Abstract
An integrated transcriptomic and proteomic analysis was undertaken to determine the physiological response of Escherichia coli O157:H7 Sakai to steady-state conditions relevant to low temperature and water activity conditions experienced during meat carcass chilling in cold air. The response of E. coli during exponential growth at 25 °C aw 0.985, 14 °C aw 0.985, 25 °C aw 0.967, and 14 °C aw 0.967 was compared with that of a reference culture (35 °C aw 0.993). Gene and protein expression profiles of E. coli were more strongly affected by low water activity (aw 0.967) than by low temperature (14 °C). Predefined group enrichment analysis revealed that a universal response of E. coli to all test conditions included activation of the master stress response regulator RpoS and the Rcs phosphorelay system involved in the biosynthesis of the exopolysaccharide colanic acid, as well as down-regulation of elements involved in chemotaxis and motility. However, colanic acid-deficient mutants were shown to achieve comparable growth rates to their wild-type parents under all conditions, indicating that colanic acid is not required for growth. In contrast to the transcriptomic data, the proteomic data revealed that several processes involved in protein synthesis were down-regulated in overall expression at 14 °C aw 0.985, 25 °C aw 0.967, and 14 °C aw 0.967. This result suggests that during growth under these conditions, E. coli, although able to transcribe the required mRNA, may lack the cellular resources required for translation. Elucidating the global adaptive response of E. coli O157:H7 during exposure to chilling and water activity stress has provided a baseline of knowledge of the physiology of this pathogen.
Because enterohemorrhagic Escherichia coli (EHEC) O157:H7 was recognized as a human food-borne pathogen in an outbreak associated with undercooked ground beef in 1982 (1), its emergence has become a concern globally. It possesses several characteristics that contribute to its significance as a threat to public health. These include toxin production; causing infections that can range from mild diarrhea to the life-threatening hemolytic uremic syndrome and; possessing a low infectious dose (2–4). The primary reservoir of this pathogen is cattle, and E. coli O157:H7 can be recovered from feces, rumen contents, saliva, and the hide. Foods of bovine origin are linked most frequently to major food-borne illnesses associated with this organism (2, 4, 5). In Australia and New Zealand, carcasses are cooled by refrigerated air leading to exposure of contaminating microbes to both chilling and osmotic stresses, which have a significant impact on the growth of bacteria including E. coli (6–7).
Bacteria sense environmental stress through signal transduction systems. They respond by activating regulatory cascades or networks that, ultimately, control the expression of numerous genes and the activity of proteins associated with a response enabling fundamental changes in cell physiology and/or morphology. This response helps bacterial cells to protect vital processes and to restore cellular homeostasis under severe conditions, as well as enhancing resistance against multiple stresses (8, 9). Growth of bacteria at temperatures below their optimum and particularly below the so-called “normal range” (10), requires a number of phenotypic changes. These include alterations in cell membranes by increasing unsaturated fatty acid content, changes in the transcriptional and translational machinery, and the production of cold shock and cold acclimation proteins (11, 12). Bacteria respond to hyperosmotic stress (or low water activity, aw) by activating osmoregulatory systems that cause accumulation of charged solutes (e.g. potassium ions and glutamate), followed by accumulation of compatible solutes either through de novo biosynthesis (e.g. trehalose) or through uptake from the external environment (e.g. glycine-betaine and proline) (9, 13). Detailed understanding of the regulatory networks involved in the response of bacteria to prolonged exposure to cold and osmotic stress, as reflected by gene and protein expression, could aid in identification of potential targets for controlling or eliminating E. coli O157:H7 from carcasses and other chilled food products. The present study was undertaken to establish a baseline of knowledge of the molecular responses of E. coli O157:H7 Sakai subjected to chilling and osmotic stresses, to be able to better interpret responses under dynamic conditions as occur during carcass chilling.
To achieve this, we employed both transcriptomic and proteomic analysis in parallel for the first time, to characterize the global genome response of exponential phase E. coli O157:H7 strain Sakai grown under four steady-state conditions, relevant to the temperature and water activity stress conditions experienced during carcass chilling, as compared with a reference culture grown at near optimal conditions (35 °C aw 0.993 in a nutrient rich broth medium).
EXPERIMENTAL PROCEDURES
Division of Experimental Work
Transcriptomic and proteomic studies were conducted in parallel at the Commonwealth Scientific and Industrial Research Organization (CSIRO) Food and Nutritional Sciences (Brisbane, Australia) and Food Safety Centre, University of Tasmania (Hobart, Australia), respectively. The only difference in methods was in the extraction protocols for RNAs and proteins (see below), the use of different equipment for optical density measurements (Transcriptomic: Pharmacia Biotech Novaspec II spectrophotometer and; Proteomic: Spectronic 20, Bausch and Lomb), and water baths for incubation of cultures (Transcriptomic: Julabo SW23 water bath (80 rpm) and FL601 Re-circulating cooler and; Proteomic: Ratek SWB20D shaking water bath (80 oscillations·min−1) with Ratek RC2 refrigeration unit).
Bacterial Strains
E. coli O157:H7 strain Sakai (14, 15) obtained from Carlton Gyles (Guelph, Canada) was used in this study. A colanic acid-deficient mutant (wza−) of the strain O157:H7 Sakai was constructed according to the method of Datsenko and Wanner (16) and designated as “FNS1” in the CSIRO Food and Nutritional Sciences culture collection. The chromosomal wza gene was targeted for mutagenesis with PCR products containing a resistance cassette flanked by FLP recognition target (FRT) sites and homologies to adjacent chromosomal sequences. Briefly, 60 bp PCR primers were designed with 40 bp of 5′ homology to the Sakai wza gene and 20 bp of 3′ homology to the chloramphenicol (cat) cassette present in pKD3 (wza cat F1, atatgtcgacgatgggcaaagacgtcatcaaacagcaggagtgtaggctggagctgcttc; wza cat R1, tgattgatcagacggttccagcgggaaaccggggcggtggatgggaattagccatggtcc). Purified PCR products were used to electroporate electrocompetent cultures of Sakai carrying plasmid pKD46 (encoding the λRed recombinase functions). The presence of the Δwza::cat mutant allele was confirmed by PCR amplification, using primers flanking the sites of substitution.
Preparation of Test Broth
Test broths were prepared as appropriate to the growth conditions employed in this study (Table I). Briefly, NaCl was added to fresh Brain-Heart Infusion medium (BHI; Oxoid, CM225) to provide the required water activity (aw)1 according to the tables of Chirife and Resnik (17). This NaCl-modified BHI broth was sterilized (121 °C, 20 min) and stored at room temperature (RT) until use. The final aw of each NaCl-modified BHI broth was confirmed with an Aqualab CX-2 dew point instrument (Decagon Devices, Inc., Pullman, USA). The measured water activity of all cultures was within ± 0.003 of the required value.
Table I. Growth conditions of E. coli O157:H7 Sakai used in this study.
| Condition | Description of condition | Number of biological replicates |
|
|---|---|---|---|
| Transcriptome | Proteomea | ||
| 35 °C aw 0.993 | Optimal | 3 | 8 (6) |
| 25 °C aw 0.985 | Intermediate | 3 | 3 (2) |
| 14 °C aw 0.985 | Low temperature | 3 | 3 (2) |
| 25 °C aw 0.967 | Low water activity | 3 | 3 (2) |
| 14 °C aw 0.967 | A combination of low temperature and low water activity | 3 | 3 (2) |
a Number of replicates performed for soluble (outside brackets) and membrane (within brackets) fractions of E. coli.
Growth Conditions
Working cultures for use in transcriptomic and proteomic studies were streaked onto BHI plates and grown at 37 °C for 24 h. An individual colony was aseptically transferred into 25 ml of BHI broth. This broth culture was incubated at 37 °C for 20 h to provide stationary phase cells. The culture was then stored at 4 °C and used as a “stock” culture for up to one month.
A 1:104 dilution of the stock culture was performed in BHI and the resulting culture was grown in a shaking water bath under each of the conditions listed in Table I. Growth was monitored turbidimetrically at 600 nm until the culture reached an OD600 of 0.1 ± 0.01 (i.e. the exponential phase of growth, ∼107 CFU/ml). The primary inoculum was then diluted 1:102 in BHI broth. This “secondary” culture was then grown under the same condition as its primary culture to the mid-exponential phase of growth (OD600 of 0.1 ± 0.01). At this point, a volume of culture was harvested as appropriate to the protocol for transcriptomic or proteomic analysis (see below). The number of biological replicates for each condition conducted for the transcriptomic and proteomic analysis is described in Table I.
Determination of Growth Rate
The growth rate of a wild-type and wza− mutant strain under each condition was determined using the viable count method. Aliquots of cultures were serially diluted in 0.1% peptone water (PW; Oxoid, LP0037), and appropriate dilutions were plated (in triplicate) onto BHI agar. All plates were then incubated for 24 h at 37 °C before colonies were enumerated. To compare the rate of growth of the wild-type and mutant strains, data at each time point were analyzed by Minitab software (Minitab 15; Minitab Inc., Minneapolis, MN) using one-way analysis of variance (ANOVA) and a post hoc Tukey test. A p value of <0.05 was considered to be significant.
Preparation of Total RNA for Transcriptomic Analysis
RNA was extracted from samples of the “secondary” cultures taken at the time of incubation, corresponding to an OD600 of 0.1 ± 0.01. RNA was stabilized by addition of a phenol-ethanol solution (5:95) and was immediately transferred to ice prior to extraction. RNA samples were prepared using a Promega SV total RNA purification kit. RNA concentration was determined using a NanoPhotometer (Implen Pty). RNA quality was determined by 16S and 23S rRNA peak examination by the Bioanalyzer 2100 (Agilent, Santa Clara, CA) using an RNA nano chip. cDNA synthesis, labeling and hybridization to GeneChip® E. coli Genome 2.0 Arrays (Affymetrix, Santa Clara, CA) was performed by the Australian Genome Research Facility (Melbourne, Australia).
Analysis of Microarray Data
Gene expression analysis was carried out using GeneSpring® GX 10.0 software (Agilent Technologies, Palo Alto, CA). Array data was normalized using the Robust Multi-Array Analysis (RMA) algorithm (18). Probesets with an intensity value in the lowest 20th percentile among all the intensity values were excluded. The remaining probesets, with intensity values between the 20th and 100th percentile, were used for downstream statistical analysis. Significance analysis was conducted using one way analysis of variance (ANOVA) using the multiple correction testing method of Benjamini-Hochberg with a p value cutoff of <0.01. Elements were considered to be differentially expressed if the fold change was >2.
Preparation of Cell Extracts for Proteomic Analysis
Parallel to the transcriptomic analysis, a 25 ml sample of each secondary culture at an OD600 of 0.1 ± 0.01 was harvested by centrifugation at 4500 × g for 10 min in a Hettich Zentrifugen EBA 12 centrifuge (Tuttlingen, Germany) operating at RT. The pellet was washed twice with 1 ml of ice-cold phosphate buffered saline (PBS; Oxoid, BR0014G) with centrifugation at 4500 × g for 10 min at 4 °C (Microcentrifuge 5417R, Eppendorf, Germany). The resulting washed pellet was frozen using liquid nitrogen and stored at −80 °C until further processing to extract the soluble proteome or membrane proteins.
