Abstract
Aims
To examine the relations between genetic loci, plasma lipoprotein(a) [Lp(a)] levels, and cardiovascular disease (CVD) risk among diabetic patients and compare with the observations in the general population.
Methods and results
In two prospective cohorts of patients with type 2 diabetes (n= 2308) from the Nurses' Health Study and the Health Professional Follow-Up Study, we performed (i) genome-wide association (GWA) scans for plasma Lp(a); (ii) prospective analysis of plasma Lp(a) for CVD risk and mortality; and (iii) genetic association analysis for CVD risk and mortality. Meta-analysis of the two GWA scans yielded 71 single-nucleotide polymorphisms (SNPs) on chromosome 6q associated with plasma Lp(a) levels at a genome-wide significance level (P< 5 × 10−8). The SNP rs10455872 in LPA was most strongly associated with Lp(a) (P= 4.60 × 10−39). Forward-selection analysis indicated that rs10455872 and other five SNPs in a region encompassing LPA, PLG, SLC22A3, and LPAL2 genes were independently associated with Lp(a) levels and jointly explained ∼20% of variation in diabetic patients. In prospective analysis, we did not find any significant association between plasma levels and CVD incidence; the relative risk for coronary heart disease (CHD), CVD, and CVD death was 1.05 [95% confidence interval (CI): 0.95–1.15], 1.05 (0.96–1.15), and 1.21 (0.99–1.47) per 1-SD higher log-transformed Lp(a) levels, respectively. Consistently, none of the Lp(a) SNPs were associated with CVD risk or mortality (all P> 0.09). For the best SNP rs10455872 for plasma Lp(a) levels, the OR for CHD, CVD, and CVD death was 0.94 (95% CI: 0.69–1.28), 0.97 (0.72–1.29), and 1.23 (0.79–1.92), respectively. The genetic effect on CHD risk showed a significant heterogeneity between the diabetic and the general populations (P= 0.006).
Conclusion
Our data indicate that the effect of Lp(a) on CVD risk among diabetic patients might be different from that in the general population. Diabetes status may attenuate the relation between Lp(a) and cardiovascular risk.
Keywords: Cardiovascular disease, Genome-wide association, Lipoprotein(a), Type 2 diabetes
Introduction
Lipoprotein(a) [Lp(a)] is an LDL-like particle that consists of an apolipoprotein(a) moiety linked to one molecule of apolipoprotein B100 via a disulfide bond.1 In the general population, elevated Lp(a) levels have been recognized as a cardiovascular risk factor.2,3 Recent genome-wide association (GWA) studies identified genetic variations associated with plasma Lp(a) levels.4,5 Data from a meta-analysis of 36 prospective studies6 and Mendelian randomization analyses in two human genetic association studies7,8 provide support for a probable causal role of elevated Lp(a) in the development of cardiovascular disease (CVD) in the general population.9
Patients with type 2 diabetes are featured by a different metabolic profile from the general populations with two- to four-fold higher cardiovascular risk than the non-diabetics.10,11 Recent studies found that Lp(a) levels were lower in patients with type 2 diabetes than the non-diabetics12. Little is known about the genetic determinants for Lp(a) levels in diabetic patients, and it remains unclear whether elevated Lp(a) levels may causally affect CVD risk in the diabetic patients. Several small prospective studies among patients with type 2 diabetes have yielded conflicting results.13–18 Three of these studies found that higher plasma Lp(a) levels were associated with increased risk for coronary heart disease (CHD),17 CVD,14 and CVD-caused mortality,16 while others did not find any significant association.13,15,18
By taking advantage of the GWA scans and plasma Lp(a) levels measured in two prospectively followed cohorts of diabetic patients from the Nurses' Health Study (NHS) and Health Professional Follow-up Study (HPFS), we performed analyses to investigate the interrelationship between genetic markers, Lp(a) levels, and risk of CVD and mortality. We also compared the observations with those reported in the general population.8
Methods
Study populations
The NHS is a prospective cohort study of 121 700 female registered nurses who were 30–55 years old at study inception in 1976 when all of them completed a mailed questionnaire on their medical history and lifestyle.19 A total of 32 826 women provided blood samples between 1989 and 1990. The HPFS is a prospective cohort study of 51 529 US male health professionals who were 40–75 years old at study inception in 1986.20 Between 1993 and 1999, 18 159 men provided blood samples. In both cohorts, information about health and disease has been collected biennially by self-administered questionnaires since inception. Diabetes cases were defined as self-reported diabetes confirmed by a validated supplementary questionnaire. For cases before 1998, we used the National Diabetes Data Group criteria to define diabetes.21 The validity of this method has been confirmed.22 We used the American Diabetes Association diagnostic criteria for diabetes diagnosis from 1998 onwards.23 Cardiovascular disease cases were defined as the occurrence of CHD [fatal or non-fatal myocardial infarction (MI) or coronary artery bypass grafting] or stroke (fatal or non-fatal) during follow-up through 2006. Non-fatal MI was confirmed by reviewing medical records using the criteria of the World Health Organization of symptoms plus either typical electrocardiographic changes or elevated levels of cardiac enzymes. Non-fatal stroke was confirmed by reviewing medical records using the criteria of the National Survey of Stroke.24 Physicians who reviewed the records had no knowledge of the self-reported risk factors. Cardiovascular deaths were confirmed by review of medical records or autopsy reports with the permission of the next of kin. The study was approved by the Human Research Committee at the Brigham and Women's Hospital, Boston, MA, USA, and all participants provided written informed consent.
In the current genetic association analysis for CVD risk, 1301 diabetic women (273 CHD and 60 stroke events; and 124 CVD deaths) and 1007 diabetic men (316 CHD and 28 stroke events; and 145 CVD deaths) with GWA scans available were included. For the prospective analysis of plasma Lp(a) levels and CVD risk, where follow-up began at the time of blood collection, 926 diabetic women (248 CHD and 56 stroke events; and 84 CVD deaths during 16-year follow-up) and 760 diabetic men (227 CHD and 30 stroke events; and 72 CVD deaths during 12-year follow-up) were included. The present analysis expanded our previous study of plasma Lp(a) and CHD risk in diabetic women by having longer follow-up and more CVD events.17 In the present study, 715 women and 735 men had both GWA scans and Lp(a) measures. Participants who were diagnosed with CVD before the diagnosis of diabetes were excluded from the present analyses.25
Measurements of lipoprotein(a) and other biomarkers
Blood samples were collected in EDTA blood tubes in the men and heparin blood tubes in the women, chilled, and sent back by prepaid overnight courier. Once in the laboratory, the samples were centrifuged and aliquoted into cryotubes as plasma, buffy coat, and red blood cells. Cryotubes were then stored in liquid nitrogen freezers at −130°C or lower. Plasma Lp(a) was measured by a latex-enhanced immunoturbidimetric method from Denka Seiken (Tokyo, Japan), with coefficients of variation (CVs) of 2.6%. The kringle IV type 2 (KIV-2) repeats polymorphism is an important determinant of Lp(a) density and distribution in plasma,26 and this method is not affected by the KIV-2 repeats. Concentrations of total cholesterol, triglycerides, and HDL cholesterol were measured simultaneously on the Hitachi 911 analyzer using reagents and calibrators from Roche Diagnostics (Indianapolis, IN, USA), with CVs of <1.8%. Low-density lipoprotein cholesterol concentration was measured by a homogenous direct method from Genzyme (Cambridge, MA, USA), with CVs of <3.1%. Haemoglobin A1c (HbA1c) concentrations were based on turbidimetric immunoinhibition with haemolysed whole blood or packed red cells, with CVs of <3.0%.
