Abstract
During the past two decades, numerous disease-causing genes for different cardiomyopathies have been identified. These discoveries have led to better understanding of disease pathogenesis and initial steps in the application of mutation analysis in the evaluation of affected individuals and their family members. As knowledge of the genetic abnormalities, and insight into cellular and organ biology has grown, so has appreciation of the level of complexity of interaction between genotype and phenotype across disease states. What were initially thought to be one-to-one gene-disease correlates have turned out to display important relational plasticity dependent in large part on the genetic and environmental backgrounds into which the genes of interest express. The current state of knowledge with regard to genetics of cardiomyopathy represents a starting point to address the biology of disease, but is not yet developed sufficiently to supplant clinically based classification systems or, in most cases, to guide therapy to any significant extent. Future work will of necessity be directed towards elucidation of the biological mechanisms of both rare and common gene variants and environmental determinants of plasticity in the genotype–phenotype relationship with the ultimate goal of furthering our ability to identify, diagnose, risk stratify, and treat this group of disorders which cause heart failure and sudden death in the young.
Keywords: Cardiomyopathy, Genetics
Introduction
Cardiomyopathies are a clinically heterogeneous group of heart muscle disorders. They are defined by the presence of abnormal myocardial structure and/or function in the absence of ischaemic heart disease or abnormal loading conditions. The current classifications of the cardiomyopathies continue to be based on phenotype defined by clinical evaluation of affected individuals, incorporating genotype when possible.
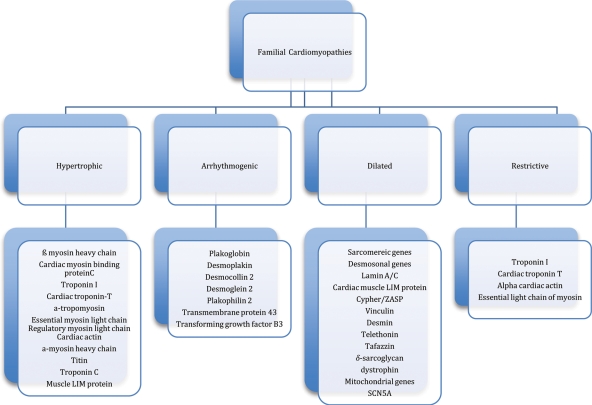
While differences exist in the classification schema of major cardiac organizations (e.g. pure channelopathies are included in the American classification system), the cardiomyopathies have historically broken down into several major phenotypic categories: hypertrophic, dilated, arrhythmogenic, and restrictive. (Figure 1).1,2 Overlap of the phenotypes is common, i.e. the patient with hypertrophic cardiomyopathy (HCM) may have restrictive physiology. With disease evolution, there also may be progression from one phenotype to another. Since the identification of the first cardiac disease-causing mutation, a point mutation in the β-myosin heavy chain gene in a French Canadian family with HCM in 1990,3,4 in excess of 600 rare genetic variants associated with cardiomyopathic disease have been recognized.5,6 Variable penetrance with incomplete expression is common in the autosomal dominant forms of cardiomyopathy, even among related individuals carrying an identical gene mutation. This combination of phenotypic and mutational heterogeneity contributes importantly to the challenges in diagnosis and prognostication, and the complexity of treatment. Additionally, genetic diagnosis represents a ‘moving target’ with new data leading to reevaluation of mutation pathogenicity. Finally, modifier gene and environmental effects are increasingly appreciated as key components of this genotype–phenotype plasticity.
Figure 1.
Classification of familial cardiomyopathies (adapted from Elliott et al.1).
The current body of knowledge on genetics of cardiomyopathy suggests a basis for understanding the pathophysiology of disease, provides potential targets for therapeutic intervention, contributes to diagnosis, allows for cascade screening, and occasionally informs prognosis. Used appropriately, genetic testing can provide important additional information for patients and their families.
This review will focus on the genetic basis of the cardiomyopathies, models of pathogenesis associated with known mutations, and the utility and yield of genetic testing. Specifically, we aim to highlight the current state-of-the-art understanding of the genetics of cardiomyopathy as it impacts the clinical presentation of these diseases. It is necessarily limited in scope but will highlight the key strengths and limitations of our current understanding of the genetics of cardiomyopathy and the determinants of genotype–phenotype plasticity (Table 1).
Table 1.
Genetic terms
| Proband: The first individual in a family who presents with clinical disease, sometimes referred to as the index case |
| Phenotype: The observable characteristics of an individual |
| Genotype: The genetic make-up of an individual |
| Mutation: A pathogenic gene variant. Mutations have a tendency to be rare, to occur in conserved or functionally important regions of the gene, to segregate with observable disease, and to occur where there is biological plausibility that the involved gene could lead to the observed phenotype |
| Modifier: Gene variants or environmental factors that are insufficient to cause observable disease on their own, but which are capable of interacting with the disease gene to alter the phenotype |
| Heterozygote: An individual who carries a single copy of a mutation |
| Homozygote: An individual who carries two copies of a mutation |
| Age-dependent expression: The tendency to develop more observable or severe phenotypes with advancing age |
| Variable penetrance: Variability in the proportion of genotypically identical individuals who express the disease phenotype |
| Variable expression: Variability in observable characteristics among carriers of an identical mutation |
| Phenotypic heterogeneity: Phenotypic variability among individuals with similar genotypes |
| Genotypic heterogeneity: Genetic variability among individuals with similar phenotypes |
| Genotype–phenotype plasticity: The concept that the link between genotype and phenotype is subject to broad variability with as yet limited predictability |
| Phenocopy: A phenotype that mimics the disease phenotype, but having a different aetiology (genetic or environmental), clinical course, and/or systemic features |
Hypertrophic cardiomyopathy
Hypertrophic cardiomyopathy is characterized by myocardial hypertrophy in the absence of clinically important abnormal loading conditions or primary valve disease. Diagnosis relies on electrocardiographic (ECG) evidence of left ventricular hypertrophy (LVH) confirmed by two-dimensional echocardiography or magnetic resonance imaging. Histological abnormalities are typified by myocyte disarray, interstitial and replacement fibrosis, and medial hypertrophy of intramyocardial small vessels.7,8 Clinical concerns in patients with HCM include development of dynamic obstruction of the left ventricular outflow tract (LVOT), atrial and ventricular arrhythmias complicated by either stroke or sudden cardiac death (SCD), and progression to diastolic and/or systolic heart failure (HF). Familial forms of HCM typically exhibit an autosomal dominant inheritance pattern, with sporadic disease and phenocopies accounting for a majority of cases of non-sarcomeric disease.
Familial HCM is characterized by marked phenotypic heterogeneity, age-dependent penetrance, and variable expression. Negligible to extreme hypertrophy, with varying degrees of fibrosis and outflow tract obstruction, can be seen within a single family. The population prevalence of clinically identifiable HCM is estimated to be 1:500 but this probably underestimates the prevalence of genetic disease.9–13 While the clinical course is relatively benign for a majority of those with HCM, the risk of sudden death is ∼1% per year for adults. A significant minority (2–5%) may develop progressive HF from a combination of pump failure and/or restrictive physiology, which can be further complicated by atrial arrhythmias and stroke.14,15
Left ventricular outflow tract obstruction is present at rest in ∼35%, may develop during exercise in another subset, and provides an important target for treatment in some symptomatic patients.16–18 The mechanisms of obstruction relate to septal thickness, LVOT dimensions, and mitral valve/papillary muscle anatomy. None of these features has a direct genetic correlate.19,20 Covariant and modifier genes not yet identified may play a role.
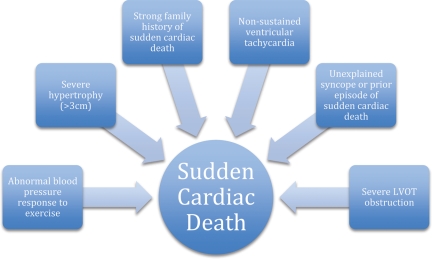
Sudden cardiac death is a primary clinical concern in the care of patients with HCM. While the absolute risk of SCD in all comers is low, it remains an important cause of SCD in the young and otherwise healthy.21 Risk factors predictive of SCD risk are relatively well established with increasing levels of risk determined by the number and severity of factors22 (Figure 2). Familial evaluation in HCM may serve to identify affected individuals who, though asymptomatic, are at risk for SCD. Risk assessment and consideration of lifestyle modification with or without an implantable cardiac defibrillator (ICD) underpins the rationale for family screening and ongoing clinical monitoring of affected individuals.10 Recommendations for lifestyle modification, such as avoidance of competitive sports, depend largely on phenotype but in specific cases (see below) may be impacted by genotype even in phenotype-negative individuals. Because of the complexity of the genotype and phenotype relationship, accurate prognostication based on genetic data is currently elusive, but remains a future goal.
Figure 2.
Risk factors for sudden cardiac death in hypertrophic cardiomyopathy.
Genetics of hypertrophic cardiomyopathy
Since the original genetic discovery, in excess of 400 mutations in nine genes encoding functionally important proteins of the cardiac sarcomere have been identified in association with HCM accounting for the majority of mutations found in clinical cohorts with this disease.4 The distribution of pathogenic sarcomeric mutations is uneven with myosin heavy chain 7 (MYH7) and myosin-binding protein C3 (MYPBC3) making up ∼25% each, and cardiac troponin T (TNNT2), cardiac troponin I (TNNI), myosin ventricular regulatory light chain (MYL2), and myosin ventricular essential light chain (MYL3) accounting for most of the remainder.23 Attempts to look beyond the sarcomeric proteins for genetic causes of HCM are driven by the recognition that 30–40% of the patients with clinical HCM do not have sarcomere mutations. Interest in a growing number of genes potentially associated with HCM broadly characterized as Z-disc/sarcomere genes and genes of calcium handling has emerged.24 The prevalence and pathogenicity of these mutations in studied populations awaits confirmation. It is possible, even likely, that some of these genes serve mainly as modifiers of primary mutations.
Five to 10% of the patients identified in cohort screenings carry multiple sarcomeric mutations and a dose–effect relationship occurs, with compound heterozygotes tending to present with more severe disease at an earlier age.23,25,26 As a result, more extensive genetic evaluation may be warranted in probands presenting with early or severe disease, especially in cases where the severity of disease significantly exceeds that seen in other family members. This speaks particularly strongly to the importance of family assessment, as most patients with multiple mutations will have inherited disease from both maternal and paternal lineages.
Although effort has gone into relating genotype and phenotype with the aim of supplementing clinically based diagnostic and risk factor assessment algorithms, at present, the data sets on specific mutations are not significant enough to be a major clinical guide. Specific mutations in MYH7 (Arg403Gln, Arg453Cys, and Arg719Trp) appear convincingly associated with adverse outcomes; however, data suggests that at-risk patients carrying these mutations also display clinical risk factors at the time of events limiting the added prognostic benefit of genetic diagnosis.27,28 An exception to this is HCM caused by mutations in cardiac troponin T which may cause ventricular arrhythmias and SCD in the absence of impressive morphological (LVH) or haemodynamic features (obstruction, diastolic dysfunction).29 In this context, knowledge that disease is caused by a TNNT mutation may influence management with a lower threshold for prophylactic ICD implantation.
The pathogenesis of HCM associated mutations is incompletely worked out but important pathogenic mechanisms have been elucidated. During normal contraction, calcium binding to α-tropomyosin and the troponin complex leads to removal of inhibitory troponin I. This allows association of the myosin head (a component of the thick filaments) with actin (a component of the thin filaments) and ATP hydrolysis with consequent conformation change in the myosin neck and sliding of thin filaments in relation to thick filaments. Basic science investigations incorporating mutated MYH7 demonstrate increased force generation and more rapid actin–myosin cross-bridge sliding.30 Similarly, mouse models of α-cardiac myosin and troponin T mutations demonstrate altered energy dynamics with increased force generation and abnormal relaxation properties.31,32 Additionally, reduced energy efficiency in sarcomere force development has been demonstrated in transgenic rat models,33 while in humans, markers of altered cellular energy dynamics (phosphocreatine to ATP ratio) have been noted in ∼30% of HCM patients independent of the degree or presence of hypertrophy.34 The pathways linking altered energy dynamics at the sarcomeric level with development of gross hypertrophy remain incompletely elucidated but alterations in intracellular calcium handling and regulation of calcium-sensitive signalling proteins appear to play a role.35 Based on these and other observations,36 clinical investigation into the use of diltiazem for prevention of hypertrophy in genotype-positive, phenotype-negative individuals is ongoing.37
Phenocopies of HCM are seen in a sizeable minority of patients presenting with idiopathic hypertrophy. Protein deposition disease (amyloidosis) and glycogen storage diseases (systemic and isolated cardiac) are most common but vary in prevalence with the population being tested. Fabry's disease, Danon's disease, Pompe's disease, and Noonan's syndrome can all present with ventricular hypertrophy that may be difficult to distinguish from sarcomere-associated HCM on a clinical basis alone, though histopathological findings are usually distinct.38–42 Hypertrophy seen in the setting of true pre-excitation is associated with 5′-amp-activated protein kinase subunit γ-2 (PRKAG2) mutations leading to cardiac-specific glycogen storage disease.39 It is important to note that the hypertrophy seen in these metabolic diseases represents true hypertrophy of the cardiomyocyte where the proportion of increased cardiac mass attributable to glycogen deposition is minimal. Thus, ECGs of these patients typically demonstrate dramatically increased QRS voltage and major repolarization changes.
