Abstract
Loss of imprinting (LOI) of IGF2 is a common event in many cancers and typically activates the maternally silenced allele. The resulting biallelic IGF2 expression correlates strongly with the hypomethylation of a differentially methylated region (DMR) near its promoter. It has also been shown that IGF2 undergoes overexpression in human malignancies; nevertheless, this phenomenon and its link to aberrant DMR methylation has not been reported in colorectal cancer (CRC). The aim of this study was to determine the relationship between IGF2 LOI, overexpression and DMR hypomethylation in CRC. By analyzing IGF2 and H19 methylation in 97 primary CRC and 64 matched normal colorectal tissues, we have shown a significant correlation between IGF2 LOI and DMR hypomethylation of IGF2 and H19. Additionally, when analyzing Affymetrix expression data of 167 primary CRC tumor and 32 normal tissues, 15% of tumors showed marked IGF2 elevation. We further investigated if substantially elevated IGF2 levels were linked to IGF2 or H19 hypomethylation, but found no significant correlation. However, we demonstrated that noticeable IGF2 overexpression, rather than LOI, negatively correlated with CRC microsatellite instability. These observations indicate that IGF2 expression, particularly when transcribed at significantly high levels, is a result of mechanisms unrelated to LOI. Our results suggest that IGF2 participates in CRC tumorigenesis through two different forms of aberrant gene expression.
Keywords: CpG methylation, LDR, colorectal cancer, IGF2, imprinting
INTRODUCTION
Epigenetic alternations describe heritable changes in gene function that is not reflective of changes in the DNA sequence. Altered genomic imprinting and aberrant DNA methylation are two examples of epigenetic events that are hallmarks of human cancers. An imprinted gene is typically expressed in a monoallelic fashion from one of the parental alleles, whereas loss of imprinting (LOI) describes mechanisms that either activates the normally silenced allele or inactivates the normally active gene copy.
LOI was initially discovered at the insulin-like growth factor 2 (IGF2) locus of Wilms tumor (WT) and has subsequently been reported to be present in a wide range of human malignancies1, 2. IGF2 is paternally expressed and is an important regulator of cell growth and apoptosis. LOI of IGF2 is generally manifested by the activation of the normally silenced maternal allele with the subsequent expression of both gene copies3. The biallelic IGF2 expression also correlates with aberrant IGF2 / H19 methylation. Studies have shown that IGF2 LOI is linked to the hypomethylation of a differentially methylated region (DMR) close to the IGF2 promoter, as well as linked to hypermethylation of a DMR upstream of the reciprocally imprinted H19 gene4–9. The H19 DMR contains six zinc finger CCCTC transcription factor (CTCF) binding sites (CBS) where their methylation levels reciprocally correlate with IGF2 DMR in various cancers, including kidney, ovarian, bladder and lung1, 2, 10–14. In a normal imprinting state, CTCF binding on the hypomethylated H19 DMR prevents a downstream enhancer from accessing the promoter of silenced maternal IGF2 allele. However, LOI of IGF2 is believed to occur through H19 DMR hypermethylation and the alleviation of CTCF shielding results in the enhancer mediated biallelic expression 11, 15–17.
In CRC, LOI of IGF2 occurs in 30% of tumors and 10% of non-cancerous tissues18. This phenomenon in CRC was originally thought to share the same enhancer competition model as described above6. However, Cui and colleagues have shown that IGF2 and H19 DMRs in CRC do not follow a reciprocal methylation pattern, suggesting LOI of IGF2 in CRC associated with H19 and IGF2 DMR hypomethylation4. These observations suggest that the enhancer competition model of IGF2 LOI may not apply to CRC, and that aberrant IGF2 promoter methylation is more likely to play a critical role in LOI mediated IGF2 expression.
The expression level of IGF2 in which the tumor has undergone LOI at this locus is typically 2–3 times higher than in corresponding normal tissues19. However, IGF2 overexpression at tumor-to-normal ratios of 10 times or higher is not unusual in human maligancies5, 20. Highly elevated IGF2 expression is commonly seen in ovarian cancer and correlated with hypermethylation of CTCF binding site at H19 DMR5. Using quantitative methods to study epigenetic and genetic abnormalities, we were interested in determining whether significant IGF2 overexpression occurs in CRC and if it is related to LOI and other genetic abnormalities. We also investigated if aberrant IGF2/H19 DMR methylation may explain these types of altered IGF2 expression in CRC.
MATERIAL AND METHODS
Study population
A detailed description of the study population was published previously21. Briefly, it included patients who presented at Memorial Sloan-Kettering Cancer Center (MSKCC) with a colonic neoplasm between 1992 and 2004. Location of the primary colon tumor varied and included right, left, and rectal cancers. Tumor samples were spread among stages I–IV (AJCC criteria) in 17%, 24%, 24% and 35%, respectively. Ages ranged from 17 years old to 86 years old with a median age of 65. Biological specimens used in the study included colorectal adenomas, primary colon adenocarcinomas and corresponding normal mucosa. All tissues were procured at the time of surgical resection and stored for future use under institutional IRB approved protocols.
Identification of IGF2 polymorphism and LOI status
Genomic DNAs extracted from CRC specimen were genotyped for two SNPs (820 G/A, refSNP ID: rs680 and 266 C/T, refSNP ID: rs2230949) located in IGF2 exon 9 using direct sequencing22. For all the tumor samples identified with heterozygous alleles, genomic DNAs from matched adjacent normal mucosas were also sequenced. Cases were scored as informative when the heterozygosity at ploymorphic sites presented in both tumor and matched normal tissues. All primer sequences and reaction conditions for Taqman assay were reported previously22. For the analysis of IGF2 LOI status, total RNA from frozen tissues was extracted and RT-PCR was carried out to amplify the IGF2 exon 9 region bearing SNP 820 G/A for LDR analysis. Complementary DNA (cDNA) was synthesized from 1µg total RNA using Multiscribe Reverse Transcriptase. A negative control reaction was prepared in parallel without reverse transcriptase to ensure the elimination of genomic DNA in cDNA synthesis. PCR was performed using the following protocol: 94 °C for 10 min, 35 cycles of 94 °C for 30 sec, 60 °C for 30 sec and 72 °C for 1 min, followed by a final extension step at 72 °C for 3 min. Inactivation of the polymerase was achieved by a thermocycled proteinase K reaction. The LDR was conducted in a final volume of 20 µl consisting of 20 mM Tris-HCl pH 7.6, 10 mM MgCl2, 100 mM KCl, 10 mM DTT, 1 mM NAD, 500 – 800 fmol each of the LDR primers, 40U of Taq DNA Ligase, and 2 µl of PCR amplicon. The reaction mixture was thermocycled with the following program: 94° C for 1.5 min followed by 20 cycles of 94° C for 1 min and 65° C for 4 min. The fluorescence intensity of LDR products was analyzed using ABI 3730 capillary DNA analyzer (Applied Biosystems, Foster City, CA). The sequences of PCR and LDR primers were shown in the Supplementary Table 1.
IGF2 and H19 DMR methylation status
DNA methylation analysis using bisulfite/PCR/LDR/Universal Array has been described previously23. Briefly, genomic DNAs were bisulfite treated using EZ DNA methylation kit (Zymo Research, CA). This method was found to yield greater than 99% C to U conversion. Two segments of the IGF2 DMR located upstream of exon 3, one segment each of H19 CBS1 and CBS6 DMRs were subsequently multiplex PCR amplified. Three CpG sites were analyzed for each amplified fragment. LDR was also conducted in a multiplex fashion allowing all 36 LDR primers interrogating 12 CpG sites in one reaction. The final LDR products were resolved and displayed on a Universal Array. The raw methylation levels of the interrogated cytosines were calculated by the ratio of Cy3/(Cy3+Cy5). The sequences of PCR and LDR primers are shown in the Supplementary Table 1.
