Abstract
Some HLA class I molecules bind a significant fraction of their constitutive peptidomes in the presence of proteasome inhibitors. In this study, A*68:01-bound peptides, and their parental proteins, were characterized through massive mass spectrometry sequencing to refine its binding motif, including the nearly exclusive preference for C-terminal basic residues. Stable isotope tagging was used to distinguish proteasome-inhibitor sensitive and resistant ligands. The latter accounted for less than 20% of the peptidome and, like in HLA-B27, arose predominantly from small and basic proteins. Under the conditions used for proteasome inhibition in vivo, epoxomicin and MG-132 incompletely inhibited the hydrolysis of fluorogenic substrates specific for the tryptic or for both the tryptic and chymotryptic subspecificities, respectively. This incomplete inhibition was also reflected in the cleavage of synthetic peptide precursors of A*68:01 ligands. For these substrates, the inhibition of the proteasome resulted in altered cleavage patterns. However these alterations did not upset the balance between cleavage at peptide bonds resulting in epitope destruction and those leading to their generation. The results indicate that inhibitor-resistant HLA class I ligands are not necessarily produced by non-proteasomal pathways. However, their generation is not simply explained by decreased epitope destruction upon incomplete proteasomal inhibition and may require additional proteolytic steps acting on incompletely processed proteasomal products.
Major Histocompatibility complex class I (MHC-I)1 molecules constitutively bind and present at the cell surface large peptide repertoires for recognition by CD8+ T cells. These peptides arise mainly from proteasomal degradation of endogenous proteins (1). Alternative pathways seem to have a limited contribution, although they generate some of these ligands (2–12). The proteasome is a complex protease located in the cytosol and nucleus. Its catalytic core, designated as the 20S proteasome, consists of a four-ring barrel structure. Each of the two external rings is formed by 7 noncatalytic α subunits, whereas the two inner rings contain seven β subunits, three of which, β1, β2, and β5 are catalytic. These three constitutive subunits can be replaced, upon γ-interferon stimulation, by homologous inducible subunits, to form the immunoproteasome (13). The 20S core binds the PA700 activator to form the 26S proteasome, which carries out the ATP-dependent degradation of ubiquitilated proteins, and can also interact with the PA28 regulator, which increases dual cleavage (14, 15). Although the proteasome can cleave virtually any peptide bond, three major subspecificities are distinguished: tryptic-like, after basic residues, chymotryptic-like, after aliphatic/aromatic residues, and caspase-like, after acidic residues, which can be assigned to β2, β5, and β1, respectively (16–20).
MHC-I molecules differ widely in their capacity to bind and present self-derived ligands in the presence of proteasome-inhibitors (21, 22). We previously reported that a significant fraction of the HLA-B27-bound peptidome was efficiently presented in cells treated with epoxomicin and MG-132 in a way essentially unaffected by the concentration of inhibitor. These inhibitor-resistant peptides predominantly arose from small basic proteins (23). This study was consistent with an undefined proteasome-independent proteolytic pathway with preference for this type of proteins. However, there were several problems with this interpretation. The inhibition of the proteasome was not directly measured in our previous study. Total inhibition of this enzyme is difficult to achieve (24), and degradation of small basic proteins might be particularly unaffected if the tryptic-like subspecificity is incompletely inhibited. Moreover, incomplete inhibition of the proteasome may lead to altered cleavage, because impairment of a catalytic site may allosterically modulate the activity of other sites (25). Because proteasomes not only produce MHC ligands and their precursors, but also degrade them by hydrolyzing their internal peptide bonds, altered cleavage by partially inhibited proteasomes may increase presentation of some ligands by favoring their production and/or decreasing their degradation (26).
To address these issues, we focused on the origin and proteasome-dependence of the HLA-A*68:01-bound peptidome. Like HLA-B27, the surface expression of HLA-A68 is relatively high in the presence of proteasome inhibitors (22). In addition, this allotype binds almost exclusively peptides with C-terminal basic residues, so that it might be particularly susceptible to incomplete inhibition of the proteasomal tryptic-like activity (24). Our study was designed as follows. First, the peptide motif of A*68:01, currently based on a limited set of ligands (http://www.syfpeithi.de), was refined through extensive sequencing of A*68:01-bound peptides, and their presumed parental proteins were identified. Second, by using stable isotope tagging, the percentage of proteasome inhibitor sensitive and resistant peptides was determined and multiple members of each subset, as well as their parental proteins, were identified. Third, the inhibition of the tryptic and chymotryptic-like proteasomal subspecificities in the conditions used for the analysis of the A*68:01 peptidome was quantified with fluorogenic substrates. Four, the effect of proteasome inhibitors on the generation and destruction of specific A*68:01 ligands and on altering cleavage patterns was analyzed with synthetic peptide substrates.
EXPERIMENTAL PROCEDURES
Cell Lines, Monoclonal Antibodies (mAb), and Inhibitors
C1R is a human lymphoid cell line with low expression of its endogenous HLA class I molecules (27). C1R-A*68:01 transfectants (22) were a kind gift of Dr. V. Engelhard (University of Virginia School of Medicine, USA). Cells were cultured in Roswell Park Memorial Institute 1640 medium supplemented with 10% fetal bovine serum (FBS) (both from Invitrogen, Paisley, UK). The mAb W6/32 (IgG2a; specific for a monomorphic HLA class I determinant) (28) was used. Epoxomicin, an irreversible and specific inhibitor of the proteasome (29) and carbobenzoxy-l-leucyl-l-leucyl-l-leucinal (MG-132) (1), a potent reversible inhibitor of the proteasome and calpains, were from Calbiochem (Schwalbach, Germany).
Electrospray-Orbitrap MS Analysis
Peptide mixtures were desalted and concentrated with Micro-Tip reverse-phase columns (C18, 200 μl, Harvard Apparatus, Holliston, MA). Each C18 tip was prepared by washing with 80% acetonitrile in 0.1% trifluoroacetic acid (TFA) followed by pre-equilibration with two washes with 150 μl of 0.1% TFA and then loaded with the peptide mixture. The C18 tips were washed twice again with 0.1% TFA, the peptides were eluted with 80% acetonitrile in 0.1% TFA, and concentrated to ∼18 μl using vacuum centrifugation.
The HLA class I peptides were analyzed by μLC-MS/MS using an Orbitrap XL mass spectrometer (Thermo Fisher, San Jose, CA) fitted with a capillary high-performance liquid chromatography (HPLC) (Eksigent, Dublin, CA). The peptides were resolved on a C18 trap column (0.3 × 5 mm, LC-Packings) connected on-line to a 75 micron I.D. fused silica capillaries (J&W, Folsom, CA) self-packed with 3.5 micron Reprosil C18 (Dr. Maisch, GmbH, Germany) as in (30), and eluted at flow rates of 0.25 μl/min, with linear gradients of 7–40% acetonitrile in 0.1% formic acid, during 90 min, followed by 15 min at 95% acetonitrile in 0.1% formic acid. The spectra were collected in the orbitrap mass analyzer using full ion scan mode over the mass-to-charge (m/z) range 400–2,000, which was set to 60,000 resolutions. The most intense seven masses from each full mass spectrum, with single, double and triple charge states, were selected for fragmentation by collision-induced disintegration in the linear ion-trap
Database Searches
Pep-Miner (31) was used for generation of the peak-lists based on the μLC-MS/MS data. The peptides were identified using multiple search engines: Pep-Miner, Sequest (Thermo-Fisher) (32) and Mascot (server 2.2, Matrix Science Inc. Boston, MA) (33) searched against the human part of the Uniprot database (http://www.uniprot.org, Jan 2009) including 20,332 proteins. The Sequest and Mascot search results were combined into one report by Proteome Discoverer 1.0 SP1 (Thermo-Fisher). The search was not limited by enzymatic specificity, the peptide tolerance was set to 0.01 Da. and the fragment ion tolerance was set to 0.5 Da. Oxidized methionine was searched as a variable modification.
Peptide identifications were accepted if their masses were below 1500 Da. and mass accuracy was better than 0.005 Da. Other required criteria were: Pep-Miner Score ≥80, Sequest Xcore ≥2.5, Sequest Probability ≥5, Mascot ionScore/Mascot high score ≥0.6, Mascot Expectation value ≥0.1, delta to next score = 0. Peptides were filtered also according to their fitness to the HLA-A68 consensus and were accepted if their HLA-A68 score was ≥20 (according to http://www-bimas.cit.nih.gov/molbio/hla_bind/) (34). Some peptides were also observed with different charge-states and as overlapping, longer or shorter peptides by one amino acid, listed in the table as variants. The false discovery rate of the A68 peptides adhering to at least three or more of these criteria was <3.9%. This was calculated by running the same data against the corresponding randomized database and calculating the ratio between the results from the random and Uniprot databases.
Matrix-assisted Laser Desorption-Ionization Time-of-Flight (MALDI-TOF) MS
This was done in a MALDI-TOF/TOF instrument (4800 Proteomics Analyzer, Applied Biosystems, Foster City, CA) as previously described (35). MS data were acquired in the mass range 800–2000 Da (or 700–4000 for synthetic substrate digests) in reflector positive mode at 25kV and analyzed using the Data Explorer software version 4.9 (Applied Biosystems). Each spectrum was externally calibrated with the Peptide Calibration Standard Mixture (Brucker, Product # 206195) to reach a typical mass measurement accuracy of <25 ppm.
