Abstract
Purpose
To determine the vaginal microbiome in women undergoing IVF-ET and investigate correlations with clinical outcomes.
Methods
Thirty patients had blood drawn for estradiol (E2) and progesterone (P4) at four time points during the IVF-ET cycle and at 4–6 weeks of gestation, if pregnant. Vaginal swabs were obtained in different hormonal milieu, and the vaginal microbiome determined by deep sequencing of the 16S ribosomal RNA gene.
Results
The vaginal microbiome underwent a transition during therapy in some but not all patients. Novel bacteria were found in 33% of women tested during the treatment cycle, but not at 6–8 weeks of gestation. Diversity of species varied across different hormonal milieu, and on the day of embryo transfer correlated with outcome (live birth/no live birth). The species diversity index distinguished women who had a live birth from those who did not.
Conclusions
This metagenomics approach has enabled discovery of novel, previously unidentified bacterial species in the human vagina in different hormonal milieu and supports a shift in the vaginal microbiome during IVF-ET therapy using standard protocols. Furthermore, the data suggest that the vaginal microbiome on the day of embryo transfer affects pregnancy outcome.
Electronic supplementary material
The online version of this article (doi:10.1007/s10815-011-9694-6) contains supplementary material, which is available to authorized users.
Keywords: Metagenomics, Vagina, Microbiome, Infertility, IVF, Pregnancy
Introduction
Hormonal status dictates the complement of microflora resident on the vaginal epithelial mucosa and susceptibility to infection. A recent study of the vaginal microflora in rats demonstrated that ovariectomy is associated with lower total bacterial load, due primarily to the absence of Lactobacillus, which was restored with estradiol replacement [1], consistent with the dependence of Lactobacilli on estrogen status in women. Furthermore, anaerobic bacteria were absent after ovariectomy, which was accompanied by a preponderance of Clostridium perfringens, Bacteroides, Staphylococcus aureus, and S. epidermis [1]. Over the past several years, metagenomics has revealed that the vast majority of microbial diversity had been missed by cultivation-based methods, and, in fact, less than 1% of bacteria grow and form colonies on agar plates ([2], review). Using solely a genome-based approach centered upon sequencing the 16S ribosomal RNA gene (rDNA), we reported on the microbiome of human vaginal epithelium in normally cycling, reproductive aged women [3]. We found that the Lactobacillus content of the vaginal epithelium was highly variable, ranging from 0 to 100%, although cycle phase and circulating hormone concentrations were not considered. Clinical treatment protocols for infertility with IVF-ET provide a unique opportunity to assess the human vaginal microbiome in defined, shifting hormonal milieu and study the potential associations of the vaginal microbiome community with cycle outcome of pregnancy. There are published data demonstrating decreased pregnancy rates in women undergoing IVF-ET with concurrent vaginal infections with cultured bacteria [4]. However, there is no information on the vaginal microbiome ascertained by technology that does not require growth of the microbes.
Herein, we have employed a genome-based methodology to identify the bacteria on the vaginal epithelium of women undergoing IVF-ET with protocols commonly encountered in clinical practice and have investigated the association of the vaginal microbiome with circulating ovarian-derived estradiol (E2) and progesterone (P4) concentrations and pregnancy outcome. This metagenomics approach also enabled the discovery of novel, as yet unidentified, bacterial species.
Materials and methods
Human subjects Patients were recruited after written informed consent under a protocol approved by the Committee on Human Research at the University of California, San Francisco (UCSF) and the Committee on the Use of Human Subjects in Research at Stanford University. All enrolled patients receiving treatment were from UCSF.Thirty IVF-ET patients were recruited and consented for this study (Table 1). Nineteen IVF-ET patients underwent the Long Luteal Protocol (LLP). The LLP is composed of ovarian suppression with a gonadotropin-releasing hormone (GnRH) agonist, followed by controlled ovarian stimulation with recombinant and urinary-derived gonadotropins, followed by administration of human chorionic gonadotropin (hCG) to induce oocyte maturation, and supplementation with progesterone in oil intramuscularly 2 days after oocyte retrieval. Five IVF-ET patients underwent the Microflare Protocol (MFP). MFP is a procedure for poor responders, using a GnRH agonist (leuprolide acetate), but in lower doses than the LLP and timed to utilize the stimulatory effect on follicular recruitment of the “flare” release of endogenous gonadotropins from the agonist effect of leuprolide acetate. Four patients underwent the Demi-Halt Protocol (DHP). DHP is a protocol for poor responders in which the GnRH agonist in a down-regulated cycle is stopped at the start of gonadotropin administration to minimize suppression of the hypothalamic pituitary axis. One patient underwent the Very Low Dose leuprolide acetate (VLDL) Protocol, which is a third protocol for poor responders. VLDL is similar to LLP except very low amounts of the GnRH agonist are used for “gentle” suppression of the hypothalamic-pituitary axis. One patient underwent an Antagonist Protocol (AP), which makes use of a short-acting GnRH antagonist instead of the long-acting GnRH agonist. In addition, all IVF-ET patients received glucocorticoids and antibiotics commonly used in practice for IVF-ET treatment cycles.Patients enrolled in this study did not have signs or symptoms of cervical, uterine, or tubal infection. The clinicians and the embryology laboratory involved in the IVF cycles followed standards to avoid transmission of infections from all possible sources established by the American Society for Reproductive Medicine/Society for Assisted Reproductive Technology, as well as the U.S. Center for Disease Control and Prevention. Follicular aspiration was performed with a 17 gauge needle at a standard aspiration pressure. Male partners provided a semen sample in a sterile container in the collection area near the embryology laboratory as per routine practice. Gametes and embryos were handled separately according to standard laboratory procedures. Oocytes were inseminated 38–42 h following time of hCG administration in Vitrolife G media (Vitrolife, Englewood, CO) supplemented with 5% human serum albumin (Sage-Cooper Surgical, Trumball, CT) or injected with a single sperm (ICSI) depending on the medical indication. Zygotes identified at the fertilization check 16–18 h post insemination or injection were cultured individually in 25 μL of growth medium (G1.2 or G1.3; Scandinavian IVF Science/Vitrolife, Gothenburg, Sweden) overlaid with 8 mL of oil in Falcon 1007 culture dishes (Becton Dickinson Labward, Franklin Lakes, NJ). All media were certified to be free of endotoxin. Embryo transfers were performed with a Wallace catheter (Irvine Scientific, CA).
Table 1.
