Abstract
Background
The genetics and pathophysiology of Alzheimer Disease (AD) and Parkinson Disease (PD) appears complex. However, mitochondrial dysfunction is a common observation in these and other neurodegenerative diseases
Scope of Review
We argue that the available data on AD and PD can be incorporated into a single integrated paradigm based on mitochondrial genetics and pathophysiology.
Major Conclusions
Rare chromosomal cases of AD and PD can be interpreted as affecting mitochondrial function, quality control, and mitochondrial DNA (mtDNA) integrity. mtDNA lineages, haplogroups, such haplogroup H5a which harbors the mtDNA tRNAGln A8336G variant, are important risk factors for AD and PD. Somatic mtDNA mutations are elevated in AD, PD, and Down Syndrome and Dementia (DSAD) both in brains and also systemically. AD, DS, and DSAD brains also have reduced mtDNA ND6 mRNA levels, altered mtDNA copy number, and perturbed Aβ metabolism. Classical AD genetic changes incorporated into the 3XTg-AD (APP, Tau, PS1) mouse result in reduced forebrain size, life-long reduced mitochondrial respiration in 3XTg-AD males, and initially elevated respiration and complex I and IV activities in 3XTg-AD females which markedly declines with age.
General Significance
Therefore, mitochondrial dysfunction provides a unifying genetic and pathophysiology explanation for AD, PD, and other neurodegenerative diseases.
Keywords: Alzheimer Disease, Parkinson Disease, Mitochondria, mtDNA, 3XTg-AD Mouse, oxidative phosphorylation
1. Mitochondrial Etiology of Alzheimer and Parkinson Disease
Evidence strongly supports the conclusion that mitochondrial dysfunction is the major factor in the etiology of Alzheimer Disease (AD) and Parkinson Disease (PD). These diseases preferentially affect the central nervous system, which is the tissue with the highest mitochondrial energy demand. They have a complex genetics which is consistent with the Mendelian and non-Mendelian components of the cellular bioenergetic systems. They exhibit a delayed-onset and progressive course consistent with the age-related accumulation of somatic mitochondrial DNA (mtDNA) mutations and they can be strongly influenced by environmental factors including diet, climate, and the effects of a broad spectrum of environmental toxins.
The original description of AD included a progressive dementia, cortical atrophy, and amyloid plaques. Most AD cases have a late-onset and a seemingly spontaneous appearance. However, less than 1% of cases have an early-onset and exhibit Mendelian inheritance [1]. Genes found to be mutated in the early-onset cases have included Amyloid Precursor Protein (APP) and the genes of the Presenilin 1 (PS1) and 2 (PS2) complexes, one function of which is to process the APP polypeptide. When processed by the β and γ-secretase activities, APP is cleaved into the amyloid peptides, Aβ1–40 and Aβ1–42. The Aβ peptides have been assumed to be toxic, with Aβ1–42 being the more so. The Aβ peptides have a tendency to aggregate, and it has been found that the toxic forms of Aβ are oligomers and to a lesser extent filamentous aggregates [2, 3].
There are now numerous studies and multiple reviews that have reported mitochondrial dysfunction in AD [4–8]. The presenilin complex proteins have been located in the mitochondrion [9], suggesting that the PS complexes are important for normal mitochondrial function. Furthermore, Aβ is imported through the mitochondrial outer membrane “TOMM” (Translocase of Outer Mitochondrial Membrane) complex and it becomes localized in the mitochondrial cristae [10, 11]. APP is also bound to the mitochondrial outer membrane and processed by mitochondrial γ-secretase to release the APP intracellular domain (AICD) within the mitochondrion. Therefore, AICD and Aβ can be generated locally within the mitochondrion as well as being imported [12]. Mitochondrial Aβ is also turned over within the mitochondrion by presequence protease [13].
AD is associated with neurofibrillary tangles, which are composed of hyperphosphorylated Tau. The N-terminal 20–22 kDa of the Tau protein is enriched in the synaptosome mitochondria of AD patients and correlates with Aβ multimeric species. Therefore, Aβ interacts with 20–22 kDa N-terminal Tau and the complex may impair mitochondrial function [14].
Dysfunctional autophagy may also contribute to the decline in mitochondrial function in AD [15]. This may be attributed in part to altered mitochondrial dynamics (fission and fusion) [16]
Aβ has been reported to enter the mitochondrion. There, it inhibits mitochondrial function by increasing mitochondrial membrane viscosity, causing a decrease in ATP/O ratio, reducing electron transport chain (ETC) activity, increasing ROS production, and facilitating cytochrome c release [17]. Incubation of cultured neurons with Aβ oligomers results in an 89% reduction in mitochondrial membrane potential within two hours of exposure, followed by mitochondrial release of cytochrome c and apoptosis initiating factor (AIF) within 8 hours [3]. Synaptic mitochondria are more sensitive to Aβ mitochondrial toxicity than are neuronal mitochondria and synaptic mitochondrial dysfunction is an early manifestation in mouse models of AD [18, 19].
Aβ has been shown to specifically inhibit cytochrome c oxidase (COX or complex IV) [20, 21]. One possible mechanism for this inhibition is that Aβ can bind heme, including the heme-a of COX, creating an Aβ-heme complex, which is also a peroxidase [22]. Chronic exposure of rats to the COX inhibitor NaN3 results in cognitive deficits and neuronal morphological changes [23].
Aβ has been reported to inhibit mitochondrial Mn superoxide dismutase (MnSOD), increasing oxidative stress, and over-expression of MnSOD is protective of AD [24]. Similarly, Aβ has been reported to inhibit cyclophilin D, a component of the mtPTP [25]. Early stage AD brains also show lipoxidation of the α-subunit of the ATP synthase, in association with reduced ATP synthase activity [26]. In addition, Aβ perturbs the mtPTP, inhibits OXPHOS complexes III and IV, and also binds and inhibits mitochondrial alcohol dehydrogenase, designated Aβ Alcohol Dehydrogenase (ABAD) [11, 27]. The interaction between Aβ and ABAD has been associated with mitochondrial dysfunction and dementia, and one hypothesis argues that alteration of ABAD increases mitochondrial aldehyde toxicity [25, 28–30].
PS1 and PS2 have been reported to interact with mitochondrial proteins. They form a complex with the outer mitochondrial membrane protein FKBP38 and Bcl-2 thus modulating mitochondrial mtPTP-mediated apoptosis [31]. PS1 also interacts with Om1/HtrA2, a pro-apoptotic protein stored in the mitochondrial intermembrane space. The C-terminus of PS1 activates the proteolytic activity of Om1/HtrA2, which degrades anti-apoptotic proteins [32].
The PS1 and PS2 complexes have recently been associated with “Mitochondrial-Associated Membranes” (MAMs). MAMs are thought to be specialized compartments of the endoplasmic reticulum (ER), which connect the ER with the mitochondrion and are important in lipid and calcium (Ca++) metabolism. Altered PS metabolism could thus alter Ca++ flux into the mitochondrion, initially activating rate-limiting mitochondrial dehydrogenases, then increasing mitochondrial reactive oxygen species (ROS) production, and ultimately activating the mitochondrial permeability transition pore (mtPTP). Activation of the mtPTP would lead to caspase activation and synaptic loss [33, 34]. Thus, the localization of PS1 & 2 in the MAMs is consistent with the observed association between ER stress, Ca++ dysregulation, and mitochondrial dysfunction in AD [35, 36].
Late-onset AD cases are commonly associated with the apolipoprotein E gene ε4 allele (ApoE ε4) [5]. This association has been linked to mitochondrial dysfunction by demonstrating that transgenic mice which over-express ApoE ε4 have reduced levels of complexes I, IV, and V [37]. The special neuronal toxicity of ApoE ε4 has been proposed to result from ApoE ε4’s unique ability to undergo an interaction between the N- and C-terminal domains, creating a compact structure. ApoE ε4 is then subject to neuron-specific cleavage and the N-terminal cleavage product becomes associated with the mitochondrion and inhibits mitochondrial function [38, 39].
The ApoE ε4 allele is linked to the mitochondrial outer membrane protein import gene, TOMM40. Recently, an association has been observed between a TOMM40 polymorphism and AD, which is independent of the ApoE ε4 association, thus further supporting a direct link between mitochondrial function and AD [40, 41].
About 20% of AD patient brains also show the neuropathologic features of PD (AD-PD). PD is a progressive movement disorder in which the brain develops protein aggregates called Lewy bodies composed of α-synuclein and ubiquitin. As is the case for AD, a small proportion of PD cases have been linked to chromosomal loci.
Of the chromosomal loci, PARK1, encodes α-synuclein, with mutations in this gene being dominant. α-synuclein has been implicated in the maintenance of the contorted mitochondrial membranes [42]. The dominant PARK8 locus encodes the leucine-rich repeat kinase 2 (LRRK2), mutations in which have been associated with OXPHOS dysfunction [43]. The PARK7 locus encodes DJ-1, which has been associated with complex I defects, increased mitochondrial ROS production, reduced mitochondrial membrane potential, and altered mitochondrial morphology [44–46]. DJ-1 defects have been shown to preferentially affect substantia nigra dopaminergic neurons over ventral tegmental neurons due to differences in Ca++ metabolism and uncoupler protein expression [47]. The PARK2 and PARK6 loci encode parkin and PTEN-induced kinase 1 (PINK1), both of which control mitochondrial turnover by mitophagy [48–55]. Thus many of the chromosomal PD genes are part of an integrated pathway for maintaining the integrity of the mitochondria.
The link between AD and PD is also supported by inclusion body myositis, which is associated with intermyofiber accumulations of Aβ. Parkin is expressed in the brain, skeletal muscle, and other tissues, and muscle and cells lacking parkin are more sensitive to intracellular Aβ toxicity. Increased levels of parkin are protective of both mitochondrial toxins and Aβ [56].
2. Mitochondrial Biology and Genetics
These observations indicate that mitochondrial dysfunction is central to the underlying pathophysiology of AD and PD. The mitochondria generate much of the cellular energy and the brain is the organ most reliant on mitochondrial energy, representing 2% of the body’s weight but consuming 20% of the oxygen. Therefore, a systemic mitochondrial defect will preferentially cause symptoms in the brain.
The mitochondria also regulate cellular REDOX state, generate most of the ROS, buffer cytosolic Ca++ levels, and regulate apoptosis through the activation of the mtPTP. Mitochondria and mtDNAs are continuously being produced within cells and the addition of mitochondria to the cell is balanced by an equal rate of degradation of the mitochondria by mitophagy. Hence, the mitochondria and mtDNAs are maintained in a dynamic steady state [5, 57].
The mitochondrial genome consists of the maternally-inherited mtDNA plus between one and two thousand nDNA-encoded genes. The mtDNA encodes key components of mitochondrial OXPHOS specifically related to electron and proton transport. Hence, the mtDNA encodes the circuit diagram for the mitochondrial power plants. The nDNA encodes all of the genes required to assemble the mitochondrion and the structural elements of the OXPHOS complexes [57, 58].
Each cell contains hundreds of mitochondria and each mitochondrion contains multiple mtDNAs packaged in nucleoids. Hence, most cells contain thousands of mtDNAs. The mtDNA encodes 13 polypeptides, 22 tRNAs, and a 12S and 16S rRNA. The 13 polypeptides are the central components of OXPHOS complexes I, III, IV and V. These four enzymes work together to generate and utilize the mitochondrial inner membrane electrochemical gradient. This inner membrane capacitance is produced by the burning of reducing equivalents (electrons) derived from the carbohydrates and fats in our diet with the oxygen that we breathe to produce water. The energy that is released as the electrons flow through the electron transport chain (ETC) complexes I, III, and IV is used to pump protons out across the mitochondrial inner membrane. The potential energy in this capacitor is then used by complex V (ATP synthase) to condense ADP + Pi into ATP. The ATP and ADP are exchanged across the mitochondrial inner membrane by the adeneine nucleotide tranlocators (ANTs) and the outer membrane by the voltage dependent anion channels (VDACs).
Complex I (NADH dehydrogenase) is composed of 45 polypeptides, seven (ND1, 2, 3, 4L, 4, 5, & 6) encoded by the mtDNA; complex III of 11 polypeptides, one (cytochrome b (cytb)) encoded by the mtDNA; complex IV of 13 polypeptides, three (COI, II, & III) encoded by the mtDNA; and complex V of approximately 15 polypeptides, two (ATP6 & ATP8) encoded by the mtDNA. All of the mtDNA mRNAs, except for ND6, are encoded by the guanosine (G)-rich heavy (H)-strand of the mtDNA and are derived from one long polycistronic transcript. This transcript also encompasses the two rRNA genes and the majority of the tRNA genes, which punctuate the rRNA and polypeptide sequences. As this transcript is generated, the tRNAs are cleaved out to generate the mature rRNA and mRNA transcripts which are then poly-adenylated. ND6, by contrast, is transcribed from the cytosine (C)-rich light (L)-strand using an independent L-strand promoter. ND6 is critical for complex I assembly.
The 22 tRNAs and two rRNAs are the structural RNAs for the bacteria-like mitochondrial protein synthesis apparatus. The mitochondrial mRNAs are translated within the mitochondrion on ribosomes that are sensitive to certain bacterial ribosome inhibitors, for example chloramphenicol [59]. Consistent with the bacterial origin of the mitochondria, the mitochondrial ribosomal translation products are initiated by a formyl-methionine, while cytosolic ribosomal translation products are not. Hence, the mitochondrial translation products have the same molecular signature as those of exogenous bacteria [5, 57].
