Abstract
A subgroup of pediatric acute T-lymphoblastic leukemia (T-ALL) was characterized by a gene expression profile comparable to that of early T-cell precursors (ETPs) with a highly unfavorable outcome. We have investigated clinical and molecular characteristics of the ETP-ALL subgroup in adult T-ALL. As ETP-ALL represents a subgroup of early T-ALL we particularly focused on this cohort and identified 178 adult patients enrolled in the German Acute Lymphoblastic Leukemia Multicenter studies (05/93–07/03). Of these, 32% (57/178) were classified as ETP-ALL based on their characteristic immunophenotype. The outcome of adults with ETP-ALL was poor with an overall survival of only 35% at 10 years, comparable to the inferior outcome of early T-ALL with 38%. The molecular characterization of adult ETP-ALL revealed distinct alterations with overexpression of stem cell-related genes (BAALC, IGFBP7, MN1, WT1). Interestingly, we found a low rate of NOTCH1 mutations and no FBXW7 mutations in adult ETP-ALL. In contrast, FLT3 mutations, rare in the overall cohort of T-ALL, were very frequent and nearly exclusively found in ETP-ALL characterized by a specific immunophenotype. These molecular characteristics provide biologic insights and implications with respect to innovative treatment strategies (for example, tyrosine kinase inhibitors) for this high-risk subgroup of adult ETP-ALL.
Keywords: acute lymphoblastic leukemia, ETP-ALL, T-ALL, molecular characteristics, FLT3, NOTCH1
Introduction
Early T-cell precursors (ETPs) are intrathymic c-Kithi double-negative (DN)1 cells, which contribute to the development of T-lymphocytes.1, 2 These cells represent immature progenitors that have recently immigrated from the bone marrow to the thymus. In addition to their T-lymphoid potential, they carry a natural killer, dendritic, and remarkably, myeloid cell differentiation potential.3
Recently, a small subgroup of pediatric acute T-lymphoblastic leukemia (T-ALL) was described showing a gene expression profile closely linked to the expression signature of ETPs.4, 5 This subtype, termed ETP-ALL, is characterized by a specific immunophenotype: CD1a−, CD8−, CD5weak and expression of stem cell or myeloid markers. Moreover, ETP-ALLs are distinguished by high expression of oncogenic transcription factors, including genes involved in the pathogenesis of T-ALL like LMO1, LYL1 and ERG. Pediatric patients with ETP-ALL showed a highly unfavorable outcome compared with typical T-ALL.4 On the basis of these results, the Italian national study Associazione Italiana Ematologia Oncologia Pediatrica (AIEOP) and St-Jude Children's hospital proposed modifications in their treatment schedules for children with ETP-ALL towards a more intensive regime including allogeneic stem cell transplantation (alloSCT).4 Up to date, the existence of a subtype with an ETP gene expression signature in adult patients has not yet been investigated.
Although several molecular risk factors with prognostic and potential therapeutic implications have been identified, such as mutations (NOTCH1) or aberrant expression (BAALC, ERG, TLX1, TLX3),6 there is still a need for more precise molecular risk stratifications to guide adapted treatment strategies. Although some study groups utilize an initial leukocyte count above 100 000/nl as an adverse prognostic factor, the German Multicenter Study Group for Adult ALL (GMALL) applies risk stratification for T-ALL based on the immunophenotype.7 On the basis of the results of earlier study protocols, early (sCD3−, CD1a−) T-ALL is regarded as a high-risk subgroup and alloSCT is recommended in first complete remission (CR).7 Therefore, patients with ETP-ALL, being exclusively found in the subgroup of early T-ALL, are already assigned towards alloSCT within the GMALL strategy. However, the multilineage ability of normal ETPs and the stem cell-like features of oncogenetically transformed ETP-ALL may translate in primary resistance to conventional chemotherapy. Thus, a standard protocol for lymphoblastic diseases might be insufficient and new molecular targets are warranted for specific targeted therapy in order to improve overall survival (OS) in this high-risk disease.
Although the implementation of specific targeted therapies have already been applied to high-risk B-ALL characterized by BCR-ABL and CRLF2/JAK activation, alterations of the tyrosine kinase pathway have only been identified in rare cases of T-ALL presenting with rearrangements NUP214-ABL1 and STRN3-JAK2.8
In this study, we have investigated the existence of ETP-ALL among adult patients and, in particular, assessed their clinical as well as molecular characteristics. We further explored the ETP-ALL as a molecular distinct subgroup of T-ALL and identified a high frequency of FLT3 mutations. Thus these findings may in future direct to innovative treatment strategies for this distinct T-ALL subgroup.
Patients and methods
Patients and treatment
We analyzed 178 patients classified as early T-ALL in the GMALL study trials between 1993 and 2008. The GMALL protocols include a combination of chemotherapy, radiation, and with the protocol 06/99, alloSCT was implemented for high-risk T-ALL-patients. Details of the protocols were previously described.9 All patients gave written informed consent to participate in the study according to the Declaration of Helsinki.10 This study was approved by the ethics board of the Johann Wolfgang Goethe-Universität Frankfurt am Main, Germany. In the GMALL study, immunophenotyping was centrally performed in the GMALL reference laboratory at the Charité University Hospital, Berlin. Immunophenotyping and subtype assignment was carried out as previously described.11, 12, 13 High-risk T-ALL was defined by an immunophenotype of an early (sCD3−, CD1−) or mature (sCD3+, CD1−) T-ALL. ETP-ALL was defined as a subgroup within early T-ALL with the following immunophenotype: CD1a−, CD8−, CD5weak with expression of stem cell (CD34, HLA-DR, CD117) and/or coexpression of myeloid antigens (CD13, CD33, CD65s). Absence, positivity and weak expression of antigens were defined according to the definitions in pediatric patients.4
Nucleic acid preparation and molecular characterization
For patients with sufficient material available, pretreatment bone marrow samples were used for DNA and total RNA extraction using TRIzol (Life Technologies, Grand Island, NY, USA) according to the manufacturer's protocol with minor modifications. Complementary DNA was synthesized using 500 ng of total RNA and avian myeloblastosis virus reverse transcriptase (RT-AMV; Roche, Mannheim, Germany) in the presence of RNase inhibitor (RNasin; Roche).
