Abstract
Currently, there is no well-accepted rating system to reliably predict which human leukocyte antigen (HLA)-mismatched (MM) unrelated donor (URD) should be selected for patients who do not have an HLA allele-matched donor. We evaluated the ability of an MM ranking system, HistoCheck, to predict the risk associated with HLA class I disparity in a population of 744 single allele or antigen HLA-A, B, or C MM myeloablative URD hematopoietic stem cell transplant (HCT) recipients with AML, ALL, CML or MDS, facilitated through the NMDP from 1988–2003. Multivariate models were used to adjust for other significant clinical risk factors. HLA MMs were scored using the HistoCheck web-based tool and the patients divided into 4 quartiles: Dissimilarity Score (DSS) 1.04–2.84 (allele MM), >2.84–13.75 (allele and antigen MM), >13.75–19.39 (antigen MM) and >19.39–36.62 (antigen MM). Using the lowest scoring quartile as the reference, the DSS groups were evaluated for associations with relapse, treatment-related mortality (TRM), acute and chronic graft-versus-host disease (GVHD), leukemia-free survival (LFS), and overall survival (OS) in the entire cohort, and in subset analyses by disease and disease stage. No significant associations were found between DSS and any outcomes in the overall cohort using the quartile categories or treating DSS as a continuous variable. Higher DSS scores were associated with decreased engraftment in early stage disease (p=0.0003) but not in other disease stages. In summary, DSS does not correlate with transplantation outcomes, and the HistoCheck scoring system does not provide an effective strategy to rank HLA class I MM. The data set used in this study is available to evaluate new algorithms proposed for donor selection.
Introduction
While HLA matching for alleles of HLA-A, -B, -C, -DRB1 (i.e. 8/8 matches) has been shown to optimize survival following hematopoietic stem cell transplantation (HCT) (1;2), 30%–40% of transplantations facilitated through the National Marrow Donor Program (NMDP) are mismatched at one or more loci (3). While mismatching for a single allele (7/8 match) results in a 10% reduction in average overall survival compared to 8/8 matched transplants, this risk may be acceptable when compared to alternative therapies.
No rating system exists to reliably predict which HLA-mismatched unrelated donor should be selected for patients who do not have an HLA allele-matched donor. Previous Center for International Blood and Marrow Transplant Research (CIBMTR) studies have evaluated mismatched donor selection based on serologically cross-reactive epitopes (i.e. CREGs) (4), amino acid triplets (i.e. HLA MatchMaker) (5), and the number of amino acid differences (6;7), and found that these selection strategies do not predict outcomes. While studies of donor-recipient pairs from the Japanese Marrow Donor Program have suggested a differential impact of specific HLA mismatches (8–10), a recent CIBMTR report discussed the difficulties in evaluating the impact of specific allele mismatches in the context of mismatches at other loci (9). Thus, it continues to be a priority to evaluate various HLA-based donor selection criteria to improve the outcomes of transplantation with HLA-mismatched donors.
In 2002, Elsner and Blasczyk suggested that a rating system based on structural data of HLA class I molecules might be used to identify acceptable mismatches (11). Their algorithm was based on the functional similarity of amino acids using a distance matrix developed by Risler et al. (12), and based on the frequency of amino acid substitutions in proteins. Risler scores were further weighted based on the position of the disparity in the HLA molecule (i.e. location in the peptide binding or T cell receptor recognition site). In 2004, Blasczyk and colleagues extended the algorithm to include evaluation of class II molecules and developed an internet-based software tool for assigning scores (13) (http://www.histocheck.de/).
In 2004, Shaw and colleagues used HistoCheck to score 26 single HLA-A allele mismatched recipients and nine single HLA-B mismatches in the Anthony Nolan clinical database (14). These recipients were matched for alleles at the other key HLA loci. The investigators compared the clinical outcomes with the HistoCheck scores. No associations were observed with neutrophil engraftment, acute or chronic graft-versus-host disease (GVHD), relapse, or survival in this small study. In 2011, Askar and colleagues evaluated the correlation between HistoCheck score and high-risk HLA allele mismatch combinations previously described by Kawase et al (15). They found that HistoCheck score distribution was no different between high- and low-risk allele combinations, and did not correlate with mismatch risk stratification in the Japanese population. We used CIBMTR data to evaluate the HistoCheck algorithm in a larger study in order to provide guidance for HLA-mismatched donor selection.
Materials and Methods
Study population
The study included patients reported to the NMDP who were transplanted from an unrelated donor between 1988 and 2003. All patients and their donors were fully HLA-typed at high resolution through the NMDP ongoing retrospective high-resolution typing project. The study included 744 donor-recipient pairs with a single HLA-A, -B or -C mismatch. All cases were matched for HLA-DRB1 and DQB1. Eligible diagnoses included acute lymphoblastic leukemia (ALL, N=199), acute myeloid leukemia (AML, N=224), chronic myeloid leukemia (CML, N=259) and myelodysplastic syndrome (MDS, N=62). Early stage disease was defined as AML or ALL in first complete remission, CML in first chronic phase, and MDS subtype refractory anemia. Intermediate stage disease was defined as AML or ALL in second or subsequent complete remission or in first relapse, and CML in accelerated phase or second chronic phase. Advanced phase disease was defined as AML in second or higher relapse or primary induction failure, CML in blast phase, MDS subtypes refractory anemia with excess blasts or in transformation, or unclassified MDS. All patients received standard myeloablative conditioning regimens. The same data set was used to evaluate another matching algorithm, HLA Matchmaker (5).
