Abstract
We previously showed that the Pituitary Adenylate Cyclase-Activating Polypeptide (PACAP) and its receptor VPAC1 are negative regulators of megakaryopoiesis and platelet function, but their downstream signaling pathway that inhibits this process still remained unknown. A combined proteomic, transcriptomic, and bioinformatic approach was here used to elucidate the molecular mechanisms underlying PACAP signaling via VPAC1 in megakaryocytes. Two-dimensional difference gel electrophoresis and tandem MS were applied to detect differentially expressed proteins in megakaryocytic CHRF cells stimulated with PACAP. The majority of the 120 proteins modulated by PACAP belong to the class of “cell cycle and apoptosis” proteins. The up- or down-regulated expression of some proteins was confirmed by immunoblot and immunohistochemical analysis. A meta-analysis of our data and 12 other published studies was performed to evaluate signaling pathways involved in different cellular models of PACAP response. From 2384 differentially expressed genes/proteins, 83 were modulated by PACAP in at least three independent studies and Ingenuity Pathway Analysis further identified apoptosis as the highest scored network with NF-κB as a key-player. PACAP inhibited serum depletion-induced apoptosis of CHRF cells via VPAC1 stimulation. In addition, PACAP switched on NF-κB dependent gene expression since higher nuclear levels of the active NF-κB p50/p65 heterodimer were found in CHRF cells treated with PACAP. Finally, a quantitative real time PCR apoptosis array was used to study RNA from in vitro differentiated megakaryocytes from a PACAP overexpressing patient, leading to the identification of 15 apoptotic genes with a 4-fold change in expression and Ingenuity Pathway Analysis again revealed NF-κB as the central player. In conclusion, our findings suggest that PACAP interferes with the regulation of apoptosis in megakaryocytes, probably via stimulation of the NF-κB pathway.
The Pituitary Adenylate Cyclase-Activating Polypeptide (PACAP)1 is a neuropeptide belonging to the secretin and glucagon family. It was first isolated from ovine hypothalamic extracts on the basis of its ability to stimulate cAMP formation in rat anterior pituitary cells (1). PACAP is widely distributed, consists of an evolutionary highly conserved sequence, and consistently mediates diverse physiological functions. It has in fact been implicated in many biological processes, including reproductive, development, growth, cardiovascular, respiratory, digestive functions, immune responses, and circadian rhythms (2). The human ADCYAP1 gene is located on chromosome 18p11.32 and encodes a 176-amino acid prepro-PACAP. In all mammalian species studied so far, the sequences of the processed and active PACAP peptides, PACAP38 and its derived form PACAP27, are located in the C-terminal domain of the prepro-PACAP precursor. The sequence of PACAP27 shares also 68% identity with the vasointestinal peptide (VIP). PACAP is a ligand for three G protein-coupled transmembrane receptors: the PACAP-specific PAC1 receptor and the PACAP/VIP-indifferent VPAC1 and VPAC2 receptors, which are primarily coupled to adenylyl cyclase (2).
Megakaryocytes and platelets express the VPAC1 receptor (3, 4). Our previous studies showed that the neuropeptide PACAP and its receptor VPAC1 are negative regulators of megakaryopoiesis and platelet function (3, 5). These studies were performed in two related trisomy 18p patients having three ADCYAP1 copies, elevated PACAP plasma concentrations and a severe bleeding tendency with thrombopathy and mild thrombocytopenia. Though these studies revealed that PACAP tempers megakaryopoiesis and platelet production, the downstream players of the PACAP/VPAC1 pathway remained unknown. A deeper understanding of the downstream pathway could also bring novel insights in this still poorly understood and complex cellular differentiation process (6).
Proteomics offers a feasible approach to explore the global protein alterations in cells after receptor stimulation. Among the most powerful proteomic technologies, difference gel electrophoresis (DIGE) has recently been implemented as a more accurate and sensitive alternative to conventional two-dimensional electrophoresis (7). The main advantage is that samples are labeled with three different spectrally resolvable fluorescent dyes, increasing sensitivity up to picogram levels. The fluorescent dyes label two samples and one internal standard, which is a pooled-mixture containing an equal aliquot of all samples, to be run together in the same two-dimensional gel. The use of an internal standard facilitates accurate matching of spots and permits data normalization among gels to minimize experimental variability (8). It has also been reported that the correlation between quantitation by DIGE and metabolic stable isotope labeling is very good (9).
In the present study, we took advantage of this technology coupled to mass spectrometry (matrix-assisted laser desorption ionization/time of flight (MALDI TOF-TOF)) to detect reproducible proteome changes in megakaryocytic CHRF cells induced by PACAP. Our findings indicate that PACAP interferes with apoptosis in CHRF cells via the VPAC1 receptor, as also found via a literature-based meta-analysis of other cellular systems after addition of PACAP. Finally, a quantitative real time PCR array performed on in vitro differentiated megakaryocytes from our previously described trisomy 18p patient with elevated PACAP plasma level (3, 5) confirmed that PACAP is a protective factor against apoptosis and a stimulator of NF-κB signaling in megakaryocytes.
EXPERIMENTAL PROCEDURES
Patient Description
Our trisomy 18p patient with elevated PACAP plasma levels has been previously described (3, 5). Briefly, the patient is a 29-year-old boy with multiple neurological (epilepsy, hypotonia, convulsions, mental retardation, tremor, psychotic, hyperactive behavior), gastro-intestinal (diarrhea, vomiting), and endocrinological (hypoplasia of the pituitary gland, hypogonadotropic hypogonadism) problems. He also has moderate thrombocytopenia (70–90 × 109 platelets/L) and obvious bleeding problems with repetitive epistaxis and a markedly prolonged Ivy bleeding time (>15 min). Informed consent for our studies was obtained from the legal representative of the patient. This study was approved by the Medical Ethical Committee of the University Hospital of Leuven.
Cell Cultures
Human megakaryocytic CHRF-288–11 cells (ATCC-CRL10107) were grown in RPMI1640 medium supplemented with 4 mm l-glutamine, 1 mm sodium pyruvate, nonessential amino acids, 1 U/ml penicillin, 1 μg/ml streptomycin and with/without 10% heat inactivated fetal bovine serum at 37 °C in a humidified atmosphere with 5% CO2. Cells were incubated with 100 nm PACAP38 (Bachem, Bubendorf, Switzerland) for 9 h or 4 days. On day 4, PACAP treated cultures received full media replacement without PACAP and were grown in fresh medium for another 4 days. The experimental design was conceived in a way that all the cells were sampled at the same stage of growth, that is day 8, to ensure that the changes in protein expression observed among the different points of the time course are really due to PACAP and not to differences in cell growth (Fig. 1). For negative control experiments, only phosphate-buffered saline (PBS) was added to cells during the same time course. After the experiment, cell pellets were washed and stored at −80 °C. Human neuroblastoma SK-N-SH cells were grown in Dulbecco's modified Eagle's medium/F12 medium, supplemented with 10% fetal bovine serum, 4 mm l-glutamine, 1 U/ml penicillin and 1 μg/ml streptomycin.
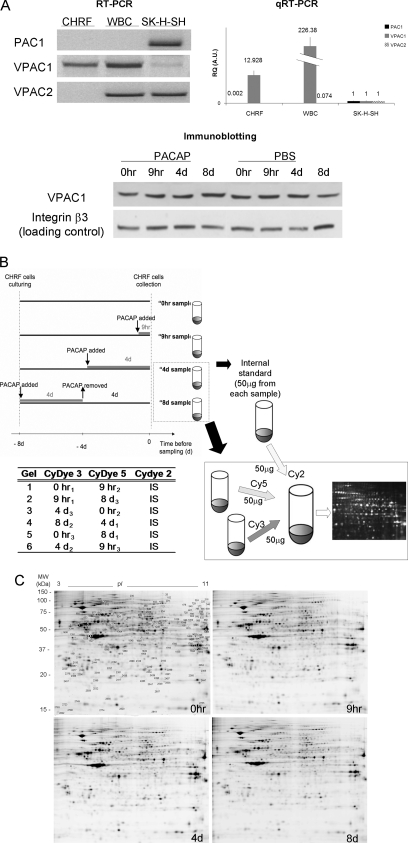
Fig. 1.
Expression of PACAP receptors in different human cell types/lines, schematic workflow of sample preparation for 2D-DIGE and representative gels for each time point of the PACAP treatment of CHRF cells. A, Total RNA from human CHRF cell line (lane 1), white blood cells (lane 2), and SK-N-SK neuroblastoma cell line (lane 3) was submitted to RT-PCR (left panel) and qRT-PCR (right panel) with primers designated to amplify PAC1, VPAC1 or VPAC2. Data reported for qRT-PCR are the average of the expression level of at least two replicates ± the standard deviation; all the RQ (Relative Quantification) expression values are normalized for the values of SK-H-SH cell line. Both experiment findings are consistent and show that CHRF cells uniquely express VPAC1 receptor. The lower panel shows the expression of VPAC1 at protein level in CHRF cells during a time course where cells were treated with PACAP38 or vehicle PBS (integrin β3 was used as loading control). The image is representative of three independent experiments. B, CHRF cells were treated with 100 mm PACAP38. Controls received fresh medium without PACAP at t = 0 h (no PACAP). PACAP was added for 9 h (early PACAP response) or 4 days (late PACAP response) and on day 4 removed and replaced with fresh medium for other 4 days (t = 8 day, PACAP washout). All CHRF cell samples were grown for the same period of time (8 days). Experiments were repeated three times to adjust for interexperimental variations. Untreated and treated cell pellets were lysed, labeled, mixed, and run in one gel with an equal amount of internal standard (IS). This standard was pooled from all 12 samples in the experiment. The lower part of the figure exemplarily depicts the procedure for time points t = 4 day and t = 8 day. The table reports the experimental design for DIGE: the samples following PACAP treatment (0 h, 9 h, 4 day, and 8 day) were analyzed in triplicate by running six gels, in order to maximize the likelihood of detecting any sample-to-sample variation. C, 2D DIGE representative image of CHRF cell lysates proteins: 0 h (control), 9 h and 4 day after PACAP treatment and 8 day (4 day after PACAP removal). Proteins were separated in 24 cm, NL pH 3–10 IPG strips for the first dimension and 12.5% polyacrylamide gel for the second dimension. The image was acquired on a Typhoon 9400 scanner. Protein spots that change in expression during the time course (p < 0.05 by the application of FDR correction methods) are indicated (protein identifications are listed in Table I).