Molecular diagnostic examinations were available from in total 297 T-ALL patients (including all immunophenotypical subgroups) from the two GMALL studies 06/99 and 07/03. As far as material was available, samples were investigated by comparative real-time PCR (RT-PCR) for expression of five genes (BAALC, ERG, IGFBP7, WT1 and MN1). mRNA expression levels for WT1,14 BAALC13 and ERG13 were determined by RT-PCR as previously described. The primers for IGFBP7 are available on request. MN1-primers were designed as reported.15 GUS was used as a housekeeping gene with the exception of ABL in the RT-PCR for MN1. In 142 patient samples, the NOTCH1 mutation status was identified by sequencing of PCR-amplified products.16, 17 For the FBWX7 mutation status, exons 8 and 9 were sequenced in 121 patients samples as previously described.18 WT1 mutations analyses was performed as recently reported.14 FLT3 mutations (internal tandem duplications (ITD)/tyrosine kinase domain (TKD)) were analyzed using a commercially available FLT3 mutation assay (InVivoScribe Technologies, San Diego, CA, USA) in 123 T-ALL patients. Because of lack of sufficient material, not all samples could be analyzed. No significant differences regarding immunophenotype, age or sex were found in the different subsets of patients analyzed in the different experiments.
Statistical analyses
Differences in the clinical characteristics as well as response to induction therapy were tested by the Pearson χ2-test. For OS and duration of remission in the different subgroups, Kaplan–Meier curves were created and compared by the log-rank test. OS was calculated from the time-point of study entry to the time-point of death or latest follow-up (censored). Remission duration was calculated in CR patients from the time-point of first CR until relapse or last follow-up. Stem cell transplantation in first CR, death in CR, withdrawal and continuous remission were censored at the respective dates.
The statistical difference of gene expression between two independent groups was tested by the nonparametric Mann–Whitney U-test. Differences in the mutation rate were analyzed by the Pearson χ2-test. For all tests, a P-value <0.05 (two-sided) was considered to indicate a significant difference. All calculations were performed using the SPSS software version 17 (SPSS Inc., Chicago, IL, USA) and GraphPad Prism software version 5 (GraphPad Software Inc., La Jolla, CA, USA).
Results
Patients' characteristics and clinical outcome
We first restricted our analysis to 178 patients diagnosed with an immunophenotype of early T-ALL who were enrolled in the GMALL trials (05/93, 06/99, 07/03) between 1993 and 2008. This cohort of early T-ALL patients represents 23% of all patients with T-ALL enrolled in the mentioned studies.7 All samples were clearly positive for cytoplasmic CD3. Applying the immunophenotypic ETP-ALL profile (CD1a−, CD8−, CD5weak with expression of stem cell or myeloid markers), 57 patients (32% of early T-ALL) were classified as ETP-ALL, corresponding to 7.4% of all adult T-ALL patients. Notably, none of the early T-ALL cases fulfilled the EGIL criteria for biphenotypic leukemia.
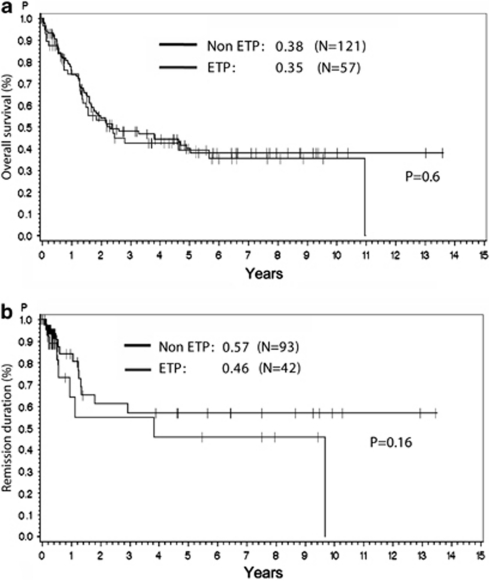
The clinical characteristics were similar between the group of ETP-ALL and non-ETP early T-ALL regarding sex, age, white blood cell count, and central nervous system involvement (Table 1). However, a lower frequency of patients with a mediastinal mass at diagnosis was found among patients with ETP-ALL compared with those with non-ETP early T-ALL (28% vs 47% P=0.02). Patients of both groups were treated equally in the three GMALL trials and the rate of patients receiving an alloSCT in first CR did not differ between ETP-ALL and non-ETP early T-ALL (59% vs 60%, respectively). Slightly fewer patients with ETP-ALL achieved a CR after induction therapy (42 of 53 patients; 79%) compared with patients with non-ETP early T-ALL (93 of 113 patients; 82% n.s.; Table 1). A total of 14 patients died during the induction therapy, 6 in the group of ETP-ALL, 8 in the group of non-ETP early T-ALL. Regarding all high-risk early T-ALL patients in the three study trials, the probability of survival at 10 years was comparably poor in patients with ETP-ALL and non-ETP early T-ALL (35% vs 38%, Figure 1a). In patients with early T-ALL, more patients remained in CR in the group of non-ETP early T-ALL than in the group of ETP-ALL (57% vs 46%, n.s., Figure 1b) with a follow-up of 9 years.
Table 1. Characteristics of patients enrolled in the three GMALL study trials 05/93, 06/99 and 07/03 with the diagnosis of early T-ALL.
| ETP-ALL (n=57) (%) | Non-ETP early T-ALL (n=121) (%) | P | |
|---|---|---|---|
| Sex | |||
| Male | 47 (82.5) | 84 (69.4) | 0.07 |
| Female | 10 (17.5) | 37 (30.6) | |
| Age | |||
| 15–35 | 27 (47.4) | 68 (56.2) | 0.27 |
| 36–65 | 30 (52.6) | 53 (43.8) | |
| WBC count (n=163) | |||
| <30/nl | 32 (62.7) | 71 (63.4) | 0.94 |
| >30/nl | 19 (37.3) | 41 (36.6) | |
| Mediastinal mass (n=162) | |||
| No | 37 (72.5) | 59 (53.2) | 0.02 |
| Yes | 14 (27.5) | 52 (46.8) | |
| CNS involvement (n=150) | |||
| No | 42 (91.3) | 100 (96.2) | 0.22 |
| Yes | 4 (8.7) | 4 (3.8) | |
| Response to induction | |||
| CR | 42 (79.2) | 93 (82.3) | 0.65 |
| PR/failure | 5 (9.4) | 12 (11.7) | |
| Death in induction | 6 (11.3) | 8 (7.8) | |
Abbreviations: CNS, central nervous system; CR, complete remission; ETP-ALL, early T-cell precursors-acute lymphoblastic leukemia; PR, partial remission; T-ALL, acute T-lymphoblastic leukemia; WBC, white blood cell.
