Abstract
Mass spectrometric sequencing of low abundance, integral membrane proteins, particularly the transmembrane domains, presents challenges that span the multiple phases of sample preparation including solubilization, purification, enzymatic digestion, peptide extraction, and chromatographic separation. We describe a method through which we have obtained high peptide coverage for 12 γ-aminobutyric acid type A receptor (GABAA receptor) subunits from 2 picomoles of affinity-purified GABAA receptors from rat brain neocortex. Focusing on the α1 subunit, we identified peptides covering 96% of the protein sequence from fragmentation spectra (MS2) using a database searching algorithm and deduced 80% of the amino acid residues in the protein from de novo sequencing of Orbitrap spectra. The workflow combined microscale membrane protein solubilization, protein delipidation, in-solution multi-enzyme digestion, multiple stationary phases for peptide extraction, and acquisition of high-resolution full scan and fragmentation spectra. For de novo sequencing of peptides containing the transmembrane domains, timed digestions with chymotrypsin were utilized to generate peptides with overlapping sequences that were then recovered by sequential solid phase extraction using a C4 followed by a porous graphitic carbon stationary phase. The specificity of peptide identifications and amino acid residue sequences was increased by high mass accuracy and charge state assignment to parent and fragment ions. Analysis of three separate brain samples demonstrated that 78% of the sequence of the α1 subunit was observed in all three replicates with an additional 13% covered in two of the three replicates, indicating a high degree of sequence coverage reproducibility. Label-free quantitative analysis was applied to the three replicates to determine the relative abundances of 11 γ-aminobutyric acid type A receptor subunits. The deep sequence MS data also revealed two N-glycosylation sites on the α1 subunit, confirmed two splice variants of the γ2 subunit (γ2L and γ2S) and resolved a database discrepancy in the sequence of the α5 subunit.
Mass spectrometric sequencing of proteins from natural sources is a valuable technique for identifying post-translational modifications (1), drug binding sites using affinity labeling techniques (2), and variations in protein sequence resulting from genomic alterations, and transcript editing and splicing. In order to fully utilize this technique, it must be: (1) applicable to the quantities of protein obtainable from small samples of native tissues, because these are the sites where sequence variation and post-translational modifications occur and; (2) capable of obtaining sequence at the amino acid residue level (rather than deducing sequence from spectral matching of database entries) to enable the identification and characterization of amino acid modifications (natural and artificial) and sequence variants. The facile application of residue level mass spectrometric sequencing to integral membrane proteins (IMPs)1 would be particularly valuable because these proteins, which play a key regulatory role in regulating communication between cells and the extracellular environment, are estimated to constitute 15–39% of the human proteome (3). To date, detailed mass spectrometric analyses of native IMPs have been limited by their low natural abundance as well as by a variety of technical issues including difficulties in solubilization, enzymatic digestion and recovery and separation of hydrophobic peptides from the transmembrane spanning domains (TMDs) (4).
Significant advances in mass spectrometry of IMPs have come from proteomic studies aimed at quantitative cataloging of the proteome of various organisms and tissues (5). These advances include improvements in membrane isolation (4), the introduction of membrane “shaving” (6), the use of multiple enzymes in digestion (7), the use of mass spectrometry (MS)-compatible surfactants in protease digestion (8), delipidation of membrane proteins (9), in-solution rather than in-gel digestion (10), and the performance of liquid chromatography at elevated temperature to facilitate resolution of hydrophobic peptides (11). These techniques have enhanced detection of TMDs from IMPs and thus increased protein identification. To date, studies have not addressed complete de novo sequencing of IMPs to define sequence variations and localize protein modifications in TMDs and other domains.
Efforts have also been directed at detailed sequencing of IMPs purified from recombinant systems. For example, Lubec and colleagues examined affinity-tagged γ-aminobutyric acid type A receptor (GABAA receptors) β3 homomers or α1β3 heteropentamers overexpressed in Sf-9 insect cells (7, 12). They isolated GABAA receptor subunits using affinity-chromatography followed by sequential electrophoretic steps beginning with blue native gel electrophoresis (rather than conventional two-dimensional gel electrophoresis) to avoid IMP precipitation during isoelectric focusing. Using multiple endoproteases (trypsin, chymotrypsin, subtilisin) and in-gel digestion, they identified peptides using a database search algorithm that covered the full sequence of the protein using liquid chromatography (LC)-electrospray ionization (ESI)-mass spectrometry (MS); however these analyses required >500 pmols of receptor. An alternative strategy to sequencing purified IMPs is the “top-down” approach in which the undigested proteins are analyzed by mass spectrometry and fragmented peptides are generated and analyzed in a collision cell. Top-down mass spectrometry has been applied to obtaining full sequence coverage of several membrane proteins and identifying post-translational modifications (13). However, this method currently requires nmol quantities of purified protein and is currently limited to proteins ∼ <50 kDa, rendering its application to many native IMPs impractical.
In this study we describe a method to obtain deep residue-level sequence from low pmol quantities of native brain GABAA receptors, a low abundance IMP that serves as the major inhibitory neurotransmitter receptor in brain. In addition to serving as a key regulator of synaptic excitability, these receptors are targets for numerous classes of drugs including benzodiazepines, barbiturates, propofol, etomidate, and the neurosteroids (14). Native GABAA receptors are pentamers composed of various combinations of nonidentical subunits, with each subunit consisting of a long N-terminal extracellular hydrophilic region, four α-helical TMDs, and a short extracellular C terminus. To date, 19 GABAA receptor subunits have been identified including α1–6, β1–3, γ1–3, δ, ε, θ, π, and ρ1–3 subunits; pentamers composed of two α1, two β2 and one γ2 subunit are the most abundant receptors in brain neocortex (14).
In this work, we affinity-purified GABAA receptors from rat neocortex to obtain ∼2 pmols of a mixture of native GABAA receptors with multiple subunit combinations. We employed timed, in-solution digestion with Lys-C/trypsin, Lys-C/chymotrypsin, or chymotrypsin alone to generate peptides of varying lengths, which were recovered using sequential solid phase extractions with two different stationary phases. High resolution survey (MS1) and fragmentation (MS2) spectra were acquired using both data-dependent and directed (e.g. accurate inclusion mass screening) mass spectrometry. Using this protocol, we obtained high peptide coverage of 12 GABAA receptor subunits. Detailed analyses of the α1 subunit identified peptides covering 96% of the sequence of the entire protein including 93% its four TMDs, based on MS2 spectra. By manual interpretation, we identified fragmentation ions in the MS2 spectra that covered 80% of the amino acid residues of the total protein sequence and 63% of the residues in the four TMDs. Analysis of replicate samples demonstrated that this method produced highly reproducible sequence coverage of the α1 subunit. Label-free quantitative analysis was applied to the replicate samples to determine the relative abundances of 11 GABAA receptor subunits. The data obtained also enabled identification of N-glycosylation sites in the α1 subunit, a splice variant in the γ2 subunit and provided de novo data to resolve different protein sequences of the α5 subunit in two protein databases.
EXPERIMENTAL PROCEDURES
Membrane Preparation
In each experiment, one adult rat was anesthetized with isoflurane and killed by decapitation. The brain was rapidly removed and the neocortex was manually separated from the cerebellum and brainstem. The isolated neocortex was placed in ice-cold 0.32 m sucrose (10 ml/gm) and homogenized using a motor-driven Teflon pestle homogenizer as previously described (15). The homogenate was centrifuged at 1000 × g for 10 min at 4 °C to obtain the pellet (P1) and supernatant (S1). The S1 supernatant was centrifuged at 100,000 × g for 45 min and the resultant pellet (P2) was resuspended in distilled water and stirred for 30 min at 4 °C. After centrifugation at 100,000 × g for 45 min at 4 °C, the membrane pellet was collected and resuspended in buffer containing 10 mm K-phosphate, pH 7.5 and 100 mm KCl. Aliquots of the membrane preparation were used for protein analysis using a bicinchoninic acid protein assay kit (Pierce, Rockford, IL) or for [3H]Ro15,4513 binding to quantify GABAA receptor number. The remainder of the membrane preparation was immediately used for extraction of membrane proteins.
[3H]Ro15,4513 Binding
[3H]Ro15,4513 binding assays were performed using previously described methods (16) with modification as described below. Briefly, aliquots of membranes (1 mg/ml) were incubated with 30 nm [3H] Ro15,4513 (25 Ci/mmol, Perkin Elmer Life Science) in a total volume of 1 ml of assay buffer (50 mm Tris-citrate, pH7.1, 150 mm NaCl). Nonspecific binding was defined as the [3H]Ro15,4513 binding observed in the presence of 100 μm flumazenil (Sigma Chemical, St. Louis, MO). The assay tubes were incubated on ice for 90 min. A Brandel (Gaithersburg, MD) cell harvester was used to collect the membranes on Whatman glass fiber (GF/B) filter paper. The filter paper was rinsed three times with 4 ml of ice-cold buffer and placed in 4 ml Biosafety II scintillation mixture (Fisher Scientific, Pittsburgh, PA). Radioactivity bound to the filters was measured by liquid scintillation spectrometry. For detergent-solubilized or purified GABAA receptors, 50 mm γ-globulin and 4% polyethylene glycol were added to the binding assays before filtration. The filter papers were washed with assay buffer containing 4% polyethylene glycol.
Solubilization and Purification of GABAA Receptors
Freshly prepared membranes from a single rat brain neocortex were solubilized at 4 °C for 2 h in 10 ml extraction buffer containing 50 mm Tris, pH 7.4, 150 mm NaCl, 1% Triton X-100, 1 mm ethylenediaminetetraacetic acid (EDTA), and a protease inhibitor mixture containing 4-(2-aminoethyl)benzenesulfonyl fluoride, pepstatinA, E-64, bestatin, leupeptin, and aprotinin (Sigma Chemical, St. Louis, MO). Insoluble membrane material was removed by centrifugation at 15,000 g for 45 min. An aliquot of the supernatant, containing solubilized membrane proteins, was saved for total protein assays and [3H]Ro15,4513 binding. The remainder of the supernatant was applied to a trivalent immunoaffinity column containing antibodies directed against the γ2, β2, and β3 subunits of the GABAA receptor (16–18) at a rate of 15 ml/h. This combination of antibodies was used because all pentameric GABAA receptors in neocortex contain at least one of these subunits and thus should be enriched on this column. The column was sequentially washed with 20 ml each of extraction buffer, buffer I (0.6 m NaCl, 50 mm Tris pH 8.3, 1 mm EDTA, and 0.5% Triton X-100), and buffer II (150 mm NaCl, 50 mm Tris pH 8.0, 1 mm EDTA, 0.2% Triton X-100). The receptor complex was eluted with 10 ml 0.1 m glycine pH 2.4, 150 mm NaCl, 0.1% Triton X-100 and collected as 1 ml/fraction into tubes with 0.1 ml 3 m Tris pH 7.4 (16). All the procedures were carried out at 4 °C. The eluted fractions were analyzed by SDS-PAGE with either SYPRO-Ruby™ protein staining or immunoblotting with an antibody to the α1 subunit of the GABAA receptor, to determine the fractions enriched in GABAA receptors. For immunoblotting, protein was transferred from the gel to a polyvinylidene fluoride membrane and probed with a primary anti-α1 antibody (goat IgG, 1:400 dilution, Santa Cruz). The α1 subunits were visualized using a horseradish peroxidase-conjugated donkey anti-goat IgG antibody and the ECL Plus detection method (GE Healthcare).