Utility of genetic testing in hypertrophic cardiomyopathy
A genetic diagnosis is obtainable in 60–70% of consecutive patients with familial HCM, though the yield is lower (∼30%) when sporadic disease is considered.23,43,44 As mentioned, troponin T mutations have been associated with SCD in the absence of traditional risk factors and identification of such a mutation should lead to consideration for early ICD placement. For other mutations, even those associated with severe disease, SCD rarely occurs in the absence of clinical risk factors making the added utility of genetic diagnosis in this group minimal. Those with special characteristics such as conduction disease, pre-excitation, or systemic disease may represent phenocopies of sarcomere-associated HCM and focused genetic testing may be helpful with diagnosis and treatment (e.g. Danon's disease, PRKAG2).
Although studies evaluating prognostic significance of mutational analysis in HCM have failed to demonstrate consistent outcomes, a family history of SCD remains an important risk factor, indicating that genetic background may yet prove helpful in risk assessment.24,29,45,46 Importantly, a large cohort study of unrelated patients with HCM demonstrated that a positive genetic diagnosis for any myofilament mutation was associated with quadruple the risk of adverse outcomes including cardiovascular death, stroke, and progression to advanced HF, in comparison with those who were found to be genotype-negative.47 However, because no specific action or change in therapy can be recommended for any individual patient based on this finding, the relevance of this observation to clinical gene testing is limited.
The potential for medical therapy to attenuate or prevent disease development in pre-clinical genotype-positive individuals has yet to be realized though early investigations in humans and animals demonstrate some promise.36,48,49 The greatest benefit of genetic testing in HCM derives from cascade screening with the ability to identify which individuals in a family are, or are not, at risk of disease development. Cascade screening is possible/feasible when a pathogenic mutation is identified in a proband and there are ‘at risk’ family members. Those carrying the mutation can be followed closely for the development of disease and counselled appropriately with regard to lifestyle, while those who do not carry the mutation may be released from follow-up (a cost-effective approach).50 For cascade screening to be safe and effective, it is critical that the pathogenicity of the mutation be certain. Because many mutations that cause HCM are private (unique to the individual family), careful family assessment demonstrating co-segregation of mutation with disease phenotype is advised.
Arrhythmogenic cardiomyopathy/arrhythmogenic right ventricular cardiomyopathy
The clinical spectrum of arrhythmogenic cardiomyopathy (ACM) is variable but is typified by electro-anatomical abnormalities, ventricular arrhythmia, and, in some cases, HF or sudden death.51–53 Disease phenotypes include the classic right-dominant form known as arrhythmogenic right ventricular cardiomyopathy (ARVC), as well as increasingly noted left-dominant and biventricular forms.54 While dysfunction of the right ventricles and/or LVs is common in later stages, the disease typically presents with ECG abnormalities and ventricular arrhythmias. Desmosomal gene mutations have been identified in association with all three subtypes leading to an appreciation for the broad spectrum of desmosomal gene expression. While familiar, the term ARVC no longer accurately reflects the breadth of phenotypes reported within this cardiomyopathy. Accordingly, the term ACM has been accepted by the HRS/EHRA Expert Consensus Statement on the State of Genetic Testing for Channelopathies and Cardiomyopathies (in press).
The clinical profile of ACM is outlined in Table 2. Updated diagnostic guidelines incorporating anatomic, histological, electrophysiological, arrhythmic, and genetic features have been proposed for arrhythmogenic ventricular cardiomyopathy (AVC).51 The updated classification system addresses the recognition that early or familial disease may be overlooked by previous criteria that were derived largely from index cases presenting with sustained ventricular arrhythmia or advanced structural disease (right ventricular failure and/or SCD).55 Importantly, early disease, manifested by subtle structural abnormalities and minor ventricular arrhythmias, may be associated with SCD, highlighting the importance of disease identification during this so-called ‘concealed’ phase.53,56
Table 2.
Clinical profile of arrhythmogenic ventricular cardiomyopathya (reproduced from Sen-Chowdhry et al.58)
| Diagnostic measure | Classic right dominant | Left dominant |
|---|---|---|
| 12-Lead ECG | Normal | |
| Poor R-wave progressionb | ||
| IVCD in V1–3 | Normal | |
| Prolongation of QRS duration in V1–3 | Leftward QRS axis (−30° < QRS axis < 0) or left-axis deviation (QRS axis < −30°) | |
| Incomplete RBBB | Early transition | |
| RBBB | LBBB | |
| Epsilon waves in V1–3 | Epsilon waves in inferior (II, III, aVF) and/or lateral leads (V5–V6 ± V4, I, aVL) | |
| Inverted/flat T-waves in V1–3, extending to V4–6 with LV involvement | Inverted/ flat T-waves in (infero)lateral leads, extending to V1–3 with RV involvement | |
| ST elevation in V1–3 | ||
| Signal-averaged ECG | Late potentials | |
| Arrhythmia | Both supraventricular tachycardia and atrial fibrillation/flutter are observed in arrhythmogenic cardiomyopathy but are not contributory to diagnosis | |
| Frequent PVCs of LBBB configuration | Frequent PVCs of RBBB configuration | |
| Sustained/non-sustained VT of LBBB configurationc | Sustained/non-sustained VT or RBBB configurationc | |
| Ventricular volumes | Normal | Normal |
| Mild, moderate, or severe RV dilation ± dysfunction | Mild, moderate, or severe LV dilation ± dysfunction | |
| RV/LV volume ratio | ≥1.2, increases with disease progression | <1, diminishes with disease progression |
| Other imaging abnormalities | Localized dilation, WMA, and/or aneurysms in RV, preferentially affecting triangle of dysplasia and mid-free wall | Localized dilation, WMA, and/or aneurysms in LV |
| Increased/abnormal trabeculation | Non-compacted appearance | |
| Fat/late enhancement in RV myocardium | Late enhancement in LV myocardium in a subepicardial/midwall distribution | |
IVCD, intraventricular conduction delay; LBBB, left bundle branch block; PVC, premature ventricular complex; RBBB, right bundle branch block; VT, ventricular tachycardia; WMA, wall motion abnormality.
aThe clinical picture in the biventricular subtype is generally a composite of right- and left-dominant features, with RV to LV volume ratio remaining ≈1 throughout the disease course.
bPoor R-wave progression is the primary ECG abnormality observed in the Newfoundland founder population, in which LV structural abnormalities are prominent, but the subtype of arrhythmogenic cardiomyopathy is still being elucidated. It has also been reported in ∼10% of patients in a cohort including all three subtypes of arrhythmogenic cardiomyopathy.
cNon-sustained VT is defined as three or more consecutive beats at a rate of >120 b.p.m.; sustained VT has a duration of >30 s.
The prevalence of ACM is thought to be ∼1:1000 though, as with the other cardiomyopathies, age-dependent penetrance and variable expression make the true prevalence difficult to ascertain.57 A minority of patients progress to clinically important ventricular dysfunction with the clinical hallmark of disease remaining ventricular arrhythmias.57–59 Inheritance patterns are mainly autosomal dominant but well-described rare recessive forms are observed and have been of significant import in the development of genetic and pathophysiological understanding in this disease.
Initial problems with the determination of the genetic basis of ACM were overcome by the recognition of recessive families with severe right-dominant disease and associated cutaneous manifestations (e.g. kinky hair, palmoplantar hyperkeratosis). Identification of a 2bp deletion in JUP-encoded plakoglobin60,61 in Naxos disease was followed by identification of a point mutation in DSP-encoded desmoplakin in the Carvajal syndrome.62,63 These discoveries provided the basis to speculate that AVC is a desmosomal disease. Using a candidate gene approach, rare variants in five genes involving structural components of the cardiac desmosome have been identified in association with autosomal dominant AVC: PKP2-encoded plakophilin-2,64 DSG2-encoded desmoglein,65 DSC2-encoded desmocollin,66 JUP-encoded plakoglobin, and DSP-encoded desmoplakin.67–70
Arrhythmic and structural abnormalities observed in AVC may be explained in part by disruption of force transmission, intercellular communication, and cell proliferation/differentiation. The mechanism behind these effects is increasingly appreciated to be dependent not only on primary abnormalities of the desmosome but also on the close association between desmosomes, gap junctions, and adherens junctions.71 Physiologically, important crosstalk exists between the cardiac desmosome, and gap and adherens junctions, with the resulting integrity of the intercalated disc and its important role in both mechanical and electrical cellular stability dependent on adequate functioning of all three subunits.72
Components of cardiac desmosomes also play an important role in direct cellular signaling and proliferation/differentiation. In the presence of cardiac-specific suppression of desmoplakin, plakoglobin undergoes nuclear translocation with consequent transdifferentiation of cardiomyocytes to adipocytes.58,73 Identification of fatty replacement on histopathological examination of cardiac tissue from affected patients may be explained by this pathway, or alternatively by cellular damage and necrosis accompanied by inflammation resulting in collagen deposition, fibrosis, and/or adipose formation.58,74 Finally, desmosomes play an important role in anchoring and function of ion channels potentially impacting cellular gradients and arrhythmogenesis.71
Non-desmosomal genes implicated in the development of AVC include transforming growth factor B3 (TGFB3) and transmembrane protein 43 (TMEM43).75–77 Mutations in TGFB3 regulatory domains have been identified in a single large cohort and one individual patient with a clinical diagnosis of AVC. Transforming growth factor B3-modulated desmosomal protein regulation is the presumed pathogenic pathway for this mutation.77 Mutation and linkage analysis have failed to yield further reports of TGFB3-associated AVC. A point mutation in TMEM43 identified in a Newfoundland founder population causes a fully penetrant form of AVC with age- and gender-dependent expression (male predominant), a high burden of SCD, and fibrofatty changes on histopathology.76 Transmembrane protein 43 is known to respond to proliferator-activated receptor-γ signaling, and dysregulation in this pathway may explain the fibrofatty changes seen on histopathology implicating a transdifferentiation pathogenesis. Mutations in RYR2 have been reported with the AVC histological phenotype.78 The pathophysiological basis for this association is unclear and the clinical phenotype of individuals carrying these mutations is more similar to catecholaminergic ventricular tachycardia (VT) than to AVC.
Genetic testing in arrhythmogenic cardiomyopathy
A number of factors limit the use of clinical genetic testing for ACM. A desmosomal variant will be found in around 50% of the patients who fulfil clinical diagnostic criteria, but interpretation may be problematic. Many of these will be single-nucleotide changes that may be found in up to 16% of healthy volunteers.79 Aside from well-described founder populations, private mutations are common and individually require determination of their pathogenicity as either sufficient to cause disease or sufficient to modify disease in those 10–15% of individuals with AVC who carry more than one variant.
While earlier presentation and more severe phenotypic characteristics have been noted in HCM patients who carry more than one variant, there appears to be a particularly strong relationship between multiple variant carrier status and disease severity in ACM. In a study of 100 families with ACM, more than one variant was present in 28.1% of probands but only 9.7% of relatives (P= 0.01) and the presence of more than one variant was associated with a nearly five-fold increase odds of penetrant disease. These data demonstrate the importance of multiple variants in clinically significant AVC and indicate that sequencing of all five desmosomal genes is required when genetic evaluation of an ACM proband and family is undertaken.80
Identification of a pathogenic mutation may enable both detection of pre-clinical disease during which the affected individual may still present with SCD and may also allow for discharge of unaffected relatives from follow-up. Therefore, in cases where clinical diagnosis in the proband is certain or highly probable, genetic testing is reasonable if cascade screening is feasible and desired. However, the data on genetic variants associated with ACM are insufficient to advocate testing in borderline or clinically uncertain cases. For these, clinical follow-up of the proband and his or her first-degree family members is recommended.
Dilated cardiomyopathy
Dilated cardiomyopathy (DCM) is characterized by LV enlargement (LVE) and systolic dysfunction. A variety of stressors can cause LV dysfunction including ischaemia, valve disease, hypertension, inflammatory disease, infections, therapeutic cytotoxic medications, and a variety of recreational and performance-enhancing drugs. When DCM occurs in the absence of an identifiable cause, the disease is referred to as idiopathic DCM (IDCM). Systematic non-invasive cardiac evaluation (to include ECG and echo) of first-degree relatives of probands with DCM will identify another affected relative in up to 50% of families.81–84 The prevalence of familial DCM (FDCM) by history alone is probably underestimated, as disease expression in family members of clinically apparent probands is often subclinical (mild LVE and minor ECG abnormalities). Accordingly, long-term serial evaluations suggest that DCM is an insidious, slowly progressive inflammatory disease that is familial in the majority of patients.81,85 In general, the clinical features of sporadic DCM and FDCM are indistinct with similar ventricular morphologies, non-specific ECG changes, and the development of clinical HF, atrial and ventricular arrhythmias, stroke, and sudden death seen in both populations. Identifiable phenotypic subsets include DCM with conduction disease often associated with LMNA-encoded lamin mutations and DCM associated with sarcomere mutations that may predispose to earlier disease onset and prominent ventricular arrhythmias.86–91
Since the identification of a mutation in cardiac actin on chromosome 15q14,92 more than 40 genes have been identified in association with non-syndromic FDCM, the majority demonstrating autosomal dominant inheritance.93 Autosomal recessive, X-linked, and mitochondrial patterns account for a minority of cases but represent important clinical subtypes of disease that are discussed in greater detail below.