Comparison between ETP-ALL and early T-ALL with a non-ETP immunophenotype.
Figure 1.
Kaplan–Meier analyses for OS (a) and remission duration (b) for patients with ETP-ALL and early T-ALL with a non-ETP immunophenotype. Patients were enrolled in the GMALL study trials GMALL 05/93, GMALL 06/99 and GMALL 07/03.
Aberrant gene expression in ETP-ALL
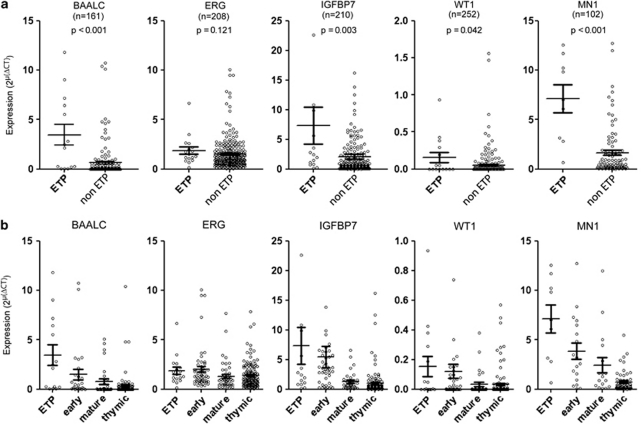
To further characterize the ETP-ALL on the molecular level, we analyzed the expression of selected genes known to be involved in the pathogenesis and with prognostic implications in T-ALL in samples with sufficient material. We first compared ETP-ALL, as a new subgroup, to all remaining T-ALL subtypes including thymic, mature and early T-ALL. As previously described, high expression of BAALC and ERG predicted an unfavorable outcome in adults with T-ALL.10, 13 Quantitative RT-PCR assays revealed that BAALC was 5.34-fold highly expressed in ETP-ALL compared with all remaining T-ALL (P<0.001, Figure 2a). ERG only showed a slight, however not significantly elevated expression (1.33-fold, P=0.12, Figure 2a). IGFBP7 is a stem cell-associated gene, which is highly associated with BAALC and overexpressed in early T-ALL19 and among the genes highly overexpressed in pediatric ETP-ALL.4 Similar to BAALC, it was significantly upregulated in ETP-ALL (3.53-fold, P=0.003, Figure 2a). The gene WT1 was of interest as it is widely overexpressed in AML20 and as its overexpression is associated with a poor outcome in thymic T-ALL patients.14 Interestingly, we found a significantly elevated WT1 expression in ETP-ALL (4.33-fold, P=0.04, Figure 2a). Because of the myeloid potential of ETPs and the frequent coexpression of myeloid surface antigens in ETP-ALL lymphoblasts, we further investigated the expression of MN1 as its overexpression was found to be associated with a shorter survival in AML without karyotypic abnormalities.15, 21 Here, we show that MN1 was significantly overexpressed in the group of ETP-ALL compared with all remaining T-ALL patients (2.86-fold, P<0.001; Figure 2a).
Figure 2.
Expression of genes known to be correlated with outcome in T-ALL and AML. Analysis of samples with sufficient RNA was carried out by RT-PCR. For each entity the median expression is shown by a horizontal bar with the s.e. of the mean. (a) Comparison between ETP-ALL and the remaining non-ETP T-ALL. (b) Expression in the different immunophenotype-defined subgroups of T-ALL. Early T-ALL refers to non-ETP early T-ALL.
Although these five genes were also upregulated in early T-ALL compared with mature or thymic T-ALL, WT1, BAALC, IGFBP7 and MN1 showed an even higher expression in the ETP-ALL group compared with the remaining group of non-ETP early T-ALL (Figure 2b).
Mutational analyses in ETP-ALL
Differences in the mutation status of candidate genes between ETP-ALL and non-ETP-ALL cases were explored (Table 2). The most frequent pathogenetic mutational event in T-ALL are NOTCH1 mutations occurring in approximately 50–70% of the cases, predominantly in thymic T-ALL22, 23, 24 Although NOTCH1 mutations have been associated with an initial good response in some studies, the prognostic impact of NOTCH1 mutations in T-ALL remains controversial.8, 25, 26, 27, 28, 29 In 142 adult T-ALL samples analyzed, we have found a low rate of NOTCH1 mutations in the immature subgroup of ETP-ALL (n=1/14, 7.1%), whereas NOTCH1 mutations were frequent (60.9%) in non-ETP T-ALL (n=78/128, P=0.001, Table 2). Similar findings were observed for the tumor-suppressor gene FBXW7 involved in NOTCH1 signaling: no FBXW7 mutations were found in the 14 ETP-ALL samples analyzed.
Table 2. Gene mutation status compared between (a) ETP-ALL and non-ETP T-ALL and (b) ETP-ALL and non-ETP early T-ALL.
| ETP-ALL (%) | Non-ETP T-ALL (%) | P | ETP-ALL (%) | Non-ETP early T-ALL (%) | P | ||
|---|---|---|---|---|---|---|---|
| (a) | (b) | ||||||
| FBXW7 (n=128) | FBXW7 (n=50) | ||||||
| wt | 14 (100) | 98 (86) | 0.13 | wt | 14 (100) | 27 (100) | — |
| mut | 0 (0) | 16 (14) | mut | 0 (0) | 0 (0) | ||
| NOTCH1 (n=142) | NOTCH1 (n=50) | ||||||
| wt | 13 (92.9) | 50 (39.1) | <0.001 | wt | 13 (92.9) | 16 (47.1) | 0.003 |
| mut | 1 (7.1) | 78 (60.9) | mut | 1 (7.1) | 18 (52.9) | ||
| FLT3 (ITD) (n=123) | FLT3 (ITD) (n=40) | ||||||
| wt | 14 (87.5) | 107 (100) | <0.001 | wt | 14 (75) | 24 (100) | 0.08 |
| mut | 2 (12.5) | 0 (0) | mut | 2 (25) | 0 (0) | ||
| FLT3 (D835) (n=116) | FLT3 (D835) (n=40) | ||||||
| wt | 12 (75) | 99 (99) | <0.001 | wt | 12 (75) | 23 (95.8) | 0.05 |
| mut | 4 (25) | 1 (1) | mut | 4 (25) | 1 (4.2) | ||
| WT1 (n=227) | WT1 (n=63) | ||||||
| wt | 12 (80) | 196 (92.5) | 0.09 | wt | 12 (80) | 42 (87.5) | 0.47 |
| mut | 3 (20) | 16 (7.5) | mut | 3 (20) | 6 (12.5) | ||
Abbreviations: ETP-ALL, early T-cell precursors-acute lymphoblastic leukemia; ITD, internal tandem duplications; mut, mutant; T-ALL, acute T-lymphoblastic leukemia; wt, wild type.
