whereas genes of unknown function and pseudogenes were similar to randomized coding sequences (fig. 2). These trends were qualitatively similar when only the noncoding region was shuffled (data not shown).
FIG. 2.—
Selection for reduced stability continues upstream of coding regions in Escherichia coli. Error bars are ± 1 standard error. (Error bars for genes of known function are smaller than the symbol size.) The dashed line is the null expectation of ZΔG for coding sequences with randomly chosen codons.
ZΔG Results Are Largely Independent of Gene Expression Level or Codon Usage Bias
We gathered information on gene expression levels to investigate whether expression levels correlate with ZΔG. As a measure of expression level, we used mRNA abundance measured by Ragavan et al. (2011). There was no overall correlation between expression level and ZΔG (Spearman’s ρ = 0.006, P = 0.68). Additionally, none of the correlations for the four class subsets were significant (known genes: Spearman’s ρ = 0.006, P = 0.74; conserved genes: Spearman’s ρ = 0.032, P = 0.59; genes of unknown function: Spearman’s ρ = 0.02, P = 0.82; pseudogenes: Spearman’s ρ = − 0.003, P = 0.98).
We also tested whether ZΔG was correlated with codon usage bias. We assessed codon usage bias with the CAI (Sharp and Li 1987). In bacteria, codon usage bias is strongly correlated with expression level, and CAI is often used as a proxy for expression level. We found a significant but weak overall correlation between ZΔG and CAI (Spearman’s ρ = 0.10, P = 4.9×10 − 10). This correlation held up only in genes of known function (known genes: Spearman’s ρ = 0.09, P = 1.3×10 − 8; conserved genes: Spearman’s ρ = 0.01, P = 0.86; genes of unknown function: Spearman’s ρ = 0.04, P = 0.63; pseudogenes: Spearman’s ρ = 0.02, P = 0.83).
Next, we examined the ZΔG values of genes with high and low expression levels. It is possible that genes with known function tend to be highly expressed compared with the other genes classes, and thus ZΔG in highly expressed genes would be higher than in lowly expressed genes solely for that reason. However, the mean ZΔG was 0.36 for lowly expressed genes and 0.37 for highly expressed genes; the difference between these two groups was not significant (two-sample t test, t = − 0.312, df = 4161, P = 0.76). Additionally, almost half (63) of genes with no known function or orthology were in the highly expressed group. By contrast, when comparing ZΔG values for genes with high and low CAI, we found a significant difference. The mean ZΔG was 0.28 for genes with low CAI and 0.45 for genes with high CAI; the difference between these two groups was highly significant (two-sample t test, t = − 5.91, df = 4,159, P = 3.6×10 − 9). Approximately, a quarter (28) of genes with no known function or orthology were in the high-CAI group.
After subsetting the E. coli genes into either genes of high or low expression level or genes of high or low CAI, we again tested whether the four gene classifications were significantly different from 0. In all cases, the prior findings remained the same (genes of known function or orthology had elevated ZΔG, pseudogenes, and genes of unknown function did not).
In summary, although there was a weak correlation between ZΔG and CAI, our conclusions were largely independent of gene expression level or codon usage bias.
Analysis of Stability Difference Yields Comparable Results
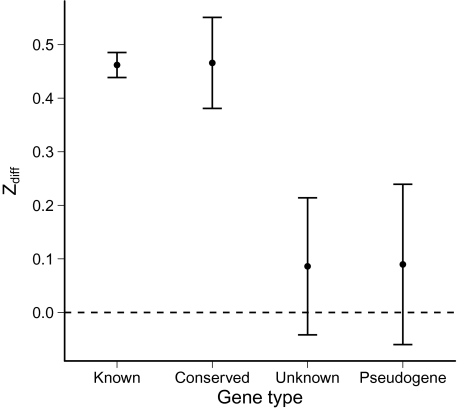
It is generally known that the majority of mRNA, outside the initial 40–50 nucleotides, is more stable than expected (Chamary et al. 2006; Gu et al. 2010). Thus, the difference between the beginning of a gene and a downstream section might also indicate whether an ORF corresponds to a functional protein. We calculated this difference between the stability of the first 30 bases and bases 101–130. We found that this difference, Zdiff, was also correlated with gene functionality in E. coli (fig. 3). The statistics were comparable to the case of considering just ZΔG: genes of known function and conserved genes had a significantly nonzero Zdiff (one-sample t test; t = 19.78, df = 3,650, P < 10 − 15 for genes of known function, t = 5.48, df = 284, P = 9.3×10 − 8 for conserved genes); genes of unknown function and pseudogenes did not (one-sample t test; t = 0.67, df = 126, P = 0.50 for genes of unknown function, t = 0.60, df = 99, P = 0.55 for pseudogenes).
