Abstract
The ubiquitin hybrid genes Uba80 and Uba52 encode ubiquitin (Ub), which is fused to the ribosomal proteins S27a (RPS27a) and L40 (RPL40), respectively. Here, we show that these genes are preferentially over-expressed during hepatoma cell apoptosis. Experiments using the tet-inducible transgenic system revealed that over-expression of the ubiquitin hybrid genes sensitized the cells to apoptosis. Further analysis suggested that Ub, and not RPS27a or RPL40, was associated with apoptotic cell death. Cleavage-resistant mutation analysis revealed that the N-terminal portion and the last two amino acids (GG) of Ub are critical for cleavage at the junction between the two protein moieties. An apoptogenic stimulus enhances the nuclear targeting and aggregation of Ub in the nucleus, resulting in histone H2A deubiquitylation followed by abnormal ubiquitylation of the nuclear envelope and the lamina. These events accompany the apoptotic nuclear morphology in the late stage of apoptosis. Each fused RP is localized in the nucleoli. These results suggest a role for Ub hybrid proteins in the altered nuclear dynamics of Ub during tumor cell apoptosis induced by apoptogenic stimuli.
Keywords: ubiquitin hybrid proteins, tumor cells, apoptosis, H2A histone, ubiquitylation, deubiquitylation
Ubiquitin (Ub) is a highly conserved protein comprised of 76 amino-acid residues with diverse intracellular activities. In many organisms, the genes encoding Ub are found in two forms.1 One structural type of the Ub gene, the polyubiquitin gene, consists of tandem repeats of the 228 bp Ub-coding unit in a head-to-tail spacerless array. A second structural type of the Ub gene (UbA) encodes a Ub hybrid protein where a single Ub and a carboxyl extension protein (CEP), which consists of either 52 or 76–80 basic amino acids, are fused.2 The human Uba80 (HUBCEP80) and Uba52 (HUBCEP52) genes encode Ub fused to the ribosomal proteins S27a (RPS27a) and L40 (RPL40), respectively.3 Both RPs have a high proportion of basic residues, a putative nuclear localization signal and a cysteine-rich motif that is common to some nucleic acid-binding proteins.4
DNA is tightly packed in chromatin, which in eukaryotic cells consists of highly organized nucleosomes. The tails of histones are subject to a variety of post-translational modifications with regulatory roles in chromatin structure and function.5 Ub targets the C-terminal tail of histone H2A, resulting in as much as 5–15% of H2A being monoubiquitylated.6 Identification of the Ub ligase Ring2 for histone H2A has revealed important roles for H2A ubiquitylation in Hox gene silencing, as well as in X-chromosome inactivation.7 The Ub ligase is also required for DNA damage-induced H2A ubiquitylation.8
USP16 (Ubp-M) deubiquitylates several critical proteins that are involved in the condensation of mitotic chromosomes, mainly on ubiquitylated proteins of the chromatin such as histones H2A and H2B9 and this deubiquitylation is also associated with both cell cycle progression and gene expression.10 USP21 also catalyzed the hydrolysis of mouse liver chromatin uH2A in nucleosome form but not that of uH2A in free form.11 Furthermore, Ub is cleaved from ubiquitylated H2A (ubH2A) during mitosis as the cells move from prophase to metaphase, and also as the chromatin condenses into chromosomes.12 The nucleosomal histones are rapidly re-ubiquitylated during anaphase.13 Besides its putative role in mitotic chromosome condensation, ubH2A deubiquitylation appears to be a feature of condensing chromatin during TGF-β1-induced apoptosis in tumor cells.14 Simultaneous decreases in ubH2A levels and apoptotic chromatin condensation occur in tumor cells that are undergoing proteasome inhibitor-induced apoptosis.15, 16 ubH2A deubiquitylation was reported to be a downstream consequence of procaspase activation. Caspase behaves as a cellular sensor of stress in situations, including apoptosis, in which cells act to preserve their genomic integrity.17 The accumulation of ubiquitylated substrate coincides with chromatin remodeling and the depletion of ubiquitylated histone H2A during proteotoxic stress, and this redistribution of Ub is a direct consequence of competition for the limited pool of free Ub.18 However, it remains unknown which Ub pool is used for histone ubiquitylation and how the dynamic Ub status of the nucleosomal histones is linked to the process of apoptogenic stress. In this study, we analyzed the behavior of Ub hybrid proteins in the dynamic changes that occur during ubiquitylation in the nucleus in response to various apoptogenic stimuli.
Results
Over-expression of ubiquitin hybrid genes during apoptosis
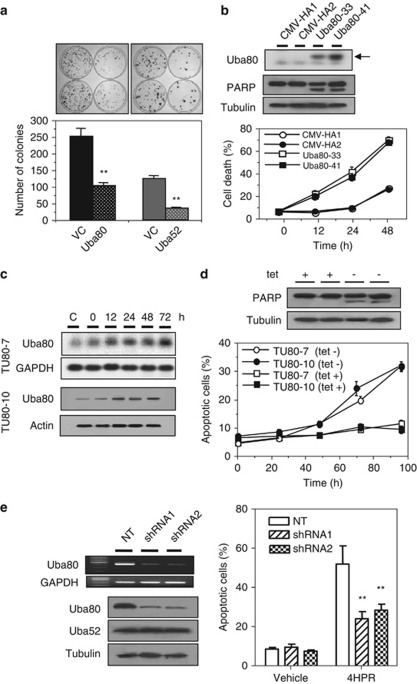
N-(4-hydroxyphenyl)retinamide (4HPR) strongly induces apoptosis through oxidative stress, and that ubiquitin hybrid genes are over-expressed during apoptotic cell death in hepatoma cells.19 Accordingly, 4HPR (10 μM) triggered apoptotic cell death (26% after 48 h of treatment) (Figure 1a) and over-expression of Uba80 and Uba52 (Figure 1c) in Hep3B cells, both in a time-dependent manner, accompanying increased poly(ADP-ribose)polymerase (PARP) cleavage. The cells were incubated under hypoxic conditions or treated with various agents. Each of these apoptogenic stimuli triggered the over-expression of Uba80 mRNA in Hep3B cells after 24–48 h of treatment, as compared with the vehicle control. The level of apoptotic cell death ranged from 45 to 56% in the treated experimental groups compared with <5% in the untreated control cells (Supplementary Figure S1a). Lung (A549), kidney (A498 and ACHN), hepatoma (SK-HEP-1), colon (HT29) and breast (MCF-7) cancer cell lines were treated with the genotoxic agents, including 5-fluorouracil (5-FU), trichostatin A (TSA), and paclitaxel (PX) for 48 h. These drugs also induced Uba80 mRNA over-expression in all of the cell lines tested (Figure 1d). 4HPR induced Uba80 and Uba52 protein expression in A549 cells in a time-dependent manner (Figure 1e). Therefore, the anticancer drug-induced over-expression of ubiquitin hybrid genes appears to be a general phenomenon that is not cell-specific.