Non-ETP T-ALL includes all thymic, mature and early T-ALL with a non-ETP immunophenotype. Analysis was performed as far as sufficient material was available.
Mutations in the WT1 gene were reported in about 8% of all T-ALL patients.14 We did not observe a significant difference in the frequency of WT1 mutations between ETP-ALL and the remaining T-ALL (Table 2).
Although FLT3 mutations are frequent in AML (∼30%) and have important prognostic and therapeutic implications, mutations for FLT3 as ITD or in the TKD in T-ALL are very rare.30, 31, 32 Interestingly, in our cohort we identified seven patients with mutations of FLT3 that were exclusive in the cohort of early T-ALL patients and predominantly found in the subgroup of ETP-ALL patients. Five cases (4.5%) had mutations in the tyrosine kinase domain of FLT3 (D835), four of them were in the group of ETP-ALL (P<0.001). Another case with an FLT3 mutation was assigned to early T-ALL, but the immunophenotype did not fulfil the criteria of an ETP-ALL due to a weak CD8 expression. This case also showed surface expression of the myeloid antigens CD13 and the stem cell markers CD34 and CD117. ITD mutations of FLT3 were only found in two of the 123 cases, both being in the group of ETP-ALL (P<0.001). In total, six of seven FLT3-mutations found were in the group of ETP-ALL, displaying a frequency of 37.5% within this subgroup. Interestingly, FLT3-mutated ETP-ALL patients showed different clinical and molecular characteristics compared with FLT3 wild-type ETP-ALL patients. The seven patients with an FLT3 mutation had a median age of 40 years and an initial white blood cell of 3800/μl. Five FLT3-mutated ETP-ALL patients were transplanted in first remission and the median OS was 31.3 months with six patients being still alive. Remarkably, patients with a FLT3 mutation showed a distinct immunophenotype with positivity for CD117 (7/7 patients), CD34 (6/7 patients), CD13 (7/7 patients), and CD2 (7/7 patients). In contrast, ETP-ALL patients with a FLT3 wild-type status had more often positivity for CD5 (7/10 patients) and CD33 (6/10 patients).
Discussion
In T-ALL, outcome has been slightly improved in the past decades, mainly because of the implementation of alloSCT for specific subgroups.7, 33 However, long-term OS only reaches rates of 30–60% depending on prognostic factors and subgroups.34 Further improvement remains warranted and may be achieved by enhanced risk stratification and development of novel treatment approaches based on the implementation of targeted therapies.
We characterized a new subgroup of adult patients with T-ALL (7.4%) with an immunophenotype of pediatric ETP-ALL.4 The frequency of ETP-ALL in adult T-ALL was slightly lower than first reported for pediatric ETP-ALL, but in the range of a recent report from the Children's Oncology Group (COG).35 Similar to the pediatric study, there were no differences in the clinical characteristics between the groups of ETP-ALL and early T-ALL with the exception of a lower frequency of a mediastinal mass in ETP-ALL.
Although many features are in common, there are relevant differences between adult and pediatric T-ALL, especially with respect to treatment. There are no immunophenotype-defined subgroups that are currently used for risk stratification in larger pediatric T-ALL trials.36, 37 In contrast, early T-ALL has been recognized as an immunophenotype-defined subtype with poor prognosis in adult T-ALL33, and since 1999, the GMALL study group in accordance with other study groups38 defines early T-ALL as high-risk T-ALL39, 40 with consecutive recommendation for alloSCT. This led to an improvement in the OS at 5 years of 40%.7 Therefore, the ETP-ALL as a subgroup of early T-ALL is already regarded as a T-ALL subgroup with poor prognosis requiring intensified therapy. In agreement with pediatric ETP-ALL, adult ETP-ALL showed an unfavorable outcome in our analysis similar to early T-ALL.7 Interestingly, a high overlap to groups with an unfavorable prognosis, characterized by negativity of CD1 and positivity for CD33, and the group of early T-ALL is evident.33 These T-ALLs overrepresented in early T-ALL are specifically contained in the subgroup of ETP-ALL.
To unravel the underlying molecular alterations of ETP-ALL as a specific T-ALL subgroup, we studied the expression of candidate genes reported to be of prognostic impact. Out of these, the expression of stem cell-associated genes (BAALC and IGFBP7) and genes known to be of prognostic significance in AML (BAALC, MN1, WT1) are underlining the immature nature of ETP-ALL. BAALC is preferentially expressed in the immature T-ALL phenotypes compared with the remaining T-ALL, and its high expression has been correlated to a subgroup with poor outcome.13 Moreover, T-ALL patients with high BAALC expression frequently showed an aberrant expression of myeloid markers. High BAALC expression is also linked to an immature phenotype in AML with normal cytogenetics,41 to a more aggressive BCR-ABL-positive acute lymphoblastic leukemia42 and is associated with an unfavorable outcome in B-precursor ALL.43 This led to the hypothesis that BAALC is a marker for acute leukemia derived from early progenitors with multi-lineage potential. Recently, IGFBP7 was identified as a lineage-independent BAALC co-expressed gene with prognostic impact and a potential link to BAALC.44 BAALC and IGFBP7 were also found as highly expressed genes within the ETP-ALL gene expression signature.4 In addition, overexpression of MN1 was recently identified to be associated with ETP-ALL, and might also provide an indirect support for the poor prognosis of ETP-ALL.45 These findings are underlined in this work as BAALC, IGFBP7 and MN1 were upregulated in ETP-ALL, probably originating from T-cell progenitors retaining myeloid differentiation potential. This multilineage potential is further strengthened by the overexpression of the molecular marker WT1 in ETP-ALL, a gene known to be of unfavorable prognosis in AML as well as in a subgroup of T-ALL46, 47, 48, 49, 50 Interestingly, a recent work identified a subgroup in adult T-ALL (∼10%) with myeloid characteristics using gene expression profiling with limited clinical data and no mutations or mutational events.51 Up to which degree this group overlaps with ETP-ALL has to be determined in further studies.