The genetics of the mitochondrion is complex because it encompasses the rules of both Mendelian and non-Mendelian genes. The mtDNA is exclusively maternally-inherited and represented by thousands of copies per cell. In the process of making energy, the mitochondrion generates much of the cellular ROS, which can damage the mitochondria and mutagenize the mtDNA. Hence, the mtDNA has a very high mutation rate. When an mtDNA mutation arises in a cell, this creates an intracellular mixture of mutant and normal mtDNAs, a state known as heteroplasmy. When the cell undergoes either mitotic or meiotic cytokinesis the mutant and normal mtDNAs are distributed to the daughter cells stochastically. Therefore, the percentage of mutant mtDNAs can drift during cell division toward either more or less mutant. As the percentage of mutant mtDNAs increases, the energy output of the cell declines until it crosses the minimum energy threshold for that tissue to function, the bioenergetic threshold, at which point symptoms appear.
Three types of mtDNA variation are relevant to neurodegenerative disease: maternally transmitted recent deleterious mutations, ancient adaptive polymorphisms that are maladaptive in the modern environment, and somatic mtDNA mutations that accumulate in tissues with age.
A large number of inherited mtDNA mutations have been described to cause neurological disease, including dementias and movement disorders [5, 60].
The mtDNA is also highly polymorphic with functional mtDNA variants having been enriched in ancient regional populations because the variants were advantageous in that local environment. As the frequency of these adaptive variants increased due to selection, additional mtDNA mutations accumulate along the radiating maternal lineage. This created groups of related mtDNA haplotypes, known as haplogroups [61–64]. Founding functional variants of the various haplogroups are now being shown to predispose to a wide range of degenerative diseases [5, 57, 65].
Finally, in post-mitotic cells, mitochondrial ROS as well as other mutagenic processes result in the accumulation of somatic mtDNA mutations in tissues with age. As more mtDNAs become damaged, the energy output of the cell declines until it falls below its bioenergetic threshold. Energy limitation initially results in a decline in cellular function, but ultimately can culminate in the activation of the mtPTP and the destruction of the faulty cell by apoptosis. This age-related accumulation of somatic mtDNA mutations provides the aging clock and can exacerbate partial OXPHOS defects resulting from inherited nuclear DNA (nDNA) or mtDNA mutations or polymorphisms [5, 57]. The common 5 kb mtDNA deletion, for example, accumulates with age in many tissues, but it reaches the highest levels in the cerebral cortex and the basal ganglion of the brain, the tissues affected by AD and PD [66, 67].
Amelioration of the effects of the age-related accumulation of somatic mtDNA mutations may be one of the main functions of the mitochondrial fusion-fission and the PINK1-parkin systems. Deleterious mtDNA mutations can complement each other in trans thus retarding mitochondrial functional decline [68], but this also masks the mutant mtDNAs. Mitochondrial fission and fusion can partition mutant mtDNA nucleoids into separate mitochondrial structures. If the mutation results in a decline in the mitochondrial membrane potential, then the PINK1-parkin system is activated to eliminate the mutant nucleoid and defective mitochondria by mitophagy [55]. Mutational inactivation of the PINK1-parkin pathway removes this mitochondrial maintenance system and permits a more rapid accumulation of mutant mtDNA.
3. Inherited mtDNA Variation in AD and PD
3.1. Haplogroups
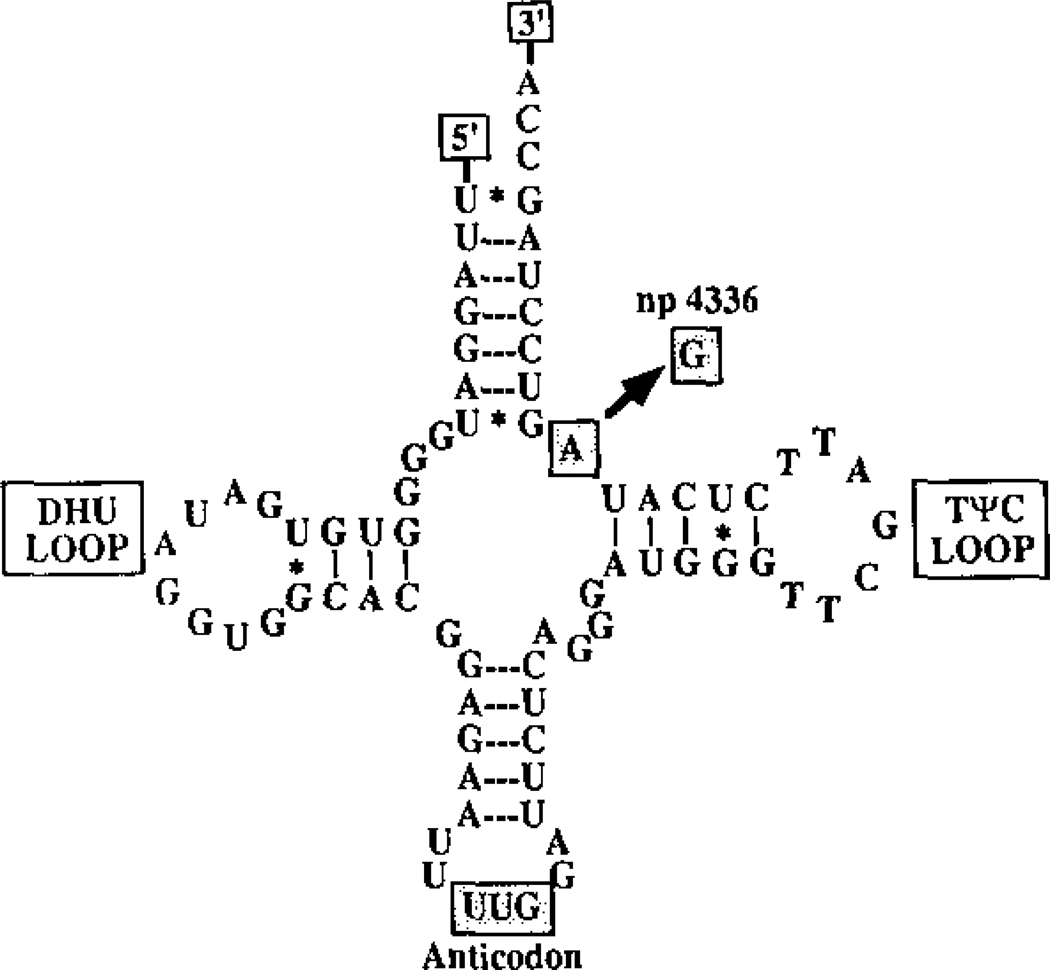
In 1993, we reported that a variant in the tRNAGln gene at nucleotide (nt) A4336G was enriched in AD and PD patients (Figure 1). In our initial survey, we found this variant in 3.2% AD, 5.3% PD and 6.8% AD+PD patients but only 0.4% controls. This variant arose about 8,500 to 17,000 years ago creating a European mtDNA lineage predisposed to neurodegenerative disease, now designated haplogroup H5a [69]. The association between the 4336 variant and AD was first confirmed in 1995 [70], has been observed multiple additional times [5], and most recently reaffirmed in a survey of 936 AD patients and 776 controls from central and northern Italy, where it was found in 2% of AD patients and 0.8% of controls [71]. Additional mtDNA variants have also been associated with AD in ours and other regional studies [5, 57, 69, 72].
Figure 1.
mtDNA tRNAGln nt A4336G Mutation Associated with Late-Onset AD and PD. Reprinted from [69].
Since this first association, multiple studies have shown that certain mtDNA haplogroups either increase or decrease the probability of neurodegenerative disease. Haplogroups U and T have been associated with decreased risk of developing AD in most contexts [69, 73–75], though Uk was found to increase AD risk in a brain imaging study [76]. Similarly, haplogroup H has been associated with increased risk, and haplogroups J and Uk with decreased risk, for developing Parkinson Disease [77–79]. The variable effects of haplogroup Uk in AD may stem from the fact that it has a relatively strong effect on mitochondrial OXPHOS and thus energy output [65]. Hence, this haplogroup may be particularly sensitive to contextual influences. For example, in a study of mtDNA haplogroups and AIDs progression, Uk had the opposite influence as did other U and J mtDNA haplogroups [80].
3.2. Somatic mtDNA Variation in AD
In addition to correlations with ancient polymorphisms, somatic mtDNA mutations, such as the common 5 kb deletion, have been observed to be increased in the frontal cortex of AD [81] and Huntingtion Disease brains [82]. Somatic mtDNA deletions have also been observed in the neurons and cells of the substantia nigra in PD patients and aged-controls. The neurons containing the mtDNA deletions also show OXPHOS enzyme defects by histochemical staining [83, 84].
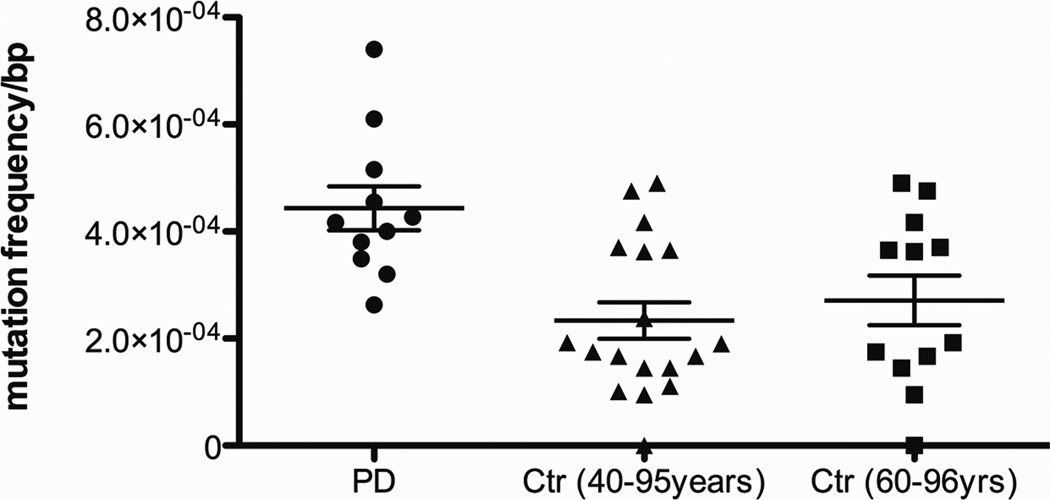
Base substitution mutations in the mtDNA are also elevated in the brains of AD patients and in Down Syndrome (DS) patients that progress to an AD-like dementia (DSAD)[85, 86]. We have found that AD brains, but not control brains, harbor a mtDNA control region mutation, T414G. This mutation alters the mitochondrial transcription factor (Tfam) binding site associated with the L-strand promoter, which transcribes the ND6 gene [87, 88]. This mutation was originally observed to accumulate with age in human skin fibroblasts [89]. Subsequent cloning and sequencing of the control regions from AD and control subjects revealed that AD brains harbored more mtDNA mutations than control brains, and that some mutations can accumulate to high levels of heteroplasmy in AD brains. Furthermore, in contrast to the less frequent control brain mutations, the AD mutations preferentially altered known mtDNA control region transcription and replication regulatory sites. An analogous significant increase in somatic mtDNA control region mutations have also been observed in PD brain samples (Figure 2).
Figure 2.
Increased mtDNA Control Region Mutation Levels in PD Brains. These data were collected as in [85] and analyzed as in [86] and found to be significant (p=0.002 ANOVA).
The elevated mtDNA control region mutation levels in AD brains were correlated with a 50% reduction in the L-strand ND6 transcript levels relative to the H-strand ND2 gene transcript levels and a comparable reduction in the mtDNA/nDNA copy number [85]. That control region mutations could affect both L-strand transcript levels and mtDNA copy number was confirmed by our demonstration that a single nucleotide change in the haplogroup J control region was associated with a doubling of mtDNA L-strand transcription and mtDNA copy number [90]. However, the reduced ND6 mtDNA levels might also be the product of an epigenetic response to the systemic mitochondrial defect. Recently, it has been found that the myocyte enhancer factor 2 (MEF2) family member, MEF2D, is imported into the mitochondrion and that it binds to the mtDNA at a site within the ND6 gene. MEF2D has been found to be required for the maintenance of ND6, and loss of MEF2D results in the decline of ND6 mRNA levels, loss of complex I activity, reduced ATP production, and increased H2O2 production. MEF2D is located in neurons and is required for neuronal development, synaptic plasticity, and survival. Moreover, brains of animals treated with the complex I inhibitors MPTP and rotenone and autopsy brains of PD patients have been found to be deficient in MEF2D [91]. If altered MEF2D expression also occurs in AD brains, it might contribute to lower ND6 levels in both neurodegenerative diseases.
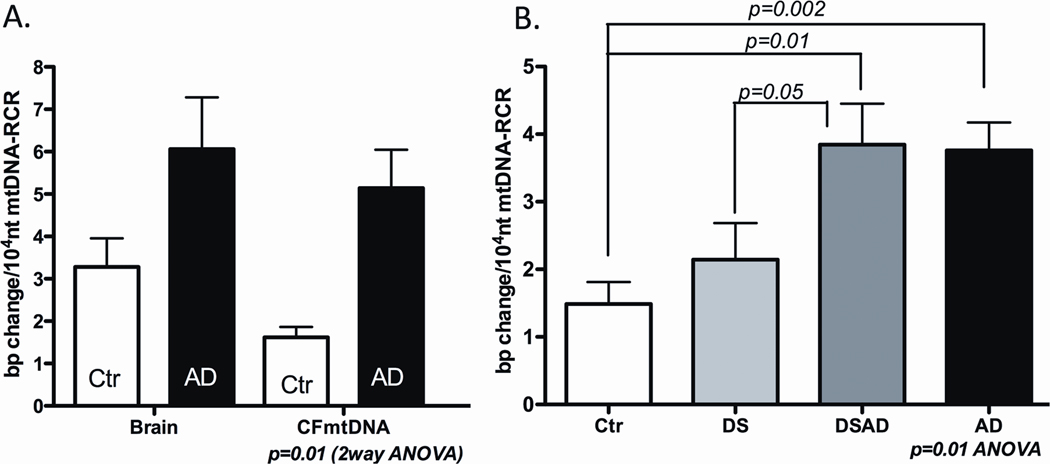
We have confirmed and expanded our studies on brain somatic mtDNA mutation rates by cloning and sequencing the entire mtDNA control region from AD, DS, DSAD, and control subjects (Figure 3). Again, the somatic mutation levels of the mtDNA control region were found to be elevated in the AD and DSAD brains relative to DS and control brains, and the DSAD and DS somatic mutations preferentially affected functional nucleotides and were occasionally found at high heteroplasmy levels [86].