Figure 1.
Induction of ubiquitin hybrid genes during apoptosis. (a) Cell apoptosis assay of Hep3B cells continuously treated with 10 μM 4HPR for 72 h. FACScan analysis of apoptotic Hep3B cells stained with Annexin V-FITC and PI (1 μg/ml). The fraction of apoptotic cells was estimated (right upper quadrant of the histogram) at the indicated times. The values are reported as the mean±S.E.M. of two independent experiments performed in triplicate (lower panel). (b) In the linear map of the ubiquitin hybrid genes, the Uba80 and Uba52 genes contain the amino-terminal Ub and the carboxy-terminal RPS27a or RPL40. (c) Time courses of Uba80 or Uba52 mRNA (upper panels) and protein induction (middle panels) accompanying PARP cleavage (lower panels). The results shown are representative of three independent experiments. (d) Uba80 mRNA induction by various anticancer drugs. Hep3B cells were incubated under hypoxia or were treated with H2O2 (0.5 mM), 5-FU (2 μg/ml), PX (100 nM), TSA (0.5 μM) or etoposide (50 μM) at their IC50 concentrations for 48 h. (e) The expression of Uba80 and Uba52 proteins over time was analyzed in A549 cells treated with 10 μM 4HPR for 48 h. (f) Luciferase activity was measured in Hep3B cells transiently transfected with Uba80-Luc and Uba52-Luc and treated with 10 μM 4HPR for the indicated time intervals. The vertical bars represent the mean±S.E.M. of three experiments performed in duplicate
Next, we examined whether 4HPR affects Uba80 mRNA transcription or its stability. In Hep3B cells, a 12 h treatment with 4HPR induced a sustained increase in Uba80 mRNA. In the presence of actinomycin D, an inhibitor of transcription, 4HPR increased the Uba80 mRNA level (Supplementary Figure S1b), showing that 4HPR stabilized RPS27a mRNA levels in Hep3B cells. The effect of 4HPR on Uba80 and Uba52 promoter activity was tested further by transiently transfecting Hep3B cells with Uba80-Luc and Uba52-Luc, which contain the human Uba80 and Uba52 promoters, respectively, linked to a luciferase reporter gene (Figure 1f).20, 21 The decrease in luciferase activity relative to the level in untreated control cells indicates that 4HPR decreased the level of Uba80 and Uba52 promoter activation in the transfected Hep3B cells. This suggests that the apoptogenic drugs increased Uba80 and Uba52 mRNA at the post-transcriptional level.
Ub, not RP, is associated with apoptotic cell death
We measured the clonogenicity of Hep3B cells transfected with Uba80, Uba52, or an empty vector and found that the number of Uba80 or Uba52 stable transfectants obtained was always lower than that of the controls (approximately 40% or 28% of the controls, respectively) (Figure 2a). The inhibition of colony formation in this type of assay can be explained by either a block in the cell cycle or induction of cell death. To distinguish these mechanisms, the Uba80 expression system was introduced into Hep3B cells, which constitutively express low levels of Uba80 mRNA. Two Hep3B sublines (Uba80-33 and Uba80-41) that stably expressed human Uba80 mRNA were isolated (Figure 2b) and treated with 10 μM 4HPR. Within 48 h of treatment, the Uba80-33 and Uba80-41 cells became apoptotic (72.2% and 70.7%, respectively) compared with the control cells (27.1% and 25.9%, respectively), which had been transfected with the empty vector. This type of apoptotic cell death was further confirmed by PARP cleavage. Similar results were obtained for the stable Uba52 transfectants (data not shown). This was tested further using an Uba80 expression construct created by placing Uba80 under the control of a tetracycline (tet)-repressible promoter.22 In both TUba80-7 and TUba80-10 clones, tetracycline depletion induced Uba80 over-expression in a time-dependent manner (Figure 2c). The level of apoptotic cell death was even slightly increased in the TUba80-7 and TUba80-10 cells cultured in the absence of 4HPR (Figure 2d). In contrast, a Uba80 gene-specific short hairpin RNA (shRNA) markedly knocked down the Uba80 transcript and protein level, but not Uba52 levels (Figure 2e). Apoptosis analysis indicated that while Uba80 silencing effectively inhibited 4HPR-mediated cell death, it did not do so completely in the Uba80 shRNA-expressing Hep3B cells compared with the cells expressing the non-target control (24.0% and 28.4% versus 56.1%, respectively, P<0.01). This finding suggests that the sensitivity of tumor cells to apoptotic cell death is modulated by the level of Uba80 expression, which is downstream of apoptotic initiation.
Figure 2.