IVF-ET patients and outcomes
| Patient | Age | Self-identified ethnicity | IVF-ET protocol | Live birth |
|---|---|---|---|---|
| A01 | 38 | Caucasian | LLP | Y |
| A02 | 34 | Caucasian | LLP | N |
| A03 | 40 | Latina | MFP | N |
| A04 | 33 | Caucasian | LLP | Y |
| A05 | 45 | Caucasian | LLP | N |
| A07 | 36 | Hispanic* | MFP | N |
| A08 | 39 | Caucasian | LLP | N |
| A09 | 38 | Vietnamese | LLP | N |
| A10 | 34 | Caucasian | LLP | Y |
| A11 | 42 | Chinese | DHP | N |
| A12 | 33 | Chinese | LLP | Y |
| A13 | 36 | Caucasian | LLP | N |
| A16 | 29 | White | LLP | Y |
| A17 | 31 | Mediterranean** | DHP | N |
| A19 | 41 | Caucasian | MFP | Y |
| A20 | 42 | Caucasian | LLP | N |
| A21 | 31 | Caucasian | AP | Y |
| A22 | 38 | Asian | LLP | N |
| A23 | 34 | Caucasian | LLP | Y |
| A24 | 35 | Caucasian | VLDL | Y |
| A25 | 35 | Caucasian | LLP | N |
| A26 | 32 | Caucasian | LLP | N |
| A27 | 44 | Japanese | LLP | N |
| A29 | 43 | Caucasian | MFP | N |
| A30 | 43 | Caucasian | DHP | N |
| A31 | 34 | Hispanic | LLP | N |
| A33 | 41 | Caucasian | LLP | N |
| A34 | 39 | Caucasian | DHP | N |
| A40 | 28 | Caucasian | LLP | Y |
| A44 | 41 | Caucasian | MFP | Y |
*Hispanic/Caucasian; ** 50% Italian, 25% Basque, 25% Spanish. AP, Antagonist Protocol; DH, Demi-Halt Protocol; LLP, Long Luteal Protocol; MFP, Microflare Protocol; VLDL, Very Low Dose leuprolide acetate Protocol
Serum hormone concentration measurements Blood was drawn, and serum prepared, at four time points during each treatment cycle plus pregnancy: at baseline (B); at the (hyperestrogenic) late follicular stage (LF) at the day close to or on the morning before hCG administration; on the day of hCG administration; at embryo transfer (TR); and after 6-to-8 weeks of gestation (GE).
Total DNA from vaginal swabs Swabs of the posterior fornix of the vagina were performed, as described [3]. The first swab was taken at the B ultrasound on the first or second day of spontaneous or induced menses (low E2, low P4). Swab 2 was taken at oocyte retrieval (36 h after hCG administration; high E2, low P4). Swab 3 was taken at TR (high E2, moderate P4). Swab 4 was taken at GE (high E2, high P4). The frozen, de-identified vaginal swabs were transferred to the Stanford Genome Technology Center (SGTC), in a blinded fashion. Total DNA was purified from each vaginal swab employing a DNeasy Tissue and Blood Kit (Qiagen, Valencia, CA). The final step was dialysis and concentration with Amicon Ultra Centrifugal Filters (0.5 ml, 100 K; EMD Millipore, Billerica, MA). Each total DNA preparation for each swab was frozen in aliquots until use.
BigDye-terminator sequencing of the 16S ribosomal RNA gene (rDNA) An aliquot of total DNA from each swab was employed as template in an individual PCR reaction. The rDNA amplification primers were 8f and 1492r and employed as described previously [3]. The ~1.4 Kb product was purified by gel electrophoresis and cloned into a plasmid vector using a TA cloning kit from Invitrogen (Carlsbad, CA). BigDye-terminator chemistry was employed for sequencing (ABI, Foster City, CA), as described previously [3]. The sequencing reactions were run on an ABI 3730xl DNA Analyzer. Because our current average good quality read length is >700 bases with some good quality read lengths of >800 bases, each insert was sequenced from only one end to avoid unproductive overlap. This is a change from our previous procedure. Nevertheless, because the cloning procedure was somewhat asymmetric, sequences for the entire rDNA were achieved.
BigDye-terminator sequence reads From each vaginal swab, we achieved four 96-well plates of individual sequences (called “reads”) employing BigDye-terminator chemistry (Sanger sequence). Individual sequence reads were trimmed, assembled into contigs, and the contigs hand edited as described previously [3], except that the ABI BaseCaller software was employed to identify the bases instead of phred [5].
From contig consensus sequence to microbe Each contig consensus sequence was compared to the data in the Ribosomal Database Project (RDP) to identify the bacterium [6]. When the sequence match was >97% identity, the contig was given the name of the bacterium with the best sequence match, even when that name was “uncultured bacterium”. (As more environments have been investigated, the category of uncultured bacterium has increased in membership.) When the best sequence match was <97%, that bacterium was deemed “novel” [7–9].
GenBank accession numbers The novel rDNA sequences reported in this manuscript have been deposited in GenBank. Their accession numbers are HQ293151-HQ293203.
Changes in the vaginal microbiome When the reads supporting the presence of a particular bacterium changed by more than 10% of the total reads between swabs of the same IVF-ET patient, that change was deemed significant. A change was scored as “1”. No change was scored as “0”.
Analyses The Shannon Diversity Index (SDI) was calculated for the micro-biome of each vaginal swab (http://math.hws.edu/javamath/ryan/DiversityTest.html). Chao1, Principal Component, and SDI analyses employed the QIIME software [10] with UniFrac distances [11]. Parametric and nonparametric testing were used to determine statistical significance of patient age and live birth and the statistical significance of diversity of the vaginal microbiome on the outcome (live birth/no live birth).
Results
IVF-ET patients and their outcomes The thirty IVF-ET patients enrolled in this study are presented in (Table 1). Eleven patients (37%) had a live birth. In addition, patient A13 became pregnant, but had a miscarriage. Patients A02 and A31 demonstrated very low human chorionic gonadotropin (hCG) concentrations, likely representing a biochemical pregnancy. The mean age of the eleven women who had a live birth was 34.2 years with a standard deviation of 4.1 years (http://www.csgnetwork.com/stddeviationcalc.html). The mean age of the nineteen women who did not have a live birth was 38.5 years with a standard deviation of 4.0 years. By the two sample Student’s t-test (http://www.usablestats.com/calcs/2samplet&summary=1), this difference was significant (p = 0.0088). That the age of the woman is inversely related to the success of the IVF-ET procedure had been established previously (see, for example, [12]).