Genome-wide association genotyping, imputation, and quality control
Genome-wide association genotyping, imputation, and quality control have been described in detail elsewhere (the NHS and HPFS T2D GWA scans).27 Briefly, samples were genotyped and analysed using the Affymetrix Genome-Wide Human 6.0 array (Santa Clara, CA, USA) and the Birdseed calling algorithm. All samples used in the present study achieved a call rate of ≥98%. Individual single-nucleotide polymorphisms (SNPs) were excluded if they were monomorphic, had a missing call rate of ≥2%, more than one discordance, a Hardy–Weinberg equilibrium P-value of <1 × 10−4 or a minor allelic frequency (MAF) of <0.02. We used MACH (http://www.sph.umich.edu/csg/abecasis/MACH) to impute SNPs on chromosomes 1–22 with NCBI build 36 of Phase II HapMap CEU data (release 22) as the reference panel. Imputation results were expressed as ‘allele dosages’ (fractional values between 0 and 2). Imputed SNPs with MAF < 0.02 and/or with poor imputation quality scores (MACH, r2 ≤ 0.30) were filtered from the analysis. Finally, ∼2.5 million directly genotyped or imputed SNPs were included for analysis.
Statistical analysis
Two GWA scans for plasma Lp(a) levels across ∼2.5 million SNPs (imputed data were expressed as allele dosage) were performed using linear regression under an additive genetic model adjusting for age and body mass index (BMI) in ProbABEL package.28 Meta-analysis of these two GWA scans was conducted using inverse variance weights under a fixed-effect model in METAL (http://www.sph.umich.edu/csg/abecasis/Metal). Lipoprotein(a) levels were first log-transformed and then scaled as SD units before analysis.
We used a forward-selection regression procedure to identify genome-wide significant SNPs that were associated with Lp(a) levels independent of other SNPs. Age and BMI were forced into the linear regression model and the selection criterion was P< 0.05 for an SNP to enter the final model. A genotype score was calculated by counting the number of higher-Lp(a) alleles from the selected SNPs, assuming that each SNP was independently associated with Lp(a) levels under an additive inheritance model. The SNPs selected were also further analysed for haplotype associations by using the THESIAS program, which is based on the Stochastic-EM algorithm (SEM).29 Logistic regression was used to test the association of the selected SNPs or the genotype score with risk of CHD, CVD, and CVD death, adjusting for age and BMI. We used Cox's proportional hazards analysis to estimate relative risks (RRs) and 95% confidence interval (CI) for CVD incidence and mortality per 1-SD higher log-transformed Lp(a) levels and by comparing the quintiles of Lp(a) levels. Participants who were diagnosed with CHD or stroke or died during follow-up were censored at the date of diagnosis or death. Combined results between men and women were calculated by using inverse variance weights under a fixed model. In addition, the regression coefficients of the SNP rs10455872 for Lp(a) levels and CHD risk in our study of diabetic patients and in the general population8 were compared by the t-test. Power calculations for genetic association analysis were performed using Quanto software (http://hydra.usc.edu/gxe/). The analyses (otherwise indicated) were performed in SAS 9.1 (SAS Institute, Inc., Cary, NC, USA).
Results
Characteristics of study participants
Table 1 presents the characteristics of study participants at the year of blood sample collection. Participants who developed CVD were more likely to be older, use insulin and cholesterol-lowering medication, have a family history of MI, have a history of hypertension, and have lower HDL cholesterol and higher HbA1c than those who remained free of CVD among both men and women. Among women, case patients had a longer duration of diabetes and had higher levels of LDL cholesterol and triglycerides than controls. Among men, case patients had higher Lp(a) levels, consumed less alcohol, and had a more frequency of aspirin use than controls.
Table 1.
Characteristics of the participantsa
| Women (NHS) |
Men (HPFS) |
|||
|---|---|---|---|---|
| CVD free | CVD cases | CVD free | CVD cases | |
| n | 998 | 333 | 663 | 344 |
| Lp(a) [median (range)] (mg/dL) | 7.4 (0.2–172.0) | 7.8 (0.2–112.8) | 7.5 (0.1–106.0) | 10.0 (0.1–119.7) |
| Age (years) | 56 ± 7 | 60 ± 6 | 61 ± 8 | 65 ± 8 |
| BMI (kg/m2) | 30.0 ± 5.6 | 30.2 ± 6.0 | 24.4 ± 10.7 | 23.0 ± 11.4 |
| Current smokers (%) | 11.4 | 16.0 | 6.8 | 5.8 |
| Alcohol (g/day) | 2.9 ± 7.0 | 2.6 ± 7.1 | 10.9 ± 15.2 | 8.3 ± 13.6 |
| Physical activity (MET-hours/week) | 13.0 ± 15.7 | 12.6 ± 16.0 | 27.9 ± 31.8 | 27.4 ± 30.5 |
| Aspirin use (%) | 46.9 | 48.7 | 38.1 | 53.0 |
| Insulin use (%) | 3.9 | 21.9 | 11.6 | 17.7 |
| Cholesterol-lowering medication use (%) | 3.2 | 9.0 | 9.8 | 18.4 |
| Postmenopausal hormone use (%) | 22.2 | 24.6 | — | — |
| Duration of type 2 diabetes (years) | 8.9 ± 8.7 | 10.9 ± 8.0 | 3.8 ± 3.0 | 4.2 ± 4.5 |
| History of hypertension (%) | 32.2 | 51.7 | 35.5 | 46.4 |
| Parental history of MI (%) | 23.9 | 30.9 | 34.4 | 40.9 |
| Fasting status (%) | 69.4 | 71.1 | 59.1 | 55.9 |
| Biomarkers | ||||
| LDL cholesterol (mg/dL) | 137 ± 38 | 148 ± 43 | 125 ± 38 | 126 ± 37 |
| HDL cholesterol (mg/dL) | 51 ± 15 | 48 ± 13 | 41 ± 11 | 38 ± 10 |
| Triglycerides (mg/dL) | 204 ± 164 | 236 ± 158 | 192 ± 99 | 202 ± 101 |
| HbA1c (%) | 6.6 ± 1.7 | 7.2 ± 1.7 | 7.1 ± 1.5 | 7.5 ± 1.7 |
aCharacteristics at 1990 for NHS and at 1994 for HPFS. Data are mean ± SD or %, unless indicated otherwise.