In-Solution Protein Digestion and Peptide Preparation
The fractions containing GABAA receptors were pooled and concentrated to a volume of 200 μl using a 30-kDa nominal molecular weight limit Centricon concentrator (Millipore). In selected experiments, an aliquot of the concentrated material was saved for [3H]Ro15,4513 assay to determine receptor recovery. The protein delipidation and peptide preparations were conducted using modifications of previously described methods (9). Briefly, the proteins were first precipitated using an SDS-PAGE Clean-up Kit (GE Healthcare Life Sciences) according to the manufacturer's protocol. The precipitated proteins were solubilized in 20 μl of 8 m urea, 5% RapiGest™ (Waters, Milford MA), 100 mm Tris, pH 8.5 for 30 min at 37 °C with agitation. The proteins were then precipitated again with the SDS-cleanup kit and resuspended in 20 μl of 8 m Urea, 0.1% RapiGest™, 100 mm Tris pH 8.5. Proteins were first reduced by adding tris (2-carboxyethyl) phosphine to 5 mm and incubating at room temperature for 30 min and then alkylated with 10 mm iodoacetamide in the dark at room temperature for 30 min. The tris (2-carboxyethyl) phosphine and iodoacetamide were quenched with 5 mm dithiothreitol at room temperature for 10 min. Twenty percent of the reduced and alkylated protein was then removed and diluted with 100 mm Tris pH 8.5 to a concentration of 1 m urea, and CaCl2 was added to a final concentration of 10 mm. The proteins were then digested with 4 μg of chymotrypsin (Roche, sequencing grade) at room temperature overnight. The remaining 80% of the sample was digested with 1 μg of Lys-C (Roche, sequencing grade) at 37 °C overnight. Lys-C digestion was the initial step in the digestion procedures, because Lys-C is active in 8 m urea. Lys-C digests the proteins into large fragments that can remain soluble in 1 or 2 m urea, concentrations required for chymotrypsin and trypsin activity, respectively. After Lys-C incubation, the sample was divided into four aliquots. One aliquot was diluted to reduce the concentration of urea to 2 m with 100 mm Tris pH 8.5 and digested with 4 μg of sequencing grade trypsin (Sigma Chemical, St. Louis MO) at 37 °C overnight. The other three aliquots were diluted with 100 mm Tris pH 8.5 to a concentration of 1 m urea and CaCl2 was added to a final concentration of 10 mm. These samples were digested with 4 μg of chymotrypsin (Roche, sequencing grade) at room temperature for 4 h, 6 h or overnight. At the conclusion of the incubations, all samples were acidified with 1% formic acid (FA) for 30 min at 37 °C.
For deglycosylation, immunopurified GABAA receptors were precipitated using the SDS-cleanup kit. The precipitated proteins were dissolved in 20 μl deglycosylation reaction buffer (0.5% SDS, 0.04 m DTT, 0.05 m Na-phosphate, pH 7.5, 1% Nonidet P-40) and incubated with PNGase F (New England Biolabs, Ipswich, MA) at 37 °C for 1 h. The samples were then digested according to the protocol described above.
Solid Phase Extraction (SPE)
The protein digests were sequentially extracted with NuTip C4 and porous graphitic carbon (PGC) wedge tips (Glygen) using a BRAVO robot (Agilent, Santa Clara, CA). Both the C4 and PGC tips had bed volumes of ∼25 μl in a 200 μl pipet tip and were conditioned with 125 μl of a mixture of 60% acetonitrile (ACN) and 1% FA. Both tips were then equilibrated with 125 μl of a solution of aqueous 1% ACN and 1% FA. Peptides from each digestion condition were initially loaded on the C4 tips by 50 repetitive pipettings, and were then washed with 125 μl of aqueous 1% ACN and 1% FA. The peptides were eluted with 20 μl of a mixture of 60% ACN and 1% FA. The digestions were extracted sequentially with three C4 tips in order to maximize peptide recovery. The C4 extracted digests were then loaded onto a conditioned PGC tip and eluted following the same protocol as with the C4 tips. The SPE with PGC tips was also performed in triplicate for each digest. The triplicate elutions from each SPE step were pooled for subsequent analysis by nano-LC-MS (nanoliter-liquid chromatograph interfaced to a hybrid linear quadrupole ion trap-Orbitrap mass spectrometer).
In-Gel Protein Digestion
Bands detected by SYPRO-Ruby™ protein staining on a SDS-PAGE gel were excised using a gel excision robot (ProPic; Genomics Solutions, Ann Arbor, MI) and transferred into 96-well plates as 1.8 mm diameter cores. The excised gel plugs were then digested in situ with trypsin using a modified accelerated digestion protocol (19). Briefly, the gel plugs were sequentially washed with water and 100% ACN, each for 10 min. The ACN was changed twice to completely dehydrate the gel plugs. The gel plugs were dried in a SpeedVac, rehydrated with 1 mm triethyl ammonium bicarbonate pH 8.3 and incubated with 1 μg trypsin at 37 °C overnight. The gel plugs were further digested with trypsin after the addition of 20 μl of aqueous 1% ACN/1% FA to each gel plug followed by incubation at 37 °C 1 h. The resulting peptides were analyzed using nano-LC-MS on a hybrid quadrupole linear ion trap-Fourier transform ion cyclotron resonance mass spectrometer as previously described (20).
High Resolution MS1 and MS2 Mass Spectrometry
Peptide mixtures were analyzed using high-resolution nano-LC-MS on a hybrid mass spectrometer consisting of a linear quadrupole ion-trap and an Orbitrap (LTQ-Orbitrap XL, Thermo Fisher Scientific). Chromatographic separations were performed using a nanoLC 1D Plus™ (Eksigent) for gradient delivery and a cHiPLC-nanoflex (Eksigent) equipped with a 15 cm × 75 μm C18 column (ChromXP C18-CL, 3 μm, 120 Å, Eksigent). The liquid chromatograph was interfaced to the mass spectrometer with a nanospray source (PicoView PV550; New Objective). Mobile phases were 1% FA in water (A) and 1% FA in 60% ACN (B). After equilibrating the column in 98% solvent A (aqueous 1% FA) and 2% of solvent B (ACN containing 1% FA), the samples (5 μl) were injected from autosampler vials using the LC-system's autosampler at a flow rate of 500 nL/min followed by gradient elution (250 nL/min) with solvent B: isocratic at 2% B, 0–5 min; 2% B to 25% B, 5–110 min; 25% to 80%, 110–170 min; 80% to 2%, 170–175; and isocratic at 2% B, 175–190 min. Total run time, including column equilibration, sample loading, and analysis was 217 min. For high resolution data directed analysis, the survey scans (m/z 350–2000) (MS1) were acquired at high resolution (60,000 at m/z = 400) in the Orbitrap in profile mode and the MS/MS spectra (MS2) were acquired at 7500 resolution in the Orbitrap in profile mode after fragmentation in the linear ion trap. The maximum injection times for the MS1 scan in the Orbitrap and the LTQ were both 500 ms, and the maximum injection times for the MSn scan in the Orbitrap and the LTQ were 800 ms and 5000 ms, respectively. The automatic gain control targets for the Orbitrap and the LTQ were 5 × 105 and 3 × 104, respectively for the MS1 scans and 2 × 105 and 1 × 104, respectively for the MSn scans. The MS1 scans were followed by three MS2 events in the linear ion trap with collision activation in the ion trap (parent threshold = 10,000; isolation width = 4.0 Da; normalized collision energy = 30%; activation Q = 0.250; activation time = 30 ms). Dynamic exclusion was used to remove selected precursor ions (–0.20/+1.0 Da) for 90 s after MS2 acquisition. A repeat count of 3, a repeat duration of 45 s, and a maximum exclusion list size of 500 was used. The following ion source parameters were used: capillary temperature 200 °C, source voltage 2.5 kV, source current 100 μA, and the tube lens at 79 V. The data were acquired using Xcalibur, version 2.0.7 (Thermo Fisher). For directed MS2 spectral acquisition (DMSA) and FT-MS, see supplemental Materials and Methods.
Data Processing and Analysis of Gel Plug Samples
Data files from the LC-MS analysis of tryptic peptides generated by in situ tryptic digestion of SDS-PAGE gel plugs were processed using MASCOT Distiller (Matrix Science, version 2.3.0.0) with the settings previously described (21). The data were searched with Mascot (Matrix Science, London, UK; version 2.1.6) using both a UniprotKB/Swiss-Prot rat database (31051 sequence entries) and a customized database that contained the sequence of all GABAA receptor subunits (FASTA files for this database containing 215 proteins are included in Supplemental database). The search was conducted using trypsin as the endoprotease allowing 9 missed cleavages, specifying carbamidomethylation of Cys residues as a fixed modification and oxidation of Met residues as a variable modification and setting a fragment ion mass tolerance of 0.800 Da and a parent ion tolerance of 20 ppm. The Protein and Peptide Prophet algorithms in Scaffold (version 3_00_03, Proteome Software Inc., Portland, OR) were used to qualify the database results. A probability of ≥50% was used to assign peptide sequences to MS2 spectra using the Peptide Prophet algorithm (22). A probability of ≥95% was used to assign proteins with the Protein Prophet algorithm (23). Proteins that contained similar peptides and could not be differentiated based on MS/MS analysis alone were grouped to satisfy the principles of parsimony.
Database Searches of In-Solution Digests
The LC-MS data files for both low resolution and high resolution MS2 were processed using MASCOT Distiller (Matrix Science, version 2.3.0.0) with previously described settings for both low resolution (21) and high resolution (24) MS2. Settings for high resolution spectra acquired in the Orbitrap were as follows: (1) MS processing: 200 data points per Da; sum aggregation method; maximum charge state, 3+; minimum number of peaks, 1; (2) MS/MS processing: 200 data points per Da; time domain aggregation method; minimum number of peaks, 10; use precursor charge as maximum; precursor charge and m/z, “try to re-determine charge from the parent scan (tolerance, 1.2 Da)”; charge defaults, 1; maximum charge state, (3) time domain parameters: minimum precursor mass, 300; maximum precursor mass, 16,000; precursor m/z tolerance for grouping, 0.01; maximum number of intermediate scans, 0; minimum number of scans in a group, 1; (4) peak picking: maximum iterations, 500; correlation threshold, 0.60; minimum signal to noise, 2; minimum peak m/z, 50; maximum peak m/z, 100,000; minimum peak width, 0.002; maximum peak width, 0.2; expected peak width, 0.02. The resulting MS2 centroided files were used for database searching with MASCOT (Matrix Science, version 2.1.6) against both a Uniprot-Rat database (see previous section) and a custom, in-house database containing the sequence of all GABAA receptor subunits (see previous section). For low resolution MS2 spectra, the search was conducted using trypsin as the specified protease, allowing nine missed cleavages, specifying carbamidomethylation of Cys residues as a fixed modification and oxidation of Met residues as a variable modification and setting a fragment ion mass tolerance of 0.800 Da and a parent ion tolerance of 20 ppm. For high resolution MS2 spectra, the search was conducted with no enzyme specificity and a fragment ion mass tolerance of 100 absolute milli-mass units. All other settings were the same as those used for searching the low resolution MS2 spectra. Scaffold (version Scaffold_3_00_03, Proteome Software Inc., Portland, OR) was used to validate MS/MS based peptide identifications. Peptide identifications were accepted if they could be established at greater than 50.0% probability as specified by the Peptide Prophet algorithm (22).