Determination of KRAS, BRAF mutation and MSI status
Methods to measure DNA mutations and MSI status were performed as previously reported21, 24. Briefly, KRAS and BRAF mutations were detected using PCR/LDR approaches. Fluorescently labeled LDR primers were designed to detect the 7 common KRAS (Val, Asp, Ala, Arg, Ser, and Cys at codon 12 and Asp at codon 13) and BRAF V600E mutations. For MSI study, fluorescently labeled oligonucleotide primers were designed to analyze microsatellite loci of BAT25, BAT26, D2S123, D5S346, and D17S250. MSI was scored as present when at least two of the five markers showed size instability.
Statistical analysis
The COPA analysis was performed using a R package25. Statistical comparisons were performed by either Student’s t-test or Wilcoxon signed-rank test. The correlation studies were performed by computing the chi-square or Fisher’s exact (two-sided) tests of the n × m contingency tables. P values of less than 0.05 were considered significant. Multiple testing correction was performed using the Benjamini-Hochberg method to find those tests with a false discovery rate (FDR) of at most 5%.
RESULTS
IGF2 and H19 were hypomethylated in CRC
We measured DNA methylation status in the CBS1 and CBS6 regions of H19 and the imprinting control region of IGF2 using a quantitative bisulfite/PCR/LDR/Universal Array assay (Figure 1A and 1B)23. This approach provides unbiased amplification of a given genomic region with both methylated and unmethylated DNA sequences. Loci of IGF2 and H19 DMRs were amplified and the methylation levels of a total of 12 CpG sites were analyzed. As shown in Figure 1C and Supplementary Figure 1, the calibration curves for each CpG methylation level were established by mixing synthetic DNA templates with methylated and unmethylated sequences in the ratios of 0:5, 1:4, 2:3, 3:2, 4:1 and 5:0 followed by bisulfite conversion, PCR amplification and LDR/Universal microarray analysis. The linearity of these curves demonstrates the quantitative ability of our assay. The overall methylation status of a DMR was determined by averaging the methylation level of individual cytosines detected at that locus. By equally weighting the methylation levels of the queried cytosines, the overall DMR or promoter methylation status of a candidate gene may be represented by multiple CpG sites across a relatively larger region of DNA sequences. The cytosine methylation level of each interrogated CG dinucleotide was also confirmed using bisulfite sequencing in a subset of samples (data not shown).
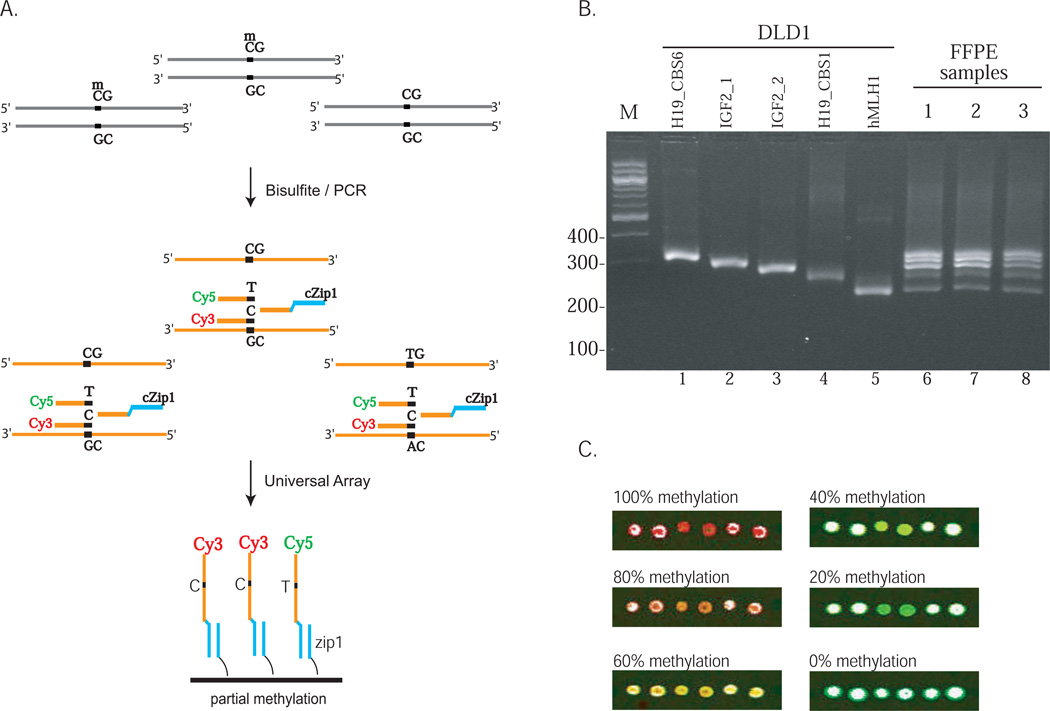
Figure 1.
Quantitative analysis of aberrant DNA methylation. (A) Schematic diagram of the bisulfite/PCR/LDR/Universal Array assay. The diagram depicts a hypothetical scenario in which the methylation levels of three copies of the same genomic locus were analyzed. After bisulfite treatment, only top-strand DNAs were PCR amplified and shown. Three LDR primers were designed to analyze the methylation status of each cytosine. Two upstream primers were fluorescently labeled and the downstream primer was tagged with a zipcode complementary sequence. LDR products with the same complementary zipcode tags were captured onto a specific array address. LDR/Universal Array is able to identify the 2:1 ratio of the methylated vs. unmethylated DNA fragments. (B) Multiplex amplification of the interrogated genomic loci. Genomic DNAs extracted from CRC cell line DLD1 (lanes 1–5) and formalin-fixed, paraffin-embedded (FFPE) tissues (lanes 6–8) were PCR amplified after bisulfite treatment and resolved on an agarose gel. The primer and assay designs were suitable for multiplex analysis of genomic DNAs extracted from FFPE tissues. The hMLH1 was shown for the demonstration of multiplex PCR and was not further investigated in this study. (C) Examples of the quantitative determination of IGF2 DMR methylation. Different amount of synthetic methylated and unmethylated DNAs were mixed and subjected to bisulfite/PCR/LDR/Universal Array analysis. The red-green color scale represents the percentage of methylation levels at each CpG dinucleotide from high to low determined by the fluorescence intensity ratio of LDR products23. The zipcode oligos corresponding to each interrogated CpG sites were double spotted to increase the detection accuracy.