Serum hormone concentrations Serum concentrations of estradiol (E2) were measured at baseline (B) and at three intervals during the IVF-ET procedure (Table S1 and Fig. S1; Supplementary Information). For those women who conceived, E2 concentrations were measured at 6-to-8 weeks of gestation. Progesterone concentrations were measured at embryo transfer and at 6–8 weeks of gestation (Fig. S2).The percent change in E2 concentration between stages of the IVF-ET procedure was calculated (Table 2). Between the baseline (B) and the late follicular stage (LF), the E2 concentration increased greatly, as anticipated (Table 2). Of the 26 values between LF and the administration of hCG, seven E2 concentrations decreased and nineteen increased. Of those patients who had a live birth, three E2 concentrations decreased and five increased (Fig. S2, Supplementary Information).Between hCG and embryo transfer (TR) (i.e., between swab 2 and swab 3), all 24 E2 concentrations decreased (Table S1, Supplementary Information). We calculated the percent decrease for each patient (Table 2). For the eight patients who had a live birth, the average decrease was 62.2 +/− 8.1% with a range of 47.3–74.5%. The average percent decrease for those patients who did not have a live birth was bimodal: 9.6 +/− 6.5% with a range of 0.6–19.9% (n = 5) and 56.5 +/− 10.0% with a range of 41.8–73.0% (n = 11) (p < 0.00001). The average decrease for the eight patients who had a live birth is statistically significantly different from the average of the sub-set of the five patients who did not have a live birth and who had only a small change in E2 concentration (p < 0.00001). These latter five patients are A05, A08, A09, A22, and A26. However, average decrease for the eight patients who had a live birth is not statistically different from the average of the eleven patients who did not have a live birth and who had a large change in E2 concentration (p = 0.19).We had measurements of E2 concentrations at TR and at 6-to-8 weeks of gestation (GE) (i.e., between swabs 3 and 4) for six patients who had a live birth (Table S1, Supplementary Information). For four of these patients, the E2 concentration increased and for two it decreased (Table 2).At embryo transfer (ET), the average serum progesterone concentration for those patients who had a live birth was 36.9 +/− 6.0 ng/ml (n = 7; Fig. S2, Supplementary Information). The average serum progesterone concentration for those who patients who did not have a live birth was 36.3 +/− 9.1 ng/ml (n = 16). These numbers are not statistically significantly different (p = 0.85). For those patients who had a live birth, at 4-to-6 weeks of gestation (GE), the average serum progesterone concentration was 37.4 +/− 9.4 ng/ml (n = 6), not different from the value at ET (p = 0.91).
Table 2.
Percent change in estradiol concentration: between baseline (B) and the late follicular stage (LF); between LF and the day of human chorionic gonadotropin (hCG) administration; between hCG and embryo transfer (TR); and between TR and 6-to-8 weeks of gestation (GE)
| Patient | B to LF (%) | LF to hCG (%) | hCG to TR (%) | TR to GE (%) |
|---|---|---|---|---|
| A01 | >3280 | 75.0 | −66.6 | 66.8 |
| A02 | >2580 | 75.5 | ||
| A03 | >6260 | 38.7 | −41.8 | |
| A04 | >8280 | 57.6 | −57.2 | −30.6 |
| A05 | >9110 | 23.1 | −11.2 | |
| A07 | >1970 | 92.7 | −65.9 | |
| A08 | >13450 | 31.8 | −19.9 | |
| A09 | >2630 | 54.5 | −11.4 | |
| A10 | >8920 | −76.0 | −54.1 | 366.6 |
| A11 | >4460 | 46.6 | −49.9 | |
| A13 | >3930 | −0.1 | −70.7 | |
| A16 | 175900 | 83.4 | −67.3 | |
| A17 | >9530 | −40.0 | −63.3 | |
| A19 | >12120 | −28.1 | −74.5 | 32.8 |
| A20 | 1341 | 14.4 | −47.4 | |
| A21 | 954 | 123.1 | −65.9 | −27.6 |
| A22 | >6690 | 84.5 | −0.6 | |
| A23 | >8280 | −44.0 | −64.9 | 194.8 |
| A24 | >9330 | 15.7 | −47.3 | |
| A25 | >5220 | 173.7 | −47.5 | |
| A26 | >6040 | 67.9 | −4.9 | |
| A27 | >9380 | 73.4 | −73.0 | |
| A29 | >16510 | −60.2 | −54.0 | |
| A30 | >11670 | −44.1 | −59.4 | |
| A31 | >10520 | 81.2 | −48.6 | |
| A33 | >3660 | 107.7 |
Vaginal microbes identified by BigDye-terminator sequencing of PCR amplified and cloned rDNAs From the 30 IVF-ET patients, we had a total of 99 vaginal swabs. The detailed vaginal bacteria results for all 30 IVF-ET patients and all swabs are presented in Table S2 (Supplementary Information). Lactobacillus species were supported by more than half of the sequence reads for each of 85 swabs (85/99 = 86%). In some cases, a single Lactobacillus species dominated. For example, for patient A01 swab 1 (A01-1), L. crispatus was supported by 96% of the sequence reads. In other cases, more than one Lactobacillus species was present. For example, for patient A03 swab 1 (A03-1), L. iners was supported by 47%, L. gasseri by 26%, and L. jensenii by 25% of the reads. Fourteen swabs evidenced little (<10% of reads) or no Lactobacillus. As an example, none of the three swabs for patient A05 contained any Lactobacillus. Six swabs evidenced >50% Lactobacillus sequence reads but, additionally, had a substantial number of reads supporting other bacteria. For example, for patient A07 swab 3 (A07-3), 55% of the reads supported Lactobacillus, while 41% of the reads supported Prevotella.We had a swab 1 for 29 of the 30 IVF-ET patients. Only three swabs were deficient in Lactobacillus: A05-1, A09-1, and A16-1 (Table S2, Supplementary Information). For A05-1, 81% of the sequence reads supported Streptococcus. For A09-1, 30% of the sequence reads supported Flavobacterium and 20% of the reads supported Acidovorax. For A16-1, 29% of the reads supported Anaerococcus and 49% of the reads supported “uncultured bacterium”. Patient A16 had a live birth, while patients A05 and A09 did not.Lactobacillus was supported by the most sequence reads on swab 2 for 27 of the 29 patients for whom we have a swab 2: 11 (41%) L. crispatus; 5 (19%) L. iners; 5 (19%) L. jensenii; 3 (11%) L. fornicalis; 2 (7%) L. gasseri; and one (4%) L. sp. A single Lactobacillus species dominated on 19 (70%), while, on eight (30%) swab 2 s, more than one Lactobacillus species was supported by, at least,100 sequence reads (Table S2, Supplementary Information). For the two swab 2 s where Lactobacillus was not supported by the most reads, Streptococcus (92%) dominated for patient A05, and Flavobacterium (73%) dominated for patient A30 (Table S2, Supplementary Information). Neither patient A05 nor patient A30 had a live birth.We had a swab 3 for all 30 patients. The vaginal microbiome was composed of virtually only Lactobacillus for 18 of the swabs: 14 L. crispatus, 3 L. gasseri, and 2 L. iners. In addition, L. crispatus plus Flavobacterium were found on A30-3, as well as L. jensenii, L. gasseri, plus Prevotella on A07-3 and L. jensenii plus Flavobacterium on A12-3. Enterococcus dominated on two swabs (A04-3, A26-3). Enterococcus plus Anaerococcus dominated on A05-3. The combination of Finegoldia plus Flavobacterium dominated on two swabs (A09-3 and A31-3). Escherichia and Prevotella each dominated on one swab (A34-3 and A25-3, respectively). Novel bacteria dominated on one swab (A13-3).All eleven swab 4 s were overwhelmingly dominated by Lactobacillus: six by L. crispatus, two by L. jensenii (including patient A13, who had a miscarriage), and one each L. fornicalis, L. gasseri, and a mixture of L. iners plus L. jensenii.