Data from in-solution digests
The data flow for high-resolution MS1 peptide mapping onto the GABAA receptor subunits is shown in supplemental Fig. S1. Briefly, the unprocessed LC-MS data were imported into Rosetta Elucidator™ (Rosetta Biosoftware, version 3.3) for retention time and m/z alignment of the peptide ion chromatograms using previously described parameters (25). To determine the mass accuracy tolerance for mapping the MS1 data, we first identified a standard set of peptides with high quality MS2 spectra (ion score ≥ 30) by database searching with Mascot (Matrix Science, London, UK; version 2.1.6) as described above and in supplemental Fig. S1. The experimentally determined mass tolerance is shown in supplemental Fig. S2. The theoretical m/z of the parent ion for each of these peptides was calculated using mMass (26) and compared with the observed m/z. The mass accuracy tolerance was used to match observed and theoretical m/z values using the mass-matching utility in Rosetta Elucidator™. Results were filtered after matching to ensure that all matches were correct by both mass and charge. The in silico digestion was conducted using mMass with trypsin, Lys-C/chymotrypsin, or chymotrypsin as the proteolytic enzyme, five missed cleavage sites, carbamidomethylation of Cys residues as a fixed modification and oxidation of Met residues as a variable modification.
Based on the operating resolution (7500), a mass accuracy of ± 20 ppm was used as a guideline for interpretation of MS2 spectra. In addition, we determined the mass accuracy tolerance using fragment ions from high quality database-annotated spectra with determinable charge state. The latter mass accuracy tolerance was determined for three analytical blocks (supplemental Figs. S5B, S5D, and S5F). In brief, to determine peptide and residue sequence coverage for the α1 subunit, all α1 peptides with Peptide Prophet probability ≥50% were exported from the Scaffold file generated for the LC-MS analysis with MS2 spectral acquisition in the Orbitrap. In those instances where duplicate peptides were present, the peptide with the highest probability and Mascot ion score was selected. For the resulting list of peptides, each spectrum was manually verified in a stepwise manner by comparing theoretical parent and fragment values from MS-Product (prospector.ucsf.edu) with observed values from the spectrum as observed in XCalibur. MS1 scans were checked to confirm that the observed mass and charge values were consistent with the theoretical values for the peptide assigned to the spectrum. If this requirement was not met, the peptide and corresponding spectrum were removed from consideration for sequence and residue coverage. In those cases where MS1 value was determined to be correct, the peaks in the MS2 spectrum were manually assigned based upon theoretical m/z values obtained from MS-Product, and the presence or absence of a 13C isotope signal was noted. Standardization sets for peptides were generated by using all accepted MS1 values, and an acceptance range (mean ± 2 SD) was determined by calculating the error for each peptide based upon theoretical m/z values obtained from MS-Product. All peptides whose error fell within the acceptance range were included in the peptide coverage map. Standardization sets for fragment ions were generated by using all b and y ions with assigned charge states, and an acceptance range (mean ± 2 SD) was determined by calculating the error for each fragment based upon theoretical m/z values obtained from MS-Product. All b and y ions whose error fell within the acceptance range were used to determine residue coverage, regardless of the presence of a 13C isotope signal. In addition, the b and y ions with determinable charge states and within 20 ppm mass tolerance were also included. These peptide and fragment acceptance ranges were used to verify spectra for α 1 peptides in the DDA set that were unassigned by Mascot (see spectra 60, 62, and 92 in supplemental spectra). Peptides from DMSA were verified using peptide and fragment acceptance ranges determined from a BSA benchmark sample in a similar manner (see spectra 54, 59, and 61). Flow charts and graphs representing acceptance criteria can be found in supplemental Figs. S4 and S5. Sequencing was based on assigned sequential, paired b or y signals that agreed within the mass accuracy.
Analysis of Reproducibility
To determine the reproducibility of our deep sequencing method, two additional samples were prepared from separate rat brains and analyzed with the original sample. The two replicate samples were control samples from a separate photoaffinity labeling study (manuscript in preparation) and were prepared using the same strategy shown in Fig. 8. The samples differed slightly, with respect to chymotryptic digestion times; specifically replicate #2 had a 2-hour rather than a 6-hour chymotryptic digestion time and did not have a “chymotrypsin alone” digest. Deglycosylation with PNGase was not performed and the glycosylated peptides were thus not considered in this analysis. For simplicity, the eluates of C4 and PGC SPE from tryptic digests of each single brain were pooled. Similarly, the C4 and PGC SPE eluates from the timed chymotryptic digests of each brain were pooled. The resulting six endoprotease digests from three rat brains were run sequentially in a single block analysis by LC-MS, as previously described. The consistency of sequence coverage of the α1 subunit was determined for the three replicates. Sequence coverage was selected as the critical metric rather than the consistent detection of individual peptides because the approach was based on both tryptic and chymotryptic digestion conditions, the latter designed to produce missed cleavages with the aim of obtaining amino acid sequence from overlapping peptides. Sequence coverage was determined by high-resolution mass matching of MS1 spectra in the replicates to the peptide m/z values that had undergone MS2 sequencing from the original sample (Supplemental ion table). The inclusion criteria that were used to assign MS1 features to peptides in the three replicates, were accurate mass tolerance, isotope distribution, chromatographic retention time similar to the original analysis, and intensity >1 × 105. Mass tolerance (mean ± 2 SD of the parent ions) was established for each chromatographic run using MS2 spectra of α1 subunit peptides with Peptide Prophet probability ≥50% and MASCOT ion score ≥30. The few sequences that were not identified in the original sample (Fig. 9) were included in the sequence coverage for the new replicates based on diagnostic MS2 spectra (accurate mass tolerance of parent ion, correct isotope distribution and manual interpretation of MS2 spectrum).
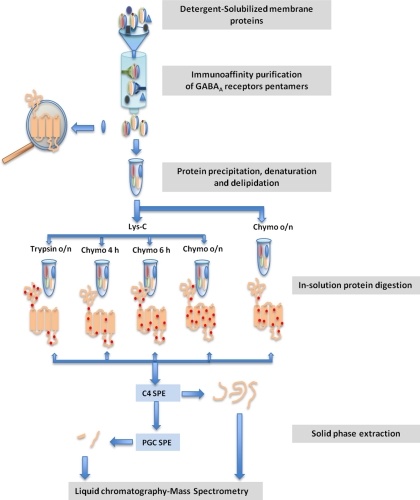
Fig. 8.
High-resolution LC-MS workflow for sequencing picomole quantities of transmembrane proteins from biological samples. Pentameric GABAA receptors were purified from an immunoaffinity column. The proteins were denatured and delipidated followed by timed, in-solution digestion with several proteolytic enzymes. Peptide recovery was enhanced using sequential solid phase extraction. Peptide identification and sequencing was optimized using nano-LC-ESI-MS with high resolution in both MS1 and MS2 spectra.
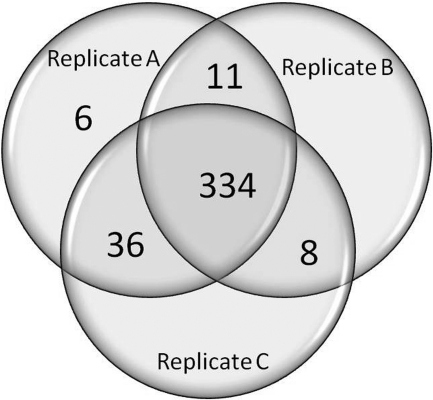
Fig. 9.
Venn diagram showing overlapping amino acid sequence coverage of the α1 subunit from three replicate brain samples. Peptides covering 392 (Replicate A; original sample), 353 (Replicate B), and 378 (Replicate C) amino acids were identified, representing 97%, 88%, and 94% of the detectable amino acid sequence of the α1 subunit.
Label-free Quantitative Analysis of Replicate Samples
The relative abundances of the 11 identified GABAA receptor subunit isoforms were determined using label-free quantitative analysis of the triplicate sample preparations. The spectrum count method was used with normalization for protein length and replicates. Peptides with identical sequences shared by multiple isoforms were assigned based on the unique spectrum count of each isoform as described by Zhang and colleagues (27). Briefly, unique spectra and spectra shared by multiple GABAA isoforms were identified using Scaffold Version 3.0 with a peptide probability ≥50% and a protein probability ≥95%. The unique spectrum counts were normalized across the three replicates and the normalized unique spectrum counts were used to partition shared spectra between isoforms (each spectrum was counted once and modified peptides were included). The assigned shared spectrum counts and the unique spectrum counts were summed and normalized for protein length. The total spectrum counts were then normalized for replicates.
The related spectra are available for download as .RAW files at the Proteome Commons Tranche data repository at https://proteomecommons.org/(TrancheHash: 9lyRCAuo77es83CipuVZGvuJ/W/nW5 PFSZ+jUn6vnjveh19TIIefb0t9uhBNqbsDqe5kGa5BNX5iGd6XqDgN 0ORGakwAAAAAAAADKw==).
RESULTS
Purification of GABAA Receptors From Brain
The low abundance of GABAA receptors in brain required that the receptors be solubilized and partially purified prior to mass spectrometric sequencing. The use of antibodies to affinity purify the receptors further necessitated that solubilization be performed with a nondenaturing detergent. Multiple detergents were tested for solubilization efficiency, including CHAPS and C12E9 combination (28), n-dodecyl-β-d-maltopyranoside (29), Triton X-100, RIPA and deoxycholate. Triton X-100 had the highest efficiency of extraction while not interfering with antibody affinity purification. Triton X-100 extracted ∼20% of the GABAA receptors present in P2 brain membranes, as assessed by densitometric comparison of Western blots (using an antibody against the α1 subunit of the GABAA receptor) of SDS-extracted P2 membranes and Triton X-100 membrane lysates. The tissue sample used for receptor protein purification was the neocortex (1 g) from a single rat brain; P2 membranes from this sample contained ∼20 picomoles of GABAA receptor as assessed by [3H]Ro15,4513 binding at a specific activity of ∼1.5 picomoles/mg protein. The Triton X-100 lysate of these membranes contained ∼4 picomoles of GABAA receptor. The lysate was applied to an immunoaffinity column containing antibodies directed against the β2, β3, and γ2 subunits of the GABAA receptor. Following washing with high concentration salt buffers, 40–50% of the GABAA receptors were eluted, yielding ∼2 picomoles of receptor (based on [3H]Ro15,4513 binding). The specific activity was >1500 picomoles/mg protein, indicating a purification of >1000-fold. At this stage of the enrichment procedure the GABAA receptor is preserved as an intact pentamer; in subsequent steps the receptor is denatured and the subunits are dissociated.