MALDI-TOF/TOF MS/MS spectra were acquired with the same software at 1Kv, using collision-induced dissociation with atmospheric air, and a precursor mass window of ±2.5 Da. A signal-to-noise ratio of three was used for processing data. Interpretation of the MS/MS spectra was done manually, but assisted by various tools as follows. Manual inspection of the spectrum, which was facilitated by using a simple tool implemented as an Excel macro, usually allowed us to derive a tentative sequence. This was used to screen the human proteome in the human protein entries of the Uniprot/Swiss-prot database (Release 14.0: 2008/07/22, with 19329 entries), using a window of 0.5 m/z units for both precursor and fragment ions, for a possible match using the Mascot server 2.2 software. For those sequences showing the highest scores in this preliminary search, the MS-product tool (version 5.1.8) at http://prospector.ucsf.edu (University of California, San Francisco), which generates a list of theoretical fragment ions from a given peptide sequence, was used to match the candidate sequences to our experimental MS/MS spectra. Alternatively, a new software (designated as MSgene) was used for de novo generation of candidate sequences. This tool is based on a genetic algorithm in which an initial population of random peptide sequences encoded in 5-bit codon units, is subjected to an iterative process of optimization, by means of recombination and mutation, leading to convergence toward candidate sequences with improved fit to the experimental MS/MS spectrum (supplemental Fig. S1). These candidate sequences were subsequently analyzed with the MS-product tool as above. Some of these sequences were confirmed with the corresponding synthetic peptides.
Isotopic Labeling
This was done as previously described (23) with minor modifications. Briefly, C1R-A*68:01 transfectants (about 5 × 108 cells per flask) were incubated for 4 h in Roswell Park Memorial Institute 1640 medium without Arg or, in some experiments, Lys, supplemented with 10% dialyzed FBS. One flask was then supplemented with standard (14N) Arg or Lys (100 μg/ml), a second one with 100 μg/ml of l-Arg-guanido[15N2] ·HCl (Cambridge Isotope Laboratories, Andover, MA), or l-Lys-[15N2]·HCl, both reagents containing two 15N atoms, and the third flask was treated with 20 μm MG-132 for 30 min prior to the addition of the 15N-tagged amino acid, and left for the entire labeling period. After 5 h, the cells were washed twice in 20 mm Tris-HCl, 150 mm NaCl, pH 7.5. Pellets were stored at −70 °C in the presence of PMSF (50 μl of a 70 mg/ml solution) for further processing. In other experiments 2.5 μm epoxomicin was used in the same conditions, except that starving of cells in the absence of Arg before labeling was carried out for 12 h. All incubations were done at 37 °C. Peptide labeling was quantified by the labeling ratio, which was defined as follows. Ratio = [(15N + inh.)- 14N]/(15N-14N), where 14N, 15N, and (15N + inh.) are the percent intensities of the relevant isotopic peak, relative to the monoisotopic one, in the MALDI-TOF spectrum of the peptide, in the absence of labeling, upon labeling in the absence of inhibitor, and in its presence, respectively.
Isolation of HLA-A*68:01-bound Peptides
This was carried out as previously described (36). Briefly, cells were lysed in 1% Igepal CA-630 (Sigma-Aldrich, St Louis, MO) in the presence of a mixture of protease inhibitors. The soluble fraction was subjected to affinity chromatography using the W6/32 mAb. HLA-A68 -bound peptides were eluted with 0.1% TFA at room temperature, filtered through Centricon 3 (Amicon, Beverly, MA), concentrated and either used as a peptide pool for sequencing, or subjected to HPLC fractionation as previously described (37). The peptides to be sequenced from individual HPLC fractions were identified as those with the same monoisotopic mass and retention time as the labeled peptides in a parallel chromatography of an unlabeled peptide pool obtained from 1010 cells, by comparing the MALDI-TOF spectra of correlative and highly matched HPLC fractions from this peptide pool and those from the labeling experiments (23).
Analysis of Residue Frequencies Among A*68:01 Ligands
This was automatically done using an updated version of the previously described MSearcher software (38).
Assignment and Analysis of the Parental Proteins of HLA-A68 Ligands
This was done on the basis of unambiguous matching with a single human protein in the UniProtKB database release 14.0: 2008/07/22 using the Fasta 3 software (htpp://www.ebi.ac.uk/fasta), after taking into account the database redundancy because of multiple entries for the same protein. When a peptide ligand matched several closely related members of a protein family, a single entry for a representative member was chosen, with the understanding that the same ligand can arise from more than one member of such families.
The molecular mass (MW) and theoretical isoelectric point (pI) of the assigned proteins was calculated with the Compute pI/Mw tool (htpp://www.expasy.org/tools/pi_tool.html). Subcellular localization of the proteins was obtained from the UniProtKB release 14.0: 2008/07/22 database using the Protein Information and Knowledge Extractor software (39), available at http://proteo.cnb.csic.es:8080/pike. Human proteome analysis was performed with the 19329 entries in the UniProtKB/Swiss-Prot database using the Compute pI/Mw tool as above. Statistical analyses were carried out using the χ2 test.
Proteasome Purification and Hydrolysis of Fluorogenic Substrates
Preparation of cell extracts containing 26S proteasome was performed as previously described (40). Briefly, about 2 × 106 cells were cultured for 1 h in phenol red-free Ivecco's modified eagle's medium (Invitrogen), supplemented with 10% FBS, in the presence or absence of proteasome inhibitors. Cells were washed twice with phosphate-buffered saline, permeabilized with 1.4 ml of 50 mm Tris-HCl, pH 7.5, 250 mm sucrose, 5 mm MgCl2, 0.5 mm EDTA, 1 mm dithiothreitol, 1 mm ATP, 0.025% digitonin (homogenization buffer), for 5 min at 4 °C, and centrifuged (2 × 104 g, 10 min). Supernatants containing the cytosol were further centrifuged (3 × 105 g, 2 h) to separate large protein complexes, including the 26S proteasome, and these pellets were resuspended in 1 ml of digitonin-free homogenization buffer. Analysis of the tryptic and chymotryptic activity was performed with 100 μm Boc-LRR-amc or Suc-LLVY-amc (both from Bachem, St. Helens, UK), respectively. The cell extracts were incubated with the fluorogenic substrates at 37 °C for 1 h, and aliquots were taken every 10 min. The reaction was stopped by adding 0.33% TFA. Fluorescence was measured in an Aminco-Bowman series 2 fluorimeter (Sim-Aminco Spectronic Instruments, Rochester, NY) with excitation and emission wavelengths of 380 and 460 nm, respectively. The hydrolysis rate was calculated as the ratio between the slope of the reaction progress curves in the presence and absence of inhibitor. 20S proteasomes were purified from C1R cells by ion exchange chromatography and centrifugation in glycerol gradient as previously described (37).
In-gel Digestion Assays
Cell extracts containing 26S proteasome from 2 × 106 cells were loaded into a 3% polyacrylamide gel, in 90 mm Tris/90 mm boric acid pH 8.3, 5 mm MgCl2, 0.5 mm EDTA, 1 mm ATP-MgCl2 (resolving buffer), and run for 3 h at 110 V and 4 °C as previously described (41). Other samples were run in a 3–14% gradient gel in the same buffer. This process was also carried out changing the electrophoretic polarity in some experiments. After electrophoresis, the gels were washed with 50 mm Tris-HCl, pH 7.4, 5 mm MgCl2, 1 mm ATP (developing buffer), and incubated in 1 ml of this buffer with 50 μm of the fluorogenic substrate for 30 min at 30 °C. Gels were visualized in a UV Transiluminator.
In Vitro Digestions of Synthetic Precursors
Two synthetic peptide substrates, a 31-mer and a 30-mer, were used. They were obtained using N-(fluorenyl)-methocarbonyl chemistry and purified by reverse phase HPLC. Their purity and correct molecular mass was established by MALDI-TOF MS. About 20 μg of the substrate were incubated for 8h at 37 °C with 2 μg of purified 20S proteasome with and without inhibitors, in 25 mm HEPES buffer, pH 7.6. The proteasome was incubated with the inhibitors for 2 h, previous to the addition of the substrate. The reaction was stopped with 0.067% TFA. For the 31-mer the digestion products were separated from the proteasome by molecular filtrating using 10 kDa pore size Centricon and fractionated by HPLC as described (37). The HPLC fractions were analyzed by MALDI-TOF/TOF MS. For the 30-mer the digestion mixture was purified with OMIX C18 tips (Agilent technologies, Santa Clara. CA), after washing with 0.2% TFA, then with 40% acetonitrile in 0.1% TFA, and finally eluting the peptides with 70% acetonitrile in 0.1% TFA. The digestion mixture was directly analyzed by MALDI-TOF MS/MS. The digestion products were identified using a software tool, designated as Digestor, developed as a Windows application. This tool assigns each ion peak to possible internal sequences in the precursor substrate on the basis of its molecular mass. If an ion peak matched several possible internal sequences, the ambiguity was either resolved by sequencing or not taken into account. The yield of individual digestion products was estimated as follows. The percentage of the total absorbance at 210 nm corresponding to a given chromatographic peak was determined. When multiple digestion products co-eluted in a same peak a percentage of its absorbance was assigned to each peptide on the basis of the relative intensity of the ion peaks in the MALDI-TOF spectrum of the corresponding HPLC fractions. Alternatively, the relative yields of individual digestion products were calculated directly from the relative intensities of the ion peaks in the MALDI-TOF MS of the unfractionated digestion mixture.
RESULTS
The A*68:01 Peptide Motif
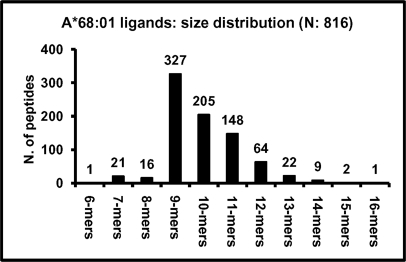
This was determined from a database of natural self-ligands, built from 3 sources. First, 35 sequences were available from the SYFPEITHI database of MHC ligands (http://www.syfpeithi.de). Second, 772 sequences were obtained from the A*68:01-bound peptide pool isolated from C1R transfectants by electrospray-Orbitrap MS/MS. The false discovery rate of this data set was 3.9%. Third, 70 sequences were obtained from the same peptide pool by MALDI-TOF/TOF MS/MS. Since some peptides were sequenced by more than one procedure, their total number was 816 (Table 1S), with a size range of 6 to 16 amino acid residues (Fig. 1). The A*68:01 peptide motif was determined from the 778 nonamer and longer peptides. The analysis was performed with this total set for the 5 anchor positions (P)1, P2, P3, PC-2, and PC (Fig. 2), because these can be properly aligned among peptides with different sizes. In addition, residue usage at all positions was analyzed separately for 9-mers, 10-mers, 11-mers and 12-mers (supplemental Fig. S2 and supplemental Table S2).