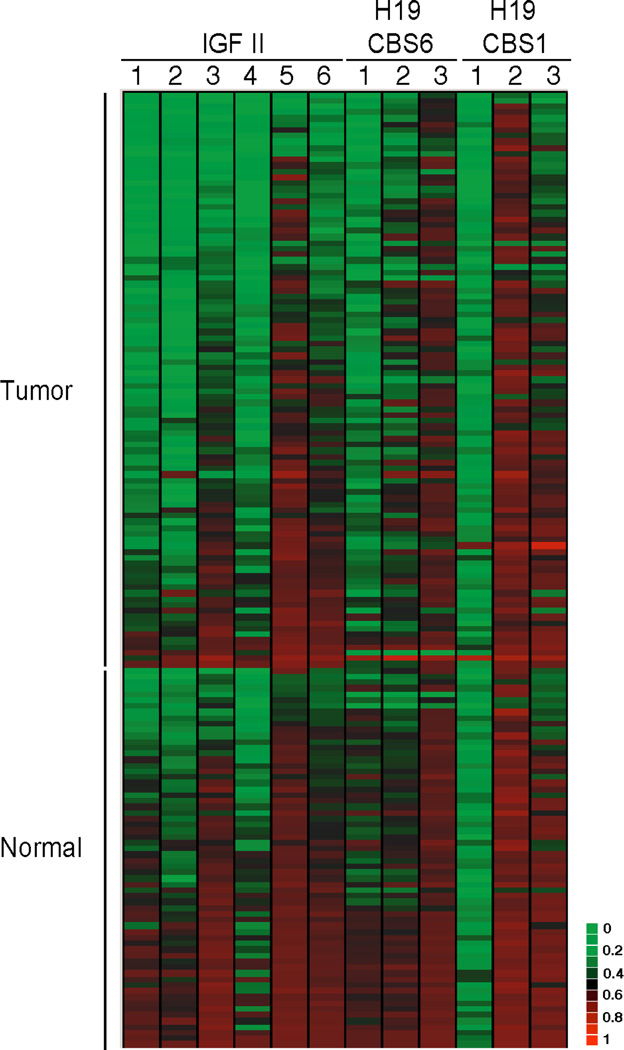
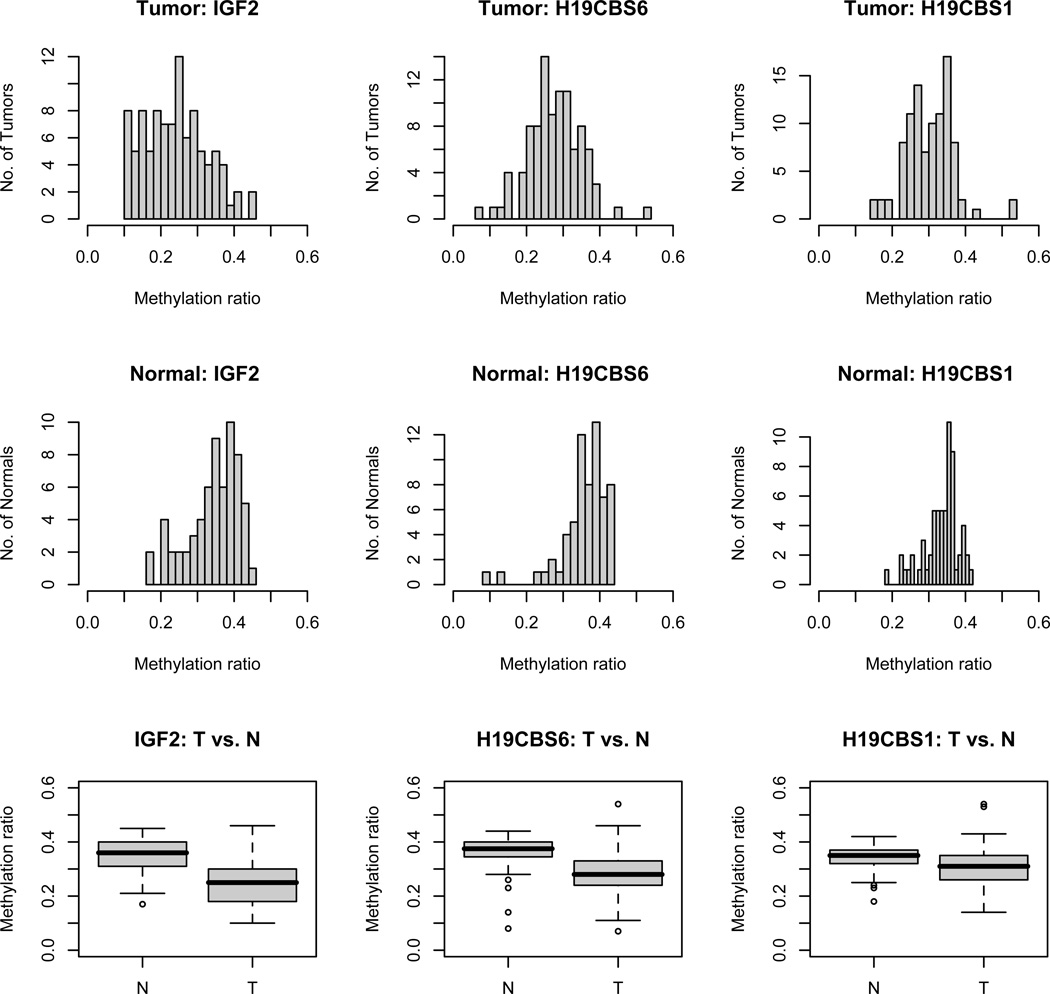
The methylation status of IGF2 and H19 DMRs was profiled in 97 primary colorectal tumors and 64 matched normal colonic tissues (Figure 2). In all three DMRs, methylation levels were significantly lower in the tumors compared to the adjacent normal tissues (p<0.0001) (Figures 2 and 3). One of the CG dinucleotide (CpG 1) in H19CBS1 appeared to be hypomethylated in the majority of tumor and normal tissues. To validate the bislufite/PCR/LDR/Universal array results, we performed bisulfite sequencing in four tumor and four normal colorectal DNA samples (Supplementary Figure 2). In general, the sequencing data indicate that CG sites at IGF2 and H19 DMRs in tumors underwent hypomethylation, while sites in normal tissues remain hemimethylated. For CG sites measured at H19 CBS1, relatively low level (around 20%) methylation was seen at site 1 in both normal and tumor samples. The majority of CG sites at H19 CBS1 have medium level methylation (30–60%) in both tissues, nevertheless, higher methylation was found in normal tissues than in tumors with statistically significant differences. The bisulfite sequencing data are consistent with bisulfite/PCR/LDR/Universal array results and support our previous study suggesting that the overall promoter methylation status of a gene is optimally determined by averaging multiple cytosine methylation levels rather than basing methylation status on a single cytosine21. The IGF2 DMR exhibited average methylation levels of 0.25±0.085 and 0.35±0.07 in the tumor and normal tissues, respectively. Hypomethylation was scored (<0.21) when the average methylation level of a given sample was two standard deviations (2SDs) below the mean of normal tissue methylation levels. Based on this criterion, 35% (n=34) tumors and 8% of normal tissues showed IGF2 hypomethylation, which is consistent with a previous report indicating comparable percentages of IGF2 LOI in tumor and normal colonic tissue18. Similarly, the average methylation levels were 0.31±0.065 and 0.34±0.048 at H19CBS1, and were 0.28±0.073 and 0.36±0.064 at H19CBS6 in the tumor and normal tissues, respectively. Thus, hypomethylation at CBS1 and CBS6 was found in 14% (n=14) and 21% (n=20) of CRC when scored using the 2SDs cut-off. Over 75% of tumor samples with H19CBS1 or H19CBS6 hypomethylation were also hypomethylated at the IGF2 DMR (p=0.0004 and 0.0001, respectively). This observation suggests that the methylation at H19 and IGF2 DMRs in CRC did not follow a reciprocal imprinting pattern.
Figure 2.
The cytosine methylation levels of 97 primary CRC and 64 matched normal mucosas were profiled using bisulfite/PCR/LDR/Universal Array assay. Six, three and three CpG sites at the IGF2 DMR, H19CBS1 and H19CBS6 were analyzed, respectively. A heat map diagram depictes the percentage of methylation at each CpG dinucleotide site. The methylation levels of these sites were calibrated using the standard curve shown in supplementary figure 1. Matched normal tissues were used as controls to calculate the statistical significance of tumor-specific aberrant methylation at each locus.
