Abstract
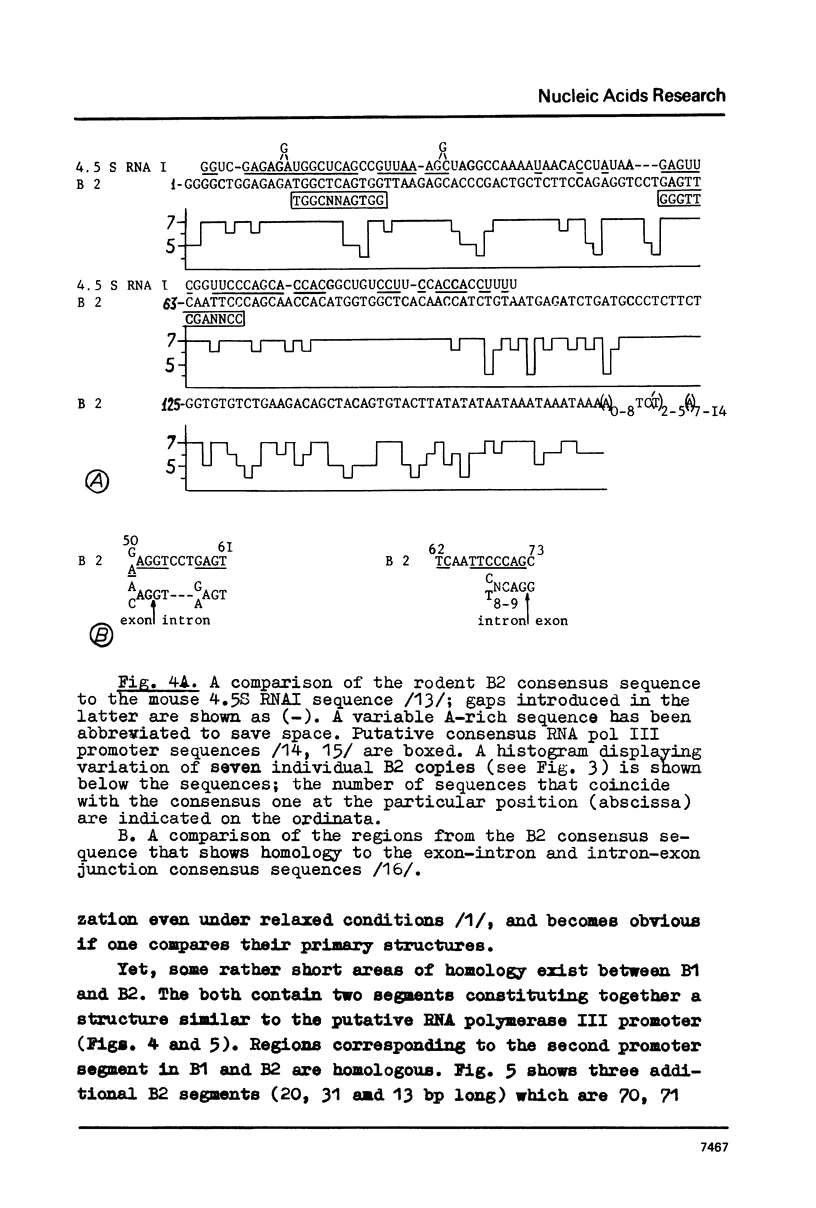
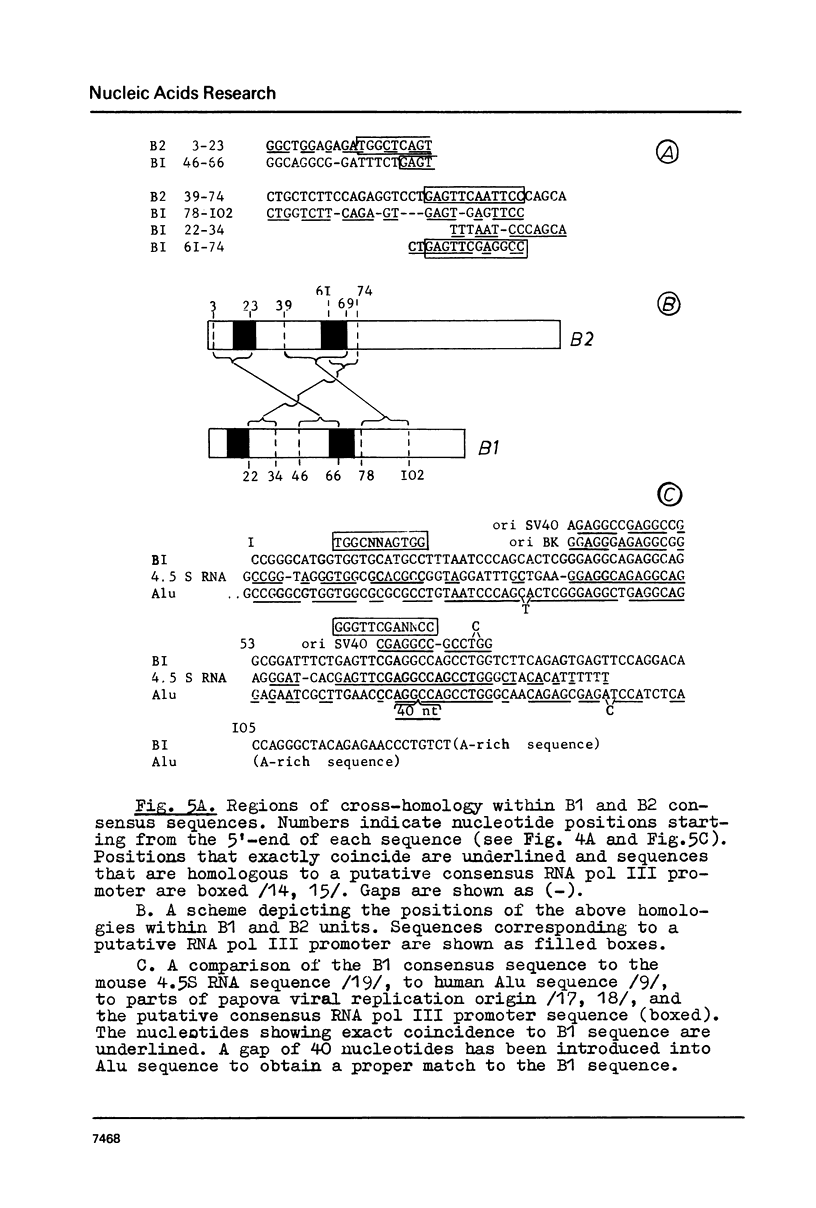
Mouse genome contains two major families of short interspersed repeats in more than 10(5) copies scattered throughout the whole genome. They are referred to as B1 and B2 sequences since they were first isolated from the genome library by means of a dsRNA-B probe /1/. In this work, two copies of the B2 family were sequenced and compared with the previously sequenced B1 repeat /2/. A B2 ubiquitous repeat is ca. 190 bp long. The members of the family deviate in 3-5% of nucleotides from the consensus sequence. B2 contains regions of homology to the RNA polymerase III split promoter and to 4.5S snRNA I. Both B1 and B2 contain regions which resemble junctions between exons and introns. In contrast to B1, B2 does not contain apparent homologies to papova viral replication origins and a human Alu sequence. One side of the B2 repeat is represented by a very AT-rich sequence (ca. 30 bp long) followed with an oligo (dA) stretch 10-15 nucleotides long. This region of the repeat is the most variable one. The whole unit is flanked with 15-16 bp direct repeats different in sequenced copies of B2. The same is true of some copies of the B1 family. The properties of B1 and B2 repeats suggest that they may represent a novel class of transposon-like elements in eukaryotic genome. A possible role of B-type repeats in genome reorganization, DNA replication and pre-mRNA processing is discussed.