Fig. 1.
Size distribution among natural A*68:01 peptide ligands. The total of 816 A*68:01 ligands, includes 35 previously available (htpp://www.syfpeithi.de) and 781 newly determined sequences.
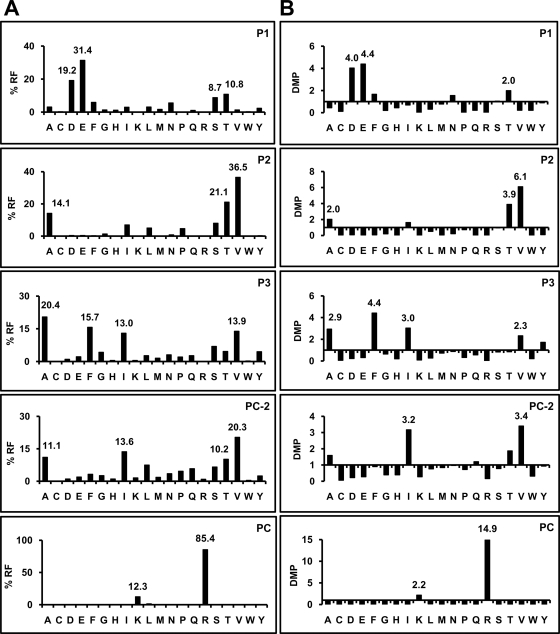
Fig. 2.
Residue usage among A*68:01 ligands longer than octamers. A, percent residue frequencies (% RF) and B, deviation relative to the mean frequency in the human proteome (DMP) among 778 A*68:01 ligands ranging from 9 to 16 residues were determined at positions (P)1, P2, P3, PC-2, and PC. At each position the % RF and DMP values of the predominant residues is highlighted.
At all anchor positions some residues were predominant (Fig. 2). At P1, acidic residues were found in 50.6% of the ligands, followed by T and S. P2 was restricted by size rather than polarity, since residues in the range of 71–103 Da accounted for 84.4%, but both polar (T) and aliphatic residues (A, V) were tolerated. At P3, aliphatic residues and F were predominant. PC-2 showed preference for aliphatic residues (54.4%), but uncharged polar ones were also frequent (26.1%). At PC, K and R jointly accounted for 97.7% of the ligands, with a minor allowance for nonpolar residues. When residue frequencies were made relative to the human proteome a good correlation was found between the frequency of a given residue at any given anchor position and an increase or decrease relative to the proteome, indicating positive or negative selection of that residue among A68 ligands. The few peptides with nonpolar C-terminal residues (2.3%) are unlikely to be contaminant HLA-C*04 ligands, expressed on C1R cells, since they have features typical of A*68:01, but not of C*04 ligands, such as acidic P1 residues, and lack the P2 F/Y motif of C*04 (supplemental Table S1). Residue frequencies among peptides of different length were small and not statistically significant (supplemental Fig. S2). At non-anchor positions residue restrictions were low (supplemental Table S2).
One hexamer, 21 heptamers and 16 octamers were sequenced from the A*68:01-bound pool. Residue usage among these peptides deviated significantly from longer ones at P1-P3, but not at PC or PC-2 (supplemental Fig. S3). Longer variants, frequently consisting of N-terminal extensions, were found for many of these ligands.
Parental Proteins of A*68:01 Ligands
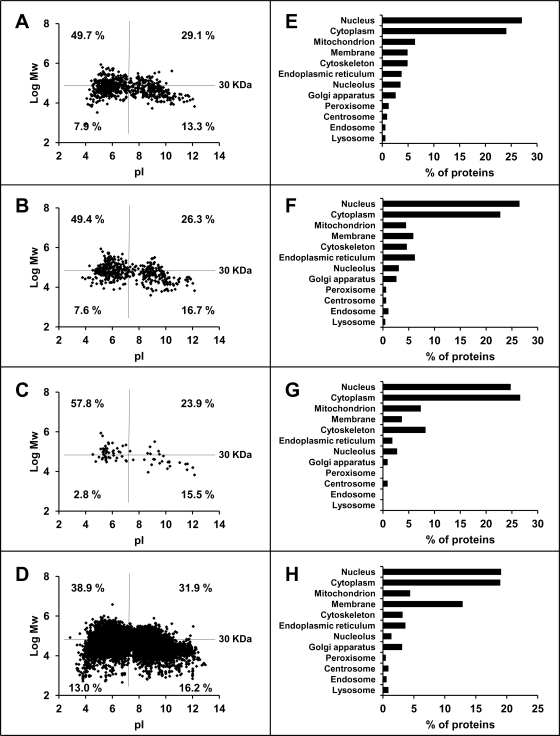
The 816 A*68:01 ligands arose from 672 parental proteins whose MW and pI distribution was similar as in HLA-B*27:05 (42) and showed, relative to the human proteome, a higher percentage of large (>30 KD) acidic proteins and a decreased percentage of small (<30 KD) acidic ones (Figs. 3A–3D). These differences were statistically significant. The majority of proteins were from the cytoplasm and nucleus and much lower percentages were from other subcellular compartments (Fig. 3E). This was similar to B*27:05 (Fig. 3F) and other MHC class I molecules (43).
Fig. 3.
Parental proteins of A*68:01 ligands and their relationship to those of HLA-B27 and to the human proteome. A–D, the molecular mass (log Mw) was plotted versus the isoelectric point (pI) for (A) the parental proteins (n = 672) of A*68:01 ligands, (B) those of B*27:05 ligands (n = 471) (42), (C) parental proteins (n = 74) common to A*68:01 and B*27:05 ligands, and (D) the human proteome (UniProtKB database release 14.0: 2008/07/22). In each panel, the proteins are classified by size (using a 30KD threshold) and pI, and the percentage of each subset is specified. E–H, subcellular distribution of (E) the parental proteins of A*68:01 ligands, (F) those of B*27:05 ligands, (G) parental proteins common to A*68:01 and B*27:05 ligands, and (H) the human proteome. The analysis was carried out using the Protein Information and Knowledge Extractor software (39) at http://proteo.cnb.csic.es/pike/analysis.html.
A total of 74 proteins were the source of both A*68:01 or B*27:05 ligands (11% and 15.7%, respectively). This is about fourfold higher than expected if each set of parental proteins were independently selected in an unbiased way, suggesting that the ligands of both MHC molecules arise from a subset of the proteome. For example, membrane proteins were under-represented, relative to the whole proteome, among the sources of A68 and B27 ligands (Figs 3E–3H). Other factors such as abundance, turnover rate, degree of cotranslational degradation of defective ribosomal products, etc., may influence the contribution of a given protein to the MHC-I-bound peptidomes. The parental proteins common to A68 and B27 showed similar distribution of MW and pI as the total sets, except for a lower percentage of small acidic proteins (Fig. 3C).
Susceptibility and Resistance of A*68:01 Ligands to Proteasome Inhibitors
The expression of A*68:01 ligands was analyzed by stable isotope labeling, using 15N-tagged Arg or Lys, in the absence or presence of MG-132 or epoxomicin. The A*68:01-bound peptide pool was fractionated by HPLC, and each fraction was analyzed by MALDI-TOF MS. The peptides isolated from cells treated with 15N-tagged amino acids showed a different isotopic distribution compared with the same unlabeled peptides. As the tagged residues are 2 Da heavier than their nontagged counterpart, labeling was detected as an increase of the intensity of the A+2, A+4, etc. peaks (A being the monoisotopic peak), depending on the number of tagged peptide residues. The labeling of inhibitor-resistant ligands should be unaltered or only slightly decreased in the presence of inhibitor, because their generation would not be abrogated by inhibition of the proteasome. In contrast, little or no labeling should be detected in inhibitor-sensitive ligands isolated from cells treated with inhibitor plus 15N-tagged amino acids.
In a first set of experiments cells were labeled with 15N-tagged Arg or Lys in the presence of 20 μm MG-132. A total of 132 ion peaks (supplemental Table S3) were amenable to analysis on the following basis: (1) they showed sufficient intensity for good detection of the isotopic envelope, and (2) the relevant isotopic peak increased at least 20% upon 15N labeling in the absence of inhibitor, relative to unlabeled cells. A given ligand was assigned as inhibitor-resistant if its labeling ratio was >0.6, that is, if the increased intensity of the corresponding isotopic peak in the presence of the inhibitor was more than 60% of the increase obtained in its absence. This threshold was adopted because decreased label in the presence of inhibitor may be because of indirect effects, such as down-regulation of protein synthesis (44). The inhibitor-sensitive ligands were those showing labeling ratios <0.35. This threshold was adopted to allow for residual proteasome activity in the presence of inhibitors, as well as for inherent experimental error. On this basis 95 peptides (71%) were sensitive to proteasome inhibition, 24 (19%) were inhibitor-resistant, and 13 (10%) were not assigned, either because they showed intermediate labeling ratios, were more inhibited with epoxomicin, or both.
In a second experiment the cells were labeled with 15N-tagged Arg in the presence of 2.5 μm epoxomicin. Of 63 peptides amenable to this analysis (supplemental Table S4), 46 (73%) were inhibitor-sensitive, 8 (13%) were inhibitor-resistant and 9 (14%) were not assigned because of intermediate labeling ratios, less inhibition with MG-132 or both.