Figure 3.
The distributions of IGF2 and H19 DMR methylation status in 97 primary CRC and 64 matched normal colorectal tissues. The overall promoter methylation status of IGF2, H19CBS1 and H19CBS6 in each sample was determined by averaging the methylation levels of six, three and three interrogated cytosines in that particular locus, respectively. The Student’s t-test was conducted to determine the significance of differential methylation between tumor and normal tissues (p<0.0001 at all three loci). T: tumor and N: normal tissues.
Hypomethylation of IGF2 / H19 correlated with IGF2 LOI status in CRC
To study IGF2 LOI, the heterozygous or informative samples were identified by analyzing a genomic polymorphism in IGF2 exon 922. A total of 38 informative cases were identified and the IGF2 allelic expression levels were measured using LDR. Biallelic expression of IGF2 was defined as the fluorescence intensity ratio of LDR products less than 3:1 between the more-abundant and less-abundant alleles. Of the 38 analyzed cases, 24 of them exhibited IGF2 hypomethylation and biallelic gene expression (Table 1, p<0.0001). Correlation between H19 hypomethylation and IGF2 LOI was also seen at the CBS1 and CBS6 loci (Table 1, p<0.05). Only two of the hypomethylated IGF2 samples showed allelically imbalanced expression. Our data is consistent with previous reports suggesting that IGF2 and H19 promoter hypomethylation correlated with IGF2 LOI4.
Table 1.
The relationship between IGF2 expression and the DMR methylation status.
| Methylation status |
||||||
|---|---|---|---|---|---|---|
| Sample ID |
IGF2 LOI | IGF2 | H19CBS1 | H19CBS6 | marked IGF2 elevation |
MSI |
| 1 | Yes | Half | Half | Half | No | No |
| 2 | Yes | Half | Half | Half | No | No |
| 3 | Yes | Hypo | Half | Hypo | No | No |
| 4 | Yes | Hypo | Half | Hypo | No | No |
| 5 | Yes | Hypo | Half | Hypo | No | No |
| 6 | Yes | Hypo | Half | Half | No | Yes |
| 7 | Yes | Hypo | Half | Half | No | Yes |
| 8 | Yes | Hypo | Half | Half | No | Yes |
| 9 | Yes | Hypo | Half | Hypo | No | No |
| 10 | Yes | Hypo | Hypo | Hypo | No | No |
| 11 | Yes | Hypo | Half | Hypo | No | No |
| 12 | Yes | Hypo | Half | Half | No | No |
| 13 | Yes | Hypo | Half | Hypo | No | No |
| 14 | Yes | Hypo | Hypo | Hypo | No | No |
| 15 | Yes | Hypo | Hypo | Hypo | No | No |
| 16 | Yes | Hypo | Hypo | Hypo | No | No |
| 17 | Yes | Hypo | Hypo | Hypo | No | No |
| 18 | Yes | Hypo | Hypo | Hypo | No | No |
| 19 | Yes | Hypo | Hypo | Hypo | No | No |
| 20 | Yes | Hypo | Hypo | Hypo | No | No |
| 21 | Yes | Hypo | Hypo | Hypo | No | No |
| 22 | Yes | Hypo | Hypo | Hypo | No | No |
| 23 | Yes | Hypo | Hypo | Hypo | No | No |
| 24 | Yes | Hypo | Hypo | Half | Yes | No |
| 25 | Yes | Hypo | Half | Half | Yes | No |
| 26 | Yes | Hypo | Half | Half | Yes | No |
| 27 | No | Half | Hypo | Half | No | No |
| 28 | No | Half | Half | Half | No | Yes |
| 29 | No | Half | Half | Half | No | Yes |
| 30 | No | Half | Half | Half | No | No |
| 31 | No | Half | Half | Half | No | No |
| 32 | No | Half | Half | Half | No | No |
| 33 | No | Half | Half | Half | No | No |
| 34 | No | Half | Half | Half | No | No |
| 35 | No | Hypo | Half | Hypo | No | No |
| 36 | No | Hypo | Half | Hypo | No | No |
| 37 | No | Half | Half | Hypo | Yes | No |
| 38 | No | Half | Half | Half | Yes | No |
| Summary |
IGF2 DMR methylation |
H19CBS1 DMR methylation |
H19CBS6 DMR methylation |
marked IGF2 elevation |
MSI | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| hypo | hemi | hypo | hemi | hypo | hemi | yes | no | yes | no | ||
| IGF2LOI | yes no |
24 2 |
2 10 |
12 1 |
14 11 |
17 3 |
9 9 |
3 2 |
23 10 |
3 2 |
23 10 |
| p=0.000008 | p=0.03 | p=0.035 | p=0.643 | p=0.643 | |||||||
IGF2 overexpression correlated with microsatellite instability but not with its promoter hypomethylation
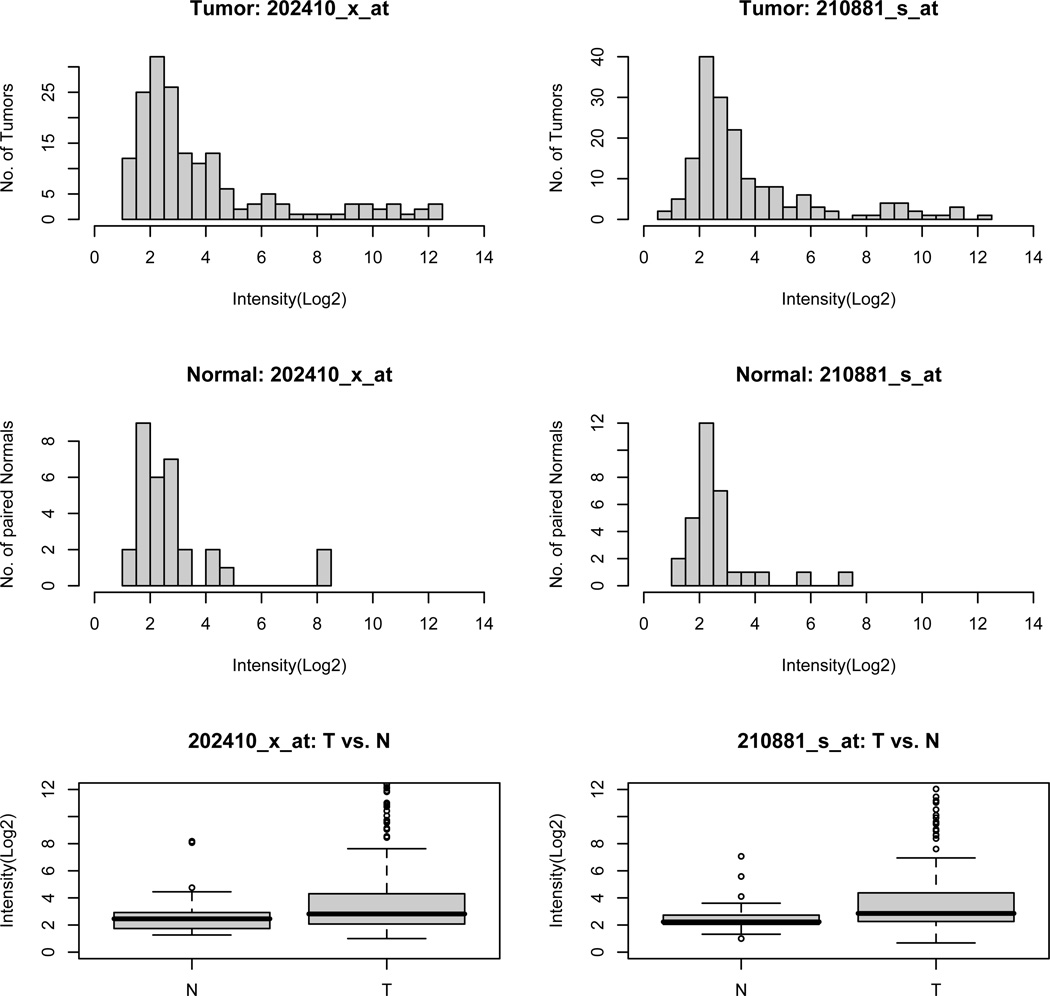
To determine if substantially elevated IGF2 is a common event in CRC, the IGF2 expression profiles of 167 primary colorectal tumors and 32 matched adjacent normal colonic tissues were obtained using Affymetrix HG-U133A2 array26. Tumor-specific IGF2 expression was seen in the signals of probe sets 202410_x_at and 210881_s_at (p=0.029 and 0.0017, respectively, Figure 4). The microarray-based expression results were subsequently confirmed using real-time reverse transcription-PCR assay (Supplementary Figure 3). Histograms of these data revealed IGF2 RNA levels skewed to the right, suggesting that the tumor-specific IGF2 expression was heavily influenced by outlier samples with expression levels greater than 1.5 fold of the interquartil range (>6.4). By this criterion, 23 colorectal tumors (13.4%) were determined as having noticeable, outlier IGF2 overexpression.