For soluble protein extraction, cell pellets maintained at −80 °C were left to thaw on ice for ∼15 min. The soluble fraction of the E. coli proteome was extracted using the Qproteome bacterial protein prep kit (Qiagen, Victoria, AUS, 37900). The supernatant; containing the soluble protein fraction, was collected after centrifugation of the homogenate at 14,000 × g for 30 min at 4 °C to pellet the cell debris containing insoluble proteins. Protein concentration was estimated by using the Quick Start Bradford protein assay with bovine plasma γ-globulin as standard (Bio-Rad, Hercules, CA). An aliquot of each supernatant containing ∼80 μg of soluble protein was then lyophilized for 2 h using a Mini Ultra Cold freeze-drier (Dynavac Australia), according to the manufacturer's instructions. The precipitated proteins were subsequently denatured in 20 μl of 100 mm ammonium bicarbonate containing 8 m urea prior to tryptic digestion.
In separate sets of the same experiment, independent samples were harvested as described above, and membrane proteins were extracted by performing additional steps following the removal of the soluble proteome. The pellets containing insoluble proteins, obtained after the centrifugation as described above, were washed twice with 1 ml of milliQ water (with centrifugation at 18,000 × g for 25 min at 4 °C) and solublized in 0.5 ml of Protein Extraction Reagent Type 4 (Sigma-Aldrich; C0356). Suspensions were ultrasonicated with four 15 s pulses (Microson XL2000 ultrasonic cell disrupter, Farmingdale, NY, USA) on ice with 10 s cooling between pulses. After centrifugation at 14,000 × g for 45 min at 15 °C, the enriched membrane protein containing supernatant was collected. An aliquot containing ∼80 μg of total protein, as determined by the Quick Start Bradford protein assay (Bio-Rad), was subjected to tryptic digestion.
Tryptic Digestion
Both sample solutions, i.e. containing soluble proteins or enriched membrane proteins were reduced with reducing agent (50 mm dithiothreitol, 100 mm ammonium bicarbonate; incubated for 1 h at RT), alkylated with alkylating reagent (200 mm iodoacetamide, 100 mm ammonium bicarbonate; for 1 h at RT) and reduced again with the reducing agent (for 1 h at RT). After dithiothreitol reduction and iodoacetamide alkylation of cysteine residues, all solutions were diluted to a final volume of 220 μl with digestion buffer containing 50 mm ammonium bicarbonate, 1 mm calcium chloride solution (CaCl2·2H2O), and digested with 0.1 μg/μl sequencing grade modified trypsin (Promega, Madison, WI; V5111) at a ratio of 1:80 (trypsin/protein). After incubation at 37 °C for 18–20 h, the digest reactions were quenched by acidification with 10 μl of 10% formic acid and stored at −20 °C for further analysis.
2D-LC/MS/MS Analysis
All tryptic digests were analyzed using multidimensional protein identification technology (MudPIT), based on the modified methodology of Delahunty and YatesIII (19). In this study, a nanoflow triphasic MudPIT system was used, which consisted of a C18 capillary trap (Peptide CapTrap, Michrom BioResources, Auburn, CA), a strong cation exchange (SCX) material (IntegraFrit Column, 100 μm i.d., 2.5 cm, New Objective, Woburn, MA), and an analytical C18 nano-column (PicoFrit Column, 15 μm i.d. pulled tip, 10 cm, New Objective). This triphasic microcapillary column was directly coupled in-line with an electrospray ionization ion trap tandem mass spectrometer (LTQ Orbitrap, ThermoElectron, San Jose, CA). A 5-step MudPIT gradient was performed with a real time data-dependent MS/MS acquisition. This 2D-LC-MS/MS analysis typically produced between 10,000 and 20,000 MS/MS spectra to identify. A detailed description of the MudPIT analysis is provided in supplemental material S1.
Analysis of MS/MS Data
Acquired MS/MS spectra generated from each sample of soluble and membrane fractions were automatically processed by the Computational Proteomics Analysis System (CPAS), a web-based system built on the LabKey Server (version 9.1, released 02.04.2009) for managing, analyzing, and sharing high volumes of tandem mass spectrometry data (20). The experimental mass spectra produced were subjected to a semitryptic search against the complete database of E. coli O157:H7 Sakai (5318 entries in total), consisting of the E. coli O157:H7 Sakai database (5230 entries, NC_002695.fasta) and two E. coli O157:H7 Sakai plasmid databases, plasmid pO157 (85 entries, NC_002128.fasta) and plasmid pOSAK1 (three entries, NC_002127.fasta) downloaded from the National Center for Biotechnology Information (NCBI, https://www.ncbi.nlm.nih.gov/, downloaded 25.11.2008) using X!Tandem version 2007.07.01 (21). The search parameters used were as follows: mass tolerance for precursor and fragment ions: 10 ppm and 0.5 Da, respectively; fixed modification: cysteine carbamidomethylation (+ 57 Da); and no variable modifications. The search results were then analyzed using the PeptideProphet and ProteinProphet algorithms from the Trans Proteomic Pipeline v3.4.2 (22–23). Peptide identifications with a PeptideProphet probability ≥0.9 were selected and subsequently submitted to ProteinProphet, where a probability of each protein was assigned based on the combined peptide probabilities (22). The derived protein identifications were then accepted at a ProteinProphet of ≥0.9, which corresponded to an error rate of ≤2%, according to the probability that the best match obtained would fall in the distribution of random database matches (24). Finally, to estimate a false-positive discovery rate in each dataset, the MS/MS spectra were searched against the target-reversed (decoy) database using the same search criteria as described above.
All protein identifications that passed the above criteria were further assessed based on the confidence level of protein identifications across biological replicates of each fraction of E. coli cells grown under a given condition. The confidence level was based on the number of unique peptides identified from one sample and the number of replicates in which the protein was detected. Specifically, proteins identified by more than one unique peptide in at least one of the replicates were considered to have a high confidence score. At the intermediate confidence level, proteins with a single peptide hit were detected in more than one replicate. Low-confidence proteins were those identified by a single unique peptide and found in only one replicate. Only protein identifications with a high and intermediate confidence level (referred as having a “high confidence score”) were accepted for further analysis.
Protein Expression Ratio and Its Significance
Spectral count (SpC), a sampling statistic output of ProteinProphet, was used to measure relative protein abundance (22, 25). The abundance of each protein in a sample was obtained by averaging SpC values detected for a given protein across replicates of each fraction. Differences in the expression level of proteins and their significance was then determined independently for membrane and soluble fractions. Log2 fold changes in protein expression under each treatment condition were calculated based on the average SpC, according to the method described by Old et al. (26). The log2 ratio of each protein was also normalized by subtraction of the log2 ratio calculated from a “simulated protein” between corresponding pairs of comparison. Significant differences in protein expression levels were assessed by an empiric test of Fu et al. (27) known as the spectral index (SpI) together with permutation analysis. Typically, the SpI method is based on the SpC data derived from each set of replicates (i.e. control and treatment) and the number of replicates for detectable peptides (i.e. number of replicates in which the peptides are detected). These features employ the robust statistical power of SpI to account for the variation in absolute number of SpC obtained in different 2D-LC/MS/MS runs, and those proteins with low spectral counts, which exhibit greater relative variability (27). All proteins with a spectral index significant at the 95% confidence level were considered to be differentially expressed.
After evaluating proteins independently from each fraction as described above, we further used the spectral index to determine which fraction was relatively more enriched with the protein identifications, and thus should be considered for the subsequent analysis. This was to avoid confusion or misinterpretation of the proteins and their relative abundance. The spectral index calculated in this study was based on the average spectral count of a given protein among all the replicates of all conditions for membrane and soluble fractions within which the peptides were detected. The proteins with a positive spectral index suggest enrichment of proteins in the E. coli membrane proteome, whereas a negative value indicates enrichment in the soluble fraction (27).
Transcriptomic and Proteomic Data Mining
Information on protein identifications with a high confidence score were obtained from the UniProt knowledgebase (http://www.uniprot.org/) and the NCBI database (accessed on 18.02.2010). These included protein and gene names, ECs numbers (locus tag), GI numbers, NCBI Reference Sequence (RefSeq), protein sizes, and molecular masses. Preliminary functions and properties of the proteins were searched in the BioCyc database collection (http://www.ecocyc.org/, accessed on 20.02.2010). Throughout the present work, protein or gene names in conjunction with ECs numbers were used where appropriate as a unique identifier for genes and/or proteins.
Predefined Group Enrichment Analysis
A predefined group enrichment analysis of Boorsma et al. (28), known as T-profiler, was conducted on the normalized log2 expression ratios to score the changes in expression of a predefined set of genes and proteins under test conditions compared with the reference culture. This type of analysis has been successfully used to interpret genome-wide expression data, and provides a more system level and informative view of biological states (compared with individual gene and protein-based interpretation). However, it is important to note that the T-profiler method of analysis uses average log2 ratios only, and therefore does not take into account any variability present within a set of replicates.
In the present study, two predefined sets of genes and proteins were used for the T-profiler analysis: the first set was based on the E. coli O157:H7 Sakai role categories obtained from the JCVI Comprehensive Microbial Resource (JCVI CMR) database (http://cmr.jcvi.org/cgi-bin/CMR/CmrHomePage.cgi, downloaded 5.12.2009); and the second set was based on genes/proteins previously found to be positively controlled by CpxRA (29), RpoE (30), RpoH (31), RpoS (32–34), and the Rcs phosphorelay system (35). The score (“T-value”) from the T-profiler analysis was determined only for sets that contained at least five members, and their significance was established by using the associated two-tailed p value. All predefined sets with a p value less than 0.1 were considered to be statistically significant.
Hierarchical Clustering Analysis
To determine similarities in the overall expression pattern of groups of genes and proteins and to investigate overall changes in both the transcriptome and proteome under the test conditions, hierarchical clustering analysis was performed on the T-values calculated for JCVI CMR functional categories across test conditions. The degree of similarity was calculated with the Euclidean distance metric and complete linkage as a clustering method, using Cluster 3.0 software (36). The clustering result was then visualized in TreeView version 1.1.3 (37).
Colanic Acid Purification and Quantification
The method used to extract colanic acid was based on procedures previously described (38–39). Briefly, 50 ml of cell culture was heated for 15 min at 100 °C to denature EPS-degrading enzymes. After cooling, the heated culture was centrifuged at 13,200 × g at 4 °C for 30 min. Then 40 ml of the supernatant fraction was precipitated by addition of three volumes of ethanol. The mixture was maintained at 4 °C overnight and centrifuged under the same conditions as described above. The resulting pellet was dissolved in 5 ml of distilled water, dialyzed for 48 h against distilled water (membrane MWCO, 3,500 Da) and dried. Residual polypeptides were removed by precipitation with 5 ml of 10% (v/v) trichloroacetic acid and centrifuged again at 13,200 × g at 4 °C for 30 min. The supernatant was dialyzed again for 5 days against distilled water and stored until quantification.
Quantification of colanic acid was carried out, according to a previously described protocol (40) by measuring the nondialyzable methylpentose (fucose), which is a specific component of this exopolysaccharide. Ten to 100 μl of the colanic acid preparation were diluted to 1 ml with distilled water, and mixed with 4.5 ml of H2SO4/H2O (6:1; v/v). The mixture was prepared at RT, then heated at 100 °C for 20 min, and finally cooled down to RT. For each sample, absorbance at 396 nm and 427 nm was measured either directly (control sample (A-co)) or after addition of 100 μl of cysteine hydrochloride (cysteine sample (A-cy)).
Transcriptomic and Proteomic Data Accession Numbers
The transcriptomic data are deposited at Array Express, accession number E-MTAB-623 (http://www.ebi.ac.uk/miamexpress/). The proteomic data are available in the PRIDE database under accession numbers 18544 - 18577 (http://www.ebi.ac.uk/pride/) (41).