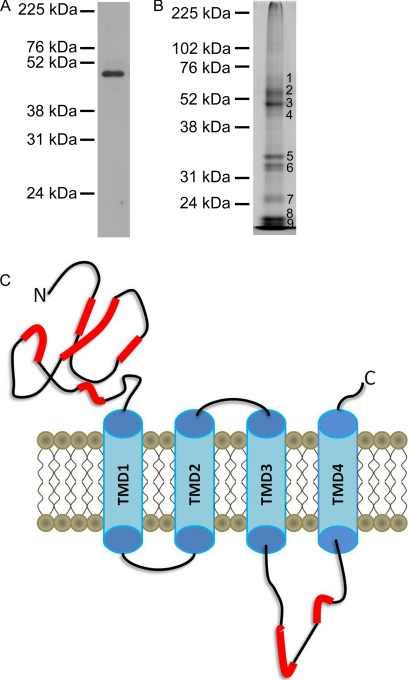
To assess the purity of the GABAA receptors, an aliquot of the purified protein was analyzed by SDS-PAGE with SYPRO-Ruby™ staining and immunoblotting with an antibody directed against the α1 subunit (Figs. 1A, 1B). The immunoblot demonstrated a single band for the α1 subunit (52 kDa). SYPRO-Ruby™ staining showed that four major bands between 48-kDa and 55-kDa, two bands near 31-kDa, and three bands at or below 24-kDa were observed (Fig. 1B). These bands were excised and digested in situ with trypsin, and the extracted peptides were analyzed by nano-LC-FT-MS with data-dependent MS2 acquisition as previously described (20). From a search of the Uniprot-Rat database with the MS2 spectra, the most abundant proteins in band 1 were the α4 subunit of the GABAA receptor with lesser amounts of the β2 subunit and trace amounts of synaptotagmin; band 2 contained the β2 subunit with lesser amounts of the β1, β3, α1, and α3 subunits as well as tubulin; band 3 contained mostly the α1 and some α2, β1, and β2 subunits; band 4 contained the α1 subunit as well as trace amounts of actin and integrin; bands 5 and 6 contained antibody from the immunoaffinity column; bands 7–9 contained degraded myelin basic protein and ATP synthase. Analysis of the tryptic peptides eluted from band 3 of the SDS-PAGE gel identified peptides covering 22% of the sequence of the α1 subunit of the GABAA receptor; none of the identified sequence was within the TMDs (Fig. 1C). These data indicate that a large amount of the proteins on the gel represented intact or fragmented GABAA receptor subunits. Because the sample was highly enriched in GABAA receptor subunits, we used in-solution digestion of the unfractionated affinity-purified receptor for further studies.
Fig. 1.
SDS-PAGE of immunopurified GABAA receptors. A, Immunoblot with antibody to the α1 subunit of the GABAA receptor. B, SYPRO-Ruby™ protein staining. C, Peptide level sequence coverage of the α1 subunit of the GABAA receptor based on MS2 spectra obtained from nano-LC-MS analysis of the in-gel tryptic digest of band 3 from panel B. The red highlights represent regions with MS2 identified peptide sequences.
Chymotrypsin Digestions of Varying Duration Enhance Sequence Coverage of the TMDs of the GABAA Receptor α1 Subunit
Trypsin digestion, usually the first step in protein identification and sequencing by MS, is of limited utility in sequencing TMDs because the tryptic cleavage sites in these domains results in large, insoluble hydrophobic peptides. In silico proteolytic digestion of the α1 subunit of the GABAA receptor with trypsin yielded hydrophobic peptides of the TMDs that were 20–33 amino acids in length with GRAVY (Grand average of hydropathicity (30)) scores >1. Preliminary work with synthetic versions of these peptides revealed that they were not soluble in LC-MS compatible solutions, e.g. 10–90% ACN (data not shown). Chymotrypsin, which cleaves on the C-terminal side of hydrophobic amino acids such as Tyr, Trp, Phe, Leu, and Met, was tested as an alternative endoprotease to produce shorter, soluble peptides of the TMDs. Since TMD's contain multiple hydrophobic residues, we designed a protocol that would allow variable degrees of chymotryptic digestion, thus producing overlapping peptides for de novo sequencing. The purified GABAA receptor was either first digested overnight with Lys-C, followed by chymotryptic digestion for 4 h, 6 h, or ∼18 h or was digested with chymotrypsin alone for ∼18 h.
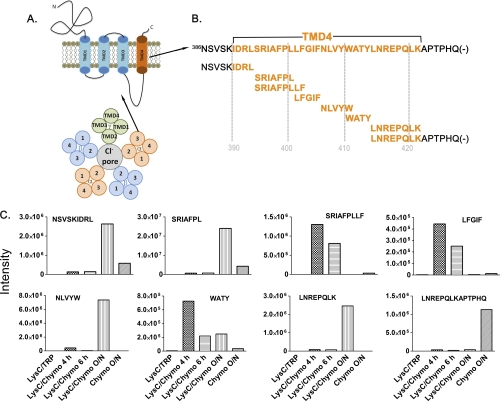
Using this digestion time course, we obtained peptides of various lengths with overlapping sequences that covered the four TMDs of the GABAA receptor. This is illustrated in Fig. 2 for the fourth TMD (TMD4) and the C terminus of the GABAA receptor α1 subunit. As shown in Fig. 2B, the timed digestion protocol yielded peptides that provided complete sequence coverage of TM4 and the C terminus of the α1 subunit. This region was sequenced by eight overlapping chymotryptic peptides recovered at different digestion times. The intensities of each peptide as the parent ions in the MS1 spectra were compared among different digestion conditions after time and m/z alignment (Fig. 2C). Shorter digestion times tended to yield longer peptides because of multiple missed cleavages, whereas longer digestion times yielded shorter peptides because of relatively complete chymotryptic cleavage. For example, the peptide SRIAFPLLF was only detected with 4 h and 6 h of chymotrypsin digestion. In contrast, the peptide SRIAFPL was only detected with overnight chymotrypsin digestion. Multiple time course samplings were needed to obtain full sequence coverage of this C-terminal region. As expected, however, Lys-C/trypsin digestion produced no detectable peptides of this region (Fig. 2C). Similar results were obtained for the other three TMDs of the α1 subunit. Using this timed LysC/chymotryptic protocol, 93% of the sequence of the four TMDs of the α1 subunit was determined from database searching of the MS2 spectra.
Fig. 2.
Time course of chymotryptic digestion to obtain increased sequence coverage of the fourth transmembrane domain of the GABAA receptor α1 subunit using LC-MS. A, Diagram of a GABAA receptor subunit composed of four transmembrane spanning regions with extracellular N- and C termini and the assembly of five such subunits to form a pentameric chloride channel. B, Overlapping peptides of the entire fourth TMD of the α1 subunit of the GABAA receptor from the sequential digestion with Lys-C and chymotrypsin for varying lengths of time. C, Peptide intensities from MS1 spectra of the indicated TMD4 peptides that were produced during the chymotryptic digestion time course.
Lys-C/Trypsin Digestion is Useful to Yield Peptides of Hydrophilic Regions
Although Lys-C/trypsin digestion produced no detectable TMD peptides, it did yield peptides that were essential for complete sequencing of the hydrophilic domains of the α1 subunit. This is illustrated in Fig. 3, for a 59 amino acid portion of the protein (residues, 15T - R73) that was completely sequenced by three tryptic peptides and six chymotryptic peptides from different digestion times. Although timed chymotrypsin digestion provided most of the sequence coverage for this region, two peptides were missing because they were too small to be detected. Three peptides from this region were observed in the Lys-C/trypsin digest, including the sequences missing from the timed chymotrypsin digests. These data illustrate that digestion with multiple enzymes significantly increases the peptide sequence coverage for hydrophilic regions of the α1 subunit of the GABAA receptor.
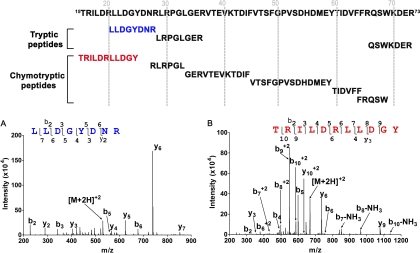
Fig. 3.
Multi-enzyme digestion of the N-terminal region of the α1 subunit of the GABAA receptor. (Top) Overlapping peptides from tryptic and chymotryptic digestions. A, MS2 spectrum of a Lys-C/tryptic peptide, 21LLDGYDNR28. B, MS2 spectrum of a Lys-C/chymotryptic peptide, 15TRILDRLLDGY25.
Additional Peptides from the N-Terminal Portion of the Receptor are Detected Following Deglycosylation
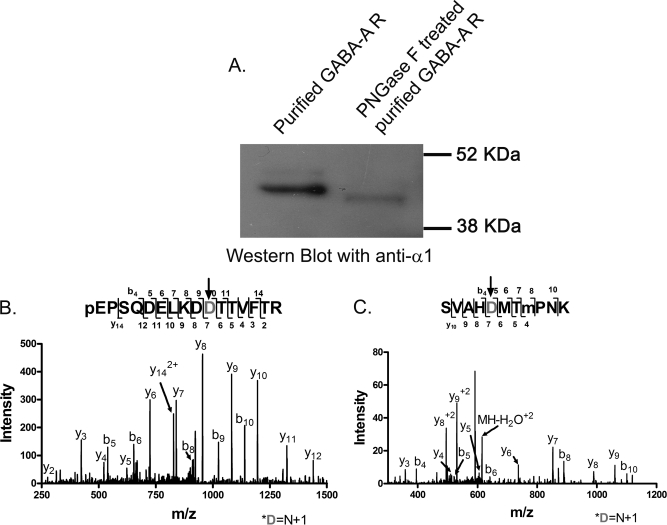
Two sequences in the N-terminal extracellular domain, N′-QPSQDELKDNTTVF14and 106SVAHNMTMPNK116 were not detected using our multi-endoprotease protocol. These sequences contain consensus N-glycosylation sequons N-X-S/T (X can be any amino acid except P). Previous studies using mutagenesis have suggested that these two sites, 10N and 110N, are glycosylated in recombinant mammalian expression systems (31). To determine if these sites are glycosylated in native tissue GABAA receptors, a purified sample was digested with PNGase F. A GABAA Western blot (α1 antibody) comparing the untreated and PNGase F treated receptors showed that the treated α1 subunit migrates faster than the native subunit, consistent with deglycosylation (Fig. 4A); the mass change was about 5-kDa. The deglycosylated sample was digested with Lys-C and trypsin and analyzed with LC-MS. PNGase F cleaves glycans at the amide bond converting Asn to Asp residues; this change can be detected by a 1 Da mass increase in the deglycosylated peptide (32). Data analysis that accounted for PNGase F catalyzed deamidation of glycosylated Asn residues resulted in the identification of a tryptic peptide (m/z = 931.936, z = 2) corresponding to 1QPSQDELKDDTTVFTR16 (The N to D change produced by deamidation is denoted by the BOLD D; the N-terminal Q is modified to pyro-E.) The peptide sequence was deduced from the MS2 spectrum (Fig. 4B). The conversion of Asn → Asp was determined from the flanking b9 and b10 and y6 and y7 ions. The signal peptide of the unprocessed form of the α1 subunit contains a G residue before this tryptic peptide (QPS…), indicating that Q is the N-terminal amino acid residue of the mature α1 subunit in rat neocortex.