Effects of ectopic Uba80 over-expression. (a) The clonogenicity of Hep3B cells transfected with an Uba80 gene (CMV-HA-Uba80), an Uba52 gene (CMV-FLAG-Uba52), or an empty vector (VC) was determined as plating efficiency in a colony-forming assay. Colonies were visualized by crystal violet staining of the cultures after 14 days of G418 selection (upper). The vertical bars represent the mean±S.E.M. from two independent experiments, each performed in duplicate (lower). **P<0.01 versus the vector control. (b) mRNA expression of Uba80 in the stably transfected cells (Uba80–33 and Uba80–41) compared with the vector control cells (CMV-HA1 and CMV-HA2). Apoptotic cell death in stably transfected Hep3B cells treated with 10 μM 4HPR for 48 h was assessed by PARP cleavage (middle panels) and DAPI staining (lower panel). The experiments were performed twice using duplicate samples. (c) The accumulation of Uba80 mRNA and protein was induced in the tet-regulated transfectants (TUb80-7 and TUb80-10, respectively) in a time-dependent manner. The transfectants were cultured in the absence of tetracycline for 72 h. (d) The TUb80-7 and TUb80-10 cells were cultured in the presence or absence of 2 μg/ml tetracycline for 96 h. The level of apoptotic cell death was determined by PARP cleavage and staining with DAPI. Each value represents the mean of triplicate experiments±S.E.M. (e) The effect of Uba80 shRNA on apoptotic cell death in Hep3B cells transduced with lentiviral Uba80 shRNA or the lentiviral nontarget control (NT, SHC002V) at an MOI=1. Semi-quantitative RT-PCR was performed to confirm the knockdown of Uba80 mRNA (top left). Immunoblot analysis of Uba80 shRNA specific for Uba80, not for Uba52 protein (bottom left). The level of cell apoptosis was evaluated further by DAPI staining (right). Each value represents the mean of triplicate experiments±S.E.M. **P<0.001
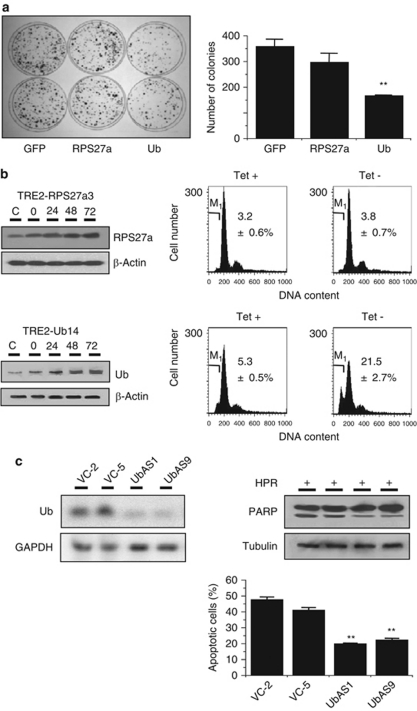
Next, colony generation assays were performed for the Hep3B cells that over-expressed either Ub or RPS27a to determine which moiety from the Ub-ribosomal hybrid proteins was essential for the 4HPR-induced apoptotic cell death. The number of Ub transfectants obtained was always significantly lower than the number of either RPS27a transfectants or green fluorescence protein (GFP)-transfected controls (Figure 3a). In TRE2-RPS27a3 or TRE2-Ub14 cells, tetracycline depletion was used to induce RPS27a or Ub over-expression in a time-dependent manner (Figure 3b). The level of apoptotic cell death increased in the TRE2-Ub14 cells, not in the TRE2-RPS27a3 cells cultured without 4HPR. During 4HPR-induced apoptosis, the inhibition of Ub mRNA by antisense Ub cDNA (the UbAS1and UbAS9 cells) seemed to be resistant to 4HPR (27% and 22.2% apoptosis versus 44.3% and 39.3%, respectively), as compared with the vector control (Figure 3c). These results indicate that Ub, not RPS27a, over-expression may be related to apoptosis.
Figure 3.
Effects of ectopic over-expression of Ub. (a) The clonogenicity of Hep3B cells after transfection with an expression vector for Ub (pIRES2-EGFP-Ub), a vector for RPS27a (pIRES2-EGFP-RPS27a) or an empty vector (pIRES2-EGFP), as assessed by plating efficiency in a colony-forming assay visualized by crystal violet staining of the cultures after 14 days of G418 selection (left). The vertical bars represent the mean±S.E.M. from two independent experiments each performed in duplicate (right). **P<0.01 versus the vector control. (b) The accumulation of RPS27a or Ub protein (left) in the tet-regulated transfectants in a time-dependent manner. Both transfectants were cultured in the presence or absence of tetracycline for 72 h. The apoptotic fractions were quantified using FACScan. The sub-G1 fraction was estimated by gating the hypodiploid cells in the histogram using a C-30 program. The DNA content was plotted on the linear abscissa (M1, apoptotic fraction) (right). Each value represents the mean of triplicate experiments±S.E.M. (**P<0.01 compared with the control). (c) Antisense Ub cDNA inhibits 4HPR-induced apoptosis. Ub mRNA expression was analyzed in Hep3B cells transfected with antisense Ub cDNA or the empty vector (VC). Northern blot analysis was performed with GAPDH as the loading control (left). The Hep3B cells transfected with the antisense Ub cDNA were treated with 10 μM 4HPR for 72 h. The level of apoptotic cell death was determined by PARP cleavage and DAPI staining (right). The vertical bars represent the mean±S.E.M. of quadruplicate experiments (**P<0.01)
Cellular localization of Ub or RPs (RPS27 and RPS40) during apoptosis
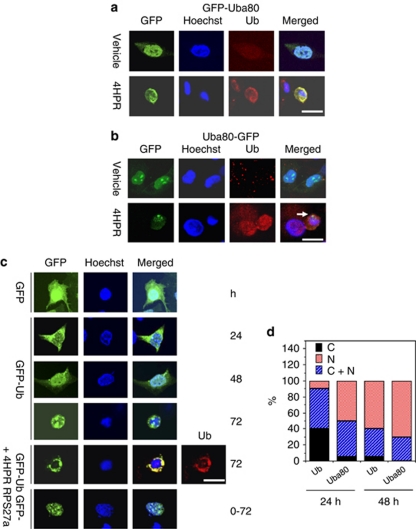
Vectors containing the GFP fused to the amino (GFP-Uba80) or carboxy terminus (Uba80-GFP) of the Uba80 gene were used to determine where the fusion protein was cleaved and to identify the cellular target of each protein during drug-induced apoptosis. The ectopic expression of the GFP-Uba80 fusion protein was prominent early in the cytoplasm and late in the nucleus, which completely overlapped with the expression of Ub. Treatment with 4HPR accelerated the perinuclear aggregation of the GFP fusion protein, which was recognized by the Ub antibody (FL-6) in the transfected cells (Figure 4a). In contrast, the Uba80-GFP transfected cells showed persistent localization of GFP in the nucleoli (Figure 4b). Similar results were obtained in the cells transfected with the GFP-Uba52a expression plasmid, and this suggests that the Ub hybrid protein is cleaved into two fragments, which then target the nucleus or nucleoli. The GFP-Ub fusion protein was mainly localized in the cytoplasm of the cells 24 h after transfection and it was then partially translocated to the nucleus (Figure 4c). At 72 h after transfection, a ring-like fluorescent aggregate appeared along the perinuclear region, surrounding the condensed and fragmented chromatin. This fluorescence overlapped perfectly with the results of immunostaining for Ub. In contrast, the GFP-RPS27a fusion protein was mainly located in the nucleoli of the cells during an early stage of apoptosis. We then compared the quantitative fractions of cells in which the Uba80 or Ub protein was localized in the cytoplasm and/or the nucleus. At 24 h after transfection, 95% of cells showed Uba80 mainly in the nucleus, whereas only 60% of cells showed Ub in the nucleus (Figure 4d). This finding suggests that although monoubiquitin itself can target the nucleus over time, either RPS27a or RPL40 can facilitate this transport.
