Abstract
Poor data have been previously reported about the mutation rates in K-RAS, BRAF, and PIK3CA genes among patients with hepatocellular carcinoma (HCC). Here we further elucidated the role of these genes in pathogenesis of primary hepatic malignancies. Archival tumour tissue from 65 HCC patients originating from South Italy were screened for mutations in these candidate genes by direct sequencing. Overall, oncogenic mutations were detected in 15 (23%) patients for BRAF gene, 18 (28%) for PIK3CA gene, and 1 (2%) for K-RAS gene. Using statistical analysis, BRAF mutations were significantly correlated with the presence of either multiple HCC nodules (P=0.021) or higher proliferation rates (P=0.034). Although further extensive screenings are awaited in HCC patients among different populations, our findings clearly indicated that mutational activation of both BRAF and PIK3CA genes does contribute to hepatocellular tumorigenesis at somatic level in Southern Italian population.
Keywords: hepatocellular carcinoma, cancer genes, mutation analysis, patients' molecular classification
Hepatocellular carcinoma (HCC) is among the most common malignancies worldwide, with approximately more than 500 000 new patients diagnosed each year; however, significant regional differences in the incidence of HCC are observed because of the frequency of the different aetiological factors involved in HCC development.1
Despite major efforts to improve diagnosis and treatment of HCC, therapeutic options remain limited; treatment options depend on the stage of the disease and the extent of liver dysfunction and cirrhosis.2 Therefore, targeting the key-role genes and pathways have increasingly gained attention. Actually, molecular mechanisms leading to malignant transformation of hepatocytes as well as development and progression of HCC still remain largely unclear. The mitogen-activated protein kinase (MAPK) pathway (including the cascade of RAS, BRAF, MEK1/2, and extracellular-signal-regulated kinase (ERK)1/2 proteins) has emerged as a major signalling cascade, involved in the control of cell growth, proliferation, and migration in majority of the cancers. Recently, sorafenib, an oral multikinase inhibitor targeting BRAF among other cellular kinases, has been demonstrated to be effective in HCC.3
Besides a well-documented overexpression of RAS in human HCC,4 activating mutations of the RAS oncogenes (particularly those affecting the K-RAS gene) have been classified as rare events during hepatocarcinogenesis.5 On the other hand, RAS mutations have been frequently identified in HCC induced by various chemical agents in animals.6 From the same signalling cascade, somatic mutations of the BRAF serine/threonine kinase gene have been reported at various frequencies among different types of human cancers.7 Similar to the RAS alterations, BRAF mutations seem to poorly participate in the pathogenesis of human HCC.8 However, a clear statement about the prevalence of both RAS and BRAF mutations in HCC is yet to be definitely assessed, and differences may exist because of the geographical origins of the patients' populations and probably to the different aetiological factors that have been involved in hepatocellular carcinogenesis.
A higher amount of reports has been published revealing that the phosphoinositide-3-kinase-catalytic-alpha (PIK3CA) gene may be somatically mutated in several types of human cancer, including HCC.9, 10 In particular, PIK3CA is an effector of the phosphatase and tensin homologue (PTEN)–AKT pathway, which is involved in the inhibition of focal adhesion formation, cell spreading, and migration, as well as in the inhibition of growth factor-stimulated MAPK signalling (alterations in the RAS–BRAF pathway are frequently associated with the PTEN–PIK3CA impairments).11
In the present study, we analysed the mutational status of the K-RAS, BRAF, and PIK3CA genes in a series of HCC tissues from patients originating from South Italy, to further elucidate the possible role of these genes in primary hepatic malignancies.
Results
Genomic DNA from 65 consecutively collected HCC patients was screened for somatic mutations in BRAF, K-RAS, and PIK3CA genes. The study population consisted of 48 (74%) males and 17 (26%) females, with a median age of 69 years (range, 58–81 years). Histological patterns included trabecular (41 cases; 63%), solid (21; 32%), mixed (1; 2%), and unclassified (2; 3%) types.