Figure 3.
Elevated Somatic mtDNA Mutation Rates AD and DSAD. Panel A: Elevated somatic mtDNA mutation levels in the brains and blood samples of the same patients and controls, Ctr = control and CFmtDNA = cell free mtDNA sample. Panel B: Elevated somatic mtDNA mutation levels in lymphoblastoid cell lines from AD and DSAD patients. Reprinted from [86].
Analyzing the mtDNA control region frequencies in blood and brain samples from the same AD and control subjects revealed that mtDNA control region mutation levels were elevated in both tissues (Figure 3A). Hence, the increased mtDNA somatic mutation levels are not brain-specific, but are likely to be elevated throughout the body. Moreover, in some cases the same somatic mutation was found at high heteroplasmy levels in both blood and brain. In one case, a control region mutation, G185A, was present in 100% of the brain mtDNA and 91% of the blood mtDNAs. This patient’s mtDNA was haplogroup U5a1, and this haplogroup does not normally harbor the G185A mutation. In another case, the mutation G207A was found in 75% of the brain mtDNAs and 94% of the blood mtDNAs. This same G207A mutation was also found in 88% of the brain mtDNAs in another case. These observations indicate that the G185A and G207A mutations arose early in development and became selectively enriched to high levels in multiple organs over the lifetime of the patient [86].
Further evidence that an increased somatic mtDNA mutation rate is a systemic feature of AD patients was obtained by evaluating the mtDNA control region mutation levels in lymphoblastoid cell lines derived from AD, DS, DSAD, and control blood samples (Figure 3B). Cell lines derived from both AD and DSAD patients retained elevated mtDNA control region somatic mutation levels relative to the DS and control lymphoblastoid cell lines. Therefore, either the mutant mtDNAs are preferentially replicated in the lymphoblastoid cell lines or the high mtDNA mutation rate is retained in these cells and is continuously adding new mtDNA mutations as the old ones are lost [86].
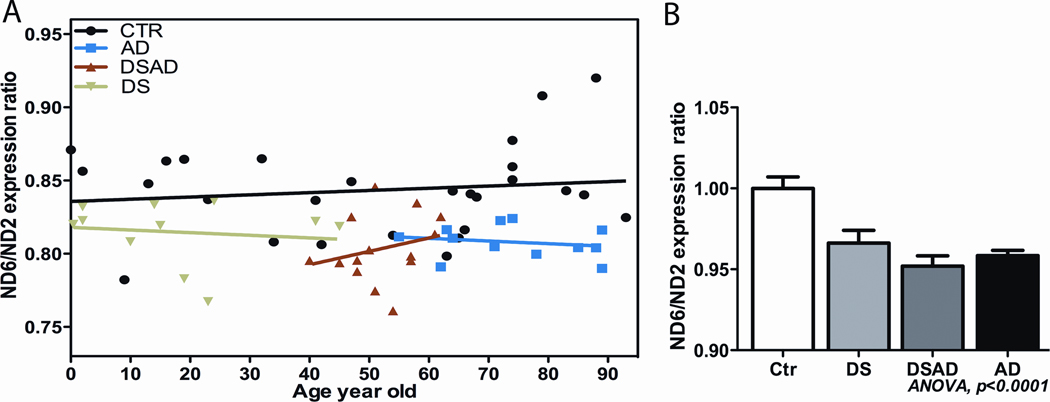
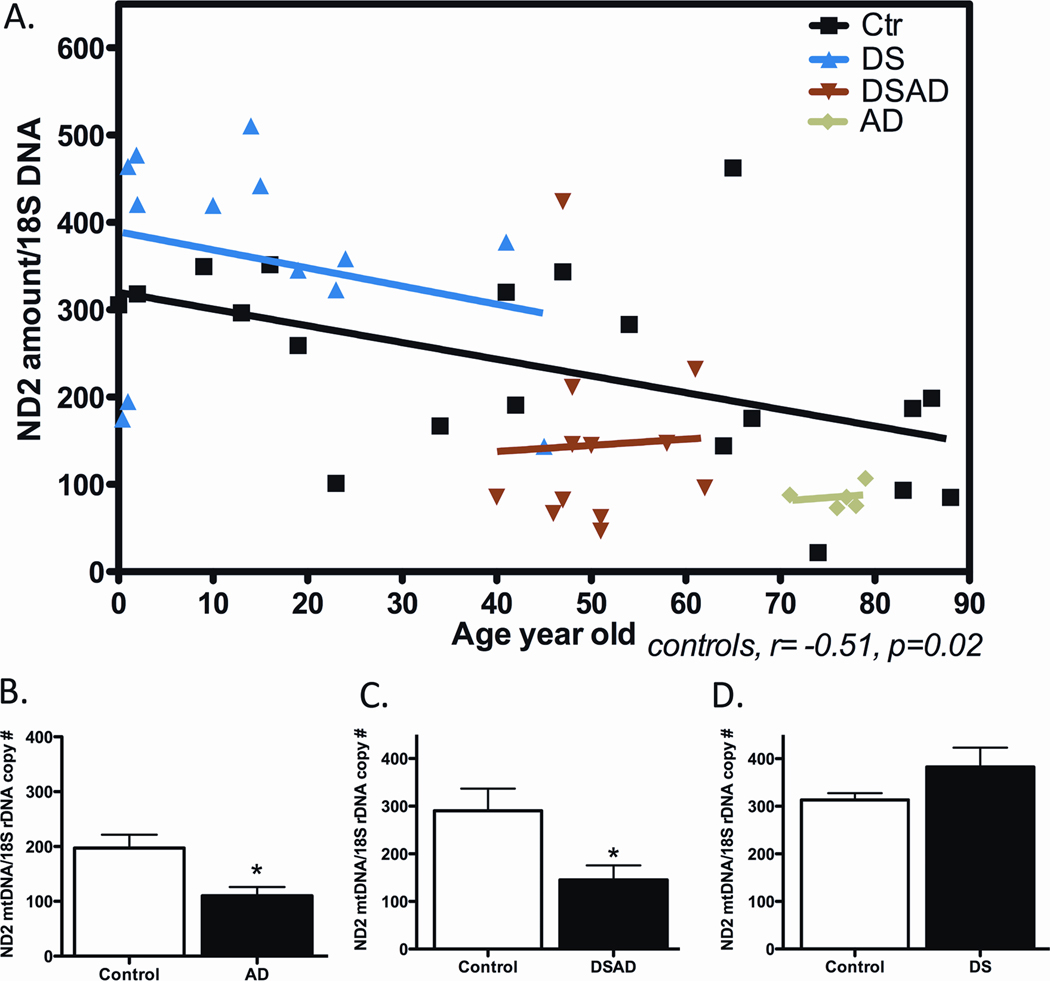
To determine if the increased mtDNA somatic mutations affected mtDNA function, we again examined the ratio of ND6 L-strand mRNAs to ND2 H-strand mRNA levels and the mtDNA/nDNA ratio in AD, DS, DSAD, and control brains. Consistent with our original AD report, we observed that the ND6 transcript levels were reduced in AD, DS, and DSAD brains relative to controls (Figure 4).
Figure 4.
Reduced mtDNA L-Strand ND6 Transcript in DS, DSAD and AD Brains. Data presented as ratio of ND6 to ND2 mRNA levels, determined by real-time quantitative PCR. Panel A: Data from individual subjects and controls plotted according to their age. Panel B: Mean and standard deviation values for ND6/ND2 ratios. Data reprinted from [86].
The cellular mtDNA/nDNA ratio was also reduced in AD brains as well as DSAD brains relative to controls (Figure 5). Interestingly, the DS mtDNA/nDNA ratio proved to be elevated (Figure 5). Given that all of these cell lines had reduced mtDNA ND6 transcript levels and presumable partial complex I defects, then the elevated DS mtDNA levels might represent a compensatory up-regulation of mitochondrial biogenesis in response to a mitochondrial defect caused by trisomy 21.
Figure 5.
Reduction in mtDNA Copy Number in AD and DSAD Brains. The mtDNA/nDNA copy number was determined by quantitative real-time PCR of purified brain genomic DNA. Panel A: The mtDNA/nDNA ratio on patients and controls plotted by age. Panel B: The mean and standard deviation of each category of patient. Reprinted from [92].
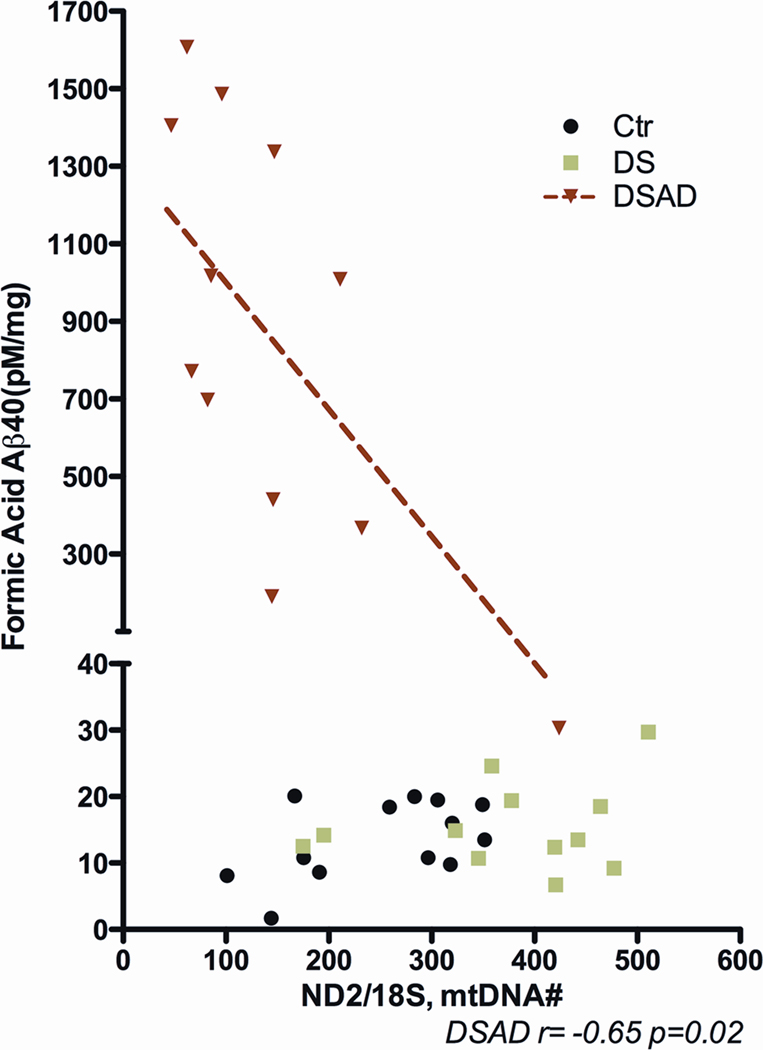
These mtDNA genetic alterations also correlate with altered Aβ metabolism. In DSAD brains, β secretase activity was observed to increase as the somatic mtDNA control region mutation levels increased in brain. Furthermore, the β secretase activity increased as the brain mtDNA/nDNA ratio declined. While soluble Aβ42 accumulated with mtDNA/nDNA ratio, insoluble Aβ40 increased as the mtDNA/nDNA ratio declined (Figure 6). Hence, mitochondrial somatic mutations and dysfunction correlate with alterations in Aβ metabolism [86].
Figure 6.
Insoluble Aβ Aggregates Increase as the mtDNA/nDNA Ratio Decreases in DSAD Brains. Results reprinted from [86].
Since somatic mtDNA mutations are found to arise early in development, long before Aβ accumulation in the brain, Aβ accumulation can not account for the increased mtDNA somatic mutation levels. Rather, the increased mtDNA somatic mutation levels can account for the increased Aβ accumulation.
3.3. Physiological Consequences of Increased mtDNA Mutations in AD & PD
Genetic evidence that mtDNA mutations can institute AD-like changes comes from cybrid studies [93, 94]. AD cybrids are generated by fusion of blood platelets from AD patients to established neuroblastoma cell lines which lack mtDNA (ρo) [95]. Extensive analysis of such cybrid cell lines has revealed that their bioenergetic function declines with increasing time in cell culture [96]. AD-neuroblastoma cybrids have been shown to have reduced complex IV specific activity, increased oxidative stress, Aβ peptide secretion, and flickering of the mitochondrial ΔψM linked to ATP-synthase activity [97]. Differentiated neuroblastoma cybrids have elongated mitochondria, reduced mitochondrial movement along cybrid neurites [98], and increased autophagic vesicles consistent with reduced microtubule networks [99]. Differentiated AD cybrids also have ROS-mediated alterations in tyrosine and serine-threonine kinase activity associated with reduced tropomyocin receptor (TrkA) signaling and p75 neurotropin receptor expression. This is associated with altered nerve growth factor signaling, generation of Aβ, and reduced cell viability [100].
Therefore, all of the above studies support the conclusion that AD is the product of the decline in mitochondrial function, to which the brain responds by altering APP processing to produce Aβ. Initially, Aβ is non-deleterious, perhaps even protective. However, as the level of Aβ accumulates and enters the mitochondrion, it ultimately oligomerizes and becomes toxic to the mitochondria disrupting energy homeostasis [101].