Figure 4.
Cellular localization of the Ub hybrid proteins. (a) Hep3B cells were transfected with 2 μg of an expression vector for the GFP- or GFP-Uba52 fusion proteins (GFP fused to the amino terminus). The cells were fixed and permeabilized after 72 h treatment with 4HPR. The cells were processed for staining with 0.1 μg/ml Hoechst 33258 to visualize the nuclei (blue) and for immunostaining of Ub (red). The GFP fluorescence was then analyzed by confocal microscopy (Zeiss LSM 510) (scale bar, 20 μm). (b) The Hep3B cells were transfected with 2 μg of the expression vector for the GFP or Uba52-GFP fusion protein (GFP fused to the carboxy terminus). The arrow indicates a nucleolus extruded from the condensed or fragmented chromatin. (c) Hep3B cells were transfected with 2 μg of the expression vector for GFP-Ub or GFP-RPS27a for the indicated amounts of time (scale bar, 20 μm). (d) The quantitative fraction of cells in which the ectopic ubiquitin hybrid gene was localized. The Hep3B cells were transfected with 2 μg of the expression vector for GFP-Uba80 or GFP-Ub (GFP fused to the amino terminus of Uba80 or Ub, respectively) for the indicated times (markers C, C+N and N represent cytoplasm, cytoplasm and nucleus, and the nucleus, respectively)
Cleavage of Ub hybrid proteins
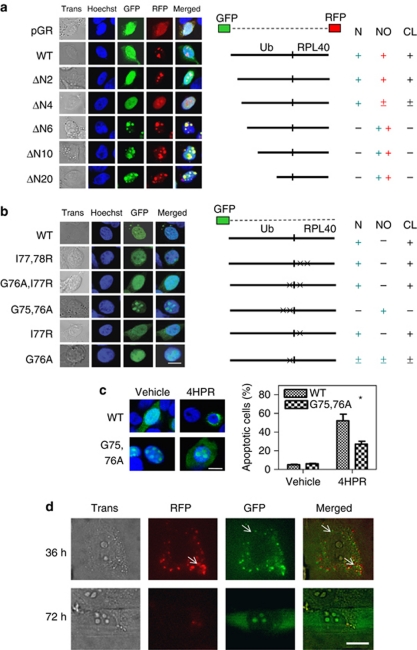
Using Ub hybrid proteins tagged with N-terminal-GFP and C-terminal-RFP, we examined the cleavage of Uba80 and Uba52 by Ub C-terminal hydrolase.23 GFP-tagged Ub was localized in the nucleus and the RFP-tagged RP was localized in the nucleoli in Hep3B and A549 cells (Supplementary Figure S2). We confirmed these cleavage sites using cleavage-resistant mutants. Ub functions as a cis-acting cleavage signal when located at the N terminus of fusion proteins.24 Mutation analysis showed that both end-tagged moieties from the N-terminal deletion mutants, ΔN6 through to ΔN20, were located in the nucleoli without cleavage. After cleavage of the minimal deletion mutants (∼ΔN4) and the wild-type Uba52, the GFP-tagged Ub moiety was localized in the nucleus and the RFP-tagged RP moiety was localized in the nucleoli (Figure 5a). This suggests that the N-terminal-deletion (Ub-deletion) mutants resist cleavage and that Ub has an important role in the cleavage of Uba into two moieties. However, the various C-terminal deletion mutants were subjected to cleavage (data not shown). The same results were observed in lung cancer A549 cells (Supplementary Figure S3a).
Figure 5.
Cleavage of the Ub hybrid proteins. (a) The Hep3B cells grown on coverslips were transiently transfected with 2 μg of the plasmid encoding the wild-type Uba52 or the deletion mutant Uba52 that was end-tagged with both GFP (N-terminal) and RFP (C-terminal), or with a vector control (pGR). GFP and RFP fluorescence was analyzed using a Zeiss LSM 510 confocal microscope. ΔN2, ΔN4, ΔN6, ΔN10 and ΔN20 indicate the deletion mutants of Uba52 lacking the N-terminal 2, 4, 6, 10 and 20 amino acids, respectively. Nuclear (N) or nucleolus (NO) indicates localization of Ub or RPL40, respectively, which was derived from the Ub hybrid protein Uba52 by cleavage (CL) Trans, transmission. (b) The Hep3B cells grown on coverslips were transiently transfected with 2 μg of plasmid encoding the wild-type or mutant GFP-Uba52 or with a vector control (GFP). I77,78 R indicates a mutant in which I77 and I78 are mutated to R. In G76A/I77R, G76 and I77 are mutated to A and R, respectively. In G75,76A, G75 and G76 are mutated to A. In I77R, I77 is mutated to R. In G76A, G76 is mutated to A (scale bar, 10 μm). (c) Effect of the G75,76A (cleavage-resistant) mutation on drug-induced apoptosis in Hep3B cells. Cells grown on coverslips were transiently transfected with 2 μg of the plasmid encoding the wild-type or mutant GFP-Uba52 (G75,76A) and treated with 10 μM 4HPR for 3 days. The cells with apoptotic morphology were identified by confocal microscopy (left panels), and the apoptotic cells among the GFP-positive cells were quantified (right panels). Each bar represents the mean of triplicate experiments±S.E.M. (*P< 0.05); scale bar, 10 μm. (d) Visualization of wild-type Uba80 in living Hep3B cells using a TIRF microscope with a transmitted all-side polished dove prism. Hep3B cells grown on coverslips were transiently transfected with 10 ng of the plasmid encoding wild-type Uba80 tagged with both RFP (N-terminal) and GFP (C-terminal). The TIRF image was examined 36 and 72 h after transfection; bar, 10 μm
Ub and Ub-like proteins bear a conserved diglycine motif at their carboxyl termini for isopeptide linkage to conjugated proteins.25 Point mutation analysis in Hep3B cells revealed that GFP-tagged Uba52 is mainly located in the nucleoli of cells that overexpress the G75,76A mutant protein (Figure 5b). In the cells that overexpress G76A, half of the GFP-tagged protein was detected in the nucleoli, whereas the other half was found in the nucleus. This indicates that the cleavage site is located at the junction between the Ub and the RP, and that the last two amino acids (GG) of Ub are essential for this cleavage. The same results were obtained in A549 cells (Supplementary Figure S3b). Accordingly, western blot analysis revealed that GFP localized to both the cytoplasm and nuclei of wild-type and cleavable mutants, whereas GFP mainly localized to the nuclei of the uncleavable mutants (Supplementary Figure S3c). We consistently observed that Ub localized to the nucleoli without perinuclear aggregation during apoptosis in cells that were transfected with the GFP-tagged G75,76A mutant, and that these cells showed more resistance to apoptosis than did the cells transfected with the GFP-tagged wild-type after 4HPR treatment (Figure 5c).