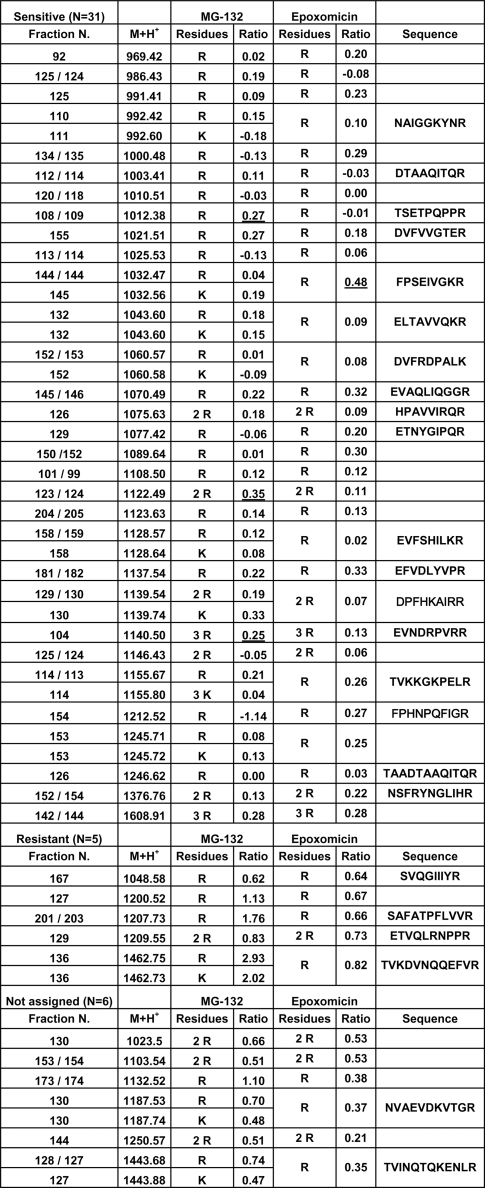
Of 42 peptides analyzed with both inhibitors 31 (74%) were sensitive, 5 (12%) were resistant and 6 (14%) were not assigned (Fig. 4). In the latter subset, 5 of the 6 peptides showed higher inhibition with epoxomicin.
Fig. 4.
Isotopic labeling of HLA-A*68:01 ligands in the presence of proteasome inhibitors. A total of 42 ion peaks were analyzed with both 20 μm MG-132 and 2.5 μm epoxomicin. Their elution position in HPLC (Fraction N), monoisotopic mass (M+H+), the number of Arg (R) or Lys (K) residues, which was inferred from the isotopic peak that increased upon 15N-labeling, and the labeling ratio (Ratio: see Exp erimental Procedures), are indicated. Peptides were classified as inhibitor-sensitive or -resistant when the labeling ratio was <0.35 or >0.6, respectively, with both inhibitors. Underlined labeling ratios for some inhibitor-sensitive ligands indicate distorted values due to inhibition of labeling close to background levels. When the labeling ratio showed intermediate values or was significantly different with both inhibitors, the peptides were not assigned. Within each set, the peptides are ordered by their M+H+. Ion peaks that were sequenced are indicated (see Tables 3S and 4S for full labeling data).
These results indicate that both inhibitors have a similar effect on most A*68:01 ligands. However, a higher inhibition by epoxomicin was suggested by the smaller percentage of resistant ligands, relative to MG-132, and by its more drastic effect on some peptides (Fig. 4).
The average intensity of the monoisotopic (M+H+) MALDI-TOF ion peaks corresponding to the peptides in Fig. 4 was significantly higher for inhibitor-sensitive than for inhibitor-resistant ligands (Table I), indicating that the resistance was not an artifact due to high peptide abundance. Indeed, many inhibitor-sensitive peptides showed ion peak intensities much higher than any of the resistant peptides. The lower intensity values observed in the presence of inhibitors affected equally to inhibitor-sensitive and -resistant peptides. It results from nonspecific global effects affecting peptide yields, such as decreased cell viability in the presence of inhibitors, and perhaps also from down-regulation of multiple cell biological processes following proteasome inhibition, including MHC-I maturation and export.
Table I. Mean intensity of the monoisotopic (M+H+) ion peaks of inhibitor-sensitive and resistant peptides. This analysis was performed for the peptides in Figure 4.
| MG-132 | Epoxomicin | |||||
|---|---|---|---|---|---|---|
| 14N | 15N | 15N+Inh. | 14N | 15N | 15N+Inh. | |
| Sensitive (n = 31) | 13461 | 11092 | 7026 (63%)c | 3001 | 2489 | 1644 (66%)c |
| Resistant (n = 5) | 2818 | 2364a | 1657 (70%)c | 551 | 356b | 176 (49%)c |
| Ratio | 4.8 | 4.7 | 4.2 | 5.4 | 7.0 | 9.3 |
a Range: 537 to 5030. As many as 20 inhibitor-sensitive peptides (64.5%) had higher intensity.
b Range: 56 to 602. As many as 17 inhibitor-sensitive peptides (54.8%) had higher intensity.
c Percent values are relative to peptides labeled in the absence of inhibitor (15N).
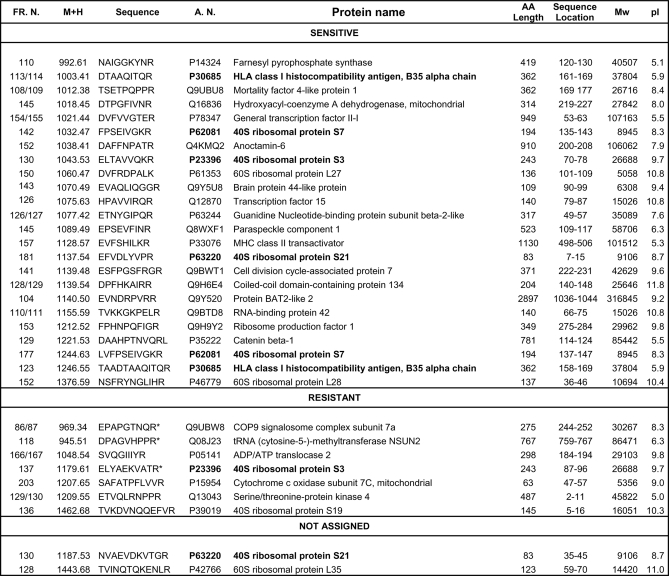
Identification of Inhibitor-resistant and Inhibitor-sensitive A*68:01 Ligands and Their Parental Proteins
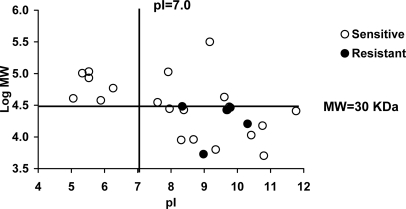
The sequence of 24 inhibitor-sensitive, seven inhibitor-resistant, and two peptides showing different susceptibility with both inhibitors was determined. All of the inhibitor-sensitive, the two nonassigned and five inhibitor-resistant peptides corresponded to internal sequences of their presumed parental proteins. Two inhibitor-resistant peptides corresponded to N-terminal (ETVQLRNPPR) or C-terminal (DPAGVHPPR) protein sequences (Fig. 5). There were no obvious differences between inhibitor-sensitive and resistant peptides in size, sequence motifs, or frequency of flanking residues (data not shown). In addition, abundance of the source protein was unrelated to inhibitor-resistance. For instance, abundant ribosomal proteins were the source of both types of ligands (Fig. 5). However, all 5 inhibitor-resistant peptides corresponding to internal protein sequences arose from small (30 kDa or smaller) and basic proteins. The two peptides arising from N- or C-terminal protein ends did not follow this rule. For inhibitor-sensitive ligands the parental proteins with MW<30 kDa and pI>7 accounted for 54.5% (Fig. 6).
Fig. 5.
Amino acid sequences of inhibitor-sensitive and inhibitor-resistant HLA-A*68:01 ligands. Peptides are classified as in Fig. 4 and ordered by their monoisotopic mass (M+H+) within each set. The corresponding parental proteins, their accession numbers (A.N.) in the Swiss-Prot database, amino acid (AA) length, location of the ligand in their sequence (sequence location), MW and theoretical pI are indicated. Polypeptides giving rise to more than one ligand are indicated in boldface. Resistant peptides marked with an asterisk (*) were assigned only on the basis of their lack of inhibition with MG-132.
Fig. 6.
Inhibitor-resistant A*68:01 ligands predominantly arise from small and basic proteins. The molecular mass (log MW) of the parental proteins from inhibitor-sensitive and -resistant A*68:01 ligands was plotted versus their theoretical pI. The value of 30 kDa corresponds to the highest molecular mass (30267 Da) observed among parental proteins of inhibitor-resistant ligands derived from internal sequences. The two parental proteins of ligands matching N- or C-terminal sequences were not included.
We found two examples of ligands derived from the same protein but showing differential sensitivity to proteasome inhibitors. First, two peptides (EFVDLYVPR and NVAEVDKVTGR) arising from the 40S ribosomal protein S21 behaved as inhibitor-sensitive and not assigned, respectively. A previously reported B27 ligand (GRFNGQFKTY) derived from the same protein was inhibitor-resistant (23). Second, ELTAVVQKR and ELYAEKVATR, from the 40S ribosomal protein S3 were inhibitor-sensitive and -resistant, respectively (Fig. 5).