Patients undergoing conditioning regimens of lower intensity, second or subsequent transplantation, or surviving patients who did not provide signed, informed consent to allow analysis of their clinical data or HLA typing of stored NMDP Research Repository samples were excluded. All surviving recipients included in this analysis were retrospectively contacted and provided informed consent for participation in the NMDP research program. To adjust for the potential bias introduced by exclusion of non-consenting surviving patients, a modeling process randomly excluded the same percentage of deceased patients using a biased coin randomization with exclusion probabilities based on characteristics associated with not providing consent for use of the data in survivors (1).
Evaluation of HLA disparity
HLA class I mismatches were scored by HistoCheck using a web-based tool (13) (http://www.histocheck.de/). Patients were divided into four quartiles based on a dissimilarity score (DSS): DSS 1.04–2.84 (Group 1), >2.84–13.75 (Group 2), >13.75–19.39 (Group 3) and >19.39–36.62 (Group 4) for analysis. In addition, all analyses included an evaluation of DSS score as a continuous variable. All cases were matched for HLA-DRB1 and DQB1. HLA-DPB1 matching was available for all cases, but was not considered in the analysis.
Clinical endpoints
The study evaluated the association between the HistoCheck score on outcomes, with disease-free survival (DFS) as the primary endpoint, and acute GVHD grades II-IV, chronic GVHD, treatment-related mortality (TRM), relapse, overall survival (OS) and neutrophil engraftment as secondary endpoints. DFS was defined as relapse or death from any cause, with patients who were alive and in complete remission censored at the time of last follow-up. The incidences of grades II-IV acute GVHD were determined during the first 100 days post transplant and defined according to the Glucksberg scale (16). Chronic GVHD was defined according to the Seattle criteria (17). TRM was defined as death during a continuous complete remission. Relapse was defined as leukemia relapse or MDS recurrence. OS considered death from any cause as the event, and surviving patients were censored at the date of last contact. Neutrophil engraftment was defined as achieving an absolute neutrophil count greater than 500 for three consecutive measurements. Events were summarized by the cumulative incidence estimate, with death as the competing risk.
Statistical methods
To compare pre-transplant characteristics for discrete factors, the number of cases and their respective percentages were calculated and chi-square tests performed to compare the HistoCheck-defined HLA-mismatched groups. For continuous factors, the medians and ranges were calculated and the Kruskal-Wallis test used to analyze differences between the HistoCheck quartile groups. Probabilities of disease-free survival were calculated using the Kaplan-Meier estimator. Estimated cumulative incidence was used to describe the probabilities for events with competing risks. These included engraftment, GVHD, relapse and TRM. Comparisons of survival curves were done with the log-rank test.
Multivariate analyses were performed using the Cox proportional hazards model to compare the HistoCheck quartile groups. Models were fit to determine which risk factors were related to a given outcome. All variables were tested for affirmation of the proportional hazards assumption. Neutrophil recovery at day 28 was modeled using a logistic regression approach. Subset analyses were conducted by disease and disease stage. Due to multiple comparisons, a p-value <0.01 was used to determine statistical significance.
Results
The characteristics of the patients in this study and their HLA mismatches are described in Table 1. Patients were grouped into quartiles based on the dissimilarity score received for their specific HLA class I mismatch. Analysis of the specific HLA mismatches found in each quartile correlated with the type of mismatches found in each category: DSS 1.04–2.84 (allele MM), >2.84–13.75 (allele and antigen MM), >13.75–19.39 (antigen MM) and >19.39–36.62 (antigen MM). Table 1 shows the distribution of mismatches at HLA-A, HLA-B and HLA-C loci. The disease stage distribution was significantly different (p=0.04) across the quartiles, with patients in the highest DSS quartile exhibiting more intermediate and advanced disease than the other quartiles. No other characteristics were significantly different between the quartile groups. While donors and recipients were all matched at HLA-DRB1 and DQB1 loci, the majority of them were HLA-DPB1 mismatched. The distribution of HLA-DPB1 mismatching did not differ across the quartile groups, with 83%, 87%, 88%, and 88% mismatch (p=0.47) for groups 1–4, respectively.
Table 1.