Changes in the vaginal microbiome for each patient Of the 30 IVF-ET patients, five evidenced no change in the microbe mix across all of their swabs (A01, A02, A27, A29, A33; Table S2, Supplementary Information). L. crispatus was the very dominant bacterium on all swabs for these five patients. An additional nine patients experienced only changes in the mix of Lactobacillus species (A03, A10, A11, A17, A19, A22, A23, A24, A44; Table S2, Supplementary Information). For example, A11 switched from L. iners (80% of the sequence reads) on swab 2 to L. crispatus (96% of the reads) on swab 3. Uniquely, patient A05 began with a vagina dominated by Streptococcus (92% of the reads) and switched to Enterococcus (68% of the reads) and Anaerococcus (21% of the reads). For each IVF-ET patient, the change in the vaginal microbiome was determined (Table 3). For all genera except Lactobacillus, all species were subsumed into the genus. For Lactobacillus, species were retained.
Table III.
Change in bacterial content between swabs, including changes in Lactobacillus species.
| Patient ID | 1-to-2 | 2-to-3 | 3-to-4 |
|---|---|---|---|
| A01 | 0 | 0 | 0 |
| A02 | 0 | 0 | |
| A03 | 1 | 1 | |
| A04 | 0 | 1 | 0 |
| A05 | 0 | 1 | |
| A07 | 0 | 1 | |
| A08 | 0 | 1 | |
| A09 | 1 | 1 | |
| A10 | 1 | 0 | 0 |
| A11 | 1 | 1 | |
| A12 | NA | 1 | 1 |
| A13 | 1 | 1 | 1 |
| A16 | 1 | 0 | 0 |
| A17 | 1 | 1 | |
| A19 | 0 | 0 | 0 |
| A20 | 0 | 1 | |
| A21 | 0 | 1 | 0 |
| A22 | 1 | 1 | |
| A23 | 1 | 1 | 0 |
| A24 | 1 | 1 | |
| A25 | 0 | 1 | |
| A26 | 1 | 1 | |
| A27 | 0 | 0 | |
| A29 | 0 | 0 | |
| A30 | 1 | 1 | |
| A31 | 1 | 1 | |
| A33 | NA | NA | |
| A34 | 0 | 1 | |
| A40 | 1 | 1 | 0 |
| A44 | 1 | 1 | 1 |
“0” means “no change”. “1” means “change”. NA means a swab was not available.
Correlation between changes in E2 concentration and changes in the vaginal microbiome Between B/swab 1 and LF/swab 2, the vaginal microbiome changed for 15/28 (54%; Table 3) of the IVF-ET patients, while all serum E2 concentrations rose dramatically (Table 2).For the five IVF-ET patients (A05, A08, A09, A22, A26) whose serum E2 concentrations decreased only modestly between hCG/swab 2 and TR/swab 3 (Table 2), all five vaginal microbiomes changed (100%; Table 3). For the remainder of the IVF-ET patients, 17/24 (71%; Table 3) of the vaginal microbiomes changed, while the E2 concentration decreased substantially.There were six measurements of the change in serum E2 concentration between TR/swab3 and GE/swab4. Four increased, and two decreased (Table 2). For those same six patients, the vaginal microbiome did not change (Table 3). For the remaining five swab 3 to swab 4 transitions, two vaginal microbiomes (including that of the patient who miscarried) changed and three did not (Table 3). Thus, it appeared that the vaginal microflora was not a simple function of circulating E2 concentration.The Wilcoxon rank sum test was employed to make several comparisons among patients with a change in vaginal microbiome to those with no change (Table 3 and Table S1). The tests and their p-values are summarized in Table S3 (Supplementary information). With one exception, these statistical tests demonstrated no significant difference. The exception was the comparison of the relative E2 changes from LF to TR between patients who have a vaginal microbiome change from swab 2 to swab 3 and patients who have no change. In this case, the difference is statistically significant (p-value = 0.003).
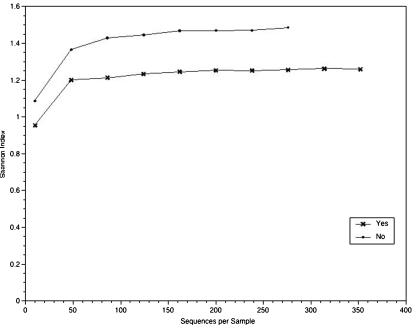
Microbiome diversity For the simplest test of vaginal microbiome diversity, as compared between those patients who had a live birth and those who did not, the number of bacterial genera on each swab was counted. The average number (+/− standard deviation) of bacteria per swab for the 42 swabs from patients who had a live birth was 6.4 +/− 4.7 (range from 1 to 31). The average number (+/− standard deviation) of bacteria per swab for the 57 swabs from patients who did not have a live birth was 8.6 +/− 5.3 (range from 1 to 24). By the two-sided t-test, these two numbers are statistically significantly different (p = 0.034). By comparing swabs from patients who had a live birth to swabs from patients who did not have a live birth, the overall difference was tracked to swab 3. There, the average number (+/− standard deviation) of bacteria per swab 3 for the eleven swabs from patients who had a live birth was 6.1 +/− 3.2 (range from 1 to 14). The average number (+/− standard deviation) of bacteria per swab 3 for the nineteen swabs from patients who did not have a live birth was 10.5 +/− 5.8 (range from 5 to 25). By the two-sided t-test, these two numbers for swab 3 s are statistically significantly different (p = 0.028).For a more sophisticated comparison of bacterial diversity between swabs, the Shannon Diversity Index (SDI) was calculated for all 99 swabs ([13]; http://math.hws.edu/javamath/ryan/DiversityTest.html). The results of the calculations are compiled in Table 4. The range was 0.0 to 3.3. For the three instances where the SDI was 0.0, the bacteria were all Lactobacillus: L. gasseri (A16-3 and A16-4) or L. crispatus (A29-1). To compare the SDI for different swabs, the Wilcoxon signed rank test (i.e., the one sample Wilcoxon test) was employed. (For these comparisons, the swabs for patients A12 and A33 were excluded because swabs were missing from the set.) The SDIs were not statistically different between swabs 1 and 2 and between swabs 2 and 3. To compare the SDI for the analogous swab between patients with different outcomes (live birth/no live birth), the Wilcoxon rank sum test (i.e., the two sample Wilcoxon test) was employed. The SDIs for swabs 1 and 2 were not statistically different between those patients who had a live birth and those who did not. The SDIs for swab 3 of patients who had a live birth differed significantly from the SDIs for swab 3 of those patients who did not have a live birth (p-value = 0.01). Thus, the diversity of the vaginal microbiome on swab 3 (i.e., at embryo transfer) may correlate with outcome (live birth/no live birth).The QIIME software [10] was employed to calculate the SDI as a function of the number of sequence reads. These results are shown in Fig. 1. In agreement with the calculations of diversity, the SDI curves distinguished those women who had a live birth from those who did not (Fig. 1). The QIIME software was also employed to calculate the SDI as a function of the number of sequence reads for each IVF-ET protocol. These results, shown in Fig. S3 (Supplementary Information), are difficult to interpret because of the different numbers of patients in each group.