RESULTS AND DISCUSSION
The present study employed both transcriptomic and proteomic analysis to investigate the physiology of E. coli O157:H7 Sakai grown under steady-state conditions of chill and/or osmotic stress, with the aim to obtain a comprehensive insight into the adaptation of this pathogen under the conditions mimicking that experienced on meat carcases during cold-air chilling. E. coli were cultivated in nutrient rich BHI media under five different conditions (Table I). The growth rate of E. coli was consistent with the predictive model of Ross et al. (42) at each condition (Table II). Although acknowledging that growth rate affects gene expression (43–44), monitoring these changes was intrinsic to gaining insight into the molecular mechanisms enabling growth and/or survival under conditions experienced during carcass chilling.
Table II. The number of valid protein identifications and significantly differentially expressed genes and proteins under each steady-state condition.
| Condition | Growth rate (h−1) | Number of proteinsa |
Number of differentially expressed elements |
|||
|---|---|---|---|---|---|---|
| Membrane fraction | Soluble fraction | Transcriptome (total) | Proteome (total) | Transcriptome vs. Proteomeb | ||
| 35 °C aw 0.993 | 2.05 | 284 | 1,180 | NAc | NA | NA |
| 25 °C aw 0.985 | 0.82 | 218 | 763 | 317 | 176 | 40 |
| 14 °C aw 0.985 | 0.18 | 232 | 815 | 788 | 301 | 75 |
| 25 °C aw 0.967 | 0.35 | 265 | 722 | 875 | 322 | 90 |
| 14 °C aw 0.967 | 0.08 | 213 | 615 | 1203 | 447 | 143 |
a Number of proteins identified in membrane and soluble fractions of the E. coli proteome that have passed the filtering criteria.
b Number of differentially expressed elements found in both the transcriptomic and proteomic analysis.
c NA, not applicable.
Integrated Transcriptomic and Proteomic Analysis of O157:H7 Sakai
During growth at the exponential phase (an OD600 of 0.1 ± 0.01), cultures were harvested and processed for RNA, and membrane and soluble protein extractions as appropriate for analysis of cDNA microarray and 2D-LC-MS/MS, respectively. The proteomic analysis identified several proteins with high confidence in membrane and soluble fractions of E. coli under each condition (Table II and supplemental material S2). To estimate reproducibility of the label-free quantitative shotgun proteomic analysis, linear regression analysis based on spectral counts was performed for each pair-wise comparison of the replicates. The observed R2 values revealed a relatively strong linear correlation between spectral counts for all possible pair-wise comparisons of each set of replicates of soluble fractions (average R2 = 0.80 ± 0.11 across all conditions tested), although the correlation observed for membrane fractions was more moderate (average R2 = 0.60 ± 0.07 across all conditions tested). These findings indicate a high level of reproducibility between replicated data sets. All proteomic data sets were further analyzed to estimate the false-positive rate of peptide identifications. The peptide false-positive discovery rate, calculated by dividing the number of spectra matching decoy peptides with the total number of spectra (45), was less than 5% for all MudPIT runs (supplementary Table S2). To enable comparison of transcriptomic and proteomic data, changes in gene and protein expression profiles of O157:H7 Sakai under four steady-state conditions of 25 °C aw 0.985, 14 °C aw 0.985, 25 °C aw 0.967, and 14 °C aw 0.967 were determined relative to the baseline response of a reference culture grown at near-optimal conditions for rapid growth (35 °C aw 0.993).
The number of genes and proteins observed to be differentially expressed by O157:H7 Sakai with respect to the reference culture is shown in Table II. Supplementary Table S13–S16 present a complete and annotated list of these genes and proteins with a comparison to previously published reports. It is worthwhile noting that the protein identifications with an intermediate confidence level (i.e. those proteins that were identified by a single unique peptide and detected in more than one replicate) contribute a small proportion (6.4% ± 1.9%) of differentially expressed proteins across test conditions (see supplementary Table S13–S16). These proteins are discussed in the present study only when transcript levels of the corresponding gene display differential expression in the transcriptomic study. Furthermore, comparison of the transcriptomic and proteomic profiles as determined by calculating the percentage of differentially expressed genes and proteins matched under each condition, revealed a low level of correlation with an average of only 8.3% ± 1.1% of elements matched across test conditions (see Venn diagrams in the supplementary Table S13–S16). The low level of correlation observed is consistent with previous studies, reporting poor correlation between mRNA and protein profiles (46–49). This suggests that different control mechanisms are present at the transcriptomic and proteomic levels, and that a combination of mRNA and protein expression patterns are required to fully develop an understanding of the functional architecture of genomes and gene networks (50, 51). Additional reasons for discrepancies between mRNA and protein expressions have been reported, and should not be discounted; including posttranscriptional mechanisms controlling the protein translation rate, the half-lives of specific proteins or mRNAs, and the intracellular location and molecular association of the protein products of expressed genes (52).
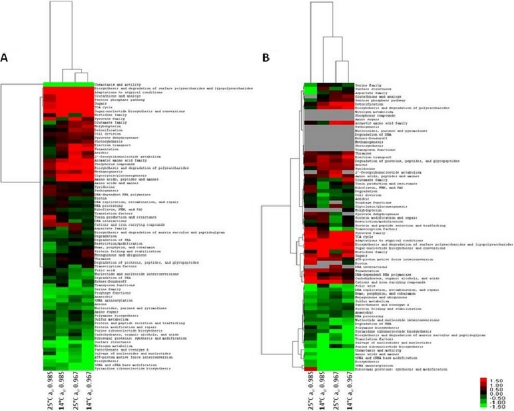
Despite the low level of correlation observed in the current study, a number of trends were observed. E. coli O157:H7 Sakai significantly altered the overall expression of several JCVI CMR functional categories and subcategories during growth under the steady-state conditions of chilling and/or osmotic stress, as determined by the T-profiler analysis (Table III). Clustering of the T-profiler results revealed that expression profiles of gene and protein were clustered by low water activity (aw 0.967) rather than low temperature (14 °C) (Fig. 1), with the global response of cultures grown at 25 °C aw 0.967 and 14 °C aw 0.967 most similar. This suggests that water activity stress has a greater effect on the physiology of E. coli than cold stress.
Table III. T-profiler analysis of transcriptomic and proteomic data based on JCVI CMR functional categories.
| Protein groups | 25 °C aw 0.985 |
14 °C aw 0.985 |
25 °C aw 0.967 |
14 °C aw 0.967 |
||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Transcriptome |
Proteome |
Transcriptome |
Proteome |
Transcriptome |
Proteome |
Transcriptome |
Proteome |
|||||||||
| T-valuea | p valueb | T-value | p value | T-value | p value | T-value | p value | T-value | p value | T-value | p value | T-value | p value | T-value | p value | |
| Amino acid biosynthesis | ||||||||||||||||
| Aromatic amino acid family | 0.63 | 0.17 | 1.15 | −0.36 | 1.42 | 1.45 | 1.76 | 0.096 | 1.35 | |||||||
| Aspartate family | −0.16 | −1.91 | 0.077 | −0.73 | −0.37 | 0.53 | 0.46 | −0.16 | 0.20 | |||||||
| Glutamate family | −0.85 | −0.08 | 0.21 | −0.41 | 0.81 | −0.52 | 0.86 | 1.23 | ||||||||
| Pyruvate family | 1.34 | 1.35 | −0.47 | 0.53 | 3.21 | 0.008 | 3.29 | 0.013 | 0.46 | 1.74 | ||||||
| Serine family | −1.25 | −1.01 | −1.25 | 0.16 | −1.06 | −0.12 | −1.17 | −0.52 | ||||||||
| Histidine family | 0.26 | 0.15 | 2.72 | 0.035 | 3.39 | 0.015 | −0.47 | −0.52 | 2.15 | 0.075 | 0.97 | |||||
| Biosynthesis of cofactors, prosthetic groups, and carriers | ||||||||||||||||
| Biotin | −0.16 | 1.25 | 0.13 | NDc | −0.43 | ND | −0.36 | ND | ||||||||
| Folic acid | 0.08 | −1.58 | −0.44 | −0.20 | −0.70 | −1.46 | −0.92 | −1.25 | ||||||||
| Heme, porphyrin, and cobalamin | −1.08 | −1.14 | −0.40 | −0.88 | 0.03 | −0.50 | −0.03 | −0.97 | ||||||||
| Menaquinone and ubiquinone | −0.34 | −0.96 | 0.03 | −0.85 | 0.05 | −0.33 | 0.48 | −0.82 | ||||||||
| Molybdopterin | −0.16 | ND | 0.42 | ND | 0.76 | ND | 1.15 | 0.09 | ||||||||
| Pantothenate and coenzyme A | −0.72 | −0.91 | −1.34 | −0.52 | −0.97 | −0.58 | −1.81 | −0.46 | ||||||||
| Pyridoxine | 0.54 | −0.14 | 0.32 | 0.90 | 0.17 | 0.79 | −0.02 | 0.81 | ||||||||
| Riboflavin, FMN, and FAD | 0.29 | 0.62 | −0.29 | −0.93 | −0.45 | −0.17 | −0.36 | −0.35 | ||||||||
| Glutathione and analogs | 1.02 | ND | 2.11 | 0.080 | −0.03 | 2.66 | 0.037 | 0.51 | 2.81 | 0.031 | 0.80 | |||||
| Thiamine | −0.45 | 0.58 | 0.28 | 0.30 | −0.04 | 0.21 | 0.12 | 0.87 | ||||||||
| Cell envelope | ||||||||||||||||
| Surface structures | −1.70 | 0.099 | −1.85 | −3.61 | 0.001 | 0.55 | −2.68 | 0.012 | −0.72 | −3.33 | 0.002 | −0.09 | ||||
| Biosynthesis and degradation of murein sacculus and peptidoglycan | −0.64 | −0.32 | −0.57 | −2.46 | 0.020 | 0.21 | −0.93 | −0.50 | −2.12 | 0.042 | ||||||
| Biosynthesis and degradation of surface polysaccharides and lipopolysaccharides | 10.86 | <0.001 | 2.19 | 0.033 | 8.30 | <0.001 | 5.11 | <0.001 | 6.74 | <0.001 | 1.53 | 5.12 | <0.001 | 1.23 | ||