Figure 4.
The distribution of IGF2 gene expression in 167 primary CRC tumors and 32 matched normal colorectal tissues. The IGF2 transcription level of each sample was measured by two corresponding probe sets on Affymetrix HG-U133A2 array. The Wilcoxon signed-rank test was conducted to determine the significance of differential IGF2 transcription between tumor and normal tissues (p=0.029 for 202410_x_at and 0.0017 for 210881_s_at). T: tumor and N: normal tissues.
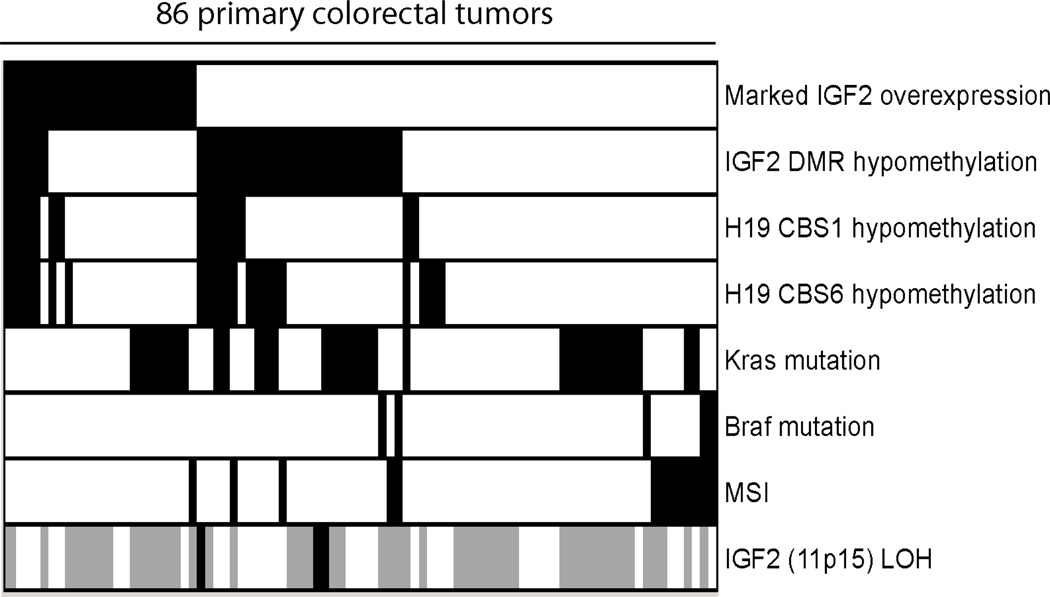
IGF2 overexpression has been shown to associate with uniparental disomy (UPD) at IGF2 locus in Wilms tumors27. It is interesting to investigate if a similar correlation may exist in CRC. We have previously performed genome-wide copy number variation (CNV) study in a subset of colorectal tumors, which allows us to determine copy number neutral LOH or paternal UPD at 11p15 (IGF2 locus)21. Among the 86 tumors in which the IGF2 expression and methylation data were analyzed, CNV were measured in 37 samples using Affymetrix 50K SNP array (Figure 5 and Supplementary Figure 4). Among them, paternal UPD at 11p15 was revealed in only three tumor samples. Although, all three tumors possessed IGF2 DMR hypomethylation and duplication of the hypomethylated allele, none of them associated with marked IGF2 overexpression.
Figure 5.
A dichotomous heat map of IGF2/H19 methylation, IGF2 expression, selected mutation and genomic instability status in 86 primary CRC. The IGF2 overexpression status was determined by averaging signals from two probe sets (202410_x_at and 210881_s_at) and scored by the criteria described in the result section. For each DMR, the scoring of hypomethylation was determined by the two standard deviation criteria described in the result section. The MSI, BRAF and KRAS mutation statuses, and copy number neutral LOH at IGF2 locus were also determined for each tumor sample. The alignment of each tumor was maintained across. The presence of noticeable IGF2 overexpression, DMR hypomethylation, MSI, BRAF and KRAS mutations, and copy number neutral IGF2 LOH were indicated in black. The presence of normal IGF2 expression, DMR hemimethylation, MSS, BRAF and KRAS wild-type alleles, and IGF2 locus not undergone copy number neutral LOH were indicated in white. Tumor samples lack of SNP array experimental results were indicated in gray.
We further investigated the association between significantly elevated IGF2 expression, promoter hypomethylation and some common genetic alterations in CRC. As shown in figure 5, among the 86 tumors where the gene expression and methylation data were both available, only five samples exhibited marked IGF2 elevation and promoter hypomethylation (p=0.20). IGF2 overexpression did not correlate with either H19 DMRs hypomethylation (p=0.64 for CBS1 and 0.27 for CBS6), KRAS (p=0.43) or BRAF mutations (p=0.32). Nevertheless, a significant negative correlation was found between samples of markedly elevated IGF2 expression and microsatellite instability (MSI) phenotype (p=0.031, Figure 5). These results indicate that marked IGF2 elevation did not arise through IGF2 and H19 promoter hypomethylation, but instead may involve other oncogenic event such as chromosomal instability.
DISCUSSIONS
Three probe sets were fabricated on Affymetrix HG-U133A2 array to study IGF2 transcription levels. Our data indicated that CRC-specific IGF2 expression was detectable by two probe sets (202410_x_at and 210881_s_at) located in the IGF2 coding region and capable of capturing multiple IGF2 isoform transcripts. However, the third probe set (202409_at) that hybridizes to loci near the end of IGF2 3’ UTR did not reveal signals of tumor-specific expression (Supplementary Figure 5) and was not employed in the data analysis. It is worth noting that IGF2 transcription may be controlled by several promoters, including a biallelically expressed P1 promoter and the imprinted P2-P4 promoters28. To assure our study primarily focused on measuring IGF2 imprinting rather than P1 derived transcription, the determination of IGF2 methylation status was performed at exon 3 and the analysis of allelic expression ratio was assayed according to a SNP at exon 94, 22.