Reverse Transcription Polymerase Chain Reactions (RT-PCR)
Total RNA extraction from white blood cells, CHRF or SK-N-SH cells using TriZol (Life sciences), cDNA conversion and RT-PCR analysis were performed as previously described (5). Real time RT-PCR was carried out by using the SYBR Green detection and the 7500 Fast Real-Time PCR system (Applied Biosystems, Foster City, CA). Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was used as a control gene for normalization. Primer sequences to amplify VPAC1, VPAC2, PAC1, and GAPDH are available upon request.
2D-DIGE and Image Analysis
CHRF cell pellets from three independent biological replicates for each point of the PACAP time-course were lysed by sonication in sample buffer (7 m urea, 2 m thiourea, 4% 3-[(3-cholamidopropyl)dimethylammonio]propanesulfonate (CHAPS), 30 mm Tris, pH 8.5) containing a mixture of protease inhibitors (Complete protease inhibitor, Roche Diagnostics). The protein concentration was determined by the Bio-Rad Dc protein assay (BioRad, Hercules, CA). DIGE analysis was done as described before (10, 11). Briefly, protein samples (50 μg) were labeled with 400 pmol of Cy3 or Cy5 (GE Healthcare) and the internal standard was labeled with Cy2 for 30 min on ice and quenched with 10 mm lysine for 10 min. The internal standard contained equivalent amounts of all the 12 samples for each time point. Labeled proteins were pooled and loading buffer was added (7 m urea, 2 m thiourea, 4% (w/v) CHAPS, 0.5% (v/v) IPG buffer and 1.2% (v/v) Destreak). IPG strips (NL pH 3–11, 24 cm) were cup-loaded with labeled samples in 450 μl of rehydration buffer (7 m urea, 2 m thiourea, 4% (w/v) CHAPS, 0.5% (v/v) IPG buffer, 1.2% (v/v) Destreak and 0.05% (w/v) OrangeG). First dimension separation was done via the IPGphor system (GE Healthcare) with the following conditions: 1.5 h at 150 V, 1 h rump up to 500 V, 2 h rump up to 1000 V, 3 h rump up to 8000 V and 8 h at 8000 V. IPG strips were incubated for 20 min in a buffer containing 6 m urea, 30% glycerol, 2% SDS, 50 mm Tris-Cl pH 8.8 and 1% DTT and 20 min in the same buffer with 4% iodoacetamide and 0.02% bromphenol blue. Second dimension separation was performed in 12.5% SDS gels at 12 mA/gel on the Ettan DaltSix system (GE Healthcare).
Labeled spots were visualized using the Typhoon Trio imager (GE Healthcare) and images were processed via ImageQuant software (GE Healthcare) and analyzed by DeCyder 2-D Differential Analysis Software version 6.5 (Amersham Biosciences GE Healthcare). For each spot, the software reported the standardized abundance as the ratio of the volume in the Cy3 (or Cy5) sample to the volume of Cy2-labeled standard. Fold change was calculated as the ratio of the average standardized abundance in pairwise comparisons among the four groups. The Extended Data Analysis module carried out intra- and intergel statistical analyses, based on PCA and ANOVA. Each comparison was filtered to find spots reaching a p value <0.05 (t test) by applying the false discovery rate (FDR) correction method (12). Differentially expressed spots between pairwise comparisons among the different time points following PACAP treatment were identified by mass spectrometry. To this aim, after image acquisition, gels were fixed in 50% methanol, 7% acetic acid for 1 h and then visualized with silver staining to pick spots of interest (13).
In-Gel Digestion and Mass Spectrometry
Gel pieces were manually cut out of a preparative gel run with 300 μg of proteins, destained by washing twice with 6 mm potassium ferricyanide in 100 g/L sodium thiosulfate, three times with MilliQ water and three times with acetonitrile (ACN) and subsequently rehydrated in a 100 mm ammonium bicarbonate solution for 10 min followed by a rehydration step in ACN for 10 min. This last step was repeated twice prior to dehydrating the gel pieces in a SpeedVac. Gel pieces were incubated overnight in digestion buffer (50 mm ammonium bicarbonate, 5 mm CaCl2) containing modified trypsin (Promega, Madison, WI) at 37 °C. The resulting peptides were extracted with 50 mm ammonium bicarbonate and then 60% (v/v) ACN and 0.1% TFA for 30 min each. Pooled supernatants were dried in a SpeedVac. Upon concentrating and desalting the tryptic fragments using Millipore C18 ZipTips (Millipore, Bedford, MA), the samples were mixed in a 1:1 (v/v) ratio with α-cyano-4-hydroxycinnamic acid matrix (saturated solution in 50% ACN and 2.5% trifluoroacetic acid) and spotted onto the MALDI target plate. MS/MS analyses were performed on a 4800 MALDI-TOF/TOF instrument (Applied Biosystems, Foster City, CA) after calibration with Applied Biosystems Calibration Mixture 1. Measurements were taken in the positive ion mode between 900 and 3000 m/z. Sequences were automatically acquired by scanning first in MS mode and selecting the 15 most intense ions for MS/MS, using an exclusion list of peaks arising from tryptic autodigestion.
Data interpretation was carried out using the GPS Explorer software version 3.5, with the following parameters for MS: mass range of 900–3500Da; peak density of max 10 peaks per 200 Da; minimum S/N of 10; minimum area of 100; maximum 50 peaks/spot. The same settings have been used for MS/MS, except the mass range of 60 Da and 20 precursors. Database searching was performed using the Mascot program version 2.0.00 by using a target/decoy approach (14) and considering a FDR of 0% for peptide matches above the identity threshold. MS/MS searches were conducted with the following settings: taxonomy set on humans, MSDB (release date: 31.08.2006, locally installed 19.07.2007; number of all entries: 3239079; number of entries for Homo sapiens: 148148) or SwissProt (UniProt_SwissProt, release date: 04.2010; number of all entries: 517000; number of entries for Homo sapiens: 20367) as database, peptide and fragment tolerance equal or lower than 0.4 Da, carbamidomethylation of cysteine as fixed modification, methionine oxidation as variable modification, one missed cleavage allowed in case of incomplete trypsin hydrolysis, precursor charge +1. Using these parameters the probability-based Molecular Weight Search (MOWSE) scores greater than the given cutoff value for MS/MS fragmentation data were taken as significant (p < 0.05). The significant score calculated by Mascot software for a threshold of p < 0.05 was 35 for the majority of protein identifications, but the lowest score was 24.
Meta-analysis and Bioinformatics Analysis
A literature search in PubMed till July 1st, 2010 was performed for the meta-analysis using a search strategy designed to identify studies evaluating gene or protein expression alterations induced by PACAP. PubMed search parameters were: ((“PACAP”[MeSH Terms] OR (“PACAP”[All Fields]) AND (“proteomics”[All Fields] OR “2D”[All Fields] OR “mass spectrometry”[All Fields] OR “microarray”[All Fields] OR “array”[All Fields]). PACAP regulated genes/proteins were analyzed by Ingenuity Pathway Analysis (IPA; Ingenuity Systems, Mountain View, CA; www.ingenuity.com). Networks with scores of 10 or higher were retained.
Real Time PCR Apoptosis Array
Human CD34+ cells were isolated from peripheral blood of the trisomy 18p patient and a healthy volunteer by magnetic cell sorting (Miltenyi Biotec, Auburn, CA) (5). Isolated CD34+ cells were cultured in StemSpan SFEM medium for 13 days (Stem cell technologies, Vancouver, Canada), supplemented with 20 ng/ml TPO, 10 ng/ml SCF, 10 ng/ml IL-6 and 10 ng/ml FLT-3 (Peprotech, London, United Kingdom). The following cytokines were added at day 6: 10 ng/ml TPO, 10 ng/ml IL-6 and 10 ng/ml IL-1β (5).
On day 13, in vitro differentiated megakaryocytes were costained with FITC-conjugated CD41a (HIP8) (BD Biosciences Pharmingen, Heidelberg, Germany) and PE-conjugated CD61 (Y2/51) (Miltenyi Biotec, Auburn, CA) and matched isotype controls. We used Cell Quest software for two-color immunofluorescence acquisition on a FACSCalibur flow cytometer (both from BD Biosciences Pharmingen) and data analysis.
RNA was isolated on day 13 using the RNeasy mini kit according to the manufacturer's instructions (Qiagen, Venlo, Netherlands) and residual DNA was removed using the Turbo DNA-free kit (Ambion, Austin, TX, USA). Corresponding cDNA was made using the RT2 First Strand Kit (SA Biosciences, Frederick, MD, USA) and analyzed for the expressed quantity of apoptosis related genes. We used the Human Apoptosis RT2 Profiler™ PCR Array (SA Biosciences) that profiles the expression of 84 key genes involved in apoptosis following the manufacturer's instructions.
cAMP Measurements
The cAMP levels for the CHRF cells at the four points of the PACAP time course were measured using a cAMP enzyme immunoassay (GE Healthcare Life Sciences, Uppsala, Sweden), as described (5).
Preparation of Nuclear and Cellular Extracts
Nuclear and cytoplasmic extracts were obtained from CHRF cells using the NE-PER kit (Thermo, Rockford, IL) according to the manufacturer's instructions.