To determine the cleavage site of the Ub hybrid protein, we used a combination of differential interference contrast (DIC) and total internal reflection fluorescence (TIRF) microscopy, using an all-side polished dove prism-type microscope. Early in the expression of the double-tagged Ub hybrid proteins, most RFPs were aligned with GFPs side by side in the cytoplasm (Figure 5d), but some fluorescent singlets were observed (white arrows). At later times, the GFP was localized in the nucleoli of the cells. These results imply that the Ub hybrid protein is cleaved into two protein moieties in the cytoplasm.
Changes in endogenous Ub during apoptosis
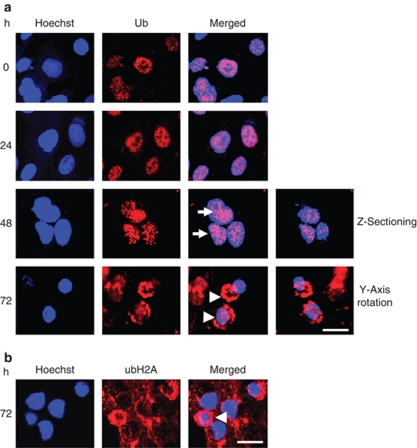
In the absence of 4HPR, most of the cells showed nuclear accumulation of the ubiquitylated proteins (Figure 6a). However, treatment with 4HPR enhanced the nuclear staining of Ub, and this caused some aggregation in the nuclei over time (white arrows). Different levels of Z-sectioning of the cells revealed additional levels of aggregation, suggesting that those aggregations were localized within the nucleus. Finally, the apoptotic cells showed a ring-like perinuclear aggregation of ubiquitylated proteins, which was accompanied by decreases in nuclear size and by nuclear condensation (white arrowheads). Rotation of the cells on the Y-axis showed a ring-like aggregation surrounding the nucleus. Results using the E6C5 monoclonal anti-uH2A antibody were similar to those using the FL-76 polyclonal anti-Ub antibody (Figure 6b).
Figure 6.
Nuclear targeting and perinuclear aggregation of endogenous ubiquitylated proteins during apoptosis. (a) Hep3B cells treated with 10 μM 4HPR for the indicated times were fixed and processed for indirect immunofluorescence with the Ub-specific antibody, FL-76. The nuclei were stained with Hoechst 33258 (blue), and the cells were examined by confocal microscopy. Z-sectioning revealed Ub aggregates in the nucleus, and Y-axial rotation showed the perinuclear aggregates of the ubiquitylated proteins. (b) Indirect immunofluorescence with the uH2A-specific antibody E6C5 in Hep3B cells treated with 10 μM 4HPR for 72 h (red). The arrows indicate multiple stained aggregates in the nuclei. The arrowheads indicate the aggregated ring-like staining in the perinuclear regions; scale bar, 20 μm
Decreased ubiquitylation of nucleosomal histone H2A during apoptosis
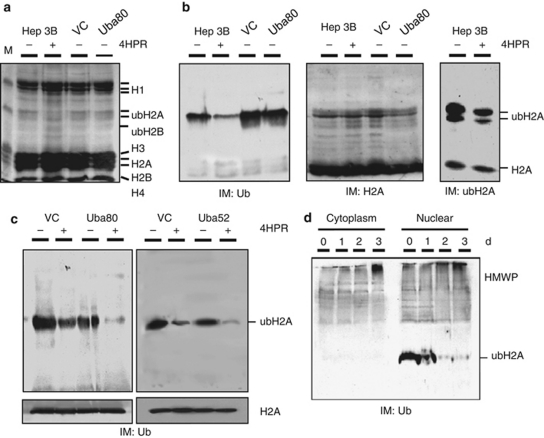
The kinetics of ubiquitylation in the cells was determined by examining the effect of 4HPR on the quantity of ubH2A (Figures 7a and b). Using three types of antibodies, FL-76 against Ub, C-20 against H2A and E6C5 against ubH2A, it was found that E6C5 detected ubH2A and H2A, with ubH2A being the main ubiquitylated band, as described elsewhere.17, 26 4HPR effectively decreased the ubH2A level in the parental cells. In the absence of 4HPR, the stable transfectants that expressed Uba80 showed a small decrease in ubiquitylated histone levels compared with the vector control cells. However, in the presence of 4HPR, the decrease in ubH2A was significantly greater than in the vector control cells (Figure 7c); this suggests that ectopic Uba80 or Uba52 expression may sensitize the cells to apoptosis and subsequently contribute to H2A deubiquitylation. In this study, as ubiquitylated proteins accumulated in the cells undergoing apoptosis,27 the abundant (poly)ubiquitylated proteins (high molecular weight proteins: HMWPs) were easily detected on immunoblots of the cytoplasmic lysates from cells treated with 4HPR using an Ub antibody (Figure 7d). This finding is consistent with results of a previous report.17 A time-dependent increase in ubiquitylated HMWPs in the nuclear lysates of cells treated with 4HPR was also observed.
Figure 7.