Inhibition of the 26S Proteasome Subspecificities
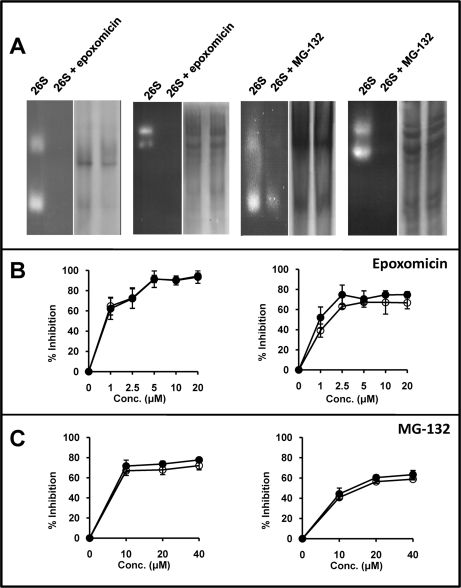
The effect of inhibitors on the chymotryptic- and tryptic-like activities of the proteasome from C1R cells was tested with specific fluorogenic substrates. When fractionated in native gels the 26S proteasome samples (Fig. 7A) contained a mixture of 26S and 20S proteasome, and some additional bands that were best observed in gradient electrophoresis. Hydrolysis of both substrates was observed in the 20S and 26S proteasome bands, but not elsewhere. Upon inverting the electrophoretic polarity, additional contaminants or hydrolytic activity were not observed (data not shown). These results suggest that the hydrolysis of the fluorogenic substrates is due to the proteasome. In the presence of 2.5 μm epoxomicin, all detectable hydrolysis of both substrates was inhibited. In contrast, with MG-132 the hydrolysis of the tryptic substrate was only partially inhibited and residual chymotryptic activity was also observed (Fig. 7A).
Fig. 7.
Activity of the 26S proteasome in the presence or absence of proteasome-inhibitors. A, In-gel hydrolysis of site-specific fluorogenic substrates by partially purified 26S proteasomes, which include significant amounts of 20S proteasomes, incubated or not with 2.5 μm epoxomicin or 20 μm MG-132. The experiments were performed in 3% polyacrylamide gel or 3–15% polyacrylamide gradient gels for the tryptic (Boc-LRR-amc) and chymotryptic (Suc-LLVY-amc) substrates, respectively. For each experiment the in-gel digestion analysis is shown on the left side. The same gel, washed and stained with Coomassie blue, is shown at right. B, Inhibition of the hydrolysis of fluorogenic substrates specific for the chymotryptic (left) or tryptic (right) activity by partially purified 26S proteasomes in the presence of various concentrations of epoxomicin. Empty circles (o) show the results obtained with proteasome purified from cells treated with the inhibitor. Black circles (●) show results obtained with proteasome treated with the inhibitor after its purification from cells. C, Inhibition of the hydrolysis of fluorogenic substrates specific for the chymotryptic (left) or tryptic (right) activity by partially purified 26S proteasomes in the presence of various concentrations of MG-132. Conventions are as in panel B.
In other experiments C1R-A*68:01 cells were incubated with or without inhibitor at various concentrations. After 1h, the cytosolic 26S proteasome was partially purified and its hydrolytic activity for the tryptic and chymotryptic substrates was determined by measuring the generation of fluorescence as a function of time. In parallel experiments, the 26S proteasome was purified from untreated C1R cells, incubated with inhibitors and tested in the same way. The results in both conditions were very similar, indicating that proteasomal inhibition was not significantly affected by incubating either whole cells or purified proteasome with MG-132 or epoxomicin (Figs. 7B–7C). The chymotryptic-like activity was totally inhibited (94 ± 5%) with 20 μm epoxomicin. At the 2.5 μm concentration used for the analysis of A*68:01 ligands, the inhibition of this subspecificity was high (72 ± 7%), but incomplete. The tryptic-like activity was inhibited to a maximum of about 67% at 5 μm or higher concentrations of epoxomicin, and nearly as much (63 ± 2%) at 2.5 μm. With MG-132 the maximal inhibition of the chymotryptic-and tryptic-like activities was obtained at 40 μm (72 ± 4% and 59 ± 1%, respectively), but similar values (68 ± 5% and 56 ± 2%, respectively) were observed at 20 μm, used for A*68:01 ligands.
These results indicate that: (1) inhibition of the tryptic- and chymotryptic-like specificities of the proteasome was higher with epoxomicin than with MG-132, (2) this inhibition was not complete, but was close to 100% with epoxomicin for the chymotryptic-like activity, (3) the residual hydrolysis of fluorogenic substrates is probably because of incomplete inhibition of the proteasome, rather than to non-proteasomal activity (Fig. 7A), and (4) in the conditions used for A*68:01 ligands both the tryptic- and the chymotryptic-like activities of the proteasome were incompletely inhibited.
Effect of Proteasome Inhibition on the Generation of Inhibitor-resistant Ligands In Vitro
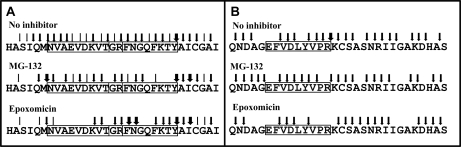
Incomplete inhibition may alter proteasomal cleavage patterns, resulting in decreased destruction and/or increased production of some MHC-I ligands, which might explain their resistance to inhibitors (26). To explore this possibility, a synthetic 31-mer spanning residues 29–59 from the 40S ribosomal protein S21 was digested in vitro with purified 20S proteasome in the absence or presence of inhibitors (Fig. 8A, Table II and supplemental Table S5). This substrate includes two overlapping sequences (NVAEVDKVTGR and GRFNGQFKTY) that correspond to an A*68:01 ligand showing partial resistance to proteasome inhibitors (Fig. 5) and to an inhibitor-resistant HLA-B27 ligand (23), respectively. In the absence of inhibitors, the substrate was totally degraded and cleavage occurred at virtually all peptide bonds. A total of 80 peptides, 66 of them requiring dual cleavage, were recovered with yields ≥ 0.1% (supplemental Table S5). Despite extensive cleavage within their sequences, the A68 (residues 7–17) and B27 (residues 16–25) epitopes were generated by the proteasome with 6.4 and 1% yield, respectively. Multiple N-terminally extended species of these ligands were also found.
Fig. 8.
Digestion of synthetic precursors of inhibitor-resistant A*68:01 and B*27:05 ligands by purified 20S proteasome. A, Cleavage of a 31-mer substrate, including the overlapping sequences of an A68 and a B27 ligand (boxed) in the absence or in the presence of the indicated inhibitors. Thin, medium and thick arrows indicate peptide bonds whose cleavage result in a joint yield of digestion products of 0.1 to 1%, 1.1 to 10% and >10%, respectively (see supplemental Table S6). B, Cleavage of a 30-mer substrate, containing the sequence of an inhibitor sensitive A68 ligand. The arrows follow the same convention as in panel A (see supplemental Table S7).
Table II. Effect of proteasome inhibitors on the digestion of the 31-mer substrate.
| No Inhibitor | MG-132 | Epoxomicin | |
|---|---|---|---|
| Amount digested | 100% | 28.9% | 54.3% |
| N. of digestion products | 80 | 28 | 16 |
| Peptides involving dual cleavagea | 66 (82.5%) | 20 (71.4%) | 7 (43.8%) |
| N. of cleaved peptide bondsb | 29 (96.7%) | 22 (73.3%) | 19 (63.3%) |
| N. of productive species for the A68 ligandc | 4 (5.1%) | 2 (7.1%) | 1 (6.3%) |
| Relative yield of productive cleavagesd | 6.9% | 5.5% | 2.6% |
| N. of destructive cleavages for the A68 ligande | 10 (34.5%) | 8 (36.4%) | 4 (21.1%) |
| Relative yield of the destructive cleavagesd | 20.3% | 26.3% | 7.8% |
| N. of productive species for the B27 ligandc | 12 (15.2%) | 2 (7.1%) | 2 (12.5%) |
| Relative yield of productive cleavagesd | 28.2% | 14.6% | 17.0% |
| N. of destructive cleavages for the B27 ligande | 9 (31%) | 6 (27.3%) | 7 (36.8%) |
| Relative yield of the destructive cleavagesd | 25.9% | 18.3% | 45.8% |
| Destruction/production ratio for the A68 ligandf | 2.9 | 4.8 | 3.0 |
| Destruction/production ratio for the B27 ligandf | 0.9 | 1.3 | 2.7 |
a Percent relative to the total number of digestion products.
b Percent relative to the total number of peptide bonds in the substrate.
c Peptides with the right C-terminus and right or extended N-terminus. Percent values are relative to the total number of digestion products (see Table VS).
d Added values from Table VIS. For productive cleavages the yields correspond to the relative contribution of cleavage after R17 or Y25 for the A68 and B27 ligands, respectively.
e Cleaved peptide bonds implying destruction of the epitope. Percent values are relative to the total number of cleaved peptide bonds.
f Ratio between the joint relative yield of cleavages implying destruction of the natural ligand and that of the potentially productive peptides.
With MG-132 only 28.9% of the substrate was cleaved. The number of digestion products, excluding the undigested substrate, was reduced to 28 peptides (Table 5S), which arose from cleavage at 22 peptide bonds. The A68 ligand was generated in very low amounts (0.3% of the digested material) and the B27 ligand was not produced, but N-terminally extended precursors of both ligands were observed (supplemental Table S5). Yet the ratio between the relative contribution of the cleavages implying epitope destruction and potential production was increased for the A68 (from 2.9 to 4.8) and the B27 ligand (from 0.9 to 1.3) in the presence of this inhibitor, relative to its absence (Table II). With epoxomicin 54.3% of the substrate was digested, and only 16 digestion products (supplemental Table S5), arising from cleavage at 19 peptide bonds, were obtained. None of the natural ligands was generated, but potential precursors with the right C-terminal residues were observed. The ratio between the relative yield of destructive and productive cleavages was unaltered (from 2.9 to 3) or increased (from 0.9 to 2.7) for the A68 and B27 ligands, respectively.