| Cellular processes | ||||||||||||||||
| Cell division | 0.36 | −0.13 | 0.46 | −1.23 | 0.68 | −0.58 | 0.68 | −0.48 | ||||||||
| Chemotaxis and motility | −42.25 | <0.001 | −4.62 | <0.001 | −28.03 | <0.001 | −2.47 | 0.024 | −28.94 | <0.001 | −2.91 | 0.010 | −22.63 | <0.001 | −1.89 | 0.077 |
| Detoxification | −0.02 | 0.18 | 0.10 | 0.82 | 1.05 | 1.82 | 0.88 | 1.43 | ||||||||
| DNA transformation | ND | ND | ND | ND | ND | ND | ND | ND | ||||||||
| Toxin production and resistance | 1.38 | 0.18 | 0.11 | −0.95 | −0.26 | 0.01 | −0.36 | 0.43 | ||||||||
| Pathogenesis | 0.60 | ND | 0.20 | ND | 0.12 | ND | −0.23 | ND | ||||||||
| Adaptations to atypical conditions | 2.32 | 0.033 | 1.04 | 1.88 | 0.077 | 2.35 | 0.043 | 1.67 | 3.60 | 0.005 | 2.59 | 0.019 | 3.99 | 0.003 | ||
| Central intermediary metabolism | ||||||||||||||||
| Amino sugars | −0.22 | ND | −0.71 | ND | −0.07 | ND | −0.58 | ND | ||||||||
| Phosphorus compounds | 0.84 | ND | 1.62 | ND | 1.40 | ND | 1.79 | ND | ||||||||
| Polyamine biosynthesis | −0.44 | −0.14 | −0.71 | −0.91 | −0.18 | −0.34 | −0.72 | −2.06 | ||||||||
| Sulfur metabolism | −0.35 | −1.16 | −0.91 | −0.74 | −0.16 | −0.64 | −0.80 | −1.26 | ||||||||
| Nitrogen metabolism | −1.36 | ND | −1.59 | ND | −0.73 | ND | −2.68 | 0.032 | ND | |||||||
| DNA metabolism | ||||||||||||||||
| DNA replication, recombination, and repair | −0.15 | −1.98 | 0.053 | 0.26 | −0.93 | −0.20 | −0.83 | −0.22 | −1.35 | |||||||
| Restriction/modification | −0.99 | −0.30 | −0.24 | 0.60 | −0.46 | −0.92 | −0.25 | 0.10 | ||||||||
| Degradation of DNA | −0.58 | ND | −0.16 | ND | −0.81 | ND | −0.83 | ND | ||||||||
| Energy metabolism | ||||||||||||||||
| Aerobic | −0.38 | −0.21 | 0.28 | −0.75 | 1.30 | −0.33 | 0.49 | −0.30 | ||||||||
| Amino acids and amines | 0.12 | −1.06 | −0.13 | −2.16 | 0.059 | 0.17 | −2.21 | 0.055 | −0.09 | −2.22 | 0.053 | |||||
| Anaerobic | −1.40 | −0.54 | −1.03 | −0.53 | −0.56 | −0.38 | −0.75 | −0.69 | ||||||||
| ATP-proton motive force interconversion | −2.05 | 0.067 | 1.39 | −1.58 | 0.87 | −1.21 | 0.89 | −1.77 | −0.17 | |||||||
| Electron transport | −0.17 | 0.47 | 0.67 | 1.06 | 2.35 | 0.020 | 0.83 | 1.18 | 1.46 | |||||||
| Entner-Doudoroff | −0.24 | ND | −0.24 | ND | −1.21 | ND | −0.45 | ND | ||||||||
| Fermentation | −0.40 | 1.14 | 0.45 | 1.13 | 1.70 | 0.097 | 0.70 | 1.56 | 0.31 | |||||||
| Glycolysis/gluconeogenesis | 0.63 | 0.14 | 1.42 | −0.80 | 2.42 | 0.021 | 0.64 | 2.11 | 0.042 | −0.35 | ||||||
| Pentose phosphate pathway | 1.02 | −0.68 | 2.16 | 0.051 | 0.13 | 2.50 | 0.028 | 0.54 | 2.49 | 0.029 | 0.53 | |||||
| Pyruvate dehydrogenase | 0.13 | 0.71 | 0.36 | 0.48 | 0.60 | −0.59 | 0.34 | ND | ||||||||
| Sugars | 1.31 | 1.07 | 2.62 | 0.011 | 2.21 | 0.039 | 2.89 | 0.006 | 0.61 | 2.71 | 0.009 | 1.43 | ||||
| TCA cycle | 1.25 | 2.08 | 0.055 | 2.68 | 0.012 | 0.77 | 4.33 | <0.001 | 3.44 | 0.004 | 3.34 | 0.002 | 1.83 | 0.087 | ||
| Methanogenesis | 0.38 | ND | 1.03 | ND | 0.78 | ND | 1.36 | ND | ||||||||
| Biosynthesis and degradation of polysaccharides | 0.86 | −0.26 | 1.53 | 1.21 | 1.66 | 1.20 | 2.06 | 0.056 | 2.17 | 0.067 | ||||||
| Photosynthesis | −0.09 | ND | 0.30 | ND | 0.38 | ND | 0.75 | ND | ||||||||
| Fatty acids and phospholipids metabolism | ||||||||||||||||
| Biosynthesis | −1.21 | −1.04 | −1.68 | −3.22 | 0.004 | −1.12 | −2.99 | 0.007 | −1.70 | 0.098 | −2.68 | 0.015 | ||||
| Degradation | −0.46 | 0.65 | −0.50 | −0.53 | −0.21 | −0.52 | −0.29 | 0.07 | ||||||||
| Mobile and extrachromosomal element functions | ||||||||||||||||
| Prophage functions | −1.27 | −0.20 | −1.21 | −0.92 | −1.14 | ND | −0.97 | −0.39 | ||||||||
| Transposon functions | −1.06 | ND | 0.09 | ND | −1.26 | ND | −0.63 | ND | ||||||||
| Protein fate | ||||||||||||||||
| Protein and peptide secretion and trafficking | −0.61 | −0.08 | −0.81 | 0.01 | −0.55 | −0.59 | −0.66 | 0.18 | ||||||||
| Protein modification and repair | −0.52 | 0.38 | −0.75 | 0.52 | −0.76 | −0.94 | −0.83 | 0.03 | ||||||||
| Protein folding and stabilization | −0.87 | −1.07 | −0.46 | −0.79 | −0.04 | −0.54 | 0.46 | −0.30 | ||||||||
| Degradation of proteins, peptides, and glycopeptides | −0.42 | 0.39 | −0.04 | 1.04 | 0.10 | 0.53 | 0.11 | 0.97 | ||||||||
| Protein synthesis | ||||||||||||||||
| tRNA aminoacylation | −1.02 | −1.08 | −1.18 | −3.14 | 0.005 | −0.47 | −4.67 | <0.001 | −1.10 | −6.07 | <0.001 | |||||
| Ribosomal proteins: synthesis and modification | −0.40 | 1.20 | −1.43 | −3.54 | 0.001 | −0.69 | −2.72 | 0.009 | −1.13 | −5.26 | <0.001 | |||||
| tRNA and rRNA base modification | −1.14 | −1.41 | −1.02 | −2.13 | 0.044 | −2.16 | 0.039 | −2.58 | 0.016 | −2.17 | 0.038 | −2.74 | 0.011 | |||
| Translation factors | 0.13 | −0.42 | −0.04 | −1.68 | −0.25 | −0.78 | −0.26 | −1.92 | 0.081 | |||||||
| Purines, pyrimidines, nucleosides, and nucleotides | ||||||||||||||||
| 2′-Deoxyribonucleotide metabolism | −0.25 | ND | 0.08 | 0.48 | 1.91 | 0.092 | 1.23 | 0.87 | ND | |||||||
| Nucleotide and nucleoside interconversions | −0.10 | −0.58 | −0.26 | −0.57 | −0.79 | −1.36 | −1.08 | −1.16 | ||||||||
| Purine ribonucleotide biosynthesis | −0.54 | −0.91 | −0.92 | −2.16 | 0.049 | −0.43 | −0.16 | −1.00 | −0.85 | |||||||
| Pyrimidine ribonucleotide biosynthesis | −0.82 | −0.50 | −1.11 | −0.62 | −1.76 | −1.86 | −1.66 | −2.65 | 0.045 | |||||||
| Salvage of nucleoside and nucleotide | −0.35 | −1.13 | −1.60 | −1.86 | 0.082 | −1.08 | −1.25 | −2.17 | 0.042 | −1.98 | 0.066 | |||||
| Sugar-nucleotide biosynthesis and conversions | 4.33 | 0.008 | 3.45 | 0.026 | 4.17 | 0.009 | 4.48 | 0.011 | 3.65 | 0.015 | 2.70 | 0.054 | 3.21 | 0.024 | 2.07 | |
| Regulatory functions | ||||||||||||||||
| DNA interactions | 0.93 | ND | 0.12 | ND | −0.78 | 0.93 | −0.20 | ND | ||||||||
| Protein interactions | ND | ND | ND | ND | ND | ND | ND | ND | ||||||||
| Transcription | ||||||||||||||||
| Degradation of RNA | −0.30 | −0.90 | −0.22 | −0.89 | 0.06 | −1.39 | −0.17 | −1.31 | ||||||||
| DNA-dependent RNA polymerase | 0.52 | 1.31 | 0.31 | −0.05 | 0.32 | 0.31 | 0.22 | 0.15 | ||||||||
| Transcription factors | −0.37 | 1.14 | −0.02 | 0.19 | −0.92 | −1.84 | 0.099 | 0.19 | −0.97 | |||||||
| RNA processing | 0.12 | −0.63 | 0.23 | −0.98 | −0.09 | −0.37 | −0.18 | −0.76 | ||||||||
| Transport and binding proteins | ||||||||||||||||
| Amino acids, peptides and amines | −0.28 | 0.44 | 1.58 | 0.63 | 2.45 | 0.015 | 1.40 | 2.33 | 0.021 | 0.60 | ||||||
| Anions | −1.13 | 0.88 | −0.55 | 0.96 | −0.74 | 0.85 | −0.74 | 1.01 | ||||||||
| Carbohydrates, organic alcohols, and acids | −0.63 | 1.87 | 0.080 | −0.76 | 0.57 | −0.16 | 0.96 | −1.06 | 0.19 | |||||||
| Cations and iron carrying compounds | 0.86 | 3.52 | 0.002 | 0.98 | 1.89 | 0.074 | 0.55 | 1.76 | 0.094 | −0.28 | 0.71 | |||||
| Nucleosides, purines and pyrimidines | −1.00 | ND | −0.64 | ND | −0.82 | ND | −1.08 | ND | ||||||||
a T-values were determined from log2 expression data (see Materials and Methods). Boldface type indicates significant T-values.
b p values <0.1 are shown only.
c ND, not determined.
Fig. 1.
Hierarchical clustering analysis of transcriptomic (A) and proteomic data (B), based on the T-profiler results. The heat map is linked by a dendrogram representing clustering of different experimental conditions (top), and T-values for JCVI CMR functional categories (side). The color code is as follows: red indicates positive T-values; green indicates negative T-values; and gray indicates not determined.
Transcription and Protein Synthesis
Gene expression and protein production depend not only on specific regulatory mechanisms, but also vary with growth rate as dictated by environmental conditions. The optimal level of RNA polymerases and ribosomes required by cells often adjust accordingly (43, 44). In the present study, differences in the growth rate of E. coli under each of the test conditions were observed to affect transcription machinery and protein synthesis. The proteomic analysis revealed a significant decrease in the production of poly(A) polymerase I (PcnB) (3.9 log2-fold on average) during growth at all conditions, and DNA-directed RNA polymerase subunit omega (RpoZ) was significantly down-regulated at 14 °C aw 0.967. In respect of translation machinery, the T-profiler analysis indicated that the overall abundance of ribosomal proteins was significantly reduced under the conditions of 14 °C aw 0.985, 25 °C aw 0.967 and 14 °C aw 0.967 (Table III). Furthermore, a number of growth-rate dependent proteins were observed to be significantly reduced in abundance. Production of a tRNA synthase (ThrS) was down-regulated under the osmotic stress conditions of 25 °C aw 0.967 and 14 °C aw 0.967, consistent with the notion that synthesis of tRNA synthetases is coupled to the synthesis of other ribosomal components (53). A decrease in cell growth was probably related to down-regulation of the cell division protein FtsZ at 14 °C aw 0.985, implying delayed cell division (53). Finally, abundance of the housekeeping sigma factor (RpoD) was reduced under high osmolarity conditions (25 °C aw 0.967 and 14 °C aw 0.967), which was most likely because of the decreased growth rate (54).
