Abstract
Lysine acetylation is a well-studied post-translational modification on both histone and nonhistone proteins. More than 2000 acetylated proteins and 4000 lysine acetylation sites have been identified by large scale mass spectrometry or traditional experimental methods. Although over 20 lysine (K)-acetyl-transferases (KATs) have been characterized, which KAT is responsible for a given protein or lysine site acetylation is mostly unknown. In this work, we collected KAT-specific acetylation sites manually and analyzed sequence features surrounding the acetylated lysine of substrates from three main KAT families (CBP/p300, GCN5/PCAF, and the MYST family). We found that each of the three KAT families acetylates lysines with different sequence features. Based on these differences, we developed a computer program, Acetylation Set Enrichment Based method to predict which KAT-families are responsible for acetylation of a given protein or lysine site. Finally, we evaluated the efficiency of our method, and experimentally detected four proteins that were predicted to be acetylated by two KAT families when one representative member of the KAT family is over expressed. We conclude that our approach, combined with more traditional experimental methods, may be useful for identifying KAT families responsible for acetylated substrates proteome-wide.
In order to function properly, natural proteins suffer from various post-translational modifications, among which acetylation plays critical roles in protein stability, gene expression regulation, protein-protein interactions, and cellular metabolism (1–5). Acetylation occurs mainly on lysine residues and has been extensively studied on histone N-terminal lysine (6, 7). Further study shows that lysine residues of nonhistone proteins, such as p53 and ER can also be acetylated (8, 9). Although the number of nonhistone protein acetylation reports is rapidly expanding, it is difficult to detect the acetylation of all lysine residues in the whole proteome by traditional experiments because of the huge number of lysine residues. There are over 600,000 lysine residues in the human proteome according to UniProtKB/Swiss-Prot (version 188).
To find more acetylated proteins new technologies have been developed, such as immune-precipitation combined with mass spectrometric analysis. At least three groups successfully used this technology to get precise acetylation information from about two thousand proteins (3, 5, 10). Their research greatly increased the number of known acetylated proteins and expanded our knowledge of acetylation. However, the number of discovered acetylation sites is still small compared with the huge amount of lysine residues in the whole proteome. In addition, acetylation is a dynamic and condition-dependent process, as different cells and the same cells in different conditions have various acetylation profiles. Because it is impossible to determine the acetylation status in every cell and condition, researchers took full advantage of known information and developed in silico methods to predict acetylation. The most widely used method is the support vector machine method which can classify new sequences by learning the features of sequence contexts surrounding the acetylated lysines. Several acetylation predict tools were based on this method and each achieved a good performance (11–13). Besides support vector machine, other methods such as meta-analysis and sequence clustering have also been used to predict acetylation sites (14, 15). All these predictions provided valuable insights for further acetylation identification.
These previous efforts, both mass spectrometry and in silico prediction, focus on the acetylation itself, and no methods give any information about which (K)-acetyl-transferases (KAT)1 is responsible for one given protein or lysine site. Like kinases which catalyze phosphorylation of a specific subset of substrates, KATs are substrate-specific enzymes. Over 20 human KATs have been identified (supplemental Table S1). Although it is not ease to categorize all of these KATs because of their variety, 9 KATs are divided into three families by sequence and structural similarity (supplemental Table S1, supplemental Fig. S1) (16–18). They are the CBP/p300 family, the General control of amino acid synthesis protein 5-like 2 (GCN5)/p300/CBP-associated factor (PCAF) family and the MYST family. Each of the three families catalyzes a special subgroup of substrates. For example, WRN is acetylated by CBP or p300, but not by PCAF or TIP60 (19); PARP-2 is acetylated by PCAF, but not by CBP or p300 (20). Even though different KAT families can target the same protein, they acetylate different lysine sites. For instance, PCAF acetylates p53 at site K320 (8), whereas TIP60 acetylates it at K120 (21); CBP acetylates HMGI(Y) at site K65, whereas PCAF acetylates it at K71 (22). Hundreds of acetylated sites in human proteins with identified KATs from the three families have been reported (supplemental Table S1). We have previously provided evidence about the acetylation of nonhistone proteins, such as p53 and Forkhead box protein O1, and their functional roles in cells (23–25). However, the KATs responsible for most non-histone proteins are not clear. Although we have predicted kinase-specific phosphorylation sites (26, 27), there is no KAT-specific acetylation site prediction tool available.
Here, making full use of the manually collected acetylated sites with known KAT information, we found that acetylation sites of different KATs have varied characters. We then developed an Acetylation Set Enrichment Based (ASEB) method to predict acetylated proteins or sites and the KAT families responsible for the acetylation. We predicted that proteins methyl-CpG-binding domain protein 1 (MBD1) and MTA1 are acetylated by the CBP/p300 family and that DNA polymerase β and DDB1 are acetylated by the GCN5/PCAF family. Using an immunoprecipitation assay combined with Western blotting we evaluated the acetylation of these proteins. To our knowledge, this is the first time that KAT-specific acetylation sites have been characterized and the initial data presented in this work should bring important information to bioinformatics and experimental studies on acetylation.