Familial DCM demonstrates age-dependent penetrance with disease developing in childhood, adolescence, and middle age, but rarely in the elderly.94 Most patients are unaware of the diagnosis until HF symptoms or arrhythmia develops, or abnormalities are detected during routine evaluation. This highlights the important role for active family screening once a proband has been identified. Among the cardiomyopathies, FDCM stands out, in that nearly all disease-causing gene mutations are unique to that family (‘private’ mutations) though examples of variants identified in multiple families exist (TNNT Lys210del; LMNA 203 variants; RNA-binding motif 20—R636).90,95
Familial DCM demonstrates marked genetic heterogeneity with mutations identified in genes encoding sarcomeric proteins (β-myosin heavy chain, cardiac troponin T, troponin C, and troponin I), cardiac muscle LIM protein (CLP), cypher/ZASP (LBD3) and vinculin (VCL), desmoplakin (DSP), desmin (DES), telethonin (TCAP), tafazzin (G4.5), as well as those encoding proteins of the dystrophin-associated complex including δ-sarcoglycan (SGCD) and dystrophin (DMD).63,89,91,96–106 Disruption of sarcomere–cytoskeletal interactions, myocyte architecture, aggresomal amyloid deposition, desmosomal abnormalities, calcium handling, ion channel function, mitochondrial energy dynamics, and nuclear membrane-cytoskeletal integrity have been described. Because this profound genetic heterogeneity exists in the context of a common phenotype, a final common pathway for disease development has been proposed in which abnormal proteins adversely affect force transmission in the cardiomyocyte leading to cellular injury, inflammation, collagen deposition, remodelling, dilation, and systolic failure.41,107,108 However, this relatively clean approach to understanding the pathophysiology of FDCM does not allow for ready incorporation of several rather unique and increasingly well-described theories of pathogenesis outlined below.
Desmin is an intermediate filament that forms a scaffold around the Z-disc of the sarcomere and connects the Z-disc to the subsarcolemmal cytoskeleton.109 Mutations in desmin have been demonstrated to lead to pre-amyloid deposition in aggresomal bodies in transgenic mice designed to model desmin-associated cardiomyopathy.110 Interestingly, voluntary exercise reduces deposition of pathogenic aggresomal bodies and dramatically improved survival in this DCM model.111,112
Genes encoding desmosomal proteins, classically associated with AVC, have been recently recognized as a cause of DCM.54,90 The initial report on this topic documented a higher burden of ventricular arrhythmias in DCM patients carrying desmosomal gene mutations. Because this initial report was limited to two families, the familial phenotypic diversity of desmosomal-associated DCM requires further investigation. Isolated reports of AVC masquerading as DCM have emerged, indicating that considerable phenotypic overlap exists between DCM caused by desmosomal gene mutations, DCM caused by other genetic mechanisms, and AVC.113–115 Clinical presentation with arrhythmia and/or sudden death is more typical of disease caused by a mutation in a desmosomal gene or lamin A/C, where the LV phenotype may have only mild structural and/or functional abnormalities at the time of arrhythmic disease presentations.
Familial DCM with conduction disease secondary to disruption in the nuclear cytoskeleton by mutations in lamin A/C deserves special attention. Nuclear lamins A and C are highly conserved proteins critical in nuclear cytoskeletal integrity. Mutations in these proteins account for 5–8% of FDCM and are particularly notable for the heterogeneity of disease expression with which they are associated. Premature conduction disease, DCM with HF, mild DCM with prominent arrhythmias, partial lipodystrophy, Emery–Dreifuss muscular dystrophy, and Hutchinson–Gilford progeria syndrome are all seen in relation to LMNA mutations.116,117 Lamins A and C play important structural and regulatory roles in the nuclear cytosol. Structural abnormalities of myocyte nuclei associated with LMNA mutations have been observed consistent with the architectural role played by the lamins.118 Abnormally cleaved and processed LMNA-encoded proteins have been associated with diverse phenomena at the nuclear level.119 Reversibility of pathogenic end products and clinical phenotype has been observed in murine models of Hutchinson–Gilford progeria.120–122 The extent to which the underlying disease mechanisms and associated therapies will be applicable to LMNA-associated cardiomyopathy is unclear.
In those affected by LMNA-associated cardiomyopathy, conduction disease can precede development of DCM in some families while in other families DCM occurs first. The practical significance of this is that individuals who may have mild DCM caused by LMNA may be at risk of SCD, while this scenario is highly unlikely with most sarcomere and all cytoskeletal abnormalities. Therefore, when SCD is seen in a family with mild DCM, testing for LMNA mutations may be helpful and lead to early consideration for ICD therapy. Reports of increased arrhythmogenicity in SCN5A-associated123,124 and desmosomal-associated DCM indicate that a similar approach may be taken when these mutations are identified.54
Rare variants in sarcomere genes may be associated with either DCM or HCM depending on the effect of the mutation. Functional analyses of representative sarcomere mutations indicate that disorders of force transmission and of force generation can both lead to development of DCM. Divergent alterations in both calcium regulation (currents and concentration at the level of the sarcolemma) and in thin filament calcium-binding affinity appear to yield different phenotypes with reduced binding affinity favouring development of DCM.88,89,125,126
Dilated cardiomyopathy is also seen as a common component of the muscular dystrophies, including myotonic dystrophy, Friedreich's ataxia, myofibrillar myopathy, and several limb girdle muscular dystrophies. Mutations for these disease states are well characterized for the most part and, although outside the breadth of this review, may ultimately provide insight into the pathogenesis of IDCM.
Utility of genetic testing in dilated cardiomyopathy
The yield of genetic testing in FDCM is low, ∼30%. Given the genetic heterogeneity in DCM, the majority of mutations demonstrate extremely low prevalence necessitating the sequencing of large numbers of genes to enable effective genetic testing. Difficulties in interpreting the results of mutation analysis arise from the high prevalence of private mutations among individual families, and the need to individually assess the pathogenicity of previously unreported mutations that are deemed pathogenic based on structure–function models and evidence of inter-species conservation.
As noted, DCM with conduction disease and/or arrhythmia represents a special subset of FDCM in which focused testing for LMNA, desmosomal, and SCN5A mutations may have a substantial clinical impact. When there is a strong family history of important ventricular arrhythmias, heart block, or SCD, practitioners may consider recommending early prophylactic ICD implantation for genotype-positive relatives, even in the presence of mild or no phenotype.
Identification of a definitively pathogenic mutation in the setting of clinical disease allows for cascade screening which can be a relief for family members who test negative and can then be discharged from follow-up. Likewise, for family members who test positive, appropriate monitoring and interventions can be initiated to prevent disease progression and adverse events. Data supporting the prophylactic use of angiotensin-converting enzyme (ACE)-inhibitors in genotype-positive, phenotype-negative patients with Duchene's muscular dystrophy for prevention or delay of DCM development are promising,127 and the use of ACE-inhibitors in asymptomatic LV dysfunction is supported by similar data.128
Other cardiomyopathies
Restrictive cardiomyopathy (RCM) and LV non-compaction (LVNC) have been subclassified individually but evidence exists for considerable overlap between these syndromes and HCM and DCM. Familial RCM is increasingly recognized as a specific phenotype within the HCM spectrum and can be seen in those who share mutations expressed as classic hypertrophic cardiomyopathy in other family members.129,130 Similarly, LVNC is an imaging diagnosis with profound overlap with both DCM and HCM phenotypes and their disease-causing mutations.131
The prevalence of pure familial vs. sporadic RCM and LVNC (in the absence of HCM and/or DCM within the family) is not known. For LVNC, the definition of the clinical phenotype remains under debate and population prevalence varies widely depending on the cohort examined and the diagnostic criteria utilized.132,133 Likewise, the clinical course of LVNC remains unclear with some reporting a high incidence of adverse events and others reporting a relatively benign prognosis apparently with the use of the same diagnostic criteria.132,134,135 Perhaps in part because the clinical syndrome remains under debate, genotypes associated with LVNC span a wide spectrum (cytoskeletal to sarcomeric to ion channel encoding genes).99,136–140 Genetic testing for LVNC should be reserved for those with syndromic presentation and those with clear familial disease. Because of the overlap with DCM and HCM, active family assessment is essential in evaluation of these patients.
Familial RCM is the rarest of the primary myocardial diseases and is increasingly recognized as an inherited disease seen in association with sarcomere mutations.129,141–143 The population prevalence remains unknown. Cardinal clinical features include atrial enlargement with normal sized ventricles with a high burden of atrial arrhythmias, progression to advanced HF, and death either related to HF or ventricular arrhythmias.144 Some RCM-associated troponin I mutations alter troponin I inhibition of actin–myosin ATPase leading to increased calcium sensitivity at the actin–myosin bridge. Increased calcium sensitivity at this site promotes myocardial stiffness by altering sarcomere response to calcium homeostasis. This mechanism may therefore be important in the differential development of RCM vs. HCM phenotypes.130
Modifier genes and environmental effects
Modifier genes are defined by their effect on expression of primary mutations. The term implies a secondary role in disease development. Modifier genes fail to consistently co-segregate with disease, but when present can significantly alter the phenotypic expression of the primary mutation. Because modifier genes do not co-segregate and are not sufficient to cause disease independently, their identification is a cumbersome task. Nevertheless, because genotype–phenotype plasticity is increasingly appreciated, a growing body of literature has arisen identifying potential modifiers.145–160
The impact of diet, fitness, and psychological stress on outcomes in patients with cardiac disease are well documented.161–163 Among patients with HCM, psychological stress has been found to be a trigger of ventricular arrhythmic events.164 There are scant but intriguing data that mental stress may impact development of ventricular arrhythmias in AVC.165,166 Endurance training/sport in patients with AVC is thought to confer increased risk of progression to functional and anatomic abnormalities of the RV and increased risk of arrhythmic death. Risk of SCD in patients with AVC is 5.4-fold higher among competitive athletes than among non-athletes.58,167
Genotype–phenotype plasticity and variance component analysis
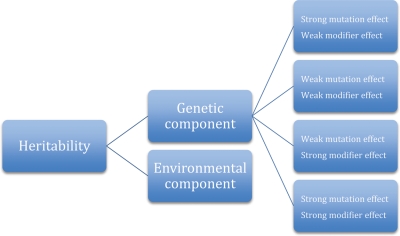
The degree of plasticity in the genotype–phenotype relationship defies explanation by identification of pathogenic rare variants alone. Influences on primary mutation expression include location and type of mutation, number of variants, modifier genes, and environmental factors. Variance component analysis (VCA) allows comparison of phenotypic variability within and between families carrying the same primary mutation. Segregation of the relative impact of genetic (primary and modifier mutations) and environmental factors may be assessed using this technique.157 In VCA, the proportion of phenotypic variance that can be attributed to summed genetic effects (primary mutation + modifier genes) is termed heritability. Environmental effects must perforce account for the remainder of phenotypic variability. Figure 3 expresses the relationship between these effects.
Figure 3.
Variance component analysis allows for breakdown of a disease state into its core components. Heritability quantifies the likelihood of familial transmission of a given trait. Transmission of a trait is dependent on both genetic and environmental components. Genetic components of transmission are determined by the variable effects of a primary mutation and associated modifier genes (modified from Sen-Chowdhry et al.157).
Variance component analysis of AVC suggests that modifier genes and environmental effects contribute significantly to phenotypic heterogeneity seen in family members carrying the same mutation, including susceptibility to arrhythmogenesis. Similar analyses have not been systematically undertaken in HCM or DCM cohorts, but the principle maps well onto these diseases and illustrates the complexity of the relationship between pathogenic rare variants and observed phenotype.
A brief note on the drawbacks of genetic testing
This review presents a conservative perspective on the utility of genetic testing. The disadvantages described in the literature mainly focus on the psychological impact of cascade screening, particularly among children and adolescents. Much of the data on the impact of genetic diagnosis and pre/post-test counselling, however, come from the non-cardiac literature.168,169 A genetic diagnosis leading to inappropriate device therapy and/or lifestyle restrictions are recognized clinical scenarios. In addition, the financial impact of the broad use of genetic testing is another important factor, though appropriate use of mutation analysis has been shown to be a cost-effective strategy, in that it can free up patients from unnecessary follow-up.50 In all cases, testing is most useful and least problematic when administered in the context of a multidisciplinary speciality clinic with expertise in the inherited cardiomyopathies.170
Conclusions
Clinical and genetic characterization of the inherited cardiomyopathies has lead to novel pathophysiological insights and a new real-time approach to genetic diagnosis. The complexity of genotype–phenotype interaction lends itself to careful clinical observation and judicious use of genetic testing. Caution with regard to application of genetic testing is warranted, in particular with regard to AVC and DCM as interpretation of genetic tests may be limited by phenotypic and genetic heterogeneity as well as prognostic utility. Ongoing efforts to expand our understanding of both pathogenesis of disease and the complex interplay between the factors involved in disease expression will offer continued opportunities for improved care.