Using quantitative assays, we have shown that the LOI of IGF2 significantly correlated with IGF2 and H19 DMR hypomethylation in CRC. This mechanism is unique to CRC and differs from the H19 DMR hypermethylation observed in other cancer types4. In fact, hypermethylation of H19 CBS1 and CBS6 was not observed in our study and the biallelic IGF2 expression correlated only with H19 CBS hypomethylation. These results suggest that the model of IGF2/H19 competing for a common downstream enhancer may not be entirely applicable to CRC. Moreover, although IGF2 promoter hypomethylation resulted in LOI, neither IGF2 nor H19 aberrant methylation correlated with the substantially elevated IGF2 expression. The observation that noticeable IGF2 overexpression was unlikely to occur in MSI or tumors with a near-diploid genome, supporting the notion that aneuploidy or chromosomal instability may cause significantly elevated IGF2 expression.
Gene fusion due to chromosomal translocation has been reported in several type of human cancers29. A well-known example is the BCR-ABL fusion protein in chronic myelogenous leukemia where the ABL proto-oncogene was activated by the 5’ activation region of BCR30, 31. Recently, gene fusion has also been identified in epithelial tumors32. In prostate cancer, fusion between TMPRSS2 and oncogenomic ETS family transcription factors (ERG or ETV1) has been found in many instances (>50%), accompanied with high ETS gene expression. This phenomenon has been reasoned that oncogenes driving tumorigenesis are drastically activated by the fusion of a strong promoter element translocated from other genes upstream genomic region. Applicable methods of cancer outlier profile analysis (COPA) were developed to identify chromosomal rearrangement events and their corresponding overexpressed candidates25, 32. We analyzed the gene expression profiles of 167 primary CRC and 32 matched normal tissues using the open source COPA package25. This method was developed on the assumption that gene translocation may involve the fusion of an activating domain to multiple downstream candidates, nevertheless only one fusion event or the highest activated candidate is likely to be found in a given tumor sample. Namely, the fusion induced gene activation is likely to be seen in a mutually exclusive fashion in a sample if there were multiple fusion products from a same activating domain. Unexpectedly, our result indicates that IGF2 was ranked the first in the outlier analysis (Supplementary Figure 6). This finding suggests that the substantially elevated IGF2 expression may involve a previously uncharacterized molecular event. This preliminary data may potentially explain why the highly expressed IGF2 does not correlate with its DMR hypomethylation.
Our study of CRC did not show a correlation between IGF2 UPD and marked overexpression like those observed in Wilms tumors. The discrepancy may be explained in several ways. First, the correlation of IGF2 UPD and overexpression may exist in a tumor type specific manner. WT1 is an IGF2 repressor and its mutation facilitates IGF2 up-regulation. Since paternal UPD duplicates the expressed IGF2 allele and is commonly found in Wilms tumors with a WT1 mutation, it has been suggested that IGF2 overexpression results from a WT1 abnormality and IGF2 paternal UPD. However, despite a WT1 mutation that occurs in 15–25% of Wilms tumors, it has not been reported in CRC. Additionally, UPD at 11p15 is neither a frequent event in colorectal tumors (relative to marked IGF2 overexpression). A similar example of tumor-specific abnormality is the presence of reciprocal methylation patterns between IGF2 and H19 DMRs in Wilms tumors, but not in CRC. Thus, one may not expect to see the correlation between IGF2 UPD and overexpression in CRC. Second, change of IGF2 expression in CRC may involve mechanisms other than LOI. An example is the discovery of a long-range interchromosomal association between one allele of IGF2 imprinting control region and Wsb1/f1 gene, mediated by a transcription factor CTCF33. The participation of cis-acting elements and trans-acting factors may explain the alteration of transcription levels at these loci. It is likely that the colocalization of looped chromosomes bring regulatory elements such as enhancers to allow mutual influence of transcription from separated genes34–36. Alternatively, multiple gene expression may be activated by shared transcription factors in which the confined chromosomal geometry permits a transcription machinery interacting with several juxtaposed promoter regions37, 38. These examples are consistent with our preliminary result suggesting that chromosomal instability, including gene fusion, might be a mechanism for marked IGF2 overexpression. Third, UPD at IGF2 locus may not be adequately identified using 50k SNP array. The interrogated SNPs in Affymetrix 50k array has a mean spacing of 47kb, which does not support a detailed mapping of chromosomal aberrations at IGF2 locus spanning 20.5kb region. To avoid the underestimation of IGF2 UPD in tumors, platforms with higher resolution (e.g. a mean spacing at 5kb) will be employed in future studies to accurately identify short genomic sequence alterations at the IGF2 locus.
A recent study demonstrated that LOI of IGF2 enhances IGF2 signaling by increasing the proliferation-related gene expression (Akt/PKB signaling)39. Interestingly, the signaling enhancement does not correlate with the increase of overall IGF2 levels. Markedly sustained Akt activation was seen in LOI cells treated with a low dose of IGF2, compared to those treated with four-fold higher IGF2 dosage. The detection of up-regulated IGF2 signaling components, IGF1 and insulin receptors (igf1r and insr) in LOI cells provided further evidence indicating that hypersensitivity of the signaling pathway, rather than IGF2 levels, accounts for the proliferation-related gene activation of Akt/PKB signaling. These observations suggest a plausible mechanism of IGF2 LOI involvement in early tumor development. The strong biological impact of LOI in responding to low IGF2 has been reasoned to facilitate the proliferation of tumorigenic cells at early cancer stages in which the density of IGF2 producing cells was low. While the above study suggested that increasing IGF2 level has little impact on IGF2 signaling, it is also likely that the marked IGF2 overexpression (typically expressed at 10-fold or greater, than the average CRC IGF2 level), especially in non-LOI tumor cells, may affect tumorigenesis through mechanisms other than LOI enhanced signal sensitivity, including the interaction with H19 gene, transcription factor mediated interchromosomal association, and other signaling pathways33, 40.
Clinically and pathologically it has been suggested that the IGF2 LOI is linked to MSI tumors41, 42. Since adult MSI patients have a favorable prognosis, the correlation between LOI and MSI may represent a valuable clinical application of IGF2 as a biomarker. However, in a similar study performed by Sasaki et al. using 95 informative IGF2 LOI, no significant correlation was found between LOI and MSI43. The authors argued that those earlier reports were conducted in a relatively small sample size or patients were recruited with a bias that resulted in a high prevalence of MSI cases (30–40%). In contrast, the number of MSI cases in our study cohort is comparable with a typical MSI prevalence (15%) in sporadic CRC. Our data showed that genetic abnormalities (e.g. MSI and KRAS/BRAF mutations) in CRC did not correlate with either IGF2 LOI or DMR hypomethylation. The results are consistent with those reported by Sasaki et al. indicating no significant correlation between MSI and biallelic IGF2 expression. The fact that IGF2 LOI was not linked to MSI may represent a unique opportunity to subclassify colorectal carcinogenesis. We conclude that the aberrant IGF2 expression in CRC consists of at least LOI and other mechanisms resulting in elevated IGF2 expression.
Supplementary Material
ACKNOWLEDGEMENTS
This work was supported by grants from Clinical Nutrition Research Unit (CNRU) P30 CA29502 (YWC) and National Cancer Institute P01-CA65930 (FB). We also thank the Gilbert Family Foundation, and the Ludwig Institute for Cancer Research / Conrad N. Hilton Foundation joint Hilton-Ludwig Cancer Metastasis Initiative for their generous funding in part of this work. The authors thank Owen Parker and Philip Feinberg for critical reading of the manuscript. Francis Barany is an Affiliate of the Ludwig Institute for Cancer Research.