EXPERIMENTAL PROCEDURES
Data Preparation
We collected human proteins acetylated by each of the three families by searching the PubMed literature with key words (supplemental Table S2). We also searched substrates of other KATs, but because of the small number of substrates, we did not include them in any further analysis. After examining these papers and related references, the papers with identified acetylation sites and KAT information were picked out. The acetylated proteins were extracted and mapped to the UniProt Database and we retrieved their exact UniProt ID. The acetylated sites were carefully checked to make sure the acetylated position was exactly the position mentioned in the literature.
The ASEB Method
Gene Set Enrichment Analysis (GSEA) was developed and used on DNA microarray data to detect coordinated expression changes in a group of functionally related genes and then was applied to find the putative functions of the long noncoding RNAs (28–30). Taking advantage of the idea of GSEA, we proposed a new method called ASEB to detect new sites acetylated by a specific KAT family. For each family, we focused on finding sites which were similar in sequence with discovered ones. We treated the acetylated lysine sites and their surrounding amino acids (eight on each side) as acetylated peptides. Acetylated peptides from two KAT families formed two acetylated peptides sets (the CBP/p300 set and the GCN5/PCAF set). Because of the small number of peptides in the MYST family, we just analyzed the sequence features of this family and omitted it from the prediction processes. The following calculation was based on the conception of the 17 amino acid long peptides. To determine whether a given peptide could be acetylated by one of the two families, we just needed to know whether the given peptide was similar to the acetylated peptides in that set. The ASEB method was developed to estimate this similarity and the significance of the similarity. Compared with the original GSEA method several necessary changes have been made. The details of the ASEB method are described as follows:
Input
Predefined KAT special peptide set Sk containing N peptides. Here, Sk (CBP/p300) containing 267 peptides (the other 13 peptides which had less than six surrounding amino acids on a side were excluded); Sk (GCN5/PCAF) containing 82 peptides (the other two peptides which had less than six surrounding amino acids on a side were excluded). Predefined background peptide set Sb containing 10,000 randomly selected peptides.
Step 1: Calculate Similarity Scores
At first, similarity scores between the query peptide (denoted as pquery) and each peptide in Sk ∪Sb were calculated according to the BLOSUM 62 matrix. Then, these similarity scores were normalized to [0, 1] and [-1,0] for positive and negative scores separately. Third, all similarity scores were mixed together and ranked from high to low. According to the above steps, we know that if pquery is similar to the peptides in Sk for a specific KAT family, these peptides in Sk should be enriched at the top of the ranked similarity score list, and pquery is likely a novel substrate for that KAT family.
Step 2: Calculate Enrichment Score (ES)
To determine how enriched the peptides in Sk were at the top of the ranked list, a running sum score was calculated by walking down the list. Let us denote ri as the similarity scores between peptide pquery and peptide pi ϵSk, and R as the sum of |ri| for all pi ϵSk. While walking down the list, the running sum statistic increased |rj|/R when encountering a peptide pj in Sk and decreased 1/10,000 when encountering a peptide in Sb. The enrichment score (ES) was defined as the maximum of the running sum statistic.
Step 3: Estimate Significance of ES
To estimate the significance of the ES for a given peptide, a total of 9999 peptide sets with the same size as Sk were randomly generated from the background peptides, and denoted as Snull1 to Snull 9999. Then the ES for each set was calculated by treating each as predefined peptide sets. Finally, these 10,000 ES (ES(Snull1), ES(Snull2),..ESnull9999, plus ES(Sk)) were ranked from high to low. Suppose the rank of ES(Sk) is L, the nominal p value of the given peptide was defined as L/10000. The p value should be between 0.0001 and 1, with the minimum interval of 0.0001. The smaller the p value, the more significant the chance that the given peptides were acetylated by the KAT family. In practice, the number of randomly generated peptide sets can be changed according to different needs.
The ASEB method has several advantages. It did not require the balanced number of positive and negative datasets, as support vector machine did. It directly calculates the similarity between two single peptides. In most of the cases, peptides in the KAT set can be divided into several subsets which share little variation with each other. The ASEB method is particularly fit to these cases, because the ES of a given peptide would be significant as soon as it is similar to some but not all peptides in the KAT set.
Cell Culture and Transfection Experiments
The human cervical carcinoma Hela cell line used in this study was purchased from the American Type Culture Collection. The cell line was maintained in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum. For transfection, cells were seeded into 10 cm tissue culture plates and grown overnight to 80% confluency. Typically, 8 μg of human p300 and PCAF expression plasmids and 16 μl Lipofectamine 2000 (Invitrogen, Carlsbad, CA) were used for each transfection. Cells were harvested 48 h after transfection.
Co-immunoprecipitation and Western blot
Cell extracts were prepared by lysing cells in Nonidet P-40 buffer (50 mm Tris-Cl, ph 7.5/300 mm NaCl/1% Nonidet P-40/0.1% SDS/2 mm EDTA/1 mm PMSF/2.5 μg/ml aprotinin/2.5 μg/ml leupeptin/10 mm sodium butyrate). MBD1, MTA1, DNA polymerase β, and DDB1 were immunopurified from clarified supernatant with the relevant antibodies. Immunocomplexes were pulled down by Protein-A beads and washed three times in Nonidet P-40 buffer, diluted in 2× SDS loading buffer, and resolved by SDS-PAGE. Pan lysine acetylation antibody was used to detect acetylation of immunoprecipitated proteins.