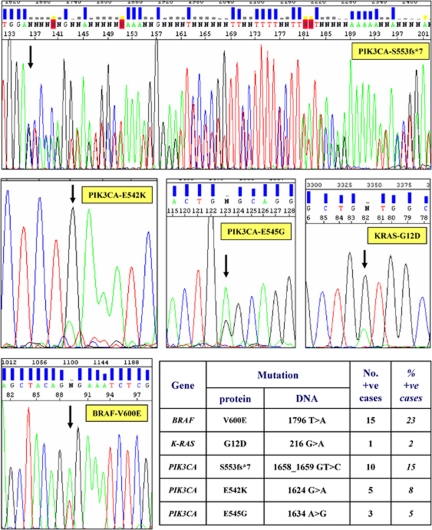
The full coding sequence and intron–exon junctions of the candidate exons (see Materials and Methods) were assessed in such different HCC samples. Figure 1 shows the nucleotide sequences for the somatic mutations identified in our series. Overall, mutations were detected in 15 (23%) HCCs for the BRAF gene, 18 (28%) cases for the PIK3CA gene, and 1 (2%) patient for the K-RAS gene (the rates of each specific mutation are shown at bottom of Figure 1). Four cases presented co-existence of BRAF and PIK3CA mutations; altogether, 30/65 (46%) patients carried a somatic mutation in at least one of the above-mentioned genes. All BRAF mutations were represented by the most common substitution of valine by glutamic acid at position 600 (V600E; Figure 1). None of the sequence changes identified was present in normal adjacent tissues from the same HCC cases, indicating that these variants are tumour-specific and somatically acquired mutations.
Figure 1.
Sequencing results for identified somatic mutations. Electropherograms show the nucleotide sequences of the genomic DNA from positive HCC samples; arrows indicate the mutation position within the sequence. Bottom right: prevalence of all mutations, designed for both DNA and amino acid changes
Using statistical tests, BRAF and PIK3CA mutations were evaluated for association with several pathological parameters: sex, age at diagnosis, tumour grading, pathological tumour size (pT; according to the TNM classification13), total number of tumour lesions within hepatic parenchyma (single or multiple HCC nodules), and rate of proliferations (as inferred by the levels of tumour mitosis; Table 1). In our series, the only significant correlation was found between the occurrence of BRAF mutations and the presence of either multiple HCC nodules (P=0.021) or higher proliferation rates (P=0.034; Table 1). No significant association was observed between the BRAF or PIK3CA mutations and other parameters (though a trend for BRAF mutations to be associated with an older age of onset, a higher tumour grade, and a larger primary tumour, as well as for PIK3CA mutations to be conversely associated with earlier primary HCC was inferred; Table 1).
Table 1. Distribution of mutations according to the characteristics of HCC patients.
|
BRAF |
PIK3CA |
|||||
|---|---|---|---|---|---|---|
| Characteristics (patients) | Mutated cases | % | P | Mutated cases | % | P |
| Sex | ||||||
| Male (N=48) | 11 | 23 | 0.972 | 13 | 27 | 0.875 |
| Female (N=17) | 4 | 24 | 5 | 29 | ||
| Age | ||||||
| ≤60 (N=13) | 2 | 15 | 0.219 | 4 | 31 | 0.822 |
| >60 (N=52) | 13 | 25 | 14 | 27 | ||
| Grading | ||||||
| G2 (N=52) | 11 | 21 | 0.211 | 15 | 29 | 0.427 |
| G3 (N=13) | 4 | 31 | 3 | 23 | ||
| pT | ||||||
| pT1 (N=20) | 4 | 20 | 8 | 40 | ||
| pT2 (N=34) | 7 | 21 | 0.187 | 8 | 24 | 0.106 |
| pT3 (N=11) | 4 | 36 | 2 | 18 | ||
| No. of tumour sites | ||||||
| Single (N=43) | 4 | 9 | 0.021 | 13 | 30 | 0.353 |
| Multiple (N=22) | 11 | 50 | 5 | 23 | ||
| No. of tumour mitoses | ||||||
| Low (N=47) | 6 | 13 | 0.034 | 14 | 30 | 0.321 |
| High (N=18) | 9 | 50 | 4 | 22 | ||
Abbreviations: HCC, hepatocellular carcinoma; PIK3CA, phosphoinositide-3-kinase-catalytic-alpha
Discussion
In this study, we examined the frequency of activating mutations of the BRAF, K-RAS, and PIK3CA genes in a series of 65 human HCC samples from patients originating from South Italy. In our series, K-RAS mutations were quite absent (only one patient presented an oncogenic mutation in this gene) confirming data from literature, indicating that mutations in K-RAS are rare and likely not a key event in hepatocarcinogenesis. On the other hand, a higher-than-expected prevalence of somatic mutations was observed for the BRAF gene (15/65; 23%). In the unique previous study on this issue, no BRAF mutation was indeed observed in a small subset of human HCC patients.8 One could speculate that the higher frequency detected in our series may be somehow due to the patients' origin or, in other words, to the different ‘genetic background' of populations from the two studies. Moreover, it cannot be excluded that different aetiological factors could be involved in the HCC development among different populations, and therefore, different mechanisms of transformation could occur. The most common mutation in BRAF gene (nearly, 90% of cases) is a substitution of valine with glutamic acid at position 600 (V600E).7 This mutation, which is present in exon 15 within the kinase domain, represented the only variant identified in our HCC tissues.