Genome-wide association scans for lipoprotein(a) levels in women and men with type 2 diabetes
The median of Lp(a) was 7.8 and 8.2 mg/dL in diabetic women and men, respectively. The SD of log-transformed Lp(a) was 1.4 in both men and women, which corresponds to about a 4.0-fold difference (e1.4) on the original scale of Lp(a) in mg/dL.
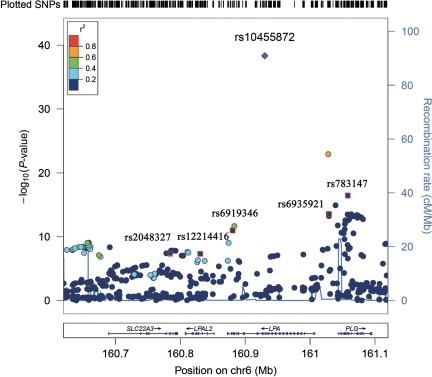
The combined analysis of GWA scans yielded 71 SNPs within or flanking 4 genes on chromosome 6 associated with plasma Lp(a) levels at a genome-wide significance level (P< 5 × 10−8) (see Supplementary material online, Figure S1 and Table S1). All top SNPs clustered within a ∼500 kb region on chromosome 6q, which harbours four genes including SLC22A, LPAL2, LPA, and PLG (Figure 1). The quantile–quantile plot (see Supplementary material online, Figure S2) suggests no evidence of systematic bias in the distribution of P-values for the analysis in both women (λ= 1.00) and men (λ= 0.999). The SNP rs10455872 in the LPA gene was most strongly associated with Lp(a) levels (P= 4.60 × 10−39), with an effect size (β) of 1.28 (95% CI: 1.08–1.48) for 1-SD log-transformed Lp(a) (Table 2), corresponding to approximately 6.0-fold higher (4.01.28) of usual Lp(a) levels per minor allele. Among these 71 SNPs, 16 SNPs (Table 2) representing distinct linkage disequilibrium (LD) blocks (r2 ≥ 0.80) were selected for further analyses on their independent associations with Lp(a) levels.
Figure 1.
Association signals in and adjacent to the LPA gene region on chromosome 6q. Single-nucleotide polymorphisms include all single-nucleotide polymorphisms (161 genotyped and 343 imputed single-nucleotide polymorphisms) within the region from 160 620 to 161 120 kb. The vertical axis representing the –log10 P-values. Recombination rates in this region are plotted in the background in blue. A pair-wise linkage disequilibrium between rs10455872 and other single-nucleotide polymorphisms was estimated using HapMap LD data. Six single-nucleotide polymorphisms labelled were selected from the forward-selection regression model.
Table 2.
Top single-nucleotide polymorphisms for plasma lipoprotein(a) levels among type 2 diabetes patientsa
| SNPs | Gene | Function | Position (kb)b | Alleles (+/−)c | NHS (n = 715) |
HPFS (n = 735) |
Combinedd |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| MAF | r2 e | βf | SE | P-valuef | MAF | r2 e | βf | SE | P-valuef | β | SE | P-value | |||||
| rs3120139 | SLC22A3 | Upstream | 160661.6 | G/A | 0.14 | 1 | 0.30 | 0.08 | 7.33E−05 | 0.13 | 1 | 0.33 | 0.07 | 1.38E−05 | 0.32 | 0.05 | 3.67E−09 |
| rs3120137 | SLC22A3 | Intron | 160691.2 | G/A | 0.15 | 1 | 0.27 | 0.08 | 0.00047 | 0.13 | 1 | 0.37 | 0.08 | 1.83E−06 | 0.32 | 0.05 | 4.37E−09 |
| rs2048327 | SLC22A3 | Intron | 160783.5 | T/C | 0.37 | 1 | 0.23 | 0.06 | 4.95E−05 | 0.34 | 1 | 0.20 | 0.06 | 0.00021 | 0.22 | 0.04 | 3.71E−08 |
| rs12214416 | LPAL2 | Intron | 160830.5 | T/A | 0.04 | 1 | −0.42 | 0.13 | 0.00148 | 0.04 | 0.998 | −0.58 | 0.13 | 6.36E−06 | −0.50 | 0.09 | 4.62E−08 |
| rs3124785 | LPA | Intron | 160874.5 | G/A | 0.28 | 0.993 | 0.17 | 0.06 | 0.00486 | 0.28 | 0.993 | 0.33 | 0.06 | 8.46E−09 | 0.25 | 0.04 | 9.24E−10 |
| rs6919346 | LPA | Intron | 160880.3 | C/T | 0.18 | 1 | −0.27 | 0.07 | 7.54E−05 | 0.18 | 1 | −0.36 | 0.06 | 2.31E−08 | −0.32 | 0.05 | 9.78E−12 |
| rs11751605 | LPA | Intron | 160883.2 | T/C | 0.16 | 0.945 | 0.31 | 0.08 | 8.24E−05 | 0.14 | 0.941 | 0.45 | 0.08 | 3.57E−09 | 0.38 | 0.05 | 2.17E−12 |
| rs10455872 | LPA | Intron | 160930.1 | A/G | 0.07 | 0.539 | 1.18 | 0.14 | 1.14E−15 | 0.07 | 0.545 | 1.36 | 0.13 | 3.00E−23 | 1.28 | 0.10 | 4.60E−39 |
| rs5014650 | LPA | Upstream | 161019.5 | G/A | 0.17 | 1 | 0.30 | 0.07 | 4.56E−05 | 0.16 | 1 | 0.27 | 0.07 | 6.68E−05 | 0.28 | 0.05 | 1.11E−08 |
| rs2315065 | LPA | Upstream | 161028.1 | C/A | 0.09 | 0.482 | 0.92 | 0.14 | 1.62E−10 | 0.08 | 0.451 | 1.08 | 0.14 | 3.64E−14 | 1.00 | 0.10 | 1.15E−23 |
| rs6935921 | PLG | Upstream | 161028.5 | T/C | 0.30 | 0.949 | −0.29 | 0.06 | 3.59E−07 | 0.30 | 0.935 | −0.33 | 0.06 | 1.99E−08 | −0.31 | 0.04 | 2.70E−14 |
| rs2314852 | PLG | Upstream | 161042.8 | A/G | 0.20 | 0.893 | −0.25 | 0.07 | 0.00032 | 0.21 | 0.877 | −0.43 | 0.07 | 3.72E−10 | −0.34 | 0.05 | 1.74E−12 |
| rs14224 | PLG | Synonymous | 161057.8 | T/C | 0.43 | 1 | 0.21 | 0.05 | 5.87E−05 | 0.43 | 1 | 0.22 | 0.05 | 9.07E−06 | 0.22 | 0.04 | 1.90E−09 |
| rs783147 | PLG | Intron | 161058.0 | G/A | 0.44 | 1 | −0.29 | 0.05 | 3.83E−08 | 0.45 | 1 | −0.31 | 0.05 | 3.10E−10 | −0.30 | 0.04 | 3.44E−17 |
| rs4252120 | PLG | Intron | 161063.6 | T/C | 0.29 | 1 | −0.28 | 0.06 | 6.37E−07 | 0.29 | 1 | −0.32 | 0.06 | 1.24E−08 | −0.30 | 0.04 | 2.95E−14 |
| rs1782627 | PLG | Downstream | 161116.8 | A/G | 0.44 | 0.978 | −0.22 | 0.05 | 2.24E−05 | 0.46 | 0.974 | −0.22 | 0.05 | 1.56E−05 | −0.22 | 0.04 | 1.24E−09 |
aLp(a) levels were first log-transformed and then scaled as SD units before analysis.