FIG. 3.—
Average Zdiff for different gene categories in Escherichia coli. Error bars are ± 1 standard error. The dashed line is the null expectation of Zdiff for coding sequences with randomly chosen codons. Zdiff is significantly different from 0 for known and conserved genes but not for unknown genes or pseudogenes.
ZΔG Is Consistently Higher for Annotated Versus Unknown Proteins in Prokaryotes
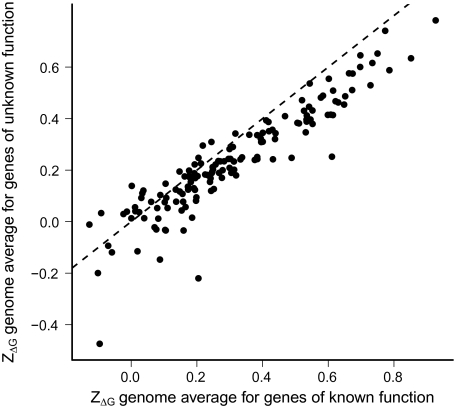
Although we found multiple lines of evidence suggesting that E. coli genes lacking orthologs or functional annotation are not under selection for reduced mRNA secondary-structure stability, the generality of this finding was unclear. Therefore, we performed similar comparisons in 126 bacterial and 19 archaeal genomes. Given our previous finding of similar ZΔG values for ORFs with a known function and for conserved ORFs, we binned these two categories together and compared them with genes of unknown function; pseudogenes were not consistently marked in most genomes and thus were excluded from this analysis. ZΔG was generally higher for genes of known or predicted function compared with genes of unknown function (fig. 4). Note that most of the overall variation in ZΔG is associated with genomic GC content (Gu et al. 2010).
FIG. 4.—
Comparison of ZΔG for genes of known function versus genes of unknown function in 145 prokaryote genomes. Each point represents a genome; 126 bacterial and 19 archeal genomes were used. The dashed line is the 1:1 null expectation of equal ZΔG values for the two gene types. The mean ZΔG for unknown genes tends to be lower than for known genes, especially for genomes with high mean ZΔG.
However, this analysis failed to consider phylogenetic relationships in the comparative analysis, which may confound interpretation because species cannot be assumed to be independent data points (Felsenstein 1985, 2003). Therefore, we constructed a BPMM to account for relatedness between species (Hadfield 2010; Hadfield and Nakagawa 2010). BPMMs control for phylogeny by incorporating into the regression model, the covariance structure given by the input tree, and the branch lengths between species. Although this type of analysis does not appear to be widely used in genomic analyses (but see Naya et al. 2006), it is a powerful method for analyzing variation within and between species.
Using an alignment of 16S ribosomal RNA sequences from the Comparative RNA website (Cannone et al. 2002), we constructed a maximum-likelihood tree for the prokaryotes used in this study (see Materials and Methods). The estimated relationships between species were then used as a random effect in a phylogenetic mixed model. After controlling for phylogeny and species identity, there was still a large difference between the average ZΔG of genes with a known or predicted function versus genes where function has not been identified (table 1). The ZΔG of genes with a known or predicted function (ZΔG = 0.201) was on average twice as large as the ZΔG of genes with unknown function (ZΔG = 0.201 − 0.105 = 0.096). This difference was highly significant (table 1).