Funding
D.J. is supported in part by NIH - 1R21 NR011387 (PI=Redeker) 8/13/09-7/31/11 Cognitive Behavioral Therapy in Stable Heart Failure Role: Co-Investigator. W.J.M.'s work undertaken at UCLH/UCL is supported by a proportion of funding from the Department of Health's NIHR Biomedical Research Centres funding scheme.
Conflict of interest: none declared.
References
- 1.Elliott P, Anderson B, Arbustini E, Bilinska Z, Cecchi F, Charron P, Dubourg O, Kuhl U, Maisch B, McKenna WJ, Monserrat L, Pankuweit S, Rapezzi C, Seferovic P, Tavazzi L, Keren A. Classification of cardiomyopathies: a position statement from the European working group on myocardial and pericardial diseases. Eur Heart J. 2008;29:270–276. doi: 10.1093/eurheartj/ehm342. [DOI] [PubMed] [Google Scholar]
- 2.Maron BJ, Towbin JA, Thiene G, Antzelevitch C, Corrado D, Arnett D, Moss AJ, Seidman CE, Young JB. Contemporary definitions and classification of the cardiomyopathies: an American Heart Association Scientific Statement from the Council on Clinical Cardiology, Heart Failure and Transplantation Committee; Quality of Care and Outcomes Research and Functional Genomics and Translational Biology Interdisciplinary Working Groups; and Council on Epidemiology and Prevention. Circulation. 2006;113:1807–1816. doi: 10.1161/CIRCULATIONAHA.106.174287. [DOI] [PubMed] [Google Scholar]
- 3.Jarcho JA, McKenna W, Pare JA, Solomon SD, Holcombe RF, Dickie S, Levi T, Donis-Keller H, Seidman JG, Seidman CE. Mapping a gene for familial hypertrophic cardiomyopathy to chromosome 14q1. N Engl J Med. 1989;321:1372–1378. doi: 10.1056/NEJM198911163212005. [DOI] [PubMed] [Google Scholar]
- 4.Geisterfer-Lowrance AA, Kass S, Tanigawa G, Vosberg HP, McKenna W, Seidman CE, Seidman JG. A molecular basis for familial hypertrophic cardiomyopathy: a beta cardiac myosin heavy chain gene missense mutation. Cell. 1990;62:999–1006. doi: 10.1016/0092-8674(90)90274-i. [DOI] [PubMed] [Google Scholar]
- 5.ARVC Database List of Pathogenic Variants. 2011. as of March 29.
- 6.Number of Sarcomere Gene Variants. Harvard Partners; Updated from 2006. [Google Scholar]
- 7.Maron BJ, McKenna WJ, Danielson GK, Kappenberger LJ, Kuhn HJ, Seidman CE, Shah PM, Spencer WH, III, Spirito P, Ten Cate FJ. American College of Cardiology/European Society of Cardiology clinical expert consensus document on hypertrophic cardiomyopathy: a report of the American College of Cardiology Foundation Task Force on Clinical Expert Consensus Documents and the European Society of Cardiology Committee for Practice Guidelines. J Am Coll Cardiol. 2003;42:1687–1713. doi: 10.1016/s0735-1097(03)00941-0. [DOI] [PubMed] [Google Scholar]
- 8.Varnava AM, Elliott PM, Sharma S, McKenna WJ, Davies MJ. Hypertrophic cardiomyopathy: the interrelation of disarray, fibrosis, and small vessel disease. Heart. 2000;84:476–482. doi: 10.1136/heart.84.5.476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pinamonti B, Di Lenarda A, Sinagra G, Camerini F. Restrictive left ventricular filling pattern in dilated cardiomyopathy assessed by Doppler echocardiography: clinical, echocardiographic and hemodynamic correlations and prognostic implications. Heart Muscle Disease Study Group. J Am Coll Cardiol. 1993;22:808–815. doi: 10.1016/0735-1097(93)90195-7. [DOI] [PubMed] [Google Scholar]
- 10.Elliott P, McKenna W. Hypertrophic cardiomyopathy. Lancet. 2004;363:1881–1891. doi: 10.1016/S0140-6736(04)16358-7. [DOI] [PubMed] [Google Scholar]
- 11.Marian AJ. Phenotypic Plasticity of Sarcomeric Protein Mutations. J Am Coll Cardiol. 2007;49:2427–2429. doi: 10.1016/j.jacc.2007.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Maron BJ, Peterson EE, Maron MS, Peterson JE. Prevalence of hypertrophic cardiomyopathy in an outpatient population referred for echocardiographic study. Am J Cardiol. 1994;73:577–580. doi: 10.1016/0002-9149(94)90337-9. [DOI] [PubMed] [Google Scholar]
- 13.Maron B, Gardin JM, Flack JM, Gidding SS, Kurosaki TT, Bild DE. Prevalence of hypertrophic cardiomyopathy in a general population of young adults. Echocardiographic analysis of 4111 subjects in the CARDIA Study. Coronary Artery Risk Development in (Young) Adults. Circulation. 1995;92:785–789. doi: 10.1161/01.cir.92.4.785. [DOI] [PubMed] [Google Scholar]
- 14.Spirito P, Maron BJ, Bonow RO, Epstein SE. Occurrence and significance of progressive left-ventricular wall thinning and relative cavity dilatation in hypertrophic cardiomyopathy. Am J Cardiol. 1987;60:123–129. doi: 10.1016/0002-9149(87)90998-2. [DOI] [PubMed] [Google Scholar]
- 15.Maron BJ, Harris KM, Spirito P, Maron MS, Zenovich AG, Formisano F, Lesser JR, Mackey-Bojack S, Manning WJ, Udelson JE. Prevalence, clinical profile, and significance of left ventricular remodeling in the end-stage phase of hypertrophic cardiomyopathy. Circulation. 2006;114:216–225. doi: 10.1161/CIRCULATIONAHA.105.583500. [DOI] [PubMed] [Google Scholar]
- 16.Geier C, Perrot A, Ozcelik C. Letter by Geier et al. regarding article, ‘Hypertrophic cardiomyopathy is predominantly a disease of left ventricular outflow tract obstruction’. Circulation. 2007;115:E622–E622. doi: 10.1161/CIRCULATIONAHA.107.690057. [DOI] [PubMed] [Google Scholar]
- 17.Maron MS, Link MS, Udelson JE, Kuvin JT, Pandian NG, Olivotto I, Nistri S, Cecchi F, Maron BJ. Response to letter regarding article, ‘Hypertrophic cardiomyopathy is predominantly a disease of left ventricular outflow tract obstruction’. Circulation. 2007;115:E623–E623. doi: 10.1161/CIRCULATIONAHA.106.644682. [DOI] [PubMed] [Google Scholar]
- 18.Maron BJ, Maron MS, Olivotto I, Zenovich AG, Link MS, Pandian NG, Kuvin JT, Nistri S, Cecchi F, Udelson JE. Hypertrophic cardiomyopathy is predominantly a disease of left ventricular outflow tract obstruction. Circulation. 2006;114:2232–2239. doi: 10.1161/CIRCULATIONAHA.106.644682. [DOI] [PubMed] [Google Scholar]
- 19.Levine RA, Vlahakes GJ, Lefebvre X, Guerrero JL, Cape EG, Yoganathan AP, Weyman AE. Papillary-muscle displacement causes systolic anterior motion of the mitral-valve—experimental validation and insights into the mechanism of subaortic obstruction. Circulation. 1995;91:1189–1195. doi: 10.1161/01.cir.91.4.1189. [DOI] [PubMed] [Google Scholar]
- 20.Henry WL, Clark CE, Griffith JM, Epstein SE. Mechanism of left-ventricular outflow obstruction in patients with obstructive asymmetric septal hypertrophy (idiopathic hypertrophic subaortic stenosis) Am J Cardiol. 1975;35:337–345. doi: 10.1016/0002-9149(75)90025-9. [DOI] [PubMed] [Google Scholar]
- 21.Maron BJ, Doerer JJ, Haas TS, Tierney DM, Mueller FO. Sudden deaths in young competitive athletes: analysis of 1866 deaths in the United States, 1980–2006. Circulation. 2009;119:1085–1092. doi: 10.1161/CIRCULATIONAHA.108.804617. [DOI] [PubMed] [Google Scholar]
- 22.Elliott PM, Poloniecki J, Dickie S, Sharma S, Monserrat L, Varnava A, Mahon NG, McKenna WJ. Sudden death in hypertrophic cardiomyopathy: identification of high risk patients. J Am Coll Cardiol. 2000;36:2212–2218. doi: 10.1016/s0735-1097(00)01003-2. [DOI] [PubMed] [Google Scholar]
- 23.Richard P. Hypertrophic cardiomyopathy: distribution of disease genes, spectrum of mutations, and implications for a molecular diagnosis strategy. Circulation. 2003;107:2227–2232. doi: 10.1161/01.CIR.0000066323.15244.54. [DOI] [PubMed] [Google Scholar]
- 24.Bos JM, Towbin JA, Ackerman MJ. Diagnostic, prognostic, and therapeutic implications of genetic testing for hypertrophic cardiomyopathy. J Am Coll Cardiol. 2009;54:201–211. doi: 10.1016/j.jacc.2009.02.075. [DOI] [PubMed] [Google Scholar]
- 25.Girolami F, Ho CY, Semsarian C, Baldi M, Will ML, Baldini K, Torricelli F, Yeates L, Cecchi F, Ackerman MJ, Olivotto I. Clinical features and outcome of hypertrophic cardiomyopathy associated with triple sarcomere protein gene mutations. J Am Coll Cardiol. 2010;55:1444–1453. doi: 10.1016/j.jacc.2009.11.062. [DOI] [PubMed] [Google Scholar]
- 26.Millat G, Bouvagnet P, Chevalier P, Dauphin C, Jouk PS, Da Costa A, Prieur F, Bresson JL, Faivre L, Eicher JC, Chassaing N, Crehalet H, Porcher R, Rodriguez-Lafrasse C, Rousson R. Prevalence and spectrum of mutations in a cohort of 192 unrelated patients with hypertrophic cardiomyopathy. Eur J Med Genet. 2010;53:261–267. doi: 10.1016/j.ejmg.2010.07.007. [DOI] [PubMed] [Google Scholar]
- 27.Watkins H, Rosenzweig A, Hwang DS, Levi T, McKenna W, Seidman CE, Seidman JG. Characteristics and prognostic implications of myosin missense mutations in familial hypertrophic cardiomyopathy. N Engl J Med. 1992;326:1108–1114. doi: 10.1056/NEJM199204233261703. [DOI] [PubMed] [Google Scholar]
- 28.Saltzman AJ, Mancini-DiNardo D, Li C, Chung WK, Ho CY, Hurst S, Wynn J, Care M, Hamilton RM, Seidman GW, Gorham J, McDonough B, Sparks E, Seidman JG, Seidman CE, Rehm HL. Short communication: the cardiac myosin binding protein C Arg502Trp mutation: a common cause of hypertrophic cardiomyopathy. Circ Res. 2010;106:1549–1552. doi: 10.1161/CIRCRESAHA.109.216291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Watkins H, McKenna WJ, Thierfelder L, Suk HJ, Anan R, O'Donoghue A, Spirito P, Matsumori A, Moravec CS, Seidman JG, Seidman CE. Mutations in the genes for cardiac troponin T and alpha-tropomyosin in hypertrophic cardiomyopathy. N Engl J Med. 1995;332:1058–1064. doi: 10.1056/NEJM199504203321603. [DOI] [PubMed] [Google Scholar]
- 30.Palmiter KA, Tyska MJ, Haeberle JR, Alpert NR, Fananapazir L, Warshaw DM. R403Q and L908V mutant beta-cardiac myosin from patients with familial hypertrophic cardiomyopathy exhibit enhanced mechanical performance at the single molecule level. J Muscle Res Cell Motil. 2000;21:609–620. doi: 10.1023/a:1005678905119. [DOI] [PubMed] [Google Scholar]
- 31.Georgakopoulos D, Christe ME, Giewat M, Seidman CM, Seidman JG, Kass DA. The pathogenesis of familial hypertrophic cardiomyopathy: early and evolving effects from an alpha-cardiac myosin heavy chain missense mutation. Nat Med. 1999;5:327–330. doi: 10.1038/6549. [DOI] [PubMed] [Google Scholar]
- 32.Harada K, Potter JD. Familial hypertrophic cardiomyopathy mutations from different functional regions of troponin T result in different effects on the pH and Ca2+ sensitivity of cardiac muscle contraction. J Biol Chem. 2004;279:14488–14495. doi: 10.1074/jbc.M309355200. [DOI] [PubMed] [Google Scholar]
- 33.Frey N, Brixius K, Schwinger RH, Benis T, Karpowski A, Lorenzen HP, Luedde M, Katus HA, Franz WM. Alterations of tension-dependent ATP utilization in a transgenic rat model of hypertrophic cardiomyopathy. J Biol Chem. 2006;281:29575–29582. doi: 10.1074/jbc.M507740200. [DOI] [PubMed] [Google Scholar]