Fig. 4.
Identification of deglycosylated N-terminal peptides from the α1 subunit of the GABAA receptor following treatment with PNGase F. A, Western blot with an anti-α1 antibody comparing untreated and PNGase-treated receptors. B, The MS2 spectrum of a tryptic peptide corresponding to 1QPSQDELKDDTTVFTR16 (m/z = 931.936, z = 2). The N-terminal Q was modified to pyro-E, labeled as pE. C, The MS2 spectrum of a tryptic peptide corresponding to 106SVAHDMTMoxidationKPNK116 (m/z = 623.779, z = 2). The N to D change produced by PNGase F catalyzed deamidation is denoted by the gray D and arrow.
The other sequence stretch in the N-terminal domain, 106SVAHDMTMPNK116 (m/z = 623.778, z = 2), was determined from the MS2 spectrum shown in Fig. 4C. A 1-Da increment was observed in the fragmentation ions b5-b10 and y7-y9. The localization of the 1 Da change to a deamidation of N was confirmed from the flanking b4 and b5 and the y6 and y7 ions. The non-glycosylated forms of the N-terminal peptides were not observed in the LC-MS analysis of samples that were not treated with PNGase F, suggesting that the receptor is completely glycosylated in the native tissue. There is another consensus glycosylation sequence of the α1 subunit at 364N in the TM3-TM4 loop. In both deglycosylated and native samples, however, this amino acid residue was observed as N without modification, indicating this site is not glycosylated in rat neocortex. It should be noted that LC-MS analyses of PNGaseF-treated samples were performed using DDA with high and low resolution acquisition of the MS1 and MS2 spectra, respectively.
Sequential Solid Phase Extraction With C4 and Porous Graphitic Carbon Increases Recovery of Peptides from Digests of the GABAA Receptor α1 Subunit
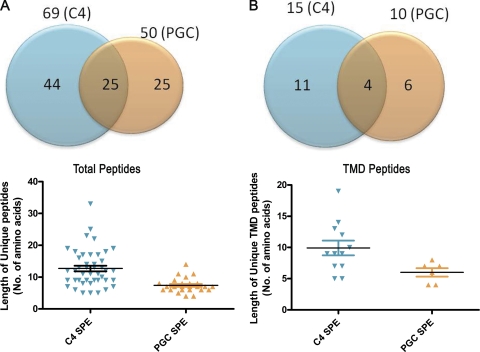
We investigated the recovery of peptides from the high-salt digests (e.g. 2 m urea) using sequential SPE with two stationary phases, C4 for hydrophobic peptides and PGC for smaller and hydrophilic peptides. A total of 94 unique peptides from the α1 subunit of the GABAA receptor were recovered by sequential C4 and PGC SPE. As shown in Fig. 5A, 44 peptides were uniquely extracted with C4 SPE, 25 peptides were uniquely extracted with PGC SPE and 25 peptides were extracted with either stationary phase. Peptides greater than 14 amino acids in length were observed following C4 but not PGC SPE (Fig. 5A lower panel). PGC SPE, in contrast, yielded several shorter peptides that were not detected following C4 extraction. The sequential SPE protocol was particularly useful in identifying the TMD sequences (Fig. 5B). Twenty-one unique peptides from the TMD regions of the α1 subunit were recovered by sequential C4 and PGC SPE. 10 peptides were recovered by PGC SPE with 6/10 uniquely recovered from PGC SPE; these peptides were all peptides <14 amino acids in length. The extraction with C4 yielded 15 TMD peptides of which 11 were uniquely recovered; TMD peptides greater than 9 amino acids in length were recovered from C4, but not PGC SPE. These results indicate that sequential SPE is superior to either C4 or PGC SPE alone and is particularly important for recovering TMD peptides. Supplemental Table I lists all of the GABAA receptor α1 subunit peptides recovered by C4 or PGC SPE. Based on these results we adopted the protocol in which samples were first extracted with C4 and then extracted with PGC.
Fig. 5.
Comparison of total and TMD peptides from the α1 subunit of the GABAA receptor recovered by sequential solid phase extraction with C4 and porous graphitic carbon. A, Peptides recovered using C4 and PGC SPE from the total peptide pool (top); peptide size distribution of the total peptides recovered by C4 and PGC SPE (lower). B, TMD peptides recovered by C4 and PGC SPE (top); size distribution of TMD peptides recovered by C4 and PGC SPE (lower). Bars indicate mean ± S.E.
High Resolution MS2 Analysis of the GABAA Receptor α1 subunit
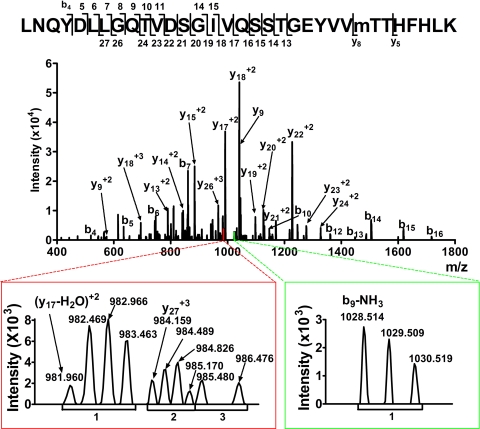
The availability of mass spectrometry for high mass accuracy measurements of both the parent ions and fragment ions in MS1 and MS2 spectra, respectively, affords the opportunity to sequence peptides de novo with a higher degree of confidence. High-resolution Orbitrap measurements of peptide fragments in MS2 spectra enable a clear distinction from noise in isotope clusters and, with the ∼100-fold increase in mass accuracy, reduce the chances that flanking residue signals are coincidental. The resolution (7500) in the MS2 spectra enabled the determination of fragment ion charge state for a majority of the MS signals that were used for peptide sequencing, minimizing erroneous assignment of fragment ion species in these spectra. Fig. 6 shows a high resolution MS2 spectrum for the tryptic peptide 187LNQYDLLGQTVDSGIVQSSTGEYVVMTTHFHLK219 (observed m/z = 924.962+4, theoretical m/z = 924.961) of the α1 subunit in which the single methionine residue is oxidized. The two lower panels show zoomed views of the low abundance fragment ions y17-H2O+2, y27+3, and b9-NH3+1. The left lower panel displays the m/z range between 981 and 986. The doubly charged fragment with an average m/z separation of 0.502 Da (cluster 1) is distinguishable from the adjacent triply charged fragments ion, with a charge separation of 0.330 (cluster 2), and the singly charged fragment ions (cluster 3). The doubly charged fragment ion with monoisotopic peak m/z = 981.960 was identified as y17-H2O+2 and the triply charged fragment ion (m/z = 984.159) as y27+3. The right inset shows the m/z range between 1027 and 1032. The intensity of these ions was just above baseline noise. However, the charge assignment indicated that fragment ion m/z = 1028.514 was the singly charged b9-NH3+1.
Fig. 6.
High resolution mass spectrometry improves the identification of peptides in the fragmentation spectra (MS2) of the tryptic peptide 187LNQYDLLGQTVDSGIVQSSTGEYVVMoxidationTTHFHLK219 (m/z = 924.962+4). Signals from peptide fragmentation are assigned using both enhanced mass accuracy and charge state determination. Accurate charge state assignment from isotopic resolution is illustrated in bottom spectra.
Sequence Coverage of the GABAA Receptor
We identified all 12 of the GABAA receptor subunit isoforms expressed in neocortex from database searching of MS2 spectra. To determine the sequence coverage of these identified subunits from MS1 data, we first determined the mean mass accuracy and standard deviation for the individual blocks of LC-MS analyses. We used the observed values for the 940 parent ions of GABAA receptor peptides that had MASCOT score ≥30. The distribution is shown in supplemental Fig. S2. The m/z and retention time aligned LC-MS analyses of all tryptic and chymotryptic digests were next queried for all observed values within the mass accuracy range (2.2 ppm) of the theoretical values of the all the GABAA receptor isoforms. As shown in Table I, the peptides from all the digestion conditions within 2.2 ppm mass tolerance on the MS1 spectra gave 100% sequence coverage for the α1 subunit, 98% for the β2 subunit, and 95% for the γ2 subunit, which represent the most abundant subunits in rat neocortex. The low abundance α4, α5, and δ subunits also had high MS1 coverage of 95%, 98%, and 89%, respectively. The MS1 coverage of other GABAA receptor subunits is also listed in Table I. Sequence coverage maps of all the GABAA receptor subunits based on high accuracy MS1 are presented in supplemental Figs. 3A–3D.