Controversial data have been published about the PIK3CA mutation rates, ranging from none14 to one-third9, 10 of the HCC cases presenting a mutated PIK3CA (interestingly, the two main contrasting reports are based on patients from the Eastern populations). In the present study, mutations were detected in quite a high fraction of the HCC tissues (18/65; 28%).
Overall, our study provides clear indications that mutational activation of both the BRAF and PIK3CA genes indeed participates to hepatocellular tumorigenesis at the somatic level in the southern Italian population. From the functional point of view, BRAF and RAS are members of the MAPK (RAS–BRAF–MEK–ERK) pathway, which mediates cellular response to growth signals. In particular, activated BRAF or RAS (mutation in encoding two genes have been demonstrated to be mutually exclusive15, 16) participate in the sustained induction of cell cycle progression. More in general, the MAPK and the PIK3CA signalling pathways both have a key role in cell proliferation and survival; their oncogenic activation deeply contributes to pathogenesis of different solid tumours, including HCC.
Nevertheless, BRAF and PIK3CA mutations were more or less equally distributed among the different patients' subsets in our series, and no statistically significant correlation with sex, onset age, histological tumour type, primary tumour size, and grading were observed. However, patients with HCC carrying a mutated BRAF are more likely to significantly present with multifocal or more aggressive disease (see Table 1).
More extensive screenings should be carried out to confirm the prevalence and distribution of BRAF- and PIK3CA-activating mutations in HCC among different populations. Actually, this need is particularly strong, as inhibition of the RAS–BRAF–MEK–ERK pathway may be an important new therapeutic strategy in HCC. More specifically, BRAF mutations have become a favoured target for the treatment of advanced HCC, because of the clinical development of the RAF-kinase inhibitor sorafenib that has found indication in the treatment of HCC in the United States, Europe, and Asia.17, 18 The different response rates observed in the clinical trials that led to the registration of sorafenib in HCC treatment could be, on the basis of our data, not only due to the different status of accompanying cirrhotic disease, but also to a different distribution of candidate gene mutations. Therefore, it should be interesting to correlate such a mutational status to the clinical responsiveness. In this view, we have recently demonstrated that the decrease of the phosphorylation of downstream ERK (a surrogate marker of the RAF kinase activity) is a predictive marker of response to a combination between sorafenib and octreotide in advanced HCC.19, 20 Trying to point out such issues, we already started a new study in which HCC patients are being prospectively enrolled and adequately followed-up, with registration of all the main clinical information (type of treatment(s), clinical behaviour of the disease, date of last control or death) and inclusion of the expression analysis by immunohistochemistry for several effectors of the BRAF–PI3K pathways (K-RAS, MEK, phospoERK, PTEN, AKT).
Finally, our findings may provide new therapeutic options for the subset of patients carrying the V600EBRAF mutations, as selective and potent BRAF inhibitors have been recently developed.21, 22, 23 Vemurafenib, a specific inhibitor of the BRAFV600E variant, has indeed showed a very promising activity in patients positive for such a mutation.23 Nevertheless, the identification of a mutated PIK3CA gene, which is becoming a promising target for newly discovered anticancer drugs,24 potentially gives an additional opportunity for identifying HCC patients who may benefit from alternative treatment strategies. One could hypothesize that new therapies, aimed at blocking the key molecular pathways, which are differently altered during hepatocellular tumorigenesis, in combination with the molecular classification of such distinct patients' subsets may become the gold standard for future treatment approaches in the HCC disease.
Materials and Methods
Samples
A total of 65 patients with histologically proven diagnosis of HCC were included into the study. For tumour typing, the classification according the AJCC/UICC guidelines was adopted.12 Tumour size was based on the largest dimension of the tumour specimen. Multiple lesions were defined by the presence of two or more nodules of tumours including intrahepatic metastasis.
Neoplastic tissue samples were obtained from pathological archives using appropriate informed consent procedures. The study was reviewed and approved by the ethical review board of the University of Sassari.