These results indicate that: (1) the 20S proteasome generated both natural ligands and their N-terminally extended precursors, (2) neither inhibitor abolished the proteasomal activity on this substrate or the generation of productive species, and (3) incomplete proteasomal inhibition did not result in increased production of the natural ligands or their precursors.
Both inhibitors showed a comparable global effect on the various subspecificities, altering only slightly their relative contribution to cleavage (Table III). However, the effect on individual peptide bonds within each subspecificity, was inhibitor-dependent (supplemental Table S6). For instance, MG-132 inhibited more efficiently cleavage after K13 than after R17, whereas epoxomicin showed higher inhibition on cleavage after K13 and R17 than after K23. For residues assigned to the chymotryptic-like activity the relative inhibition of cleavage after M6 was more efficient with epoxomicin, whereas after F18 and F22 was more efficient with MG-132. For residues not assigned to a particular subspecificity the inhibition was generally more efficient with epoxomicin than with MG-132, but the opposite was true for N19. Thus, incomplete inhibition of the proteasome resulted in an inhibitor-dependent alteration of cleavage patterns.
Table III. Effect of inhibitors on the cleavage of synthetic peptide precursors by proteasome 20S.
| Activity | No inhibitor | MG-132 | Epoxomicin | |||
|---|---|---|---|---|---|---|
| 31-mer | N. of Cleavages | % Contributiona | N. of Cleavages | % Contributiona | N. of Cleavages | % Contributiona |
| Tryptic | 3 | 15.4 | 2 | 8.3 | 3 | 11.6 |
| Chymotryptic | 13 | 65.1 | 11 | 70.3 | 9 | 70.1 |
| Caspase | 2 | 1.2 | 2 | 0.9 | 0 | 0 |
| Other | 11 | 18.3 | 7 | 20.5 | 7 | 18.4 |
| Total | 29 | 100 | 23 | 100 | 20 | 100 |
| 30-mer | N. of Cleavages | % Contributionb | N. of Cleavages | % Contributionb | N. of Cleavages | % Contributionb |
| Tryptic | 4 | 27.6 | 4 | 17.3 | 3 | 12.7 |
| Chymotryptic | 8 | 25.4 | 9 | 35.2 | 6 | 37.5 |
| Caspase | 3 | 21.1 | 2 | 12.1 | 2 | 12.5 |
| Other | 7 | 25.9 | 8 | 35.4 | 7 | 37.2 |
| Total | 22 | 100 | 23 | 100 | 18 | 100 |
a Added values from Table VIS.
b Added values from Table VIIS.
The ligands analyzed above derived from the same protein as an inhibitor-sensitive A68 ligand: EFVDLYVPR (Fig. 5). Thus, we analyzed the effect of proteasome inhibitors on the in vitro generation of this epitope from a synthetic 30-mer with the sequence of its parental protein (residues 2–31) in this region (Fig. 8B, Table IV, supplemental Table S7). The natural ligand was not detected in the digest, but a productive cleavage, generating a single digestion product corresponding to residues 1–14 of the substrate, occurred with a relative yield of 16.8% in the absence of inhibitor, at lower yield (9%), with MG-132, and not with epoxomicin. The joint relative yield of cleavages implying epitope destruction was similar in the absence or in the presence of inhibitors (Table IV). Compared with the non-assigned one, the inhibitor-sensitive A68 ligand from the same protein showed a lower destruction/production ratio without inhibitor and with MG-132 and was not generated in the presence of epoxomicin.
Table IV. Effect of proteasome inhibitors on the digestion of the 30-mer substrate.
| No Inhibitor | MG-132 | Epoxomicin | |
|---|---|---|---|
| Amount digested | 75.8% | 56.8% | 49.9% |
| N. of digestion products | 21 | 18 | 13 |
| Peptides involving dual cleavagea | 13 (61.9%) | 8 (44.4%) | 8 (61.5%) |
| N. of cleaved peptide bondsb | 22 (75.9%) | 23 (79.3%) | 18 (62.1%) |
| N. of productive species for the ligandc | 1 | 1 | 0 |
| Relative yield of productive cleavagesd | 16.8% | 9.0% | 0 |
| N. of destructive cleavages for the ligande | 6 (27.3%) | 6 (26.1%) | 4 (22.2%) |
| Relative yield of the destructive cleavagesd | 19.8% | 21.0% | 25.9% |
| Destruction/production ratio for the ligandf | 1.2 | 2.3 | – |
a Percent relative to the total number of digestion products.
b Percent relative to the total number of peptide bonds in the substrate.
c Peptides with the right C-terminus and right or extended N-terminus. Percent values are relative to the total number of digestion products.
d Added values from Table VIIS. Productive cleavages correspond to cleavage after R14.
e Cleaved peptide bonds implying destruction of the epitope. Percent values are relative to the total number of cleaved peptide bonds.
f Ratio between the joint relative yield of cleavages implying destruction of the natural ligand and that of the potentially productive peptides.
Alteration of cleavage patterns in the presence of inhibitors was also observed with this substrate. Both MG-132 and epoxomicin had a similar global effect, consisting in the preferential inhibition of the tryptic and caspase subspecificities, at the expense of the chymotryptic and non-assigned ones (Table III). However, inhibitor-dependent differences on cleavage of individual peptide bonds were observed, most notably after R14, but also at other positions (Fig. 8B and supplemental Table S7).
Although extrapolating in vitro results to proteasomal processing and inhibition in vivo must be done with great caution, the main differences between the non-assigned and the sensitive A68 ligand were that MG-132 had a larger inhibitory effect on the generation of the productive species of the second ligand and that epoxomicin abrogated its generation. This is consistent with the sensitivity of this ligand to inhibitors observed in vivo.
DISCUSSION
This study was undertaken to address whether the observation that proteasome inhibitor-resistant HLA-B27 ligands predominantly arose from small and basic proteins (23) could be generalized to other MHC-I molecules and whether this results from proteasome-independent processing or incomplete proteasomal inhibition.
The canonical A*68:01 motif can be described by significant restrictions at four anchor positions: a preference for acidic residues, T and S at P1 (joint frequency: 70.1%), V, T, and A at P2 (joint frequency: 71.7%), nonpolar residues, mainly A, F, I, and V (joint frequency: 63%) at P3 and an almost absolute restriction for C-terminal basic residues. Short peptides deviated significantly from this pattern at P1-P3. Some, especially 8-mers, are probably canonical ligands or may be bound in noncanonic ways (45). Yet, we cannot rule out that, despite the protease inhibitors used during purification, some of the shortest species might result from residual amino peptidase activity.
The A*68:01 motif is very different from that of B*27:05, where basic residues and G are favored, and acidic residues are very rare at P1. The R2 motif of HLA-B27 is absent in A68, and C-terminal basic residues account only for 42% of B*27:05 ligands (42). Thus, the two molecules bind largely divergent peptidomes. However, that 11% of the parental proteins of A68 ligands were also the source of B27-bound peptides indicates that both MHC molecules sample for the immune system both distinct subsets of the proteome and distinct epitopes of the same protein.
The rationale to distinguish between proteasome inhibitor sensitive and resistant A68 ligands was based on their metabolic labeling and the quantization of its inhibition. Since proteasomal inhibition severely impairs cell viability, the labeling times were relatively short, precluding uniform labeling. The observation that most of the inhibitor-resistant-peptides arose from small and basic proteins suggests that our previous observations on HLA-B27 (23) have a more general significance. Yet, the following facts hinted at an incomplete inhibition of the proteasome. First, the percentage of inhibitor-resistant ligands was smaller with epoxomicin than with MG-132. Second, inhibitor-resistant ligands from proteins other than small and basic ones corresponded to N- or C-terminal protein ends, which do not require dual cleavage. Third, five peptides were more inhibited with epoxomicin than with MG-132. Fourth, the intermediate inhibition of labeling observed on some A68 ligands might reflect down-regulation of protein synthesis (44), but also incomplete proteasomal inhibition. Fifth, the differential susceptibility to inhibitors of some peptides from the same parental protein could be explained by two mechanisms. Altered cleavage by a partially inhibited proteasome might result in increased production of only one of the ligands. Alternatively, the inhibitor-resistant ligand might be generated from protein fragments produced by a partially inhibited proteasome by nardylisin and/or thimet oligopeptidase, as recently reported (46).
Because replicate isotope-labeling experiments were not performed, we cannot rule out some inaccuracy in the assignments of inhibitor-sensitive and resistant ligands. However, the consistency in the assignment of most peptides amenable to analysis with both inhibitors, or with the same inhibitor upon K and R tagging, underlines the reliability of our analysis.
Incomplete inhibition of the proteasome was directly demonstrated both with fluorogenic substrates and synthetic peptide precursors. At the concentrations used in vivo both inhibitors impaired the hydrolysis of the chymotryptic substrate in vitro by about 70%. This value was lower than in HeLa cells (47), and reflects incomplete inhibition of the proteasome in C1R, because higher concentrations of epoxomicin, inhibited the hydrolysis of this substrate by >90%. Epoxomicin inhibited the tryptic activity by a maximum of about 67%, also less than in HeLa cells (47). With MG-132 the inhibition of the tryptic activity was still lower and partial inhibition of the proteasome was clear in gel experiments. The caspase activity was not tested with fluorogenic substrates, but it was reported that epoxomicin and MG-262, a boronate analog of MG-132, have lower effect on this than on the chymotryptic and tryptic subspecificities (47). In the same study the efficacy of inhibitors was protein substrate dependent, presumably as a function of the amino acid composition. Thus, incomplete inhibition of the proteasome might explain the predominance of inhibitor-resistant ligands arising from small basic proteins, because these are comparatively rich in basic residues and might be particularly susceptible to a residual tryptic-like activity.