Characteristics of patients by Histocheck (DSS) score quartiles of mismatched cases (7/8 HLA class I mismatches)
| Variable | DSS Quartile | P-valueb | |||
|---|---|---|---|---|---|
| 1.04 – 2.84 N (%) |
> 2.84 – 13.75 N (%) |
> 13.75 – 19.39 N (%) |
> 19.39 – 36.62 N (%) |
||
| Number of patients | 187 | 189 | 182 | 186 | |
| Number of centers | 62 | 69 | 55 | 66 | |
| Matching of locia | <0.001 | ||||
| HLA-A allele mismatched | 49 (26) | 49 (26) | 0 | 0 | |
| HLA-A antigen mismatched | 0 | 38 (20) | 32 (18) | 74 (40) | |
| HLA-B allele mismatched | 63 (34) | 22 (12) | 0 | 0 | |
| HLA-B antigen mismatched | 0 | 4 ( 2) | 2 ( 1) | 6 ( 3) | |
| HLA-C allele mismatched | 75 (40) | 5 ( 3) | 0 | 0 | |
| HLA-C antigen mismatched | 0 | 71 (37) | 148 (81) | 106 (57) | |
| Age, median (range), years | 33 (<1–60) | 32 (<1–60) | 32 (<1–65) | 30 (1–59) | 0.42 |
| Age at transplant | 0.22 | ||||
| ≤ 10 y | 19 (10) | 26 (14) | 16 ( 9) | 27 (15) | |
| 10–19 y | 33 (18) | 19 (10) | 30 (17) | 38 (20) | |
| 20–29 y | 27 (14) | 40 (21) | 35 (19) | 27 (15) | |
| 30–39 y | 44 (24) | 35 (18) | 39 (21) | 36 (19) | |
| 40–49 y | 43 (23) | 51 (27) | 47 (26) | 37 (20) | |
| 50 y and older | 21 (11) | 18 (10) | 15 ( 8) | 21 (11) | |
| Male sex | 93 (50) | 98 (52) | 107 (59) | 99 (53) | 0.34 |
| Karnofsky prior to TX ≥ 90 | 124 (66) | 143 (76) | 130 (71) | 137 (74) | 0.12 |
| Disease at transplant | 0.10 | ||||
| AML | 57 (31) | 52 (28) | 49 (27) | 66 (35) | |
| ALL | 47 (25) | 49 (26) | 48 (26) | 55 (30) | |
| CML | 73 (39) | 74 (39) | 64 (35) | 48 (26) | |
| MDS | 10 ( 5) | 14 ( 7) | 21 (12) | 17 ( 9) | |
| Disease status at transplant | 0.04 | ||||
| Early | 71 (38) | 84 (44) | 72 (39) | 58 (31) | |
| Intermediate | 89 (48) | 67 (35) | 74 (41) | 76 (41) | |
| Advanced | 27 (14) | 37 (20) | 36 (20) | 51 (27) | |
| Other | 0 | 1 ( 1) | 0 | 1 ( 1) | |
| Graft type | 0.008 | ||||
| Marrow | 164 (88) | 183 (97) | 169 (93) | 167 (90) | |
| Peripheral blood | 23 (12) | 6 ( 3) | 13 ( 7) | 19 (10) | |
| Conditioning Regimen | 0.71 | ||||
| Traditional myeloablative | 180 (96) | 185 (98) | 176 (97) | 182 (98) | |
| Nontraditional ablative | 7 ( 4) | 4 ( 2) | 6 ( 3) | 4 ( 2) | |
| Donor/recipient sex match | 0.26 | ||||
| Male/Male | 64 (34) | 66 (35) | 63 (35) | 53 (28) | |
| Male/Female | 51 (27) | 47 (25) | 45 (25) | 44 (24) | |
| Female/Male | 29 (16) | 32 (17) | 44 (24) | 46 (25) | |
| Female/Female | 43 (23) | 44 (23) | 30 (16) | 43 (23) | |
| GVHD prophylaxis | 0.20 | ||||
| Tacrolimus + (MTX or MMF or Steroids) ± other | 35 (19) | 34 (18) | 31 (17) | 38 (20) | |
| Tacrolimus ± other | 2 ( 1) | 0 | 1 (<1) | 1 (<1) | |
| CsA + MTX ± other | 114 (61) | 109 (58) | 98 (54) | 87 (47) | |
| CsA ± MMF ± Steroids ± other (No MTX) | 2 ( 1) | 7 ( 4) | 6 ( 3) | 3 ( 2) | |
| MMF +/− other | 1 (<1) | 0 | 0 | 0 | |
| MTX +/− other (No CsA) | 0 | 1 (<1) | 2 ( 1) | 0 | |
| T-cell depletion | 33 (18) | 38 (20) | 43 (24) | 56 (30) | |
| Other | 0 | 0 | 1 (<1) | 1 (<1) | |
| Donor/recipient CMV match | 0.84 | ||||
| Negative/Negative | 62 (33) | 58 (31) | 59 (32) | 73 (39) | |
| Negative/Positive | 56 (30) | 56 (30) | 49 (27) | 46 (25) | |
| Positive/Negative | 30 (16) | 35 (18) | 28 (15) | 32 (17) | |