Table 1.
BPMM Fit of ZΔG to Gene Function (Known/Predicted or Unknown) While Controlling for Phylogeny and Species (126 Bacterial and 19 Archeal Genomes)
| Fixed Effect | Parameter Estimatea | 95% Credible Interval | P Value |
| Known/predicted function | 0.201 | 0.049–0.329 | 0.006 |
| Unknown function | −0.105 | −0.112 to −0.098 | <5×10−4 |
| Random Effect | Estimated Variance | 95% Credible Interval | |
| Phylogeny | 0.029 | 0.013–0.049 | |
| Species | 0.018 | 0.011–0.027 | |
| Residual | 1.016 | 1.011–1.020 |
The parameter estimate for known/predicted function is the mean ZΔG for genes in this category. The parameter estimate for unknown function is the change in mean ZΔG relative to known/predicted function.
ZΔG Can Serve as Predictor of Gene Functionality
Finally, we wanted to determine to what extent ZΔG could actually serve as a predictor of gene functionality. To address this question, we developed logistic regression models that predicted gene functionality from ZΔG and other predictor variables. We fitted these models to the E. coli data. We classified genes of known function or conserved genes as functional and all other genes (i.e., pseudogenes and genes of unknown function) as nonfunctional.
For the simplest model, we considered only ZΔG as a predictor variable; both ZΔG and the intercept were significant (Model I in table 2). In the second model, we used CAI and log-transformed mRNA expression levels as predictor variables. In this model, CAI and expression were significant, whereas the intercept was not (Model II in table 2). Finally, we fitted a model using ZΔG, CAI, and expression levels. In this model, all three predictor variables were significant, whereas the intercept was not (Model III in table 2). We fit identical models to a reduced E. coli data set that had the core genome removed. The results were very similar to those obtained for the whole genome. The main difference was that mRNA expression level was not significant for any model on the reduced genome (table 3). In aggregate, these results show that ZΔG is a significant predictor of gene functionality, even when used jointly with other predictors and that it performs better than mRNA expression level.
Table 2.
Logistic Regression of Gene Functionality Against Predictor Variables, Using the Full Escherichia coli Genome
| Predictor | Estimate | Standard Error | z Value | P Value |
| Model I | ||||
| ZΔG | 0.315 | 0.065 | 4.83 | 1.35×10 − 6 |
| Intercept | 2.79 | 0.069 | 40.3 | < 2×10 − 16 |
| Model II | ||||
| CAI | 10.9 | 1.12 | 9.79 | < 2×10 − 16 |
| Expression | 0.099 | 0.045 | 2.19 | 0.028 |
| Intercept | −0.60 | 0.32 | −1.90 | 0.056 |
| Model III | ||||
| ZΔG | 0.245 | 0.067 | 3.64 | 2.7×10 − 4 |
| CAI | 10.5 | 1.12 | 9.42 | < 2×10 − 16 |
| Expression | 0.099 | 0.045 | 2.19 | 0.029 |
| Intercept | −0.546 | 0.32 | –1.72 | 0.085 |
Table 3.
