Abstract
Purpose
This study sought to determine whether morphology of simultaneously biopsied polar bodies is predictive of their cell division origin.
Methods
Levels of heterozygosity were measured using single nucleotide polymorphism (SNP) microarrays in sequentially biopsied polar bodies to establish the predictive value on samples with known cell division origins. The validated method of predicting cell division origin of polar bodies using heterozygosity rates was then applied to simultaneously biopsied polar bodies which had origin predictions made by morphological assessment.
Results
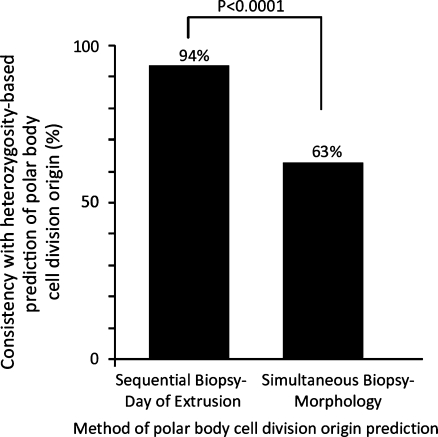
SNP microarray heterozygosity analysis was proven to be 94% predictive when tested against the known origin of sequentially biopsied polar bodies (n = 133). This methodology subsequently demonstrated that morphology was only 63% consistent when tested on simultaneously biopsied polar bodies (n = 455; P < 0.0001). Predictions of the origins of aneuploidy using morphology assignment were also significantly different than with heterozygosity assignment of polar body cell division origin (P = 0.0003).
Conclusions
Studies of the origin of aneuploidy utilizing morphological assignment of polar body cell division origin without heterozygosity analysis may be inaccurate and should be interpreted with caution.
Electronic supplementary material
The online version of this article (doi:10.1007/s10815-011-9683-9) contains supplementary material, which is available to authorized users.
Keywords: Polar body, SNP microarray, Meiosis, Heterozygosity
Introduction
Aneuploidy represents the leading genetic cause of miscarriage and developmental delay in humans and increases in prevalence as a function of advanced maternal age [1]. The primary origin of aneuploidy based upon analysis of products of conception is maternal meiosis [2]. However, controversy remains over whether chromosome segregation errors primarily arise during maternal meiosis I or II and whether meiosis I errors are primarily due to premature separation of sister chromatids or chromosome non-disjunction. Recent literature has evaluated 1st and 2nd polar bodies obtained by simultaneous biopsy at the time of fertilization check [3]. In this setting, polar body morphology was used to designate one as the 1st polar body and the other as the 2nd polar body—a technique which has not been validated.
One alternative to morphology based prediction of the cell division origin of polar bodies might involve characterization of heterozygosity. At a given single nucleotide polymorphism (SNP) locus, a diploid cell will be either homozygous (identical) or heterozygous (different). Second polar bodies have only 1 copy of each chromosome and thus are homozygous for every SNP. In spite of having sister-sister homologues, 1st polar bodies have a biological potential for heterozygosity due to crossover. Thus, when comparing polar bodies, the one with greatest heterozygosity should be the 1st polar body. This concept was tested and applied in the present study in order to assess the validity of heterozygosity and morphology based predictions of the cell division origin of polar bodies.
Materials and methods
This study was conducted in 2 phases. First, polar bodies sequentially biopsied and therefore with known meiotic cell division origin (1st or 2nd) were evaluated for consistency with heterozygosity predictions. Second, the validated method of predicting cell division origin of each polar body using heterozygosity analysis was applied to simultaneously biopsied polar bodies with morphology based origin assignments. All materials were collected and evaluated with institutional review board approval. In addition, all oocytes included in this study were suitable for clinical use and for polar body biopsy for preimplantation genetic screening of aneuploidy based on standard morphological criteria [4].
In order to obtain polar bodies with known cell division origin (positive control), the 1st polar body was biopsied at the time of intracytoplasmic sperm injection and the second polar body was biopsied when fertilization success was assessed the next day (+1). The biopsy procedure was performed as previously described [5]. In addition, a second set of polar bodies were obtained simultaneously when fertilization success was assessed on day +1. In order to assess the validity of assignment of polar body cell division origin based on morphology, the larger polar body was assigned to be the 1st polar body and the smaller was assigned to be the 2nd polar body.
Each polar body was subject to whole genome amplification and single nucleotide polymorphism (SNP) microarray analysis performed as previously described [6]. The percentage of SNPs given a heterozygous genotype call was obtained for each polar body. Each pair of polar bodies was then ranked by rate of heterozygosity and the polar body with the highest rate of heterozygosity was assigned as the 1st polar body and the lowest as the 2nd polar body. Heterozygosity based assignments were made without knowledge of the known or predicted cell division origin of the polar bodies (blinded).
Predictions of the cell division origin of polar bodies using heterozygosity were compared to either the known origin (sequential biopsy) or the morphology predicted origin (simultaneous biopsy). The rates of consistency for sequentially and simultaneously biopsied polar body data sets, and maternal meiosis cell division origin of aneuploidy from morphology or heterozygosity based assignment of which polar body was 1st or 2nd were compared using a chi square test of proportions. Aneuploidy was identified as previously described [6].