Carbohydrate Metabolism and Energy Generation
The alteration in metabolic pathways caused by stress typically disturbs the balance of metabolic flux, which leads to metabolic burden during cellular growth. An increased metabolic burden is connected to metabolic readjustments to meet an energy requirement (55). The T-profiler analysis indicated an increase in the activity of major processes associated with carbon and energy metabolism, and metabolic activity varied between different test conditions (Table III). Particularly, cultures grown at low aw (25 °C aw 0.967 and 14 °C aw 0.967) exhibited different metabolic activities from those grown at intermediate aw (25 °C aw 0.985 and 14 °C aw 0.985). Several genes encoding key enzymes in the glycolysis/gluconeogenesis pathway were up-regulated at low aw, as indicated by significant positive T-values. Of which, transcript levels of fbaB, pfkB, and yihT were significantly induced by up to 5.5 log2-fold. Furthermore, it was observed that growth under the stress conditions of 14 °C aw 0.985, 25 °C aw 0.967, and 14 °C aw 0.967 significantly induced the overall expression of genes encoding enzymes of the pentose phosphate pathway (Table III). Among these genes, transaldolase A (talA) and transketolase B (tktB), encoding the enzymes in the nonoxidative branch of the pentose phosphate pathway, were commonly found to be significantly up-regulated. Both talA and tktB are positively regulated by RpoS at their transcriptional level (34), and up-regulation of these genes has also been described in previous studies of low temperature (15 °C) (54) and high osmolarity (4.5% and 0.4 m NaCl) stress (54, 56). Taken together, our findings suggest that the pentose phosphate pathway serves as a more general response to the stress conditions. The differences in metabolic activities might also reflect the different requirements in energy generation for growth under each condition tested.
E. coli is capable of utilizing three different modes for energy generation including aerobic respiration, anaerobic respiration and fermentation (57). During growth under all test conditions, cultures increased aerobic respiration with positive T-values observed for the TCA cycle (Table III). Genes of the main operon encoding the enzymatic complexes involved in aerobic oxidative phosphorylation were significantly up-regulated during growth at 14 °C aw 0.985, 25 °C aw 0.967 and 14 °C aw 0.967, including those genes (sdhABCD) encoding succinate/quinine oxidoreductase (1.7 log2-fold on average). This was despite that in the proteomic analysis, only SdhA exhibited a significant up-regulation (1.9 log2-fold on average) under all test conditions except 14 °C aw 0.967. Regarding anaerobic respiration, the formate dehydrogenase-O subunit FdoG involved in rapid adaptation during anaerobiosis (58) was significantly down-regulated during growth at all conditions, although the transcriptomic analysis revealed a significant decrease in the transcript level of fdo operon only at 14 °C aw 0.967. In addition, the production of ArcA, a component of the ArcAB two-component signal transduction pathway that responds to growth under microaerobiosis was significantly reduced under osmotic stress conditions (25 °C aw 0.967 and 14 °C aw 0.967).
Fatty Acid and Phospholipids Metabolism
A number of proteins involved in fatty acid biosynthesis exhibited a significant decrease in their abundance under the stress conditions of 14 °C aw 0.985, 25 °C aw 0.967 and 14 °C aw 0.967 (Table III). These included the key enzymes in fatty acid biosynthesis initiation (ECs4341, AccC and AccD), and elongation of saturated (FabA and ECs4341) and unsaturated fatty acids (FabA). Consistently, the T-profiler analysis also revealed significant negative T-values for proteins involved in the biosynthesis of fatty acids and phospholipids under these conditions, whereas this was only observed in the transcriptomic data at 14 °C aw 0.967.
Cell Envelope Components
During growth at all conditions, genes and proteins previously known to be induced by the Rcs phosphorelay system, which controls the biosynthesis of extracellular polysaccharide capsule or colanic acid (35), were among the most highly up-regulated (Table IV). Expression levels of gmd/Gmd, wcaI/WcaI, wza/Wza, and wzc/Wzc were commonly observed to increase significantly by up to 6.9 log2-fold. Although the physiological role of colanic acid remains unclear, several studies have demonstrated that colanic acid protects bacterial cells against a variety of environmental stresses, such as acid, osmotic, oxidative, and desiccation stress (59–61), and is essential for biofilm formation (62). An increase in colanic acid production has previously been reported to occur in response to membrane instability originating from a damaged cell envelope (63). Here, we observed that enzymes of the peptidoglycan biosynthetic pathway such as MurIF and DdlA were significantly reduced by up to 4.0 log2-fold under all test conditions. This was despite the fact that the T-profiler analysis revealed a significant decrease in the overall abundance of proteins related to the biosynthesis and degradation of murein sacculus and peptidoglycan only under the cold stress conditions of 14 °C aw 0.985 and 14 °C aw 0.967 (Table III). Down-regulation of proteins involved in peptidoglycan synthesis may lead to envelope stress, resulting in an increase in the biosynthesis of colanic acid. Indeed, a recent study has also demonstrated that peptidoglycan stress caused by antibiotics increases colanic acid synthesis (64).
Table IV. T-profiler analysis of transcriptomic and proteomic data based on major regulons.
| Major regulons | 25 °C aw 0.985 |
14 °C aw 0.985 |
25 °C aw 0.967 |
14 °C aw 0.967 |
||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Transcriptome |
Proteome |
Transcriptome |
Proteome |
Transcriptome |
Proteome |
Transcriptome |
Proteome |
|||||||||
| T-valuea | p valueb | T-value | p value | T-value | p value | T-value | p value | T-value | p value | T-value | p value | T-value | p value | T-value | p value | |
| CpxRA regulon | 0.86 | −2.07 | 0.072 | 0.87 | −0.33 | 0.63 | −1.87 | 0.098 | 1.04 | −1.50 | ||||||
| Rcs regulon | 24.29 | <0.001 | 8.44 | <0.001 | 21.18 | <0.001 | 12.73 | <0.001 | 20.43 | <0.001 | 10.28 | <0.001 | 17.99 | <0.001 | 8.63 | <0.001 |
| RpoE regulon | 1.63 | 0.78 | 1.11 | −0.41 | 1.29 | −1.20 | 0.98 | −0.37 | ||||||||
| RpoH regulon | 0.11 | −0.66 | 0.80 | −1.05 | 0.54 | −1.74 | 0.088 | 1.47 | −0.63 | |||||||
| RpoS regulon | 10.25 | <0.001 | 0.57 | 14.56 | <0.001 | 7.51 | 5.98E-09 | 17.35 | <0.001 | 10.80 | <0.001 | 18.79 | <0.001 | 13.82 | <0.001 | |
a T-values were determined from log2 expression data (see Materials and Methods). Boldface type indicates significant T-values.
b p values <0.1 only are shown.
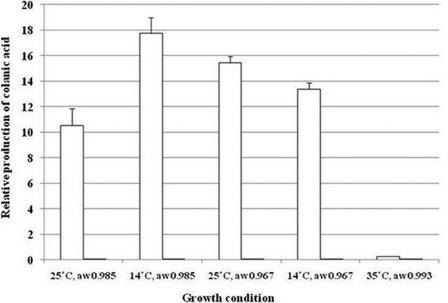
To further investigate the critical role of colanic acid in E. coli, a colanic acid assay was undertaken with a wild-type and colanic acid-deficient mutant (wca−) grown under each condition. Our results indicated that colanic acid synthesis occurred under all test conditions with the highest level of synthesis observed during growth at 14 °C aw 0.985 (Fig. 2), aligning with the results of both transcriptomic and proteomic analyses. However, the growth rate of the colanic acid mutant was not significantly different (p values >0.05) to that of the wild-type parent under each condition, indicating that colanic acid biosynthesis is not required for growth and/or survival under the conditions tested. This result is in keeping with a previously published study in which a colanic acid-deficient mutant still induced an acid adaptation response (61). Ionescu and Belkin (65) have reported that the colanic acid synthesis operon is negatively regulated by RpoS, and that the production of colanic acid may have simply evolved to cope with the low levels of RpoS or rpoS mutations in order to improve nutrient scavenging. In their report, it is predicted that colanic acid production may allow cells that have become rpoS deficient to grow and cope with osmotic upshifts while enjoying a better nutrient-scavenging capability because of the overexpression of RpoD-dependent uptake and nutrient utilization systems. In the current experiments, nutrient rich BHI media were used to support the growth of cultures, and this may have masked the benefits of colanic acid production under conditions requiring nutrient scavenging. It also should be noted that the observation of combined expression of colanic acid and the RpoS regulon under all test conditions is inconsistent with the reported negative regulation of the colanic acid synthesis regulon by RpoS (65). Synthesis of colanic acid in E. coli is regulated by a complex network of regulatory proteins (66), and it is possible that other modes of regulation may come in to play under conditions requiring expression of both regulons.
Fig. 2.
Production of total colanic acid by an E. coli O157:H7 Sakai wild-type (solid white bars) and colanic acid mutant (solid black bars) during growth under different culture conditions. Error bars represent standard errors of the means based on assays from three replicate populations. In most cases, the value obtained for the colanic acid mutant was barely visible above the x axis.
E. coli grown under all test conditions had reduced mRNA levels of several genes involved in surface structures, as indicated by significant negative T-values (Table III). The majority of these genes, especially those commonly found under the conditions of 14 °C aw 0.985, 25 °C aw 0.967 and 14 °C aw 0.967 (ECs4431, ECs2112, ydeT, ydeR, and ydeS) are known to encode pilus proteins. However, in the proteomic analysis only growth at 25 °C aw 0.985 and 25 °C aw 0.967 exhibited a significant decrease in overall abundance of proteins involved in the CpxRA-signaling pathway, which controls the biogenesis of adhesive organelles such as pili (29, 67) (Table IV). This observation contradicts previous studies, reporting that the CpxRA system is activated by structural damage resulting from a variety of stress conditions, including high osmolarity (67, 68). However, recent studies have indicated the involvement of the Cpx-controlled adhesive mechanism and Rcs-regulated colanic acid production in the final stage of biofilm development (62, 69). The positive regulation of the Rcs pathways together with down-regulation of genes encoding pilus proteins and the negative regulation of the CpxRA system might be a feature to prepare cells for the transition from attached cells to mature biofilm under all test conditions (69).
Amino Acid Biosynthesis
Growth under cold stress conditions (14 °C aw 0.985 and 14 °C aw 0.967) was uniquely found to increase expression level of several genes and proteins involved in histidine biosynthesis. The T-profiler analysis of the transcriptomic data revealed up-regulation of several histidine biosynthetic genes at 14 °C aw 0.985 (hisG, hisI, hisA, hisF, hisB, hisC, and hisD; 1.1 to 1.4 log2-fold) and at 14 °C aw 0.967 (hisA, hisF, hisB, hisC and hisD; 1.1 to 1.2 log2-fold), whereas the proteomic analysis only found a significant positive T-value for histidine biosynthesis under the condition of 14 °C aw 0.985 (Table III). The histidine biosynthetic pathway is an energetically expensive process, with 41 ATP molecules sacrificed for each histidine molecule made (70). It is possible that transport of this amino acid is a rate-limiting step during growth at reduced temperature, as shown for the transport of tryptophan in yeast (71). Indeed, O'Donovan and Ingraham (72) have shown that the first enzyme in histidine biosynthesis was more sensitive to feedback inhibition by histidine at low temperatures. The up-regulation of histidine biosynthesis observed here is also in line with a previously published report demonstrating an increase in transcript levels of genes (hisHI) encoding enzymes in histidine biosynthesis at 15 °C (54). However, Kim et al. (55) observed that E. coli down-regulated key proteins (HisACH) for histidine biosynthesis after prolonged incubation at 20 °C following temperature downshift from 37 °C. This may be because of the fact that, in contrast to our study, cultures were not completely cold-adapted. In support of this, a reduction in expression of hisF has been observed in response to cold shock from 37 °C to 15 °C (73). In addition, constitutive expression of the his operon has been reported to cause a number of changes in E. coli including growth inhibition in high-salt media (74–75), although this response was not evident under low water activity conditions in the present study.