Logistic Regression of Gene Functionality Against Predictor Variables, Using a Reduced Escherichia coli Data Set in Which the Core Genome Has Been Removed
| Predictor | Estimate | Standard Error | z Value | P Value |
| Model I | ||||
| ZΔG | 0.385 | 0.092 | 4.18 | 2.92×10 − 5 |
| Intercept | 2.89 | 0.098 | 29.4 | < 2×10 − 16 |
| Model II | ||||
| CAI | 10.7 | 1.62 | 6.62 | 3.56×10 − 11 |
| Expression | 0.116 | 0.067 | 1.74 | 0.083 |
| Intercept | −0.436 | 0.46 | −0.95 | 0.342 |
| Model III | ||||
| ZΔG | 0.305 | 0.094 | 3.24 | 1.21×10 − 3 |
| CAI | 10.2 | 1.63 | 6.27 | 3.73×10 − 10 |
| Expression | 0.112 | 0.067 | 1.68 | 0.093 |
| Intercept | −0.327 | 0.46 | –0.71 | 0.476 |
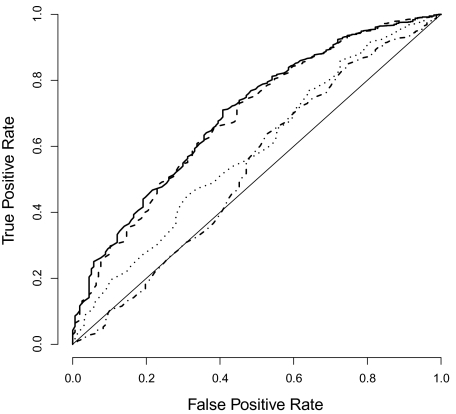
However, the statistical significance in a regression model does not quantify the predictive power of a given variable. To quantify predictive power, we used the logistic regression models to predict gene functionality and then calculated ROC curves for these predictors. We used the E. coli core genome plus pseudogenes as the training data set and all other genes as the test data set. In the test data set, we considered genes of known function and conserved genes as functional and genes of unknown function as nonfunctional. As our previous logistic regression models would suggest, gene expression level as measured by mRNA abundance was a poor predictor of gene functionality. In fact, for false-positive rates below approximately 0.4, it performed worse than random guessing (fig. 5). We therefore did not consider it any further. By contrast, ZΔG performed somewhat better than random guessing (AUC = 0.594), and CAI performed substantially better than random guessing (AUC = 0.689, fig. 5). The combined predictor of ZΔG and CAI performed approximately 1 percentage point better than CAI alone (AUC = 0.699). Note that most of the improvement was obtained in the region of interest, at low false-positive rates (fig. 5). In summary, these results recapitulated the earlier logistic-regression models: CAI by itself is the best individual predictor of gene functionality but ZΔG by itself also has significant predictive power. In combination, ZΔG and CAI perform slightly better than CAI alone.
FIG. 5.—
ROC curves for gene-functionality prediction. We trained logistic regression models on the Escherichia coli core genome plus pseudogenes and tested the predictors on genes outside the core genome (excluding pseudogenes). We considered genes of known function and conserved genes as functional and genes of unknown function as nonfunctional. The solid line corresponds to a model with ZΔG and CAI as predictors. The AUC is 0.699. The dashed line corresponds to a model where CAI is the only predictor (AUC = 0.689). The dotted line corresponds to the model where ZΔG is the only predictor (AUC = 0.594). The dot-dashed line corresponds to a model where expression level is the only predictor (AUC = 0.540).
Discussion
We have compared the level of mRNA secondary-structure stability near the start codon for genes with different functional annotations. In E. coli, we found that two broad classes (genes with known function and genes with known orthologs in other species) had similar levels of reduced mRNA secondary-structure stability. There was no evidence that the remaining genes of unknown function were under selection for reduced mRNA stability. Indeed, their ZΔG scores were similar to those of pseudogenes, suggesting that many of the remaining unannotated ORFs are nonfunctional.
We then extended our analysis to include 144 other prokaryote genomes. We found that genes with a known function have generally higher ZΔG than genes with no predicted function. Thus, there seems to be a general trend in prokaryotes that lower ZΔG indicates reduced probability of gene functionality. However, since few organisms have been studied as extensively as E. coli, we expect that many of the unknown genes in other organisms will ultimately turn out to be functional. In fact, in 2002 nearly one-third of E. coli ORFs lacked functional annotation or orthology in other genomes (Jackson et al. 2002). As of 2010, only 5% of ORFs remain unidentified at any level. Likewise, although our analysis suggests that in E. coli the majority of these 5% of ORFs are nonfunctional, we cannot exclude the possibility that some of the genes that we currently classify as being of unknown function will eventually be found to have a specific function as well. For this reason, our analyses both of E. coli and of other prokaryotes are possibly biased, since we may have included functional genes in the nonfunctional category. However, this bias can only weaken our conclusions, making our study conservative.