Immunoblot Analysis
Equal amounts of protein extracts (30 μg) from CHRF cells were resolved by SDS-PAGE on 10% gels and transferred onto Hybond ECL-nitrocellulose membrane (GE Healthcare). Membranes were blocked in 5% milk powder in Tris-buffered saline-Tween 20 (TBST; 10 mm Tris-HCl pH 8.0, 150 mm sodium cloride, 0,1% Tween 20) for 2 h and then incubated overnight at 4 °C with one of the following primary antibodies (1:500): mouse rabbit superoxide dismutase, goat calpain, rabbit NF-κB p50 and p65, mouse adenylyl cyclase associated protein 1, B-tubulin (all from Santa Cruz Biotechnology; Santa Cruz, CA), rabbit integrin β3 (Cell Signaling Technology, Danvers, MA) and VPAC1 produced in our laboratory as previously described (5). Subsequently, membranes were incubated with HRP-conjugated secondary antibody (1:1000). Staining was performed with the Western blotting ECL detection reagent (GE Healthcare). Expression was quantified by measuring the density of the bands using the Java image processing program ImageJ version 1.34 (NIH Image software).
Immunostaining
CHRF cells or fibroblasts from the patient (3, 5) and two normal controls were plated for 1 h at 37 °C on glass cover slips coated with human fibrinogen (100 μg/ml), previously blocked with 1% bovine serum albumin for 1h at 37 °C. Cells were washed with PBS, fixed with 3% PFA and 300 mm sucrose for 20 min and permeabilized with 0.5% Triton X-100 for 15 min. Cells were stained with phalloidin-rhodamine (Sigma) for 30 min, washed five times with PBS and incubated with an adenylyl cyclase associated protein 1, superoxide dismutase, NF-κB p50, NF-κB p65 or gelsolin antibody (1:50) for 1 h. After five washing steps with PBS, cells were incubated with a 1:200 diluted specific secondary antibody (Alexa Fluor 488 conjugated; Invitrogen) for 45 min at 37 °C. Cover slips were analyzed on a Zeiss CLSM510 confocal microscope (Carl Zeiss Inc., Germany) at 63 × (for CHRF cells) or 20 × (for fibroblasts) magnification. Images were analyzed using the KS 300 version 3.0 program (Carl Zeiss).
Immunoprecipitation Assay
Five hundred micrograms of CHRF lysate proteins were diluted in 1 ml of binding buffer (20 mm Tris pH 7.5, 150 mm sodium chloride, 1% Nonidet P-40, 1 mm DTT with protease inhibitors) and incubated with 100 μl of protein G-Sepharose beads for 1 h at 4 °C with rotation. The samples were next immunoprecipitated using 1 μg of NF-κB p50 or p65 antibody (Santa Cruz) at 4 °C overnight with rotation. A no-antibody control was included for each experiment. Beads were washed three times with TBS-Tween and immune complexes were collected in sample buffer by heating at 90 °C, loaded on 10% SDS-PAGE and visualized by silver stain. Protein bands of interest were cut from the gel and identified by mass spectrometry as described above.
Apoptosis Assay
Serum starved CHRF cells were grown in presence or absence of 100 nm PACAP38 and with or without 250 nm PG 97–269 (15), a VPAC1 selective antagonist (kind gift from Dr. Robberecht, Université Libre de Bruxelles, Belgium) for 24 h. APC-conjugated recombinant Annexin V (ImmuniTools, Friesoythe, Germany) was used for the detection of apoptotic cells. FACSDiva software was used for immunofluorescence acquisition on the FACSCalibur flow cytometer and data analysis.
RESULTS
Proteomic Analysis of Megakaryocytic CHRF Cells After PACAP Treatment
CHRF cells are widely used as a model for human megakaryocytes and express VPAC1 but not PAC1 or VPAC2 receptors, as shown by regular and real time RT-PCR analysis (Fig. 1A). RT-PCR analysis of white blood cells and SK-N-SH cells were included as positive control for PAC1 and VPAC2 amplification. Proteomic profile changes in PACAP-treated CHRF cells were determined at the following time points: 0 h without addition of PACAP38, 9 h and 4 days after the addition of PACAP to monitor early and late response, and 8 days after a PACAP wash out of 4 days (Fig. 1B). Immunoblot analysis showed no changes in VPAC1 expression during the time course of the experiment or due to PACAP treatment (Fig. 1A). Equal amounts (50 ug) of protein extracts from CHRF cells at the different time points were labeled with Cy3 or Cy5 dyes. Replicate samples from each condition were never labeled all with Cy3 or Cy5 to avoid expression differences based on labeling efficiency. A representative DIGE gel for each time point is shown (Fig. 1C). Replicates performed for each condition revealed about 1700 spots with high reproducibility.
Identification of Proteins Regulated by PACAP in CHRF Cells
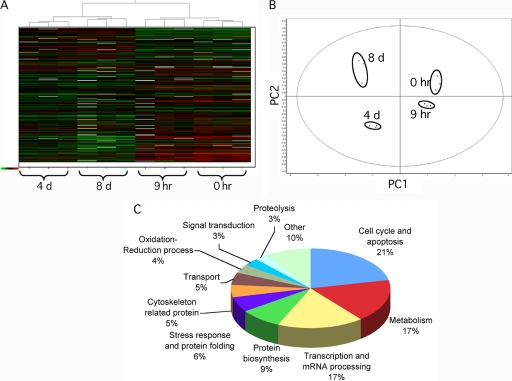
Decyder software was used to detect differentially expressed proteins among the four conditions (p < 0.05 by the application of FDR correction method). The following additional selection criteria were used: (1) proteins with a threshold expression ratio >1.5 (above the average plus standard deviation of the absolute values of the expression ratios of the statistically significant modulated spots) and present in at least 2 out of 3 gel replicates were considered, (2) all spots found to be differentially expressed by Decyder software were manually checked to have the 3D profile characteristics of a protein spot, and (3) spots with volume measurements close to background values were excluded. ANOVA was performed on the normalized abundances of matched spots to compare similarity according to the expression patterns among CHRF cells during PACAP addition and removal. Fig. 2A reports the heat map resulting from hierarchical clustering analysis in which each vertical column represents one proteomic map of the 12 samples (three replicates for each time point) and each horizontal row represents one protein with relative expression values ranging from green (decreased expression) to red (increased expression). Hierarchical clustering analysis classified the 12 samples in four major clusters, each corresponding to a different time point during PACAP treatment. These grouping assignments were reiterated in an unsupervised PCA of the protein expression patterns within each sample (Fig. 2B). The first and second principal component, which distinguished 88% of the variance, correctly assigned the different experimental groups during PACAP treatment. Moreover, the proteomic profile of cells after 9 h of PACAP treatment more closely resembled the initial condition than after 4 days of treatment, indicating that most protein changes are related to late stages of PACAP response. After PACAP removal, the proteomic profile of CHRF cells returned closer to the initial condition though not completely, suggesting that several changes induced by PACAP were irreversible. PCA and hierarchical cluster analysis indicated a complete separation of the four groups, demonstrating high reproducibility among the replicate DIGE samples. It is noteworthy to highlight that statistical analysis is performed without a priori assigning the samples to the different experimental conditions.
Fig. 2.
Multivariate analysis of DIGE results. A, Hierarchical clustering of the CHRF cell lysates based on the global expression patterns of modulated proteins detailed in supplemental Table S1. In the heat map, each vertical column represents one proteomic map of the 12 samples, whereas each horizontal row represents one protein, with relative expression values displayed as an expression matrix (heat map) using a relative scale ranging from –0.5 (green) to +0.5 (red). The green and red color mean that the expression decreased or increased compared with the standard, respectively: the brighter the color, the stronger the change. B, Each dot in the scatter plots represents a proteomic map, which describes the global expression values for the subset of proteins listed in supplemental Table S1. The PCA and hierarchical clustering analysis were consistent between them and indicated a complete and clear separation of the 12 individual Cy3- and Cy5-labeled DIGE expression maps into the four points of the time course following PACAP treatment. PC1: principal component analysis 1; PC2: principal component analysis 2. C, Functional classification of PACAP modulated proteins in CHRF cells identified by DIGE and MS. Proteins were classified using the information provided by the Gene Ontology project (http://amigo.geneontology.org/cgi-bin/amigo/go.cgi).
Pairwise comparisons between the different time point gels revealed that 159 protein spots were modulated by PACAP (p < 0.05, FDR) (Table I). No significant protein changes were found at the early 9 h response. The majority of proteins found to be differentially expressed were detected at 8 days, i.e. after 4 days of PACAP removal. The fold change (increase or decrease) in protein expression among differentially expressed spots was 1.5–1.7 for 70%, 1.5–1.7 for 22%, and >2.0 for 8% of the spots. Out of the 159 differentially expressed proteins with a foldchange >1.5, 120 could be identified by MALDI TOF-TOF MS (supplemental Table S1 and S2). Some protein spots consist of multiple proteins (supplemental Table S1B); in this cases, the top-hit identification by MALDI-TOF/TOF were considered, assuming that the hit rank, as well as the sequence coverage and the number of matching peptides correlate with protein abundance (16). In addition, more than one spot was found to correspond to the same protein, probably indicating the presence of posttranslational modifications of the same protein: therefore the total number of uniquely identified proteins was 95. These could be divided into four groups according to their expression kinetics during stimulation with PACAP: (1) proteins up-regulated throughout the experiment (irreversible up-regulation); (2) proteins whose expression increased after PACAP treatment, but decreased or remained unchanged after PACAP removal (transient up-regulation); (3) proteins down-regulated throughout the experiment (irreversible down-regulation); (4) proteins whose expression decreased after PACAP treatment, but increased or remained unchanged after PACAP removal (transient down-regulation) (supplemental Fig. S1). Proteins could also be classified according to their biological function (Fig. 2C). The most represented classes were those of cell cycle and apoptosis (21%), metabolism (17%), transcription and mRNA processing (17%) and protein biosynthesis (9%).