That Aβ is produced as a soluble monomer and only becomes toxic after it reaches a sufficient concentration to form oligomers implies that it has two different functional phases. Phase one is either innocuous or benefical. Phase two is lethal.
Proteomic analysis of AD brain mitochondria has revealed an induction of proteins associated with mitochondrial energy metabolism, including ANT, VDAC, hexokinase, and creatine kinase [102]. This might reflect a compensatory up-regulation of mitochondrial energy production in response to a chronic mitochondrial dysfunction or an initially protective aspect of Aβ and/or Tau for mitochondrial function.
We have observed a similar biphasic consequence of the induction of the heart-muscle-brain adenine nucleotide translocase, isoform 1 (Ant1) in response to brain injury. The Ant proteins not only export mitochondrial ATP into the cytosol in exchange for cytosolic ADP, they also regulate the mtPTP and thus the initiation of apoptosis. In damaged brain, Ant1 expression is induced in astrocytes by elevated TGFβ, acting through the SMAD pathway [103, 104]. Presumably, the increased Ant1 increases ATP export to facilitate repair of the cellular damage as well as facilitate the uptake of excitotoxic glutamate. While knockout of Ant1 has no effect on unchallenged brain function, when Ant1−/− mice are challenged with the kainate, they prove to be substantially more resistant to excitotoxic death. This correlates with an increased resistance of the neuronal mtPTP to Ca++ activation [105]. These data indicate that initially, the Ant1 induction increases ATP export to aid in the repair of damaged cells and neurons. However, if the cells are sufficiently damaged that the increased ATP does not resolve the cellular problem, the Ant continues to be induced, eventually reaching a sufficiently high level that it activates the mtPTP and destroys the damaged cell by apoptosis. Early in life when cells are plentiful, this strategy would rid tissues of faulty cells which could degrade neurological function. However, later in life, as cells become limiting, the continued loss of cells would impair the computational power of the brain.
By analogy, induction of Aβ by mitochondrial dysfunction might initially serve a protective function for the cell and its mitochondria. For example, Aβ monomers may function as anti-oxidants [5]. However, if continued mitochondrial dysfunction persists, then induction of Aβ will continue until the Aβ begins to aggregate into oligomers. At this point, the Aβ begins to inhibit mitochondrial function, ultimately leading to bioenergetic failure, activation of the mtPTP, and the destruction of the faulty neurons. Again, such a system would be advantageous early in life with abundant cells, but leads to cortical and basal ganglion neuronal depletion later in life resulting in AD and PD, respectively. By this model, Ant1 and Aβ induction exhibits antagonistic pleiotrophy.
4. Mitochondrial Functional Changes with Age in 3XTg-AD Mice
To test this biphasic hypothesis of Aβ action, we characterized mitochondrial OXPHOS function in the forebrains of a mouse AD model throughout their life span. We used the triple transgenic-AD mouse model (3XTg-AD), which is homozygous for three transgenes, PS1M146V, tauP301L, and APPSwe(KM670/671NL), maintained on a 129/C57BL6 hybrid background. This model develops the plaques and tangles seen in AD neuropathology and develops behavior abnormalities later in life [106].
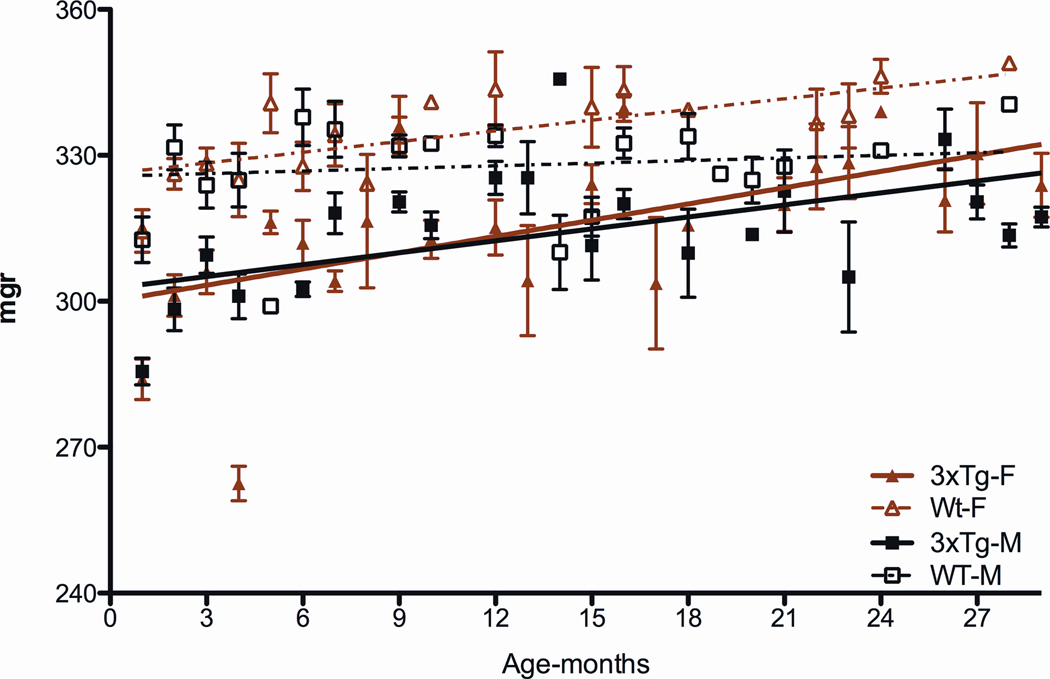
Analysis of the 3XTg-AD mouse forebrain weights revealed that both male and female 3XTg mice have reduced brain sizes at birth and throughout life (Figures 7). We hypothesize that this brain size reduction may be the result of some developmental abnormality.
Figure 7.
Reduced Forebrain Weight in 3XTg-AD Mice Throughout Life.
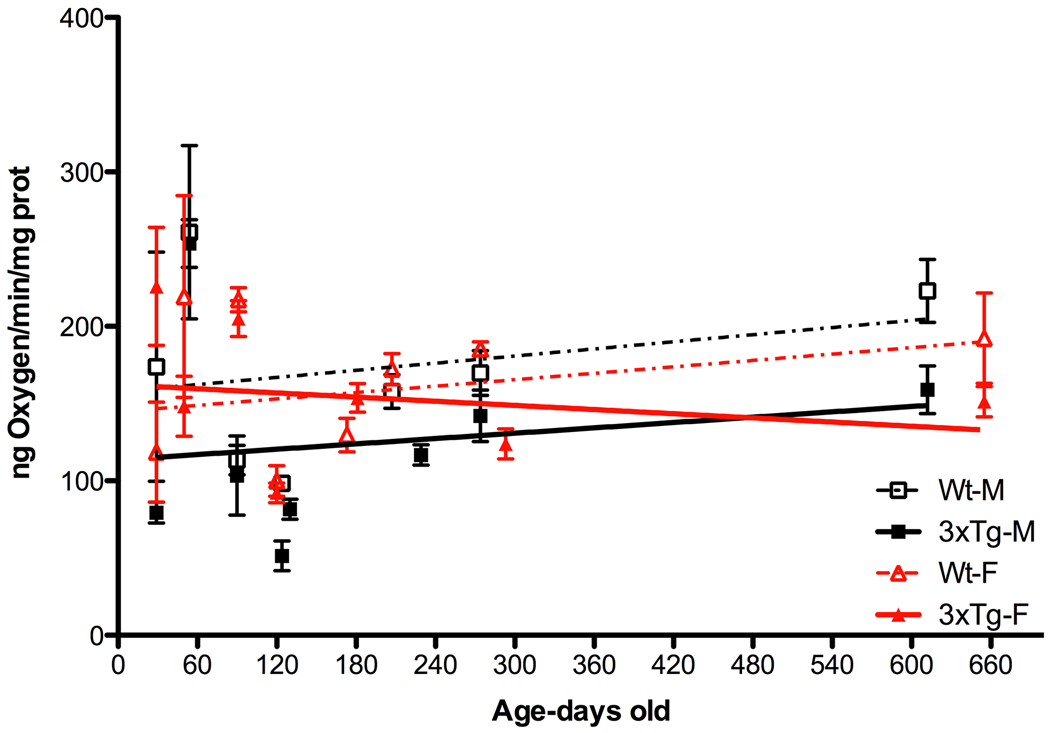
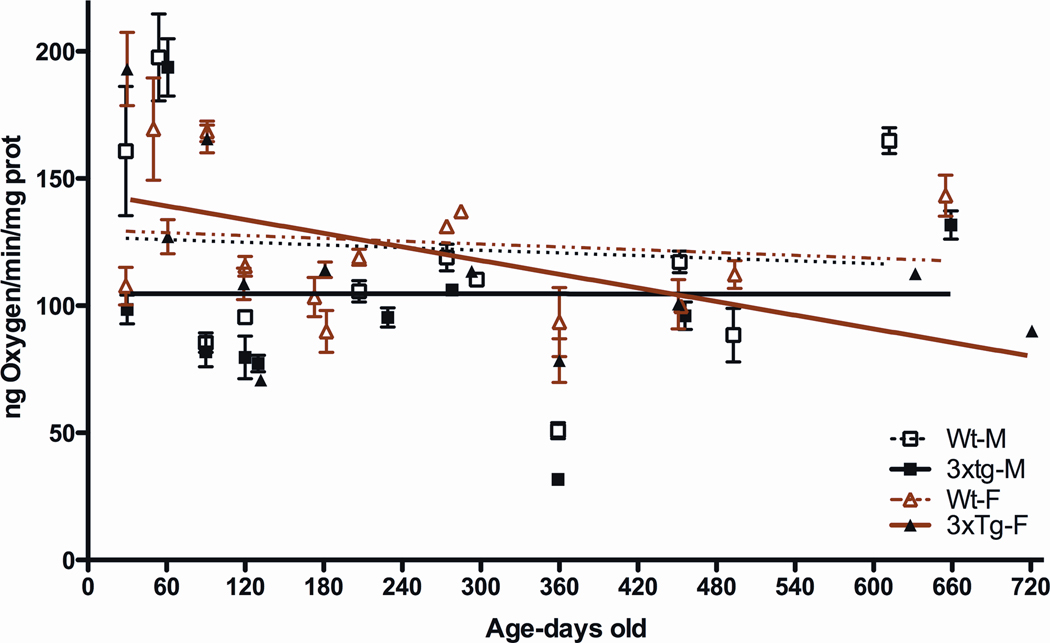
We next analyzed the mitochondrial respiration of isolated forebrain mitochondria. We first examined the electron flux down the ETC by measuring mitochondrial oxygen consumption, driven by the site I substrates, pyruvate and malate, which feed electrons into complex I (Figure 8). These studies were performed in the presence of the OXPHOS uncoupler (FCCP) to release the constraints on respiration imposed by ATP synthase (complex V). The male 3XTg-AD mouse mitochondria had consistently lower uncoupled respiration rates than male controls. The female 3XTg-AD mouse mitochondria initially had an uncoupled mitochondrial respiration rate similar to that of the female control mitochondria, but this declined throughout life descending to the same final level as seen for the male 3XTg-AD male mitochondria. Thus, both male and female 3XTg-AD mice had impaired mitochondrial ETC, but the males appear to be affected much earlier than the females.
Figure 8.
Consistently Reduced Mitochondrial Uncoupled Respiration in Male 3XTg-AD Mice and Declining Respiration in 3XTg-AD Female Mice
To further investigate the sex difference in mitochondrial responses of the 3XTg-AD mouse mitochondria, we analyzed the ADP-stimulated (State III) respiration when metabolizing the site I substrates, pyruvate and malate (Figure 9). The state III respiration rates of the male 3XTg-AD mice were again consistently below that of control mice throughout life. The female 3XTg-AD mice had initial state III respiration rates that were greater than those of female control mice. However, these female state III respiration rates declined as the mice aged, ultimately falling below the control level by about 420 days of age and continuing to decline until the end of life.
Figure 9.
Consistently Reduced Mitochondrial State III Respiration in Male 3XTg-AD Mice and Declining Respiration in 3XTg-AD Female Mice
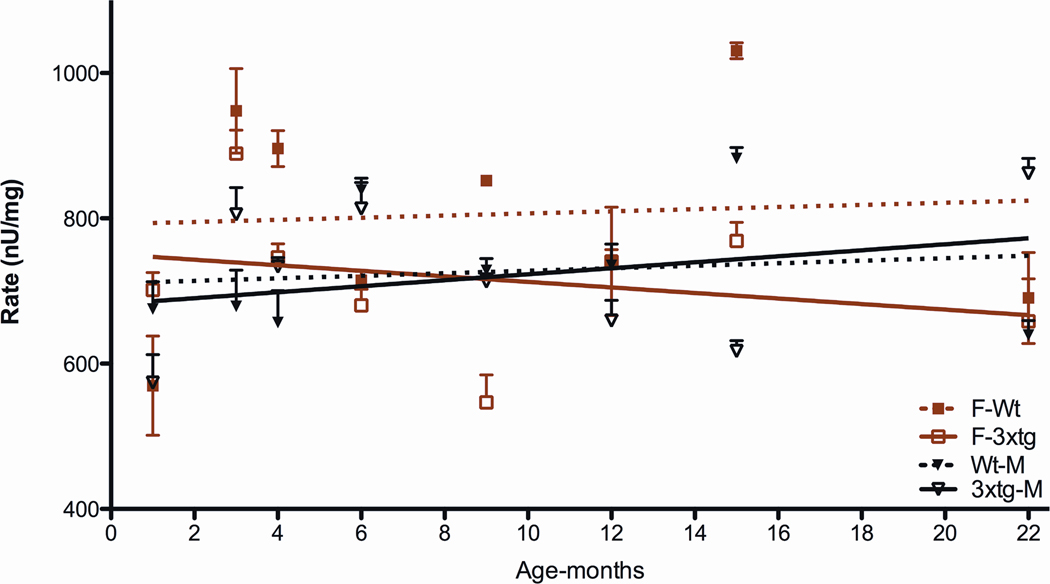
To determine which component of the ETC might be responsible for the biphasic response in the female 3XTg-AD mice, we analyzed the specific activity of each of the ETC enzymes. This revealed that the complex IV activity in the female control mice was consistently higher than that of the male control mice. Furthermore, the complex IV activity of the female 3XTg-AD mice started just below that of the control female mouse level, but then declined throughout life (Figure 10).