In addition to these particular gene expression patterns, we found a clear difference in mutation events of genes known to be involved in the pathogenesis of T-ALL or AML. The high rate of NOTCH1 mutations in T-ALL with a frequency of about 50%26, 27, 52, 53 makes the NOTCH1 signaling pathway an interesting candidate for targeted therapies by the implementation of ã-secretase inhibitors.54, 55 Whether NOTCH1 mutations could initiate leukemia alone56, 57, 58 or are mainly secondary effects in human T-ALL59 is discussed controversially.60, 61 Interestingly, only one NOTCH1 mutation was found in the small group of analyzed ETP-ALL, thus demonstrating a clear pathogenetic difference to non-ETP T-ALL (61% NOTCH1 mutations) as ETP-ALL is yet the only subgroup that lacks NOTCH1 mutations. Additionally, in the group of ETP-ALL, mutations of the less frequent mutated tumor suppressor gene FBXW7 were not found, again supporting a different pathogenesis for the group of ETP-ALL, likely independent from the activated NOTCH1 pathway. Therefore, targeted therapies implementing γ-secretase inhibitors would presumably be less effective in ETP-ALL lacking mutational activated NOTCH1 signaling.
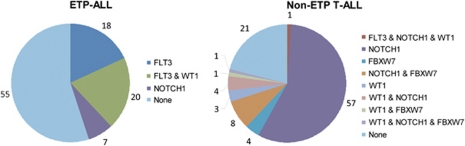
Most remarkably, we found a high rate of FLT3 mutations in ETP-ALL with a not yet reported high incidence of 37.5%, albeit in a small cohort (16 patients with ETP-ALL). Thus, with respect to the FLT3, NOTCH1/FBXW7 and WT1 mutation status, ETP-ALL shows a clearly different mutational profile compared with non-ETP T-ALL (Figure 3) indicating a distinct biological entity.
Figure 3.
Distribution of mutations (FBXW7, NOTCH1, FLT3, WT1) in percent in ETP-ALL (n=16 on the left) and non-ETP T-ALL (n=212 on the right). Not all T-ALL samples tested for WT1 were also tested for NOTCH1 and FLT3 mutations in the cohort of non-ETP T-ALL.
With respect to FLT3 mutations, we found in contrast to AML more frequently TKD than ITD mutations. This finding is in line with data from a murine bone marrow transplantation model, where mice that received a transplant of FLT3-ITD-transduced bone marrow cells developed myeloproliferative diseases, whereas a transplant of FLT3-TKD-transduced bone marrow cells induced lymphoid disorders.62 More recently, it was shown that a subset of common myeloid progenitors (FLT3+CD150−) has the potential to develop into T cells.63 Although FLT3 mutations in T-ALL are generally very rare (1–3%),30 FLT3 mutations were proposed to be frequently found in CD117+ T-ALL patients,32, 64 an antigen frequently expressed on ETP-ALL lymphoblasts. Although FLT3 mutation screening is clinically not indicated in unselected newly diagnosed T-ALL, ETP-ALL as a distinct subgroup defined by immunocytology should now be prospectively tested for FLT3 mutations. Moreover, these findings result in enlarged treatment options for this group with poor outcome65 including tyrosine kinase inhibitors (TKI), which are currently investigated in trials for the treatment in AML. Importantly, similar to myeloid cells, T-ALL cell lines transfected with an FLT3 ITD expression construct showed particular sensitivity to tyrosine kinase inhibition further justifying the use of TKI in FLT3-mutated ETP-ALL (data not shown). As ETP-ALL in pediatric patients is associated with high minimal residual disease levels after induction therapy,32 the use of TKI may be efficient to lower the leukemic blast burden before alloSCT in minimal residual disease-positive patients. Furthermore, TKI could be implemented in the treatment of a relapse after alloSCT.
We identified the specific subgroup of ETP-ALL based on the definition of a certain immunophenotype in adult T-ALL. Overall, the outcome of ETP-ALL is comparably poor to early T-ALL. Therefore, intensified treatment protocols including alloSCT, as already implemented in the therapy of adult T-ALL, seems warranted for these high-risk patients. Moreover, as ETP-ALL has distinct molecular features with a high rate of FLT3 mutations, these data point to new targeted therapy options including TKI for these high-risk T-ALL patients.
Acknowledgments
This work was supported by grants from the Deutsche Krebshilfe (Max Eder Nachwuchsförderung) to CD Baldus, from Deutsche Krebshilfe 70–2657-Ho2 to Dieter Hoelzer and partly BMBF 01GI 9971 to Dieter Hoelzer and Nicola Gökbuget.
The authors declare no conflict of interest.