Table 4.
Shannon Diversity Index for each swab
| Patient | Swab 1 | Swab 2 | Swab 3 | Swab 4 |
|---|---|---|---|---|
| A01 | 0.312 | 0.187 | 0.286 | 0.428 |
| A02 | 1.030 | 0.211 | 1.148 | |
| A03 | 1.660 | 1.694 | 0.725 | |
| A04 | 0.740 | 0.714 | 1.558 | 0.394 |
| A05 | 1.192 | 0.605 | 1.553 | |
| A07 | 1.389 | 1.051 | 1.782 | |
| A08 | 1.382 | 0.410 | 2.604 | |
| A09 | 3.232 | 0.101 | 3.317 | |
| A10 | 0.831 | 0.423 | 0.392 | 0.595 |
| A11 | 1.926 | 1.071 | 0.329 | |
| A12 | NA | 1.452 | 1.294 | 0.262 |
| A13 | 1.606 | 0.523 | 2.080 | 0.872 |
| A16 | 2.008 | 0.026 | 0.0 | 0.0 |
| A17 | 0.674 | 1.155 | 1.400 | |
| A19 | 0.382 | 0.751 | 0.027 | 0.237 |
| A20 | 0.053 | 0.193 | 0.917 | |
| A21 | 0.191 | 0.712 | 0.676 | 0.026 |
| A22 | 0.668 | 1.711 | 1.772 | |
| A23 | 0.331 | 2.021 | 0.747 | 0.544 |
| A24 | 0.377 | 1.874 | 0.887 | |
| A25 | 0.276 | 0.980 | 1.104 | |
| A26 | 0.318 | 1.011 | 0.718 | |
| A27 | 0.342 | 0.361 | 0.106 | |
| A29 | 0.0 | 0.027 | 0.207 | |
| A30 | 1.410 | 1.279 | 1.496 | |
| A31 | 0.456 | 0.049 | 0.795 | |
| A33 | 0.927 | NA | 1.267 | |
| A34 | 1.043 | 0.757 | 0.893 | |
| A40 | 1.699 | 1.190 | 0.147 | 0.379 |
| A44 | 1.261 | 1.695 | 0.770 | 1.264 |
NA means that the swab was not available
Fig. 1.
Shannon Diversity Index as a function of the number of sequences
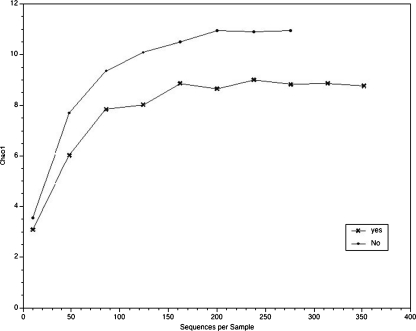
Estimates of species richness In every study of microbes in a given ecological niche (in this case, human vagina), time and money limit the amount of sampling and analysis accomplished. Therefore, statistical methods have been derived and employed to calculate species richness. Chao1 is one such statistical method [14]. The QIIME software was employed to calculate Chao1 as a function of the number of sequence reads [10]. These results are shown in Fig. 2. The Chao1 curves distinguished those women who had a live birth from those who did not (Fig. 2). The QIIME software was also employed to calculate Chao1 as a function of the number of sequence reads for each IVF-ET protocol. These results are shown in Fig. S4 (Supplementary Information). The very different number of patients in each group made interpretation of those curves difficult.
Fig. 2.
Chao1 analysis of the data
Principal Component Analysis (PCA) PCA is a method of extracting relationship information from groups of data. PCA reduces the observed variables into a smaller number of “principal components” (which are artificial variables) that account for all/most/some of the variance in the data. (PCA cannot identify what the principal components actually are.) The QIIME software was employed for PCA [10]. The results are shown in Fig. S5 (Supplementary Information). Where the variable was live birth/no live birth in one case and the particular IVF-ET procedure in the other, the first principal component accounted for 52% of the variance.
Novel Bacteria During the course of this study, we identified novel bacteria on the vaginal epithelium (Table S1, Supplementary Information). This finding was not surprising as novel bacteria have been found previously in the vagina [3, 15, 16]. Table 5 reports those novel bacteria supported by at least ten reads. Ten (33%) of the 30 IVF-ET patients were found to harbor novel bacteria on their vaginal epithelium: three patients (A16, A33, A40) on swab 1 (Baseline); five patients (A10, A12, A13, A24, A25) on swab 2 (oocyte retrieval); five patients (A08, A12, A13, A20, A33) on swab 3 (embryo transfer); and none on swab 4 (gestation). In particular, 63% of the reads for swab 3 for patient A13 supported a novel bacterium. For swab 2 of patient A25, 38% of the reads supported a novel bacterium.
Table 5.