- 34.Crilley JG, Boehm EA, Blair E, Rajagopalan B, Blamire AM, Styles P, McKenna WJ, Ostman-Smith I, Clarke K, Watkins H. Hypertrophic cardiomyopathy due to sarcomeric gene mutations is characterized by impaired energy metabolism irrespective of the degree of hypertrophy. J Am Coll Cardiol. 2003;41:1776–1782. doi: 10.1016/s0735-1097(02)03009-7. [DOI] [PubMed] [Google Scholar]
- 35.Bers DM. Calcium cycling and signaling in cardiac myocytes. Annu Rev Physiol. 2008;70:23–49. doi: 10.1146/annurev.physiol.70.113006.100455. [DOI] [PubMed] [Google Scholar]
- 36.Semsarian C, Ahmad I, Giewat M, Georgakopoulos D, Schmitt JP, McConnell BK, Reiken S, Mende U, Marks AR, Kass DA, Seidman CE, Seidman JG. The L-type calcium channel inhibitor diltiazem prevents cardiomyopathy in a mouse model. J Clin Invest. 2002;109:1013–1020. doi: 10.1172/JCI14677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang L, Seidman JG, Seidman CE. Narrative review: harnessing molecular genetics for the diagnosis and management of hypertrophic cardiomyopathy. Ann Intern Med. 2010;152:513–520. doi: 10.1059/0003-4819-152-8-201004200-00008. W181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Eishi Y, Takemura T, Sone R, Yamamura H, Narisawa K, Ichinohasama R, Tanaka M, Hatakeyama S. Glycogen storage disease confined to the heart with deficient activity of cardiac phosphorylase kinase: a new type of glycogen storage disease. Hum Pathol. 1985;16:193–197. doi: 10.1016/s0046-8177(85)80071-x. [DOI] [PubMed] [Google Scholar]
- 39.Arad M, Benson DW, Perez-Atayde AR, McKenna WJ, Sparks EA, Kanter RJ, McGarry K, Seidman JG, Seidman CE. Constitutively active AMP kinase mutations cause glycogen storage disease mimicking hypertrophic cardiomyopathy. J Clin Invest. 2002;109:357–362. doi: 10.1172/JCI14571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Arad M, Moskowitz IP, Patel VV, Ahmad F, Perez-Atayde AR, Sawyer DB, Walter M, Li GH, Burgon PG, Maguire CT, Stapleton D, Schmitt JP, Guo XX, Pizard A, Kupershmidt S, Roden DM, Berul CI, Seidman CE, Seidman JG. Transgenic mice overexpressing mutant PRKAG2 define the cause of Wolff–Parkinson–White syndrome in glycogen storage cardiomyopathy. Circulation. 2003;107:2850–2856. doi: 10.1161/01.CIR.0000075270.13497.2B. [DOI] [PubMed] [Google Scholar]
- 41.Ahmad F, Seidman JG, Seidman CE. The genetic basis for cardiac remodeling. Annu Rev Genomics Hum Genet. 2005;6:185–216. doi: 10.1146/annurev.genom.6.080604.162132. [DOI] [PubMed] [Google Scholar]
- 42.Elleder M, Bradova V, Smid F, Budesinsky M, Harzer K, Kustermann-Kuhn B, Ledvinova J, Belohlavek M, Kral V, Dorazilova V. Cardiocyte storage and hypertrophy as a sole manifestation of Fabry's disease. Report on a case simulating hypertrophic non-obstructive cardiomyopathy. Virchows Archiv A Pathol Anat Histopathol. 1990;417:449–455. doi: 10.1007/BF01606034. [DOI] [PubMed] [Google Scholar]
- 43.Van Driest SL, Ommen SR, Tajik AJ, Gersh BJ, Ackerman MJ. Yield of genetic testing in hypertrophic cardiomyopathy. Mayo Clin Proc. 2005;80:739–744. doi: 10.1016/S0025-6196(11)61527-9. [DOI] [PubMed] [Google Scholar]
- 44.Andersen PS, Havndrup O, Hougs L, Sorensen KM, Jensen M, Larsen LA, Hedley P, Thomsen AR, Moolman-Smook J, Christiansen M, Bundgaard H. Diagnostic yield, interpretation, and clinical utility of mutation screening of sarcomere encoding genes in Danish hypertrophic cardiomyopathy patients and relatives. Hum Mutat. 2009;30:363–370. doi: 10.1002/humu.20862. [DOI] [PubMed] [Google Scholar]
- 45.Niimura H, Bachinski LL, Sangwatanaroj S, Watkins H, Chudley AE, McKenna W, Kristinsson A, Roberts R, Sole M, Maron BJ, Seidman JG, Seidman CE. Mutations in the gene for cardiac myosin-binding protein C and late-onset familial hypertrophic cardiomyopathy. N Engl J Med. 1998;338:1248–1257. doi: 10.1056/NEJM199804303381802. [DOI] [PubMed] [Google Scholar]
- 46.Van Driest SL, Ackerman MJ, Ommen SR, Shakur R, Will ML, Nishimura RA, Tajik AJ, Gersh BJ. Prevalence and severity of ‘benign’ mutations in the beta-myosin heavy chain, cardiac troponin T, and alpha-tropomyosin genes in hypertrophic cardiomyopathy. Circulation. 2002;106:3085–3090. doi: 10.1161/01.cir.0000042675.59901.14. [DOI] [PubMed] [Google Scholar]
- 47.Olivotto I, Girolami F, Ackerman MJ, Nistri S, Bos JM, Zachara E, Ommen SR, Theis JL, Vaubel RA, Re F, Armentano C, Poggesi C, Torricelli F, Cecchi F. Myofilament protein gene mutation screening and outcome of patients with hypertrophic cardiomyopathy. Mayo Clin Proc. 2008;83:630–638. doi: 10.4065/83.6.630. [DOI] [PubMed] [Google Scholar]
- 48.Abozguia K, Elliott P, McKenna W, Phan TT, Nallur-Shivu G, Ahmed I, Maher AR, Kaur K, Taylor J, Henning A, Ashrafian H, Watkins H, Frenneaux M. Metabolic modulator perhexiline corrects energy deficiency and improves exercise capacity in symptomatic hypertrophic cardiomyopathy. Circulation. 2010;122:1562–1569. doi: 10.1161/CIRCULATIONAHA.109.934059. [DOI] [PubMed] [Google Scholar]
- 49.Penicka M, Gregor P, Kerekes R, Marek D, Curila K, Krupicka J. The effects of candesartan on left ventricular hypertrophy and function in nonobstructive hypertrophic cardiomyopathy: a pilot, randomized study. J Mol Diagn. 2009;11:35–41. doi: 10.2353/jmoldx.2009.080082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wordsworth S, Leal J, Blair E, Legood R, Thomson K, Seller A, Taylor J, Watkins H. DNA testing for hypertrophic cardiomyopathy: a cost-effectiveness model. Eur Heart J. 2010;31:926–935. doi: 10.1093/eurheartj/ehq067. [DOI] [PubMed] [Google Scholar]
- 51.Marcus FI, McKenna WJ, Sherrill D, Basso C, Bauce B, Bluemke DA, Calkins H, Corrado D, Cox MG, Daubert JP, Fontaine G, Gear K, Hauer R, Nava A, Picard MH, Protonotarios N, Saffitz JE, Sanborn DM, Steinberg JS, Tandri H, Thiene G, Towbin JA, Tsatsopoulou A, Wichter T, Zareba W. Diagnosis of arrhythmogenic right ventricular cardiomyopathy/dysplasia: proposed modification of the Task Force Criteria. Eur Heart J. 2010;31:806–814. doi: 10.1093/eurheartj/ehq025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Basso C, Corrado D, Thiene G. Cardiovascular causes of sudden death in young individuals including athletes. Cardiol Rev. 1999;7:127–135. doi: 10.1097/00045415-199905000-00009. [DOI] [PubMed] [Google Scholar]
- 53.Tabib A, Loire R, Chalabreysse L, Meyronnet D, Miras A, Malicier D, Thivolet F, Chevalier P, Bouvagnet P. Circumstances of death and gross and microscopic observations in a series of 200 cases of sudden death associated with arrhythmogenic right ventricular cardiomyopathy and/or dysplasia. Circulation. 2003;108:3000–3005. doi: 10.1161/01.CIR.0000108396.65446.21. [DOI] [PubMed] [Google Scholar]
- 54.Elliott P, O'Mahony C, Syrris P, Evans A, Rivera Sorensen C, Sheppard MN, Carr-White G, Pantazis A, McKenna WJ. Prevalence of desmosomal protein gene mutations in patients with dilated cardiomyopathy. Circ Cardiovasc Genet. 2010;3:314–322. doi: 10.1161/CIRCGENETICS.110.937805. [DOI] [PubMed] [Google Scholar]
- 55.Hamid MS, Norman M, Quraishi A, Firoozi S, Thaman R, Gimeno JR, Sachdev B, Rowland E, Elliott PM, McKenna WJ. Prospective evaluation of relatives for familial arrhythmogenic right ventricular cardiomyopathy/dysplasia reveals a need to broaden diagnostic criteria. J Am Coll Cardiol. 2002;40:1445–1450. doi: 10.1016/s0735-1097(02)02307-0. [DOI] [PubMed] [Google Scholar]
- 56.Sen-Chowdhry S, Syrris P, Ward D, Asimaki A, Sevdalis E, McKenna WJ. Clinical and genetic characterization of families with arrhythmogenic right ventricular dysplasia/cardiomyopathy provides novel insights into patterns of disease expression. Circulation. 2007;115:1710–1720. doi: 10.1161/CIRCULATIONAHA.106.660241. [DOI] [PubMed] [Google Scholar]
- 57.Peters S, Trummel M, Meyners W. Prevalence of right ventricular dysplasia-cardiomyopathy in a non-referral hospital. Int J Cardiol. 2004;97:499–501. doi: 10.1016/j.ijcard.2003.10.037. [DOI] [PubMed] [Google Scholar]
- 58.Sen-Chowdhry S, Morgan RD, Chambers JC, McKenna WJ. Arrhythmogenic cardiomyopathy: etiology, diagnosis, and treatment. Annu Rev Med. 2010;61:233–253. doi: 10.1146/annurev.med.052208.130419. [DOI] [PubMed] [Google Scholar]
- 59.Dalal D, Nasir K, Bomma C, Prakasa K, Tandri H, Piccini J, Roguin A, Tichnell C, James C, Russell SD, Judge DP, Abraham T, Spevak PJ, Bluemke DA, Calkins H. Arrhythmogenic right ventricular dysplasia: a United States experience. Circulation. 2005;112:3823–3832. doi: 10.1161/CIRCULATIONAHA.105.542266. [DOI] [PubMed] [Google Scholar]
- 60.Sen-Chowdhry S, Syrris P, McKenna WJ. Genetics of right ventricular cardiomyopathy. J Cardiovasc Electrophysiol. 2005;16:927–935. doi: 10.1111/j.1540-8167.2005.40842.x. [DOI] [PubMed] [Google Scholar]
- 61.McKoy G, Protonotarios N, Crosby A, Tsatsopoulou A, Anastasakis A, Coonar A, Norman M, Baboonian C, Jeffery S, McKenna WJ. Identification of a deletion in plakoglobin in arrhythmogenic right ventricular cardiomyopathy with palmoplantar keratoderma and woolly hair (Naxos disease) Lancet. 2000;355:2119–2124. doi: 10.1016/S0140-6736(00)02379-5. [DOI] [PubMed] [Google Scholar]
- 62.Protonotarios N, Tsatsopoulou A. Naxos disease and Carvajal syndrome: cardiocutaneous disorders that highlight the pathogenesis and broaden the spectrum of arrhythmogenic right ventricular cardiomyopathy. Cardiovasc Pathol. 2004;13:185–194. doi: 10.1016/j.carpath.2004.03.609. [DOI] [PubMed] [Google Scholar]
- 63.Norgett EE, Hatsell SJ, Carvajal-Huerta L, Cabezas JC, Common J, Purkis PE, Whittock N, Leigh IM, Stevens HP, Kelsell DP. Recessive mutation in desmoplakin disrupts desmoplakin-intermediate filament interactions and causes dilated cardiomyopathy, woolly hair and keratoderma. Hum Mol Genet. 2000;9:2761–2766. doi: 10.1093/hmg/9.18.2761. [DOI] [PubMed] [Google Scholar]
- 64.Syrris P, Ward D, Asimaki A, Sen-Chowdhry S, Ebrahim HY, Evans A, Hitomi N, Norman M, Pantazis A, Shaw AL, Elliott PM, McKenna WJ. Clinical expression of plakophilin-2 mutations in familial arrhythmogenic right ventricular cardiomyopathy. Circulation. 2006;113:356–364. doi: 10.1161/CIRCULATIONAHA.105.561654. [DOI] [PubMed] [Google Scholar]
- 65.Pilichou K, Nava A, Basso C, Beffagna G, Bauce B, Lorenzon A, Frigo G, Vettori A, Valente M, Towbin J, Thiene G, Danieli GA, Rampazzo A. Mutations in desmoglein-2 gene are associated with arrhythmogenic right ventricular cardiomyopathy. Circulation. 2006;113:1171–1179. doi: 10.1161/CIRCULATIONAHA.105.583674. [DOI] [PubMed] [Google Scholar]