Table I. Distribution of the spots differentially expressed (p < 0.05, FDR) between pairwise comparisons of the different points of the time course following PACAP treatment (0 hr, 9 hr, 4 day, and 8 day), as detected by DIGE.
| N° of protein spots | |||
|---|---|---|---|
| total | ↑ | ↓ | |
| 0hr - 9hr | 0 | 0 | 0 |
| 0hr - 4d | 47 | 15 | 32 |
| 0hr - 8d | 119 | 51 | 68 |
| 9hr - 4d | 27 | 7 | 20 |
| 9hr - 8d | 99 | 35 | 64 |
| 4d - 8d | 4 | 2 | 2 |
Validation of Proteomic Results by Immunoblot Analysis and Immunohistochemistry
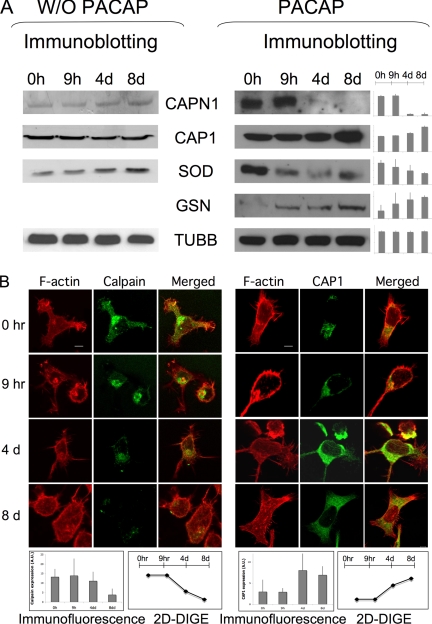
To confirm these protein expression differences observed in CHRF cells during PACAP treatment, some of the proteins were subsequently studied via immunoblot analysis. These include calpain (CAPN1), adenylyl cyclase-associated protein 1 (CAP1), superoxide dismutase (SOD) and gelsolin (GSN), and Beta-tubulin (TUBB) was used as loading control (Fig. 3A). Densitometric quantitation of the blots revealed similar expression profile changes as observed by DIGE data for all studied proteins (Fig. 3A and supplemental Table S1).
Fig. 3.
Validation of the DIGE/MS results by immunoblot and immunofluorescence analysis. A, Conventional 1D SDS-PAGE gels were run with proteins from CHRF cells treated with PACAP at several time points. Proteins were transferred onto nitrocellulose membranes and incubated with antibodies against some of the proteins identified in DIGE/MS analysis: calpain (CAPN1, 28kDa), adenylyl cyclase-associated protein 1 (CAP1, 52kDa), superoxide dismutase (SOD1, 16kDa) and gelsolin (GSN, 86kDa). Beta-tubulin (TUBB, 50kDa) was used as the internal loading control. The densitometric quantification of protein patterns showed similar trends with DIGE data for all proteins analyzed. Data obtained by performing the same experiment with vehicle PBS indicate that the change in protein expression observed after PACAP treatment are PACAP related and not due to CHRF cell growth. B, CHRF cells at the different points following PACAP treatment were plated on coverslips precoated with fibrinogen (100 μg/ml) 1 h at 37 °C. Cells were stained with rhodamin-phalloidin to visualize F-actin and after with Alexa468 anti-CAPN1 or -CAP1 antibody. The degrees of differential expression in the different time points for both DIGE (supplemental Table S1) and immunohistochemical analysis are highly consistent and are shown in the graphics as average of three experiment replicates ± standard deviation. Bar = 5 μm.
We also performed immunoblot analysis of samples collected at the same time points during CHRF growth, but with addition of vehicle PBS instead of PACAP. The level of CAPN1 and CAP1, which showed respectively a decreased and increased expression after PACAP treatment, remained unchanged without PACAP addition (Fig. 3A). SOD1, down-regulated during PACAP treatment, was up-regulated at 4 and 8 days without PACAP (Fig. 3A). These data indicate that the changes in protein expression observed after PACAP treatment by DIGE are actually because of a PACAP effect and not just induced by cellular growth.
CHRF cells were also analyzed by immunohistochemistry as validation assay to study protein expression differences for CAP1 and CAPN1. Cells were seeded on fibrinogen-coated coverslips and double stained with fluorescent phalloidin and an anti-CAPN1 or -CAP1 antibody. A similar change in expression was again found (up- and down-regulation for CAP1 and CAPN1, respectively), as detected by DIGE (Fig. 3B). Moreover, after PACAP treatment CAPN1 was detected in the cell membrane and nucleus, while its initial cellular localization is mainly the cytosol (Fig. 3B). Interestingly, it was already previously shown that PACAP induced CAPN1 translocation from the cytosol to the membrane of rat pituitary cells (17).
Bioinformatic Analysis Revealed Apoptosis as Major PACAP Regulatory Pathway
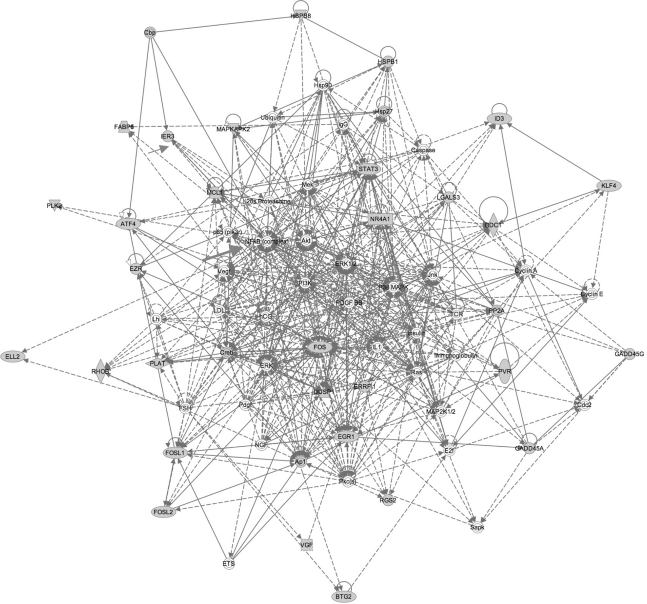
DIGE identified 120 differentially expressed proteins after PACAP treatment of CHRF cells that seemed to be mainly involved in cell cycling and apoptosis. IPA was then used to explore the presence of relevant networks among these proteins (listed in supplemental Table S1). Highly interconnected networks are likely to represent significant biological functions and the score is calculated as the likelihood that proteins are in the network because of random chance. The highest ranked network resulting from our study consists of 35 proteins of which 34 were actually identified by our DIGE approach and one, namely NF-κB, was present based upon reported interactions with the other DIGE-based identified proteins (supplemental Fig. S2).
A literature-based meta-analysis was subsequently performed to evaluate the main pathways involved in other cellular models of PACAP response. From a total of 60 PubMed papers, 12 unrelated studies (listed in supplemental Table S3) were selected based on the availability of incorporated raw data of all genes or proteins that were obtained from transcriptomic or proteomic platforms and used to identify genes or proteins modulated by PACAP in diverse biological models: bovine chromaffin, rat PC12 cells, primary sympathetic neuronal cells, chick embryo ciliary ganglion neurons, rat tear, chicken endolymph and adrenal glands and brain cortex of PACAP knockout versus wild-type mice. The complete list with potential biomarkers of PACAP response reported in these studies and our DIGE analysis on CHRF cells was used for an additional IPA analysis. From the 2384 differentially expressed genes and/or proteins, 83 were further selected for IPA as being modified by PACAP in at least 3 different studies (Table II). The highest scored network was implicated in apoptosis, namely the “Cell Death, Endocrine System Disorders and Metabolic Disease” cluster, with an overall score of 59 (Fig. 4). Interestingly, NF-κB was again an important player of this network. Three other important genes modulated by PACAP in different models are IER3, HSPA5 and FABP5, all previously described with anti-apoptotic properties (Table II) (18–20).
Table II. PACAP-responsive genes/proteins identified at least in three of the data sets derived from the 13 studies (12 papers selected from literature and our study) used for meta-analysis. The genes/proteins found also in our study are listed in bold.
| N° studies | Gene symbol |
|---|---|
| 7 | Ier3 |
| 6 | Azin1, Odc1, Ptp4a1 |
| 5 | Anxa2, Egr1, Fos, Gas1, Klf4, Lgals3, Lmo1, OLFM3, Plat, Rgs2, Snap25, Tubb2b |
| 4 | Akr1b3, ATP11A, Btg2, Cd63, Coq10b, Ezr, Fosl2, Herc4, Hspb1, Hspb8, Mapkapk2, Obfc2a, Por, Stk40 |
| 3 | 2810407C02Rik, Actg1, Agfg1, Agl, Akr1b7, ALCAM, Ap1s2, Arf4, ARL4A, Arpc3, As3mt, Atf4, B4galt3, Cited1, Creb3l2, Csrnp1, Csrp1, Dgat2, Dusp1, Ehd4, Eif1a, Ell2, Errfi1, Fabp5, Fosl1, Gadd45a, Gadd45g, Hmgb2, HspA5, Id3, Lama5, Mcl1, Nr4a1, Pdk3, Plk2, Plxna2, Ppp2r2b, Ptges3, PTPRO, Pvr, RHOB, Serpinb1a, Slc7a8, SMAD1, Stat3, Syt4, Tead1, Tpm4, Tsc22d1, Tuba1, Vgf, Zdbf2, Znhit6 |
Fig. 4.