Several genes and proteins related to the biosynthesis of amino acids in the pyruvate family were up-regulated during growth at 25 °C aw 0.967, as indicated by significant positive T-values (Table III). These included ilvB/IlvB and ilvD/IlvD that have functions related to the biosynthesis of isoleucine and valine. The transcriptomic analysis also indicated a significant increase in the expression of genes involved in trytophan biosynthesis (trpABC; 1.4 log2-fold on average) under all stress conditions (14 °C aw 0.985, 25 °C aw 0.967 and 14 °C aw 0.967). Up-regulation of the trp operon has previously been reported during exposure to cold (15 °C) and osmotic (4.5% NaCl) stress (54).
Taken together, activation of the amino acid biosynthetic pathways (i.e. histidine, valine, isoleucine, and tryptophan), as already mentioned above seems to be varied during growth under each of the different stress conditions. This suggests that an increase in the intracellular content of these amino acids may be part of the adaptive response enabling growth and/or survival of E. coli under these stress conditions. In support of this hypothesis, Horinouchi et al. (76) demonstrated that supplementing culture media with isoleucine, tryptophan or histidine increased the growth rate under ethanol stress by enabling the culture to develop ethanol tolerance, whereas the growth enhancement was not observed at all in the absence of ethanol stress.
Transport Systems
A significant up-regulation of several genes encoding transporters was observed under low water activity conditions (25 °C aw 0.967 and 14 °C aw 0.967) (supplementary Table S13–S16). These included those responsible for transporting lysine/arginine/ornithine (argT and hisMQ; 25 °C aw 0.967), glutamate/aspartate (gltLJKI; 25 °C aw 0.967 and 14 °C aw 0.967), peptide/nickel (oppADC; 25 °C aw 0.967), and putrescine (potFG; 14 °C aw 0.985, 25 °C aw 0.967 and 14 °C aw 0.967.) Other transport systems that were significantly up-regulated under all test conditions in both transcriptomic and proteomic analyses included those responsible for the uptake of glycine-betaine and proline and osmoprotectant. It has also been reported that transport systems for histidine are strongly impaired under high salt conditions (77). However, our transcriptomic data revealed a significant up-regulation of genes (hisMQ) involved in histidine transporter at 25 °C aw 0.967, whereas a significant decrease in abundance of a subunit (HisJ) of this transporter was observed under all test conditions, except the condition of 14 °C aw 0.985. Furthermore, genes involved in the transport proteins for glutamine (glnPQ) were significantly down-regulated during growth at 25 °C aw 0.985, 14 °C aw 0.985 and 25 °C aw 0.967. The expression of genes encoding the transporters for methionine (metQIN) was significantly reduced at 14 °C aw 0.967.
In the proteomic analysis, many proteins involved in the transport of cations and iron compounds were significantly up-regulated during growth at 25 °C aw 0.985, 14 °C aw 0.985 and 25 °C aw 0.967 (Table III). Among these proteins, only YncD (putative outer membrane receptor for iron transport) was commonly up-regulated (∼1.5 log2-fold on average) under these conditions. The abundance of CirA and Fiu, which act as receptors for siderophores and colicin increased under the conditions of 25 °C aw 0.985 and 14 °C aw 0.985, whereas the iron uptake protein Iha was up-regulated at 25 °C aw 0.985 and 25 °C aw 0.967. It has previously been suggested that the enhancement of iron uptake is caused by changes in the intracellular redox state, originating from alterations in the respiratory system. These changes typically lead to increased levels of intracellular hydrogen peroxide (H2O2) and superoxide (O2−), which invoke the oxidative stress response (76). Previous studies have also shown that high levels of these reactive oxygen species result in increased transcript levels of genes involved in the iron uptake through the inactivation of Fur, a repressor for iron ion transport-related genes (78, 79). This hypothesis is consistent with the fact that several oxidative stress-responsive genes were significantly up-regulated in the present study (see below).
Bacterial Chemotaxis and Flagellar Assembly
The T-profiler analysis showed a significant down-regulation of genes and proteins involved in chemotaxis and motility during growth at all conditions (Table III). It was also observed that these genes and proteins were present as the most heavily down-regulated elements under all conditions (supplementary Table S13–S16). This implies that chemotaxis and motility are the most dispensable functions under these conditions. Consistent with our findings, Moen et al. (54) demonstrated reduced expression of flagella biosynthetic genes in response to cold (15 °C) and osmotic (4.5% NaCl) stress. A similar observation was also found in another study on the response of E. coli to osmotic shock (0.8 m NaCl) (80). The observed down-regulation of chemotaxis and motility is in keeping with the fact that expression of genes encoding flagella are negatively regulated by the master stress regulator RpoS (32) and the Rcs system-activated colanic acid biosynthesis (81).
Stress Response
Both transcriptomic and proteomic analyses revealed activation of the master stress response regulator RpoS (32–34) during growth at all stress conditions (14 °C aw 0.985, 25 °C aw 0.967, and 14 °C aw 0.967) with the greatest T-value at 14 °C aw 0.967 (Table IV). Surprisingly, the transcriptomic analysis also revealed a significant positive T-value for the RpoS regulon under the intermediate growth condition of 25 °C aw 0.985 (T-value 10.25). This was accompanied by a significant up-regulation of many RpoS-dependent elements (32–34), including yciF/YciF, osmB/OsmB, and elaB/ElaB (see supplementary Table S13–S16).
Recent studies have shown that cold (82) and osmotic stress (83) typically induce a defense response mechanism against oxidative stress. Consistent with these observations, the transcriptomic data revealed that cells grown at 14 °C aw 0.985, 25 °C aw 0.967, and 14 °C aw 0.967 exhibited increased expression of various genes involved in oxidative stress resistance, although this trend was not obvious in the proteomic analysis. These genes included those encoding aconitate hydratase (acnA), DNA protection starvation protein (dps), glutaredoxin II (grxB), hydroperoxidase II (katE), osmotically inducible protein (osmC), superoxide dismutase (sodC), and components of a secondary pathway of iron-sulfur cluster assembly (sufABCDES) that have been found to be induced by osmotic and/or heat stress (83). The T-profiler analysis also indicated a significant increase in overall expression of genes involved in the biosynthesis of glutathione and analogs (ggt, grxB, nrdH, and ybdK; up to 3.2 log2-fold) under the stress conditions of 14 °C aw 0.985, 25 °C aw 0.967, and 14 °C aw 0.967 (Table III). Glutathione is the most abundant intracellular low-molecular-weight thiol and has been shown to protect cells against oxidative stress (84).
In general, both transcriptomic and proteomic response of E. coli grown under all conditions correlated well with previously published transcriptomic and proteomic studies of the cold shock response (73, 85), cold adaptation response (54, 73), osmotic shock response (53, 56, 86), osmotic adaptation response (54, 83), and oxidative shock response (87) (see supplementary Table S13–S16). Particularly, we observed up-regulation of most genes/proteins under all test conditions, including imposition of cold temperature without osmotic stress, that have previously been described to be involved in osmotic adaptation (54). These included groups of the glycine-betaine and proline transporter (pro operon), osmotically inducible proteins (osm operon), trehalose biosynthetic enzymes (otsAB/OtsAB), and the yehWXYZ operon of the predicted ATP-dependant transporter for osmoprotectant. These findings support previous reports, suggesting that many of the genes and proteins typically induced by osmotic stress play a more general role in cross-protection against diverse stress conditions (e.g. cold stress in this study) (34, 56). Furthermore, genes of the glutamate-dependent gad system involved in acid resistance (88), were significantly up-regulated at 14 °C aw 0.985 (gadBEX), 25 °C aw 0.967 (gadBX), and 14 °C aw 0.967 (gadBCX). This is in keeping with the fact that gad genes have previously been shown to be induced by cold and osmotic stress (54), and osmotic shock (56).
The results of the current analysis were also evaluated by a direct comparison of the data, in which changes exclusively because of low temperature (14 °C aw 0.985 versus 25 °C aw 0.985 and 14 °C aw 0.967 versus 25 °C aw 0.967) and those exclusively because of low water activity (25 °C aw 0.967 versus 25 °C aw 0.985 and 14 °C aw 0.967 versus. 14 °C aw 0.985) were investigated (results not shown). The T-profiler analysis revealed that E. coli specifically responded to low temperature by up-regulating several genes and proteins involved in the biosynthetic pathway of histidine, while reducing the overall abundance of ribosomal proteins and those proteins related to the biosynthesis and degradation of murein sacculus and peptidoglycan. For the response exclusively associated with low water activity, it was found that E. coli up-regulated a number of groups of genes or proteins involved in the TCA cycle, detoxication and the biosynthesis of glutathione and analogs. It was observed that this was also accompanied by down-regulation of several ribosomal proteins and elements of biosynthesis and degradation of surface polysaccharides and lipopolysaccharides. In addition, activation of the RpoS general stress response regulon and the Rcs regulon involved in colanic acid biosynthesis was observed as a specific response to both low temperature and low aw.
CONCLUDING REMARKS
In this study, we explored the response of E. coli O157:H7 Sakai under steady-state conditions relevant to low temperature and water activity conditions experienced during meat carcass chilling. E. coli O157:H7 responded to the conditions of chill temperature and osmotic stress by increasing the expression of genes and proteins controlled by the RpoS general stress response regulon and the Rcs regulon involved in colanic acid biosynthesis, as well as down-regulating the elements involved in chemotaxis and motility. In the colanic acid study, it was shown that colanic acid biosynthesis was not necessary for enabling E. coli to maintain their exponential growth under conditions of cold and/or osmotic stress. Although the present study was descriptive, the data generated here will be analyzed in combination with recently generated data monitoring the time-dependent changes in the global pattern of gene and protein expression of E. coli in response to a sudden downshift in temperature and water activity stress. The information garnered may enable us to exploit the physiology of E. coli to enable the selection and/or development of potential strategies to inactivate this pathogen.
Acknowledgments
Their support, in particular that of Ian Jenson, Manager of the Food Safety and Strategic Science program, is gratefully acknowledged. We also thank Edwin Lowe and Richard Wilson from Central Science Laboratory (University of Tasmania, Hobart, AUS) for technical support in 2D-LC/MS/MS analysis.
Footnotes
* This study was entirely funded by Meat and Livestock Australia.
 This article contains supplemental Tables S1 to S16 and Material S1 and S2.
This article contains supplemental Tables S1 to S16 and Material S1 and S2.
1 The abbreviations used are:
- aw
- water activity
- 2D-LC-MS/MS analysis
- Two-dimensional liquid chromatography tandem mass spectrometry
- FRT FLP
- recognition target
- MS/MS
- Tandem mass spectrometry
- MudPIT
- Multidimensional protein identification technology
- SpC
- Spectral count
- SpI
- Spectral index.