Table I. Coverage of the subunits. Peptide coverage of GABAA receptor subunit sequences using different mass spectrometric methods of analysis. MS1 coverage indicates the percentage of sequence identified from accurate mass determination of survey spectra with mass matching to in silico enzymatic digests of known subunit sequences. MS2 coverage indicates the sequence coverage obtained by confirming MS1 identifications with MS2 spectra collected in data-driven acquisition mode. Directed MS2 spectral acquisition (DMSA) increases the peptide coverage of the α1 subunit to 96%.
| Subunit | MS1 coverage | Peptide coverage based on DDA | Peptide coverage based on DDA and AIMS |
|---|---|---|---|
| α1 | 100% | 90% | 96% |
| α2 | 97% | 77% | |
| α3 | 99% | 67% | |
| α4 | 95% | 42% | |
| α5 | 98% | 59% | |
| β1 | 95% | 67% | |
| β2 | 98% | 90% | |
| β3 | 94% | 80% | |
| γ1 | 81% | 31% | |
| γ2S | 95% | 57% | |
| γ2L | 93% | 57% | |
| γ3 | 92% | 20% | |
| δ | 89% | 20% |
MS2 MASCOT search results from DDA were validated using the Peptide Prophet algorithm in Scaffold. The peptides with more than 50% probability in Scaffold and manually verified spectra were accepted. These peptides provided 85% sequence coverage of the α1 subunit and 86% of the β2 subunit, which are the most abundant GABAA receptor subunits in neocortex. In this paper, we focused on sequencing the α1 subunit. The α1 subunit peptides from the MS1 spectra for which a diagnostic MS2 was not obtained using DDA were re-analyzed using DMSA (note, the DMSA analysis was performed on a separate brain sample). To do the DMSA, we first determined peptide retention times by searching selective ion chromatograms for theoretical m/z of the peptide with single, double, and triple charges. The features from the MS1 spectra within 5 ppm mass tolerance deviation from the theoretical m/z with correct isotopic distribution, charge assignment, and intensity >1e5 were selected for DMSA. The MS2 spectra obtained from DMSA were validated using acceptance criteria as described in Materials and Methods. An additional three α1 subunit peptides were identified using this approach. One of these peptides, 231LPCIMTVIL239 (Supplemental spectra, #54), was not included in the α1 sequence coverage because manual interpretation of the spectrum was ambiguous. Inclusion of the two peptides from DMSA increased the peptide sequence coverage of the α1 subunit to 90% overall with 88% peptide coverage of the TMDs. Including the peptides identified following deglycosylation, our analyses provide 96% peptide coverage of the sequence of the α1 subunit and 90% of the β2 subunit of the GABAA receptor (Fig. 7 and Table I). Peptide sequence coverage maps for the other GABAA receptor subunits based on MS2 spectra are shown in supplemental Figs. S6A–S6D.
Fig. 7.
Residue level sequencing of the α1 subunit of the GABAA receptor, purified from rat brain neocortex. Amino acid residues definitively identified from analysis of fragment ions of the MS2 spectra are in red and BOLD. The residue coverage of the whole protein is 80%. Frames indicate the TMDs of the protein. The residue coverage of the TMDs is 63%. The sequencing coverage of this protein at the peptide level is 96%. The missing peptides/residues are 74LK75, 132L, 274N, 231LPCIM235, 289IAVCY293, and 310TK311. 289IAVCY293 was observed in both new replicates.
Peptide fragmentation spectra were also analyzed to determine the level of residue coverage by de novo sequencing. Identification of a specific residue was based on identification of two adjacent b ions or y ions, or an adjacent N-terminal y ion and C-terminal b ion with an accurate mass difference corresponding to a specific amino acid. These identifications were assisted by the increased mass accuracy and the determination of fragment ion charge state from these high-resolution MS2 spectra, particularly for signals with a low signal to noise ratio, as illustrated above in Fig. 6. From the interpretation of 94 MS2 spectra (Supplemental data spectra), 80% of the amino acid residues of the α1 subunit were identified including 63% of the TMD residues as shown in red and BOLD in Fig. 7. (The 80% residue coverage includes the amino acids deduced from the HR-LR deglycosylated peptides shown in Figs. 4B and 4C.) Supplemental ion tables report all the individual α1 subunit peptides and their corresponding ion fragments. The corresponding spectra are presented in (supplemental spectra).
Reproducibility of α1 Sequence Coverage
Reproducibility of the method was determined based on sequence coverage of the α1 subunit from the three replicates. In order to test the reproducibility of this method, brain samples from two additional rats were analyzed in the same block as the original sample. Peptides covering 392 (replicate A; original sample), 353 (replicate B) and 378 (replicate C) amino acids were detected. This represents 96%, 88 and 94% of the detectable amino acid sequence of the α1 subunit (not including the signal peptide or glycosylated peptides). The Venn diagram in Fig. 9 shows that the same 78% of the α1 subunit sequence was observed in all three replicates with an additional 13% observed in two of the three replicates. Only 1% of the α1 subunit sequence (6 amino acids) was uniquely observed in the original sample. As expected, the two glycosylated peptides (Figs. 4B and 4C) were absent in all three replicates, confirming the complete glycosylation of these two sites in native tissue. Interestingly, the TM3 peptide (289IAVCY293), not observed in the original sample, was found in both new replicates. The interpreted MS2 spectrum of this peptide is included as (supplemental Spectra (#95)).
Relative Abundance of GABAA Receptor Subunits Determined by Label-free Quantitative Analysis
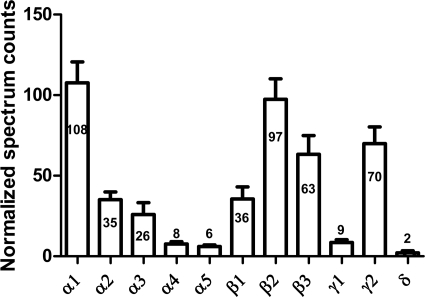
The relative abundance of the 11 identified GABAA receptor subunits was determined using label-free quantitative analysis of the triplicate sample preparations (Fig. 10). Each of the 11 GABAA receptor subunits was detected with Protein Prophet Probability ≥95% in all three samples, and the normalized spectrum counts of each subunit (640 spectra in total) were consistent across the three samples. Statistical analysis (one-way ANOVA with Tukey's post-hoc multiple comparison test) indicated that the α1 subunit spectrum counts were significantly higher than any of the other α subunits (p < 0.001); β2 counts were significantly higher than β1 (p < 0.001), but not significantly different from β3; and γ2 subunit counts were significantly higher than γ1 subunit. These results are in agreement with previous reports, based on RNA expression and quantitative immunoblotting (33, 34), that α1, β2, and γ2 are the most abundant subunits in brain neocortex. Interestingly, the sum of the normalized spectrum counts of all the α subunits (548), all the β subunits (589), and all the γ and δ subunits (242), yields a ratio of 2.3: 2.4: 1, consistent with the known stoichiometry (2α: 2β: 1γ or δ) of individual heteropentameric GABAA receptors (33, 34). To our knowledge, this work provides the first direct measurement of the relative abundance of GABAA receptors subunits at the protein level in native tissue.
Fig. 10.
Relative abundance of GABAA receptor subunits determined by label-free quantitative analysis. The mean ± S.E. of the normalized spectrum counts for three replicate samples are shown on the graph, with the mean value labeled for each subunit. The stoichiometry of α: β: (γ+δ) = 2.3: 2.4: 1, consistent with the expected 2: 2: 1 stoichiometry.
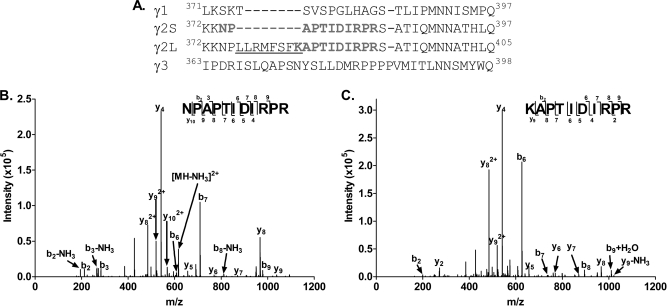
Identification of Splice Variants of GABAA Receptor γ2 Subunits
Residue level sequence coverage affords the opportunity to identify sequence variants such as splice variant isoforms. Two distinct splice variant isoforms of the γ2 subunit, γ2Short (γ2S) and γ2Long (γ2L), have previously been identified from cDNA sequences (35). γ2S and γ2L differ by 8 amino acids (LLRMFSF) in the intracellular loop between TMD3 and TMD4 (Fig. 11A). In our MS2 data, a chymotryptic peptide (m/z = 583.853, z = 2) was identified as KNAPTIDIRPR, a specific γ2L peptide. A specific γ2S peptide, NPAPTIDIRPR, was also identified in the tryptic digest samples (m/z = 625.3539, z = 2). The diagnostic MS2 spectra of these two peptides are shown in Figs. 11B and 11C. These data provide the first direct protein-level evidence that both γ2S and γ2L isoforms exist in brain tissue and provide a practical methodology for future investigation of the relative abundance of each isoform in different brain regions and developmental stages.
Fig. 11.
Identification of γ2 subunit splice variant isoforms of the GABAA receptor. A, The γ2S and γ2L splice variants differ by 8 amino acids (LLRMFSF; underlined). B, An MS2 spectrum of the tryptic peptide, NPAPTIDIRPR (m/z = 625.3542+) was observed, identifying the γ2S variant isoform. C, An MS2 spectrum of the chymotryptic peptide KNAPTIDIRPR (m/z = 583.8532+) was observed, identifying the γ2L variant isoform.
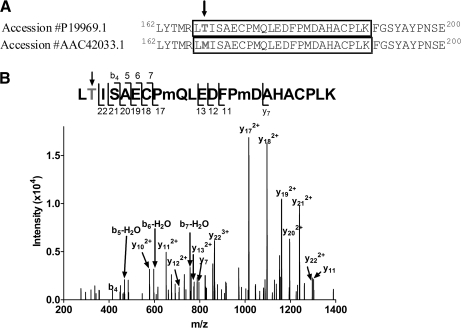
Resolution of a Single Amino Acid Discrepancy in a Protein Sequence Database of the α5 Subunit
There are several single amino acid differences in GABAA receptor subunit sequences between different protein sequence databases. For example, the sequence of the α5 subunit is different in residue 168 (the sequence numbered with the signal peptide included) between accession #P19969.1 (168Thr) and accession #AAC42033.1 (168Met) in the PubMed/protein database (Fig. 12A). A search of our LC-MS data against a database containing both sequences detected a peptide (m/z = 936.091, z = 3) consistent with 167LTISAEC*PmQLEDFPmDAHAC*PLK190 (theoretical m/z = 936.090, z = 3; * carbamidomethylation of Cys). The high resolution MS2 spectrum of this peptide (Fig. 12B) contains 4 sequential b ions (b4-b7), supporting the identification of this peptide with Thr as residue 168. The adjacent 6 sequential y ions (y17–y22) further support the assignment of 168Thr. No peptides consistent with oxidized or nonoxidized 168Met were detected in the MS2 or MS1 spectra of either the tryptic or chymotryptic digests. These results suggest that 168Thr is the correct residue in the α5 subunit.
Fig. 12.
Resolution of a single amino acid database discrepancy in the sequence of the α5 subunit. A, Amino acid differences in residue 168 of the α5 subunit are indicated by the arrow. B, The MS2 spectrum of a peptide (m/z = 936.0913+) consistent with 167LTISAEC*PmQLEDFPmDAHAC*PLK190 (theoretical m/z = 936.0903+, * indicates carbamidomethylation of Cys) was observed, suggesting that 168Thr is the correct residue in the α5 subunit from our biological source.