References
- Rothenberg EV, Moore JE, Yui MA. Launching the T-cell-lineage developmental programme. Nat Rev Immunol. 2008;8:9–21. doi: 10.1038/nri2232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tremblay CS, Hoang T, Hoang T. Early T cell differentiation lessons from T-cell acute lymphoblastic leukemia. Prog Mol Biol Transl Sci. 2010;92:121–156. doi: 10.1016/S1877-1173(10)92006-1. [DOI] [PubMed] [Google Scholar]
- Bell JJ, Bhandoola A. The earliest thymic progenitors for T cells possess myeloid lineage potential. Nature. 2008;452:764–767. doi: 10.1038/nature06840. [DOI] [PubMed] [Google Scholar]
- Coustan-Smith E, Mullighan CG, Onciu M, Behm FG, Raimondi SC, Pei D, et al. Early T-cell precursor leukaemia: a subtype of very high-risk acute lymphoblastic leukaemia. Lancet Oncol. 2009;10:147–156. doi: 10.1016/S1470-2045(08)70314-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meijerink JP. Genetic rearrangements in relation to immunophenotype and outcome in T-cell acute lymphoblastic leukaemia. Best Pract Res Clin Haematol. 2010;23:307–318. doi: 10.1016/j.beha.2010.08.002. [DOI] [PubMed] [Google Scholar]
- Chiaretti S, Foa R. T-cell acute lymphoblastic leukemia. Haematologica. 2009;94:160–162. doi: 10.3324/haematol.2008.004150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoelzer D, Thiel E, Arnold R, Beck J, Beelen D, Bornhäuser M, et al. Sucessful subtype oriented treatment strategies in adult T-ALL: results of 744 patients treated in three consecutive GMALL studies. Blood. 2009;144:324. [Google Scholar]
- Mullighan CG. New strategies in acute lymphoblastic leukemia: translating advances in genomics into clinical practice. Clin Cancer Res. 2011;17:396–400. doi: 10.1158/1078-0432.CCR-10-1203. [DOI] [PubMed] [Google Scholar]
- Bruggemann M, Raff T, Flohr T, Gökbuget N, Nakao M, Droese J, et al. Clinical significance of minimal residual disease quantification in adult patients with standard-risk acute lymphoblastic leukemia. Blood. 2006;107:1116–1123. doi: 10.1182/blood-2005-07-2708. [DOI] [PubMed] [Google Scholar]
- Baldus CD, Burmeister T, Martus P, Schwartz S, Gökbuget N, Bloomfield CD, et al. High expression of the ETS transcription factor ERG predicts adverse outcome in acute T-lymphoblastic leukemia in adults. J Clin Oncol. 2006;24:4714–4720. doi: 10.1200/JCO.2006.06.1580. [DOI] [PubMed] [Google Scholar]
- Ludwig WD, Rieder H, Bartram CR, Heinze B, Schwartz S, Gassmann W, et al. Immunophenotypic and genotypic features, clinical characteristics, and treatment outcome of adult pro-B acute lymphoblastic leukemia: results of the German multicenter trials GMALL 03/87 and 04/89. Blood. 1998;92:1898–1909. [PubMed] [Google Scholar]
- Schwartz S, Rieder H, Schlager B, Burmeister T, Fischer L, Thiel E. Expression of the human homologue of rat NG2 in adult acute lymphoblastic leukemia: close association with MLL rearrangement and a CD10(-)/CD24(-)/CD65s(+)/CD15(+) B-cell phenotype. Leukemia. 2003;17:1589–1595. doi: 10.1038/sj.leu.2402989. [DOI] [PubMed] [Google Scholar]
- Baldus CD, Martus P, Burmeister T, Schwartz S, Gökbuget N, Bloomfield CD, et al. Low ERG and BAALC expression identifies a new subgroup of adult acute T-lymphoblastic leukemia with a highly favorable outcome. J Clin Oncol. 2007;25:3739–3745. doi: 10.1200/JCO.2007.11.5253. [DOI] [PubMed] [Google Scholar]
- Heesch S, Goekbuget N, Stroux A, Tanchez JO, Schlee C, Burmeister T, et al. Prognostic implications of mutations and expression of the Wilms tumor 1 (WT1) gene in adult acute T-lymphoblastic leukemia. Haematologica. 2010;95:942–949. doi: 10.3324/haematol.2009.016386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heuser M, Beutel G, Krauter J, Döhner K, von Neuhoff N, Schlegelberger B, et al. High meningioma 1 (MN1) expression as a predictor for poor outcome in acute myeloid leukemia with normal cytogenetics. Blood. 2006;108:3898–3905. doi: 10.1182/blood-2006-04-014845. [DOI] [PubMed] [Google Scholar]
- Weng AP, Nam Y, Wolfe MS, Pear WS, Griffin JD, Blacklow SC, et al. Growth suppression of pre-T acute lymphoblastic leukemia cells by inhibition of notch signaling. Mol Cell Biol. 2003;23:655–664. doi: 10.1128/MCB.23.2.655-664.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baldus CD, Thibaut J, Goekbuget N, Stroux A, Schlee C, Mossner M, et al. Prognostic implications of NOTCH1 and FBXW7 mutations in adult acute T-lymphoblastic leukemia. Haematologica. 2009;94:1383–1390. doi: 10.3324/haematol.2008.005272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nowak D, Mossner M, Baldus CD, Hopfer O, Thiel E, Hofmann WK. Mutation analysis of hCDC4 in AML cells identifies a new intronic polymorphism. Int J Med Sci. 2006;3:148–151. doi: 10.7150/ijms.3.148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heesch S, Schlee C, Neumann M, Stroux A, Kühnl A, Schwartz S, et al. BAALC-associated gene expression profiles define IGFBP7 as a novel molecular marker in acute leukemia. Leukemia. 2010;24:1429–1436. doi: 10.1038/leu.2010.130. [DOI] [PubMed] [Google Scholar]
- Menssen HD, Renkl HJ, Rodeck U, Maurer J, Notter M, Schwartz S, et al. Presence of Wilms' tumor gene (wt1) transcripts and the WT1 nuclear protein in the majority of human acute leukemias. Leukemia. 1995;9:1060–1067. [PubMed] [Google Scholar]
- Grosveld GC. MN1, a novel player in human AML. Blood Cells Mol Dis. 2007;39:336–339. doi: 10.1016/j.bcmd.2007.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malyukova A, Dohda T, von der LN, Akhoondi S, Corcoran M, Heyman M, et al. The tumor suppressor gene hCDC4 is frequently mutated in human T-cell acute lymphoblastic leukemia with functional consequences for Notch signaling. Cancer Res. 2007;67:5611–5616. doi: 10.1158/0008-5472.CAN-06-4381. [DOI] [PubMed] [Google Scholar]