Abbreviations
- CRC
colorectal cancer
- DMR
differentially methylated region
- IGF2
insulin-like growth factor 2
- LDR
ligase detection reaction
- LOI
Loss of imprinting
- MSI
microsatellite instability
REFERENCES
- 1.Rainier S, Johnson LA, Dobry CJ, Ping AJ, Grundy PE, Feinberg AP. Relaxation of imprinted genes in human cancer. Nature. 1993;362:747–749. doi: 10.1038/362747a0. [DOI] [PubMed] [Google Scholar]
- 2.Ogawa O, Eccles MR, Szeto J, McNoe LA, Yun K, Maw MA, Smith PJ, Reeve AE. Relaxation of insulin-like growth factor II gene imprinting implicated in Wilms' tumour. Nature. 1993;362:749–751. doi: 10.1038/362749a0. [DOI] [PubMed] [Google Scholar]
- 3.Ferguson-Smith AC. Genetic imprinting: silencing elements have their say. Curr Biol. 2000;10:R872–R875. doi: 10.1016/s0960-9822(00)00817-4. [DOI] [PubMed] [Google Scholar]
- 4.Cui H, Onyango P, Brandenburg S, Wu Y, Hsieh CL, Feinberg AP. Loss of imprinting in colorectal cancer linked to hypomethylation of H19 and IGF2. Cancer Res. 2002;62:6442–6446. [PubMed] [Google Scholar]
- 5.Murphy SK, Huang Z, Wen Y, Spillman MA, Whitaker RS, Simel LR, Nichols TD, Marks JR, Berchuck A. Frequent IGF2/H19 domain epigenetic alterations and elevated IGF2 expression in epithelial ovarian cancer. Mol Cancer Res. 2006;4:283–292. doi: 10.1158/1541-7786.MCR-05-0138. [DOI] [PubMed] [Google Scholar]
- 6.Nakagawa H, Chadwick RB, Peltomaki P, Plass C, Nakamura Y, de La Chapelle A. Loss of imprinting of the insulin-like growth factor II gene occurs by biallelic methylation in a core region of H19-associated CTCF-binding sites in colorectal cancer. Proc Natl Acad Sci U S A. 2001;98:591–596. doi: 10.1073/pnas.011528698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sullivan MJ, Taniguchi T, Jhee A, Kerr N, Reeve AE. Relaxation of IGF2 imprinting in Wilms tumours associated with specific changes in IGF2 methylation. Oncogene. 1999;18:7527–7534. doi: 10.1038/sj.onc.1203096. [DOI] [PubMed] [Google Scholar]
- 8.Steenman MJ, Rainier S, Dobry CJ, Grundy P, Horon IL, Feinberg AP. Loss of imprinting of IGF2 is linked to reduced expression and abnormal methylation of H19 in Wilms' tumour. Nat Genet. 1994;7:433–439. doi: 10.1038/ng0794-433. [DOI] [PubMed] [Google Scholar]
- 9.Moulton T, Crenshaw T, Hao Y, Moosikasuwan J, Lin N, Dembitzer F, Hensle T, Weiss L, McMorrow L, Loew T, et al. Epigenetic lesions at the H19 locus in Wilms' tumour patients. Nat Genet. 1994;7:440–447. doi: 10.1038/ng0794-440. [DOI] [PubMed] [Google Scholar]
- 10.Bell AC, Felsenfeld G. Methylation of a CTCF-dependent boundary controls imprinted expression of the IGF2 gene. Nature. 2000;405:482–485. doi: 10.1038/35013100. [DOI] [PubMed] [Google Scholar]
- 11.Hark AT, Schoenherr CJ, Katz DJ, Ingram RS, Levorse JM, Tilghman SM. CTCF mediates methylation-sensitive enhancer-blocking activity at the H19/IGF2 locus. Nature. 2000;405:486–489. doi: 10.1038/35013106. [DOI] [PubMed] [Google Scholar]
- 12.Kim HT, Choi BH, Niikawa N, Lee TS, Chang SI. Frequent loss of imprinting of the H19 and IGF-II genes in ovarian tumors. Am J Med Genet. 1998;80:391–395. doi: 10.1002/(sici)1096-8628(19981204)80:4<391::aid-ajmg16>3.0.co;2-h. [DOI] [PubMed] [Google Scholar]
- 13.Elkin M, Shevelev A, Schulze E, Tykocinsky M, Cooper M, Ariel I, Pode D, Kopf E, de Groot N, Hochberg A. The expression of the imprinted H19 and IGF-2 genes in human bladder carcinoma. FEBS Lett. 1995;374:57–61. doi: 10.1016/0014-5793(95)01074-o. [DOI] [PubMed] [Google Scholar]
- 14.Kondo M, Suzuki H, Ueda R, Osada H, Takagi K, Takahashi T, Takahashi T. Frequent loss of imprinting of the H19 gene is often associated with its overexpression in human lung cancers. Oncogene. 1995;10:1193–1198. [PubMed] [Google Scholar]
- 15.Leighton PA, Ingram RS, Eggenschwiler J, Efstratiadis A, Tilghman SM. Disruption of imprinting caused by deletion of the H19 gene region in mice. Nature. 1995;375:34–39. doi: 10.1038/375034a0. [DOI] [PubMed] [Google Scholar]
- 16.Ohlsson R, Renkawitz R, Lobanenkov V. CTCF is a uniquely versatile transcription regulator linked to epigenetics and disease. Trends Genet. 2001;17:520–527. doi: 10.1016/s0168-9525(01)02366-6. [DOI] [PubMed] [Google Scholar]
- 17.Reik W, Murrell A. Genomic imprinting. Silence across the border. Nature. 2000;405:408–409. doi: 10.1038/35013178. [DOI] [PubMed] [Google Scholar]
- 18.Cui H, Cruz-Correa M, Giardiello FM, Hutcheon DF, Kafonek DR, Brandenburg S, Wu Y, He X, Powe NR, Feinberg AP. Loss of IGF2 imprinting: a potential marker of colorectal cancer risk. Science. 2003;299:1753–1755. doi: 10.1126/science.1080902. [DOI] [PubMed] [Google Scholar]
- 19.Ravenel JD, Broman KW, Perlman EJ, Niemitz EL, Jayawardena TM, Bell DW, Haber DA, Uejima H, Feinberg AP. Loss of imprinting of insulin-like growth factor-II (IGF2) gene in distinguishing specific biologic subtypes of Wilms tumor. J Natl Cancer Inst. 2001;93:1698–1703. doi: 10.1093/jnci/93.22.1698. [DOI] [PubMed] [Google Scholar]
- 20.Fottner C, Hoeflich A, Wolf E, Weber MM. Role of the insulin-like growth factor system in adrenocortical growth control and carcinogenesis. Horm Metab Res. 2004;36:397–405. doi: 10.1055/s-2004-814563. [DOI] [PubMed] [Google Scholar]
- 21.Cheng YW, Pincas H, Bacolod MD, Schemmann G, Giardina SF, Huang J, Barral S, Idrees K, Khan SA, Zeng Z, Rosenberg S, Notterman DA, et al. CpG island methylator phenotype associates with low-degree chromosomal abnormalities in colorectal cancer. Clin Cancer Res. 2008;14:6005–6013. doi: 10.1158/1078-0432.CCR-08-0216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Woodson K, Flood A, Green L, Tangrea JA, Hanson J, Cash B, Schatzkin A, Schoenfeld P. Loss of insulin-like growth factor-II imprinting and the presence of screen-detected colorectal adenomas in women. J Natl Cancer Inst. 2004;96:407–410. doi: 10.1093/jnci/djh042. [DOI] [PubMed] [Google Scholar]