bPosition based on NCBI build 36.3.
c‘+’, major allele; ‘−’, minor allele.
dResults were combined using inverse variance weights (all P for heterogeneity > 0.05).
eMeasurement of SNP imputation quality.
fRegression coefficients and P-values were estimated as every one copy effect of the minor allele adjusted for age and BMI.
Forward-selection regression analysis yielded six SNPs that were independently associated with Lp(a) levels in both women and men (all P< 0.02): rs10455872 and rs6919346 (LPA), rs783147 and rs6919346 (PLG), rs2048327 (SLC22A3), and rs12214416 (LPAL2) (Table 3 and Figure 1). The best-associated SNP rs10455872 explained 3.33 and 3.74%, and the six SNPs jointly explained 18.3 and 21.2% of the total variation of plasma Lp(a) levels in women and men, respectively.
Table 3.
Multiple linear regression analysis of selected single-nucleotide polymorphisms in relation to plasma lipoprotein(a) levelsa
| Variable | Women (NHS) |
Men (HPFS) |
||||||
|---|---|---|---|---|---|---|---|---|
| β | SE | P-value | Variance explained (%) | β | SE | P-value | Variance explained (%) | |
| Age | 0.00 | 0.01 | 0.69 | 0.02 | 0.01 | 0.01 | 0.12 | 0.34 |
| BMI | −0.01 | 0.01 | 0.39 | 0.10 | −0.02 | 0.01 | 0.10 | 0.38 |
| rs10455872 | 0.81 | 0.17 | 1.04E−06 | 3.33 | 0.88 | 0.17 | 1.45E−07 | 3.74 |
| rs783147 | −0.40 | 0.07 | 1.22E−08 | 4.51 | −0.37 | 0.07 | 4.78E−08 | 4.03 |
| rs6935921 | −0.47 | 0.08 | 2.03E−09 | 4.89 | −0.49 | 0.08 | 2.72E−09 | 4.76 |
| rs2048327 | 0.28 | 0.09 | 0.001 | 1.51 | 0.22 | 0.08 | 0.011 | 0.89 |
| rs6919346 | −0.23 | 0.09 | 0.012 | 0.90 | −0.37 | 0.09 | 7.13E−05 | 2.15 |
| rs12214416 | −0.44 | 0.18 | 0.014 | 0.86 | −0.65 | 0.18 | 0.0002 | 1.85 |
aLp(a) levels were log-transformed before analysis.
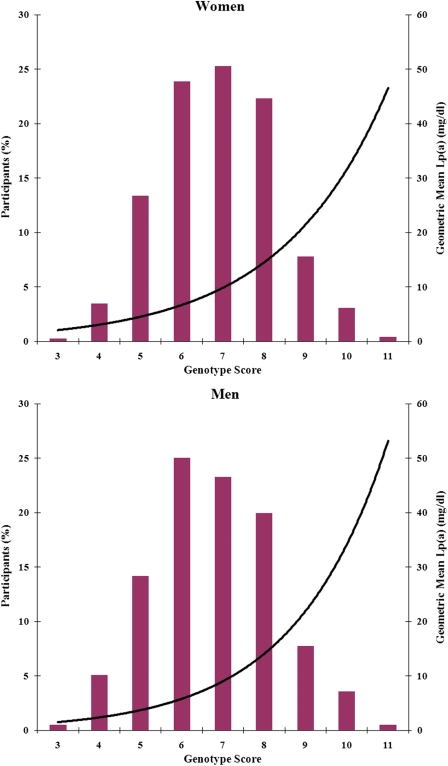
The genotype score calculated from these six SNPs ranged from 3 to 11, and plasma Lp(a) levels increased in a geometrical progression across the genotype score in both women and men (Figure 2). The estimated effect size (β) for 1-SD log-transformed Lp(a) was 0.29 (95% CI: 0.25–0.32), corresponding to approximately 1.5-fold higher (4.00.29) of usual Lp(a) levels per allele. Based on these SNPs, we also performed haplotype analysis for Lp(a) levels. None of these haplotypes showed additional stronger association with Lp(a) levels beyond the individual SNPs (see Supplementary material online, Table S2).
Figure 2.
Distribution of the genotype score and its association with plasma lipoprotein(a) levels in women and men with type 2 diabetes. Bars represent the percentage of participants. Black lines are trend lines of geometric means of plasma lipoprotein(a) across the genotype score.
Prospective analyses on plasma lipoprotein(a) levels and risk of cardiovascular disease and mortality
In the two cohorts of women and men with type 2 diabetes, we only found a marginal association between plasma Lp(a) levels and CVD incidence and mortality in age-adjusted models (Table 4). After further adjustment for lipids and other conventional risk factors, the RRs for CHD and CVD were reduced to 1.05 (95% CI: 0.95–1.15) and1.05 (0.96–1.15), respectively, while that for CVD death slightly increased to 1.21 (0.99–1.47) per 1-SD higher log-transformed Lp(a) levels. We also examined the associations by comparing extreme quintiles of Lp(a) levels and did not find any significant results. The corresponding RRs for CHD and CVD were 1.19 (0.88–1.62) and 1.15 (0.87–1.53), respectively.
Table 4.