- 66.Syrris P, Ward D, Evans A, Asimaki A, Gandjbakhch E, Sen-Chowdhry S, McKenna WJ. Arrhythmogenic right ventricular dysplasia/cardiomyopathy associated with mutations in the desmosomal gene desmocollin-2. Am J Hum Genet. 2006;79:978–984. doi: 10.1086/509122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Corrado D, Basso C, Thiene G. Arrhythmogenic right ventricular cardiomyopathy: an update. Heart. 2009;95:766–773. doi: 10.1136/hrt.2008.149823. [DOI] [PubMed] [Google Scholar]
- 68.Gerull B, Heuser A, Wichter T, Paul M, Basson CT, McDermott DA, Lerman BB, Markowitz SM, Ellinor PT, MacRae CA, Peters S, Grossmann KS, Michely B, Sasse-Klaassen S, Birchmeier W, Dietz R, Breithardt G, Schulze-Bahr E, Thierfelder L. Mutations in the desmosomal protein plakophilin-2 are common in arrhythmogenic right ventricular cardiomyopathy. Nat Genet. 2004;36:1162–1164. doi: 10.1038/ng1461. [DOI] [PubMed] [Google Scholar]
- 69.van der Zwaag PA, Jongbloed JDH, van den Berg MP, van der Smagt JJ, Jongbloed R, Bikker H, Hofstra RMW, van Tintelen JP. A genetic variants database for arrhythmogenic right ventricular dysplasia/cardiomyopathy. Hum Mutat. 2009;30:1278–1283. doi: 10.1002/humu.21064. [DOI] [PubMed] [Google Scholar]
- 70.Rampazzo A, Nava A, Malacrida S, Beffagna G, Bauce B, Rossi V, Zimbello R, Simionati B, Basso C, Thiene G, Towbin JA, Danieli GA. Mutation in human desmoplakin domain binding to plakoglobin causes a dominant form of arrhythmogenic right ventricular cardiomyopathy. Am J Hum Genet. 2002;71:1200–1206. doi: 10.1086/344208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Delmar M, McKenna WJ. The cardiac desmosome and arrhythmogenic cardiomyopathies: from gene to disease. Circ Res. 2010;107:700–714. doi: 10.1161/CIRCRESAHA.110.223412. [DOI] [PubMed] [Google Scholar]
- 72.Delmar M. The intercalated disk as a single functional unit. Heart Rhythm. 2004;1:12–13. doi: 10.1016/j.hrthm.2004.02.003. [DOI] [PubMed] [Google Scholar]
- 73.Garcia-Gras E, Lombardi R, Giocondo MJ, Willerson JT, Schneider MD, Khoury DS, Marian AJ. Suppression of canonical Wnt/beta-catenin signaling by nuclear plakoglobin recapitulates phenotype of arrhythmogenic right ventricular cardiomyopathy. J Clin Invest. 2006;116:2012–2021. doi: 10.1172/JCI27751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Pilichou K, Remme CA, Basso C, Campian ME, Rizzo S, Barnett P, Scicluna BP, Bauce B, van den Hoff MJ, de Bakker JM, Tan HL, Valente M, Nava A, Wilde AA, Moorman AF, Thiene G, Bezzina CR. Myocyte necrosis underlies progressive myocardial dystrophy in mouse dsg2-related arrhythmogenic right ventricular cardiomyopathy. J Exp Med. 2009;206:1787–1802. doi: 10.1084/jem.20090641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Christensen AH, Andersen CB, Tybjaerg-Hansen A, Haunso S, Svendsen JH. Mutation analysis and evaluation of the cardiac localization of TMEM43 in arrhythmogenic right ventricular cardiomyopathy. Clin Genet. doi: 10.1111/j.1399-0004.2011.01623.x. doi:10.1111/j.1399-0004.2011.01623.x. Published online ahead of print 2011. [DOI] [PubMed] [Google Scholar]
- 76.Merner ND, Hodgkinson KA, Haywood AF, Connors S, French VM, Drenckhahn JD, Kupprion C, Ramadanova K, Thierfelder L, McKenna W, Gallagher B, Morris-Larkin L, Bassett AS, Parfrey PS, Young TL. Arrhythmogenic right ventricular cardiomyopathy type 5 is a fully penetrant, lethal arrhythmic disorder caused by a missense mutation in the TMEM43 gene. Am J Hum Genet. 2008;82:809–821. doi: 10.1016/j.ajhg.2008.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Beffagna G, Occhi G, Nava A, Vitiello L, Ditadi A, Basso C, Bauce B, Carraro G, Thiene G, Towbin JA, Danieli GA, Rampazzo A. Regulatory mutations in transforming growth factor-beta3 gene cause arrhythmogenic right ventricular cardiomyopathy type 1. Cardiovasc Res. 2005;65:366–373. doi: 10.1016/j.cardiores.2004.10.005. [DOI] [PubMed] [Google Scholar]
- 78.Tiso N, Stephan DA, Nava A, Bagattin A, Devaney JM, Stanchi F, Larderet G, Brahmbhatt B, Brown K, Bauce B, Muriago M, Basso C, Thiene G, Danieli GA, Rampazzo A. Identification of mutations in the cardiac ryanodine receptor gene in families affected with arrhythmogenic right ventricular cardiomyopathy type 2 (ARVD2) Hum Mol Genet. 2001;10:189–194. doi: 10.1093/hmg/10.3.189. [DOI] [PubMed] [Google Scholar]
- 79.Kapplinger JD, Landstrom AP, Salisbury BA, Callis TE, Pollevick GD, Tester DJ, Cox MG, Bhuiyan Z, Bikker H, Wiesfeld AC, Hauer RN, van Tintelen JP, Jongbloed JD, Calkins H, Judge DP, Wilde AA, Ackerman MJ. Distinguishing arrhythmogenic right ventricular cardiomyopathy/dysplasia-associated mutations from background genetic noise. J Am Coll Cardiol. 2011;57:2317–2327. doi: 10.1016/j.jacc.2010.12.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Quarta G, Muir A, Pantazis A, Syrris P, Gehmlich K, Garcia-Pavia P, Ward D, Sen-Chowdhry S, Elliott PM, McKenna WJ. Familial evaluation in arrhythmogenic right ventricular cardiomyopathy: impact of genetics and revised Task Force Criteria. Circulation. 2011;123:2701–2709. doi: 10.1161/CIRCULATIONAHA.110.976936. [DOI] [PubMed] [Google Scholar]
- 81.Mahon NG, Murphy RT, MacRae CA, Caforio AL, Elliott PM, McKenna WJ. Echocardiographic evaluation in asymptomatic relatives of patients with dilated cardiomyopathy reveals preclinical disease. Ann Intern Med. 2005;143:108–115. doi: 10.7326/0003-4819-143-2-200507190-00009. [DOI] [PubMed] [Google Scholar]
- 82.Pankuweit S, Richter A, Ruppert V, Gelbrich G, Maisch B. Prevalence of different etiologies in dilated cardiomyopathy. J. Am. Coll. Cardiol. 2010;55:A35.E342. [Google Scholar]
- 83.Grunig E, Tasman JA, Kucherer H, Franz W, Kubler W, Katus HA. Frequency and phenotypes of familial dilated cardiomyopathy. J Am Coll Cardiol. 1998;31:186–194. doi: 10.1016/s0735-1097(97)00434-8. [DOI] [PubMed] [Google Scholar]
- 84.Baig MK, Goldman JH, Caforio AL, Coonar AS, Keeling PJ, McKenna WJ. Familial dilated cardiomyopathy: cardiac abnormalities are common in asymptomatic relatives and may represent early disease. J Am Coll Cardiol. 1998;31:195–201. doi: 10.1016/s0735-1097(97)00433-6. [DOI] [PubMed] [Google Scholar]
- 85.Caforio AL, Mahon NG, Baig MK, Tona F, Murphy RT, Elliott PM, McKenna WJ. Prospective familial assessment in dilated cardiomyopathy: cardiac autoantibodies predict disease development in asymptomatic relatives. Circulation. 2007;115:76–83. doi: 10.1161/CIRCULATIONAHA.106.641472. [DOI] [PubMed] [Google Scholar]
- 86.Rankin J, Ellard S. The laminopathies: a clinical review. Clin Genet. 2006;70:261–274. doi: 10.1111/j.1399-0004.2006.00677.x. [DOI] [PubMed] [Google Scholar]
- 87.Tardiff JC. Tropomyosin and dilated cardiomyopathy: revenge of the actinomyosin ‘gatekeeper’. J Am Coll Cardiol. 2010;55:330–332. doi: 10.1016/j.jacc.2009.11.018. [DOI] [PubMed] [Google Scholar]
- 88.Lakdawala NK, Dellefave L, Redwood CS, Sparks E, Cirino AL, Depalma S, Colan SD, Funke B, Zimmerman RS, Robinson P, Watkins H, Seidman CE, Seidman JG, McNally EM, Ho CY. Familial dilated cardiomyopathy caused by an alpha-tropomyosin mutation: the distinctive natural history of sarcomeric dilated cardiomyopathy. J Am Coll Cardiol. 2010;55:320–329. doi: 10.1016/j.jacc.2009.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Mogensen J, Murphy RT, Shaw T, Bahl A, Redwood C, Watkins H, Burke M, Elliott PM, McKenna WJ. Severe disease expression of cardiac troponin C and T mutations in patients with idiopathic dilated cardiomyopathy. J Am Coll Cardiol. 2004;44:2033–2040. doi: 10.1016/j.jacc.2004.08.027. [DOI] [PubMed] [Google Scholar]
- 90.Kamisago M, Sharma SD, DePalma SR, Solomon S, Sharma P, McDonough B, Smoot L, Mullen MP, Woolf PK, Wigle ED, Seidman JG, Seidman CE. Mutations in sarcomere protein genes as a cause of dilated cardiomyopathy. N Engl J Med. 2000;343:1688–1696. doi: 10.1056/NEJM200012073432304. [DOI] [PubMed] [Google Scholar]
- 91.Villard E, Duboscq-Bidot L, Charron P, Benaiche A, Conraads V, Sylvius N, Komajda M. Mutation screening in dilated cardiomyopathy: prominent role of the beta myosin heavy chain gene. Eur Heart J. 2005;26:794–803. doi: 10.1093/eurheartj/ehi193. [DOI] [PubMed] [Google Scholar]
- 92.Olson TM, Michels VV, Thibodeau SN, Tai YS, Keating MT. Actin mutations in dilated cardiomyopathy, a heritable form of heart failure. Science. 1998;280:750–752. doi: 10.1126/science.280.5364.750. [DOI] [PubMed] [Google Scholar]
- 93.Hershberger RE, Morales A, Siegfried JD. Clinical and genetic issues in dilated cardiomyopathy: a review for genetics professionals. Genet Med. 2010;12:655–667. doi: 10.1097/GIM.0b013e3181f2481f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Mestroni L, Rocco C, Gregori D, Sinagra G, Di Lenarda A, Miocic S, Vatta M, Pinamonti B, Muntoni F, Caforio AL, McKenna WJ, Falaschi A, Giacca M, Camerini F. Familial dilated cardiomyopathy: evidence for genetic and phenotypic heterogeneity. Heart Muscle Disease Study Group . J Am Coll Cardiol. 1999;34:181–190. doi: 10.1016/s0735-1097(99)00172-2. [DOI] [PubMed] [Google Scholar]
- 95.Li D, Morales A, Gonzalez-Quintana J, Norton N, Siegfried JD, Hofmeyer M, Hershberger RE. Identification of novel mutations in RBM20 in patients with dilated cardiomyopathy. Clin Transl Sci. 2010;3:90–97. doi: 10.1111/j.1752-8062.2010.00198.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Li D, Czernuszewicz GZ, Gonzalez O, Tapscott T, Karibe A, Durand JB, Brugada R, Hill R, Gregoritch JM, Anderson JL, Quinones M, Bachinski LL, Roberts R. Novel cardiac troponin T mutation as a cause of familial dilated cardiomyopathy. Circulation. 2001;104:2188–2193. doi: 10.1161/hc4301.098285. [DOI] [PubMed] [Google Scholar]
- 97.Murphy RT, Mogensen J, Shaw A, Kubo T, Hughes S, McKenna WJ. Novel mutation in cardiac troponin I in recessive idiopathic dilated cardiomyopathy. Lancet. 2004;363:371–372. doi: 10.1016/S0140-6736(04)15468-8. [DOI] [PubMed] [Google Scholar]
- 98.Mohapatra B, Jimenez S, Lin JH, Bowles KR, Coveler KJ, Marx JG, Chrisco MA, Murphy RT, Lurie PR, Schwartz RJ, Elliott PM, Vatta M, McKenna W, Towbin JA, Bowles NE. Mutations in the muscle LIM protein and alpha-actinin-2 genes in dilated cardiomyopathy and endocardial fibroelastosis. Mol Genet Metab. 2003;80:207–215. doi: 10.1016/s1096-7192(03)00142-2. [DOI] [PubMed] [Google Scholar]
- 99.Vatta M, Mohapatra B, Jimenez S, Sanchez X, Faulkner G, Perles Z, Sinagra G, Lin JH, Vu TM, Zhou Q, Bowles KR, Di Lenarda A, Schimmenti L, Fox M, Chrisco MA, Murphy RT, McKenna W, Elliott P, Bowles NE, Chen J, Valle G, Towbin JA. Mutations in Cypher/ZASP in patients with dilated cardiomyopathy and left ventricular non-compaction. J Am Coll Cardiol. 2003;42:2014–2027. doi: 10.1016/j.jacc.2003.10.021. [DOI] [PubMed] [Google Scholar]