DISCUSSION
We have described a method for using LC-ESI-MS to obtain complete sequencing of GABAA receptor subunits from picomole quantities of isolated brain receptors. A workflow diagram of the method is shown in Fig. 8. The analysis identified peptides, using a database searching algorithm of MS2 spectra, which cover 96% of the sequence of the α1 subunit including N-terminal domain glycosylation sites after enzymatic removal of the carbohydrate moieties. Further analysis using de novo sequencing gave 80% of the amino acid residues of the α1 subunit. Using a combination of database searching and peptide mass mapping, our analyses also delineated most of the sequence, at the peptide level, of the other 11 GABAA receptor subunits expressed in neocortex. Sequence coverage of the α1 subunit was highly reproducible in three samples, enabling us to use label-free quantitative analysis to provide the first direct measurement of the relative amounts of GABAA receptor subunits in brain. Analysis of our data also identified peptides diagnostic for splice variants of the γ2 subunit and corrected a database discrepancy in the sequence of the α5 subunit. The methods and strategy developed and exploited here should be applicable to identifying additional post-translational modifications and sequence variants of GABAA receptors, as well as other IMPs, in specific brain regions under a variety of physiological and pharmacological conditions.
Several previously described methods were incorporated into our microscale workflow to achieve a high level of sequence coverage from a small quantity of IMP. In-solution digestion was used to avoid protein losses associated with gel electrophoresis and peptide losses during gel extraction, thereby optimizing both protease access to the protein and peptide recovery. In-solution digestion also allowed us to delipidate the protein, facilitating digestion of the TMDs (9). The use of multiple proteases (Lys-C, trypsin, and chymotrypsin) contributed to our ability to obtain overlapping peptides covering both the hydrophilic and TMD portions of the protein (7, 12). Finally, the specificity of peptide and residue identification was enhanced by using high resolution mass spectrometry for the acquisition of both the MS1 and MS2 spectra.
Two new techniques for sequencing TMDs also contributed to the success of our method: Variable duration chymotryptic digestions and sequential solid phase extraction of peptides with multiple stationary phases. The major obstacle to mass spectrometric sequencing of IMPs has been the generation, isolation and sequencing of peptides covering the TMDs. Our initial strategy was to generate long tryptic peptides in which TMDs were tethered to adjacent hydrophilic regions of the protein to maintain their solubility in the solvents used for LC-MS. This strategy was unsuccessful. We found that long tryptic peptides were not recovered from PGC SPE. Although some long tryptic peptides could be recovered using C4 SPE, these peptides provided minimal sequence coverage of the TMDs (Fig. 2). The lack of TMD sequence coverage from long tryptic peptides may be explained, in part, by experiments conducted with a 24 amino acid synthetic tryptic peptide of the TM4 region of the α1 subunit (LSRIAFPLLFGIFNLVYWATYLNR; data not shown). This peptide was not soluble in ACN, methanol, isopropanol, FA or in nondenaturing or denaturing (SDS, RapiGest™) detergents, suggesting that long tryptic peptides containing TMD sequences are likely to wax out of solution and thus not be detected (Fig. 2). Chymotrypsin, which cleaves peptide bonds adjacent to aromatic and other hydrophobic amino acids, in contrast, yields shorter, more soluble hydrophobic peptides. To produce TMD hydrophobic peptides of varying length and with overlapping sequences, proteins were digested with either Lys-C followed by chymotrypsin for varying lengths of time or chymotrypsin alone. Digestion for different times was essential to improving the coverage of the TMDs. Shorter digestion times yielded longer peptides because of missed cleavages, whereas longer digestion times produced short peptides. This approach generated overlapping chymotryptic peptides that provided complete sequence coverage of the TMD regions of the α1 subunit. Recovery of the short chymotryptic peptides was markedly better with PGC than with C4 SPE. Ensembles of both long and short peptides were produced using sequential SPE with C4 followed by PGC.
The methods developed here should be generally applicable to sequencing low abundance IMPs from native tissues and could also readily be applied to IMPs expressed in recombinant systems. To exploit this approach, it is necessary to have reliable methods for solubilizing and enriching and purifying the target protein. The selected enrichment method will dictate the solubilization methods that are applicable. For example, to utilize immunoaffinity or ligand-protein affinity purification (12), nondenaturing solvents or detergents are required. Other purification methods such as immobilized metal ion affinity chromatography can be used with denaturing detergents or solvents, allowing for more efficient protein solubilization. This can be an important consideration since nondenaturing detergents often selectively extract membrane proteins from specific environments (See below). The steps in our method subsequent to solubilization should require limited modification for sequencing other IMPs. The timed Lys-C/chymotrypsin and chymotrypsin digestions should be generally applicable to all TMDs. The remaining hydrophilic sequences may require use of proteases in addition to trypsin to maximize sequence coverage (12), but the need for, and identity of, additional proteases can be readily discerned from the consensus proteolytic sites in the predicted protein sequence.
A significant limitation of the method, as exploited here, is incomplete solubilization of GABAA receptors from tissue with a bias to solubilizing receptors from specific subcellular environments. Incomplete solubilization has been an issue in all of the described purifications/enrichments of GABAA receptors from native tissue (36). Triton X-100 was used in our study because it was the most efficient solubilizing agent that did not interfere with immunoaffinity purification. However, it has been previously reported that extrasynaptic GABAA receptors containing the δ-subunit are found predominantly in TX-100 insoluble lipid raft fractions of the membrane. Similarly, synaptic GABAA receptors (containing the γ subunit) are principally located in high density Triton X-100 insoluble membrane fractions that are thought to correspond to the post-synaptic density (36). Our study focused on the α1 subunit, the most abundant GABAA receptor subunit, which is known to be present in both Triton X-100 soluble and insoluble membrane fractions. Although we detected and sequenced all of the GABAA receptor subunits expressed in brain, the receptors we studied may be selectively enriched in intracellular GABAA receptors (endoplasmic reticulum and golgi) and receptors from non-specialized portions of the neuronal plasma membrane. This selective solubilization is important because GABAA receptors (and other IMPs) in specific cellular and subcellular environments, may have specific post-translational modifications and sequence variants. It is noteworthy that other signaling proteins including G-protein coupled receptors (37) and excitatory neurotransmitter receptors (36, 38), are also selectively localized in lipid rafts and synapses and may be subject to the same selective solubilization phenomena. Some of the issues of selective solubilization can, however, be addressed. For example, denaturing detergents and high salt conditions can also enhance solubilization of membrane proteins without interrupting purification when methods such as immobilized metal ion affinity chromatography are used for affinity purification.
There are several ways in which the sensitivity and efficiency of the methods described here could be further improved. First, we performed separate mass spectrometric analyses of the C4 and PGC extracts of each digestion and of the various timed chymotryptic digests. Alternatively, the various chymotryptic digests and the C4 and PGC solid phase extracts could be pooled. This would reduce the amount of starting protein needed, the number of mass spectrometric runs required and the complexity of the data analysis. This approach was successfully implemented in our analysis of replicate samples to validate reproducible sequence coverage. Although we did not attempt to optimize the reverse phase packing material used in the chromatographic separations, the use of C4 improves recovery and resolution of long hydrophobic peptides (data not shown) and could be employed to further optimize the detection of hydrophobic peptides. Finally, performing chromatography at elevated temperatures has been reported to improve resolution of hydrophobic peptides (11).
To test the utility of our method for addressing biological questions, we applied label-free quantitative analysis to determine the relative abundance of the eleven GABAA receptor subunit isoforms and searched our data to identify examples of PTMs, splice variants and database discrepancies. Label-free quantitative analysis provided the first direct measurement of the relative amount of GABAA receptor subunits in brain tissue. Our results demonstrate that the aggregate amount of all α isoforms is roughly equal to the aggregate of all β isoforms and is double that of the δ + γ subunits, consistent with the known 2: 2: 1 ratio of α: β: (δ or γ) in a single pentameric receptor. The relative abundance of the various isoforms is consistent with immunoprecipitation/immunoblotting studies estimating the prevalence of various heteropentameric receptor combinations (33, 34). Samples of purified GABAA receptors treated with PNGase allowed us to identify which N-glycosylation sequons on the α1 subunit were glycosylated in vivo. We confirmed one α1 N-glycosylation site (10N) that has recently been identified using shotgun analysis of the brain glycoproteome (39). We also identified an additional N-glycosylation site (110N) and demonstrated that a third sequon (364N) is not detectably glycosylated in vivo. A splice variant of the γ2 subunit of the GABAA receptor has been identified at the mRNA level, but not confirmed at the protein sequence level. As illustrated in Fig. 11, our data contained diagnostic MS2 spectra for peptides covering the spliced regions of both the γ2L and γ2S sequences, confirming the existence of both splice variants. Analysis of GABAA receptor subunit sequences from several databases detected a single amino acid discrepancy in the sequence of the α5 receptor. This discrepancy was definitively resolved by the residue level sequencing obtained with our method (Fig. 12). Collectively, these examples demonstrate the utility of our microscale, residue-level sequencing methodology for quantifying GABAA receptors and identifying sequence variants and modifications.
Although glycosylation sites and the γ2S/γ2L splice variants were readily detected in a 2 pmol sample of GABAA receptor, it may be more challenging to identify modifications (e.g. phosphorylation) and splice variants that affect a small percentage of total receptor protein. Enrichment techniques such as IMAC are routinely used to identify these “trace” PTMs. Application of enrichment techniques to the peptides generated in our workflow should facilitate the identification of low abundance PTMs such as phosphorylation, palmitoylation etc., and should provide more complete identification of PTMs on GABAA receptors than the whole tissue shotgun methods currently in use (40). Depending on the abundance of the IMP being studied, it may also be necessary to begin sample preparation with larger amounts of tissue for rare splice variants and low prevalence PTMs.
In summary, we report a method for obtaining virtually complete, residue-level mass spectrometric sequence coverage of low picomole quantities of native GABAA receptor subunits. This method should be applicable to other low abundance, native IMPs and will be useful for identifying new post-translational modifications as well as sequence variants that occur in vivo.
Acknowledgments
We thank Natalia Akentieva for her contributions to the preliminary phases of this work and Cheryl Lichti for assistance with spectral interpretation. We also thank Alan Davis, James Malone, Petra Erdmann-Gilmore, Randy Hastings, and Brad Manion for their technical assistance, Joe Henry Steinbach for his advice and support and Jeanne Nerbonne and Nick Franks for their suggestions about the manuscript.
Footnotes
* This work was supported by a grant from the National Institute of General Medical Sciences PO1-GM37846 and NIH National Center for Research Resources P41 RR00954 and UL1 RR024992.
 This article contains two supplemental Tables and supplemental Fig. S1–S6.
This article contains two supplemental Tables and supplemental Fig. S1–S6.
1 The abbreviations used are:
- IMP
- integral membrane protein
- LC
- liquid chromatography
- ESI
- electrospray ionization
- TMD
- transmembrane domain
- FA
- formic acid
- PGC
- porous graphitic carbon
- ACN
- acetonitrile
- SPE
- solid phase extraction
- MS1
- Full scan spectral acquisition
- MS2
- tandem MS to acquire peptide fragmentation mass spectra
- DDA
- data-dependent acquisition
- DMS
- directed MS2 spectral
- nano-LC-MS
- nanoliter-flow liquid chromatograph interfaced to a hybrid linear quadrupole ion trap-Orbitrap mass spectrometer.