Full text
PDF












Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Avvedimento V. E., Vogeli G., Yamada Y., Maizel J. V., Jr, Pastan I., de Crombrugghe B. Correlation between splicing sites within an intron and their sequence complementarity with U1 RNA. Cell. 1980 Oct;21(3):689–696. doi: 10.1016/0092-8674(80)90432-8. [DOI] [PubMed] [Google Scholar]
- Bell G. I., Pictet R., Rutter W. J. Analysis of the regions flanking the human insulin gene and sequence of an Alu family member. Nucleic Acids Res. 1980 Sep 25;8(18):4091–4109. doi: 10.1093/nar/8.18.4091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson E. H., Britten R. J. Regulation of gene expression: possible role of repetitive sequences. Science. 1979 Jun 8;204(4397):1052–1059. doi: 10.1126/science.451548. [DOI] [PubMed] [Google Scholar]
- Deininger P. L., Jolly D. J., Rubin C. M., Friedmann T., Schmid C. W. Base sequence studies of 300 nucleotide renatured repeated human DNA clones. J Mol Biol. 1981 Sep 5;151(1):17–33. doi: 10.1016/0022-2836(81)90219-9. [DOI] [PubMed] [Google Scholar]
- Duncan C. H., Jagadeeswaran P., Wang R. R., Weissman S. M. Structural analysis of templates and RNA polymerase III transcripts of Alu family sequences interspersed among the human beta-like globin genes. Gene. 1981 Mar;13(2):185–196. doi: 10.1016/0378-1119(81)90007-x. [DOI] [PubMed] [Google Scholar]
- Fowlkes D. M., Shenk T. Transcriptional control regions of the adenovirus VAI RNA gene. Cell. 1980 Nov;22(2 Pt 2):405–413. doi: 10.1016/0092-8674(80)90351-7. [DOI] [PubMed] [Google Scholar]
- Galli G., Hofstetter H., Birnstiel M. L. Two conserved sequence blocks within eukaryotic tRNA genes are major promoter elements. Nature. 1981 Dec 17;294(5842):626–631. doi: 10.1038/294626a0. [DOI] [PubMed] [Google Scholar]
- Georgiev G. P., Ilyin Y. V., Chmeliauskaite V. G., Ryskov A. P., Kramerov D. A., Skryabin K. G., Krayev A. S., Lukanidin E. M., Grigoryan M. S. Mobile dispersed genetic elements and other middle repetitive DNA sequences in the genomes of Drosophila and mouse: transcription and biological significance. Cold Spring Harb Symp Quant Biol. 1981;45(Pt 2):641–654. doi: 10.1101/sqb.1981.045.01.082. [DOI] [PubMed] [Google Scholar]
- Grimaldi G., Queen C., Singer M. F. Interspersed repeated sequences in the African green monkey genome that are homologous to the human Alu family. Nucleic Acids Res. 1981 Nov 11;9(21):5553–5568. doi: 10.1093/nar/9.21.5553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harada F., Kato N. Nucleotide sequences of 4.5S RNAs associated with poly(A)-containing RNAs of mouse and hamster cells. Nucleic Acids Res. 1980 Mar 25;8(6):1273–1285. doi: 10.1093/nar/8.6.1273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haynes S. R., Jelinek W. R. Low molecular weight RNAs transcribed in vitro by RNA polymerase III from Alu-type dispersed repeats in Chinese hamster DNA are also found in vivo. Proc Natl Acad Sci U S A. 1981 Oct;78(10):6130–6134. doi: 10.1073/pnas.78.10.6130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haynes S. R., Toomey T. P., Leinwand L., Jelinek W. R. The Chinese hamster Alu-equivalent sequence: a conserved highly repetitious, interspersed deoxyribonucleic acid sequence in mammals has a structure suggestive of a transposable element. Mol Cell Biol. 1981 Jul;1(7):573–583. doi: 10.1128/mcb.1.7.573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higuchi R., Stang H. D., Browne J. K., Martin M. O., Huot M., Lipeles J., Salser W. Human ribosomal RNA gene spacer sequences are found interspersed elsewhere in the genome. Gene. 1981 Nov;15(2-3):177–186. doi: 10.1016/0378-1119(81)90127-x. [DOI] [PubMed] [Google Scholar]
- Jagadeeswaran P., Forget B. G., Weissman S. M. Short interspersed repetitive DNA elements in eucaryotes: transposable DNA elements generated by reverse transcription of RNA pol III transcripts? Cell. 1981 Oct;26(2 Pt 2):141–142. doi: 10.1016/0092-8674(81)90296-8. [DOI] [PubMed] [Google Scholar]