Association of plasma lipoprotein(a) and genetic variants with cardiovascular disease incidence and mortality
| CHD |
CVD |
CVD death |
||||
|---|---|---|---|---|---|---|
| RR (95% CI) | P-value | RR (95% CI) | P-value | RR (95% CI) | P-value | |
| Plasma Lp(a)a | ||||||
| Age adjusted | ||||||
| NHS | 1.12 (0.98–1.26) | 0.09 | 1.10 (0.98–1.23) | 0.10 | 1.18 (0.95–1.47) | 0.14 |
| HPFS | 1.04 (0.90–1.20) | 0.63 | 1.04 (0.91–1.20) | 0.56 | 1.15 (0.83–1.58) | 0.40 |
| Combinedb | 1.08 (0.98–1.19) | 0.12 | 1.08 (0.98–1.18) | 0.10 | 1.17 (0.98–1.40) | 0.09 |
| Multivariate adjustedc | ||||||
| NHS | 1.12 (0.98–1.28) | 0.10 | 1.09 (0.97–1.23) | 0.14 | 1.25 (0.99–1.57) | 0.06 |
| HPFS | 0.97 (0.83–1.12) | 0.65 | 0.99 (0.85–1.14) | 0.85 | 1.09 (0.74–1.63) | 0.66 |
| Combinedb | 1.05 (0.95–1.16) | 0.35 | 1.05 (0.96–1.15) | 0.32 | 1.21 (0.99–1.47) | 0.06 |
| Genetic variantsd | ||||||
| SNP rs10455872 | ||||||
| NHS | 0.95 (0.61–1.47) | 0.81 | 0.99 (0.66–1.47) | 0.94 | 0.76 (0.39–1.51) | 0.43 |
| HPFS | 0.94 (0.61–1.46) | 0.79 | 0.95 (0.63–1.45) | 0.82 | 1.75 (0.98–3.13) | 0.06 |
| Combinedb | 0.94 (0.69–1.28) | 0.71 | 0.97 (0.73–1.30) | 0.84 | 1.23 (0.79–1.92) | 0.36 |
| Genotype score | ||||||
| NHS | 0.97 (0.88–1.07) | 0.51 | 0.97 (0.89–1.06) | 0.49 | 0.99 (0.87–1.14) | 0.96 |
| HPFS | 0.98 (0.89–1.08) | 0.67 | 1.00 (0.91–1.09) | 0.96 | 0.95 (0.83–1.08) | 0.42 |
| Combinedb | 0.98 (0.91–1.04) | 0.48 | 0.98 (0.91–1.06) | 0.69 | 0.97 (0.88–1.07) | 0.53 |
aThe RRs for plasma Lp(a) were estimated using Cox's regression.
bResults were combined between women and men using inverse variance weights under fixed model, as there was no heterogeneity between women and men (all P for heterogeneity > 0.16).
cMultivariate RR adjusted for age, fasting status, smoking, alcohol intake, physical activity, duration of diabetes, insulin use, aspirin use, cholesterol-lowering medication use, family history of MI, history of hypertension, BMI, LDL cholesterol, HDL cholesterol, triglycerides, A1C, and hormone replacement therapy use (women only).
dData are odds ratios for the genetic association analysis after adjustment for age and BMI.
Genetic variants and cardiovascular disease risk and mortality
Combined results in women and men showed that the most strongly Lp(a)-associated SNP rs10455872, as well as the genotype score, was not significantly associated with CHD, CVD, or CVD death (all P> 0.36) (Table 4). The other selected five SNPs were also not significantly associated with CVD risk or mortality (all P> 0.09) (see Supplementary material online, Table S3). Since a previous randomized trial reported that the use of low-dose aspirin attenuated the effect of LPA genetic variation on cardiovascular risk,30 we further examined the potential interactions in our study samples. However, we did not observed a significant interaction between aspirin use and individual LPA genetic variants or the genotype score on CHD or CVD risk (all P for interaction >0.07). In addition, we did not find any haplotypes that were significantly associated with these outcomes.
Comparison of the results in patients with type 2 diabetes and in the general populations
The regression coefficient of the SNP rs10455872 for 1-SD log Lp(a) levels in our study of diabetic patients was higher but not significantly different from that estimated in the general populations8 [1.28 (95% CI: 1.08–1.48) vs. 1.08 (1.02–1.13), P= 0.06]. However, the associations between this SNP and CHD risk showed a significant heterogeneity between the diabetic [OR= 0.94 (0.69–1.28)] and the general population9 [OR= 1.47 (1.35–1.60), P for heterogeneity= 0.006]. Assuming there was no difference in the association between genetic variants and CHD risk between the diabetic patients and general populations, our study had 89% power to detect the reported OR of 1.47.
Discussion
We performed comprehensive analyses combining GWA scans, plasma Lp(a) levels, and CVD risk in two prospective cohorts of type 2 diabetic patients. The present GWA scans identified the associations of common genetic variants in the SLC22A, LPAL2, LPA, and PLG loci with plasma Lp(a) levels. We found that different from the general population, the Lp(a) loci and plasma Lp(a) levels were not associated with CVD risk or mortality in patients with type 2 diabetes.
In our meta-analysis of two GWA scans in men and women with type 2 diabetes, SNP rs10455872 showed the strongest association with plasma Lp(a), consistent with the previous observations in the general populations.8 The SNP rs10455872 had similar effect size [1.28 vs. 1.08 for 1-SD log-transformed Lp(a)] on plasma Lp(a) levels in the diabetic and general populations. We found that other five SNPs in LPA or nearby genes (SLC22A, LPAL2, and PLG), which were not in LD with rs10455872, were also independently associated with plasma Lp(a) levels; and the genotype score calculated based on these six SNPs showed a cumulative effect on plasma Lp(a) levels. Consistent with our results, a recent GWA analysis in 386 Hutterites identified multiple SNPs with weak LD (r2< 0.3) in at least six loci including LPA and PLG gene independently associated with plasma Lp(a) levels.4 In addition, Trégouët et al.5 reported that the haplotypes in the SLC22A3–LPAL2–LPA gene cluster were associated with plasma Lp(a) levels. These findings suggest that besides the LPA locus, several additional loci close to the LPA locus may confer independent genetic effects on plasma Lp(a) levels.