Mutation analysis
For mutation analysis, genomic DNA was isolated from tumour tissue samples, using standard methods. Tumour tissues were estimated to contain at least 70% neoplastic cells by light microscopy.
The complete coding sequences and intron–exon boundaries of the K-RAS exons 2–3, BRAF exon 15, and PIK3CA exons 9–20, were screened for mutations by direct sequencing, using an automated fluorescence-based cycle sequencer (ABIPRISM3100, Applied Biosystems, Foster City, CA, USA). Primer sequences were as reported in Genome DataBase (http://www.gdb.org).
Statistical analysis
Fisher's exact test and χ2-test were used to evaluate possible associations between covariates (mutated and wild-type genotypes) and clinical or pathological parameters. Variables were also evaluated for independent correlations by Cox regression analysis. Statistical comparisons were performed using the SPSS statistical software package, version 15.0 (SPSS Inc., Chicago, IL, USA). All tests were two-tailed and P-values of less than 0.05 were considered to be statistically significant.
Acknowledgments
We are grateful to the patients for their important contribution to this study. We like to thank Dr Mario Budroni for the statistical analyses. This work was supported by the Ricerca Finalizzata Ministero della Salute and Sardinian Regional Government (Regione Autonoma Sardegna). AB was supported by a grant from MIUR (PRIN 2008).
Author Contributions
MC carried out mutation analysis; PC performed the research; FI participated in patients' collection; FT and GB performed pathological review; AL and MA contributed to data analysis; LT participated in patients' collection; IS and MA participated in performing the research; AA contributed to the study design; MC participated in both designing the research study and writing the paper; GP wrote the paper
Glossary
- HCC
hepatocellular carcinoma
- MAPK
mitogen-activated protein kinase
- ERK
extracellular-signal-regulated kinase
- PIK3CA
phosphoinositide-3-kinase-catalytic-alpha
- PTEN
phosphatase and tensin homologue
The authors declare no conflict of interest.
Footnotes
Edited by G Melino
References
- Parkin DM, Bray F, Ferlay J, Pisani P. Global cancer statistics. CA Cancer J Clin. 2005;55:74–108. doi: 10.3322/canjclin.55.2.74. [DOI] [PubMed] [Google Scholar]
- Llovet JM. Updated treatment approach to hepatocellular carcinoma. J Gastroenterol. 2005;40:225–235. doi: 10.1007/s00535-005-1566-3. [DOI] [PubMed] [Google Scholar]
- Llovet JM, Ricci S, Mazzaferro V, Hilgard P, Gane E, Blanc JF, et al. Sorafenib in advanced hepatocellular carcinoma. N Engl J Med. 2008;359:378–390. doi: 10.1056/NEJMoa0708857. [DOI] [PubMed] [Google Scholar]
- Bose S, Sakhuja P, Bezawada L, Agarwal AK, Kazim SN, Khan LA, et al. Hepatocellular carcinoma with persistent hepatitis B virus infection shows unusual downregulation of Ras expression and differential response to Ras mediated signaling. J Gastroenterol Hepatol. 2011;26:135–144. doi: 10.1111/j.1440-1746.2010.06305.x. [DOI] [PubMed] [Google Scholar]
- Teufel A, Staib F, Kanzler S, Weinmann A, Schulze-Bergkamen H, Galle PR. Genetics of hepatocellular carcinoma. World J Gastroenterol. 2007;13:2271–2282. doi: 10.3748/wjg.v13.i16.2271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai F, Nakanishi Y, Takayama K, Pei XH, Inoue K, Harada T, et al. Codon 64 of K-ras gene mutation pattern in hepatocellular carcinomas induced by bleomycin and 1-nitropyrene in A/J mice. Teratog Carcinog Mutagen. 2003. pp. 161–170. [DOI] [PubMed]
- Davies H, Bignell G, Cox C, Stephens P, Edkins S, Clegg S, et al. Mutations of the BRAF gene in human cancer. Nature. 2002;417:949–954. doi: 10.1038/nature00766. [DOI] [PubMed] [Google Scholar]