Ubiquitylation of the nucleoprotein during apoptosis. (a) Hep3B cells were treated with either 4HPR or the vehicle for 72 h, and then lysed with TNESV buffer plus protease inhibitors. The stable transfectants of Uba80 or the vector control cells (VC) were also lysed. Equal amounts of the acid–soluble proteins from each fraction were electrophoretically resolved on 15% SDS-PAGE gels. The gels were stained with coomassie brilliant blue. ubH2A and ubH2B represent the Ub adducts of H2A and H2B, respectively. (b) Immunoblots (IM) for Ub (left), H2A (middle) and ubH2A (right). (c) The stable transfectants of the Uba80, Uba52 or vector control cells (VC) were treated with either 4HPR or the vehicle for 72 h. The nucleoprotein fractions were electrophoretically resolved on 15% SDS-PAGE gels, transferred to membranes and then immunoblotted to detect ubiquitylated proteins. (d) Ubiquitylated HMWP during 4HPR-induced apoptosis. The cells treated with 4HPR for the indicated times were lysed. The nucleoprotein and soluble lysate (cytoplasmic) fractions were separated by SDS-PAGE and the gels were immunoblotted to detect ubiquitylated proteins
Ubiquitylation of the nuclear envelope and the lamina instead of histones during apoptosis
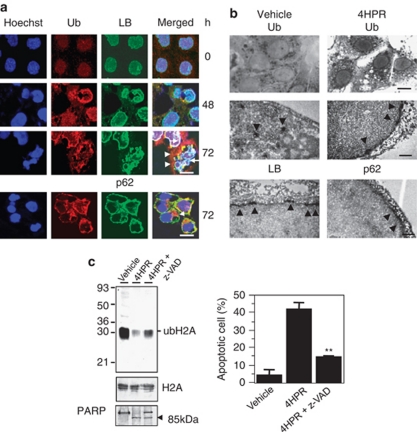
In the nuclei, apoptosis is manifested by chromatin condensation, nucleosomal fragmentation and perturbation of the nucleus.28 The nuclear envelope proteins include those of the nuclear pore (NUP153), the inner nuclear membrane (LBR and LAP2) and the nuclear lamina (lamin B, LB), and specific executioner caspases target these proteins.29, 30, 31 Immunocytochemistry revealed that the LB protein was restricted to the periphery of the fragmented chromatin. In some areas, the linear ubiquitylated protein overlapped with the linear LB protein. However, the ubiquitylated protein aggregates were mainly located outside of the lamina protein (Figure 8a). The patchy regions of Ub immunoreactivity were localized mainly inside of p62, which is one of the major nuclear pore complex proteins. This suggests that the deubiquitylation of nuclear histones is accompanied by ubiquitylation of the nuclear envelope and the lamina.
Figure 8.
Ubiquitylation of the nuclear envelope and the lamina during apoptosis. (a) Hep3B cells treated with 10 μM 4HPR for the indicated times were fixed and processed for indirect immunofluorescence with antibodies against Ub (red), LB (green) or p62 (green). The nuclei were stained with Hoechst 33258 (blue). The white arrows indicate the lamina in close contact with chromatin. The white arrowheads indicate the aggregated staining of the ubiquitylated proteins in the perinuclear region; scale bar, 5 μm. (b) Optical (upper panels) and electron (middle and lower panels) micrographs of the Hep3B cells. Dark Ub-immunoreactivity can be seen in the cytoplasmic granules of the Hep3B cells treated with the vehicle (upper left). Dark linear Ub-immunopositivity can be seen along the nuclear membrane of the Hep3B cells treated with 10 μM 4HPR (upper right); scale bar, 10 μm. Ub-immunoreactivity (arrowheads) can be seen as electron-dense patches in the nucleus of the Hep3B cells treated with the vehicle (middle left). Electron dense linear Ub-immunoreactivity can be seen along the nuclear membrane in Hep3B cells treated with 10 μM 4HPR (middle right). LB-(lower left) or p62–immunoreactivity (lower right) can be seen along the nuclear membrane in Hep3B cells treated with 10 μM 4HPR. The electron-dense immunoreactive product (arrowheads) is localized at the nuclear membrane of the cells; scale bar, 1 μm. (c) Caspase-dependent inhibition of histone ubiquitylation. Hep3B cells were treated with 10 μM 4HPR in the presence or absence of z-VAD-FMK for 72 h and then lysed. Equal amounts of the acid-soluble proteins of each fraction were resolved electrophoretically on 15% SDS-PAGE gels and the ubiquitylated proteins or H2A as an internal control were detected by immunoblotting. Concurrently, the soluble lysate (cytoplasmic) fractions were immunoblotted for PARP (left). The apoptotic fractions were quantified using a FACScan system (right). Each bar represents the mean of triplicate experiments±S.E.M. (**P<0.01 compared with 4HPR treatment alone)
To confirm these observations, the precise localization of Ub relative to the nuclear lamina and envelope during apoptosis was determined by immuno–electron microscopic analysis (Figure 8b). The optical microscopic examination showed that the dark linear Ub-immunoreactivity occurred along the nuclear membrane of the Hep3B cells treated with 4HPR, but not in the cells treated with the vehicle. Therefore, the electron-dense immunoreaction against Ub in apoptotic cells is consistent with that against the lamina and p62. Furthermore, the electron-dense patch in the nucleus disappeared in cells treated with the apoptogenic drug.
Next, changes in ubH2A and PARP cleavage were measured to determine whether this alternative ubiquitylation of the nuclear envelope depends on caspase activation. The pan-caspase inhibitor, zVAD-fmk, partially blocked the 4HPR-induced apoptosis (15% versus 42%, respectively). The PARP cleavage was proportionally related to the decrease in ubH2A. In other words, the quantity of ubH2A decreased in accordance with levels of PARP cleavage and apoptotic cell death (Figure 8c), showing that the ubiquitylation of the nuclear envelope and lamina depends, at least in part, on activation of the pro-apoptotic caspase. Thus, the caspase-mediated cleavage of nuclear envelope proteins may precede their ubiquitylation.