Figure 10.
Cytochrome c Oxidase (Complex IV) Activities of 3XTg-AD Mice Throughout Life. Male 3XTg-AD and male control activities are comparable. However, the female 3XTg-AD levels start comparable to control females but then decline with age.
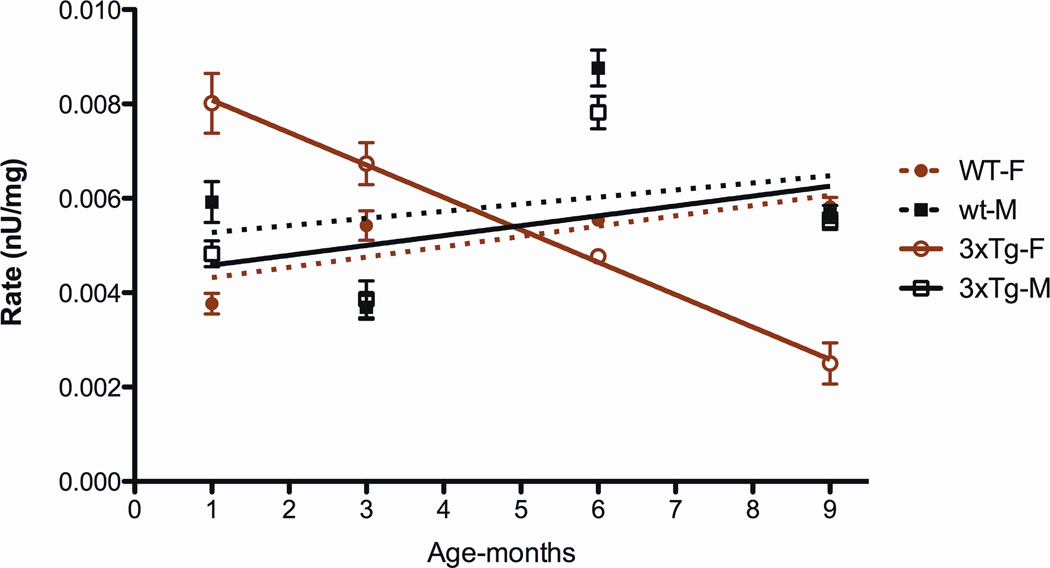
Finally, while the complex I specific activities of the male and female control and male 3XTg-AD activities were quite similar, the complex I activity of the female 3XTg-AD mice was strikingly different (Figure 11). At one month after birth, the 3XTg-AD female complex I activity was almost twice that of the female controls. However, it then declined sharply throughout life, falling below that of the control animals by five months of age.
Figure 11.
Striking Biphasic Complex I Activity in 3XTg-AD Female Mice Throughout Life
Thus, there is a striking sexual dimorphism in response to over-expression of mutant APP, tau, and PS1. The male 3XTg-AD mice show a consistently reduced forebrain mitochondrial respiration rate, both when coupled and uncoupled to ATP synthesis. By contrast, the females, show a striking biphasic response with an initially elevated brain state III respiration rate followed by progressive decline. This biphasic response seems to be a primary consequence of complex I activity with a secondary contribution by complex IV.
Interestingly, in an independent study, it was reported that the respiratory control ratio (RCR) was reduced in female 3XTg-AD female mice at 3, 6, 9 and 12 months relative to controls, while in our studies the RCR was similar to matched control mice, averaging about 4. These investigators also observed reduced pyruvate dehydrogenase at all four ages and reduced complex IV at 6, 9 and 12 months. However, they did not report on the activity of complex I [107, 108].
In an effort to determine the biochemical factor(s) that cause this decline in mitochondrial function in the 3XTg-AD mice, we examined mitochondrial Δψ, calcium sensitivity, mitochondrial H2O2 production, and mitochondria O2˙− production by Mitosox fluorescence. The only significant difference was found in two to three month old male 3XTg-AD mice, whose mitochondria produced more H202 than age matched controls (p = 0.02). Perhaps, this elevated ROS production inhibits electron transfer between the OXPHOS complexes in the male 3XTg-AD mouse mitochondria explaining the reduced electron transport rate. By contrast, the elevated state III respiration and complex I and IV activities of the female 3XTg-AD mice suggest that the initial increased Aβ and tau levels may stabilize complexes I and IV or increase their biogenesis. However, Aβ subsequently forms Aβ oligomers, which inhibit the enzyme function.
The difference in mitochondrial response of the male and female 3XTg-AD mice would seem surprising from a classical Mendelian genetics perspective. However, such sex biased expression of mitochondrial defects is seen in mitochondrial disease. Leber Hereditary Optic Neuropathy (LHON) is a form of blindness that is caused by mild mtDNA mutations. In most LHON families, all maternal relatives have high levels of the mutant mtDNA, yet there is a 3 to 1 male bias in developing blindness [5]. Since all maternal relatives in LHON pedigrees have high levels of mutant mtDNAs, females must have a greater resistance to the deleterious effects of the mtDNA mutations than males. One possibility is that estrogen is protective of mitochondrial dysfunction. We have shown that the estrogen receptor is present in the mitochondrial matrix and that exposure of mitochondria to estrogen activates MnSOD antioxidant activity [109]. A similar mechanism might explain the greater resistance of females to the adverse effects of Aβ toxicity early in life, which they lose as estrogen production declines during the post-reproductive period.
5. Conclusion: A Mitochondrial Pathophysiology of AD and PD
From this accumulate evidence, we can now formulate a mitochondrial etiology for AD and PD that integrates all of the observations to date. The core concept is that the underlying pathophysiology of AD, PD, DS, and DSAD is bioenergetic decline due to mitochondrial dysfunction. The neuronal dysfunction is the result of the age-related progressive decline in mitochondrial function, which ultimately becomes insufficient for optimal neurological function. The increasing mitochondrial dysfunction stimulates the expression of Aβ in AD and DSAD, initially as a compensatory response, which ultimately reaches sufficient concentrations to form aggregates and the plaques ad tangles. Perhaps, in PD, α-synuclein is also initially induced as a protective function, but later reaches sufficient concentration to aggregate into Lewy bodies.
Individual predisposition to AD and PD can result from partial defects in mitochondrial function resulting from mild variants in the over a thousand nuclear DNA-encoded mitochondrial genes, recent relatively deleterious mtDNA mutations, or more ancient functional mtDNA variants that are now maladaptive. These inherited mitochondrial defects are insufficient in themselves to drop mitochondrial energy production below the minimum bioenergetic threshold for normal brain function. However, they increase the rate at which somatic mtDNA mutations accumulate in the cortex and/or the basal ganglion. This age-associated accumulation of somatic mutations further erodes brain cell energetics until the combination of the inherited and the acquired mitochondrial defects become so severe that there is insufficient mitochondrial energy production to sustain normal neuronal function.
Nuclear gene mutations which contribute to this process include partial defects in OXPHOS genes, and mutations in genes that compromise the nuclear genetic functions that counteract the inherent age-related decline in mitochondrial function. In the case of PD, such nuclear DNA mutations affect mitochondrial structural integrity (α-synuclein), antioxidant defenses (DJ-1, LRRK2), and mitochondrial and mtDNA quality control through compromising mitophagy (PINK1 and parkin). In the case of AD, rare mutations in APP gene and its PS processing enzymes result in an overly aggressive production of Aβ. Initially, this excess phase 1 response is strongly protective of female mitochondrial function, particularly of complex I. However, consistently high levels of Aβ early in life prematurely initiate Aβ aggregation. The resulting toxic oligmeric form begins the destructive Aβ phase 2, resulting in premature neuronal death and dementia. In DS, with an extra copy of the APP gene on chromosome 21, the protective aspect of soluble Aβ might account for the increased mtDNA levels in younger DS subjects, but also for the premature aggregation of Aβ resulting in the early initiation of the destructive phase 2 leading to DSAD.
These considerations leave one persistent observation unaccounted for. In both AD and PD, the decline in mental function and the increase in neuronal loss is associated in increased local inflammation. Increased mitochondrial ROS is likely to contribute to this state. However, there is an additional possibility. The mitochondrially translated polypeptides contain a formyl methionine, making them similar to foreign bacterial antigenic polypeptides. Indeed, it has recently been shown that both mitochondrially translated polypeptides and mtDNA have the properties necessary for the cell and the immune system to see them as bacterial invaders [110].
The polypeptides with the mtDNA sequence present during development would be protected from immune rejection by the development of tolerance during thymic maturation of T cells. However, as the mtDNA accumulates somatic mtDNA mutations, the mutant polypeptides would no longer be protected by tolerance and thus seen as foreign. That mtDNA polypeptides with altered amino acid sequences can initiate a T-cell response has been demonstrated by generating mice that are isogenic for all nDNA genes, but differ in their mtDNA sequence. Injecting the cells with one mtDNA type into mice with another mtDNA type results in T cells that can kill the injected cells in a mixed lymphocyte assay. This has been shown to be driven by amino acid polymorphisms that occur toward the N-terminal end of the mtDNA encoded polypeptides [111].
Therefore, as somatic mtDNA mutations accumulate throughout the body, mitochondrial function progressively declines. The cerebral cortex and basal ganglion are especially sensitivity to this decline, because they both already have the highest somatic mtDNA mutation rate of any tissue in the body [66] and because they are the most reliant on mitochondrial energy production for normal function [5]. As the energetics of the brain cells declines, the apoptotic process fails due to energetic insufficiency. Mitochondria with the mutant formyl-methionine peptides are then released into the intercellular space, where they can be recognized as foreign and instigate an inflammatory response. These summed mitochondrial factors then combine to produce the observed features of AD, PD and DSAD.
6. Materials and Methods
6.1. Transgenic mice and handling
3xTg AD mice on C57Bl/6J and 129S4hybrid background were generated as previously described [106]. For body and brain weight assessment, 3 to 6 mice in each sex and each genotype (3xTg and wild type) were used starting from the ages of one month to 28 months with one month intervals except for the ages of 11, 14, 19, 20 and 25 months. For mitochondrial respiration and complex IV activity, 3 to 6 mice in each sex and each genotype were used from the ages of 1, 2, 3, 4, 6, 9/10, 15/16 and 21 months old and repeated the analysis twice. In mitochondrial respiration, uncoupler respiration rate was not acquired from 12 and 15 months group due to the limited samples. For complex I, 4 to 6 mice in each sex and each genotype were used from the ages of 1, 3, 4, 6 and 9 months. For Complex II activity, same animals were used at same time point except 9 months old. Mitochondrial membrane potential and Calcium sensitivity were measured at the ages of 2, 4, 11, 15 and 21 months using 3 to 6 mice in each sex and genotype. For H2O2 assay, 3 to 6 mice in each sex and genotype were used at the ages of 2, 3, 4, 6/7, 9, 12, 15 and 21 months, while for Mito-Sox assay, 3 to 6 mice in each sex and genotype used at 5/6, 9 and 21 months. Mice were maintained at 20–23 °C on a 13-hour light/11-hour dark cycle using the procedures mandated by the National Institutes of Health guidelines and the University of California Institutional Animal Care and Use Committee. Animals were sacrificed by cervical dislocation, and body weight was measured before dissection. Brain weight was measured prior to dissecting out brain stem and cerebellum.
6.2. Mitochondrial isolation and respiration
The forebrain tissue is dissected and placed in ice-cold homogenization buffer (10 ml homogenization buffer consisting of 225 mM mannitol, 75 mM sucrose, 10 mM MOPS, 1 mM EGTA and 0.5% BSA (pH 7.2)). The rest of the steps are performed at 4 °C unless otherwise indicated. Mitochondria are isolated by gentle mincing using Potter-Elvehjem grinder with Teflon pestle with seven strokes of the pestle in homogenization buffer. Homogenate centrifuged at 1000xg for 5 mins twice and supernatant collected to spin at 8000xg for 15 mins. Pellets were treated with 0.02 % final concentration digitonin in 10mL homogenization buffer and centrifuged at 8000xg for 15 mins. The pellet was washed with 10 mL homogenization buffer and centrifuged at 8000xg for 15 mins. The pellet was resuspended in 400uL homogenization buffer and protein amount was measured using Bradford method using Bio-Rad reagents and correcting for the BSA content in the homogenization buffer. For respiration, mitochondrial membrane potential, Ca++ sensitivity and mitochondrial oxidative stress measures were performed on freshly prepared mitochondria within first 2 hours after isolation. For complex activities, mitochondria were frozen in liquid nitrogen and stored at −80°C until the experiments were executed. Respiration rates were determined polarographically by oxygen consumption using a Clark-type electrode by Hansatech Instruments Ltd Oxygen Electrode Measurement Systems. For each experiment, we used 500ugr mitochondrial protein. State 3 and 4 oxygen consumption rates were determined using complex I substrates Glutamate and malate. FCCP (p-trifluoromethoxy carbonyl cyanide phenyl hydrazone) was used to measure uncoupling rate for the maximum respiration rate and oligomycin was used to inhibit the ATP production.