That a C-terminal basic residue is not a pre-requisite for the observed pattern of parental proteins among inhibitor-resistant ligands was clear from our previous study on HLA-B27 (23) because a significant portion of B27 ligands lack a basic C-terminal motif. However, B27 ligands have R2. Whether the same pattern holds for allotypes lacking basic residues at any anchor position is not known.
The digestion of synthetic peptide substrates addressed the following issues. First, whether inhibitor-resistant ligands or their N-terminal precursors were directly produced by the proteasome in vitro. Second, whether the inhibitors abrogated cleavage and the generation of the MHC ligands or their precursors. Third, whether incomplete inhibition of the proteasome resulted in altered cleavage and, if so, whether this would lead to increased production of the natural ligand or their precursors. That significant amounts of productive species for the A68 and B27 ligands were generated from the 31-mer in the absence of inhibitors suggests that they might be produced by the proteasome in vivo. Incomplete inhibition of the proteasome altered the cleavage patterns, but these alterations, although failing to abrogate the generation of productive species for the 31-mer, did not result in increased production of the inhibitor-resistant or non-assigned ligands, in contrast to a previously reported example (26).
Some recent advances on the role of cytosolic peptidases in the generation of MHC-bound peptides might provide a plausible explanation for the presentation of inhibitor-resistant ligands under conditions of decreased proteasomal activity. Bleomycin hydrolase and puromycine-sensitive amino peptidase promote optimal peptide binding and enhanced surface expression of some allotypes (48). In addition, it was recently suggested that some inhibitor-resistant ligands, including those from HLA-B27, could be generated by post-proteasomal processing of protein fragments by the combined action of nardilysin and thimet oligopeptidase, and that the former enzyme could generate the basic C-terminal end of HLA-A3 and -A11 ligands from synthetic precursors in vitro (46). The cleavage specificity of nardilysin at dibasic sequences (49, 50) could be particularly suited to generate the C terminus of peptides ending at basic residues. Of the A68 ligands reported in this study (supplemental Table S1) 168 (20.7%) had a basic C+1 residue in the corresponding parental protein (data not shown). However, only 1 of 7 (14.3%) of the inhibitor-resistant ligands showed a dibasic C/C+1 motif (ETVQLRNPPR-RQL), as opposed to 8 of 24 (33.3%) inhibitor-sensitive ligands. Yet this does not exclude a role of nardilysin, because this enzyme can also cleave at monobasic sites (51).
In conclusion, because of incomplete inhibition of the proteasome, inhibitor-resistant ligands are not necessarily generated by non-proteasomal pathways. Yet, their presence is not explained by decreased epitope destruction. It may require further processing of intermediate proteasomal products by other cytosolic enzymes, whose contribution would become more prominent under conditions of proteasome impairment.
Acknowledgments
We thank the staff of the Proteomics facilities at the Centro Nacional de Biotecnología, Madrid (member of the ProteoRed network) for help in MS.
Footnotes
* This work was supported by grants SAF2008/00461 and RD08/0075 from the Ministry of Science and Innovation, and an institutional grant of the Fundación Ramón Areces to the Centro de Biología Molecular Severo Ochoa and by the Israel Science Foundation (ISF 916/05 to AA).
 This article contains supplemental Figs. S1 to S3 and Tables S1 to S7.
This article contains supplemental Figs. S1 to S3 and Tables S1 to S7.
1 The abbreviations used are:
- MHC-I
- Major Histocompatibility Complex class I
- mAb
- monoclonal antibody
- FBS
- fetal bovine serum
- MG-132
- carbobenzoxy-L-leucyl-L-leucyl-L-leucinal
- TFA
- trifluoroacetic acid
- m/z
- mass-to-charge
- MALDI-TOF
- Matrix-assisted laser desorption/ionization time-of-flight
- MW
- molecular mass
- pI
- Isoelectric point
- P
- position.
REFERENCES
- 1. Rock K. L., Gramm C., Rothstein L., Clark K., Stein R., Dick L., Hwang D., Goldberg A. L. (1994) Inhibitors of the proteasome block the degradation of most cell proteins and the generation of peptides presented on MHC class I molecules. Cell 78, 761–771 [DOI] [PubMed] [Google Scholar]
- 2. Henderson R. A., Michel H., Sakaguchi K., Shabanowitz J., Appella E., Hunt D. F., Engelhard V. H. (1992) HLA-A2.1-associated peptides from a mutant cell line: a second pathway of antigen presentation. Science 255, 1264–1266 [DOI] [PubMed] [Google Scholar]
- 3. Wei M. L., Cresswell P. (1992) HLA-A2 molecules in an antigen-processing mutant cell contain signal sequence-derived peptides. Nature 356, 443–446 [DOI] [PubMed] [Google Scholar]
- 4. Snyder H. L., Bacik I., Bennink J. R., Kearns G., Behrens T. W., Bächi T., Orlowski M., Yewdell J. W. (1997) Two novel routes of transporter associated with antigen processing (TAP)-independent major histocompatibility complex class I antigen processing. J. Exp. Med. 186, 1087–1098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Gil-Torregrosa B. C., Raúl Castano A., Del Val M. (1998) Major histocompatibility complex class I viral antigen processing in the secretory pathway defined by the trans-Golgi network protease furin. J. Exp. Med. 188, 1105–1116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Gil-Torregrosa B. C., Castaño A. R., López D., Del Val M. (2000) Generation of MHC class I peptide antigens by protein processing in the secretory route by furin. Traffic. 1, 641–651 [DOI] [PubMed] [Google Scholar]
- 7. Zhang Y., Kida Y., Kuwano K., Misumi Y., Ikehara Y., Arai S. (2001) Role of furin in delivery of a CTL epitope of an anthrax toxin-fusion protein. Microbiol. Immunol. 45, 119–125 [DOI] [PubMed] [Google Scholar]
- 8. Seifert U., Marañón C., Shmueli A., Desoutter J. F., Wesoloski L., Janek K., Henklein P., Diescher S., Andrieu M., de la Salle H., Weinschenk T., Schild H., Laderach D., Galy A., Haas G., Kloetzel P. M., Reiss Y., Hosmalin A. (2003) An essential role for tripeptidyl peptidase in the generation of an MHC class I epitope. Nature Immunology 4, 375–379 [DOI] [PubMed] [Google Scholar]
- 9. Shen L., Sigal L. J., Boes M., Rock K. L. (2004) Important role of cathepsin S in generating peptides for TAP-independent MHC class I crosspresentation in vivo. Immunity. 21, 155–165 [DOI] [PubMed] [Google Scholar]
- 10. Leonhardt R. M., Keusekotten K., Bekpen C., Knittler M. R. (2005) Critical role for the tapasin-docking site of TAP2 in the functional integrity of the MHC class I-peptide-loading complex. J. Immunol. 175, 5104–5114 [DOI] [PubMed] [Google Scholar]
- 11. Guil S., Rodríguez-Castro M., Aguilar F., Villasevil E. M., Antón L. C., Del Val M. (2006) Need for tripeptidyl-peptidase II in major histocompatibility complex class I viral antigen processing when proteasomes are detrimental. J. Biol. Chem. 281, 39925–39934 [DOI] [PubMed] [Google Scholar]
- 12. Parmentier N., Stroobant V., Colau D., de Diesbach P., Morel S., Chapiro J., van Endert P., Van den Eynde B. J. (2010) Production of an antigenic peptide by insulin-degrading enzyme. Nat. Immunol. 11, 449–454 [DOI] [PubMed] [Google Scholar]
- 13. Griffin T. A., Nandi D., Cruz M., Fehling H. J., Kaer L. V., Monaco J. J., Colbert R. A. (1998) Immunoproteasome assembly: cooperative incorporation of interferon gamma (IFN-gamma)-inducible subunits. J. Exp. Med. 187, 97–104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Dick T. P., Ruppert T., Groettrup M., Kloetzel P. M., Kuehn L., Koszinowski U. H., Stevanović S., Schild H., Rammensee H. G. (1996) Coordinated dual cleavages induced by the proteasome regulator PA28 lead to dominant MHC ligands. Cell 86, 253–262 [DOI] [PubMed] [Google Scholar]
- 15. Shimbara N., Nakajima H., Tanahashi N., Ogawa K., Niwa S., Uenaka A., Nakayama E., Tanaka K. (1997) Double-cleavage production of the CTL epitope by proteasomes and PA28: role of the flanking region. Genes Cells 2, 785–800 [DOI] [PubMed] [Google Scholar]
- 16. Heinemeyer W., Fischer M., Krimmer T., Stachon U., Wolf D. H. (1997) The active sites of the eukaryotic 20 S proteasome and their involvement in subunit precursor processing. J. Biol. Chem. 272, 25200–25209 [DOI] [PubMed] [Google Scholar]
- 17. Dick T. P., Nussbaum A. K., Deeg M., Heinemeyer W., Groll M., Schirle M., Keilholz W., Stevanović S., Wolf D. H., Huber R., Rammensee H. G., Schild H. (1998) Contribution of proteasomal beta-subunits to the cleavage of peptide substrates analyzed with yeast mutants. J. Biol. Chem. 273, 25637–25646 [DOI] [PubMed] [Google Scholar]