REFERENCES
- 1. Riley L. W., Remis R. S., Helgerson S. D., McGee H. B., Wells J. G., Davis B. R., Hebert R. J., Olcott E. S., Johnson L. M., Hargrett N. T., Blake P. A., Cohen M. L. (1983) Hemorrhagic colitis associated with a rare Escherichia coli serotype. N. Engl. J. Med. 308, 681–685 [DOI] [PubMed] [Google Scholar]
- 2. Cagney C., Crowley H., Duffy G., Sheridan J. J., Brien S. O., Carney E., Anderson W., McDowell D. A., Blair I. S., Bishop R. H. (2004) Prevalence and numbers of Escherichia coli O157:H7 in minced beef and beef burgers from butcher shops and supermarkets in the Republic of Ireland. Food Microbiol. 21, 203–212 [Google Scholar]
- 3. Hussein H. S., Bollinger L. M. (2005) Prevalence of Shiga toxin-producing Escherichia coli in beef. Meat Sci. 71, 676–689 [DOI] [PubMed] [Google Scholar]
- 4. McClure P. (2000) The impact of E. coli O157 on the food industry. World J. Microbiol. Biotechnol. 16, 749–755 [Google Scholar]
- 5. Elder R. O., Keen J. E., Siragusa G. R., Barkocy-Gallagher G. A., Koohmaraie M., Laegreid W. W. (2000) Correlation of enterohemorrhagic Escherichia coli O157 prevalence in feces, hides, and carcasses of beef cattle during processing. Proc. Natl. Acad. Sci. U.S.A. 97, 2999–3003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Gill C. O., Bryant J. (1997) Assessment of the hygienic performances of two beef carcass cooling processes from product temperature history data or enumeration of bacteria on carcass surfaces. Food Microbiol. 14, 593–602 [Google Scholar]
- 7. Lenahan M., Crowley H., O'Brien S. B., Byrne C., Sweeney T., Sheridan J. J. (2009) The potential use of chilling to control the growth of Enterobacteriaceae on porcine carcasses and the incidence of E. coli O157:H7 in pigs. J. Appl. Microbiol. 106, 1512–1520 [DOI] [PubMed] [Google Scholar]
- 8. Aertsen A., Michiels C. W. (2004) Stress and how bacteria cope with death and survival. Crit. Rev. Microbiol. 30, 263–273 [DOI] [PubMed] [Google Scholar]
- 9. Chung H. J., Bang W., Drake M. A. (2006) Stress response of Escherichia coli. Compr. Rev. Food Sci. Food Saf. 5, 52–64 [Google Scholar]
- 10. Ingraham J. L., Marr A. G. (1996) Effect of temperature, pressure, pH, and osmotic stress on growth. In: Neidhardt F. C., Curtiss R., III, Ingraham J. L., Lin E. C. C., Low K. B., Magasanik B., Reznikoff W. S., Riley M., Schaechter M., Umbarger H. E., eds. Escherichia coli and Salmonella: cellular and molecular biology, pp. 1570–1578, American Society for Microbiology, Washington, D.C. [Google Scholar]
- 11. Cao-Hoang L., Dumont F., Marechal P. A., Le-Thanh M., Gervais P. (2008) Rates of chilling to 0°C: implications for the survival of microorganisms and relationship with membrane fluidity modifications. Appl. Microbiol. Biotechnol. 77, 1379–1387 [DOI] [PubMed] [Google Scholar]
- 12. Yamanaka K. (1999) Cold shock response in Escherichia coli. J. Mol. Microbiol. Biotechnol. 1, 193–202 [PubMed] [Google Scholar]
- 13. O'Byrne C. P., Booth I. R. (2002) Osmoregulation and its importance to foodborne microorganisms. Int. J. Food Microbiol. 74, 203–216 [DOI] [PubMed] [Google Scholar]
- 14. Hayashi T., Makino K., Ohnishi M., Kurokawa K., Ishii K., Yokoyama K., Han C. G., Ohtsubo E., Nakayama K., Murata T., Tanaka M., Tobe T., Iida T., Takami H., Honda T., Sasakawa C., Ogasawara N., Yasunaga T., Kuhara S., Shiba T., Hattori M., Shinagawa H. (2001) Complete Genome Sequence of enterohemorrphagic Escherichia coli O157:H7 and genomic comparison with a laboratory strain K-12. DNA Res. 8, 11–22 [DOI] [PubMed] [Google Scholar]
- 15. Watanabe H., Wada A., Inagaki Y., Itoh K., Tamura K. (1996) Outbreaks of enterohaemorrhagic Escherichia coli O157:H7 infection by two different genotype strains in Japan. Lancet 348, 831–832 [DOI] [PubMed] [Google Scholar]
- 16. Datsenko K. A., Wanner B. L. (2000) One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. U.S.A. 97, 6640–6645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Chirife J., Resnik S. L. (1984) Unsaturated solutions of sodium chloride as reference sources of water activity at various temperatures. J. Food Sci. 49, 1486–1488 [Google Scholar]
- 18. Irizarry R. A., Hobbs B., Collin F., Beazer-Barclay Y. D., Antonellis K. J., Scherf U., Speed T. P. (2003) Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics 4, 249–264 [DOI] [PubMed] [Google Scholar]
- 19. Delahunty C., Yates J. R., 3rd (2005) Protein identification using 2D-LC-MS/MS. Methods 35, 248–255 [DOI] [PubMed] [Google Scholar]
- 20. Rauch A., Bellew M., Eng J., Fitzgibbon M., Holzman T., Hussey P., Igra M., Maclean B., Lin C. W., Detter A., Fang R., Faca V., Gafken P., Zhang H., Whiteaker J., States D., Hanash S., Paulovich A., McIntosh M. W. (2006) Computational proteomics analysis system (CPAS): an extensible, open-source analytic system for evaluating and publishing proteomic data and high throughput biological experiments. J. Proteome Res. 5, 112–121 [DOI] [PubMed] [Google Scholar]
- 21. Craig R., Beavis R. C. (2004) TANDEM: matching proteins with tandem mass spectra. Bioinformatics 20, 1466–1467 [DOI] [PubMed] [Google Scholar]
- 22. Nesvizhskii A. I., Keller A., Kolker E., Aebersold R. (2003) A statistical model for identifying proteins by tandem mass spectrometry. Anal. Chem. 75, 4646–4658 [DOI] [PubMed] [Google Scholar]
- 23. Keller A., Nesvizhskii A. I., Kolker E., Aebersold R. (2002) Empirical statistical model to estimate the accuracy of peptide identifications made by MS/MS and database search. Anal. Chem. 74, 5383–5392 [DOI] [PubMed] [Google Scholar]
- 24. Choi H., Nesvizhskii A. I. (2008) False discovery rates and related statistical concepts in mass spectrometry-based proteomics. J. Proteome Res. 7, 47–50 [DOI] [PubMed] [Google Scholar]
- 25. Liu H., Sadygov R. G., Yates J. R., 3rd (2004) A model for random sampling and estimation of relative protein abundance in shotgun proteomics. Anal. Chem. 76, 4193–4201 [DOI] [PubMed] [Google Scholar]
- 26. Old W. M., Meyer-Arendt K., Aveline-Wolf L., Pierce K. G., Mendoza A., Sevinsky J. R., Resing K. A., Ahn N. G. (2005) Comparison of label-free methods for quantifying human proteins by shotgun proteomics. Mol. Cell Proteomics 4, 1487–1502 [DOI] [PubMed] [Google Scholar]
- 27. Fu X., Gharib S. A., Green P. S., Aitken M. L., Frazer D. A., Park D. R., Vaisar T., Heinecke J. W. (2008) Spectral index for assessment of differential protein expression in shotgun proteomics. J. Proteome Res. 7, 845–854 [DOI] [PubMed] [Google Scholar]
- 28. Boorsma A., Foat B. C., Vis D., Klis F., Bussemaker H. J. (2005) T-profiler: scoring the activity of predefined groups of genes using gene expression data. Nucleic Acids Res. 33, W592–W595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Price N. L., Raivio T. L. (2009) Characterization of the Cpx regulon in Escherichia coli strain MC4100. J. Bacteriol. 191, 1798–1815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Rhodius V. A., Suh W. C., Nonaka G., West J., Gross C. A. (2006) Conserved and variable functions of the σE stress response in related genomes. PLoS Biol. 4, e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Nonaka G., Blankschien M., Herman C., Gross C. A., Rhodius V. A. (2006) Regulon and promoter analysis of the E. coli heat-shock factor, σ32, reveals a multifaceted cellular response to heat stress. Genes Dev. 20, 1776–1789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Patten C. L., Kirchhof M. G., Schertzberg M. R., Morton R. A., Schellhorn H. E. (2004) Microarray analysis of RpoS-mediated gene expression in Escherichia coli K-12. Mol. Genet. Genomics 272, 580–591 [DOI] [PubMed] [Google Scholar]
- 33. Lacour S., Landini P. (2004) σS-dependent gene expression at the onset of stationary phase in Escherichia coli: function of σS-dependent genes and identification of their promoter sequences. J. Bacteriol. 186, 7186–7195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Weber H., Polen T., Heuveling J., Wendisch V. F., Hengge R. (2005) Genome-wide analysis of the general stress response network in Escherichia coli: σS-dependent genes, promoters, and sigma factor selectivity. J. Bacteriol. 187, 1591–1603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Hagiwara D., Sugiura M., Oshima T., Mori H., Aiba H., Yamashino T., Mizuno T. (2003) Genome-wide analyses revealing a signaling network of the RcsC-YojN-RcsB phosphorelay system in Escherichia coli. J. Bacteriol. 185, 5735–5746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. de Hoon M. J., Imoto S., Nolan J., Miyano S. (2004) Open source clustering software. Bioinformatics 20, 1453–1454 [DOI] [PubMed] [Google Scholar]
- 37. Saldanha A. J. (2004) Java Treeview–extensible visualization of microarray data. Bioinformatics 20, 3246–3248 [DOI] [PubMed] [Google Scholar]
- 38. Bergmaier D., Lacroix C., Guadalupe Macedo M., Champagne C. P. (2001) New method for exopolysaccharide determination in culture broth using stirred ultrafiltration cells. Appl. Microbiol. Biotechnol. 57, 401–406 [DOI] [PubMed] [Google Scholar]
- 39. Obadia B., Lacour S., Doublet P., Baubichon-Cortay H., Cozzone A. J., Grangeasse C. (2007) Influence of tyrosine-kinase Wzc activity on colanic acid production in Escherichia coli K12 Cells. J. Mol. Biol. 367, 42–53 [DOI] [PubMed] [Google Scholar]
- 40. Dische Z., Shettles L. B. (1951) A new spectrophotometric test for the detection of methylpentose. J. Biol. Chem. 192, 579–582 [PubMed] [Google Scholar]
- 41. Vizcaino J. A., Cote R., Reisinger F., Barsnes H., Foster J. M., Rameseder J., Hermjakob H., Martens L. (2010) The proteomics identifications database: 2010 update. Nucleic Acids Res. 38(Database issue), D736–D742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Ross T., Ratkowsky D. A., Mellefont L. A., McMeekin T. A. (2003) Modelling the effects of temperature, water activity, pH and lactic acid concentration on the growth rate of Escherichia coli. Int. J. Food Microbiol. 82, 33–43 [DOI] [PubMed] [Google Scholar]
- 43. Gyaneshwar P., Paliy O., McAuliffe J., Jones A., Jordan M. I., Kustu S. (2005) Lessons from Escherichia coli genes similarly regulated in response to nitrogen and sulfur limitation. Proc. Natl. Acad. Sci. U.S.A. 102, 3453–3458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Klumpp S., Zhang Z., Hwa T. (2009) Growth rate-dependent global effects on gene expression in bacteria. Cell 139, 1366–1375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Elias J. E., Haas W., Faherty B. K., Gygi S. P. (2005) Comparative evaluation of mass spectrometry platforms used in large-scale proteomics investigations. Nat. Methods 2, 667–675 [DOI] [PubMed] [Google Scholar]