Regulatory network for PACAP-responsive genes. PACAP-regulated transcripts or proteins common to CHRF, PC12 and chromaffin primary sympathetic neuronal cells, ciliary ganglion neurons or PACAP knock out and wild-type mice (listed in Table II) were used as input for analysis of regulatory networks using IPA. The pathway showed is the highest-ranked network (score = 59). Abbreviations: Akt: Protein Kinase B; ATF4: activating transcription factor 4 (tax-responsive enhancer element B67); ATF4: activating transcription factor 4 (tax-responsive enhancer element B67); BTG2: BTG family, member 2; Caspase: caspase-1; Cbp: carboxypeptidase B1; Cdc2: cell cycle controller CDC2; Creb: Creb protein; DUSP1: dual specificity phosphatase 1; EGR1: early growth response 1; ELL2: elongation factor, RNA polymerase II, 2; ERRFI1: ERBB receptor feedback inhibitor 1; EZR: ezrin; FABP5: fatty acid binding protein 5 (psoriasis-associated); FOS: FBJ murine osteosarcoma viral oncogene homolog; FOSL1: FOS-like antigen 1; FOSL2: FOS-like antigen 2; FSH: follitropin; GADD45A: growth arrest and DNA-damage-inducible, alpha; GADD45G: growth arrest and DNA-damage-inducible, gamma; hCG: CG, Chorionic Gonadotropin; Hsp27: heat shock 27kDa protein; Hsp90: heat shock 90kDa protein; HSPB1: heat shock 27kDa protein 1; HSPB8: heat shock 22kDa protein 8; ID3: inhibitor of DNA binding 3, dominant negative helix-loop-helix protein; IER3: immediate early response 3; IL1: Interleukin 1; KLF4: Kruppel-like factor 4 (gut); LDL: Beta Lipoprotein, low density lipoprotein; LGALS3: lectin, galactoside-binding, soluble, 3; Lh: Luteinizing hormone; MAPKAPK2: mitogen-activated protein kinase-activated protein kinase 2; MCL1: myeloid cell leukemia sequence 1 (BCL2-related); Mek: mitogen-activated protein kinase kinase; NGF: nerve growth factor; NR4A1: nuclear receptor subfamily 4, group A, member 1; ODC1: ornithine decarboxylase 1; Pdgf: Platelet-Derived Growth Factor; PDGF BB: Pdgfb dimer; PI3K: Pi 3-kinase; Pkc(s): Protein kinase c; PLAT: plasminogen activator, tissue; PLK2: polo-like kinase 2 (Drosophila); PP2A: Protein Phosphatase Type2a; PVR: poliovirus receptor; RGS2: regulator of G-protein signaling 2, 24kDa; RHOB: ras homolog gene family, member B; Sapk: Stress-responsive protein kinase; STAT3: signal transducer and activator of transcription 3 (acute-phase response factor); TCR: T cell receptor, TCR protein; VGF: VGF nerve growth factor inducible.
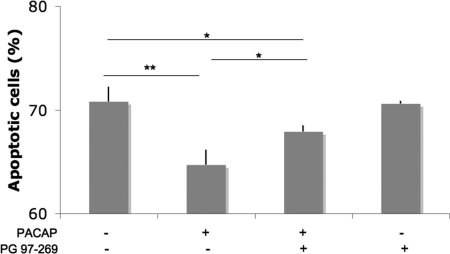
The degree of apoptosis was also assessed by measuring the level of expression of Annexin V by flow cytometry analysis of CHRF cells grown in the absence of serum for 24 h. The addition of PACAP decreased apoptosis, whereas this effect could be abolished by co-adding PG 97–269, a selective VPAC1 antagonist (Fig. 5C). This indicates that PACAP could protect CHRF cells via its receptor VPAC1 from defects in cell proliferation, cell cycling and/or the apoptotic regulation induced by serum starvation (21). However, further detailed studies are needed to define the precise mechanism for this process.
Fig. 5.
PACAP protects CHRF cells from serum starvation via its VPAC1 receptor. CHRF cells were grown in serum free medium for 24h in presence or absence of PACAP 100 nm and the VPAC1 selective antagonist PG 97–269 (250 nm) and apoptotic cells were measured by Annexin V flow cytometric analysis (mean of three independent experiments + standard deviation). The presence of PG 97–269 reduces the apoptotic inhibition induced by PACAP. *p < 0.05; **p < 0.01.
PACAP Modulates NF-κB Signaling in Megakaryocytes
We previously found that PACAP increases intracellular cAMP levels in CACO-2 cells, platelets, and megakaryocytes via the VPAC1 receptor (3, 5). Our initial hypothesis consisted of cAMP as secondary messenger activating some cAMP responsive element binding (CREB)-dependent genes and thereby being an inhibitor of megakaryopoiesis. However, this hypothesis failed as (1) intracellular cAMP levels in CHRF cells were not significantly different during the time points of PACAP treatment chosen for this study and (2) the increase of cAMP after the addition of forskolin, the direct activator of adenylyl cyclase, could not completely mimic the PACAP-induced inhibition of megakaryocyte proliferation and differentiation using an in vitro megakaryocyte differentiation assay (data not shown).
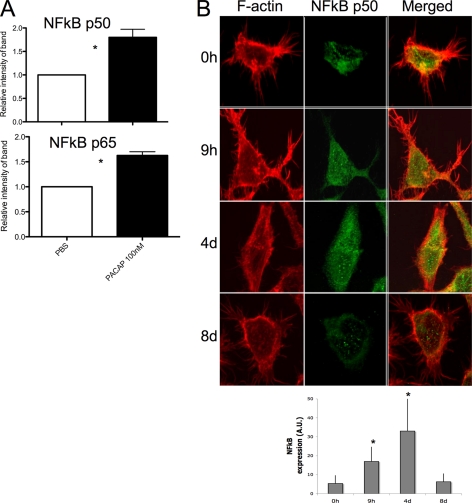
Therefore, based on the proteomic and bioinformatic data obtained from this study, we further investigated NF-κB signaling in megakaryocytes after PACAP treatment. The relative protein expression of the active NF-κB p50/p65 heterodimer was measured in nuclear extracts from CHRF cells treated with PACAP in three independent experiments. It is known that upon activation of this pathway, the p50/p65 heterodimer translocates to the nucleus to activate some target genes (22). An increased level of nuclear p50/p65 NF-κB was observed in CHRF cells after 1 h of PACAP treatment by immunoblot analysis (Fig. 6A). Moreover, immunohistochemical analysis of p50 NF-κB showed a different subcellular localization in CHRF cells spread on fibrinogen. PACAP treatment (9 h and 4 days) induced p50 NF-κB expression, whereas a decrease was observed after PACAP removal (Fig. 6B). In addition, p50 NF-κB seemed to accumulate in the nucleus after PACAP treatment (Fig. 6B). PACAP induced NF-κB expression could also be observed in CACO-2 cells having high VPAC1 expression levels (data not shown).
Fig. 6.
PACAP stimulates NF-κB signaling in CHRF cells. A, CHRF cells were stimulated for 1 h with 100 nm PACAP38 or vehicle PBS. Nuclear extracts were analyzed by immunoblotting for NF-κB subunits p50 and p65. Higher nuclear levels of p50 and p65 (p < 0.05) were observed when PACAP was added to the cells. B, CHRF cells at the different time points following PACAP treatment were plated on coverslips precoated with fibrinogen (100 μg/ml) for 1 h at 37 °C. Cells were stained with rhodamin-phalloidin to visualize F-actin and after with Alexa468 anti-NF-κB p50. The increased expression of NF-kB subunits after PACAP treatment were consistently detected by both immunoblot and immunofluorescence analysis. Figures show the average of three independent experiments + standard deviation. *p < 0.05.
Immunoprecipitation experiments coupled to mass spectrometry were carried out using CHRF cell lysates in order to identify the molecular binding partners of the p50/p60 NF-κB heterodimer components. Some of these interacting proteins (listed in supplemental Table S4 and S2B) are also known to be involved in regulation of apoptosis.
Validation of the Apoptosis Pathway in In Vitro Differentiated Megakaryocytes from the Trisomy 18p Patient
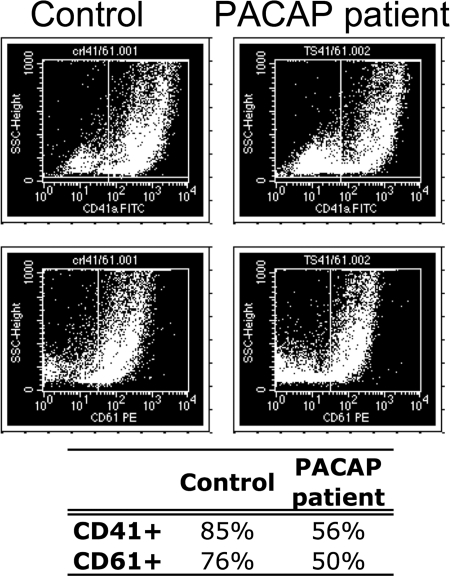
All data from this study showed that PACAP induces changes in the apoptotic pathway in megakaryocytic CHRF cells. Therefore, an apoptosis gene expression array was carried out using RNA extracted from in vitro differentiated megakaryocytes from the trisomy 18p patient with PACAP overexpression (3, 5) and from a normal control subject. Peripheral blood isolated CD34+ hematopoietic stem cells were in vitro differentiated into megakaryocytes for 13 days. The inhibitory role of PACAP during a late stage of megakaryocytic differentiation was obvious since flow cytometry showed a reduced number of differentiated megakaryocytes in the culture from the PACAP overexpressing patient compared with the control (Fig. 7A). Indeed, about 85% CD41+ and 76% CD61+ megakaryocytic cells for the control versus 56 and 50% for the PACAP overexpressing patient were observed. Total RNA was extracted from these megakaryocytic cells for a real time PCR apoptosis array. Of the 84 apoptotic regulatory genes present in the array, 15 were found to be increased or decreased by at least fourfold in the patient versus control megakaryocytes (p < 0.05, FDR) (Table III).
Fig. 7.