- 100.Olson TM, Illenberger S, Kishimoto NY, Huttelmaier S, Keating MT, Jockusch BM. Metavinculin mutations alter actin interaction in dilated cardiomyopathy. Circulation. 2002;105:431–437. doi: 10.1161/hc0402.102930. [DOI] [PubMed] [Google Scholar]
- 101.Taylor MR, Slavov D, Ku L, Di Lenarda A, Sinagra G, Carniel E, Haubold K, Boucek MM, Ferguson D, Graw SL, Zhu X, Cavanaugh J, Sucharov CC, Long CS, Bristow MR, Lavori P, Mestroni L. Prevalence of desmin mutations in dilated cardiomyopathy. Circulation. 2007;115:1244–1251. doi: 10.1161/CIRCULATIONAHA.106.646778. [DOI] [PubMed] [Google Scholar]
- 102.Hayashi T, Arimura T, Itoh-Satoh M, Ueda K, Hohda S, Inagaki N, Takahashi M, Hori H, Yasunami M, Nishi H, Koga Y, Nakamura H, Matsuzaki M, Choi BY, Bae SW, You CW, Han KH, Park JE, Knoll R, Hoshijima M, Chien KR, Kimura A. Tcap gene mutations in hypertrophic cardiomyopathy and dilated cardiomyopathy. J Am Coll Cardiol. 2004;44:2192–2201. doi: 10.1016/j.jacc.2004.08.058. [DOI] [PubMed] [Google Scholar]
- 103.Bione S, D'Adamo P, Maestrini E, Gedeon AK, Bolhuis PA, Toniolo D. A novel X-linked gene, G4.5, is responsible for Barth syndrome. Nat Genet. 1996;12:385–389. doi: 10.1038/ng0496-385. [DOI] [PubMed] [Google Scholar]
- 104.Barresi R, Di Blasi C, Negri T, Brugnoni R, Vitali A, Felisari G, Salandi A, Daniel S, Cornelio F, Morandi L, Mora M. Disruption of heart sarcoglycan complex and severe cardiomyopathy caused by beta sarcoglycan mutations. J Med Genet. 2000;37:102–107. doi: 10.1136/jmg.37.2.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Feng J, Yan J, Buzin CH, Towbin JA, Sommer SS. Mutations in the dystrophin gene are associated with sporadic dilated cardiomyopathy. Mol Genet Metab. 2002;77:119–126. doi: 10.1016/s1096-7192(02)00153-1. [DOI] [PubMed] [Google Scholar]
- 106.Homayoun H, Khavandgar S, Hoover JM, Mohsen AW, Vockley J, Lacomis D, Clemens PR. Novel mutation in MYH7 gene associated with distal myopathy and cardiomyopathy. Neuromuscul Disord. 2011;21:219–222. doi: 10.1016/j.nmd.2010.12.005. [DOI] [PubMed] [Google Scholar]
- 107.Jefferies JL, Towbin JA. Dilated cardiomyopathy. Lancet. 2010;375:752–762. doi: 10.1016/S0140-6736(09)62023-7. [DOI] [PubMed] [Google Scholar]
- 108.Bowles NE, Bowles KR, Towbin JA. The ‘final common pathway’ hypothesis and inherited cardiovascular disease. The role of cytoskeletal proteins in dilated cardiomyopathy. Herz. 2000;25:168–175. doi: 10.1007/s000590050003. [DOI] [PubMed] [Google Scholar]
- 109.Paulin D, Li Z. Desmin: a major intermediate filament protein essential for the structural integrity and function of muscle. Exp Cell Res. 2004;301:1–7. doi: 10.1016/j.yexcr.2004.08.004. [DOI] [PubMed] [Google Scholar]
- 110.Sanbe A, Osinska H, Saffitz JE, Glabe CG, Kayed R, Maloyan A, Robbins J. Desmin-related cardiomyopathy in transgenic mice: a cardiac amyloidosis. Proc Natl Acad Sci USA. 2004;101:10132–10136. doi: 10.1073/pnas.0401900101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Maloyan A, Gulick J, Glabe CG, Kayed R, Robbins J. Exercise reverses preamyloid oligomer and prolongs survival in alphaB-crystallin-based desmin-related cardiomyopathy. Proc Natl Acad Sci USA. 2007;104:5995–6000. doi: 10.1073/pnas.0609202104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Calkins H. Use of mouse models to evaluate novel therapeutic approaches to treatment of arrhythmogenic right ventricular cardiomyopathy: the future is now. J Am Coll Cardiol. 2011;57:751–752. doi: 10.1016/j.jacc.2010.09.043. [DOI] [PubMed] [Google Scholar]
- 113.Fontaine G, Fontaliran F. Arrhythmogenic right ventricular dysplasia masquerading as dilated cardiomyopathy. Am J Cardiol. 1999;84:1143. [PubMed] [Google Scholar]
- 114.Nemec J, Edwards BS, Osborn MJ, Edwards WD. Arrhythmogenic right ventricular dysplasia masquerading as dilated cardiomyopathy. Am J Cardiol. 1999;84:237–239. doi: 10.1016/s0002-9149(99)00244-1. A9. [DOI] [PubMed] [Google Scholar]
- 115.Lui CY, Marcus FI, Sobonya RE. Arrhythmogenic right ventricular dysplasia masquerading as peripartum cardiomyopathy with atrial flutter, advanced atrioventricular block and embolic stroke. Cardiology. 2002;97:49–50. doi: 10.1159/000047419. [DOI] [PubMed] [Google Scholar]
- 116.Parks SB, Kushner JD, Nauman D, Burgess D, Ludwigsen S, Peterson A, Li D, Jakobs P, Litt M, Porter CB, Rahko PS, Hershberger RE. Lamin A/C mutation analysis in a cohort of 324 unrelated patients with idiopathic or familial dilated cardiomyopathy. Am Heart J. 2008;156:161–169. doi: 10.1016/j.ahj.2008.01.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Perrot A, Hussein S, Ruppert V, Schmidt HH, Wehnert MS, Duong NT, Posch MG, Panek A, Dietz R, Kindermann I, Bohm M, Michalewska-Wludarczyk A, Richter A, Maisch B, Pankuweit S, Ozcelik C. Identification of mutational hot spots in LMNA encoding lamin A/C in patients with familial dilated cardiomyopathy. Basic Res Cardiol. 2009;104:90–99. doi: 10.1007/s00395-008-0748-6. [DOI] [PubMed] [Google Scholar]
- 118.Muchir A, Medioni J, Laluc M, Massart C, Arimura T, van der Kooi AJ, Desguerre I, Mayer M, Ferrer X, Briault S, Hirano M, Worman HJ, Mallet A, Wehnert M, Schwartz K, Bonne G. Nuclear envelope alterations in fibroblasts from patients with muscular dystrophy, cardiomyopathy, and partial lipodystrophy carrying lamin A/C gene mutations. Muscle Nerve. 2004;30:444–450. doi: 10.1002/mus.20122. [DOI] [PubMed] [Google Scholar]
- 119.Maraldi NM, Lattanzi G, Capanni C, Columbaro M, Merlini L, Mattioli E, Sabatelli P, Squarzoni S, Manzoli FA. Nuclear envelope proteins and chromatin arrangement: a pathogenic mechanism for laminopathies. Eur J Histochem. 2006;50:1–8. [PubMed] [Google Scholar]
- 120.Eriksson M, Sagelius H, Rosengardten Y, Schmidt E, Sonnabend C, Rozell B. Reversible phenotype in a mouse model of Hutchinson–Gilford progeria syndrome. J Med Genet. 2008;45:794–801. doi: 10.1136/jmg.2008.060772. [DOI] [PubMed] [Google Scholar]
- 121.Fong LG, Frost D, Meta M, Qiao X, Yang SH, Coffinier C, Young SG. A protein farnesyltransferase inhibitor ameliorates disease in a mouse model of progeria. Science. 2006;311:1621–1623. doi: 10.1126/science.1124875. [DOI] [PubMed] [Google Scholar]
- 122.Young SG, Yang SH, Meta M, Qiao X, Frost D, Bauch J, Coffinier C, Majumdar S, Bergo MO, Fong LG. A farnesyltransferase inhibitor improves disease phenotypes in mice with a Hutchinson-Gilford progeria syndrome mutation. J Clin Invest. 2006;116:2115–2121. doi: 10.1172/JCI28968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.McNair WP, Ku L, Taylor MR, Fain PR, Dao D, Wolfel E, Mestroni L. SCN5A mutation associated with dilated cardiomyopathy, conduction disorder, and arrhythmia. Circulation. 2004;110:2163–2167. doi: 10.1161/01.CIR.0000144458.58660.BB. [DOI] [PubMed] [Google Scholar]
- 124.Olson TM, Michels VV, Ballew JD, Reyna SP, Karst ML, Herron KJ, Horton SC, Rodeheffer RJ, Anderson JL. Sodium channel mutations and susceptibility to heart failure and atrial fibrillation. JAMA. 2005;293:447–454. doi: 10.1001/jama.293.4.447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Robinson P, Griffiths PJ, Watkins H, Redwood CS. Dilated and hypertrophic cardiomyopathy mutations in troponin and alpha-tropomyosin have opposing effects on the calcium affinity of cardiac thin filaments. Circ Res. 2007;101:1266–1273. doi: 10.1161/CIRCRESAHA.107.156380. [DOI] [PubMed] [Google Scholar]
- 126.Chang AN, Potter JD. Sarcomeric protein mutations in dilated cardiomyopathy. Heart Fail Rev. 2005;10:225–235. doi: 10.1007/s10741-005-5252-6. [DOI] [PubMed] [Google Scholar]
- 127.Duboc D, Meune C, Lerebours G, Devaux JY, Vaksmann G, Becane HM. Effect of perindopril on the onset and progression of left ventricular dysfunction in Duchenne muscular dystrophy. J Am Coll Cardiol. 2005;45:855–857. doi: 10.1016/j.jacc.2004.09.078. [DOI] [PubMed] [Google Scholar]
- 128.Effect of enalapril on mortality and the development of heart failure in asymptomatic patients with reduced left ventricular ejection fractions. The SOLVD Investigators. N Engl J Med. 1992;327:685–691. doi: 10.1056/NEJM199209033271003. [DOI] [PubMed] [Google Scholar]
- 129.Sen-Chowdhry S, Syrris P, McKenna WJ. Genetics of restrictive cardiomyopathy. Heart Fail Clin. 2010;6:179–186. doi: 10.1016/j.hfc.2009.11.005. [DOI] [PubMed] [Google Scholar]
- 130.Gomes AV. Mutations in human cardiac troponin i that are associated with restrictive cardiomyopathy affect basal ATPase activity and the calcium sensitivity of force development. J Biol Chem. 2005;280:30909–30915. doi: 10.1074/jbc.M500287200. [DOI] [PubMed] [Google Scholar]
- 131.Pantazis AA, Elliott PM. Left ventricular noncompaction. Curr Opin Cardiol. 2009;24:209–213. doi: 10.1097/HCO.0b013e32832a11e7. [DOI] [PubMed] [Google Scholar]
- 132.Kohli SK, Pantazis AA, Shah JS, Adeyemi B, Jackson G, McKenna WJ, Sharma S, Elliott PM. Diagnosis of left-ventricular non-compaction in patients with left-ventricular systolic dysfunction: time for a reappraisal of diagnostic criteria? Eur Heart J. 2008;29:89–95. doi: 10.1093/eurheartj/ehm481. [DOI] [PubMed] [Google Scholar]
- 133.Sandhu R, Finkelhor RS, Gunawardena DR, Bahler RC. Prevalence and characteristics of left ventricular noncompaction in a community hospital cohort of patients with systolic dysfunction. Echocardiography. 2008;25:8–12. doi: 10.1111/j.1540-8175.2007.00560.x. [DOI] [PubMed] [Google Scholar]
- 134.Aras D, Tufekcioglu O, Ergun K, Ozeke O, Yildiz A, Topaloglu S, Deveci B, Sahin O, Kisacik HL, Korkmaz S. Clinical features of isolated ventricular noncompaction in adults long-term clinical course, echocardiographic properties, and predictors of left ventricular failure. J Card Fail. 2006;12:726–733. doi: 10.1016/j.cardfail.2006.08.002. [DOI] [PubMed] [Google Scholar]
- 135.Oechslin EN, Attenhofer Jost CH, Rojas JR, Kaufmann PA, Jenni R. Long-term follow-up of 34 adults with isolated left ventricular noncompaction: a distinct cardiomyopathy with poor prognosis. J Am Coll Cardiol. 2000;36:493–500. doi: 10.1016/s0735-1097(00)00755-5. [DOI] [PubMed] [Google Scholar]
- 136.Captur G, Nihoyannopoulos P. Left ventricular non-compaction: genetic heterogeneity, diagnosis and clinical course. Int J Cardiol. 2010;140:145–153. doi: 10.1016/j.ijcard.2009.07.003. [DOI] [PubMed] [Google Scholar]
- 137.Neustein HB, Lurie PR, Dahms B, Takahashi M. An X-linked recessive cardiomyopathy with abnormal mitochondria. Pediatrics. 1979;64:24–29. [PubMed] [Google Scholar]