6.3. Electron chain enzyme activities
Mitochondrial electron chain enzyme complex I, II, IV and citrate synthase activities were assayed as previously discussed [112]. All the activities were measured on a 96 well plate set up, and specific wavelength absorption was measured using spectrophotometer, SpectroMax Molecular Devices. Complex I and II assays were tested using 20ug mitochondrial protein and Complex IV and citrate synthase were tested using 5ug mitochondrial protein. Complex I, II and IV assays were run with proper inhibitors to subtract the background activity.
6.4. Mitochondrial membrane potential and Ca++ sensitivity
Mitochondrial membrane potential and permeability transition were measured using the fluorescent probe rhodamine 123 with a Perkin Elmer LS50B luminescent spectrophotometer set at 490 nm excitation and 535 nm emission with stirring according to the protocol described by Lee et al [105]. Half a milligram of mitochondrial protein was added to 3 ml of reaction buffer.
6.5. Mitochondrial oxidative stress measures
Mitochondrial H2O2 production
H2O2 production in 0.5 mg forebrain mitochondrial protein was measured by increase in fluorescence by oxidation of PHPA (p-hydroxyphenylacetate) in the presence of horseradish peroxidase (HRP) as described by Hyslop et al [113].
Measuring mitochondrial superoxide production
Generated superoxide anion production in 0.5ug mitochondrial protein was measured using Mito-Sox (Invitrogen) fluorescence at 510 nm excitation/580 nm emission as described in detail by Robinson et al [114].
Highlights.
> mtDNA haplogroup polymorphisms such as the tRNAGln nt 4336 have been established as risk factors for AD and PD. > Somatic mtDNA mutations accumulate with age and are elevated in brains & systemically in AD, PD, and DSAD. > AD, DS, and DSAD brains showed reduced mtDNA ND6 transcript levels and altered mtDNA/nDNA ratios. >Triple transgenic-AD mouse males have reduced mitochondrial respiration rates throughout life. > 3XTg-AD female mice initially have elevated respiration and complex I & IV activities which declines with age.
Acknowledgements
This work was supported by NIH grants NS21328, AG24373, DK73691, and AG13154, an ADRC Grant (AG-16573) sub-project, and CIRM a Comprehensive Grant RC1-00353-1 awarded to Douglas C. Wallace.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Alzheimer's Association. Alzheimer’s Disease Facts and Figures, vol. 2011, 2011, pp. An annual report released by the Alzheimer’s Association, reveals the burden of Alzheimer's and dementia on individuals, caregivers, government, and the nation's healthcare system. 2011
- 2.Glabe CG, Kayed R. Common structure and toxic function of amyloid oligomers implies a common mechanism of pathogenesis. Neurology. 2006;66:S74–S78. doi: 10.1212/01.wnl.0000192103.24796.42. [DOI] [PubMed] [Google Scholar]
- 3.Deshpande A, Mina E, Glabe C, Busciglio J. Different conformations of amyloid beta induce neurotoxicity by distinct mechanisms in human cortical neurons. Journal of Neuroscience. 2006;26:6011–6018. doi: 10.1523/JNEUROSCI.1189-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Reddy PH, Beal MF. Are mitochondria critical in the pathogenesis of Alzheimer's disease? Brain Research. Brain Research Reviews. 2005;49:618–632. doi: 10.1016/j.brainresrev.2005.03.004. [DOI] [PubMed] [Google Scholar]
- 5.Wallace DC. A mitochondrial paradigm of metabolic and degenerative diseases, aging, and cancer: a dawn for evolutionary medicine. Annual Review of Genetics. 2005;39:359–407. doi: 10.1146/annurev.genet.39.110304.095751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sullivan PG, Brown MR. Mitochondrial aging and dysfunction in Alzheimer's disease. Progress in Neuro-Psychopharmacology and Biological Psychiatry. 2005;29:407–410. doi: 10.1016/j.pnpbp.2004.12.007. [DOI] [PubMed] [Google Scholar]
- 7.Diana FF, Silva Esteves AR, Oliveira CR, Cardoso SM. Mitochondria: the common upstream driver of a-beta and tau pathology in Alzheimer's Disease. Current Alzheimer Research. 2011 doi: 10.2174/156720511796391872. [ePub ahead of print] http://1.usa.gov/euaROo. [DOI] [PubMed] [Google Scholar]
- 8.Bonda DJ, Lee HP, Lee HG, Friedlich AL, Perry G, Zhu X, Smith MA. Novel therapeutics for Alzheimer's disease: an update. Current Opinion in Drug Discovery and Development. 2010;13:235–246. [PMC free article] [PubMed] [Google Scholar]
- 9.Hansson CA, Frykman S, Farmery MR, Tjernberg LO, Nilsberth C, Pursglove SE, Ito A, Winblad B, Cowburn RF, Thyberg J, Ankarcrona M. Nicastrin, presenilin, APH-1 and PEN-2 form active gamma -secretase complexes in mitochondria. The Journal of Biological Chemistry. 2004;279:51654–51660. doi: 10.1074/jbc.M404500200. [DOI] [PubMed] [Google Scholar]
- 10.Hansson Petersen CA, Alikhani N, Behbahani H, Wiehager B, Pavlov PF, Alafuzoff I, Leinonen V, Ito A, Winblad B, Glaser E, Ankarcrona M. The amyloid beta-peptide is imported into mitochondria via the TOM import machinery and localized to mitochondrial cristae. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:13145–13150. doi: 10.1073/pnas.0806192105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Devi L, Anandatheerthavarada HK. Mitochondrial trafficking of APP and alpha synuclein: Relevance to mitochondrial dysfunction in Alzheimer's and Parkinson's diseases. Biochimica et Biophysica acta. 2010;1802:11–19. doi: 10.1016/j.bbadis.2009.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pavlov PF, Wiehager B, Sakai J, Frykman S, Behbahani H, Winblad B, Ankarcrona M. Mitochondrial gamma-secretase participates in the metabolism of mitochondria-associated amyloid precursor protein. FASEB Journal. 2011;25:78–88. doi: 10.1096/fj.10-157230. [DOI] [PubMed] [Google Scholar]
- 13.Glaser E, Alikhani N. The organellar peptidasome, PreP: a journey from Arabidopsis to Alzheimer's disease. Biochimica et Biophysica Acta. 2010;1797:1076–1080. doi: 10.1016/j.bbabio.2009.12.016. [DOI] [PubMed] [Google Scholar]
- 14.Amadoro G, Corsetti V, Stringaro A, Colone M, D'Aguanno S, Meli G, Ciotti M, Sancesario G, Cattaneo A, Bussani R, Mercanti D, Calissano P. A NH2 tau fragment targets neuronal mitochondria at AD synapses: possible implications for neurodegeneration. Journal of Alzheimers Disease. 2010;21:445–470. doi: 10.3233/JAD-2010-100120. [DOI] [PubMed] [Google Scholar]
- 15.Moreira PI, Carvalho C, Zhu X, Smith MA, Perry G. Mitochondrial dysfunction is a trigger of Alzheimer's disease pathophysiology. Biochimica et Biophysica Acta. 2010;1802:2–10. doi: 10.1016/j.bbadis.2009.10.006. [DOI] [PubMed] [Google Scholar]
- 16.Su B, Wang X, Zheng L, Perry G, Smith MA, Zhu X. Abnormal mitochondrial dynamics and neurodegenerative diseases. Biochimica et Biophysica Acta. 2010;1802:135–142. doi: 10.1016/j.bbadis.2009.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Aleardi AM, Benard G, Augereau O, Malgat M, Talbot JC, Mazat JP, Letellier T, Dachary-Prigent J, Solaini GC, Rossignol R. Gradual alteration of mitochondrial structure and function by beta-myloids: importance of membrane viscosity changes, energy deprivation, reactive oxygen species production, and cytochrome c release. Journal of Bioenergetics and Biomembranes. 2005;37:207–225. doi: 10.1007/s10863-005-6631-3. [DOI] [PubMed] [Google Scholar]
- 18.Du H, Guo L, Yan S, Sosunov AA, McKhann GM, Yan SS. Early deficits in synaptic mitochondria in an Alzheimer's disease mouse model. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:18670–18675. doi: 10.1073/pnas.1006586107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dragicevic N, Mamcarz M, Zhu Y, Buzzeo R, Tan J, Arendash GW, Bradshaw PC. Mitochondrial amyloid-beta levels are associated with the extent of mitochondrial dysfunction in different brain regions and the degree of cognitive impairment in Alzheimer's transgenic mice. Journal of Alzheimers Disease. 2010;20 Suppl 2:S535–S550. doi: 10.3233/JAD-2010-100342. [DOI] [PubMed] [Google Scholar]
- 20.Canevari L, Clark JB, Bates TE. beta-Amyloid fragment 25–35 selectively decreases complex IV activity in isolated mitochondria. FEBS Letters. 1999;457:131–134. doi: 10.1016/s0014-5793(99)01028-5. [DOI] [PubMed] [Google Scholar]
- 21.Crouch PJ, Blake R, Duce JA, Ciccotosto GD, Li QX, Barnham KJ, Curtain CC, Cherny RA, Cappai R, Dyrks T, Masters CL, Trounce IA. Copper-dependent inhibition of human cytochrome c oxidase by a dimeric conformer of amyloid-beta1-42. Journal of Neuroscience. 2005;25:672–679. doi: 10.1523/JNEUROSCI.4276-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Atamna H, Boyle K. Amyloid-beta peptide binds with heme to form a peroxidase: relationship to the cytopathologies of Alzheimer's disease. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:3381–3386. doi: 10.1073/pnas.0600134103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Szabados T, Dul C, Majtenyi K, Hargitai J, Penzes Z, Urbanics R. A chronic Alzheimer's model evoked by mitochondrial poison sodium azide for pharmacological investigations. Behavioural Brain Research. 2004;154:31–40. doi: 10.1016/j.bbr.2004.01.016. [DOI] [PubMed] [Google Scholar]
- 24.Massaad CA, Amin SK, Hu L, Mei Y, Klann E, Pautler RG. Mitochondrial superoxide contributes to blood flow and axonal transport deficits in the Tg2576 mouse model of Alzheimer's disease. PLoS One. 2010;5:e10561. doi: 10.1371/journal.pone.0010561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Muirhead KE, Borger E, Aitken L, Conway SJ, Gunn-Moore FJ. The consequences of mitochondrial amyloid beta-peptide in Alzheimer's disease. The Biochemical Journal. 2010;426:255–270. doi: 10.1042/BJ20091941. [DOI] [PubMed] [Google Scholar]
- 26.Terni B, Boada J, Portero-Otin M, Pamplona R, Ferrer I. Mitochondrial ATP-synthase in the entorhinal cortex is a target of oxidative stress at stages I/II of Alzheimer's disease pathology. Brain Pathology. 2010;20:222–233. doi: 10.1111/j.1750-3639.2009.00266.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lustbader JW, Cirilli M, Lin C, Xu HW, Takuma K, Wang N, Caspersen C, Chen X, Pollak S, Chaney M, Trinchese F, Liu S, Gunn-Moore F, Lue LF, Walker DG, Kuppusamy P, Zewier ZL, Arancio O, Stern D, Yan SS, Wu H. ABAD directly links Abeta to mitochondrial toxicity in Alzheimer's disease. Science. 2004;304:448–452. doi: 10.1126/science.1091230. [DOI] [PubMed] [Google Scholar]
- 28.Michel TM, Gsell W, Kasbauer L, Tatschner T, Sheldrick AJ, Neuner I, Schneider F, Grunblatt E, Riederer P. Increased activity of mitochondrial aldehyde dehydrogenase (ALDH) in the putamen of individuals with Alzheimer's disease: a human postmortem study. Journal of Alzheimers Disease. 2010;19:1295–1301. doi: 10.3233/JAD-2010-1326. [DOI] [PubMed] [Google Scholar]
- 29.Yan SD, Stern DM. Mitochondrial dysfunction and Alzheimer's disease: role of amyloid-beta peptide alcohol dehydrogenase (ABAD) International Journal of Experimental Pathology. 2005;86:161–171. doi: 10.1111/j.0959-9673.2005.00427.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yao J, Du H, Yan S, Fang F, Wang C, Lue LF, Guo L, Chen D, Stern DM, Gunn Moore FJ, Xi Chen J, Arancio O, Yan SS. Inhibition of amyloid-beta (Abeta) peptide-binding alcohol dehydrogenase-Abeta interaction reduces Abeta accumulation and improves mitochondrial function in a mouse model of Alzheimer's disease. The Journal of Neuroscience. 2011;31:2313–2320. doi: 10.1523/JNEUROSCI.4717-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang HQ, Nakaya Y, Du Z, Yamane T, Shirane M, Kudo T, Takeda M, Takebayashi K, Noda Y, Nakayama KI, Nishimura M. Interaction of presenilins with FKBP38 promotes apoptosis by reducing mitochondrial Bcl-2. Human Molecular Genetics. 2005;14:1889–1902. doi: 10.1093/hmg/ddi195. [DOI] [PubMed] [Google Scholar]
- 32.Gupta S, Singh R, Datta P, Zhang Z, Orr C, Lu Z, Dubois G, Zervos AS, Meisler MH, Srinivasula SM, Fernandes-Alnemri T, Alnemri ES. The C-terminal tail of presenilin regulates Omi/HtrA2 protease activity. The Journal of Biological Chemistry. 2004;279:45844–45854. doi: 10.1074/jbc.M404940200. [DOI] [PubMed] [Google Scholar]
- 33.Schon EA, Area-Gomez E. Is Alzheimer's disease a disorder of mitochondria-associated membranes? Journal of Alzheimers Disease. 2010;20 Suppl 2:S281–S292. doi: 10.3233/JAD-2010-100495. [DOI] [PubMed] [Google Scholar]