RESULTS
We began the project by manually collecting and reading the related literature. We then extracted acetylated proteins with identified sites and KATs, and summarized them in Table I. In total, we found 280 unique peptides acetylated by the CBP/p300 family; 84 by the GCN5/PCAF family; and 20 by the MYST family. Large scale acetylation analysis data with mass spectrometric data but with no KAT information were also collected. All the validated acetylation sites (with or without KAT information) can be downloaded from (http://cmbi.bjmu.edu.cn/huac). Using this data set, we first analyzed the function, secondary structure and amino acid frequency of substrates (or acetylated sites) from each of the three KAT families, then predicted and validated substrates of two KAT families (prediction of the MYST family was omitted because of the small number of known acetylated unique peptides).
Table I. Collected acetylated proteins or sites.
| Proteins | Sites | |
|---|---|---|
| All validated | 2114 | 4483 |
| CBP/p300 family | 83 | 280 |
| GCN5/PCAF family | 33 | 84 |
| MYST family | 10 | 20 |
Functional and Secondary Structure Analysis of Substrates From the Three KAT Families
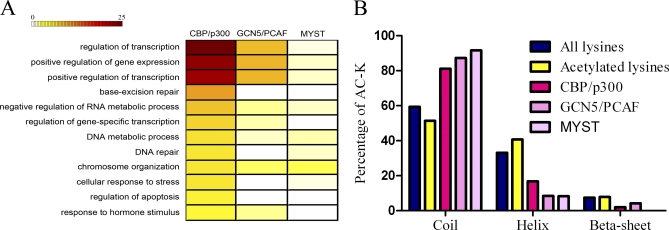
Because the three KAT families have differential substrate specificity, we first intended to find out whether substrates of each of the three families participate in different biological processes. Analysis of biological process enrichment was performed by the online DAVID program (Fig. 1A) (31, 32). Transcription related processes were enriched in all three families, which is consistent with the previous reports that KATs are transcription cofactors (7, 33–35). Substrates of the CBP/p300 family also participate in other important processes such as “stimulus response, apoptosis, chromosome organization, and DNA repair” (yellow rectangle in Fig. 1A). This indicates that CBP/p300 regulates universal biological processes, including transcription (8, 36–38). There are fewer substrates of the GCN5/PCAF family than that of the CBP/p300 family. No substrates of this family have been found enriched in the DNA repair process. Similar to the CBP/p300 family, substrates of the GCN5/PCAF family also play roles in stimulus response and chromosome organization. The most significant character of the MYST family substrates is their high enrichment in the chromosome organization process. Altogether, the three KAT families catalyze substrates which have overlapping but different functions.
Fig. 1.
A, Functional analysis of substrates from the three KAT families. Color represents the significance of functional term enrichment. A deep color means significant enrichment. Numbers on the color bar are equal to -log10P. p value is adjusted by the Benjamini method. B, Secondary structure analysis of substrates sites. All lysines: all the lysine sites in the human proteome. Acetylated lysines: all validated acetylated lysines including those without KAT information. CBP/p300: CBP/p300 family acetylated lysines. GCN5/PCAF: GCN5/PCAF family acetylated lysines. MYST: MYST family acetylated lysines.
Different enzyme structures are always associated with distinct substrate selection, which is an indication of enzyme functional preference. Our analysis of functional enrichment of substrates demonstrates that the three KAT families own different but overlapping functional spectra. For example, the CBP/p300 family is functionally enriched in apoptosis, while the other two are not. The GCN5/PCAF family substrates are not enriched in the DNA repair process, while the others are. We do not exclude the possibility that this functional spectrum will change as increasing number of new substrates are discovered. However, the functional differences among the enzymes should exist, even after all the substrates of the KAT families have been identified.
Secondary structure can influence post-translational modifications. Thus, we analyzed the secondary structure of acetylated lysine sites (validated by mass spectrometric data or traditional experiments), and the three KAT families acetylated lysine sites, by SABLE (39). As shown in Fig. 1B, compared with all the lysines in the human proteome, acetylated lysines were enriched in helix and reduced in coil structures (10), while no difference was found in β-sheet structures. Surprisingly, catalysis of lysine by the three KAT families increased in coil structures and decreased in helixes and β sheets. This difference may be biologically meaningful. Because genome-wide acetylation is preferential on structured sequence, it is significant that the three KAT family mediated acetylation prefer unstructured sequence. This indicates again the existence of undiscovered KATs or undiscovered substrates of known KATs.
What we found in the secondary structure analysis is interesting. For all acetylated lysine sites, structured helix is preferred. For the three KAT family acetylated lysine sites, unstructured coil is enriched. We assumed that the three KAT families tend to acetylate unstructured peptides. Weather there are other KAT families that preferentially acetylate structured peptides remains to be discovered.