- 138.Klaassen S, Probst S, Oechslin E, Gerull B, Krings G, Schuler P, Greutmann M, Hurlimann D, Yegitbasi M, Pons L, Gramlich M, Drenckhahn JD, Heuser A, Berger F, Jenni R, Thierfelder L. Mutations in sarcomere protein genes in left ventricular noncompaction. Circulation. 2008;117:2893–2901. doi: 10.1161/CIRCULATIONAHA.107.746164. [DOI] [PubMed] [Google Scholar]
- 139.Hermida-Prieto M, Monserrat L, Castro-Beiras A, Laredo R, Soler R, Peteiro J, Rodriguez E, Bouzas B, Alvarez N, Muniz J, Crespo-Leiro M. Familial dilated cardiomyopathy and isolated left ventricular noncompaction associated with lamin A/C gene mutations. Am J Cardiol. 2004;94:50–54. doi: 10.1016/j.amjcard.2004.03.029. [DOI] [PubMed] [Google Scholar]
- 140.Ichida F, Tsubata S, Bowles KR, Haneda N, Uese K, Miyawaki T, Dreyer WJ, Messina J, Li H, Bowles NE, Towbin JA. Novel gene mutations in patients with left ventricular noncompaction or Barth syndrome. Circulation. 2001;103:1256–1263. doi: 10.1161/01.cir.103.9.1256. [DOI] [PubMed] [Google Scholar]
- 141.Kaski JP, Syrris P, Burch M, Tome-Esteban MT, Fenton M, Christiansen M, Andersen PS, Sebire N, Ashworth M, Deanfield JE, McKenna WJ, Elliott PM. Idiopathic restrictive cardiomyopathy in children is caused by mutations in cardiac sarcomere protein genes. Heart. 2008;94:1478–1484. doi: 10.1136/hrt.2007.134684. [DOI] [PubMed] [Google Scholar]
- 142.Mogensen J. Idiopathic restrictive cardiomyopathy is part of the clinical expression of cardiac troponin I mutations. J Clin Invest. 2003;111:209–216. doi: 10.1172/JCI16336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Kubo T, Gimeno JR, Bahl A, Steffensen U, Steffensen M, Osman E, Thaman R, Mogensen J, Elliott PM, Doi Y. Prevalence, clinical significance, and genetic basis of hypertrophic cardiomyopathy with restrictive phenotype. J Am Coll Cardiol. 2007;49:2419–2426. doi: 10.1016/j.jacc.2007.02.061. [DOI] [PubMed] [Google Scholar]
- 144.Ammash NM, Seward JB, Bailey KR, Edwards WD, Tajik AJ. Clinical profile and outcome of idiopathic restrictive cardiomyopathy. Circulation. 2000;101:2490–2496. doi: 10.1161/01.cir.101.21.2490. [DOI] [PubMed] [Google Scholar]
- 145.Daw EW, Lu Y, Marian AJ, Shete S. Identifying modifier loci in existing genome scan data. Ann Hum Genet. 2008;72(Pt 5):670–675. doi: 10.1111/j.1469-1809.2008.00449.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Koren MJ, Devereux RB, Casale PN, Savage DD, Laragh JH. Relation of left ventricular mass and geometry to morbidity and mortality in uncomplicated essential hypertension. Ann Intern Med. 1991;114:345–352. doi: 10.7326/0003-4819-114-5-345. [DOI] [PubMed] [Google Scholar]
- 147.Haider AW, Larson MG, Benjamin EJ, Levy D. Increased left ventricular mass and hypertrophy are associated with increased risk for sudden death. J Am Coll Cardiol. 1998;32:1454–1459. doi: 10.1016/s0735-1097(98)00407-0. [DOI] [PubMed] [Google Scholar]
- 148.Olivotto I, Gistri R, Petrone P, Pedemonte E, Vargiu D, Cecchi F. Maximum left ventricular thickness and risk of sudden death in patients with hypertrophic cardiomyopathy. J Am Coll Cardiol. 2003;41:315–321. doi: 10.1016/s0735-1097(02)02713-4. [DOI] [PubMed] [Google Scholar]
- 149.Bruneau BG, Bao ZZ, Fatkin D, Xavier-Neto J, Georgakopoulos D, Maguire CT, Berul CI, Kass DA, Kuroski-de Bold ML, de Bold AJ, Conner DA, Rosenthal N, Cepko CL, Seidman CE, Seidman JG. Cardiomyopathy in Irx4-deficient mice is preceded by abnormal ventricular gene expression. Mol Cell Biol. 2001;21:1730–1736. doi: 10.1128/MCB.21.5.1730-1736.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Bayrak F, Komurcu-Bayrak E, Mutlu B, Kahveci G, Erginel-Unaltuna N. Genetic analysis of the Irx4 gene in hypertrophic cardiomyopathy. Turk Kardiyol Dern Ars. 2008;36:90–95. [PubMed] [Google Scholar]
- 151.Gottlieb PD, Pierce SA, Sims RJ, Yamagishi H, Weihe EK, Harriss JV, Maika SD, Kuziel WA, King HL, Olson EN, Nakagawa O, Srivastava D. Bop encodes a muscle-restricted protein containing MYND and SET domains and is essential for cardiac differentiation and morphogenesis. Nat Genet. 2002;31:25–32. doi: 10.1038/ng866. [DOI] [PubMed] [Google Scholar]
- 152.Costantini DL, Arruda EP, Agarwal P, Kim KH, Zhu Y, Zhu W, Lebel M, Cheng CW, Park CY, Pierce SA, Guerchicoff A, Pollevick GD, Chan TY, Kabir MG, Cheng SH, Husain M, Antzelevitch C, Srivastava D, Gross GJ, Hui CC, Backx PH, Bruneau BG. The homeodomain transcription factor Irx5 establishes the mouse cardiac ventricular repolarization gradient. Cell. 2005;123:347–358. doi: 10.1016/j.cell.2005.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Yetman AT, Hamilton RM, Benson LN, McCrindle BW. Long-term outcome and prognostic determinants in children with hypertrophic cardiomyopathy. J Am Coll Cardiol. 1998;32:1943–1950. doi: 10.1016/s0735-1097(98)00493-8. [DOI] [PubMed] [Google Scholar]
- 154.Perkins MJ, Van Driest SL, Ellsworth EG, Will ML, Gersh BJ, Ommen SR, Ackerman MJ. Gene-specific modifying effects of pro-LVH polymorphisms involving the renin–angiotensin–aldosterone system among 389 unrelated patients with hypertrophic cardiomyopathy. Eur Heart J. 2005;26:2457–2462. doi: 10.1093/eurheartj/ehi438. [DOI] [PubMed] [Google Scholar]
- 155.Ogimoto A, Okayama H, Nagai T, Ohtsuka T, Suzuki J, Inoue K, Nishimura K, Saito M, Shigematsu Y, Hamada M, Miki T, Higaki J. Pharmacogenetic interactions between angiotensin-converting enzyme insertion/deletion polymorphism and response to cibenzoline in patients with hypertrophic obstructive cardiomyopathy. J Cardiovasc Pharmacol. 2010;55:506–510. doi: 10.1097/FJC.0b013e3181d8bc4b. [DOI] [PubMed] [Google Scholar]
- 156.Coto E, Palacin M, Martin M, Castro MG, Reguero JR, Garcia C, Berrazueta JR, Moris C, Morales B, Ortega F, Corao AI, Diaz M, Tavira B, Alvarez V. Functional polymorphisms in genes of the Angiotensin and Serotonin systems and risk of hypertrophic cardiomyopathy: AT1R as a potential modifier. J Transl Med. 2010;8:64. doi: 10.1186/1479-5876-8-64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Sen-Chowdhry S, Syrris P, Pantazis A, Quarta G, McKenna WJ, Chambers JC. Mutational heterogeneity, modifier genes, and environmental influences contribute to phenotypic diversity of arrhythmogenic cardiomyopathy. Circ Cardiovasc Genet. 2010;3:323–330. doi: 10.1161/CIRCGENETICS.109.935262. [DOI] [PubMed] [Google Scholar]
- 158.Forleo C, Resta N, Sorrentino S, Guida P, Manghisi A, De Luca V, Romito R, Iacoviello M, De Tommasi E, Troisi F, Rizzon B, Guanti G, Rizzon P, Pitzalis MV. Association of beta-adrenergic receptor polymorphisms and progression to heart failure in patients with idiopathic dilated cardiomyopathy. Am J Med. 2004;117:451–458. doi: 10.1016/j.amjmed.2004.04.012. [DOI] [PubMed] [Google Scholar]
- 159.Arvanitis DA, Sanoudou D, Kolokathis F, Vafiadaki E, Papalouka V, Kontrogianni-Konstantopoulos A, Theodorakis GN, Paraskevaidis IA, Adamopoulos S, Dorn GW, 2nd, Kremastinos DT, Kranias EG. The Ser96Ala variant in histidine-rich calcium-binding protein is associated with life-threatening ventricular arrhythmias in idiopathic dilated cardiomyopathy. Eur Heart J. 2008;29:2514–2525. doi: 10.1093/eurheartj/ehn328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Wilkinson JD, Landy DC, Colan SD, Towbin JA, Sleeper LA, Orav EJ, Cox GF, Canter CE, Hsu DT, Webber SA, Lipshultz SE. The pediatric cardiomyopathy registry and heart failure: key results from the first 15 years. Heart Fail Clin. 2010;6:401–413. doi: 10.1016/j.hfc.2010.05.002. vii. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Metkus TS, Jr, Baughman KL, Thompson PD. Exercise prescription and primary prevention of cardiovascular disease. Circulation. 2010;121:2601–2604. doi: 10.1161/CIRCULATIONAHA.109.903377. [DOI] [PubMed] [Google Scholar]
- 162.Lloyd-Jones DM, Hong Y, Labarthe D, Mozaffarian D, Appel LJ, Van Horn L, Greenlund K, Daniels S, Nichol G, Tomaselli GF, Arnett DK, Fonarow GC, Ho PM, Lauer MS, Masoudi FA, Robertson RM, Roger V, Schwamm LH, Sorlie P, Yancy CW, Rosamond WD. Defining and setting national goals for cardiovascular health promotion and disease reduction: the American Heart Association's strategic Impact Goal through 2020 and beyond. Circulation. 2010;121:586–613. doi: 10.1161/CIRCULATIONAHA.109.192703. [DOI] [PubMed] [Google Scholar]
- 163.Whang W, Shimbo D, Kronish IM, Duvall WL, Julien H, Iyer P, Burg MM, Davidson KW. Depressive symptoms and all-cause mortality in unstable angina pectoris (from the Coronary Psychosocial Evaluation Studies [COPES]) Am J Cardiol. 2010;106:1104–1107. doi: 10.1016/j.amjcard.2010.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Lampert R, Salberg L, Burg M. Emotional stress triggers symptoms in hypertrophic cardiomyopathy: a survey of the hypertrophic cardiomyopathy association. Pacing Clin Electrophysiol. 2010;33:1047–1053. doi: 10.1111/j.1540-8159.2010.02770.x. [DOI] [PubMed] [Google Scholar]
- 165.Fornes P, Ratel S, Lecomte D. Pathology of arrhythmogenic right ventricular cardiomyopathy/dysplasia–an autopsy study of 20 forensic cases. J Forensic Sci. 1998;43:777–783. [PubMed] [Google Scholar]
- 166.Kolar AJ, Milroy CM, Day PF, Suvarna SK. Dilated cardiomyopathy and sudden death in a teenager with palmar-plantar keratosis (occult Carvajal syndrome) J Forensic Leg Med. 2008;15:185–188. doi: 10.1016/j.jflm.2007.09.002. [DOI] [PubMed] [Google Scholar]
- 167.Corrado D, Basso C, Rizzoli G, Schiavon M, Thiene G. Does sports activity enhance the risk of sudden death in adolescents and young adults? J Am Coll Cardiol. 2003;42:1959–1963. doi: 10.1016/j.jacc.2003.03.002. [DOI] [PubMed] [Google Scholar]
- 168.Lewis CSH, Jones R. Can we make assumptions about the psychosocial impact of living as a carrier, based on studies assessing the effects of carrier testing? J Genet Couns. 2011;20:80–97. doi: 10.1007/s10897-010-9327-8. [DOI] [PubMed] [Google Scholar]
- 169.Aatre RD, Day SM. Psychological issues in genetic testing for inherited cardiovascular diseases. Circ Cardiovasc Genet. 2011;4:81–90. doi: 10.1161/CIRCGENETICS.110.957365. [DOI] [PubMed] [Google Scholar]
- 170.Charron P, Arad M, Arbustini E, Basso C, Bilinska Z, Elliott P, Helio T, Keren A, McKenna WJ, Monserrat L, Pankuweit S, Perrot A, Rapezzi C, Ristic A, Seggewiss H, van Langen I, Tavazzi L. Genetic counselling and testing in cardiomyopathies: a position statement of the European Society of Cardiology Working Group on Myocardial and Pericardial Diseases. Eur Heart J. 2010;31:2715–2726. doi: 10.1093/eurheartj/ehq271. [DOI] [PubMed] [Google Scholar]