- 34.Contreras L, Drago I, Zampese E, Pozzan T. Mitochondria: the calcium connection. Biochimica et Biophysica Acta. 2010;1797:607–618. doi: 10.1016/j.bbabio.2010.05.005. [DOI] [PubMed] [Google Scholar]
- 35.Takuma K, Yan SS, Stern DM, Yamada K. Mitochondrial dysfunction, endoplasmic reticulum stress, and apoptosis in Alzheimer's disease. Journal of Pharmacological Sciences. 2005;97:312–316. doi: 10.1254/jphs.cpj04006x. [DOI] [PubMed] [Google Scholar]
- 36.Ferreiro E, Oliveira CR, Pereira CM. The release of calcium from the endoplasmic reticulum induced by amyloid-beta and prion peptides activates the mitochondrial apoptotic pathway. Neurobiology of Disease. 2008;30:331–342. doi: 10.1016/j.nbd.2008.02.003. [DOI] [PubMed] [Google Scholar]
- 37.Chen HK, Ji ZS, Dodson SE, Miranda RD, Rosenblum CI, Reynolds IJ, Freedman SB, Weisgraber KH, Huang Y, Mahley RW. Apolipoprotein e4 domain interaction mediates detrimental effects on mitochondria and is a potential therapeutic target for Alzheimer disease. The Journal of Biological Chemistry. 2011;286:5215–5221. doi: 10.1074/jbc.M110.151084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chang S, ran Ma T, Miranda RD, Balestra ME, Mahley RW, Huang Y. Lipid- and receptor-binding regions of apolipoprotein E4 fragments act in concert to cause mitochondrial dysfunction and neurotoxicity. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:18694–18699. doi: 10.1073/pnas.0508254102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mahley RW, Weisgraber KH, Huang Y. Apolipoprotein E4: a causative factor and therapeutic target in neuropathology, including Alzheimer's disease. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:5644–5651. doi: 10.1073/pnas.0600549103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Potkin SG, Guffanti G, Lakatos A, Turner JA, Kruggel F, Fallon JH, Saykin AJ, Orro A, Lupoli S, Salvi E, Weiner M, Macciardi F. Hippocampal atrophy as a quantitative trait in a genome-wide association study identifying novel susceptibility genes for Alzheimer's disease. PLoS One. 2009;4:e6501. doi: 10.1371/journal.pone.0006501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Roses AD. The medical and economic roles of pipeline pharmacogenetics: Alzheimer's disease as a model of efficacy and HLA-B(*)5701 as a model of safety. Neuropsychopharmacology. 2009;34:6–17. doi: 10.1038/npp.2008.153. [DOI] [PubMed] [Google Scholar]
- 42.Kamp F, Exner N, Lutz AK, Wender N, Hegermann J, Brunner B, Nuscher B, Bartels T, Giese A, Beyer K, Eimer S, Winklhofer KF, Haass C. Inhibition of mitochondrial fusion by alpha-synuclein is rescued by PINK1, Parkin and DJ-1. The EMBO Journal. 2010;29:3571–3589. doi: 10.1038/emboj.2010.223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mortiboys H, Johansen KK, Aasly JO, Bandmann O. Mitochondrial impairment in patients with Parkinson disease with the G2019S mutation in LRRK2. Neurology. 2010;75:2017–2020. doi: 10.1212/WNL.0b013e3181ff9685. [DOI] [PubMed] [Google Scholar]
- 44.Krebiehl G, Ruckerbauer S, Burbulla LF, Kieper N, Maurer B, Waak J, Wolburg H, Gizatullina Z, Gellerich FN, Woitalla D, Riess O, Kahle PJ, Proikas-Cezanne T, Kruger R. Reduced basal autophagy and impaired mitochondrial dynamics due to loss of Parkinson's disease-associated protein DJ-1. PLoS One. 2010;5:e9367. doi: 10.1371/journal.pone.0009367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.McCoy MK, Cookson MR. DJ-1 regulation of mitochondrial function and autophagy through oxidative stress. Autophagy. 2011;7 doi: 10.4161/auto.7.5.14684. ePub ahead of print, http://www.landesbioscience.com/journals/autophagy/article/14684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hao LY, Giasson BI, Bonini NM. DJ-1 is critical for mitochondrial function and rescues PINK1 loss of function. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:9747–9752. doi: 10.1073/pnas.0911175107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Guzman JN, Sanchez-Padilla J, Wokosin D, Kondapalli J, Ilijic E, Schumacker PT, Surmeier DJ. Oxidant stress evoked by pacemaking in dopaminergic neurons is attenuated by DJ-1. Nature. 2010;468:696–700. doi: 10.1038/nature09536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Narendra DP, Jin SM, Tanaka A, Suen DF, Gautier CA, Shen J, Cookson MR, Youle RJ. PINK1 is selectively stabilized on impaired mitochondria to activate Parkin. PLoS Biology. 2010;8 doi: 10.1371/journal.pbio.1000298. e1000298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Narendra D, Tanaka A, Suen DF, Youle RJ. Parkin is recruited selectively to impaired mitochondria and promotes their autophagy. The Journal of Cell Biology. 2008;183:795–803. doi: 10.1083/jcb.200809125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gegg ME, Schapira AH. PINK1-parkin-dependent mitophagy involves ubiquitination of mitofusins 1 and 2: Implications for Parkinson disease pathogenesis. Autophagy. 2011;7:243–245. doi: 10.4161/auto.7.2.14332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ziviani E, Whitworth AJ. How could Parkin-mediated ubiquitination of mitofusin promote mitophagy? Autophagy. 2010;6:660–662. doi: 10.4161/auto.6.5.12242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Guo M. What have we learned from Drosophila models of Parkinson's disease? Progress in Brain Research. 2010;184:3–16. doi: 10.1016/S0079-6123(10)84001-4. [DOI] [PubMed] [Google Scholar]
- 53.Burbulla LF, Krebiehl G, Kruger R. Balance is the challenge--the impact of mitochondrial dynamics in Parkinson's disease. European Journal of Clinical Investigation. 2010;40:1048–1060. doi: 10.1111/j.1365-2362.2010.02354.x. [DOI] [PubMed] [Google Scholar]
- 54.Zhu J, Chu CT. Mitochondrial dysfunction in Parkinson's disease. Journal of Alzheimer's Disease. 2010;20 Suppl 2:S325–S334. doi: 10.3233/JAD-2010-100363. [DOI] [PubMed] [Google Scholar]
- 55.Suen DF, Narendra DP, Tanaka A, Manfredi G, Youle RJ. Parkin overexpression selects against a deleterious mtDNA mutation in heteroplasmic cybrid cells. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:11835–11840. doi: 10.1073/pnas.0914569107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Rosen KM, Veereshwarayya V, Moussa CE, Fu Q, Goldberg MS, Schlossmacher MG, Shen J, Querfurth HW. Parkin protects against mitochondrial toxins and beta-amyloid accumulation in skeletal muscle cells. The Journal of Biological Chemistry. 2006;281:12809–12816. doi: 10.1074/jbc.M512649200. [DOI] [PubMed] [Google Scholar]
- 57.Wallace DC. Why do we have a maternally inherited mitochondrial DNA? Insights from Evolutionary Medicine. Annual Review of Biochemistry. 2007;76:781–821. doi: 10.1146/annurev.biochem.76.081205.150955. [DOI] [PubMed] [Google Scholar]
- 58.Pagliarini DJ, Calvo SE, Chang B, Sheth SA, Vafai SB, Ong SE, Walford GA, Sugiana C, Boneh A, Chen WK, Hill DE, Vidal M, Evans JG, Thorburn DR, Carr SA, Mootha VK. A mitochondrial protein compendium elucidates complex I disease biology. Cell. 2008;134:112–123. doi: 10.1016/j.cell.2008.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wallace DC. Cytoplasmic inheritance of chloramphenicol resistance in mammalian cells. Chapter 12. In: Shay JW, editor. Techniques in Somatic Cell Genetics. New York: Plenum Press; 1982. pp. 159–187. [Google Scholar]
- 60.MITOMAP, A Human Mitochondrial Genome Database. 2006 doi: 10.1093/nar/24.1.177. http://www.mitomap.org. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ruiz-Pesini E, Lott MT, Procaccio V, Poole J, Brandon MC, Mishmar D, Yi C, Kreuziger J, Baldi P, Wallace DC. An enhanced MITOMAP with a global mtDNA mutational phylogeny. Nucleic Acids Research. 2007;35:D823–D828. doi: 10.1093/nar/gkl927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Mishmar D, Ruiz-Pesini EE, Golik P, Macaulay V, Clark AG, Hosseini S, Brandon M, Easley K, Chen E, Brown MD, Sukernik RI, Olckers A, Wallace DC. Natural selection shaped regional mtDNA variation in humans. Proceedings of the National Academy of Sciences of the United States of America. 2003;100:171–176. doi: 10.1073/pnas.0136972100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ruiz-Pesini E, Mishmar D, Brandon M, Procaccio V, Wallace DC. Effects of purifying and adaptive selection on regional variation in human mtDNA. Science. 2004;303:223–226. doi: 10.1126/science.1088434. [DOI] [PubMed] [Google Scholar]
- 64.Ruiz-Pesini E, Wallace DC. Evidence for adaptive selection acting on the tRNA and rRNA genes of the human mitochondrial DNA. Human Mutation. 2006;27:1072–1081. doi: 10.1002/humu.20378. [DOI] [PubMed] [Google Scholar]
- 65.Gomez-Duran A, Pacheu-Grau D, Lopez-Gallardo E, Diez-Sanchez C, Montoya J, Lopez-Perez MJ, Ruiz-Pesini E. Unmasking the causes of multifactorial disorders: OXPHOS differences between mitochondrial haplogroups. Human Molecular Genetics. 2010;19:3343–3353. doi: 10.1093/hmg/ddq246. [DOI] [PubMed] [Google Scholar]
- 66.Corral-Debrinski M, Horton T, Lott MT, Shoffner JM, Beal MF, Wallace DC. Mitochondrial DNA deletions in human brain: regional variability and increase with advanced age. Nature Genetics. 1992;2:324–329. doi: 10.1038/ng1292-324. [DOI] [PubMed] [Google Scholar]
- 67.Soong NW, Hinton DR, Cortopassi G, Arnheim N. Mosaicism for a specific somatic mitochondrial DNA mutation in adult human brain. Nature Genetics. 1992;2:318–323. doi: 10.1038/ng1292-318. [DOI] [PubMed] [Google Scholar]
- 68.Oliver NA, Wallace DC. Assignment of two mitochondrially synthesized polypeptides to human mitochondrial DNA and their use in the study of intracellular mitochondrial interaction. Molecular and Cellular Biology. 1982;2:30–41. doi: 10.1128/mcb.2.1.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Shoffner JM, Brown MD, Torroni A, Lott MT, Cabell MR, Mirra SS, Beal MF, Yang C, Gearing M, Salvo R, Watts RL, Juncos JL, Hansen LA, Crain BJ, Fayad M, Reckord CL, Wallace DC. Mitochondrial DNA variants observed in Alzheimer disease and Parkinson disease patients. Genomics. 1993;17:171–184. doi: 10.1006/geno.1993.1299. [DOI] [PubMed] [Google Scholar]
- 70.Hutchin T, Cortopassi G. A mitochondrial DNA clone is associated with increased risk for Alzheimer disease. Proceedings of the National Academy of Sciences of the United States of America. 1995;92:6892–6895. doi: 10.1073/pnas.92.15.6892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Santoro A, Balbi V, Balducci E, Pirazzini C, Rosini F, Tavano F, Achilli A, Siviero P, Minicuci N, Bellavista E, Mishto M, Salvioli S, Marchegiani F, Cardelli M, Olivieri F, Nacmias B, Chiamenti AM, Benussi L, Ghidoni R, Rose G, Gabelli C, Binetti G, Sorbi S, Crepaldi G, Passarino G, Torroni A, Franceschi C. Evidence for sub-haplogroup h5 of mitochondrial DNA as a risk factor for late onset Alzheimer's disease. PLoS One. 2010;5:e12037. doi: 10.1371/journal.pone.0012037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Tanaka N, Goto YI, Akanuma J, Kato M, Kinoshita T, Yamashita F, Tanaka M, Asada T. Mitochondrial DNA variants in a Japanese population of patients with Alzheimer's disease. Mitochondrion. 2010;10:32–37. doi: 10.1016/j.mito.2009.08.008. [DOI] [PubMed] [Google Scholar]
- 73.Chagnon P, Gee M, Filion M, Robitaille Y, Belouchi M, Gauvreau D. Phylogenetic analysis of the mitochondrial genome indicates significant differences between patients with Alzheimer disease and controls in a French-Canadian founder population. American Journal of Medical Genetics. 1999;85:20–30. doi: 10.1002/(sici)1096-8628(19990702)85:1<20::aid-ajmg6>3.0.co;2-k. [DOI] [PubMed] [Google Scholar]
- 74.van der Walt JM, Dementieva YA, Martin ER, Scott WK, Nicodemus KK, Kroner CC, Welsh-Bohmer KA, Saunders AM, Roses AD, Small GW, Schmechel DE, Murali Doraiswamy P, Gilbert JR, Haines JL, Vance JM, Pericak-Vance MA. Analysis of European mitochondrial haplogroups with Alzheimer disease risk. Neuroscience Letters. 2004;365:28–32. doi: 10.1016/j.neulet.2004.04.051. [DOI] [PubMed] [Google Scholar]