Impaired megakaryopoiesis in a PACAP overexpressing patient. The in vitro differentiated megakaryocytes from the PACAP overexpressing patient clearly show an impaired megakaryopoiesis since less CD41+ and CD61+ megakaryocytic cells were formed (13 days).
Table III. List of dysregulated apoptotic genes in CD34+ derived megakaryocytes of one patient (P) with increased plasmatic level of PACAP compared to one healthy control (C).
| Symbol | Protein product | Definition | Fold change (P/C) |
|---|---|---|---|
| TNFRSF10A | NM_003844 | Tumor necrosis factor receptor superfamily, member 10a | 93.6 |
| TNFSF8 | NM_001244 | Tumor necrosis factor (ligand) superfamily, member 8 | 68.6 |
| TNFRSF10B | NM_003842 | Tumor necrosis factor receptor superfamily, member 10b | 33.9 |
| CD70 | NM_001252 | CD70 molecule | 26.8 |
| CD27 | NM_001242 | CD27 molecule | 18.9 |
| CASP1 | NM_033292 | Caspase 1, apoptosis-related cysteine peptidase | 16.0 |
| BCL2A1 | NM_004049 | BCL2-related protein A1 | 9.2 |
| TNFSF10 | NM_003810 | Tumor necrosis factor (ligand) superfamily, member 10 | 8.2 |
| PYCARD | NM_013258 | PYD and CARD domain containing | 7.6 |
| BCL2 | NM_000633 | B-cell CLL/lymphoma 2 | 6.7 |
| DFFA | NM_004401 | DNA fragmentation factor, 45kDa, alpha polypeptide | 6.0 |
| DAPK1 | NM_004938 | Death-associated protein kinase 1 | 5.6 |
| TRAF3 | NM_003300 | TNF receptor-associated factor 3 | 5.0 |
| TRAF4 | NM_004295 | TNF receptor-associated factor 4 | 4.4 |
| FASLG | NM_000639 | Fas ligand (TNF superfamily, member 6) | −8.0 |
Moreover, to elucidate which relevant pathways were represented among the genes identified by the real time PCR apoptosis array, we applied IPA. The pathway in supplemental Fig. S3 represents the highest score resulting network (score of 35), in which NF-κB revealed to be once more an important player.
DISCUSSION
We previously found that PACAP and its receptor VPAC1 negatively regulate megakaryopoiesis and platelet function in patients having three copies of the PACAP gene, elevated PACAP plasma concentrations and mild thrombocytopenia (3, 5), but the underlying molecular mechanisms remained to be elucidated. In this study, we used the innovative and powerful proteomic-based DIGE technology coupled to mass spectrometry to identify the proteins modulated by PACAP in megakaryocytic CHRF cells. We used the human megakaryocytic CHRF-288–11 cell line because it was not possible to obtain a sufficient amount of in vitro differentiated primary megakaryocytic cells to perform DIGE for different time points of PACAP treatment. Furthermore, the use of in vitro differentiated megakaryocytes are not suitable for this experiment because PACAP treatment inhibits megakaryocyte differentiation, which would obviously leads to different protein expression patterns at the different time points not because of the PACAP treatment itself but to differences in the degree of megakaryocyte differentiation. Therefore, CHRF cells represented the best choice, being the most mature megakaryoblast-like cell line available and expressing all important markers and signal transduction pathways characteristic of megakaryocytes and platelets (23–25). Moreover, CHRF cells are generally considered as a valuable model of a late stage megakaryocytic differentiation (26–31). However, this also implicates that this study shows the downstream effects of PACAP on a certain differentiation stage of megakaryocytes and not during megakaryopoiesis itself.
We compared the proteomic profile of CHRF cells treated with PACAP for 9 h, 4 days, and 4 days after PACAP removal at day 8, with that of untreated cells to determine the downstream signaling pathways associated with VPAC1 stimulation by PACAP. This specific treatment schedule was chosen based on other studies using different cell types, but always indicating that early (9 h) as well as late-response (4 days) genes are induced by PACAP (32–34). We did not include a DIGE analysis of the culture conditions without adding PACAP for all different time points because preliminary experiments showed that the proteomic profile of CHRF cells grown in presence of PACAP or vehicle PBS for 4 days are significantly different (data not shown). Moreover, our experimental design was conceived in a way that all CHRF cells obtained the same stage of growth before cell collection (Fig. 1B). Therefore, all changes detected in our study are specifically because of the effect of PACAP treatment. As a confirmation however, our immunoblot analysis of some of the differentially expressed proteins identified by DIGE with and without PACAP treatment showed changes in protein expression only upon addition of PACAP to the culture (Fig. 3A).
The main findings of this study lead to the hypothesis that PACAP represents a novel regulator of apoptosis in megakaryocytic CHRF cells and megakaryocytes from a PACAP overexpressing patient. Our proteomic analysis indeed revealed that the majority of proteins modulated by PACAP in CHRF cells belong to the functional class of cell cycle and apoptosis. Moreover, PACAP protects CHRF cells against apoptosis induced by serum removal and this effect could be inhibited by a VPAC1 specific antagonist. A literature-based meta-analysis including different models of PACAP response also resulted in the identification of multiple markers involved in apoptosis. Finally, several apoptotic genes were differentially expressed in in vitro differentiated megakaryocytes from a PACAP overexpressing patient compared with these from a control using a real time PCR apoptosis array.
Apoptosis is the final step in the differentiation process of mature megakaryocytes before the release of platelets (35–38). PACAP overexpressing patients have a defect in the late phase of megakaryopoiesis, as the histological examination of their bone marrow showed a normal amount of immature megakaryoblasts, but almost no fully mature megakaryocytes (3). We therefore hypothesize that their elevated PACAP levels protect megakaryocytes from undergoing normal apoptosis at the end of their differentiation and therefore prevent megakaryocytes to release platelets, resulting in the development of thrombocytopenia. A dysregulation of megakaryopoiesis hypothetically because of an enhanced activation of the apoptotic process was recently reported in patients with a heterozygous cytochrome c mutation, which showed naked megakaryocytic nuclei and a mild thrombocytopenia (39).
The knowledge of the regulation of apoptosis during megakaryopoiesis is still not well known. Major regulators of apoptosis include members of the B-cell lymphoma 2 family, such as Bcl-2, Bcl-xL (anti-apoptotic) and Bim (pro-apoptotic), which act upstream of caspases. The impact of Bcl-2 has been evaluated mainly in B and T lymphocytes (40, 41) whereas studies of these apoptotic regulators in megakaryocytes remain limited. The overexpression of Bcl-2 resulted in decreased apoptosis and a significant reduction in platelet numbers (38). Moreover, a 2-fold reduction in platelet numbers was found in transgenic mice overexpressing Bcl-2 under the control of a hematopoietic stem cell-specific promoter, although their megakaryocyte numbers remained unchanged (42). A similar observation was obtained in mice with a homozygous deletion of Bim (43).
We hypothesize that NF-κB is the major player in PACAP response in megakaryocytic cells on the basis of different findings presented in this study resulting from the: (1) analysis of modulated proteins in CHRF cells after PACAP treatment, (2) literature-based meta-analysis of PACAP-induced biomarkers, and (3) array analysis of apoptosis-related genes in megakaryocytes from the PACAP overexpressing patient. PACAP increased the nuclear level of the p50/p65 NF-κB heterodimer in CHRF cells. NF-κB is a transcription factor that promotes cell survival by inducing transcription of genes involved in cell proliferation or regulating apoptosis (44). Megakaryocytes and platelets express nearly all known NF-κB family members (45). NF-κB activation participates in thrombopoietin receptor signaling (46, 47), but the exact mechanism of NF- κB signaling in megakaryocyte differentiation and platelet release is not yet known. The ability to induce apoptosis via inhibition of NF-κB has been shown by several compounds in different cells (48–51). Moreover, NF-κB signaling decreases during megakaryopoiesis (52), suggesting that NF-κB signaling might be important to induce apoptosis in megakaryocytes and produce platelets. Several genes (such as Bcl-2, Bcl-2A1, Casp1, Pycard, Traf-3, and Tnfrsf8) that were found in a previously published study (52) were also detected in our apoptosis array as being up-regulated in megakaryocytes from the PACAP overexpressing patient. The fact that NF-κB plays an important role in apoptosis by regulating the expression of different Bcl-2 family members would explain the Bcl-2 overexpression in megakaryocytes from our patient. It is known that PACAP inhibits apoptosis in many other cells (2, 15, 53–60). The anti-apoptotic actions of PACAP have been widely studied though mainly in the nervous system (54, 56, 58, 59), but to our knowledge not in hematopoietic cells.
In conclusion, this study revealed PACAP as a regulator of apoptosis in megakaryocytes. If PACAP induces NF-κB dependent gene expression, it counteracts the physiological decrease in NF-κB signaling that is necessary for the terminal differentiation of megakaryocytes. Further studies are needed to define the specific timing of induction of NF- κB signaling during megakaryocyte differentiation and platelet release and to identify the exact downstream NF- κB candidate genes that are critical for the regulation of this extremely complex differentiation process.
Acknowledgments
We thank Prof Vermylen for critical reading of the manuscript, Dr Vinckier for helping with confocal microscopy analysis (Katholieke Universiteit Leuven, Belgium), Prof Robberecht (Université Libre de Bruxelles, Belgium) for providing PG 97–269 and the Leuven Biostatistics and Statistical Bioinformatics Centre, Dr Boonen, D'Imperio and Mantini for useful statistical discussion.
Footnotes
* This work was supported by ThromboGenics nv. and the Agency for Innovation by Science and Technology (I.W.T. Belgium). CVG is holder of the Bayer and Norbert Heimburger (CSL Behring) Chairs and of a clinical-fundamental research mandate of the FWO-Vlaanderen. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
 This article contains supplemental Figs. S1 to S3 and Tables S1 to S3.