| Positive/Positive | 37 (20) | 37 (19) | 41 (23) | 31 (17) | |
| Unknown | 2 ( 1) | 3 ( 2) | 5 ( 3) | 4 ( 2) | |
| Donor age, median (range), years | 36 (19–57) | 36 (19–54) | 36 (19–59) | 35 (19–59) | 0.74 |
| Donor age | 0.65 | ||||
| 18–29 | 45 (24) | 56 (30) | 47 (26) | 50 (27) | |
| 30–39 | 69 (37) | 63 (33) | 70 (38) | 74 (40) | |
| 40–49 | 57 (30) | 58 (31) | 45 (25) | 47 (25) | |
| 50 and older | 16 ( 9) | 12 ( 6) | 20 (11) | 15 ( 8) | |
| Year of transplant | 0.06 | ||||
| 1988 | 1 (<1) | 1 (<1) | 0 | 0 | |
| 1989 | 1 (<1) | 2 ( 1) | 2 ( 1) | 2 ( 1) | |
| 1990 | 3 ( 2) | 2 ( 1) | 2 ( 1) | 6 ( 3) | |
| 1991 | 6 ( 3) | 1 (<1) | 5 ( 3) | 8 ( 4) | |
| 1992 | 3 ( 2) | 15 ( 8) | 12 ( 7) | 9 ( 5) | |
| 1993 | 9 ( 5) | 6 ( 3) | 10 ( 5) | 9 ( 5) | |
| 1994 | 10 ( 5) | 12 ( 6) | 16 ( 9) | 14 ( 7) | |
| 1995 | 16 ( 9) | 16 ( 8) | 22 (12) | 9 ( 5) | |
| 1996 | 12 ( 6) | 15 ( 8) | 13 ( 7) | 16 ( 9) | |
| 1997 | 21 (11) | 13 ( 7) | 11 ( 6) | 11 ( 6) | |
| 1998 | 15 ( 8) | 20 (11) | 15 ( 8) | 11 ( 6) | |
| 1999 | 19 (10) | 38 (20) | 18 (10) | 18 (10) | |
| 2000 | 24 (13) | 17 ( 9) | 18 (10) | 19 (10) | |
| 2001 | 30 (16) | 19 (10) | 18 (10) | 29 (16) | |
| 2002 | 11 ( 6) | 11 ( 6) | 15 ( 8) | 19 (10) | |
| 2003 | 6 ( 3) | 1 (<1) | 5 ( 3) | 6 ( 3) | |
| Median follow-up of survivors, months | 61 (6–191) | 60 (7–179) | 74 (24–184) | 50 (12–175) | 0.36 |
Antigen mismatches were defined as mismatches differing in the first digits of the allele name (e.g., A*02 vs. A*68). Allele mismatches were defined as mismatches differing in the third and fourth digits of the allele name (i.e. digits following the first colon, e.g. A*02:01 vs A*02:05).
P-values are chi-square for descriptive variables, Kruskal-Wallis for continuous variables and log-rank P-value for median follow-up of survivors.
Graft-versus-host disease
Univariate analysis showed that the probabilities of grades II-IV acute GVHD at 100 days did not differ among the various DSS groups, with 51%, 51%, 56% and 51% (p=0.75) for groups 1–4, respectively (Table 2, Figure 1a). Chronic GVHD rates at one year did not differ between the DSS groups, with 39%, 34%, 35% and 38% (p=0.70) for groups 1–4, respectively.
Table 2.
Univariate probabilities of clinical outcomes by Histocheck (DSS) score quartiles of mismatched cases
| Outcomes | Time point | Percent Probability (95% CI) | P-valuea | |||
|---|---|---|---|---|---|---|
| Group 1 DSS 1.04 – 2.84 |
Group 2 DSS >2.84 – 13.75 |
Group 3 DSS >13.75 – 19.39 |
Group 4 DSS >13.39 – 36.62 |
|||
| Acute GVHD II-IV | 100 days | 51 (44–59) | 51 (44–58) | 56 (49–63) | 51 (44–58) | 0.75 |
| Chronic GVHD | 1 year | 39 (32–47) | 34 (27–42) | 35 (28–42) | 38 (31–46) | 0.70 |
| Treatment related mortality | 1 year | 41 (34–48) | 49 (42–56) | 44 (37–52) | 45 (38–52) | 0.48 |
| 3 years | 46 (39–53) | 51 (44–58) | 54 (46–61) | 49 (42–57) | 0.54 | |
| 5 years | 48 (41–56) | 54 (47–61) | 55 (48–62) | 52 (44–59) | 0.59 | |
| Relapse | 1 year | 14 (10–20) | 16 (11–22) | 14 (9–19) | 19 (14–25) | 0.53 |
| 3 years | 18 (13–24) | 21 (16–27) | 17 (12–22) | 25 (19–31) | 0.25 | |
| 5 years | 19 (14–25) | 21 (16–27) | 18 (13–24) | 25 (19–31) | 0.44 | |
| Overall survival | 1 year | 47 (40–54) | 41 (34–48) | 46 (39–53) | 41 (34–49) | 0.50 |