REFERENCES
- 1. Choudhary C., Mann M. (2010) Decoding signalling networks by mass spectrometry-based proteomics. Nat. Rev. Mol. Cell Biol. 11, 427–439 [DOI] [PubMed] [Google Scholar]
- 2. Hall M. A., Xi J., Lor C., Dai S., Pearce R., Dailey W. P., Eckenhoff R. G. (2010) m-Azipropofol (AziPm) a photoactive analogue of the intravenous general anesthetic propofol. J. Med. Chem. 53, 5667–5675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Almén M. S., Nordström K. J., Fredriksson R., Schiöth H. B. (2009) Mapping the human membrane proteome: a majority of the human membrane proteins can be classified according to function and evolutionary origin. BMC Biol. 7, 50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Speers A. E., Wu C. C. (2007) Proteomics of integral membrane proteins–theory and application. Chem. Rev. 107, 3687–3714 [DOI] [PubMed] [Google Scholar]
- 5. Sprenger R. R., Jensen O. N. (2010) Proteomics and the dynamic plasma membrane: Quo Vadis? Proteomics 10, 3997–4011 [DOI] [PubMed] [Google Scholar]
- 6. Wu C. C., MacCoss M. J., Howell K. E., Yates J. R., 3rd (2003) A method for the comprehensive proteomic analysis of membrane proteins. Nat. Biotechnol. 21, 532–538 [DOI] [PubMed] [Google Scholar]
- 7. Kang S. U., Fuchs K., Sieghart W., Lubec G. (2008) Gel-based mass spectrometric analysis of recombinant GABA(A) receptor subunits representing strongly hydrophobic transmembrane proteins. J. Proteome Res. 7, 3498–3506 [DOI] [PubMed] [Google Scholar]
- 8. Chen E. I., Cociorva D., Norris J. L., Yates J. R., 3rd (2007) Optimization of mass spectrometry-compatible surfactants for shotgun proteomics. J. Proteome Res. 6, 2529–2538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Zybailov B., Coleman M. K., Florens L., Washburn M. P. (2005) Correlation of relative abundance ratios derived from peptide ion chromatograms and spectrum counting for quantitative proteomic analysis using stable isotope labeling. Anal. Chem. 77, 6218–6224 [DOI] [PubMed] [Google Scholar]
- 10. Marionneau C., LeDuc R. D., Rohrs H. W., Link A. J., Townsend R. R., Nerbonne J. M. (2009) Proteomic analyses of native brain K(V)4.2 channel complexes. Channels 3, 284–294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Speers A. E., Blackler A. R., Wu C. C. (2007) Shotgun analysis of integral membrane proteins facilitated by elevated temperature. Anal. Chem. 79, 4613–4620 [DOI] [PubMed] [Google Scholar]
- 12. Kang S. U., Lubec G. (2009) Complete sequencing of GABAA receptor subunit beta 3 by a rapid technique following in-gel digestion of the protein. Electrophoresis 30, 2159–2167 [DOI] [PubMed] [Google Scholar]
- 13. Ryan C. M., Souda P., Bassilian S., Ujwal R., Zhang J., Abramson J., Ping P., Durazo A., Bowie J. U., Hasan S. S., Baniulis D., Cramer W. A., Faull K. F., Whitelegge J. P. (2010) Post-translational modifications of integral membrane proteins resolved by top-down Fourier transform mass spectrometry with collisionally activated dissociation. Mol. Cell. Proteomics 9, 791–803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Olsen R. W., Sieghart W. (2009) GABAA receptors: subtypes provide diversity of function and pharmacology. Neuropharmacology 56, 141–148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Evers A. S., Chen Z. W., Manion B. D., Han M., Jiang X., Darbandi-Tonkabon R., Kable T., Bracamontes J., Zorumski C. F., Mennerick S., Steinbach J. H., Covey D. F. (2010) A synthetic 18-norsteroid distinguishes between two neuroactive steroid binding sites on GABAA receptors. J. Pharmacol. Exp. Ther. 333, 404–413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Jechlinger M., Pelz R., Tretter V., Klausberger T., Sieghart W. (1998) Subunit composition and quantitative importance of hetero-oligomeric receptors: GABAA receptors containing alpha 6 subunits. J. Neurosci. 18, 2449–2457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Slany A., Zezula J., Tretter V., Sieghart W. (1995) Rat beta 3 subunits expressed in human embryonic kidney 293 cells form high affinity [35S]t-butylbicyclophosphorothionate binding sites modulated by several allosteric ligands of gamma-aminobutyric acid type A receptors. Mol. Pharmacol. 48, 385–391 [PubMed] [Google Scholar]
- 18. Tretter V., Ehya N., Fuchs K., Sieghart W. (1997) Stoichiometry and assembly of a recombinant GABAA receptor subtype. J. Neurosci. 17, 2728–2737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Havlis J., Thomas H., Sebela M., Shevchenko A. (2003) Fast-response proteomics by accelerated in-gel digestion of proteins. Anal. Chem. 75, 1300–1306 [DOI] [PubMed] [Google Scholar]
- 20. King J. B., Gross J., Lovly C. M., Rohrs H., Piwnica-Worms H., Townsend R. R. (2006) Accurate mass-driven analysis for the characterization of protein phosphorylation. Study of the human Chk2 protein kinase. Anal. Chem. 78, 2171–2181 [DOI] [PubMed] [Google Scholar]
- 21. Nittis T., Guittat L., LeDuc R. D., Dao B., Duxin J. P., Rohrs H., Townsend R. R., Stewart S. A. (2010) Revealing novel telomere proteins using in vivo cross-linking, tandem affinity purification, and label-free quantitative LC-FTICR-MS. Mol. Cell. Proteomics 9, 1144–1156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Keller A., Nesvizhskii A. I., Kolker E., Aebersold R. (2002) Empirical statistical model to estimate the accuracy of peptide identifications made by MS/MS and database search. Anal. Chem. 74, 5383–5392 [DOI] [PubMed] [Google Scholar]
- 23. Nesvizhskii A. I., Keller A., Kolker E., Aebersold R. (2003) A statistical model for identifying proteins by tandem mass spectrometry. Anal. Chem. 75, 4646–4658 [DOI] [PubMed] [Google Scholar]
- 24. Meyer M. R., Lichti C. F., Townsend R. R., Rao A. G. (2011) Identification of in vitro autophosphorylation sites and effects of phosphorylation on the Arabidopsis CRINKLY4 (ACR4) receptor-like kinase intracellular domain: insights into conformation, oligomerization, and activity. Biochemistry 50, 2170–2186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Neubert H., Bonnert T. P., Rumpel K., Hunt B. T., Henle E. S., James I. T. (2008) Label-free detection of differential protein expression by LC/MALDI mass spectrometry. J. Proteome Res. 7, 2270–2279 [DOI] [PubMed] [Google Scholar]
- 26. Strohalm M., Kavan D., Novak P., Volný M., Havlicek V. (2010) mMass 3: a cross-platform software environment for precise analysis of mass spectrometric data. Anal. Chem. 82, 4648–4651 [DOI] [PubMed] [Google Scholar]
- 27. Zhang Y., Wen Z., Washburn M. P., Florens L. (2010) Refinements to label free proteome quantitation: how to deal with peptides shared by multiple proteins. Anal. Chem. 82, 2272–2281 [DOI] [PubMed] [Google Scholar]
- 28. Li G. D., Chiara D. C., Sawyer G. W., Husain S. S., Olsen R. W., Cohen J. B. (2006) Identification of a GABAA receptor anesthetic binding site at subunit interfaces by photolabeling with an etomidate analog. J. Neurosci. 26, 11599–11605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Dostalova Z., Liu A., Zhou X., Farmer S. L., Krenzel E. S., Arevalo E., Desai R., Feinberg-Zadek P. L., Davies P. A., Yamodo I. H., Forman S. A., Miller K. W. (2010) High-level expression and purification of Cys-loop ligand-gated ion channels in a tetracycline-inducible stable mammalian cell line: GABAA and serotonin receptors. Protein Sci. 19, 1728–1738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Kyte J., Doolittle R. F. (1982) A simple method for displaying the hydropathic character of a protein. J. Mol. Biol. 157, 105–132 [DOI] [PubMed] [Google Scholar]
- 31. Buller A. L., Hastings G. A., Kirkness E. F., Fraser C. M. (1994) Site-directed mutagenesis of N-linked glycosylation sites on the gamma-aminobutyric acid type A receptor alpha 1 subunit. Mol. Pharmacol. 46, 858–865 [PubMed] [Google Scholar]
- 32. Carr S. A., Roberts G. D. (1986) Carbohydrate mapping by mass spectrometry: a novel method for identifying attachment sites of Asn-linked sugars in glycoproteins. Anal. Biochem. 157, 396–406 [DOI] [PubMed] [Google Scholar]
- 33. McKernan R. M., Whiting P. J. (1996) Which GABAA-receptor subtypes really occur in the brain? Trends Neurosci. 19, 139–143 [DOI] [PubMed] [Google Scholar]
- 34. Sieghart W., Sperk G. (2002) Subunit composition, distribution and function of GABA(A) receptor subtypes. Curr. Top. Med. Chem. 2, 795–816 [DOI] [PubMed] [Google Scholar]
- 35. Huntsman M. M., Tran B. V., Potkin S. G., Bunney W. E., Jr., Jones E. G. (1998) Altered ratios of alternatively spliced long and short gamma2 subunit mRNAs of the gamma-amino butyrate type A receptor in prefrontal cortex of schizophrenics. Proc. Natl. Acad. Sci. U.S.A. 95, 15066–15071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Li X., Serwanski D. R., Miralles C. P., Bahr B. A., De Blas A. L. (2007) Two pools of Triton X-100-insoluble GABA(A) receptors are present in the brain, one associated to lipid rafts and another one to the post-synaptic GABAergic complex. J. Neurochem. 102, 1329–1345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Patel H. H., Murray F., Insel P. A. (2008) G-protein-coupled receptor-signaling components in membrane raft and caveolae microdomains. Handb. Exp. Pharmacol. 167–184 [DOI] [PubMed] [Google Scholar]
- 38. Hou Q., Huang Y., Amato S., Snyder S. H., Huganir R. L., Man H. Y. (2008) Regulation of AMPA receptor localization in lipid rafts. Mol. Cell. Neurosci. 38, 213–223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Zielinska D. F., Gnad F., Wiśniewski J. R., Mann M. (2010) Precision mapping of an in vivo N-glycoproteome reveals rigid topological and sequence constraints. Cell 141, 897–907 [DOI] [PubMed] [Google Scholar]
- 40. Mohammed S., Heck A., Jr. (2011) Strong cation exchange (SCX) based analytical methods for the targeted analysis of protein post-translational modifications. Curr. Opin. Biotechnol. 22, 9–16 [DOI] [PubMed] [Google Scholar]