Results
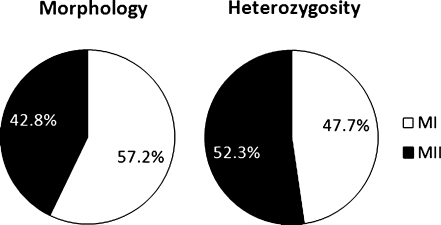
In phase 1, a total of 133 pairs of sequentially biopsied polar bodies were evaluated by microarray analysis (Supplemental Table 1). Blinded assignments made from heterozygous call rates were 94% consistent with the known cell division origin of the polar bodies (Fig. 1). In phase 2, a total of 455 pairs of simultaneously biopsied polar bodies were evaluated (Supplemental Table 2). Blinded assignments made from heterozygous call rates were 63% consistent with the morphology based prediction of the cell division origin of the polar bodies (Fig. 1). This was significantly different from the consistency observed in phase 1 (P < 0.0001). Origin of aneuploidy predictions based on either morphology or heterozygosity based assignment of which polar body was the first and which was the second were also significantly different (P = 0.0003)(Fig. 2).
Fig. 1.
The consistency of either the known cell division origin of sequentially biopsied polar bodies, or the morphology predicted origin of simultaneously biopsied polar bodies with SNP microarray heterozygosity based predictions
Fig. 2.
The relative contribution of meiosis I and meiosis II to chromosome segregation errors based upon 2 different methodologies for identifying which polar body is the 1st and which is the 2nd
Discussion
This study developed a valid method of predicting the cell division origin of polar bodies through SNP microarray based analysis of heterozygosity (phase I). There are a few possible reasons for the 6% inconsistency between heterozygosity based predictions of polar body cell division origin and the known origin based upon sequential biopsy. For example, if the 2nd polar body inherited too many copies of a chromosome (aneuploidy) the rate of heterozygosity may increase and exceed that found in the corresponding 1st polar body. While this explanation involves a biological explanation, it is also possible that errors associated with the technique could result in excessive false positive hetetrozygous genotype calls in the 2nd polar body or false negative heterozygous calls in the 1st polar body. The fact that any heterozygous calls in the 2nd polar body were observed at all (particularly in those that were euploid) is at least indicative of the former. However, this is not unexpected given the previously identified accuracy limitations of single cell genotyping after whole genome amplification and microarray analysis [7].
Nonetheless, polar body heterozygosity analysis clearly demonstrated that morphology is poorly predictive of polar body cell division origin. These results indicate that as many as one third of morphology based assignments of polar body cell division origin are likely to be incorrect. This is of particular significance to recent studies which have relied upon morphology based assignments to characterize the relative contribution of errors from meiosis I compared to meiosis II [3]. In addition, since chromatid errors in the 1st polar body would be equivalent in relative copy number change with errors in the second polar body, inaccurate assignment of the 2nd polar body as the 1st could lead to overestimation of relative contribution of premature separation of sister chromatids to the origin of meiosis I errors. Therefore, origin of aneuploidy studies utilizing simultaneously biopsied polar bodies without heterozygosity analysis may be inaccurate and should be interpreted with caution.
Electronic supplementary material
(XLS 1965 kb)
(XLS 68 kb)
Acknowledgments
Conflict of interest None.
Footnotes
Capsule SNP-microarray heterozygosity analysis demonstrates that morphology based assignment of polar body cell division origin is inaccurate and undermines conclusions from previous origin of aneuploidy studies.
References
- 1.Hassold T, Hunt P. Maternal age and chromosomally abnormal pregnancies: what we know and what we wish we knew. Curr Opin Pediatr. 2009;21:703–8. doi: 10.1097/MOP.0b013e328332c6ab. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hassold T, Hunt P. To err (meiotically) is human: the genesis of human aneuploidy. Nat Rev. 2001;2:280–91. doi: 10.1038/35066065. [DOI] [PubMed] [Google Scholar]
- 3.Fragouli E, Alfarawati S, Goodall NN, Sanchez-Garcia JF, Colls P, Wells D. The cytogenetics of polar bodies: insights into female meiosis and the diagnosis of aneuploidy. Molecular human reproduction 2011. [DOI] [PubMed]
- 4.Veeck LL. Oocyte assessment and biological performance. Ann N Y Acad Sci. 1988;541:259–74. doi: 10.1111/j.1749-6632.1988.tb22263.x. [DOI] [PubMed] [Google Scholar]
- 5.Treff NR, Su J, Kasabwala N, Tao X, Miller KA, Scott RT., Jr Robust embryo identification using first polar body single nucleotide polymorphism microarray-based DNA fingerprinting. Fertil Steril. 2010;93:2453–5. doi: 10.1016/j.fertnstert.2009.08.070. [DOI] [PubMed] [Google Scholar]
- 6.Treff NR, Su J, Tao X, Levy B, Scott RT., Jr Accurate single cell 24 chromosome aneuploidy screening using whole genome amplification and single nucleotide polymorphism microarrays. Fertil Steril. 2010;94:2017–21. doi: 10.1016/j.fertnstert.2010.01.052. [DOI] [PubMed] [Google Scholar]
- 7.Treff NR, Su J, Tao X, Northrop LE, Scott RT., Jr Single-cell whole-genome amplification technique impacts the accuracy of SNP microarray-based genotyping and copy number analyses. Mol Hum Reprod. 2011;17:335–43. doi: 10.1093/molehr/gaq103. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(XLS 1965 kb)
(XLS 68 kb)