- 18. Nussbaum A. K., Dick T. P., Keilholz W., Schirle M., Stevanović S., Dietz K., Heinemeyer W., Groll M., Wolf D. H., Huber R., Rammensee H. G., Schild H. (1998) Cleavage motifs of the yeast 20S proteasome beta subunits deduced from digests of enolase 1. Proc. Natl. Acad. Sci. U.S.A. 95, 12504–12509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Kisselev A. F., Akopian T. N., Castillo V., Goldberg A. L. (1999) Proteasome active sites allosterically regulate each other, suggesting a cyclical bite-chew mechanism for protein breakdown. Mol. Cell 4, 395–402 [DOI] [PubMed] [Google Scholar]
- 20. Kisselev A. F., Garcia-Calvo M., Overkleeft H. S., Peterson E., Pennington M. W., Ploegh H. L., Thornberry N. A., Goldberg A. L. (2003) The caspase-like sites of proteasomes, their substrate specificity, new inhibitors and substrates, and allosteric interactions with the trypsin-like sites. J. Biol. Chem. 278, 35869–35877 [DOI] [PubMed] [Google Scholar]
- 21. Benham A. M., Grommé M., Neefjes J. (1998) Allelic differences in the relationship between proteasome activity and MHC class I peptide loading. J. Immunol. 161, 83–89 [PubMed] [Google Scholar]
- 22. Luckey C. J., Marto J. A., Partridge M., Hall E., White F. M., Lippolis J. D., Shabanowitz J., Hunt D. F., Engelhard V. H. (2001) Differences in the expression of human class I MHC alleles and their associated peptides in the presence of proteasome inhibitors. J. Immunol. 167, 1212–1221 [DOI] [PubMed] [Google Scholar]
- 23. Marcilla M., Cragnolini J. J., López de Castro J. A. (2007) Proteasome-independent HLA-B27 ligands arise mainly from small basic proteins. Mol. Cell Proteomics 6, 923–938 [DOI] [PubMed] [Google Scholar]
- 24. Rock K. L., York I. A., Saric T., Goldberg A. L. (2002) Protein degradation and the generation of MHC class I-presented peptides. Adv. Immunol. 80, 1–70 [DOI] [PubMed] [Google Scholar]
- 25. Kisselev A. F., Akopian T. N., Castillo V., Goldberg A. L. (1999) Proteasome active sites allosterically regulate each other, suggesting a cyclical bite-chew mechanism for protein breakdown. Mol. Cell 4, 395–402 [DOI] [PubMed] [Google Scholar]
- 26. Wherry E. J., Golovina T. N., Morrison S. E., Sinnathamby G., McElhaugh M. J., Shockey D. C., Eisenlohr L. C. (2006) Re-evaluating the generation of a “proteasome-independent” MHC class I-restricted CD8 T cell epitope. J. Immunol. 176, 2249–2261 [DOI] [PubMed] [Google Scholar]
- 27. Zemmour J., Little A. M., Schendel D. J., Parham P. (1992) The HLA-A,B “negative” mutant cell line C1R expresses a novel HLA-B35 allele, which also has a point mutation in the translation initiation codon. J. Immunol. 148, 1941–1948 [PubMed] [Google Scholar]
- 28. Barnstable C. J., Bodmer W. F., Brown G., Galfre G., Milstein C., Williams A. F., Ziegler A. (1978) Production of monoclonal antibodies to group A erythrocytes, HLA and other human cell surface antigens. New tools for genetic analysis. Cell 14, 9–20 [DOI] [PubMed] [Google Scholar]
- 29. Kim K. B., Myung J., Sin N., Crews C. M. (1999) Proteasome inhibition by the natural products epoxomicin and dihydroeponemycin: insights into specificity and potency. Bioorg. Med. Chem. Lett. 9, 3335–3340 [DOI] [PubMed] [Google Scholar]
- 30. Ishihama Y., Rappsilber J., Andersen J. S., Mann M. (2002) Microcolumns with self-assembled particle frits for proteomics. J. Chromatogr. A 979, 233–239 [DOI] [PubMed] [Google Scholar]
- 31. Beer I., Barnea E., Ziv T., Admon A. (2004) Improving large-scale proteomics by clustering of mass spectrometry data. Proteomics 4, 950–960 [DOI] [PubMed] [Google Scholar]
- 32. Eng J. K., McCormack A. L., Yates J. R. (1994) An approach to correlate tandem mass spectral data of peptides with amino acid sequences in a protein database. J. Am. Soc. Mass. Spectrom. 5, 976–989 [DOI] [PubMed] [Google Scholar]
- 33. Perkins D. N., Pappin D. J., Creasy D. M., Cottrell J. S. (1999) Probability-based protein identification by searching sequence databases using mass spectrometry data. Electrophoresis 20, 3551–3567 [DOI] [PubMed] [Google Scholar]
- 34. Parker K. C., Bednarek M. A., Coligan J. E. (1994) Scheme for ranking potential HLA-A2 binding peptides based on independent binding of individual peptide side-chains. J. Immunol. 152, 163–175 [PubMed] [Google Scholar]
- 35. Cragnolini J. J., Garcia-Medel N., Lopez de Castro J. A. (2009) Endogenous processing and presentation of T-cell Epitopes from chlamydia trachomatis with relevance in HLA-B27-associated reactive arthritis. Mol. Cell. Proteomics 80, 1850–1859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Paradela A., Garcia-Peydro M., Vázquez J., Rognan D., López de Castro J. A. (1998) The same natural ligand is involved in allorecognition of multiple HLA-B27 subtypes by a single T cell clone: role of peptide and the MHC molecule in alloreactivity. J. Immunol. 161, 5481–5490 [PubMed] [Google Scholar]
- 37. Paradela A., Alvarez I., Garcia-Peydró M., Sesma L., Ramos M., Vázquez J., López de Castro J. A. (2000) Limited diversity of peptides related to an alloreactive T cell epitope in the HLA-B27-bound peptide repertoire results from restrictions at multiple steps along the processing-loading pathway. J. Immunol. 164, 329–337 [DOI] [PubMed] [Google Scholar]
- 38. Gómez P., Mavian C., Galocha B., Garcia-Medel N., López de Castro J. A. (2009) Presentation of cytosolically stable peptides by HLA-B27 is not dependent on the canonic interactions of N-terminal basic residues in the A pocket. J. Immunol. 182, 446–455 [DOI] [PubMed] [Google Scholar]
- 39. Medina-Aunon J. A., Paradela A., Macht M., Thiele H., Corthals G., Albar J. P. (2010) Protein Information and Knowledge Extractor: Discovering biological information from proteomics data. Proteomics 10, 3262–3271 [DOI] [PubMed] [Google Scholar]
- 40. Kisselev A. F., Goldberg A. L. (2005) Monitoring activity and inhibition of 26S proteasomes with fluorogenic peptide substrates. Methods Enzymol. 398, 364–378 [DOI] [PubMed] [Google Scholar]
- 41. Elsasser S., Schmidt M., Finley D. (2005) Characterization of the proteasome using native gel electrophoresis. Methods Enzymol. 398, 353–363 [DOI] [PubMed] [Google Scholar]
- 42. Ben Dror L., Barnea E., Beer I., Mann M., Admon A. (2010) The HLA-B*2705 peptidome. Arthritis Rheum. 62, 420–429 [DOI] [PubMed] [Google Scholar]
- 43. Hickman H. D., Luis A. D., Buchli R., Few S. R., Sathiamurthy M., VanGundy R. S., Giberson C. F., Hildebrand W. H. (2004) Toward a definition of self: proteomic evaluation of the class I peptide repertoire. J. Immunol. 172, 2944–2952 [DOI] [PubMed] [Google Scholar]
- 44. Ding Q., Dimayuga E., Markesbery W. R., Keller J. N. (2006) Proteasome inhibition induces reversible impairments in protein synthesis. FASEB J. 20, 1055–1063 [DOI] [PubMed] [Google Scholar]
- 45. Khan A. R., Baker B. M., Ghosh P., Biddison W. E., Wiley D. C. (2000) The structure and stability of an HLA-A*0201/octameric tax peptide complex with an empty conserved peptide-N-terminal binding site. J. Immunol. 164, 6398–6405 [DOI] [PubMed] [Google Scholar]
- 46. Kessler J. H., Khan S., Seifert U., Le Gall S., Chow K. M., Paschen A., Bres-Vloemans S. A., de Ru A., van Montfoort N., Franken K. L., Benckhuijsen W. E., Brooks J. M., van Hall T., Ray K., Mulder A., Doxiadis I. I., van Swieten P. F., Overkleeft H. S., Prat A., Tomkinson B., Neefjes J., Kloetzel P. M., Rodgers D. W., Hersh L. B., Drijfhout J. W., van Veelen P. A., Ossendorp F., Melief C. J. (2011) Antigen processing by nardilysin and thimet oligopeptidase generates cytotoxic T cell epitopes. Nat. Immunol. 12, 45–53 [DOI] [PubMed] [Google Scholar]
- 47. Kisselev A. F., Callard A., Goldberg A. L. (2006) Importance of the different proteolytic sites of the proteasome and the efficacy of inhibitors varies with the protein substrate. J. Biol. Chem. 281, 8582–8590 [DOI] [PubMed] [Google Scholar]
- 48. Kim E., Kwak H., Ahn K. (2009) Cytosolic aminopeptidases influence MHC class I-mediated antigen presentation in an allele-dependent manner. J. Immunol. 183, 7379–7387 [DOI] [PubMed] [Google Scholar]
- 49. Chesneau V., Pierotti A. R., Barré N., Créminon C., Tougard C., Cohen P. (1994) Isolation and characterization of a dibasic selective metalloendopeptidase from rat testes that cleaves at the amino terminus of arginine residues. J. Biol. Chem. 269, 2056–2061 [PubMed] [Google Scholar]
- 50. Chow K. M., Csuhai E., Juliano M. A., St Pyrek J., Juliano L., Hersh L. B. (2000) Studies on the subsite specificity of rat nardilysin (N-arginine dibasic convertase). J. Biol. Chem. 275, 19545–19551 [DOI] [PubMed] [Google Scholar]
- 51. Chow K. M., Oakley O., Goodman J., Ma Z., Juliano M. A., Juliano L., Hersh L. B. (2003) Nardilysin cleaves peptides at monobasic sites. Biochemistry 42, 2239–2244 [DOI] [PubMed] [Google Scholar]