- 75.Carrieri G, Bonafe M, De Luca M, Rose G, Varcasia O, Bruni A, Maletta R, Nacmias B, Sorbi S, Corsonello F, Feraco E, Andreev KF, Yashin AI, Franceschi C, De Benedictis G. Mitochondrial DNA haplogroups and APOE4 allele are non-independent variables in sporadic Alzheimer's disease. Human Genetics. 2001;108:194–198. doi: 10.1007/s004390100463. [DOI] [PubMed] [Google Scholar]
- 76.Lakatos A, Derbeneva O, Younes D, Keator D, Bakken T, Lvova M, Brandon M, Guffanti G, Reglodi D, Saykin A, Weiner M, Macciardi F, Schork N, Wallace DC, Potkin SG. Association between mitochondrial DNA variations and Alzheimer's disease in the ADNI cohort. Neurobiology of Aging. 2010;31:1355–1363. doi: 10.1016/j.neurobiolaging.2010.04.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.van der Walt JM, Nicodemus KK, Martin ER, Scott WK, Nance MA, Watts RL, Hubble JP, Haines JL, Koller WC, Lyons K, Pahwa R, Stern MB, Colcher A, Hiner BC, Jankovic J, Ondo WG, Allen FH, Jr, Goetz CG, Small GW, Mastaglia F, Stajich JM, McLaurin AC, Middleton LT, Scott BL, Schmechel DE, Pericak-Vance MA, Vance JM. Mitochondrial polymorphisms significantly reduce the risk of Parkinson disease. American Journal of Human Genetics. 2003;72:804–811. doi: 10.1086/373937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Ghezzi D, Marelli C, Achilli A, Goldwurm S, Pezzoli G, Barone P, Pellecchia MT, Stanzione P, Brusa L, Bentivoglio AR, Bonuccelli U, Petrozzi L, Abbruzzese G, Marchese R, Cortelli P, Grimaldi D, Martinelli P, Ferrarese C, Garavaglia B, Sangiorgi S, Carelli V, Torroni A, Albanese A, Zeviani M. Mitochondrial DNA haplogroup K is associated with a lower risk of Parkinson's disease in Italians. European Journal of Human Genetics. 2005;13:748–752. doi: 10.1038/sj.ejhg.5201425. [DOI] [PubMed] [Google Scholar]
- 79.Khusnutdinova E, Gilyazova I, Ruiz-Pesini E, Derbeneva O, Khusainova R, Khidiyatova I, Magzhanov R, Wallace DC. A mitochondrial etiology of neurodegenerative diseases: evidence from Parkinson's disease. Annals of the New York Academy of Sciences. 2008;1147:1–20. doi: 10.1196/annals.1427.001. [DOI] [PubMed] [Google Scholar]
- 80.Hendrickson SL, Hutcheson HB, Ruiz-Pesini E, Poole JC, Lautenberger J, Sezgin E, Kingsley L, Goedert JJ, Vlahov D, Donfield S, Wallace DC, O'Brien SJ. Mitochondrial DNA haplogroups influence AIDS progression. AIDS. 2008;22:2429–2439. doi: 10.1097/QAD.0b013e32831940bb. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Corral-Debrinski M, Horton T, Lott MT, Shoffner JM, McKee AC, Beal MF, Graham BH, Wallace DC. Marked changes in mitochondrial DNA deletion levels in Alzheimer brains. Genomics. 1994;23:471–476. doi: 10.1006/geno.1994.1525. [DOI] [PubMed] [Google Scholar]
- 82.Horton TM, Graham BH, Corral-Debrinski M, Shoffner JM, Kaufman AE, Beal BF, Wallace DC. Marked increase in mitochondrial DNA deletion levels in the cerebral cortex of Huntington's Disease patients. Neurology. 1995;45:1879–1883. doi: 10.1212/wnl.45.10.1879. [DOI] [PubMed] [Google Scholar]
- 83.Bender A, Krishnan KJ, Morris CM, Taylor GA, Reeve AK, Perry RH, Jaros E, Hersheson JS, Betts J, Klopstock T, Taylor RW, Turnbull DM. High levels of mitochondrial DNA deletions in substantia nigra neurons in aging and Parkinson disease. Nature Genetics. 2006;38:515–517. doi: 10.1038/ng1769. [DOI] [PubMed] [Google Scholar]
- 84.Kraytsberg Y, Kudryavtseva E, McKee AC, Geula C, Kowall NW, Khrapko K. Mitochondrial DNA deletions are abundant and cause functional impairment in aged human substantia nigra neurons. Nature Genetics. 2006;38:518–520. doi: 10.1038/ng1778. [DOI] [PubMed] [Google Scholar]
- 85.Coskun PE, Beal MF, Wallace DC. Alzheimer's brains harbor somatic mtDNA control-region mutations that suppress mitochondrial transcription and replication. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:10726–10731. doi: 10.1073/pnas.0403649101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Coskun PE, Wyrembak J, Derbereva O, Melkonian G, Doran E, Lott IT, Head E, Cotman CW, Wallace DC. Systemic mitochondrial dysfunction and the etiology of Alzheimer's disease and down syndrome dementia. Journal of Alzheimer's Disease. 2010;20 Suppl 2:S293–S310. doi: 10.3233/JAD-2010-100351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Murdock DG, Christacos NC, Wallace DC. The age-related accumulation of a mitochondrial DNA control region mutation in muscle, but not brain, detected by a sensitive PNA-directed PCR clamping based method. Nucleic Acids Research. 2000;28:4350–4355. doi: 10.1093/nar/28.21.4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Michikawa Y, Laderman K, Richter K, Attardi G. Role of nuclear background and in vivo environment in variable segregation behavior of the aging-dependent T414G mutation at critical control site for human fibroblast mtDNA replication. Somatic Cell and Molecular Genetics. 1999;25:333–342. doi: 10.1023/a:1019972500785. [DOI] [PubMed] [Google Scholar]
- 89.Michikawa Y, Mazzucchelli F, Bresolin N, Scarlato G, Attardi G. Aging-dependent large accumulation of point mutations in the human mtDNA control region for replication. Science. 1999;286:774–779. doi: 10.1126/science.286.5440.774. [DOI] [PubMed] [Google Scholar]
- 90.Suissa S, Wang Z, Poole J, Wittkopp S, Feder J, Shutt TE, Wallace DC, Shadel GS, Mishmar D. Ancient mtDNA genetic variants modulate mtDNA transcription and replication. PLoS Genetics. 2009;5 doi: 10.1371/journal.pgen.1000474. e1000474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.She H, Yang Q, Shepherd K, Smith Y, Miller G, Testa C, Mao Z. Direct regulation of complex I by mitochondrial MEF2D is disrupted in a mouse model of Parkinson disease and in human patients. The Journal of Clinical Investigation. 2011;121:930–940. doi: 10.1172/JCI43871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Basile AS, Bolger GT, Lueddens HW, Skolnick P. Electrophysiological actions of Ro5-4864 on cerebellar Purkinje neurons: evidence for 'peripheral' benzodiazepine receptor-mediated depression. Journal of Pharmacology and Experimental Therapeutics. 1989;248:463–469. [PubMed] [Google Scholar]
- 93.Bunn CL, Wallace DC, Eisenstadt JM. Cytoplasmic inheritance of chloramphenicol resistance in mouse tissue culture cells. Proceedings of the National Academy of Sciences of the United States of America. 1974;71:1681–1685. doi: 10.1073/pnas.71.5.1681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Wallace DC, Bunn CL, Eisenstadt JM. Cytoplasmic transfer of chloramphenicol resistance in human tissue culture cells. Journal of Cell Biology. 1975;67:174–188. doi: 10.1083/jcb.67.1.174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.King MP, Attardi G. Human cells lacking mtDNA: repopulation with exogenous mitochondria by complementation. Science. 1989;246:500–503. doi: 10.1126/science.2814477. [DOI] [PubMed] [Google Scholar]
- 96.Trimmer PA, Keeney PM, Borland MK, Simon FA, Almeida J, Swerdlow RH, Parks JP, Parker WD, Jr, Bennett JP., Jr Mitochondrial abnormalities in cybrid cell models of sporadic Alzheimer's disease worsen with passage in culture. Neurobiology of Disease. 2004;15:29–39. doi: 10.1016/j.nbd.2003.09.011. [DOI] [PubMed] [Google Scholar]
- 97.Thiffault C, Bennett JP., Jr Cyclical mitochondrial deltapsiM fluctuations linked to electron transport, F0F1 ATP-synthase and mitochondrial Na+/Ca+2 exchange are reduced in Alzheimer's disease cybrids. Mitochondrion. 2005;5:109–119. doi: 10.1016/j.mito.2004.12.002. [DOI] [PubMed] [Google Scholar]
- 98.Trimmer PA, Borland MK. Differentiated Alzheimer's disease transmitochondrial cybrid cell lines exhibit reduced organelle movement. Antioxidants and Redox Signaling. 2005;7:1101–1109. doi: 10.1089/ars.2005.7.1101. [DOI] [PubMed] [Google Scholar]
- 99.Cardoso SM, Pereira CF, Moreira PI, Arduino DM, Esteves AR, Oliveira CR. Mitochondrial control of autophagic lysosomal pathway in Alzheimer's disease. Experimental Neurology. 2010;223:294–298. doi: 10.1016/j.expneurol.2009.06.008. [DOI] [PubMed] [Google Scholar]
- 100.Onyango IG, Ahn JY, Tuttle JB, Bennett JP, Jr, Swerdlow RH. Nerve growth factor attenuates oxidant-induced beta-amyloid neurotoxicity in sporadic Alzheimer's disease cybrids. Journal of Neurochemistry. 2010;114:1605–1618. doi: 10.1111/j.1471-4159.2010.06871.x. [DOI] [PubMed] [Google Scholar]
- 101.Rhein V, Baysang G, Rao S, Meier F, Bonert A, Muller-Spahn F, Eckert A. Amyloid-beta leads to impaired cellular respiration, energy production and mitochondrial electron chain complex activities in human neuroblastoma cells. Cellular and Molecular Neurobiology. 2009;29:1063–1071. doi: 10.1007/s10571-009-9398-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Lynn BC, Wang J, Markesbery WR, Lovell MA. Quantitative changes in the mitochondrial proteome from subjects with mild cognitive impairment, early stage, and late stage Alzheimer's disease. Journal of Alzheimers Disease. 2010;19:325–339. doi: 10.3233/JAD-2010-1254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Buck CR, Jurynec MJ, Gupta DK, Law AK, Bilger J, Wallace DC, McKeon RJ. Increased adenine nucleotide translocator 1 in reactive astrocytes facilitates glutamate transport. Experimental Neurology. 2003;181:149–158. doi: 10.1016/s0014-4886(03)00043-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Law AK, Gupta D, Levy S, Wallace DC, McKeon RJ, Buck CR. TGF-beta1 induction of the adenine nucleotide translocator 1 in astrocytes occurs through Smads and Sp1 transcription factors. BMC Neuroscience. 2004;5:1. doi: 10.1186/1471-2202-5-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Lee J, Schriner SE, Wallace DC. Adenine nucleotide translocator I deficiency increases resistance of mouse brain and neurons to excitotoxic insults. Biochimica et Biophysica Acta. 2009;1787:364–370. doi: 10.1016/j.bbabio.2009.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Oddo S, Caccamo A, Shepherd JD, Murphy MP, Golde TE, Kayed R, Metherate R, Mattson MP, Akbari Y, LaFerla FM. Triple-transgenic model of Alzheimer's disease with plaques and tangles: intracellular Abeta and synaptic dysfunction. Neuron. 2003;39:409–421. doi: 10.1016/s0896-6273(03)00434-3. [DOI] [PubMed] [Google Scholar]
- 107.Yao J, Irwin RW, Zhao L, Nilsen J, Hamilton RT, Brinton RD. Mitochondrial bioenergetic deficit precedes Alzheimer's pathology in female mouse model of Alzheimer's disease. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:14670–14675. doi: 10.1073/pnas.0903563106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Yao J, Hamilton RT, Cadenas E, Brinton RD. Decline in mitochondrial bioenergetics and shift to ketogenic profile in brain during reproductive senescence. Biochimica et Biophysica Acta. 2010;1800:1121–1126. doi: 10.1016/j.bbagen.2010.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Pedram A, Razandi M, Wallace DC, Levin ER. Functional estrogen receptors in the mitochondria of breast cancer cells. Molecular Biology of the Cell. 2006;17:2125–2137. doi: 10.1091/mbc.E05-11-1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Zhang Q, Raoof M, Chen Y, Sumi Y, Sursal T, Junger W, Brohi K, Itagaki K, Hauser CJ. Circulating mitochondrial DAMPs cause inflammatory responses to injury. Nature. 2010;464:104–107. doi: 10.1038/nature08780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Lindahl KF, Hermel E, LB E, Wang CR. Maternally transmitted antigen of mice, a model transplantation antigen. Annual Review of Immunology. 1991;9:351–372. doi: 10.1146/annurev.iy.09.040191.002031. [DOI] [PubMed] [Google Scholar]
- 112.Barrientos A. In vivo and in organello assessment of OXPHOS activities. Methods. 2002;26:307–316. doi: 10.1016/S1046-2023(02)00036-1. [DOI] [PubMed] [Google Scholar]
- 113.Hyslop PA, Sklar LA. A quantitative fluorimetric assay for the determination of oxidant production by polymorphonuclear leukocytes: its use in the simultaneous fluorimetric assay of cellular activation processes. Analytical Biochemistry. 1984;141:280–286. doi: 10.1016/0003-2697(84)90457-3. [DOI] [PubMed] [Google Scholar]
- 114.Robinson KM, Janes MS, Pehar M, Monette JS, Ross MF, Hagen TM, Murphy MP, Beckman JS. Selective fluorescent imaging of superoxide in vivo using ethidium-based probes. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:15038–15043. doi: 10.1073/pnas.0601945103. [DOI] [PMC free article] [PubMed] [Google Scholar]