This article contains supplemental Figs. S1 to S3 and Tables S1 to S3.
Author contributions: MDM and KP performed experiments, participated in study design and wrote the manuscript. SL and CT participated in performing experiments. EW and LO participated in proteomic analysis. CVG studied the patient. KF, CVG and MH participated in study design and manuscript preparation.
1 The abbreviations used are:
- PACAP
- pituitary adenylate cyclase-activating polypeptide
- IPA
- ingenuity pathway analysis
- NF-κB
- nuclear transcription factor-kappaB
- FDR
- false discovery rate
- PCA
- principal component analysis
- TBST
- tris-buffered saline-Tween 20
- CAPN1
- calpain
- CAP1
- adenylyl cyclase-associated protein 1.
REFERENCES
- 1. Miyata A., Arimura A., Dahl R. R., Minamino N., Uehara A., Jiang L., Culler M. D., Coy D. H. (1989) Isolation of a novel 38 residue-hypothalamic polypeptide which stimulates adenylate cyclase in pituitary cells. Biochem. Biophys. Res. Commun. 164, 567–574 [DOI] [PubMed] [Google Scholar]
- 2. Vaudry D., Gonzalez B. J., Basille M., Yon L., Fournier A., Vaudry H. (2000) Pituitary adenylate cyclase-activating polypeptide and its receptors: from structure to functions. Pharmacol. Rev. 52, 269–324 [PubMed] [Google Scholar]
- 3. Freson K., Peeters K., De Vos R., Wittevrongel C., Thys C., Hoylaerts M. F., Vermylen J., Van Geet C. (2008) PACAP and its receptor VPAC1 regulate megakaryocyte maturation: therapeutic implications. Blood 111, 1885–1893 [DOI] [PubMed] [Google Scholar]
- 4. Nam C., Case A. J., Hostager B. S., O'Dorisio M. S. (2009) The role of vasoactive intestinal peptide (VIP) in megakaryocyte proliferation. J. Mol. Neurosci. 37, 160–167 [DOI] [PubMed] [Google Scholar]
- 5. Freson K., Hashimoto H., Thys C., Wittevrongel C., Danloy S., Morita Y., Shintani N., Tomiyama Y., Vermylen J., Hoylaerts M. F., Baba A., Van Geet C. (2004) The pituitary adenylate cyclase-activating polypeptide is a physiological inhibitor of platelet activation. J. Clin. Invest. 113, 905–912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Peeters K., Stassen J. M., Collen D., Van Geet C., Freson K. (2008) Emerging treatments for thrombocytopenia: increasing platelet production. Drug Discov. Today 13, 798–806 [DOI] [PubMed] [Google Scholar]
- 7. Unlü M., Morgan M. E., Minden J. S. (1997) Difference gel electrophoresis: a single gel method for detecting changes in protein extracts. Electrophoresis 18, 2071–2077 [DOI] [PubMed] [Google Scholar]
- 8. Marouga R., David S., Hawkins E. (2005) The development of the DIGE system: 2D fluorescence difference gel analysis technology. Anal. Bioanal. Chem. 382, 669–678 [DOI] [PubMed] [Google Scholar]
- 9. Alfonso P., Núñez A., Madoz-Gurpide J., Lombardia L., Sánchez L., Casal J. I. (2005) Proteomic expression analysis of colorectal cancer by two-dimensional differential gel electrophoresis. Proteomics 5, 2602–2611 [DOI] [PubMed] [Google Scholar]
- 10. Cicchillitti L., Di Michele M., Urbani A., Ferlini C., Donat M. B., Scambia G., Rotilio D. (2009) Comparative proteomic analysis of paclitaxel sensitive A2780 epithelial ovarian cancer cell line and its resistant counterpart A2780TC1 by 2D-DIGE: the role of ERp57. J. Proteome Res. 8, 1902–1912 [DOI] [PubMed] [Google Scholar]
- 11. Di Michele M., Della Corte A., Cicchillitti L., Del Boccio P., Urbani A., Ferlini C., Scambia G., Donati M. B., Rotilio D. (2009) A proteomic approach to paclitaxel chemoresistance in ovarian cancer cell lines. Biochim. Biophys. Acta 1794, 225–236 [DOI] [PubMed] [Google Scholar]
- 12. Benjamini Y., Hochberg Y. (2000) On the adaptive control of the false discovery rate in multiple testing with independent statistics. J. Educ. Behav. Stat. 25, 60–83 [Google Scholar]
- 13. Shevchenko A., Wilm M., Vorm O., Mann M. (1996) Mass spectrometric sequencing of proteins silver-stained polyacrylamide gels. Anal. Chem. 68, 850–858 [DOI] [PubMed] [Google Scholar]
- 14. Elias J. E., Haas W., Faherty B. K., Gygi S. P. (2005) Comparative evaluation of mass spectrometry platforms used in large-scale proteomics investigations. Nat. Methods 2, 667–675 [DOI] [PubMed] [Google Scholar]
- 15. Gourlet P., De Neef P., Cnudde J., Waelbroeck M., Robberecht P. (1997) In vitro properties of a high affinity selective antagonist of the VIP1 receptor. Peptides 18, 1555–1560 [DOI] [PubMed] [Google Scholar]
- 16. Yang Y., Thannhauser T. W., Li L., Zhang S. (2007) Development of an integrated approach for evaluation of 2-D gel image analysis: impact of multiple proteins in single spots on comparative proteomics in conventional 2-D gel/MALDI workflow. Electrophoresis 28, 2080–2094 [DOI] [PubMed] [Google Scholar]
- 17. Sato-Kusubata K., Yajima Y., Kawashima S. (2000) Persistent activation of Gsalpha through limited proteolysis by calpain. Biochem. J. 347, 733–740 [PMC free article] [PubMed] [Google Scholar]
- 18. Yu Z., Luo H., Fu W., Mattson M. P. (1999) The endoplasmic reticulum stress-responsive protein GRP78 protects neurons against excitotoxicity and apoptosis: suppression of oxidative stress and stabilization of calcium homeostasis. Exp. Neurol. 155, 302–314 [DOI] [PubMed] [Google Scholar]
- 19. Rocher G., Letourneux C., Lenormand P., Porteu F. (2007) Inhibition of B56-containing protein phosphatase 2As by the early response gene IEX-1 leads to control of Akt activity. J. Biol. Chem. 282, 5468–5477 [DOI] [PubMed] [Google Scholar]
- 20. Ma X., Ren X., Han P., Hu S., Wang J., Yin J. (2010) SiRNA against Fabp5 induces 3T3-L1 cells apoptosis during adipocytic induction. Mol. Biol. Rep. 37, 4003–4011 [DOI] [PubMed] [Google Scholar]
- 21. Schamberger C. J., Gerner C., Cerni C. (2005) Caspase-9 plays a marginal role in serum starvation-induced apoptosis. Exp. Cell Res. 302, 115–128 [DOI] [PubMed] [Google Scholar]
- 22. Pascal V., Nathan N. R., Claudio E., Siebenlist U., Anderson S. K. (2007) NF-kappa B p50/p65 affects the frequency of Ly49 gene expression by NK cells. J. Immunol. 179, 1751–1759 [DOI] [PubMed] [Google Scholar]
- 23. Fugman D. A., Witte D. P., Jones C. L., Aronow B. J., Lieberman M. A. (1990) In vitro establishment and characterization of a human megakaryoblastic cell line. Blood 75, 1252–1261 [PubMed] [Google Scholar]
- 24. Dorn G. W., 2nd, Davis M. G. (1992) Differential megakaryocytic desensitization to platelet agonists. Am. J. Physiol. 263, C864–72 [DOI] [PubMed] [Google Scholar]
- 25. Yang M., Khachigian L. M., Hicks C., Chesterman C. N., Chong B. H. (1997) Identification of PDGF receptors on human megakaryocytes and megakaryocytic cell lines. Thromb. Haemost. 78, 892–896 [PubMed] [Google Scholar]
- 26. van der Vuurst H., van Willigen G., van Spronsen A., Hendriks M., Donath J., Akkerman J. W. (1997) Signal transduction through trimeric G proteins in megakaryoblastic cell lines. Arterioscler. Thromb. Vasc. Biol. 17, 1830–1836 [DOI] [PubMed] [Google Scholar]
- 27. Conran N., Hemming F. W. (1999) Phorbol ester induces a transient increase in alpha 5 beta 1-mediated adhesion of the megakaryoblastic cell line CHRF 288–11. Platelets 10, 117–123 [DOI] [PubMed] [Google Scholar]
- 28. den Dekker E., Heemskerk J. W., Gorter G., van der Vuurst H., Donath J., Kroner C., Mikoshiba K., Akkerman J. W. (2002) Cyclic AMP raises intracellular Ca(2+) in human megakaryocytes independent of protein kinase A. Arterioscler. Thromb. Vasc. Biol. 22, 179–186 [DOI] [PubMed] [Google Scholar]
- 29. Yang M., Li K., Ng M. H., Yuen P. M., Fok T. F., Li C. K., Hogg P. J., Chong B. H. (2003) Thrombospondin-1 inhibits in vitro megakaryocytopoiesis via CD36. Thromb. Res. 109, 47–54 [DOI] [PubMed] [Google Scholar]
- 30. Chui C. M., Li K., Yang M., Chuen C. K., Fok T. F., Li C. K., Yuen P. M. (2003) Platelet-derived growth factor up-regulates the expression of transcription factors NF-E2, GATA-1 and c-Fos in megakaryocytic cell lines. Cytokine 21, 51–64 [DOI] [PubMed] [Google Scholar]
- 31. Fuhrken P. G., Chen C., Miller W. M., Papoutsakis E. T. (2007) Comparative, genome-scale transcriptional analysis of CHRF-288–11 and primary human megakaryocytic cell cultures provides novel insights into lineage-specific differentiation. Exp. Hematol. 35, 476–489 [DOI] [PubMed] [Google Scholar]