Frequency Analysis of Acetylation Motifs
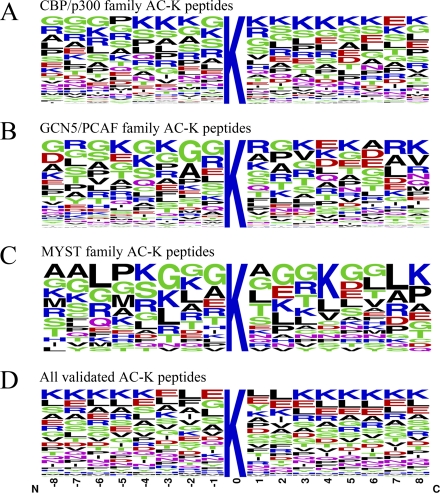
We further characterized the acetylation motifs of different families. We extracted 17 amino acid (AA) long peptide sequences with the acetylated lysines surrounded by eight residues on both sides. The length selected was based on the structural studies of published KAT domains coupled with peptide substrates (16–18). We found that Glycine (G) at –1 is the dominant residue in all three families (Fig. 2A–2C), which is consistent with previous reports that “GKXP” might be the KAT recognition motif (18, 40). The CBP/p300 family prefers lysine (K) from the –4 to +8 positions (Fig. 2A). However, this preference decreases in the GCN5/PCAF and MYST families. Compared with the CBP/p300 family, K seldom appears at –7, –6, –2, –1, +6, or +7 position in the GCN5/PCAF family and –6, –1, +1, +5, +6, or +7 position in the MYST family (Figs. 2B, 2C). The –1 and +1position is very important for KAT recognition and catalysis. The difference in K alignment in this position between the CBP/p300 and GCN5/PCAF MYST families might reflect that the CBP/p300 family prefers to acetylate two neighboring lysines, where the GCN5/PCAF and MYST families tends to accept a single lysine.
Fig. 2.
Sequence frequency analysis of the three KAT families acetylated peptides. A, B, and C, Frequency analysis of acetylated peptides of the three KAT families as indicated in the figure. D, All validated acetylated lysine (AC-K) peptides including those without KAT information. Sequence logo plots represent amino acid frequencies for 8 amino acids from both sides of the lysine acetylation site. N: amino side. C: carboxyl side. AC-K: acetylated-lysine. Number of peptides in each family: CBP/p300 280, GCN5/PCAF 84, MYST 20, All validated 4483.
We also collected all the validated acetylated lysine sites (including those without enzyme information) and analyzed the acetylated peptide frequency (Fig. 2D). A different pattern of acetylation motifs was discovered: lysine was the preferred amino acid beyond the −3 to +2 position which is different from the CBP/p300 family's preference of lysine at these positions. Also, lysine frequency decreases gradually from the –3 to –1 position and increases from the 1 to 3 position (Fig. 2D), which is consistent with a previous analysis (10). This indicates that acetylation may take place at lysine without other lysine residues in the surrounding sequence. The CBP/p300 family may be distinct from other KAT families according to this pattern. Altogether, we identified a KAT family specific pattern of acetylation motifs. All the function, secondary structure and amino acid frequency analysis showed distinct patterns between all acetylated sites and the three KAT families. We conclude that the sequence features (both the amino acid frequency and secondary structure) of acetylated lysine from different KAT families vary.
Prediction of KAT-specific Acetylation Sites with the ASEB Method
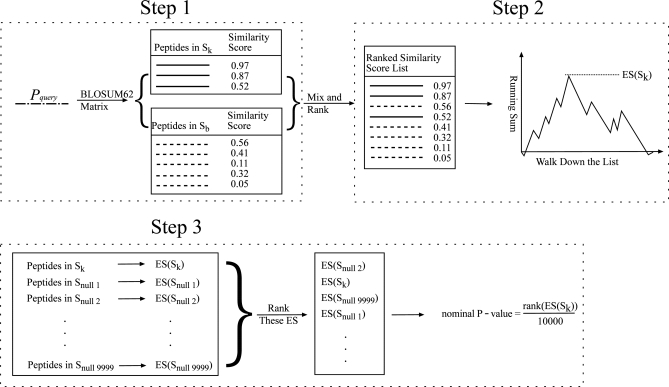
We next sought to predict novel acetylation sites in a KAT-specific way. Here we proposed an ASEB method. In this method we first define all the validated acetylated peptide sequences from one KAT family as a KAT-specific set, and define random peptides from the whole proteome as a background set. When given a new peptide sequence, scores are calculated according to the BLOSUM62 matrix between this new peptide and peptides in the KAT-specific set and background set (Fig. 3 step 1). A list is then created by ranking the scores (Fig. 3 step 2). If most of the KAT-specific set peptides are on top of the list, which means that the new given peptide is more similar to the KAT-specific set rather than the background set, the probability of this new peptide being acetylated by this KAT family is large. By defining a nominal p value, we could estimate the significance of the probability (Fig. 3 step 3). Using a threshold p value, we could predict substrate acetylation of specific KAT families.
Fig. 3.
Detailed processes of the ASEB method. Step1: Calculating similarity score. Step2: Calculating enrichment score (ES). Step3: Estimating significance of ES by calculating a nominal p value.