- Kramerov D. A., Grigoryan A. A., Ryskov A. P., Georgiev G. P. Long double-stranded sequences (dsRNA-B) of nuclear pre-mRNA consist of a few highly abundant classes of sequences: evidence from DNA cloning experiments. Nucleic Acids Res. 1979 Feb;6(2):697–713. doi: 10.1093/nar/6.2.697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramerov D. A., Lomidze N. V. "Paradoksal'naia" évoliutsiia individual'noi trnaskribiruemoi povtoriaiushcheisia posledovatel'nosti DNK (Bl) u gryzunov. Dokl Akad Nauk SSSR. 1980;255(4):1005–1009. [PubMed] [Google Scholar]
- Kramerov D. A., Ryskov A. P., Georgiev G. P. The structural organization of nuclear pre-mRNA. II. Very long double-stranded structures in nuclear pre-mRNA. Biochim Biophys Acta. 1977 Apr 4;475(3):461–475. doi: 10.1016/0005-2787(77)90062-4. [DOI] [PubMed] [Google Scholar]
- Krayev A. S., Kramerov D. A., Skryabin K. G., Ryskov A. P., Bayev A. A., Georgiev G. P. The nucleotide sequence of the ubiquitous repetitive DNA sequence B1 complementary to the most abundant class of mouse fold-back RNA. Nucleic Acids Res. 1980 Mar 25;8(6):1201–1215. doi: 10.1093/nar/8.6.1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lerner M. R., Steitz J. A. Snurps and scyrps. Cell. 1981 Aug;25(2):298–300. doi: 10.1016/0092-8674(81)90047-7. [DOI] [PubMed] [Google Scholar]
- Mount S. M. A catalogue of splice junction sequences. Nucleic Acids Res. 1982 Jan 22;10(2):459–472. doi: 10.1093/nar/10.2.459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohshima Y., Okada N., Tani T., Itoh Y., Itoh M. Nucleotide sequences of mouse genomic loci including a gene or pseudogene for U6 (4.8S) nuclear RNA. Nucleic Acids Res. 1981 Oct 10;9(19):5145–5158. doi: 10.1093/nar/9.19.5145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Page G. S., Smith S., Goodman H. M. DNA sequence of the rat growth hormone gene: location of the 5' terminus of the growth hormone mRNA and identification of an internal transposon-like element. Nucleic Acids Res. 1981 May 11;9(9):2087–2104. doi: 10.1093/nar/9.9.2087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan J., Elder J. T., Duncan C. H., Weissman S. M. Structural analysis of interspersed repetitive polymerase III transcription units in human DNA. Nucleic Acids Res. 1981 Mar 11;9(5):1151–1170. [PMC free article] [PubMed] [Google Scholar]
- Posakony J. W., Scheller R. H., Anderson D. M., Britten R. J., Davidson E. H. Repetitive sequences of the sea urchin genome. III. Nucleotide sequences of cloned repeat elements. J Mol Biol. 1981 Jun 15;149(1):41–67. doi: 10.1016/0022-2836(81)90259-x. [DOI] [PubMed] [Google Scholar]
- Reddy V. B., Thimmappaya B., Dhar R., Subramanian K. N., Zain B. S., Pan J., Ghosh P. K., Celma M. L., Weissman S. M. The genome of simian virus 40. Science. 1978 May 5;200(4341):494–502. doi: 10.1126/science.205947. [DOI] [PubMed] [Google Scholar]
- Ryskov A. P., Ivanov P. L., Kramerov D. A., Georgiev G. P. Povtoriaiushchiesia posledovatel'nosti tipa B1 i B2 v klonakh komplementarnoi DNK, poluchennoi na tsitoplazmaticheskoi poli(A)+ RNK pecheni myshi. Dokl Akad Nauk SSSR. 1982;263(4):997–1000. [PubMed] [Google Scholar]
- Ryskov A. P., Saunders G. F., Farashyan V. R., Georgiev G. P. Double-helical regions in nuclear precursor of mRNA (pre-mRNA). Biochim Biophys Acta. 1973 Jun 8;312(1):152–164. doi: 10.1016/0005-2787(73)90060-9. [DOI] [PubMed] [Google Scholar]
- Stumph W. E., Kristo P., Tsai M. J., O'Malley B. W. A chicken middle-repetitive DNA sequence which shares homology with mammalian ubiquitous repeats. Nucleic Acids Res. 1981 Oct 24;9(20):5383–5397. doi: 10.1093/nar/9.20.5383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Temin H. M. Structure, variation and synthesis of retrovirus long terminal repeat. Cell. 1981 Nov;27(1 Pt 2):1–3. doi: 10.1016/0092-8674(81)90353-6. [DOI] [PubMed] [Google Scholar]
- Yang R. C., Wu R. BK virus DNA: complete nucleotide sequence of a human tumor virus. Science. 1979 Oct 26;206(4417):456–462. doi: 10.1126/science.228391. [DOI] [PubMed] [Google Scholar]