Novel bacteria supported by ten or more reads
| swab 1 | swab 2 | swab 3 | |||||||
|---|---|---|---|---|---|---|---|---|---|
| ID | reads | closest named match | reads | % match range | closest named match | reads | % match range | closest named match | |
| 08 | 48 | 92.1–96.7 | Anaerococcus | ||||||
| 10 | 10 | 84.3–92.8 | Lactobacillus crispatus | ||||||
| 12 | no swab 1 | 33 | uncultured bacterium: order Sphingobacteriales | ||||||
| 13 | 10 | 96.2 | Flavobacterium | 219 | 96.7 | Flavobacterium | |||
| 1 | 188 | reads: | uncultured bacterium: | ||||||
| 6 | 120 | genus Anaerococcus | |||||||
| 45 | genus Prevotella | ||||||||
| 18 | genus Streptococcus | ||||||||
| 20 | 58 | 96.4 | Lactobacillus crispatus | ||||||
| 24 | 35 | uncultured bacterium: genus Lactobacillus | |||||||
| 25 | 141 | 96.7 | Lactobacillus crispatus | ||||||
| 33 | 101 | uncultured bacterium: order Sphingobacteriales | no swab 2 | 134 | uncultured bacterium: order Sphingobacteriales | ||||
| 40 | 45 | uncultured bacterium: genus Lactobacillus | |||||||
Discussion
Our data for the Lactobacillus content of the vaginal microbiome can be compared to the results of two recent studies. In our current study, Lactobacillus dominated the vaginal bacteria of swab 1 for 26 of 29 (90%) of the IVF-ET patients. For women attending a public health or sexually transmitted disease clinic, but without bacterial vaginosis by rigorous criteria, Oakley et al. [16] found that Lactobacillus accounted for 86% of all sequences. Ravel et al. [17] identified the bacteria in the vagina of reproductive age, asymptomatic women using 454 pyrosequencing technology to produce short sequence reads from two small regions of the 16S ribosomal RNA gene. Ninety-eight Caucasian women were included in their study. The vaginal microflora of these women were dominated (> 50% of the sequence reads) by L. crispatus (45.4%), L. iners (26.8%), L. jensenii (9.3%), and L. gasseri (8.2%). There were 20 Caucasian women in our study whose swab 1 was dominated (> 50% of the sequence reads) by only one species of Lactobacillus: L. crispatus (13; 65%), L. iners (4; 20%), L. jensenii (2; 10%), L. gasseri (none; 0%), and other Lactobacillus species (1; 5%). These series of numbers are in remarkable agreement. Forney et al. [18] identified the Lactobacillus species in the vagina of 19 women and found that L. crispatus was dominant for 42%, L. iners for 26%, and L. gasseri for 16%. Again, these series of numbers are in remarkable agreement.
For a quantitative comparison, the percent of reads supporting the presence of Lactobacillus was calculated for swab 1. For the ten women who had a live birth and for whom we had a swab 1, the mean value of Lactobacillus reads was 87% with a standard deviation of 29% and a range of 5% to 100% (Table S1, Supplementary Information). For the nineteen women who did not have a live birth, the mean was 80% with a standard deviation of 30% and a range of 0% to 100%. Comparing these two populations by the Wilcoxon rank sum test yields a p-value of 0.42. Therefore, the two populations are indistinguishable on the basis of swab 1 Lactobacillus content.
Of interest, 33% of patients harbored previously unidentified bacteria. The presence of novel bacteria in the vaginal mucosa may also be a contributing factor to IVF-ET outcome and warrants further investigation. Furthermore, since our subjects routinely received glucocorticoids in their IVF-ET cycles, it is biologically plausible that an inflammatory cytokine response from certain bacteria could affect implantation rates [4]. The value of such therapy warrants re-examination in view of the vaginal (and perhaps cervical) microbiome in future studies.
Protocols for IVF-ET provide a unique opportunity to evaluate hormonal influences on the vaginal microbiome, and in our study the use of different protocols could affect bacterial growth. Jakobsson and Forsum [19] characterized the normal cultivable vaginal Lactobacillus flora, using signature matching of nucleotide sequences in the V1 and V3 regions of the 16S rRNA genes in women undergoing IVF-ET. They found that L. crispatus, L. gasseri and/or L. jensenii were present in 10 of the 17 patients throughout the study period, with little variation among these three species in individual patients. Three women had dominance of L. delbrueckii, L. rhamnosus or L. vaginalis, and one had dominance of L. iners. For three women whose vaginae were initially dominated by L. rhamnosus or L. reuteri, as their E2 levels rose, their flora composition and dominance changed to one of the three species most common in the normal, healthy vagina. These observations are consistent with our data and further demonstrate the variability of vaginal communities in individuals and their hormonal dependence.
Since IVF-ET involves transfer of embryos by a catheter through the cervix into the uterus, vaginal and cervical microflora and pathogens and microbial contamination of the catheter tip have been suggested to affect implantation rates and pregnancy outcomes. It is established that detection of Chlamydia species in the endocervix of women undergoing IVF-ET is associated with decreased implantation and ongoing pregnancy rates [20], and women who have bacterial vaginosis have a higher risk of pregnancy loss in IVF-ET cycles [21]. Also, pathogenic bacteria cultured from the embryo transfer catheter tip adversely affect live birth rates [4, 22–25].
In the current study, we used a metagenomics approach to investigate the hormonal dependence of the microbiome in a typical IVF setting. During this study, we have uncovered several factors that appear to influence the success of the IVF-ET procedure. (i) All women who had a live birth also had a substantial decrease in serum E2 concentration between hCG administration and embryo transfer. This substantial decrease appears to be a necessary, but not sufficient, condition for a successful outcome of the IVF-ET procedure [26, 27]. (ii) All but one of the women who had a live birth had a swab1 dominated by Lactobacillus. (The exception, A16, had a swab1 dominated by “uncultured bacterium”.) The presence of Lactobacillus on swab 1 appears to be favorable, although not sufficient, for a successful outcome of the IVF-ET procedure. (iii) Anaerococcus, Acidovorax, Enterococcus, Escherichia, Finegoldia, Flavobacterium, Prevotella, and Streptococcus are considered noxious bacteria when found in the vagina. Despite routine antibiotic prophylaxis before the IVF-ET cycles, ten patients (A04, A05, A07, A09, A12, A25, A26, A30, A31, A34) had, at least, one of these bacteria on their vaginal swabs. Of these ten, only two had a live birth: A04 and A12. In both cases, the third swab evidenced a noxious bacterium. In the case of A12-3, Lactobacillus was the majority bacterium and Flavobacterium the minority bacterium. The ratio of sequence reads was Lactobacillus : Flavobacterium = 2.5: 1. A04-3 (Enterococcus) appears to be the only anomaly. This observation raises the question of potential negative effects of these bacteria on IVF-ET outcome of live birth, as well as whether antibiotic prophylaxis may select for pathogenic bacteria and be harmful in IVF-ET treatment cycles in select women. (iv) Unexpectedly, the diversity of the vaginal microflora at the time of embryo transfer appeared to be an important factor in the success of the IVF-ET procedure using standard-of-care protocols in clinical practice despite different stimulation protocols, use of glucocorticoids, and routine use of antibiotics. A vaginal microbiome composed solely of Lactobacillus (SDI = 0) yields the best prospect for a successful outcome of the IVF-ET procedure.