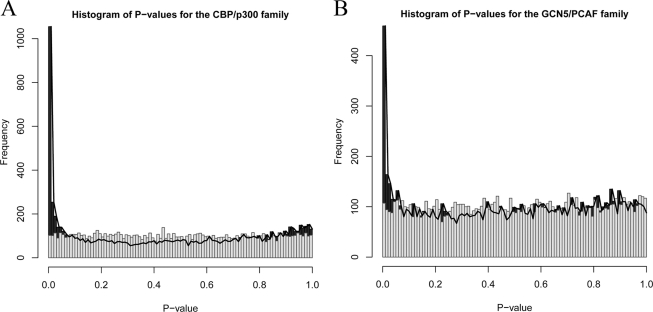
To test the power of the new method for increasing signal relative to noise, the background distribution of p values was estimated. We randomly generated 10 peptide sets for each family. Each set had 267 and 82 peptides for CBP/p300 and GCN5/PCAF respectively, and were used as predefined peptide sets to test 1000 randomly selected peptides. 10,000 p values were then determined to estimate the background distribution. A histogram for these p values was shown as gray bars in Fig. 4A and Fig. 4B for CBP/p300 and GCN5/PCAF. For comparison, we also randomly selected 10,000 peptides and tested with the original known predefined peptides sets for each family, and their p values represented the distribution of p values for our real tests. A histogram for these p values was shown as black bars in Fig. 4A and Fig. 4B for CBP/p300 and GCN5/PCAF. From these two results, we observed that the ASEB method can increase signal relative to noise and detect a lot of sites with relatively low p values.
Fig. 4.
Histogram of p values in the CBP/p300 and GCN5/PCAF families. A, Histogram of p values for the CBP/p300 family. The black bars and black lines are for p values for 10,000 sites that are randomly extracted from all sites with the pre-defined 267 peptides for CBP/p300. The gray bars are for p values of 10,000 sites that are randomly extracted from all sites with random predefined peptides (267 in size). B, Histogram of p values for the GCN5/PCAF family.
Finally a leave-one-out method was used to test the significance for known acetylated peptides from each KAT family. Each time, a known peptide was picked and the others were treated as the predefined peptide set. The p values for all known peptides were then calculated by the ASEB method. For comparison, 1000 peptides were randomly selected and tested using all known peptides as the predefined set. The results are represented in Table II. From the table we can see that, for each family, the percentage of known peptides that were picked by the method was much higher than the ones picked from the background.
Table II. Evaluation of the ASEB method. While testing a specific known peptide, the other peptides were used as the predefined peptide set. While testing the background peptides that were randomly selected, the known peptides for each family were used as the predefined peptide set.
| Total peptides | CBP/p300 |
GCN5/PCAF |
||
|---|---|---|---|---|
| Known | Background | Known | Background | |
| 267 | 1000 | 82 | 1000 | |
| Significant peptides (p value ≤1e-4) | 36 | 32 | 10 | 11 |
| Significant peptides (p value ≤1e-3) | 64 | 65 | 16 | 22 |
| Significant peptides (p value ≤1e-2) | 101 | 111 | 29 | 47 |
| Significant peptides (p value ≤1e-1) | 149 | 243 | 43 | 144 |
Experimental Validation
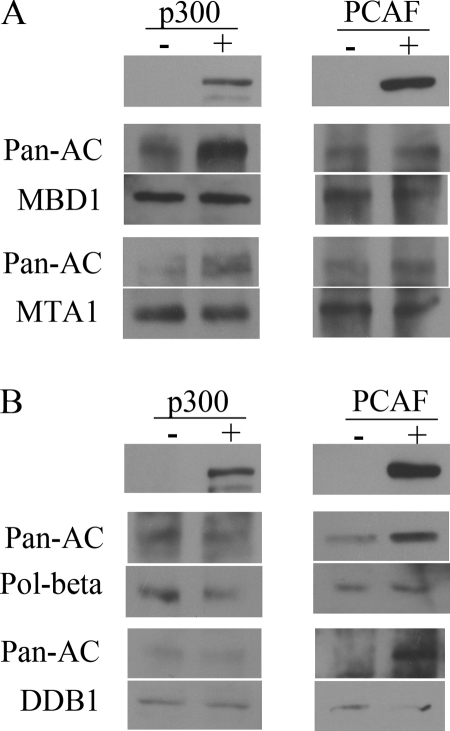
We scanned human proteins for putative KAT-specific acetylation sites by the ASEB method. The acetylation status of proteins MBD1, MTA1, DNA polymerase β and DDB1 which were predicted to be more acetylated by the CBP/p300 or GCN5/PCAF KAT families were monitored after overexpression of p300 or PCAF. When p300 expression plasmids were transfected into HeLa cells and expressed at high level, the acetylation signal of MBD1 and MTA1 increased as equal amounts of the proteins were immunoprecipitated (Fig. 5A). By contrast, when PCAF expressed at high level, acetylation of MBD1 and MTA1 increased slightly, consistent with the prediction that MBD1 and MTA1 were preferred substrates of CBP/p300 family. Similarly, for DNA polymerase beta and DDB1 which were predicted to be more acetylated by GCN5/PCAF family than by CBP/p300 family, DNA polymerase beta and DDB1 acetylation increased when PCAF plasmids were overexpressed (Fig. 5B). These experimental data indicates that the ASEB method can predict KAT families which were responsible for a given protein.
Fig. 5.