Despite the high consistency of the strongest association between the SNP rs10455872 and Lp(a) levels in our diabetic population and the general population, we did not find a significant association of this variant with CVD risk. In the general population, the SNP rs10455872 showed a significant association with CHD risk with OR of 1.47 (95% CI: 1.35–1.60).8 Our study had 89% statistical power to detect such an effect size, assuming that the association is the same between the diabetic and the general population. We noted that the SNP rs10455872 accounted for much smaller proportion of the variation of plasma Lp(a) levels in patients with type 2 diabetes (∼3–4%) compared with that in the general population (∼25%).8 This may partly explain the null association between this SNP and CVD risk in the diabetic population. However, the genotype score calculated based on the six SNPs which jointly explained ∼20% variation of plasma Lp(a) levels also was not associated with CVD risk. We posit that the diabetes-specific metabolic profile may attenuate the genetic effects on cardiovascular risk.31
A large body of evidence suggests independent association between elevated plasma Lp(a) levels and increased CVD risk in the general population.6 However, several prospective studies have examined the relationships between plasma Lp(a) levels and cardiovascular risk among patients with type 2 diabetes and reported conflicting results (see Supplementary material online, Table S4).13–18 The inconsistent results from these studies were probably due to confounding from environmental factors and differences in study design (i.e. follow-up years and case numbers), population characteristics (i.e. age, sex, and ethnicity), disease status (i.e. duration of diabetes), or the Lp(a) assay method used. Shai et al.17 previously reported a borderline association between Lp(a) levels and CHD (P for trend= 0.035) in diabetic women from the NHS. In the current analysis, the association became attenuated after inclusion of the new events from the extension of follow-up. This might be due to the weakening of this association after longer follow-up, which is supported by the similar observations in the studies of other biomarkers and CVD risk.32–34 We did not observe a significant association with CVD risk or mortality in diabetic men with 12 years of follow-up and combined analysis only yielded a marginal association with CVD mortality. Of note, compared with these observational studies which might be affected by confounding, the genetic association analysis may minimize some of the study bias.35,36 In line with the current and three previous observational studies,13,15,18 we also did not find any association between the genetic variants for Lp(a) and CHD or CVD risk in both men (HPFS) and women (NHS). Furthermore, our data indicate a significant heterogeneity in the associations between the Lp(a) associated genetic variant and CHD risk between the diabetic and the general populations.8 Such observations are in line with a previous study that reported that Lp(a) was a strong and independent predictor of future cardiovascular events in non-diabetic patients, but not in patients with type 2 diabetes, with a significant interaction between diabetes and Lp(a) (P= 0.008).18 Overall, these findings suggest that Lp(a) might contribute less to cardiovascular risk in patients with type 2 diabetes than in the general population.
Previous studies have shown that several major pathophysiological features of type 2 diabetes such as hyperglycaemia,37–39 insulin resistance,40 and dyslipidaemia (i.e. high triglycerides and low HDL cholesterol levels)15,37–39,41 are also risk factors for CHD/CVD among patients with type 2 diabetes. However, Lp(a) has been inversely associated with triglycerides42 and insulin and 2 h glucose,43 and a lower Lp(a) level was also observed in patients with type 2 diabetes.44 Very recently, Mora et al.12 reported that plasma Lp(a) was inversely associated with risk of type 2 diabetes in the Women's Health Study (26 746 healthy US women for 13-year follow-up), with confirmation of their findings in a general population of Danish men and women. The intriguing opposite effects of Lp(a) on type 2 diabetes and CVD indicate a more complicated role of Lp(a) in cardiometabolic diseases. Thus, Lp(a) might not be a relevant marker in the assessment of CVD risk among diabetic patients. Our data do not support Lp(a) lowering as a treatment for reduction in cardiovascular risk among diabetic patients.
The major strengths of our study include high-quality genotype data, careful quality control, and minimized population stratification in GWA scans,27 and simultaneous analysis of genetic variants, plasma Lp(a) levels, and cardiovascular risk in two well-established cohorts with similar study design. We acknowledged that plasma Lp(a) isoforms and KIV-2 copy number were not measured in the current study. However, previous studies have shown that LPA SNPs are in LD with the KIV-2 copy number.4,8,45 In addition, the participants included in this study are Caucasians of European ancestry, while plasma Lp(a) levels vary between different ethnic groups.46
In conclusion, our GWA scans in diabetic men and women identified common genetic variants in the SLC22A, LPAL2, LPA, and PLG loci independently associated with plasma Lp(a) levels. We found that Lp(a) genetic markers and plasma levels were not significantly associated with CVD risk in diabetic patients. Our data indicate a significant heterogeneity in the associations of Lp(a) and CVD risk between the diabetic patients and the general population and suggest that diabetes status may attenuate the effect of Lp(a) on CVD risk.
Supplementary material
Supplementary material is available at European Heart Journal online.
Funding
This study was supported by grant HL71981 from the National Institutes of Health, DK46200, from the Boston Obesity Nutrition Research Center. L.Q. was a recipient of the American Heart Association Scientist Development Award (0730094N). C.Z. was supported by the Intramural Research Program of the Eunice Kennedy Shriver National Institute of Child Health and Human Development, National Institutes of Health.
Conflict of interest: none declared.
Supplementary Material
Acknowledgements
We thank all the participants of the NHS and the HPFS for their continued cooperation.
References
- 1.Hobbs HH, White AL. Lipoprotein(a): intrigues and insights. Curr Opin Lipidol. 1999;10:225–236. doi: 10.1097/00041433-199906000-00005. [DOI] [PubMed] [Google Scholar]