Our finding that genes with unknown function generally have lower ZΔG values suggests that ZΔG may be a useful diagnostic to target ORFs with an unknown function that are likely to be functionally important. Thus, researchers interested in understanding which novel genes in a genome are functionally important might begin by selecting genes with high ZΔG scores. However, one possible problem with using ZΔG as a tool for choosing genes for further study is that individually it is a noisy statistic. Thus, while most genomes overall show reduced mRNA secondary structure stability, there are many genes (including ones with a known and important function) that have increased stability compared with null expectations. Indeed, several genes of known function had extremely high levels of mRNA secondary-structure stability (more than 3 standard deviations below null expectations). It is unclear whether these ZΔG values indicate selection for increased mRNA stability or are merely a by-product of a noisy statistic.
To assess the possibility of ZΔG as a predictor of function, we fit logistic regression models that used ZΔG alone or in conjunction with expression and CAI. We found that ZΔG alone had moderate predictive power and CAI had substantial predictive power. Expression level (as measured by mRNA abundance) performed poorly as predictor. A model that combined ZΔG and CAI performed slightly better than the model using just CAI. These results show that ZΔG is a useful predictor of gene functionality and that it provides some information not captured by CAI.
It is not surprising that CAI would be useful to predict gene functionality. After all, if a gene is functional it needs to be translated efficiently, whereas if the gene is not functional then the organism will likely benefit if translation of that gene’s transcripts is inhibited. It was more surprising that mRNA abundance was not useful at all to predict gene functionality. This finding seems to indicate that in E. coli, a substantial portion of expression regulation occurs at the translation stage, via translation initiation and/or translation efficiency, rather than at the transcription stage. It is not clear why CAI was a better predictor than ZΔG. One possibility is that CAI is simply a more precise estimator, since it averages over all codons in a transcript, whereas ZΔG is calculated from the first 10–15 codons only. Alternatively, gene-wide codon usage may be more important for overall translation efficiency than mRNA stability near the initiation site is, as argued by Welch et al. (2009).
Several recent experimental studies have shown that synonymous mutations can have dramatic effects on phenotype. Two studies found that the function of a protein can be altered due to differences at synonymous sites (Kimchi-Sarfaty et al. 2007; Zhang et al. 2010). Other studies have demonstrated that the expression level of a protein can also be affected by synonymous mutations (Kudla et al. 2009; Welch et al. 2009; Allert et al. 2010). Yet another experimental study demonstrated that the fitness of a bacterium can be altered via synonymous mutations in ribosomal proteins (Lind et al. 2010). Changes in codon usage and changes in the mRNA secondary structure are two mechanistic hypotheses that can potentially explain these experimental results. Indeed, both factors seem to contribute to these experimental findings. The synonymous mutation underlying functional differentiation in the Kimchi-Sarfaty et al. (2007) study results in a change of a frequently used codon to a rarely used codon. Kudla et al. (2009) found that mRNA stability at the beginning of genes was the primary determinant of protein expression, not codon usage; others argue that the gene constructs used exhibit more secondary structure than generally found in organisms, which may have obscured the effect of codon usage (Tuller et al. 2010). Allert et al. (2010) found that both mRNA secondary structure and codon usage were important, though secondary structure had a larger effect. Finally, Lind et al. (2010) and Zhang et al. (2010) suggested that their results were likely due to changes in mRNA secondary structure rather than codon usage.
In the age of genomics, it will become increasingly common to analyze signatures of selection over a large number of genomes (as we did here). For such analyses, we need powerful statistical tools that enable us to fit complex models while properly controlling for phylogeny and other extraneous variables. Phylogenetic mixed models (Lynch 1991) are an appropriate tool for many such analyses. However, they have been used infrequently (Housworth et al. 2004; Naya et al. 2006), likely because they were difficult to implement. The release of the R package MCMCglmm removes much of the technical obstacles to carry out such analyses (Hadfield 2010; Hadfield and Nakagawa 2010). We hope that it will lead to a more wide-spread utilization of phylogenetic mixed models in future comparative genomics studies.