Detection of predicted acetylation substrates by the CBP/p300 and GCN5/PCAF families with Western blotting. A, Acetylation of MBD1 and MTA1 significantly increased after p300 overexpression, whereas increased little after PCAF over-expression. -: pcDNA-vector transfection. +: pcDNA-p300 or pcDNA-PCAF transfection as indicated in the figure. The upper panel plots indicate expression of p300 or PCAF. Equal amounts of indicated proteins were immuniprecipitated and followed by blotting with pan-acetylation antibody after transfection with pcDNA-vector, pcDNA-p300 or pcDNA-PCAF. B, Acetylation of DNA polymerase β (pol-β) and MTA1 significantly increased after PCAF than p300 overexpression. Labels indicate the same as in (A).
The other two proteins RB and PARP1 which were predicted to be more acetylated by CBP/p300 family did not show to be acetylated by either p300 or PCAF, at least under the condition of overexpression (supplemental Fig. S2). We also tried the other two proteins LSD1 and EED for acetylation, both of them did not seem to be acetylated by the predicted KATs (data not shown). For the proteins that can't be acetylated after overexpression of p300 or PCAF, there might be several reasons. First, these proteins really were not acetylated by the indicated KAT family. Second, these proteins can be acetylated by the KAT family, but not in the cell line we used. Third, the proteins can be acetylated by the KAT family, but not under our experimental conditions.
DISCUSSION
In this paper, we first collected the experimentally identified acetylated protein and lysine sites, and integrated them into our website, which can be freely visited (http://cmbi.bjmu.edu.cn/huac). We then analyzed the sequence features surrounding the acetylated lysine of substrates from three KAT families, and developed a computer program ASEB to predict both the acetylation sites and the KAT families responsible for the acetylation. Evaluation by p value distribution, leave-one-out analysis and experimental validation of predicted proteins reveal the good performance of the method.
Because of the limited number of acetylated lysines identified in the three families (especially the MYST family), the acetylation motif patterns of all three families may change when more acetylated sites are discovered in the future. However, there are indeed distinct features of substrate peptides in the three families, such as a lysine preference near the acetylated sites in the CBP/p300 family and a lysine reduction at the –1 position in the GCN5/PCAF family. Furthermore, the frequency distribution of all validated acetylated peptides show patterns different from all three families, which might indicate that unknown KATs exist which catalyze different lysine peptides. In fact, novel KATs, such as Circadian locomoter output cycles protein kaput and MEC-17 have been found and their acetylated lysine peptides remain to be explored (41, 42). The biggest problem in acetylation prediction is the sequence heterogeneity surrounding the acetylated lysines. The reason stems from the large number of KATs which catalyze different sequence peptides. Here, we picked two main KAT families for acetylation prediction, which can efficiently reduce the sequence heterogeneity. Prediction of acetylation using KAT family specific peptides not only avoided the sequence heterogeneity, but also revealed the KAT family of the predicted acetylation sites. For validated acetylation sites, the prediction strategy directly predicts the KAT families responsible for the acetylation.
Although we predicted substrates for only two KAT families because of the limitation of known acetylation sites with KAT information, the ASEB method can be used to predict more KAT families or particular KAT substrates as soon as identified acetylation lysine sites with KAT information accumulates. The website will be updated and the ASEB method can be used to predict substrates of more KAT families or KATs.
Acknowledgments
We thank Xi Wang and Dr. Pufeng Du for useful discussions and Dr. Greg Vatcher for his kind help in English writing.
Footnotes
* This work was supported by grants from the National Basic Research Program of China (2011CBA01104) and the National Natural Science Foundation of China (60905014, 61021063 and 31030041).
 This article contains supplemental Figs. S1 and S2 and Tables S1 and S2.
This article contains supplemental Figs. S1 and S2 and Tables S1 and S2.
1 The abbreviations used are:
- KAT
- Lysine (K)-Acetyl-Transferase
- ASEB
- Acetylation Set Enrichment Based
- DDB1
- DNA damage-binding protein 1
- ES
- Enrichment score
- GCN5
- General control of amino acid synthesis protein 5-like 2
- GSEA
- Gene Set Enrichment Analysis
- MBD1
- methyl-CpG-binding domain protein 1
- MEC-17
- Alpha-tubulin N-acetyltransferase
- MTA1
- metastasis-associated protein
- MYST
- MOZ, YBF2/SAS3, SAS2, and TIP60 protein
- p300
- E1A-associated protein p300
- PCAF
- p300/CBP-associated factor
- WRN
- Werner syndrome ATP-dependent helicase
- pquery
- the query peptide
- Sb
- background peptides set
- Sk
- KAT special peptides set
- Snull
- randomly generated peptides set.