Discussion
Various forms of Ub, including free Ub and mono- and poly-ubiquitylated proteins, exist in eukaryotic cells.29 The Ub cycles between these pools are mediated by ubiquitylation and deubiquitylation enzymes.32 The dynamic exchange between Ub pools can be visually observed in living cells using a GFP-Ub fusion construct.21 This construct reflects many aspects of the behavior and localization of the endogenous protein that is involved in both mono-Ub protein modification and in the polyubiquitin chains involved in protein degradation.18, 24 Ub and several Ub-like proteins are synthesized as fusion proteins and processed to the mature domain by cleavage at the C-terminal glycine.31
Our study showed upregulation and nuclear translocation of Ub hybrid proteins in various cells exposed to apoptogenic insults. Cells showing ectopic over-expression of the Ub hybrid genes are more susceptible to apoptotic cell death. This suggests that the initial over-ubiquitylation triggered by an apoptogenic insult leading to the aggregation of ubiquitylated proteins (polyubiquitylation) in the nucleus, is harmful to cells. Subsequently, we observed the deubiquitylation of histone H2A and further chromatin condensation. Furthermore, the results were similar in non-tumor cells, including HEK293T and MEF cells (data not shown). Therefore, this altered dynamics of Ub hybrid proteins is thought to be a general phenomenon, not specific for cell types and apoptogenic insults.
Our findings are consistent with those that occur during heat shock, that is, polyubiquitylated material primarily accumulated in the nucleus.18 The proteasome inhibitor MG132 also induces a rapid cytosolic accumulation of GFP-Ub and aggresome formation in the perinuclear region. This is accompanied by a profound loss of nuclear GFP-Ub that corresponds to the redistribution of endogenous Ub. In the nuclear fractions, depletion of H2A ubiquitylation correlates with an accumulation of Ub conjugates, as well as with a shift of the conjugates to higher molecular masses.18 These results support our findings in the cells treated with 4HPR, but do not explain the nuclear redistribution of Ub to the perinuclear region.
Immunofluorescence staining and immuno–electron microscopic examination can be used to trace Ub translocation associated with histone deubiquitylation during apoptosis. The nuclear integral membrane and the nuclear pore complex proteins are ubiquitylated and then form a perichromatin ring-like aggregation. Pretreatment with a caspase inhibitor effectively inhibited histone deubiquitylation and apoptotic cell death. Therefore, cleavage of the nuclear envelope and subsequent ubiquitylation occur downstream of caspase activation.
Over-expression of Ub has been observed in various malignant tumors including astrocytoma, lung cancer, prostate cancer, and HCC.33, 34, 35, 36 Furthermore, the labeling index of Ub is positively correlated with the Ki-67 index of HCC.37 Considering that the deubiquitylation and reubiquitylation of uH2A and uH2B occurs according to the normal change in chromatin structure during cell cycling, Ub levels may reflect the proliferation of tumor cells. In the present study, we observed that most HCC-over-expressed Ub was present primarily in the nuclei (data not shown). This proliferation-related upregulation of Ub seems to increase the susceptibility of tumor cells to apoptosis, compared with nontumor cells. Apoptogenic insults further contribute to extraordinarily high levels of Ub and Ub aggregation in the nuclei of tumor cells, which subsequently results in abnormal ubiquitylation of the nuclear envelope, accompanied by deubiquitylation of nucleosomal histones. This histopathological condition may be associated with microenvironmental stresses or chemotherapy-associated genotoxic injuries.
In conclusion, we found that Ub hybrid proteins target the nucleus of a tumor cell, where they appear to mediate histone mono-ubiquitylation. Various apoptogenic stimuli induce the over-expression of these Ub hybrid proteins by stabilizing the mRNA. This results in an aggregation of ubiquitylated proteins in the nucleus, which disturbs the nucleosomal histone ubiquitylation/deubiquitylation process. Finally, Ub targets the nuclear envelope and the lamina and forms a perichromatin ring-like aggregation, which is controlled in part through events downstream of caspase activation.
Materials and Methods
Cell lines and materials
The human hepatoma (Hep3B and SK-HEP-1), lung (A549), kidney (A498 and ACHN), colon (HT29) and breast (MCF7) cancer cell lines were obtained from the American Tissue Culture Collection (ATCC, Rockville, MD, USA). 4HPR was a generous gift from the Johnson Pharmaceutical Research Institute (Spring House, PA, USA). The 5-FU, PX, and TSA agents were purchased from Sigma-Aldrich Co. (St. Louis, MO, USA). z-VAD was purchased from Calbiochem (San Diego, CA, USA), the goat polyclonal antibodies against human H2A (C-19), human nucleosporin p62 (D-20) and lamin B (C-20), and rabbit polyclonal antibodies against human PARP, H-250 and against human Ub (FL-76) were purchased from Santa Cruz Biotechnology (Santa Cruz, CA, USA). The mouse monoclonal antibody against ubiquityl-H2A (E6C5) and rabbit polyclonal antibody against trimethyl-H3 (Lys27) were purchased from Upstate (Charlottesville, VA, USA). The rabbit polyclonal antibody for human RPSL40 (Uba52) was produced by immunization with a peptide antigen (amino acid 97-109) at Labfrontier (Anyang, South Korea). The monoclonal antibody (M01) against RPS27a (Uba80) was purchased from Abnova (Taipei, Taiwan) and the shRNA lentiviral particles (shRNA Target Set against Uba52), which were transduction-ready for gene silencing, were purchased from Sigma.
Cell culture and detection of apoptotic cells
The cells were cultured in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum (FBS) in air that contained 5% CO2. The percentage of apoptotic cells was evaluated by 4′–6′-diamidino-z-phenylindole (DAPI) or Hoechst 33258 staining, as described previously.19 For flow-cytometric analysis of apoptotic cells, the hypodiploid cells in the DNA histogram were gated using the C-30 program to estimate the sub-G1 fraction. Alternatively, cells were harvested and stained with annexin-V-FITC and PI using the Annexin-V-FITC apoptosis detection kit (BD Bioscience, San Jose, CA, USA) after 4HPR treatment.
Northern blot analysis
The cells were cultured in DMEM with 10% FBS until they reached 60% confluence, and were then treated with 10 or 15 μM 4HPR for the time intervals indicated. Total RNA was extracted and northern blot analysis was performed as described previously.23 To control loading, blots were probed with GAPDH cDNA.
Luciferase assay
The DNA fragment (−945 to +84) of the Uba52 promoter generated by digestion of pUA1.1 was kindly provided by Dr. R Baker.21 The DNA fragment was subcloned into pGL3B at the 5′ SacI and 3′ XhoI sites (pGL3B-Uba52). A PCR-amplified DNA fragment of the Uba80 promoter was generated and cloned to pGL3B at the 5′ KpnI and 3′ BglII sites (pGL3B-Uba80). Hep3B or SK-HEP-1 cells were transfected with the pGL3B-Uba52 construct using lipofectin (Gibco–BRL, Grand Island, NY, USA) as described previously.20 Luciferase activity was measured using the Dual-Luciferase Reporter Assay System (Promega, Madison, WI, USA), according to the manufacturer's instructions, and was normalized by the Renilla luciferase assay.