Supplementary Material
Supplementary tables 1 and 2 are available at Genome Biology and Evolution online (http://gbe.oxfordjournals.org/).
Acknowledgments
We thank members of the Bull and Hillis labs for helpful comments on the manuscript. This work was supported in part by the National Institutes of Health grant R01 GM088344 and by the National Science Foundation under Cooperative Agreement No. DBI-0939454.
References
- Allert M, Cox JC, Hellinga HW. Multifactorial determinants of protein expression in prokaryotic open reading frames. J Mol Biol. 2010;402:905–918. doi: 10.1016/j.jmb.2010.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behnel S, et al. Cython: the best of both worlds. Comput Sci Eng. 2011;13:31–39. [Google Scholar]
- Blattner FR, et al. The complete genome sequence of Escherichia coli K-12. Science. 1997;277:1453–1474. doi: 10.1126/science.277.5331.1453. [DOI] [PubMed] [Google Scholar]
- Cannone JJ, et al. The comparative RNA web (CRW) site: an online database of comparative sequence and structure information for ribosomal, intron, and other RNAs. BMC Bioinformatics. 2002;3:2. doi: 10.1186/1471-2105-3-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chamary JV, Parmley JL, Hurst LD. Hearing silence: non-neutral evolution at synonymous sites in mammals. Nat Rev Genet. 2006;7:98–108. doi: 10.1038/nrg1770. [DOI] [PubMed] [Google Scholar]
- Cock PJA, et al. Biopython: freely available Python tools for computational molecular biology and bioinformatics. Bioinformatics. 2010;6:1422–1423. doi: 10.1093/bioinformatics/btp163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felsenstein J. Phylogenies and the comparative method. Am Nat. 1985;125:1–15. [Google Scholar]
- Felsenstein J. Inferring phylogenies. Sunderland (MA): Sinauer Associates; 2003. [Google Scholar]
- Geweke J. Bayesian statistics 4. London: Oxford University Press; 1992. Evaluating the accuracy of sampling-based approaches to the calculation of posterior moments; pp. 169–193. [Google Scholar]
- Gu W, Zhou T, Wilke CO. A universal trend of reduced mRNA stability near the translation-imitation site in prokaryotes and eukaryotes. PLoS Comput Biol. 2010;6:e1000664. doi: 10.1371/journal.pcbi.1000664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hadfield JD. MCMC methods for multi-response generalized linear mixed models: the MCMCglmm R package. J Stat Softw. 2010;33:2. [Google Scholar]
- Hadfield JD, Nakagawa S. General quantitative genetic methods for comparative biology: phylogenies, taxonomies and multi-trait models for continuous and categorical characters. J Evol Biol. 2010;23:454–508. doi: 10.1111/j.1420-9101.2009.01915.x. [DOI] [PubMed] [Google Scholar]
- Heidelberger P, Welch PD. Simulation run length control in the presence of an initial transient. Opns Res. 1983;31:1109–1144. [Google Scholar]
- Hofacker IL, et al. Fast folding and comparison of RNA secondary structures. Monatshefte f. Chemie. 1994;125:167–188. [Google Scholar]
- Housworth EA, Martins EP, Lynch M. The phylogenetic mixed model. Am Nat. 2004;163:84–96. doi: 10.1086/380570. [DOI] [PubMed] [Google Scholar]
- Jackson JH, Harrison SH, Herring PA. A theoretical limit to coding space in chromosomes of bacteria. Omics. 2002;6:115–141. doi: 10.1089/15362310252780861. [DOI] [PubMed] [Google Scholar]
- Kimchi-Sarfaty C, et al. A “silent” polymorphism in the MDR1 gene changes substrate specificity. Science. 2007;315:525–528. doi: 10.1126/science.1135308. [DOI] [PubMed] [Google Scholar]