- Ferrando AA. The role of NOTCH1 signaling in T-ALL. Hematology Am Soc Hematol Educ Program. 2009. pp. 353–361. [DOI] [PMC free article] [PubMed]
- Mansur MB, Emerenciano M, Splendore A, Brewer L, Hassan R, Pombo-de-Oliveira MS. T-cell lymphoblastic leukemia in early childhood presents NOTCH1 mutations and MLL rearrangements. Leuk Res. 2010;34:483–486. doi: 10.1016/j.leukres.2009.06.026. [DOI] [PubMed] [Google Scholar]
- Kox C, Zimmermann M, Stanulla M, Schrappe M, Ludwig WD, Koehler R. The favorable effect of activating NOTCH1 receptor mutations on long-term outcome in T-ALL patients treated on the ALL-BFM 2000 protocol can be separated from FBXW7 loss of function. Leukemia. 2010;24:2005–2013. doi: 10.1038/leu.2010.203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clappier E, Collette S, Grardel N, Suarez L, Brunie G, Kaltenbach S, et al. NOTCH1 and FBXW7 mutations have a favorable impact on early response to treatment, but not on outcome, in children with T-cell acute lymphoblastic leukemia (T-ALL) treated on EORTC trials 58881 and 58951. Leukemia. 2010;24:2023–2031. doi: 10.1038/leu.2010.205. [DOI] [PubMed] [Google Scholar]
- Zuurbier L, Homminga I, Calvert V, te Winkel ML, Buijs-Gladdines JG, Kooi C, et al. NOTCH1 and/or FBXW7 mutations predict for initial good prednisone response but not for improved outcome in pediatric T-cell acute lymphoblastic leukemia patients treated on DCOG or COALL protocols. Leukemia. 2010;24:2014–2022. doi: 10.1038/leu.2010.204. [DOI] [PubMed] [Google Scholar]
- Ferrando A. NOTCH mutations as prognostic markers in T-ALL. Leukemia. 2010;24:2003–2004. doi: 10.1038/leu.2010.237. [DOI] [PubMed] [Google Scholar]
- Breit S, Stanulla M, Flohr T, Schrappe M, Ludwig WD, Tolle G, et al. Activating NOTCH1 mutations predict favorable early treatment response and long-term outcome in childhood precursor T-cell lymphoblastic leukemia. Blood. 2006;108:1151–1157. doi: 10.1182/blood-2005-12-4956. [DOI] [PubMed] [Google Scholar]
- Gilliland DG, Griffin JD. The roles of FLT3 in hematopoiesis and leukemia. Blood. 2002;100:1532–1542. doi: 10.1182/blood-2002-02-0492. [DOI] [PubMed] [Google Scholar]
- Xu B, Li L, Tang JH, Zhou SY. Detection of FLT3 gene and FLT3/ITD mutation by polymerase chain reaction-single-strand conformation polymorphism in patients with acute lymphoblastic leukemia. Di Yi Jun Yi Da Xue Xue Bao. 2005;25:1207–1210. [PubMed] [Google Scholar]
- Paietta E, Ferrando AA, Neuberg D, Bennett JM, Racevskis J, Lazarus H, et al. Activating FLT3 mutations in CD117/KIT(+) T-cell acute lymphoblastic leukemias. Blood. 2004;104:558–560. doi: 10.1182/blood-2004-01-0168. [DOI] [PubMed] [Google Scholar]
- Marks DI, Paietta EM, Moorman AV, Richards SM, Buck G, DeWald G, et al. T-cell acute lymphoblastic leukemia in adults: clinical features, immunophenotype, cytogenetics, and outcome from the large randomized prospective trial (UKALL XII/ECOG 2993) Blood. 2009;114:5136–5145. doi: 10.1182/blood-2009-08-231217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gokbuget N, Hoelzer D. Treatment of adult acute lymphoblastic leukemia. Semin Hematol. 2009;46:64–75. doi: 10.1053/j.seminhematol.2008.09.003. [DOI] [PubMed] [Google Scholar]
- Wood B, Winter S, Dunsmore K, Raetz E, Borowitz M, Devidas M, et al. Patients with early T-cell precursor (ETP) acute lymphoblastic leukemia (ALL) have high levels of minimal residual disease (MRD) at the end of induction - a Children's Oncology Group (COG) Study. Blood. 2009;114:9. [Google Scholar]
- Moricke A, Zimmermann M, Reiter A, Henze G, Schrauder A, Gadner H, et al. Long-term results of five consecutive trials in childhood acute lymphoblastic leukemia performed by the ALL-BFM study group from 1981 to 2000. Leukemia. 2010;24:265–284. doi: 10.1038/leu.2009.257. [DOI] [PubMed] [Google Scholar]
- Pui CH, Pei D, Sandlund JT, Ribeiro RC, Rubnitz JE, Raimondi SC, et al. Long-term results of St Jude Total Therapy Studies 11, 12, 13A, 13B, and 14 for childhood acute lymphoblastic leukemia. Leukemia. 2010;24:371–382. doi: 10.1038/leu.2009.252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassan R, Hoelzer D. Modern therapy of acute lymphoblastic leukemia. J Clin Oncol. 2011;29:532–543. doi: 10.1200/JCO.2010.30.1382. [DOI] [PubMed] [Google Scholar]
- Hoelzer D, Gokbuget N, Ottmann O, Pui CH, Relling MV, Appelbaum FR, et al. Acute lymphoblastic leukemia. Hematology. Am Soc Hematol Educ Program. 2002. pp. 162–192. [DOI] [PubMed]
- Thiel E, Kranz BR, Raghavachar A, Bartram CR, Löffler H, Messerer D, et al. Prethymic phenotype and genotype of pre-T (CD7+/ER-)-cell leukemia and its clinical significance within adult acute lymphoblastic leukemia. Blood. 1989;73:1247–1258. [PubMed] [Google Scholar]
- Baldus CD, Tanner SM, Ruppert AS, Whitman SP, Archer KJ, Marcucci G, et al. BAALC expression predicts clinical outcome of de novo acute myeloid leukemia patients with normal cytogenetics: a Cancer and Leukemia Group B Study. Blood. 2003;102:1613–1618. doi: 10.1182/blood-2003-02-0359. [DOI] [PubMed] [Google Scholar]
- Juric D, Lacayo NJ, Ramsey MC, Racevskis J, Wiernik PH, Rowe JM, et al. Differential gene expression patterns and interaction networks in BCR-ABL-positive and negative adult acute lymphoblastic leukemias. J Clin Oncol. 2007;25:1341–1349. doi: 10.1200/JCO.2006.09.3534. [DOI] [PubMed] [Google Scholar]
- Kuhnl A, Gokbuget N, Stroux A, Burmeister T, Neumann M, Heesch S, et al. High BAALC expression predicts chemoresistance in adult B-precursor acute lymphoblastic leukemia. Blood. 2010;115:3737–3744. doi: 10.1182/blood-2009-09-241943. [DOI] [PubMed] [Google Scholar]
- Heesch S, Schlee C, Neumann M, Stroux A, Kuehnl A, Schwartz S, et al. Gene expression profiling identifies IGFBP-7 as a BAALC co-expressed gene with a functional role in acute leukemia. Blood. 2009;114:2629. [Google Scholar]