REFERENCES
- 1. Das C., Kundu T. K. (2005) Transcriptional regulation by the acetylation of nonhistone proteins in humans – a new target for therapeutics. IUBMB Life 57, 137–149 We were unable to verify your reference in PubMed, please confirm that it is correct or change as needed [DOI] [PubMed] [Google Scholar]
- 2. Glozak M. A., Sengupta N., Zhang X., Seto E. (2005) Acetylation and deacetylation of non-histone proteins. Gene 363, 15–23 [DOI] [PubMed] [Google Scholar]
- 3. Kim S. C., Sprung R., Chen Y., Xu Y., Ball H., Pei J., Cheng T., Kho Y., Xiao H., Xiao L., Grishin N. V., White M., Yang X. J., Zhao Y. (2006) Substrate and functional diversity of lysine acetylation revealed by a proteomics survey. Mol. Cell 23, 607–618 [DOI] [PubMed] [Google Scholar]
- 4. Spange S., Wagner T., Heinzel T., Krämer O. H. (2009) Acetylation of non-histone proteins modulates cellular signalling at multiple levels. Int. J. Biochem. Cell. Biol. 41, 185–198 [DOI] [PubMed] [Google Scholar]
- 5. Zhao S., Xu W., Jiang W., Yu W., Lin Y., Zhang T., Yao J., Zhou L., Zeng Y., Li H., Li Y., Shi J., An W., Hancock S. M., He F., Qin L., Chin J., Yang P., Chen X., Lei Q., Xiong Y., Guan K. L. (2010) Regulation of cellular metabolism by protein lysine acetylation. Science 327, 1000–1004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Allfrey V. G., Faulkner R., Mirsky A. E. (1964) Acetylation and Methylation of Histones and Their Possible Role in the Regulation of Rna Synthesis. Proc. Natl. Acad. Sci. U. S. A. 51, 786–794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. MacDonald V. E., Howe L. J. (2009) Histone acetylation: where to go and how to get there. Epigenetics 4, 139–143 [DOI] [PubMed] [Google Scholar]
- 8. Liu L., Scolnick D. M., Trievel R. C., Zhang H. B., Marmorstein R., Halazonetis T. D., Berger S. L. (1999) p53 sites acetylated in vitro by PCAF and p300 are acetylated in vivo in response to DNA damage. Mol. Cell. Biol. 19, 1202–1209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Wang C., Fu M., Angeletti R. H., Siconolfi-Baez L., Reutens A. T., Albanese C., Lisanti M. P., Katzenellenbogen B. S., Kato S., Hopp T., Fuqua S. A., Lopez G. N., Kushner P. J., Pestell R. G. (2001) Direct acetylation of the estrogen receptor alpha hinge region by p300 regulates transactivation and hormone sensitivity. J. Biol. Chem. 276, 18375–18383 [DOI] [PubMed] [Google Scholar]
- 10. Choudhary C., Kumar C., Gnad F., Nielsen M. L., Rehman M., Walther T. C., Olsen J. V., Mann M. (2009) Lysine acetylation targets protein complexes and co-regulates major cellular functions. Science 325, 834–840 [DOI] [PubMed] [Google Scholar]
- 11. Gnad F., Ren S., Choudhary C., Cox J., Mann M. (2010) Predicting post-translational lysine acetylation using support vector machines. Bioinformatics 26, 1666–1668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Xu Y., Wang X. B., Ding J., Wu L. Y., Deng N. Y. (2010) Lysine acetylation sites prediction using an ensemble of support vector machine classifiers. J. Theor. Biol. 264, 130–135 [DOI] [PubMed] [Google Scholar]
- 13. Li X., Wu L., Corsa C. A., Kunkel S., Dou Y. (2009) Two mammalian MOF complexes regulate transcription activation by distinct mechanisms. Mol. Cell 36, 290–301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Schwartz D., Chou M. F., Church G. M. (2009) Predicting protein post-translational modifications using meta-analysis of proteome scale data sets. Mol. Cell. Proteomics 8, 365–379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Basu A., Rose K. L., Zhang J., Beavis R. C., Ueberheide B., Garcia B. A., Chait B., Zhao Y., Hunt D. F., Segal E., Allis C. D., Hake S. B. (2009) Proteome-wide prediction of acetylation substrates. Proc. Natl. Acad. Sci. U. S. A. 106, 13785–13790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Marmorstein R. (2001) Structure of histone acetyltransferases. J. Mol. Biol. 311, 433–444 [DOI] [PubMed] [Google Scholar]
- 17. Marmorstein R. (2001) Structure and function of histone acetyltransferases. Cell. Mol. Life Sci. 58, 693–703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Marmorstein R., Roth S. Y. (2001) Histone acetyltransferases: function, structure, and catalysis. Curr. Opin. Genet. Dev. 11, 155–161 [DOI] [PubMed] [Google Scholar]
- 19. Li K., Wang R., Lozada E., Fan W., Orren D. K., Luo J. (2010) Acetylation of WRN protein regulates its stability by inhibiting ubiquitination. PLoS One 5, e10341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Haenni S. S., Hassa P. O., Altmeyer M., Fey M., Imhof R., Hottiger M. O. (2008) Identification of lysines 36 and 37 of PARP-2 as targets for acetylation and auto-ADP-ribosylation. Int. J. Biochem. Cell. Biol. 40, 2274–2283 [DOI] [PubMed] [Google Scholar]
- 21. Sykes S. M., Mellert H. S., Holbert M. A., Li K., Marmorstein R., Lane W. S., McMahon S. B. (2006) Acetylation of the p53 DNA-binding domain regulates apoptosis induction. Mol. Cell 24, 841–851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Munshi N., Agalioti T., Lomvardas S., Merika M., Chen G., Thanos D. (2001) Coordination of a transcriptional switch by HMGI(Y) acetylation. Science 293, 1133–1136 [DOI] [PubMed] [Google Scholar]