- Kudla G, Murray AW, Tollervey D, Plotkin JB. Coding-sequence determinants of gene expression in Escherichia coli. Science. 2009;324:255–258. doi: 10.1126/science.1170160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lind PA, Berg OG, Andersson DI. Mutational robustness of ribosomal protein genes. Science. 2010;330:825–827. doi: 10.1126/science.1194617. [DOI] [PubMed] [Google Scholar]
- Lynch M. Methods for the analysis of comparative data in evolutionary biology. Evolution. 1991;45:1065–1080. doi: 10.1111/j.1558-5646.1991.tb04375.x. [DOI] [PubMed] [Google Scholar]
- Naya H, Gianola D, Romero H, Urioste JI, Musto H. Inferring parameters shaping amino acid usage in prokaryotic genomes using Bayesian MCMC methods. Mol Biol Evol. 2006;23:203–211. doi: 10.1093/molbev/msj023. [DOI] [PubMed] [Google Scholar]
- Paradis E, Claude J, Strimmer K. APE: analyses of phylogenetics and evolution in R language. Bioinformatics. 2004;20:289–290. doi: 10.1093/bioinformatics/btg412. [DOI] [PubMed] [Google Scholar]
- Plotkin JB, Kudla G. Synonymous but not the same: the causes and consequences of codon bias. Nat Rev Genet. 2011;12:32–42. doi: 10.1038/nrg2899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plummer M, Best N, Cowles K, Vines K. Convergence diagnosis and output analysis for MCMC. R News. 2006;6:7–11. [Google Scholar]
- R Development Core Team. R: a language and environment for statistical computing. Vienna (Austria): R Foundation for Statistical Computing; 2010. [Google Scholar]
- Ragavan R, Groisman EA, Ochman H. Genome-wide detection of novel regulatory RNAs in E coli. Genome Res. Forthcoming 2011 doi: 10.1101/gr.119370.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanderson MJ. Estimating absolute rates of molecular evolution and divergence times: a penalized likelihood approach. Mol Biol Evol. 2002;19:101–109. doi: 10.1093/oxfordjournals.molbev.a003974. [DOI] [PubMed] [Google Scholar]
- Sauna ZE, Kimchi-Sarfaty C. Understanding the contribution of synonymous mutations to human disease. Nat Rev Genet. 2011;12:683–691. doi: 10.1038/nrg3051. [DOI] [PubMed] [Google Scholar]
- Sharp P, Li W. The codon adaptation index—a measure of directional synonymous codon usage bias, and its potential applications. Nucleic Acids Res. 1987;15:1281–1295. doi: 10.1093/nar/15.3.1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shine J, Dalgarno L. Determinant of cistron specificity in bacterial ribosomes. Nature. 1975;254:34–38. doi: 10.1038/254034a0. [DOI] [PubMed] [Google Scholar]
- Stamatakis A. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics. 2006;22:2688–2690. doi: 10.1093/bioinformatics/btl446. [DOI] [PubMed] [Google Scholar]
- Touchon M, et al. Organised genome dynamics in the Escherichia coli species results in highly diverse adaptive paths. PLoS Genetics. 2009;5:e1000344. doi: 10.1371/journal.pgen.1000344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuller T, Waldman YY, Kupiec M, Ruppin E. Translation efficiency is determined by both codon bias and folding energy. Proc Natl Acad Sci U S A. 2010;107:3645–3650. doi: 10.1073/pnas.0909910107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welch M, et al. Design parameters to control synthetic gene expression in Escherichia coli. PLoS One. 2009;4:e7002. doi: 10.1371/journal.pone.0007002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang F, Saha S, Shabalina S, Kashina A. Differential arginylation of actin isoforms is regulated by coding sequence-dependent degradation. Science. 2010;329:1534–1537. doi: 10.1126/science.1191701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou T, Wilke CO. Reduced stability of mRNA secondary structure near the translation-initiation site in dsDNA viruses. BMC Evol Biol. 2011;11:59. doi: 10.1186/1471-2148-11-59. [DOI] [PMC free article] [PubMed] [Google Scholar]