- 2.Berglund L, Ramakrishnan R. Lipoprotein(a): an elusive cardiovascular risk factor. Arterioscler Thromb Vasc Biol. 2004;24:2219–2226. doi: 10.1161/01.ATV.0000144010.55563.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Danesh J, Collins R, Peto R. Lipoprotein(a) and coronary heart disease: meta-analysis of prospective studies. Circulation. 2000;102:1082–1085. doi: 10.1161/01.cir.102.10.1082. [DOI] [PubMed] [Google Scholar]
- 4.Ober C, Nord AS, Thompson EE, Pan L, Tan Z, Cusanovich D, Sun Y, Nicolae R, Edelstein C, Schneider DH, Billstrand C, Pfaffinger D, Phillips N, Anderson RL, Philips B, Rajagopalan R, Hatsukami TS, Rieder MJ, Heagerty PJ, Nickerson DA, Abney M, Marcovina S, Jarvik GP, Scanu AM, Nicolae DL. Genome-wide association study of plasma lipoprotein(a) levels identifies multiple genes on chromosome 6q. J Lipid Res. 2009;50:798–806. doi: 10.1194/jlr.M800515-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Trégouët D-A, Konig IR, Erdmann J, Munteanu A, Braund PS, Hall AS, Groszhennig A, Linsel-Nitschke P, Perret C, DeSuremain M, Meitinger T, Wright BJ, Preuss M, Balmforth AJ, Ball SG, Meisinger C, Germain C, Evans A, Arveiler D, Luc G, Ruidavets J-B, Morrison C, van der Harst P, Schreiber S, Neureuther K, Schafer A, Bugert P, El Mokhtari NE, Schrezenmeir J, Stark K, Rubin D, Wichmann HE, Hengstenberg C, Ouwehand W, Ziegler A, Tiret L, Thompson JR, Cambien F, Schunkert H, Samani NJ. Genome-wide haplotype association study identifies the SLC22A3–LPAL2–LPA gene cluster as a risk locus for coronary artery disease. Nat Genet. 2009;41:283–285. doi: 10.1038/ng.314. [DOI] [PubMed] [Google Scholar]
- 6.Erqou S, Kaptoge S, Perry PL, Di AE, Thompson A, White IR, Marcovina SM, Collins R, Thompson SG, Danesh J. Lipoprotein(a) concentration and the risk of coronary heart. JAMA. 2009;302:412–423. doi: 10.1001/jama.2009.1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kamstrup PR, Tybjaerg-Hansen A, Steffensen R, Nordestgaard BG. Genetically elevated lipoprotein(a) and increased risk of myocardial infarction. JAMA. 2009;301:2331–2339. doi: 10.1001/jama.2009.801. [DOI] [PubMed] [Google Scholar]
- 8.Clarke R, Peden JF, Hopewell JC, Kyriakou T, Goel A, Heath SC, Parish S, Barlera S, Franzosi MG, Rust S, Bennett D, Silveira A, Malarstig A, Green FR, Lathrop M, Gigante B, Leander K, de Faire U, Seedorf U, Hamsten A, Collins R, Watkins H, Farrall M. Genetic variants associated with lp(a) lipoprotein level and coronary disease. N Engl J Med. 2009;361:2518–2528. doi: 10.1056/NEJMoa0902604. [DOI] [PubMed] [Google Scholar]
- 9.Nordestgaard BG, Chapman MJ, Ray K, Boren J, Andreotti F, Watts GF, Ginsberg H, Amarenco P, Catapano A, Descamps OS, Fisher E, Kovanen PT, Kuivenhoven JA, Lesnik P, Masana L, Reiner Z, Taskinen MR, Tokgozoglu L, Tybjaerg-Hansen A. Lipoprotein(a) as a cardiovascular risk factor: current status. Eur Heart J. 2010;31:2844–2853. doi: 10.1093/eurheartj/ehq386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Beckman JA, Creager MA, Libby P. Diabetes and atherosclerosis. JAMA. 2002;287:2570–2581. doi: 10.1001/jama.287.19.2570. [DOI] [PubMed] [Google Scholar]
- 11.Almdal T, Scharling H, Jensen JS, Vestergaard H. The independent effect of type 2 diabetes mellitus on ischemic heart disease, stroke, and death: a population-based study of 13 000 men and women with 20 years of follow-up. Arch Intern Med. 2004;164:1422–1426. doi: 10.1001/archinte.164.13.1422. [DOI] [PubMed] [Google Scholar]
- 12.Mora S, Kamstrup PR, Rifai N, Nordestgaard BG, Buring JE, Ridker PM. Lipoprotein(a) and risk of type 2 diabetes. Clin Chem. 2010;56:1252–1260. doi: 10.1373/clinchem.2010.146779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Haffner SM, Moss SE, Klein BE, Klein R. Lack of association between lipoprotein(a) concentrations and coronary heart disease mortality in diabetes: the Wisconsin Epidemiologic Study of Diabetic Retinopathy. Metabolism. 1992;41:194–197. doi: 10.1016/0026-0495(92)90152-z. [DOI] [PubMed] [Google Scholar]
- 14.Hiraga T, Kobayashi T, Okubo M, Nakanishi K, Sugimoto T, Ohashi Y, Murase T. Prospective study of lipoprotein(a) as a risk factor for atherosclerotic cardiovascular disease in patients with diabetes. Diabetes Care. 1995;18:241–244. doi: 10.2337/diacare.18.2.241. [DOI] [PubMed] [Google Scholar]
- 15.Abu-Lebdeh HS, Hodge DO, Nguyen TT. Predictors of macrovascular disease in patients with type 2 diabetes mellitus. Mayo Clin Proc. 2001;76:707–712. doi: 10.4065/76.7.707. [DOI] [PubMed] [Google Scholar]
- 16.Hernández C, Francisco G, Chacón P, Simó R. Lipoprotein(a) as a risk factor for cardiovascular mortality in type 2 diabetic patients. Diabetes Care. 2005;28:931–933. doi: 10.2337/diacare.28.4.931. [DOI] [PubMed] [Google Scholar]
- 17.Shai I, Schulze M, Manson J, Stampfer M, Rifai N, Hu F. A prospective study of lipoprotein(a) and risk of coronary heart disease among women with type 2 diabetes. Diabetologia. 2005;48:1469–1476. doi: 10.1007/s00125-005-1814-3. [DOI] [PubMed] [Google Scholar]
- 18.Saely CH, Koch L, Schmid F, Marte T, Aczel S, Langer P, Hoefle G, Drexel H. Lipoprotein(a), type 2 diabetes and vascular risk in coronary patients. Eur J Clin Invest. 2006;36:91–97. doi: 10.1111/j.1365-2362.2006.01604.x. [DOI] [PubMed] [Google Scholar]
- 19.Colditz GA, Manson JE, Hankinson SE. The Nurses' Health Study: 20-year contribution to the understanding of health among women. J Womens Health. 1997;6:49–62. doi: 10.1089/jwh.1997.6.49. [DOI] [PubMed] [Google Scholar]
- 20.Rimm EB, Giovannucci EL, Willett WC, Colditz GA, Ascherio A, Rosner B, Stampfer MJ. Prospective study of alcohol consumption and risk of coronary disease in men. Lancet. 1991;338:464–468. doi: 10.1016/0140-6736(91)90542-w. [DOI] [PubMed] [Google Scholar]
- 21.National Diabetes Data Group. Classification and diagnosis of diabetes mellitus and other categories of glucose intolerance. Diabetes. 1979;28:1039–1057. doi: 10.2337/diab.28.12.1039. [DOI] [PubMed] [Google Scholar]
- 22.Manson JE, Colditz GA, Stampfer MJ, Willett WC, Krolewski AS, Rosner B, Arky RA, Speizer FE, Hennekens CH. A prospective study of maturity-onset diabetes mellitus and risk of coronary heart disease and stroke in women. Arch Intern Med. 1991;151:1141–1147. [PubMed] [Google Scholar]
- 23.American Diabetes Association. Report of the expert committee on the diagnosis and classification of diabetes mellitus. Diabetes Care. 1997;20:1183–1197. doi: 10.2337/diacare.20.7.1183. [DOI] [PubMed] [Google Scholar]