| 3 years | 38 (31–45) | 32 (25–39) | 33 (27–40) | 29 (23–36) | 0.31 | |
| 5 years | 35 (28–43) | 28 (22–35) | 31 (24–38) | 26 (19–33) | 0.26 | |
| Disease free survival | 1 year | 45 (38–52) | 35 (28–42) | 42 (35–49) | 36 (29–43) | 0.16 |
| 3 years | 35 (29–43) | 27 (21–34) | 30 (23–37) | 26 (20–33) | 0.21 | |
| 5 years | 33 (26–40) | 25 (19–31) | 27 (21–34) | 24 (17–31) | 0.28 | |
| Neutrophil engraftment | 28 days | 92 (88–95) | 90 (86–94) | 87 (81–91) | 85 (79–89) | 0.09 |
P-values are pointwise
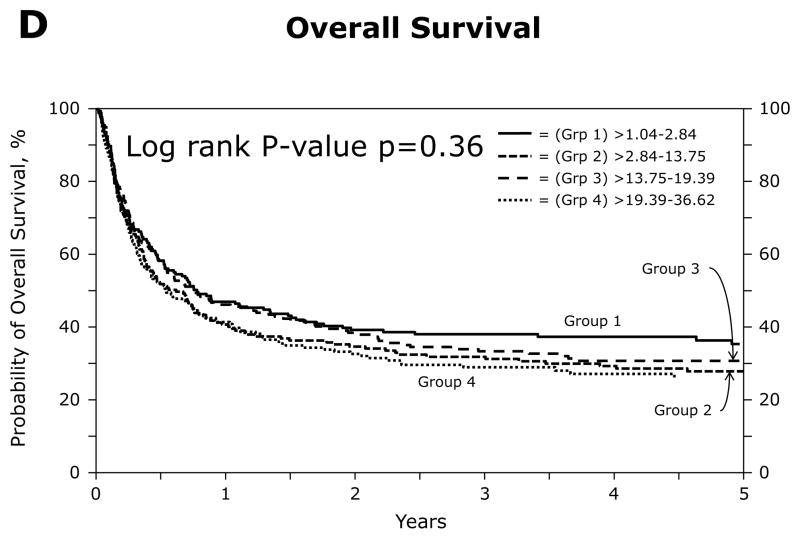
Figure 1.
Univariate analyses of outcomes by patient quartiles, based on the HistoCheck dissimilarity scores of HLA class I mismatches: a. Grades II-IV acute GVHD; b. Treatment-related mortality at five years; c. Relapse at five years; d. Overall survival at five years
Multivariate analysis found significant associations with grades II-IV acute GVHD and patient age, graft type and GVHD prophylaxis. After adjusting for the significant covariates, no associations were found with the DSS groups (overall p=0.78) or with DSS as a continuous variable (p=0.50) (Table 3). Chronic GVHD was significantly associated with donor/recipient sex match, graft type, and GVHD prophylaxis. After adjusting for significant covariates, no associations were found with the DSS groups (overall p=0.61) or with DSS as a continuous variable (p=0.89) (Table 3).
Table 3.
Multivariate analysis of clinical outcomes by HistoCheck DSS score quartile groups and treated as a continuous variable
| Outcome | Group 1 DSS 1.04 – 2.84a |
Group 2 DSS >2.84 – 13.75 RR (95% CI) |
Group 3 DSS >13.75 – 19.39 RR (95% CI) |
Group 4 DSS >19.39 – 36.62 RR (95% CI) |
Overall P-value | DSS Continuous P-value |
|---|---|---|---|---|---|---|
| Acute GVHD II-IV | 1.00 | 1.06 (0.79 – 1.41) | 1.05 (0.79 – 1.39) | 1.16 (0.88 – 1.54) | 0.78 | 0.50 |
| Chronic GVHD | 1.00 | 0.86 (0.61 – 1.21) | 0.82 (0.59 – 1.14) | 0.97 (0.69 – 1.37) | 0.61 | 0.89 |
| Treatment-related mortality | 1.00 | 1.22 (0.91 – 1.63) | 1.13 (0.84 – 1.51) | 1.32 (0.98 – 1.77) | 0.29 | 0.58 |
| Relapse | 1.00 | 1.47 (0.93 – 2.32) | 0.99 (0.61 – 1.60) | 1.44 (0.93 – 2.26) | 0.14 | 0.34 |
| Overall survival | 1.00 | 1.25 (0.98 – 1.61) | 1.07 (0.83 – 1.38) | 1.28 (1.00 – 1.64) | 0.14 | 0.57 |
| Disease-free survival | 1.00 | 1.30 (1.01 – 1.66) | 1.11 (0.86 – 1.42) | 1.35 (1.05 – 1.73) | 0.06 | 0.45 |
| Neutrophil engraftmentb | 1.00 | 0.85 (0.41 – 1.75) | 0.63 (0.32 – 1.25) | 0.48 (0.25 – 0.95) 0.13 | 0.03 |
Reference group
Data reported as an odds ratio in favor of engraftment
Treatment-related mortality
Univariate analysis found no differences in the probabilities of TRM at one (p=0.48), three (p=0.54) or five (p=0.59) years between the DSS groups (Table 2, Figure 1b). The probabilities of TRM at one year were 41%, 49%, 44% and 45% for groups 1–4, respectively.
Multivariate analysis revealed significant associations between TRM and cytomegalovirus (CMV) match, Karnofsky score, recipient age, and year of transplant. After adjusting for the significant covariates, no associations were found between TRM and the DSS groups (p=0.29) or with DSS as a continuous variable (p=0.58) (Table 3).