- 32. Vaudry D., Chen Y., Ravni A., Hamelink C., Elkahloun A. G., Eiden L. E. (2002) Analysis of the PC12 cell transcriptome after differentiation with pituitary adenylate cyclase-activating polypeptide (PACAP). J. Neurochem. 83, 1272–1284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Grumolato L., Elkahloun A. G., Ghzili H., Alexandre D., Coulouarn C., Yon L., Salier J. P., Eiden L. E., Fournier A., Vaudry H., Anouar Y. (2003) Microarray and suppression subtractive hybridization analyses of gene expression in pheochromocytoma cells reveal pleiotropic effects of pituitary adenylate cyclase-activating polypeptide on cell proliferation, survival, and adhesion. Endocrinology 144, 2368–2379 [DOI] [PubMed] [Google Scholar]
- 34. Ishido M., Masuo Y. (2004) Transcriptome of pituitary adenylate cyclase-activating polypeptide-differentiated PC12 cells. Regul. Pep.t 123, 15–21 [DOI] [PubMed] [Google Scholar]
- 35. Radley J. M., Haller C. J. (1983) Fate of senescent megakaryocytes in the bone marrow. Br. J. Haematol. 53, 277–287 [DOI] [PubMed] [Google Scholar]
- 36. Zauli G., Vitale M., Falcieri E., Gibellini D., Bassini A., Celeghini C., Columbaro M., Capitani S. (1997) In vitro senescence and apoptotic cell death of human megakaryocytes. Blood 90, 2234–2243 [PubMed] [Google Scholar]
- 37. Falcieri E., Bassini A., Pierpaoli S., Luchetti F., Zamai L., Vitale M., Guidotti L., Zauli G. (2000) Ultrastructural characterization of maturation, platelet release, and senescence of human cultured megakaryocytes. Anat. Rec. 258, 90–99 [DOI] [PubMed] [Google Scholar]
- 38. De Botton S., Sabri S., Daugas E., Zermati Y., Guidotti J. E., Hermine O., Kroemer G., Vainchenker W., Debili N. (2002) Platelet formation is the consequence of caspase activation within megakaryocytes. Blood 100, 1310–1317 [DOI] [PubMed] [Google Scholar]
- 39. Morison I. M., Cramer Bordé E. M., Cheesman E. J., Cheong P. L., Holyoake A. J., Fichelson S., Weeks R. J., Lo A., Davies S. M., Wilbanks S. M., Fagerlund R. D., Ludgate M. W., da Silva Tatley F. M., Coker M. S., Bockett N. A., Hughes G., Pippig D. A., Smith M. P., Capron C., Ledgerwood E. C. (2008) A mutation of human cytochrome c enhances the intrinsic apoptotic pathway but causes only thrombocytopenia. Nat. Genet. 40, 387–389 [DOI] [PubMed] [Google Scholar]
- 40. McDonnell T. J., Deane N., Platt F. M., Nunez G., Jaeger U., McKearn J. P., Korsmeyer S. J. (1989) bcl-2-immunoglobulin transgenic mice demonstrate extended B cell survival and follicular lymphoproliferation. Cell 57, 79–88 [DOI] [PubMed] [Google Scholar]
- 41. Sentman C. L., Shutter J. R., Hockenbery D., Kanagawa O., Korsmeyer S. J. (1991) bcl-2 inhibits multiple forms of apoptosis but not negative selection in thymocytes. Cell 67, 879–888 [DOI] [PubMed] [Google Scholar]
- 42. Ogilvy S., Metcalf D., Print C. G., Bath M. L., Harris A. W., Adams J. M. (1999) Constitutive Bcl-2 expression throughout the hematopoietic compartment affects multiple lineages and enhances progenitor cell survival. Proc. Natl. Acad. Sci. U.S.A. 96, 14943–14948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Bouillet P., Metcalf D., Huang D. C., Tarlinton D. M., Kay T. W., Köntgen F., Adams J. M., Strasser A. (1999) Proapoptotic Bcl-2 relative Bim required for certain apoptotic responses, leukocyte homeostasis, and to preclude autoimmunity. Science 286, 1735–1738 [DOI] [PubMed] [Google Scholar]
- 44. Lee H. H., Dadgostar H., Cheng Q., Shu J., Cheng G. (1999) NF-kappaB-mediated up-regulation of Bcl-x and Bfl-1/A1 is required for CD40 survival signaling in B lymphocytes. Proc. Natl. Acad. Sci. U.S.A. 96, 9136–9141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Spinelli S. L., Casey A. E., Pollock S. J., Gertz J. M., McMillan D. H., Narasipura S. D., Mody N. A., King M. R., Maggirwar S. B., Francis C. W., Taubman M. B., Blumberg N., Phipps R. P. (2010) Platelets and megakaryocytes contain functional nuclear factor-kappaB. Arterioscler. Thromb. Vasc. Biol. 30, 591–598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Kim K. W., Kim S. H., Lee E. Y., Kim N. D., Kang H. S., Kim H. D., Chung B. S., Kang C. D. (2001) Extracellular signal-regulated kinase/90-KDA ribosomal S6 kinase/nuclear factor-kappa B pathway mediates phorbol 12-myristate 13-acetate-induced megakaryocytic differentiation of K562 cells. J. Biol. Chem. 276, 13186–13191 [DOI] [PubMed] [Google Scholar]
- 47. Zhang Y., Sun S., Wang Z., Thompson A., Kaluzhny Y., Zimmet J., Ravid K. (2002) Signaling by the Mpl receptor involves IKK and NF-kappaB. J. Cell. Biochem. 85, 523–535 [DOI] [PubMed] [Google Scholar]
- 48. Ahmad N., Gupta S., Mukhtar H. (2000) Green tea polyphenol epigallocatechin-3-gallate differentially modulates nuclear factor kappaB in cancer cells versus normal cells. Arch. Biochem. Biophys. 376, 338–346 [DOI] [PubMed] [Google Scholar]
- 49. Estrov Z., Shishodia S., Faderl S., Harris D., Van Q., Kantarjian H. M., Talpaz M., Aggarwal B. B. (2003) Resveratrol blocks interleukin-1beta-induced activation of the nuclear transcription factor NF-kappaB, inhibits proliferation, causes S-phase arrest, and induces apoptosis of acute myeloid leukemia cells. Blood 102, 987–995 [DOI] [PubMed] [Google Scholar]
- 50. Chendil D., Ranga R. S., Meigooni D., Sathishkumar S., Ahmed M. M. (2004) Curcumin confers radiosensitizing effect in prostate cancer cell line PC-3. Oncogene 23, 1599–1607 [DOI] [PubMed] [Google Scholar]
- 51. Shishodia S., Aggarwal B. B. (2004) Guggulsterone inhibits NF-kappaB and IkappaBalpha kinase activation, suppresses expression of anti-apoptotic gene products, and enhances apoptosis. J. Biol. Chem. 279, 47148–47158 [DOI] [PubMed] [Google Scholar]
- 52. Chen C., Fuhrken P. G., Huang L. T., Apostolidis P., Wang M., Paredes C. J., Miller W. M., Papoutsakis E. T. (2007) A systems-biology analysis of isogenic megakaryocytic and granulocytic cultures identifies new molecular components of megakaryocytic apoptosis. BMC Genomics 8, 384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Castorina A., Tiralongo A., Giunta S., Carnazza M. L., Rasi G., D'Agata V. (2008) PACAP and VIP prevent apoptosis in schwannoma cells. Brain Res. 1241, 29–35 [DOI] [PubMed] [Google Scholar]
- 54. Brenneman D. E., Phillips T. M., Festoff B. W., Gozes I. (1997) Identity of neurotrophic molecules released from astroglia by vasoactive intestinal peptide. Ann. N.Y. Acad. Sci. 814, 167–173 [DOI] [PubMed] [Google Scholar]
- 55. Villalba M., Bockaert J., Journot L. (1997) Pituitary adenylate cyclase-activating polypeptide (PACAP-38) protects cerebellar granule neurons from apoptosis by activating the mitogen-activated protein kinase (MAP kinase) pathway. J. Neurosci. 17, 83–90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Lindholm D., Skoglösa Y., Takei N. (1998) Developmental regulation of pituitary adenylate cyclase activating polypeptide (PACAP) and its receptor 1 in rat brain: function of PACAP as a neurotrophic factor. Ann. N.Y. Acad. Sci. 865, 189–196 [DOI] [PubMed] [Google Scholar]
- 57. Vaudry D., Basille M., Anouar Y., Fournier A., Vaudry H., Gonzalez B. J. (1998) The neurotrophic activity of PACAP on rat cerebellar granule cells is associated with activation of the protein kinase A pathway and c-fos gene expression. Ann. N.Y. Acad. Sci. 865, 92–99 [DOI] [PubMed] [Google Scholar]
- 58. Onoue S., Endo K., Ohshima K., Yajima T., Kashimoto K. (2002) The neuropeptide PACAP attenuates beta-amyloid (1–42)-induced toxicity in PC12 cells. Peptides 23, 1471–1478 [DOI] [PubMed] [Google Scholar]
- 59. Vaudry D., Pamantung T. F., Basille M., Rousselle C., Fournier A., Vaudry H., Beauvillain J. C., Gonzalez B. J. (2002) PACAP protects cerebellar granule neurons against oxidative stress-induced apoptosis. Eur. J. Neurosci. 15, 1451–1460 [DOI] [PubMed] [Google Scholar]
- 60. Seaborn T., Masmoudi-Kouli O., Fournier A., Vaudry H., Vaudry D. (2011) Protective effects of pituitary adenylate cyclase-activating polypeptide (PACAP) against apoptosis. Current Pharm. Des. 17, 204–214 [DOI] [PubMed] [Google Scholar]