- 24.Walker AE, Robins M, Weinfeld FD. The National Survey of Stroke. Clinical findings. Stroke. 1981;12:I13–I44. [PubMed] [Google Scholar]
- 25.Qi L, Li T, Rimm E, Zhang C, Rifai N, Hunter D, Doria A, Hu FB. The +276 polymorphism of the apm1 gene, plasma adiponectin concentration, and cardiovascular risk in diabetic men. Diabetes. 2005;54:1607–1610. doi: 10.2337/diabetes.54.5.1607. [DOI] [PubMed] [Google Scholar]
- 26.Reblin T, Rader DJ, Beisiegel U, Greten H, Brewer HB., Jr Correlation of apolipoprotein(a) isoproteins with Lp(a) density and distribution in fasting plasma. Atherosclerosis. 1992;94:223–232. doi: 10.1016/0021-9150(92)90247-e. [DOI] [PubMed] [Google Scholar]
- 27.Qi L, Cornelis MC, Kraft P, Stanya KJ, Linda Kao WH, Pankow JS, Dupuis J, Florez JC, Fox CS, Paré G, Sun Q, Girman CJ, Laurie CC, Mirel DB, Manolio TA, Chasman DI, Boerwinkle E, Ridker PM, Hunter DJ, Meigs JB, Lee CH, Hu FB, van Dam RM. Genetic variants at 2q24 are associated with susceptibility to type 2 diabetes. Hum Mol Genet. 2010;19:2706–2715. doi: 10.1093/hmg/ddq156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Aulchenko Y, Struchalin M, van Duijn C. ProbABEL package for genome-wide association analysis of imputed data. BMC Bioinformatics. 2010;11:134. doi: 10.1186/1471-2105-11-134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tregouet DA, Escolano S, Tiret L, Mallet A, Golmard JL. A new algorithm for haplotype-based association analysis: the Stochastic-EM algorithm. Ann Hum Genet. 2004;68:165–177. doi: 10.1046/j.1529-8817.2003.00085.x. [DOI] [PubMed] [Google Scholar]
- 30.Chasman DI, Shiffman D, Zee RYL, Louie JZ, Luke MM, Rowland CM, Catanese JJ, Buring JE, Devlin JJ, Ridker PM. Polymorphism in the apolipoprotein(a) gene, plasma lipoprotein(a), cardiovascular disease, and low-dose aspirin therapy. Atherosclerosis. 2009;203:371–376. doi: 10.1016/j.atherosclerosis.2008.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Doria A, Wojcik J, Xu R, Gervino EV, Hauser TH, Johnstone MT, Nolan D, Hu FB, Warram JH. Interaction between poor glycemic control and 9p21 locus on risk of coronary artery disease in type 2 diabetes. JAMA. 2008;300:2389–2397. doi: 10.1001/jama.2008.649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hatoum IJ, Hu FB, Nelson JJ, Rimm EB. Lipoprotein-associated phospholipase a2 activity and incident coronary heart disease among men and women with type 2 diabetes. Diabetes. 2010;59:1239–1243. doi: 10.2337/db09-0730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Danesh J, Wheeler JG, Hirschfield GM, Eda S, Eiriksdottir G, Rumley A, Lowe GDO, Pepys MB, Gudnason V. C-reactive protein and other circulating markers of inflammation in the prediction of coronary heart disease. N Engl J Med. 2004;350:1387–1397. doi: 10.1056/NEJMoa032804. [DOI] [PubMed] [Google Scholar]
- 34.Tiret L, Godefroy T, Lubos E, Nicaud V, Tregouet D-A, Barbaux S, Schnabel R, Bickel C, Espinola-Klein C, Poirier O, Perret C, Munzel T, Rupprecht H-J, Lackner K, Cambien F, Blankenberg S for the AtheroGene I. Genetic analysis of the interleukin-18 system highlights the role of the interleukin-18 gene in cardiovascular disease. Circulation. 2005;112:643–650. doi: 10.1161/CIRCULATIONAHA.104.519702. [DOI] [PubMed] [Google Scholar]
- 35.Hunter DJ, Altshuler D, Rader DJ. From Darwin's finches to canaries in the coal mine—mining the genome for new biology. N Engl J Med. 2008;358:2760–2763. doi: 10.1056/NEJMp0804318. [DOI] [PubMed] [Google Scholar]
- 36.Sheehan NA, Didelez V, Burton PR, Tobin MD. Mendelian randomisation and causal inference in observational epidemiology. PLoS Med. 2008;5:e177. doi: 10.1371/journal.pmed.0050177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lehto S, Ronnemaa T, Pyorala K, Laakso M. Predictors of stroke in middle-aged patients with non-insulin-dependent diabetes. Stroke. 1996;27:63–68. doi: 10.1161/01.str.27.1.63. [DOI] [PubMed] [Google Scholar]
- 38.Lehto S, Ronnemaa T, Haffner SM, Pyorala K, Kallio V, Laakso M. Dyslipidemia and hyperglycemia predict coronary heart disease events in middle-aged patients with NIDDM. Diabetes. 1997;46:1354–1359. doi: 10.2337/diab.46.8.1354. [DOI] [PubMed] [Google Scholar]
- 39.Turner RC, Millns H, Neil HA, Stratton IM, Manley SE, Matthews DR, Holman RR. Risk factors for coronary artery disease in non-insulin dependent diabetes mellitus: United Kingdom Prospective Diabetes Study (UKPDS: 23) BMJ. 1998;316:823–828. doi: 10.1136/bmj.316.7134.823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Saely CH, Aczel S, Marte T, Langer P, Hoefle G, Drexel H. The metabolic syndrome, insulin resistance, and cardiovascular risk in diabetic and nondiabetic patients. J Clin Endocrinol Metab. 2005;90:5698–5703. doi: 10.1210/jc.2005-0799. [DOI] [PubMed] [Google Scholar]
- 41.Drexel H, Aczel S, Marte T, Benzer W, Langer P, Moll W, Saely CH. Is atherosclerosis in diabetes and impaired fasting glucose driven by elevated LDL cholesterol or by decreased HDL cholesterol? Diabetes Care. 2005;28:101–107. doi: 10.2337/diacare.28.1.101. [DOI] [PubMed] [Google Scholar]
- 42.Hernández C, Chacón P, García-Pascual L, Simó R. Differential influence of LDL cholesterol and triglycerides on lipoprotein(a) concentrations in diabetic patients. Diabetes Care. 2001;24:350–355. doi: 10.2337/diacare.24.2.350. [DOI] [PubMed] [Google Scholar]
- 43.Rainwater DL, Haffner SM. Insulin and 2-h glucose levels are inversely related to lp(a) concentrations controlled for LPA genotype. Arterioscler Thromb Vasc Biol. 1998;18:1335–1341. doi: 10.1161/01.atv.18.8.1335. [DOI] [PubMed] [Google Scholar]
- 44.Rainwater DL, MacCluer JW, Stern MP, VandeBerg JL, Haffner SM. Effects of NIDDM on lipoprotein(a) concentration and apolipoprotein(a) size. Diabetes. 1994;43:942–946. doi: 10.2337/diab.43.7.942. [DOI] [PubMed] [Google Scholar]
- 45.Lanktree MB, Anand SS, Yusuf S, Hegele RA on behalf of the SI. Comprehensive analysis of genomic variation in the LPA locus and its relationship to plasma lipoprotein(a) in South Asians, Chinese, and European Caucasians. Circ Cardiovasc Genet. 2010;3:39–46. doi: 10.1161/CIRCGENETICS.109.907642. [DOI] [PubMed] [Google Scholar]
- 46.Karen AM, Mary Fran S, Carol AD, Evan S, Heidi M-M, Sybil LC, Richard CP. Ethnic differences in cardiovascular risk factor burden among middle-aged women: Study of Women's Health Across the Nation (SWAN) Am Heart J. 2005;149:1066–1073. doi: 10.1016/j.ahj.2004.08.027. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.