Relapse
Univariate analysis revealed no differences in the probabilities of relapse at one (p=0.53), three (p=0.25) or five (p=0.44) years between the DSS groups (Table 2, Figure 1c). The probabilities of relapse at one year were 14%, 16%, 14% and 19% for groups 1–4, respectively.
Multivariate analysis found significant associations between relapse and disease status. After adjusting for the significant covariate, no associations were found between relapse and the DSS groups (p=0.14) or with DSS as a continuous variable (p=0.34) (Table 3).
Overall survival
Univariate analysis revealed no differences in the probabilities of OS at one (p=0.50), three (p=0.31) or five (p=0.26) years between the DSS groups (Table 2, Figure 1d). The probabilities of OS at one year were 47%, 41%, 46% and 41% for groups 1–4, respectively.
Multivariate analysis showed significant associations between OS and donor/recipient CMV match, disease status, Karnofsky score, recipient age and year of transplant. After adjusting for the significant covariates, no association was found between OS and the DSS groups (p=0.14) or with DSS as a continuous variable (p=0.57) (Table 3).
Disease-free survival
Univariate analysis revealed no differences in the probabilities of relapse at one (p=0.16), three (p=0.21) or five (p=0.28) years between the DSS groups (Table 2). The probabilities of relapse at one year were 45%, 35%, 42% and 36% for groups 1–4, respectively.
Multivariate analysis showed significant associations between relapse and disease status. After adjusting for the significant covariate, no association was found between relapse and the DSS groups (p=0.06) or with DSS as a continuous variable (p=0.45) (Table 3).
Neutrophil engraftment
Univariate analysis found no differences in the probabilities of neutrophil engraftment at day 28 (p=0.09) between the DSS groups (Table 2). The probabilities of neutrophil engraftment at day 28 were 92%, 90%, 87% and 85% for groups 1–4, respectively.
Multivariate analysis showed significant associations between neutrophil engraftment and year of transplant. After adjusting for the significant covariate, no associations were found between relapse and the DSS groups (p=0.13) or with DSS as a continuous variable (p=0.03) (Table 3).
Subset analyses
Previous studies suggested differing effects of HLA matching based on disease and disease status (1;18). Disease and disease status subset multivariate models were evaluated to assess association between DSS scores and all outcomes. Significant associations were noted between engraftment rates at day 28 and increasing DSS scores (p=0.0003) in early stage disease. In addition, higher DSS scores were associated with TRM, DFS and OS in the MDS population. However, the MDS population was quite small (N=10–21), which limits confidence in the relevance of this association.
Discussion
The goal of this study was to test the hypothesis that a scoring system, HistoCheck, based on the functional similarity of amino acids and their position within the HLA molecule (i.e. DSS score), could predict the outcomes of unrelated HCT using 7/8 class I mismatched donors. The results of this study of 744 single allele or antigen-mismatched transplant patients confirms and extends the results observed in a study of 35 patients by Shaw et al. (14), and an analysis of another registry by Askar et al (15). The patients were divided into quartiles based on their DSS score, and tests to detect differences between the quartiles were performed. The overall observation of equality among the quartiles suggests that the HistoCheck scoring system for HLA class I mismatches is not predictive of transplant outcomes as measured by survival, GVHD, relapse or engraftment. The HistoCheck scoring system of HLA class II mismatches was not evaluated in our study. However, a recent study showed that mean DSS scores were not significantly different and DSS distributions were overlapping among the high- and low-risk allele combinations considered responsible for severe acute GVHD within loci HLA-DRB1 and DPB1 (15). Taken together, the results of our analysis and prior studies do not support the selection of unrelated donors using the HistoCheck scoring system to improve clinical outcomes.
There were some significant observations. Higher DSS scores were associated with decreased engraftment in early stage disease (p=0.0003) but not in other disease stages. Anti-HLA antibodies in the recipient are associated with higher rates of graft failure and lower engraftment levels in HLA mismatched unrelated donor transplantation (19). Higher DSS scores do correlate with more antigen level mismatches (table 1) that could serve as targets for anti-HLA antibodies. However, the presence of anti-HLA antibodies was not evaluable in the dataset. In disease subset analyses, DSS score was associated with TRM, LFS and OS in MDS. However, the MDS subgroups were very small (n=10–21), diminishing confidence in the association.
There are multiple limitations to the HistoCheck algorithm that likely limit its ability to predict outcomes. In general, the algorithm may not adequately reflect the biological complexity inherent in the HLA/peptide/T cell receptor (TCR) complex. The amino acid substitution score by Risler may not represent functional similarities of amino acids with regard to peptide and TCR binding. The algorithm’s adjustments for impact on peptide and/or TCR binding may be too simplistic. Its structural considerations based on crystallographic data of a single allelic product, A*02:01, may not be readily extrapolated to other allelic products, particularly products of loci other than HLA-A. The influence of molecular interaction among amino acids is not considered and the differential binding of peptides, particularly in the case of minor antigens, may alter matching based on similarity. The algorithm scoring also strongly correlates with allele and antigen mismatching (table 1), however prior studies have found that both allele and antigen mismatches can be detrimental to transplant outcomes (1). It will be challenging to develop an algorithm capable of predicting the impact of HLA allelic variation on the strength of the allo-recognition response.