One of the disadvantages of our study is that the sample size (30 IVF-ET patients) is small. As such, the study could be underpowered for many statistical tests. This situation is undesirable but inevitable given that the experiments (sequencing) are expensive and are limited with regard to time and money. Parametric statistical tests might identify strong effects even with a small sample size. However, fewer assumptions need to be made when employing nonparametric statistical tests, compared to parametric statistical tests, making the nonparametric statistical tests more robust. The use of parametric statistical tests on data violating the distributional assumptions of such tests inherently includes a high risk of making false discoveries that cannot be reproduced by larger scale studies. In point of fact, we verified the statistical significance of the patient’s age and found the statistical significance of the diversity of the vaginal microbiome on the outcome (live birth/no live birth). These two results were established by the use of Student’s t-test. Nonparametric statistical tests, including the Wilcoxon rank sum test and the Wilcoxon signed rank test, were also employed when the data were of non-Gaussian distribution. This category included changes in the blood estradiol concentration, the Shannon diversity indices, and the percent of Lactobacillus reads. Thus, some statistically significant relationships were established with nonparametric statistical tests despite the small sample size.
Using different IVF-ET protocols for different patients may further reduce the sample size for testing, whether the outcome (live birth/no live birth) correlated with the measured parameters. However, the clinical protocols used were part of the patients’ routine care to maximize the probability of a successful outcome (live birth), given a patient’s individual history. The authors had no input into choosing which protocol was employed for each patient. It should be noted that the diversity of the IVF-ET protocols was probably of little relevance to the blood draws because all patients who started treatment had low estradiol levels at baseline and were hyperestrogenic during stimulation. The individual IVF-ET protocols may influence the mix of microflora of the vaginal epithelium because the vaginal epithelia of LLP patients are exposed for a longer period to estradiol and progesterone than those of patients receiving antagonist protocols. In addition, all IVF-ET patients received glucocorticoids and antibiotics commonly used in practice for IVF-T treatment cycles. These treatments could also influence the mix of microflora of the vaginal epithelium.
In conclusion, we believe that this study will serve as a pilot study, such that the discoveries made may be used to guide the design of future larger scale studies. While our patient population was heterogeneous and relatively small, this study supports the need for large, well-controlled studies on the vaginal microbiome and IVF-ET outcomes.
Electronic supplementary materials
(DOC 159 kb)
(DOC 51 kb)
(DOC 85 kb)
(DOC 86 kb)
(DOC 683 kb)
(DOC 56 kb)
(DOC 570 kb)
(DOC 30 kb)
Acknowledgements
We thank Liza Jalalian and Shehua Shen for assistance with the measurement of the serum hormone concentrations, and Monika Trebo for posting the CEL files on the Stanford Genome Technology Center website.
This work was supported by National Human Genome Research Institute grant P01 HG000205 (RWD, LCG).
Competing interests statement The authors declare no competing financial interests.
Glossary
- AP
Antagonist Protocol
- B
At baseline
- DH
Demi-Halt Protocol
- E2
Estradiol
- GE
After 6-to-8 weeks of gestation
- GnRH
Gonadotropin-releasing hormone
- hCG
Human chorionic gonadotropin
- IRB
Institutional Review Board
- IVF-ET
In vitro fertilization-embryo transfer
- LF
At late follicular stage
- LLP
Long Luteal Protocol
- MFP
Microflare Protocol
- P4
Progesterone
- PCA
Principal Component Analysis
- RDP
Ribosomal Database Project
- rDNA
The 16S ribosomal RNA gene
- SDI
Shannon Diversity Index
- SGTC
Stanford Genome Technology Center
- TR
At embryo transfer
- UCSF
University of California San Francisco
- VLDL
Very Low Dose leuprolide acetate Protocol
Footnotes
Capsule
Metagenomics was used to determine the vaginal microbiome in IVF-ET cycles. Diversity of species varied in different hormonal milieu and on the day of embryo transfer correlated with outcome (live birth/no live birth). The species diversity index distinguished women who had a live birth from those who did not.
Author contributions
RWH and LCG designed the experiments and wrote this manuscript. CNH developed the clinical database of IVF-ET patients and coordinated running the hormone assays. HJ performed several of the statistical analyses. CP applied UniFrac to the data and undertook the QIIME analyses. MF carried out the sequencing reactions, processed and assembled the sequence reads, and compared the consensus sequences to the data in the RDP. MF and RWH hand edited the contigs. KCV and DB identified appropriate patients, screened and enrolled patients, facilitated UCSF IRB compliance and sample collection, and transfer to the SGTC. ZZ measured the hormone concentrations. RWD provided the intellectual, physical, and financial milieu for the experiments at the SGTC.