- Homminga I, Pieters R, Langerak AW, de Rooi JJ, Stubbs A, Verstegen M, et al. Integrated transcript and genome analyses reveal NKX2–1 and MEF2C as potential oncogenes in T cell acute lymphoblastic leukemia. Cancer Cell. 2011;19:484–497. doi: 10.1016/j.ccr.2011.02.008. [DOI] [PubMed] [Google Scholar]
- Barragan E, Cervera J, Bolufer P, Ballester S, Martín G, Fernández P, et al. Prognostic implications of Wilms' tumor gene (WT1) expression in patients with de novo acute myeloid leukemia. Haematologica. 2004;89:926–933. [PubMed] [Google Scholar]
- Paschka P, Marcucci G, Ruppert AS, Whitman SP, Mrózek K, Maharry K, et al. Wilms' tumor 1 gene mutations independently predict poor outcome in adults with cytogenetically normal acute myeloid leukemia: a cancer and leukemia group B study. J Clin Oncol. 2008;26:4595–4602. doi: 10.1200/JCO.2007.15.2058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tosello V, Mansour MR, Barnes K, Paganin M, Sulis ML, Jenkinson S, et al. WT1 mutations in T-ALL. Blood. 2009;114:1038–1045. doi: 10.1182/blood-2008-12-192039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renneville A, Kaltenbach S, Clappier E, Collette S, Micol JB, Nelken B, et al. Wilms tumor 1 (WT1) gene mutations in pediatric T-cell malignancies. Leukemia. 2010;24:476–480. doi: 10.1038/leu.2009.221. [DOI] [PubMed] [Google Scholar]
- Virappane P, Gale R, Hills R, Kakkas I, Summers K, Stevens J, et al. Mutation of the Wilms' tumor 1 gene is a poor prognostic factor associated with chemotherapy resistance in normal karyotype acute myeloid leukemia: the United Kingdom Medical Research Council Adult Leukaemia Working Party. J Clin Oncol. 2008;26:5429–5435. doi: 10.1200/JCO.2008.16.0333. [DOI] [PubMed] [Google Scholar]
- Chiaretti S, Messina M, Tavolaro S, Zardo G, Elia L, Vitale A, et al. Gene expression profiling identifies a subset of adult T-cell acute lymphoblastic leukemia with myeloid-like gene features and over-expression of miR-223. Haematologica. 2010;95:1114–1121. doi: 10.3324/haematol.2009.015099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weng AP, Ferrando AA, Lee W, Morris JP, IV, Silverman LB, Sanchez-Irizarry C, et al. Activating mutations of NOTCH1 in human T cell acute lymphoblastic leukemia. Science. 2004;306:269–271. doi: 10.1126/science.1102160. [DOI] [PubMed] [Google Scholar]
- Grabher C, von Boehmer H, Look AT. Notch 1 activation in the molecular pathogenesis of T-cell acute lymphoblastic leukaemia. Nat Rev Cancer. 2006;6:347–359. doi: 10.1038/nrc1880. [DOI] [PubMed] [Google Scholar]
- Paganin M, Ferrando A. Molecular pathogenesis and targeted therapies for NOTCH1-induced T-cell acute lymphoblastic leukemia. Blood Rev. 2010. [DOI] [PMC free article] [PubMed]
- Palomero T, Ferrando A. Therapeutic targeting of NOTCH1 signaling in T-cell acute lymphoblastic leukemia. Clin Lymphoma Myeloma. 2009;9 (Suppl 3:S205–S210. doi: 10.3816/CLM.2009.s.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pear WS, Aster JC, Scott ML, Hasserjian RP, Soffer B, Sklar J, et al. Exclusive development of T cell neoplasms in mice transplanted with bone marrow expressing activated Notch alleles. J Exp Med. 1996;183:2283–2291. doi: 10.1084/jem.183.5.2283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiang MY, Xu L, Shestova O, Histen G, L'heureux S, Romany C, et al. Leukemia-associated NOTCH1 alleles are weak tumor initiators but accelerate K-ras-initiated leukemia. J Clin Invest. 2008;118:3181–3194. doi: 10.1172/JCI35090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Neil J, Calvo J, McKenna K, Krishnamoorthy V, Aster JC, Bassing CH, et al. Activating Notch1 mutations in mouse models of T-ALL. Blood. 2006;107:781–785. doi: 10.1182/blood-2005-06-2553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mansour MR, Duke V, Foroni L, Patel B, Allen CG, Ancliff PJ, et al. Notch-1 mutations are secondary events in some patients with T-cell acute lymphoblastic leukemia. Clin Cancer Res. 2007;13:6964–6969. doi: 10.1158/1078-0432.CCR-07-1474. [DOI] [PubMed] [Google Scholar]
- Jeannet R, Mastio J, Macias-Garcia A, Oravecz A, Ashworth T, Geimer Le Lay AS, et al. Oncogenic activation of the Notch1 gene by deletion of its promoter in Ikaros-deficient T-ALL. Blood. 2010;116:5443–5454. doi: 10.1182/blood-2010-05-286658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashworth TD, Pear WS, Chiang MY, Blacklow SC, Mastio J, Xu L, et al. Deletion-based mechanisms of Notch1 activation in T-ALL: key roles for RAG recombinase and a conserved internal translational start site in Notch1. Blood. 2010;116:5455–5464. doi: 10.1182/blood-2010-05-286328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hosoya T, Kuroha T, Moriguchi T, Cummings D, Maillard I, Lim KC, et al. GATA-3 is required for early T lineage progenitor development. J Exp Med. 2009;206:2987–3000. doi: 10.1084/jem.20090934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chi AW, Chavez A, Xu L, Weber BN, Shestova O, Schaffer A, et al. Identification of Flt3+CD150− myeloid progenitors in adult mouse bone marrow that harbor T lymphoid developmental potential. Blood. 2011;118:2723–2732. doi: 10.1182/blood-2010-09-309989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scharnhorst V, Wals J, Beverloo HB, Langerak AW, van der Velden VH. Mutation of FLT3 is not a general phenomenon in CD117-positive T-ALL. Leuk Res. 2006;30:245–246. doi: 10.1016/j.leukres.2005.06.018. [DOI] [PubMed] [Google Scholar]
- Sanz M, Burnett A, Lo-Coco F, Lowenberg B. FLT3 inhibition as a targeted therapy for acute myeloid leukemia. Curr Opin Oncol. 2009;21:594–600. doi: 10.1097/CCO.0b013e32833118fd. [DOI] [PubMed] [Google Scholar]