A major hurdle for developing an evidence-based scoring system for HLA disparities is HLA diversity. This was recently demonstrated by an NMDP study of HLA-A disparities, which showed that there were 190 different mismatch combinations in 4,226 donor-recipient pairs, and that 51% of these mismatches could be observed in only one pair (20). Since the most frequent HLA-A mismatches in the HLA-B, -C, and -DRB1, and matched pairs occurred only 2–6 times per 1,000 U.S. transplant patients, it is not feasible to directly determine associations between a particular isolated HLA-A disparity and transplant outcomes for patients transplanted in the United States.
In contrast, a study of 5,210 Japanese donor-recipient pairs showed a high incidence of recurring HLA-A disparities (9). In this study, one HLA disparity was observed in 269 pairs and several more were observed in more than 40 pairs. Three of these HLA-A disparities were associated with acute GVHD, but only one direction of each disparity reached statistical significance. This observation could provide important insights regarding factors related to T cell selection and/or recognition that might modulate allogenicity. While homogeneous populations such as the one in the Japanese study can provide sufficient numbers of a particular mismatch for isolated study, the observations might not be applicable to other populations because other factors (e.g. genetic differences, environmental exposures) could alter the alloreactive response. Further, the particular recurring HLA disparities observed in the Japanese population are not often observed in other populations that have been studied (8). The challenge intensifies if the investigation is reduced to an amino acid level, because the context of each amino acid difference will likely influence allorecognition (20).
The complexity generated by extensive HLA diversity, genetic variation and other transplant factors (e.g. disease and stage of disease, recipient age) creates a daunting challenge for developing an evidence-based scoring system for HLA disparities. Until this challenge can be resolved, it is important to exercise great caution in using untested algorithms to guide donor selection. It is likely that very complex models will likely be required to meet the challenge.
Acknowledgments
This research is supported by funding from the Office of Naval Research N00014-08-1-1078 (CKH). CIBMTR is supported by Public Health Service Grant/Cooperative Agreement U24-CA76518 from the National Cancer Institute (NCI), the National Heart, Lung and Blood Institute (NHLBI) and the National Institute of Allergy and Infectious Diseases (NIAID); a Grant/Cooperative Agreement 5U01HL069294 from NHLBI and NCI; a contract HHSH234200637015C with Health Resources and Services Administration (HRSA/DHHS); two Grants N00014-10-1-0204 and N00014-1-1-0339 from the Office of Naval Research; and grants from Allos, Inc.; Amgen, Inc.; Angioblast; Anonymous donation to the Medical College of Wisconsin; Ariad; Be the Match Foundation; Blue Cross and Blue Shield Association; Buchanan Family Foundation; CaridianBCT; Celgene Corporation; CellGenix, GmbH; Children’s Leukemia Research Association; Fresenius-Biotech North America, Inc.; Gamida Cell Teva Joint Venture Ltd.; Genentech, Inc.; Genzyme Corporation; GlaxoSmithKline; Kiadis Pharma; The Leukemia & Lymphoma Society; The Medical College of Wisconsin; Millennium Pharmaceuticals, Inc.; Milliman USA, Inc.; Miltenyi Biotec, Inc.; National Marrow Donor Program; Optum Healthcare Solutions, Inc.; Otsuka America Pharmaceutical, Inc.; Seattle Genetics; Sigma-Tau Pharmaceuticals; Soligenix, Inc.; Swedish Orphan Biovitrum; THERAKOS, Inc.; and Wellpoint, Inc. The views expressed in this article do not reflect the official policy or position of the National Institute of Health, the Department of the Navy, the Department of Defense, or any other agency of the U.S. Government.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Lee SJ, Klein J, Haagenson M, et al. High-resolution donor-recipient HLA matching contributes to the success of unrelated donor marrow transplantation. Blood. 2007;110(13):4576–4583. doi: 10.1182/blood-2007-06-097386. [DOI] [PubMed] [Google Scholar]
- 2.Woolfrey A, Klein JP, Haagenson M, et al. HLA-C Antigen mismatches are associated with worse outcomes in unrelated donor peripheral blood stem cell transplantation. Biol Blood Marrow Transplant. 2010 doi: 10.1016/j.bbmt.2010.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hurley CK, Fernandez-Vina M, Hildebrand WH, et al. A high degree of HLA disparity arises from limited allelic diversity: analysis of 1775 unrelated bone marrow transplant donor-recipient pairs. Hum Immunol. 2007;68(1):30–40. doi: 10.1016/j.humimm.2006.09.004. [DOI] [PubMed] [Google Scholar]