- 46. Chen G., Gharib T. G., Huang C. C., Taylor J. M., Misek D. E., Kardia S. L., Giordano T. J., Iannettoni M. D., Orringer M. B., Hanash S. M., Beer D. G. (2002) Discordant protein and mRNA expression in lung adenocarcinomas. Mol. Cell Proteomics 1, 304–313 [DOI] [PubMed] [Google Scholar]
- 47. Griffin T. J., Gygi S. P., Ideker T., Rist B., Eng J., Hood L., Aebersold R. (2002) Complementary profiling of gene expression at the transcriptome and proteome levels in Saccharomyces cerevisiae. Mol. Cell Proteomics 1, 323–333 [DOI] [PubMed] [Google Scholar]
- 48. Ideker T., Thorsson V., Ranish J. A., Christmas R., Buhler J., Eng J. K., Bumgarner R., Goodlett D. R., Aebersold R., Hood L. (2001) Integrated genomic and proteomic analyses of a systematically perturbed metabolic network. Science 292, 929–934 [DOI] [PubMed] [Google Scholar]
- 49. Tian Q., Stepaniants S. B., Mao M., Weng L., Feetham M. C., Doyle M. J., Yi E. C., Dai H., Thorsson V., Eng J., Goodlett D., Berger J. P., Gunter B., Linseley P. S., Stoughton R. B., Aebersold R., Collins S. J., Hanlon W. A., Hood L. E. (2004) Integrated genomic and proteomic analyses of gene expression in mammalian cells. Mol. Cell Proteomics 3, 960–969 [DOI] [PubMed] [Google Scholar]
- 50. Hatzimanikatis V., Choe L. H., Lee K. H. (1999) Proteomics: theoretical and experimental considerations. Biotechnol. Prog. 15, 312–318 [DOI] [PubMed] [Google Scholar]
- 51. Hatzimanikatis V., Lee K. H. (1999) Dynamical analysis of gene networks requires both mRNA and protein expression information. Metab. Eng. 1, 275–281 [DOI] [PubMed] [Google Scholar]
- 52. Gygi S. P., Rochon Y., Franza B. R., Aebersold R. (1999) Correlation between protein and mRNA abundance in yeast. Mol. Cell. Biol. 19, 1720–1730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Weber A., Jung K. (2002) Profiling early osmostress-dependent gene expression in Escherichia coli using DNA macroarrays. J. Bacteriol. 184, 5502–5507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Moen B., Janbu A. O., Langsrud S., Langsrud O., Hobman J. L., Constantinidou C., Kohler A., Rudi K. (2009) Global responses of Escherichia coli to adverse conditions determined by microarrays and FT-IT spectroscopy. Can. J. Microbiol. 55, 714–728 [DOI] [PubMed] [Google Scholar]
- 55. Kim Y. H., Han K. Y., Lee K., Lee J. (2005) Proteome response of Escherichia coli fed-batch culture to temperature downshift. Appl. Microbiol. Biotechnol. 68, 786–793 [DOI] [PubMed] [Google Scholar]
- 56. Weber A., Kögl S. A., Jung K. (2006) Time-dependent proteome alterations under osmotic stress during aerobic and anaerobic growth in Escherichia coli. J. Bacteriol. 188, 7165–7175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Pin C., Rolfe M. D., Munoz-Cuevas M., Hinton J. C., Peck M. W., Walton N. J., Baranyi J. (2009) Network analysis of the transcriptional pattern of young and old cells of Escherichia coli during lag phase. BMC Syst. Biol. 3, 108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Benoit S., Abaibou H., Mandrand-Berthelot M. A. (1998) Topological analysis of the aerobic membrane-bound formate dehydrogenase of Escherichia coli. J. Bacteriol. 180, 6625–6634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Ophir T., Gutnick D. L. (1994) A role for exopolysaccharides in the protection of microorgansims from desiccation. Appl. Environ. Microbiol. 60, 740–745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Chen J., Lee S. M., Mao Y. (2004) Protective effect of exopolysaccharide colanic acid of Escherichia coli O157:H7 to osmotic and oxidative stress. Int. J. Food Microbiol. 93, 281–286 [DOI] [PubMed] [Google Scholar]
- 61. Mao Y., Doyle M. P., Chen J. (2006) Role of colanic acid exopolysaccharide in the survival of enterohaemorrhagic Escherichia coli O157:H7 in simulated gastrointestinal fluids. Lett. Appl. Microbiol. 42, 642–647 [DOI] [PubMed] [Google Scholar]
- 62. Danese P. N., Pratt L. A., Kolter R. (2000) Exopolysaccharide production is required for development of Escherichia coli K-12 biofilm architecture. J. Bacteriol. 182, 3593–3596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Meredith T. C., Mamat U., Kaczynski Z., Lindner B., Holst O., Woodard R. W. (2007) Modification of lipopolysaccharide with colanic acid (M-antigen) repeats in Escherichia coli. J. Biol. Chem. 282, 7790–7798 [DOI] [PubMed] [Google Scholar]
- 64. Laubacher M. E., Ades S. E. (2008) The Rcs phosphorelay is a cell envelope stress response activated by peptidoglycan stress and contributes to intrinsic antibiotics resistance. J. Bacteriol. 190, 2065–2074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Ionescu M., Belkin S. (2009) Overproduction of exopolysaccharides by an Escherichia coli K-12 rpoS mutant in response to osmotic stress. Appl. Environ. Microbiol. 75, 483–492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Gottesman S., Stout V. (1991) Regulation of capsular polysaccharide synthesis in Escherichia coli K12. Mol. Microbiol. 5, 1599–1606 [DOI] [PubMed] [Google Scholar]
- 67. Raivio T. L., Silhavy T. J. (1999) The σE and Cpx regulatory pathways: overlapping but distinct envelope stress responses. Curr. Opin. Microbiol. 2, 159–165 [DOI] [PubMed] [Google Scholar]
- 68. Bergholz T. M., Wick L. M., Qi W., Riordan J. T., Ouellette L. M., Whittam T. S. (2007) Global transcriptional response of Escherichia coli O157:H7 to growth transitions in glucose minimal medium. BMC Microbiol. 7, 97–123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Ferrières L., Clarke D. J. (2003) The RcsC sensor kinase is required for normal biofilm formation in Escherichia coli K-12 and controls the expression of a regulon in response to growth on a solid surface. Mol. Microbiol. 50, 1665–1682 [DOI] [PubMed] [Google Scholar]
- 70. Brenner M., Ames B. N. (1971) The histidine operon and its regulation. In: Vogel H. J., ed. Metabolic pathways, Academic Press Inc, New York [Google Scholar]
- 71. Kawamura D., Yamashita I., Nimi O., Toh-e A. (1994) Cloning and nucleotide sequence of a gene conferring ability to grow at a low temperature on Saccharomyces cerevisiae tryptophan auxotrophs. J. Ferment. Bioeng. 77, 1–9 [Google Scholar]
- 72. O'Donovan G. A., Ingraham J. L. (1965) Cold-sensitive mutants of Escherichia coli resulting from increased feedback inhibition. Proc. Natl. Acad. Sci. U.S.A. 54, 451–457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Phadtare S., Inouye M. (2004) Genome-wide transcriptional analysis of the cold shock response in wild-type and cold-sensitive, quadruple-csp-deletion strains of Escherichia coli. J. Bacteriol. 186, 7007–7014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Casadesus J., Roth J. R. (1989) Absence of insertions among spontaneous mutants of Salmonella typhimurium. Mol. Gen. Genet. 216, 210–216 [DOI] [PubMed] [Google Scholar]
- 75. Frandsen N., D'Ari R. (1993) Excess histidine enzymes cause AICAR-independent filamentation in Escherichia coli. Mol. Gen. Genet. 240, 348–354 [DOI] [PubMed] [Google Scholar]
- 76. Horinouchi T., Tamaoka K., Furusawa C., Ono N., Suzuki S., Hirasawa T., Yomo T., Shimizu H. (2010) Transcriptome analysis of parallel-evolved Escherichia coli strains under ethanol stress. BMC Genomics 11, 579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Liu C. E., Ames G. F.-L. (1997) Characterization of transport through the periplasmic histidine permease using proteoliposomes reconstituted by dialysis. J. Biol. Chem. 272, 859–866 [DOI] [PubMed] [Google Scholar]
- 78. Blanchard J. L., Wholey W. Y., Conlon E. M., Pomposiello P. J. (2007) Rapid changes in gene expression dynamics in response to superoxide reveal SoxRS-depedent and independent transcriptional networks. PLoS ONE 2, e1186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Varghese S., Wu A., Park S., Imlay K. R., Imlay J. A. (2007) Submicromolar hydrogen peroxide disrupts the ability of Fur protein to control free-iron levels in Escherichia coli. Mol. Microbiol. 64, 822–830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Cheung K. J., Badarinarayana V., Selinger D. W., Janse D., Church G. M. (2003) A microarray-based antibiotic screen identifies a regulatory role for supercoiling in the osmotic stress response of Escherichia coli. Genome Res. 13, 206–215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Francez-Charlot A., Laugel B., Van Gemert A., Dubarry N., Wiorowski F., Castanié-Cornet M. P., Gutierrez C., Cam K. (2003) RcsCDB His-Asp phosphorelay system negatively regulates the flhDC operon in Escherichia coli. Mol. Microbiol. 49, 823–832 [DOI] [PubMed] [Google Scholar]
- 82. Smirnova G. V., Zakirova O. N., Oktyabrskii O. N. (2001) The role of antioxidant systems in the cold stress response of Escherichia coli. Microbiology 70, 45–5011338835 [Google Scholar]
- 83. Gunasekera T. S., Csonka L. N., Paliy O. (2008) Genome-wide transcriptional responses of Escherichia coli K-12 to continuous osmotic and heat stresses. J. Bacteriol. 190, 3712–3720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Carmel-Harel O., Storz G. (2000) Roles of the glutathione- and thioredoxin-dependent reduction systems in the Escherichia coli and Saccharomyces cerevisiae responses to oxidative stress. Annu. Rev. Microbiol. 54, 439–461 [DOI] [PubMed] [Google Scholar]
- 85. Polissi A., Laurentis W. D., Zangrossi S., Briani F., Longhi V., Pesole G., Deho G. (2003) Changes in Escherichia coli transcriptome during acclimatization at low temperature. Res. Microbiol. 154, 573–580 [DOI] [PubMed] [Google Scholar]
- 86. Shabala L., Bowman J., Brown J., Ross T., McMeekin T., Shabala S. (2009) Ion transport and osmotic adjustment in Escherichia coli in response to ionic and non-ionic osmotica. Environ. Microbiol. 11, 137–148 [DOI] [PubMed] [Google Scholar]
- 87. Zheng M., Wang X., Templeton L. J., Smulski D. R., LaRossa R. A., Storz G. (2001) DNA microarray-mediated transcriptional profiling of the Escherichia coli response to hydrogen peroxide. J. Bacteriol. 183, 4562–4570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Tramonti A., Visca P., De Canio M., Falconi M., De Biase D. (2002) Functional characterization and regulation of gadX, a gene encoding an AraC/XylS-like transcriptional activator of the Escherichia coli glutamic acid decarboxylase system. J. Bacteriol. 184, 2603–2613 [DOI] [PMC free article] [PubMed] [Google Scholar]