- 23. Yang Y., Zhao Y., Liao W., Yang J., Wu L., Zheng Z., Yu Y., Zhou W., Li L., Feng J., Wang H., Zhu W. G. (2009) Acetylation of FoxO1 activates Bim expression to induce apoptosis in response to histone deacetylase inhibitor depsipeptide treatment. Neoplasia 11, 313–324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Zhao Y., Lu S., Wu L., Chai G., Wang H., Chen Y., Sun J., Yu Y., Zhou W., Zheng Q., Wu M., Otterson G. A., Zhu W. G. (2006) Acetylation of p53 at lysine 373/382 by the histone deacetylase inhibitor depsipeptide induces expression of p21(Waf1/Cip1). Mol. Cell. Biol. 26, 2782–2790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Zhao Y., Yang J., Liao W., Liu X., Zhang H., Wang S., Wang D., Feng J., Yu L., Zhu W. G. (2010) Cytosolic FoxO1 is essential for the induction of autophagy and tumour suppressor activity. Nat. Cell. Biol. 12, 665–675 [DOI] [PubMed] [Google Scholar]
- 26. Li T., Li F., Zhang X. (2008) Prediction of kinase-specific phosphorylation sites with sequence features by a log-odds ratio approach. Proteins, 70, 404–414 We were unable to verify your reference in PubMed, please confirm that it is correct or change as needed [DOI] [PubMed] [Google Scholar]
- 27. Li T., Du P., Xu N. (2010) Identifying human kinase-specific protein phosphorylation sites by integrating heterogeneous information from various sources. PLoS One 5, e15411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Mootha V. K., Lindgren C. M., Eriksson K. F., Subramanian A., Sihag S., Lehar J., Puigserver P., Carlsson E., Ridderstrale M., Laurila E., Houstis N., Daly M. J., Patterson N., Mesirov J. P., Golub T. R., Tamayo P., Spiegelman B., Lander E. S., Hirschhorn J. N., Altshuler D., Groop L. C. (2003) PGC-1alpha-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat. Genetics 34, 267–273 We were unable to verify your reference in PubMed, please confirm that it is correct or change as needed [DOI] [PubMed] [Google Scholar]
- 29. Subramanian A., Tamayo P., Mootha V. K., Mukherjee S., Ebert B. L., Gillette M. A., Paulovich A., Pomeroy S. L., Golub T. R., Lander E. S., Mesirov J. P. (2005) Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. U. S. A. 102, 15545–15550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Guttman M., Amit I., Garber M., French C., Lin M. F., Feldser D., Huarte M., Zuk O., Carey B. W., Cassady J. P., Cabili M. N., Jaenisch R., Mikkelsen T. S., Jacks T., Hacohen N., Bernstein B. E., Kellis M., Regev A., Rinn J. L., Lander E. S. (2009) Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature 458, 223–227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Huang da W., Sherman B. T., Lempicki R. A. (2009) Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 4, 44–57 [DOI] [PubMed] [Google Scholar]
- 32. Huang da W., Sherman B. T., Lempicki R. A. (2009) Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 37, 1–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Brown C. E., Lechner T., Howe L., Workman J. L. (2000) The many HATs of transcription coactivators. Trends Biochem. Sci. 25, 15–19 [DOI] [PubMed] [Google Scholar]
- 34. Chen H., Tini M., Evans R. M. (2001) HATs on and beyond chromatin. Curr. Opin. Cell Biol. 13, 218–224 [DOI] [PubMed] [Google Scholar]
- 35. Thomas M. C., Chiang C. M. (2006) The general transcription machinery and general cofactors. Crit. Rev. Biochem. Mol. Biol. 41, 105–178 [DOI] [PubMed] [Google Scholar]
- 36. Kishimoto M., Kohno T., Okudela K., Otsuka A., Sasaki H., Tanabe C., Sakiyama T., Hirama C., Kitabayashi I., Minna J. D., Takenoshita S., Yokota J. (2005) Mutations and deletions of the CBP gene in human lung cancer. Clin. Cancer Res. 11, 512–519 [PubMed] [Google Scholar]
- 37. Szerlong H. J., Prenni J. E., Nyborg J. K., Hansen J. C. (2010) Activator-dependent p300 acetylation of chromatin in vitro: enhancement of transcription by disruption of repressive nucleosome-nucleosome interactions. J. Biol. Chem. 285, 31954–31964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Ionov Y., Matsui S., Cowell J. K. (2004) A role for p300/CREB binding protein genes in promoting cancer progression in colon cancer cell lines with microsatellite instability. Proc. Natl. Acad. Sci. U. S. A. 101, 1273–1278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Wagner M., Adamczak R., Porollo A., Meller J. (2005) Linear regression models for solvent accessibility prediction in proteins. J. Comput. Biol. 12, 355–369 [DOI] [PubMed] [Google Scholar]
- 40. Polevoda B., Sherman F. (2002) The diversity of acetylated proteins. Genome Biol. 3, reviews0006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Akella J. S., Wloga D., Kim J., Starostina N. G., Lyons-Abbott S., Morrissette N. S., Dougan S. T., Kipreos E. T., Gaertig J. (2010) MEC-17 is an alpha-tubulin acetyltransferase. Nature 467, 218–222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Doi M., Hirayama J., Sassone-Corsi P. (2006) Circadian regulator CLOCK is a histone acetyltransferase. Cell 125, 497–508 [DOI] [PubMed] [Google Scholar]