- Tannapfel A, Sommerer F, Benicke M, Katalinic A, Uhlmann D, Witzigmann H, et al. Mutations of the BRAF gene in cholangiocarcinoma but not in hepatocellular carcinoma. Gut. 2003;52:706–712. doi: 10.1136/gut.52.5.706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JW, Soung YH, Kim SY, Lee HW, Park WS, Nam SW, et al. PIK3CA gene is frequently mutated in breast carcinomas and hepatocellular carcinomas. Oncogene. 2005;24:1477–1480. doi: 10.1038/sj.onc.1208304. [DOI] [PubMed] [Google Scholar]
- Boyault S, Rickman DS, de Reyniès A, Balabaud C, Rebouissou S, Jeannot E, et al. Transcriptome classification of HCC is related to gene alterations and to new therapeutic targets. Hepatology. 2007;45:42–52. doi: 10.1002/hep.21467. [DOI] [PubMed] [Google Scholar]
- Arcaro A, Guerreiro AS. The phosphoinositide 3-kinase pathway in human cancer: genetic alterations and therapeutic implications. Curr Genomics. 2007;8:271–306. doi: 10.2174/138920207782446160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greene FL, Page DL, Fleming ID.(eds)American Joint Committee on Cancer Staging Manual6th edn,Springer: Philadelphia; 2002 [Google Scholar]
- Sobin LH, Mary K, Gospodarowicz MK, Christian Wittekind C.(eds). TNM Classification of Malignant Tumours7th edn,Wiley Blackwell: New York, USA; 2009 [Google Scholar]
- Tanaka Y, Kanai F, Tada M, Asaoka Y, Guleng B, Jazag A, et al. Absence of PIK3CA hotspot mutations in hepatocellular carcinoma in Japanese patients. Oncogene. 2006;25:2950–2952. doi: 10.1038/sj.onc.1209311. [DOI] [PubMed] [Google Scholar]
- Sensi M, Nicolini G, Petti C, Bersani I, Lozupone F, Molla A, et al. Mutually exclusive NRASQ61R and BRAFV600E mutations at the single-cell level in the same human melanoma. Oncogene. 2006;25:3357–3364. doi: 10.1038/sj.onc.1209379. [DOI] [PubMed] [Google Scholar]
- Gray-Schopfer V, Wellbrock C, Marais R. Melanoma biology and new targeted therapy. Nature. 2007;445:851–857. doi: 10.1038/nature05661. [DOI] [PubMed] [Google Scholar]
- Abou-Alfa GK, Schwartz L, Ricci S, Amadori D, Santoro A, Figer A, et al. Phase II study of sorafenib in patients with advanced hepatocellular carcinoma. J Clin Oncol. 2006;24:4293–4300. doi: 10.1200/JCO.2005.01.3441. [DOI] [PubMed] [Google Scholar]
- Yau T, Chan P, Epstein R, Poon RT. Management of advanced hepatocellular carcinoma in the era of targeted therapy. Liver Int. 2009;291:10–17. doi: 10.1111/j.1478-3231.2008.01916.x. [DOI] [PubMed] [Google Scholar]
- Del Prete S, Montella L, Caraglia M, Maiorino L, Cennamo G, Montesarchio V, et al. Sorafenib plus octreotide is an effective and safe treatment in advanced hepatocellular carcinoma: multicenter phase II So.LAR. study. Cancer Chemother Pharmacol. 2009;645:863–866. doi: 10.1007/s00280-009-1226-z. [DOI] [PubMed] [Google Scholar]
- Caraglia M, Giuberti G, Marra M, Addeo R, Montella L, Murolo M, et al. Oxidative stress and ERK1/2 phosphorylation as predictors of outcome in hepatocellular carcinoma patients treated with sorafenib plus octreotide LAR. Cell Death Dis. 2011;2:e150. doi: 10.1038/cddis.2011.34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bollag G, Hirth P, Tsai J, Zhang J, Ibrahim PN, Cho H, et al. Clinical efficacy of a RAF inhibitor needs broad target blockade in BRAF-mutant melanoma. Nature. 2010;467:596–599. doi: 10.1038/nature09454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flaherty KT, Puzanov I, Kim KB, Ribas A, McArthur GA, Sosman JA, et al. Inhibition of mutated, activated BRAF in metastatic melanoma. N Engl J Med. 2010;363:809–819. doi: 10.1056/NEJMoa1002011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman PB, Hauschild A, Robert C, Haanen JB, Ascierto P, Larkin J, et al. Improved survival with vemurafenib in melanoma with BRAF V600E mutation. N Engl J Med. 2011;364:2507–2516. doi: 10.1056/NEJMoa1103782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Echeverria C, Sellers WR. Drug discovery approaches targeting the PI3K/Akt pathway in cancer. Oncogene. 2008;27:5511–5526. doi: 10.1038/onc.2008.246. [DOI] [PubMed] [Google Scholar]