References
- 1.Bezirtzoglou E, Voidarou Ch, Papadaki A, Tsiotsias A, Kotsovolou O, et al. Hormone therapy alters the composition of the vaginal flora in ovariectomized rats. Microb Ecol. 2008;55:751–759. doi: 10.1007/s00248-007-9317-z. [DOI] [PubMed] [Google Scholar]
- 2.Schloss PD, Handelsman J. Status of the microbial census. Microbiol Mol Biol Rev. 2004;68:686–691. doi: 10.1128/MMBR.68.4.686-691.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hyman RW, Fukushima M, Diamond L, Kumm J, Giudice LC, Davis RW. Microbes on the Human Vaginal Epithelium. Proc Natl Acad Sci USA. 2005;102:7952–7957. doi: 10.1073/pnas.0503236102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Selman H, Mariani M, Barnocchi N, Mencacci A, Bistoni F, et al. Examination of bacterial contamination at the time of embryo transfer and its impact on the IVF/pregnancy outcome. J Assist Reprod Genet. 2007;24:395–399. doi: 10.1007/s10815-007-9146-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hyman RW, Jiang H, Fukushima M, Davis RW. A direct comparison of the KB Basecaller and phred for identifying the bases from DNA sequencing using chain termination chemistry. BMC Research Notes. 2010;3:257. doi: 10.1186/1756-0500-3-257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cole JR, Wang Q, Cardenas E, Fish J, Chai B, Farris RJ, Kulam-Syed-Mohideen AS, McGarrell DM, Marsh T, Garrity GM, Tiedje JM. The Ribosomal Database Project: improved alignments and new tools for rRNA analysis. Nucleic Acids Res. 2009;37:D141–5. doi: 10.1093/nar/gkn879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wayne LG, Brenner DJ, Colwell RR, Grimont PAD, Kandler O, et al. Report of the ad-hoc-committee on reconciliation of approaches to bacterial systematics. Int J Syst Bacteriol. 1987;37:463–464. doi: 10.1099/00207713-37-4-463. [DOI] [Google Scholar]
- 8.Pace NR. A molecular view of microbial diversity and the biosphere. Science. 1997;276:734–740. doi: 10.1126/science.276.5313.734. [DOI] [PubMed] [Google Scholar]
- 9.Stackebrandt E, Frederiksen W, Garrity GM, Grimont PA, Kämpfer P. Report of the ad hoc committee for the re-evaluation of the species definition in bacteriology. Int J Syst Evol Microbiol. 2002;52(3):1043–1047. doi: 10.1099/ijs.0.02360-0. [DOI] [PubMed] [Google Scholar]
- 10.Caporaso JG, Kucynski J, Stombaugh J, Bittner K, Bushman FD, Costello EK, Fierer N, Pena AG, Goodrich JK, Gordon JI, Huttley GA, Kelley ST, Knights D, Koenig JE, Ley RE, Lozupone CA, McDonald D, Muegge BD, Pirrung M, Reeder J, Sevinsky JR, Turnbaugh PJ, Walters WA, Widman J, Yatsunenko T, Zaneveld J, Knight R. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods. 2010;7:335–6. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lozupone C, Lladser ME, Knights D, Stombaugh J, Knight R. UniFrac: an effective distance metric for microbial community comparison. ISME J. 2011;5:169–172. doi: 10.1038/ismej.2010.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Roseboom TJ, Vermeiden JPW, Schoute E, Lens JW, Schats R. The probability of pregnancy after embryo transfer is affected by the age of the patient, cause of infertility, number of embryos transferred and the average morphology score, as revealed by multiple logistic regression analysis. Hum Reprod. 1995;10:3035–3041. doi: 10.1093/oxfordjournals.humrep.a135842. [DOI] [PubMed] [Google Scholar]
- 13.Shannon CE. A mathematical theory of communication. Bell System Technical Journal. 1948;27:379–423. [Google Scholar]
- 14.Hughes JB, Hellmann JJ, Ricketts TH, Bohannan BJM. Counting the Uncountable: Statistical Approaches to Estimating Microbial Diversity. Appl. Environ. Micro. 2001;67:4399–4406. doi: 10.1128/AEM.67.10.4399-4406.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fredricks DN, Fiedler TL, Marrazzo JM. Molecular identification of bacteria associated with bacterial vaginosis. N Engl J Med. 2005;353:1899–911. doi: 10.1056/NEJMoa043802. [DOI] [PubMed] [Google Scholar]
- 16.Oakley BB, Fiedler TL, Marrazzo JM, Fredricks DN. Diversity of human vaginal bacterial communities and associations with clinically defined bacterial vaginosis. Appl Environ Microbiol. 2008;74:4898–4909. doi: 10.1128/AEM.02884-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ravel J, Gajer P, Abdo Z, Schneider GM, Koenig SS, McCulle SL, Karlebach S, Gorle R, Russell J, Tacket CO, Brotman RM, Davis CC, Ault K, Peralta L, Forney LJ. Vaginal microbiome of reproductive-age women. Proc Natl Acad Sci U S A. 2011;108(Suppl 1):4680–7. doi: 10.1073/pnas.1002611107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Forney LJ, Gajer P, Williams CJ, Schneider GM, Koenig SS, et al. Comparison of self-collected and physician-collected vaginal swabs for microbiome analysis. J Clin Microbiol. 2010;48:1741–8. doi: 10.1128/JCM.01710-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jakobsson T, Forsum U. Changes in the predominant human Lactobacillus flora during in vitro fertilisation. Ann Clin Microbiol Antimicrob. 2008;7:14–21. doi: 10.1186/1476-0711-7-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Witkin SS, Kligman II, Grifo JA, Rosenwaks Z. Chlamydia trachomatis detected by polymerase chain reaction in cervices of culture-negative women correlates with adverse in vitro fertilization outcome. J Infect Dis. 1995;17:1657–659. doi: 10.1093/infdis/171.6.1657. [DOI] [PubMed] [Google Scholar]
- 21.Liversedge NH, Turner A, Horner PJ, Keay SD, Jenkins JM, et al. The influence of bacterial vaginosis on in vitro fertilization and embryo implantation during assisted reproduction treatment. Hum Reprod. 1999;14:2411–2415. doi: 10.1093/humrep/14.9.2411. [DOI] [PubMed] [Google Scholar]
- 22.Egbase PE, Al-Sharhan M, Al-Othman S, Al-Mutawa M, Udo EE, et al. Incidence of microbial growth from the tip of the embryo transfer catheter after embryo transfer in relation to the clinical pregnancy rate following in vitro fertilization and embryo transfer. Hum Reprod. 1996;11:1687–9. doi: 10.1093/oxfordjournals.humrep.a019470. [DOI] [PubMed] [Google Scholar]
- 23.Egbase PE, Udo EE, Al-Sharhan M, et al. Prophylactic antibiotics and endocervical microbial inoculation of the endometrium at embryo transfer. Lancet. 1999;354:651–2. doi: 10.1016/S0140-6736(99)02415-0. [DOI] [PubMed] [Google Scholar]
- 24.Fanchin R, Harmas A, Benaoudia F, Lundkvist U, Oliveness F, et al. (Microbials of the cervix assessed at the time of embryo transfer adversely affects in vitro fertilization outcome. Fertil Steril. 1998;70:866–70. doi: 10.1016/S0015-0282(98)00277-5. [DOI] [PubMed] [Google Scholar]
- 25.Moore DE, Soules MR, Klein NA, Fujimoto VY, Agnew KJ, et al. Bacteria in the transfer catheter tip influence the live-birth rate after in vitro fertilization. Fertil Steril. 2000;74:1118–1124. doi: 10.1016/S0015-0282(00)01624-1. [DOI] [PubMed] [Google Scholar]
- 26.Kim YJ, Ku SY, Jee BC, Suh CS, Kim SH, Choi YM, Kim JG, Moon SY. Dynamics of early estradiol production may be associated with outcomes of in vitro fertilization. Fertil Steril. 2010;94:2866–2870. doi: 10.1016/j.fertnstert.2010.06.070. [DOI] [PubMed] [Google Scholar]
- 27.Var T, Tonguc E, Dogan M, Mollamahmutoglu L. Relationship between the oestradiol/oocyte ratio and the outcome of assisted reproductive technology cycles with gonadotropin releasing hormone agonist. Gynecol Endocrinol. 2011;27(8):558–61. doi: 10.3109/09513590.2010.501887. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(DOC 159 kb)
(DOC 51 kb)
(DOC 85 kb)
(DOC 86 kb)
(DOC 683 kb)
(DOC 56 kb)
(DOC 570 kb)
(DOC 30 kb)