- 23.Cheng YW, Shawber C, Notterman D, Paty P, Barany F. Multiplexed profiling of candidate genes for CpG island methylation status using a flexible PCR/LDR/Universal Array assay. Genome Res. 2006;16:282–289. doi: 10.1101/gr.4181406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Weisenberger DJ, Siegmund KD, Campan M, Young J, Long TI, Faasse MA, Kang GH, Widschwendter M, Weener D, Buchanan D, Koh H, Simms L, et al. CpG island methylator phenotype underlies sporadic microsatellite instability and is tightly associated with BRAF mutation in colorectal cancer. Nat Genet. 2006;38:787–793. doi: 10.1038/ng1834. [DOI] [PubMed] [Google Scholar]
- 25.MacDonald JW, Ghosh D. COPA--cancer outlier profile analysis. Bioinformatics. 2006;22:2950–2951. doi: 10.1093/bioinformatics/btl433. [DOI] [PubMed] [Google Scholar]
- 26.Tsafrir D, Bacolod M, Selvanayagam Z, Tsafrir I, Shia J, Zeng Z, Liu H, Krier C, Stengel RF, Barany F, Gerald WL, Paty PB, et al. Relationship of gene expression and chromosomal abnormalities in colorectal cancer. Cancer Res. 2006;66:2129–2137. doi: 10.1158/0008-5472.CAN-05-2569. [DOI] [PubMed] [Google Scholar]
- 27.Haruta M, Arai Y, Sugawara W, Watanabe N, Honda S, Ohshima J, Soejima H, Nakadate H, Okita H, Hata J, Fukuzawa M, Kaneko Y. Duplication of paternal IGF2 or loss of maternal IGF2 imprinting occurs in half of Wilms tumors with various structural WT1 abnormalities. Genes Chromosomes Cancer. 2008;47:712–727. doi: 10.1002/gcc.20572. [DOI] [PubMed] [Google Scholar]
- 28.Issa JP, Vertino PM, Boehm CD, Newsham IF, Baylin SB. Switch from monoallelic to biallelic human IGF2 promoter methylation during aging and carcinogenesis. Proc Natl Acad Sci U S A. 1996;93:11757–11762. doi: 10.1073/pnas.93.21.11757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rowley JD. Chromosome translocations: dangerous liaisons revisited. Nat Rev Cancer. 2001;1:245–250. doi: 10.1038/35106108. [DOI] [PubMed] [Google Scholar]
- 30.Rowley JD. Letter: A new consistent chromosomal abnormality in chronic myelogenous leukaemia identified by quinacrine fluorescence and Giemsa staining. Nature. 1973;243:290–293. doi: 10.1038/243290a0. [DOI] [PubMed] [Google Scholar]
- 31.de Klein A, van Kessel AG, Grosveld G, Bartram CR, Hagemeijer A, Bootsma D, Spurr NK, Heisterkamp N, Groffen J, Stephenson JR. A cellular oncogene is translocated to the Philadelphia chromosome in chronic myelocytic leukaemia. Nature. 1982;300:765–767. doi: 10.1038/300765a0. [DOI] [PubMed] [Google Scholar]
- 32.Tomlins SA, Rhodes DR, Perner S, Dhanasekaran SM, Mehra R, Sun XW, Varambally S, Cao X, Tchinda J, Kuefer R, Lee C, Montie JE, et al. Recurrent fusion of TMPRSS2 and ETS transcription factor genes in prostate cancer. Science. 2005;310:644–648. doi: 10.1126/science.1117679. [DOI] [PubMed] [Google Scholar]
- 33.Ling JQ, Li T, Hu JF, Vu TH, Chen HL, Qiu XW, Cherry AM, Hoffman AR. CTCF mediates interchromosomal colocalization between IGF2/H19 and Wsb1/Nf1. Science. 2006;312:269–272. doi: 10.1126/science.1123191. [DOI] [PubMed] [Google Scholar]
- 34.Lopes S, Lewis A, Hajkova P, Dean W, Oswald J, Forne T, Murrell A, Constancia M, Bartolomei M, Walter J, Reik W. Epigenetic modifications in an imprinting cluster are controlled by a hierarchy of DMRs suggesting long-range chromatin interactions. Hum Mol Genet. 2003;12:295–305. doi: 10.1093/hmg/ddg022. [DOI] [PubMed] [Google Scholar]
- 35.Murrell A, Heeson S, Reik W. Interaction between differentially methylated regions partitions the imprinted genes IGF2 and H19 into parent-specific chromatin loops. Nat Genet. 2004;36:889–893. doi: 10.1038/ng1402. [DOI] [PubMed] [Google Scholar]
- 36.Sasaki H, Ishihara K, Kato R. Mechanisms of IGF2/H19 imprinting: DNA methylation, chromatin and long-distance gene regulation. J Biochem. 2000;127:711–715. doi: 10.1093/oxfordjournals.jbchem.a022661. [DOI] [PubMed] [Google Scholar]
- 37.Grande MA, van der Kraan I, de Jong L, van Driel R. Nuclear distribution of transcription factors in relation to sites of transcription and RNA polymerase II. J Cell Sci. 1997;110(Pt 15):1781–1791. doi: 10.1242/jcs.110.15.1781. [DOI] [PubMed] [Google Scholar]
- 38.Osborne CS, Chakalova L, Brown KE, Carter D, Horton A, Debrand E, Goyenechea B, Mitchell JA, Lopes S, Reik W, Fraser P. Active genes dynamically colocalize to shared sites of ongoing transcription. Nat Genet. 2004;36:1065–1071. doi: 10.1038/ng1423. [DOI] [PubMed] [Google Scholar]
- 39.Kaneda A, Wang CJ, Cheong R, Timp W, Onyango P, Wen B, Iacobuzio-Donahue CA, Ohlsson R, Andraos R, Pearson MA, Sharov AA, Longo DL, et al. Enhanced sensitivity to IGF-II signaling links loss of imprinting of IGF2 to increased cell proliferation and tumor risk. Proc Natl Acad Sci U S A. 2007;104:20926–20931. doi: 10.1073/pnas.0710359105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gabory A, Ripoche MA, Yoshimizu T, Dandolo L. The H19 gene: regulation and function of a non-coding RNA. Cytogenet Genome Res. 2006;113:188–193. doi: 10.1159/000090831. [DOI] [PubMed] [Google Scholar]
- 41.Cui H, Horon IL, Ohlsson R, Hamilton SR, Feinberg AP. Loss of imprinting in normal tissue of colorectal cancer patients with microsatellite instability. Nat Med. 1998;4:1276–1280. doi: 10.1038/3260. [DOI] [PubMed] [Google Scholar]
- 42.Nishihara S, Hayashida T, Mitsuya K, Schulz TC, Ikeguchi M, Kaibara N, Oshimura M. Multipoint imprinting analysis in sporadic colorectal cancers with and without microsatellite instability. Int J Oncol. 2000;17:317–322. doi: 10.3892/ijo.17.2.317. [DOI] [PubMed] [Google Scholar]
- 43.Sasaki J, Konishi F, Kawamura YJ, Kai T, Takata O, Tsukamoto T. Clinicopathological characteristics of colorectal cancers with loss of imprinting of insulin-like growth factor 2. Int J Cancer. 2006;119:80–83. doi: 10.1002/ijc.21741. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.