- 4.Wade JA, Hurley CK, Takemoto SK, et al. HLA mismatching within or outside of cross-reactive groups (CREGs) is associated with similar outcomes after unrelated hematopoietic stem cell transplantation. Blood. 2007;109(9):4064–4070. doi: 10.1182/blood-2006-06-032193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Duquesnoy R, Spellman S, Haagenson M, Wang T, Horowitz MM, Oudshoorn M. HLAMatchmaker-defined triplet matching is not associated with better survival rates of patients with class I HLA allele mismatched hematopoietic cell transplants from unrelated donors. Biol Blood Marrow Transplant. 2008;14(9):1064–1071. doi: 10.1016/j.bbmt.2008.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Heemskerk MB, Roelen DL, Dankers MK, et al. Allogeneic MHC class I molecules with numerous sequence differences do not elicit a CTL response. Hum Immunol. 2005;66(9):969–976. doi: 10.1016/j.humimm.2005.06.007. [DOI] [PubMed] [Google Scholar]
- 7.Heemskerk MB, Cornelissen JJ, Roelen DL, et al. Highly diverged MHC class I mismatches are acceptable for haematopoietic stem cell transplantation. Bone Marrow Transplant. 2007;40(3):193–200. doi: 10.1038/sj.bmt.1705721. [DOI] [PubMed] [Google Scholar]
- 8.Morishima Y, Kawase T, Malkki M, Petersdorf EW. Effect of HLA-A2 allele disparity on clinical outcome in hematopoietic cell transplantation from unrelated donors. Tissue Antigens. 2007;69 (Suppl 1):31–35. doi: 10.1111/j.1399-0039.2006.759_3.x. [DOI] [PubMed] [Google Scholar]
- 9.Kawase T, Morishima Y, Matsuo K, et al. High-risk HLA allele mismatch combinations responsible for severe acute graft-versus-host disease and implication for its molecular mechanism. Blood. 2007;110(7):2235–2241. doi: 10.1182/blood-2007-02-072405. [DOI] [PubMed] [Google Scholar]
- 10.Kawase T, Matsuo K, Kashiwase K, et al. HLA mismatch combinations associated with decreased risk of relapse: implications for the molecular mechanism. Blood. 2009;113(12):2851–2858. doi: 10.1182/blood-2008-08-171934. [DOI] [PubMed] [Google Scholar]
- 11.Elsner H, Blasczyk R. Sequence similarity matching: proposal of a structure-based rating system for bone marrow transplantation. European Journal of Immunogenetics. 2002;29:229–236. doi: 10.1046/j.1365-2370.2002.00301.x. [DOI] [PubMed] [Google Scholar]
- 12.Risler JL, Delorme MO, Delacroix H, Henaut A. Amino acid substitutions in structurally related proteins. A pattern recognition approach. Determination of a new and efficient scoring matrix. J Mol Biol. 1988;204(4):1019–1029. doi: 10.1016/0022-2836(88)90058-7. [DOI] [PubMed] [Google Scholar]
- 13.Elsner HA, DeLuca D, Strub J, Blasczyk R. HistoCheck: rating of HLA class I and II mismatches by an internet-based software tool. Bone Marrow Transplant. 2004;33(2):165–169. doi: 10.1038/sj.bmt.1704301. [DOI] [PubMed] [Google Scholar]
- 14.Shaw BE, Barber LD, Madrigal JA, Cleaver S, Marsh SG. Scoring for HLA matching? A clinical test of HistoCheck. Bone Marrow Transplant. 2004;34(4):367–368. doi: 10.1038/sj.bmt.1704586. [DOI] [PubMed] [Google Scholar]
- 15.Askar M, Sobecks R, Morishima YH, et al. Predictions in the Face of Clinical Reality: HistoCheck versus High-Risk HLA Allele Mismatch Combinations Responsible for Severe Acute Graft-versus-Host Disease. Biol Blood Marrow Transplant. 2011 doi: 10.1016/j.bbmt.2011.01.015. [DOI] [PubMed] [Google Scholar]
- 16.Glucksberg H, Storb R, Fefer A, et al. Clinical manifestations of graft-versus-host disease in human recipients of marrow from HL-A-matched sibling donors. Transplantation. 1974;18(4):295–304. doi: 10.1097/00007890-197410000-00001. [DOI] [PubMed] [Google Scholar]
- 17.Shulman HM, Sullivan KM, Weiden PL, et al. Chronic graft-versus-host syndrome in man. A long-term clinicopathologic study of 20 Seattle patients. Am J Med. 1980;69(2):204–217. doi: 10.1016/0002-9343(80)90380-0. [DOI] [PubMed] [Google Scholar]
- 18.Petersdorf EW, Kollman C, Hurley CK, et al. Effect of HLA class II gene disparity on clinical outcome in unrelated donor hematopoietic cell transplantation for chronic myeloid leukemia: the US National Marrow Donor Program Experience. Blood. 2001;98(10):2922–2929. doi: 10.1182/blood.v98.10.2922. [DOI] [PubMed] [Google Scholar]
- 19.Spellman S, Bray R, Rosen-Bronson S, et al. The detection of donor-directed, HLA-specific alloantibodies in recipients of unrelated hematopoietic cell transplantation is predictive of graft failure. Blood. 2010;115(13):2704–8. doi: 10.1182/blood-2009-09-244525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Baxter-Lowe LA, Maiers M, Spellman SR, et al. HLA-A disparities illustrate challenges for ranking the impact of HLA mismatches on bone marrow transplant outcomes in the United States. Biol Blood Marrow Transplant. 2009;15(8):971–981. doi: 10.1016/j.bbmt.2009.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]