Histone extraction and immunoblot analysis
The cells were harvested, resuspended in TNESV lysis buffer (50 mM Tris-HCl, pH 7.5, 1% Nonidet P-40 detergent, 2 mM EDTA, 100 mM NaCl, 10 mM orthovanadate) with added protease inhibitors (1 mM phenylmethanesulfonyl fluoride, 20 μg/ml leupeptin, 20 μg/ml aprotinin), and homogenized. The nuclei were collected by centrifugation, and the pellets were resuspended in nuclei-isolation buffer. Sulfuric acid (0.4 N) was added and the suspensions were then incubated on ice for 30 min. The acid-soluble proteins were collected in the supernatant following centrifugation.21 The protein concentrations were determined by the bicinchoninic acid method with bovine serum albumin as the standard.19
Immunofluorescence and immuno–electron microscopy
The Hep3B cells that had been grown on glass coverslips were fixed in 4% paraformaldehyde and permeabilized with 0.2% Triton-X in phosphate-buffered saline (PBS). Immunofluorescence was performed as described previously.19 For immuno–electron microscopy, cells grown on Aclar film (EMS, Hatfield, PA, USA) were coated with 0.01% poly--lysine and fixed with 4% paraformaldehyde in PBS (pH 7.4) for 30 min at room temperature. Immunohistochemistry was then performed with 3,3′-diaminobenzidine for visualization. The thin sections were processed using an osmium-embedding protocol, as described elsewhere.38 The thin sections were not counterstained with uranyl acetate or lead citrate in order to better observe the immunoreactive products in the nucleus. The grids were examined using a Hitachi H7500 electron microscope (Hitachi, Tokyo, Japan).
Combination of DIC and TIRF microscopy
A combined system of DIC and a TIRF microscope with a transmitted all-side polished dove prism was used for direct observation of the N-terminal GFP, C-terminal RFP Ub hybrid proteins in living Hep3B cells. DIC imaging was used to examine the image and the details of the living cell structure, and epifluorescence imaging was used to detect the distribution of the fluorescence as described previously.39 All of the experiments were performed on the all-side polished dove prism without moving the sample. MetaMorph 6.3 software (Universal Imaging Co., Downingtown, PA, USA) was used to collect images of individual molecules and to process the data.
Colony generation assay
The Hep3B cells were co-transfected with the indicated amounts of expression vectors (Uba80, Uba52, Ub or RPS27a) or an empty vector (pCMV-HA, pCMV2-FLAG or pIRES-EGFP) with or without a fixed amount (500 ng) of the pSV2neo selection plasmid. After 24 h of co-transfection, the cells were split and selected in G418 (600 μg/ml) (Gibco) for 2–3 weeks. The resistant colonies were counted after fixing and staining in a solution that contained crystal violet (Sigma) in 20% ethanol.
Lentiviral transduction
A lentivirus vector encoding the shRNA that targeted Uba80 and the shRNA non-target control were purchased from Sigma and were used to transduce the Hep3B cells as instructed by the manufacturer. Briefly, 5 × 104 cells were incubated overnight in a 24-well plate and transduced with the lentiviral particles at a multiplicity of infection (MOI) of 1 in the presence of 8 μg/ml hexadimethrine bromide. Semiquantitative reverse transcription-PCR (RT-PCR) was performed to confirm the knockdown of Uba80 mRNA using a primer set (forward; 5′-GCTGACGCAAACATGCAG-3′, reverse; 5′-TTTGACCTTCTTCTTG-3′).
Plasmids and transfections
The construct of the Uba80 expression vector was created by ligating rat Uba80 (a gift from Dr. Ira G Wool) with pCMV-HA (Clontech, Palo Alto, CA, USA) at each SalI/BglII site in the sense orientation. To create the GFP-Uba80 fusion expression vector, the PCR-amplified rat Uba80 DNA was ligated to pEGFP-C3 or pEGFP-N1 (Clontech) at the SalI/BamHI site in frame. For the GFP-Ub and GFP-RPS27a constructs, the rat Ub gene and the RPS27a gene fragment were PCR-amplified and then ligated to pEGFP-C3 (Clontech) at the SalI/BamHI site. For the pTRE2-Uba80 construct, the fragment of Uba80 DNA from pCMV-HA-Uba80 was ligated to the SalI/EcoRV site of pTRE2 (Clontech). For the pTRE2-Ub construct, the fragment of Ub DNA from pEGFP-Ub was ligated to the HindIII/EcoRV site of pTRE2 (Clontech). Transfections were carried out using lipofectin (Gibco-BRL) according to the manufacturer's instructions. The sequence-verified Uba80, Ub or RPS27a expression vector was co-transfected with pTK-Hyg (Clontech) and cells were selected in the presence of 50 μg/ml hygromycin for 2–3 weeks (or 600 μg/ml G418 for 2–3 weeks for neo-transfectants). Finally, the individual colonies were isolated using cloning rings, expanded and assayed for expression of the transfected gene by northern and western blotting. For the GFP-Uba52 fusion expression vector, the PCR-amplified human Uba52 cDNA was ligated to pEGFP-C3 or pEGFP-N3 (Clontech) at the EcoRI/BamHI site in frame. PCMV2-Flag-Uba52 was generated by PCR amplification of rat Uba52 cloned into the pCMV2-FLAG expression vector at the EcoRI/BamHI site in frame.
Construction of the cleavage-resistant mutants
The expression vector (pGR) with both ends tagged was constructed by ligating pEGFP-C3 (Clontech) with pDsRed2-N1 (Clontech) at each BamHI/HpaI site in the sense orientation and this was then used for the deletion mutants and the N-terminal GFP-tagged vector was used for the point mutants in these experiments. The wild-type rat Uba52 gene was amplified by PCR with the forward primer containing the EcoRI restriction enzyme site (5′-CCGGAATTCTCGCCACCATGCAGATCTTTGTGAAGACCCTC-3′) and the reverse primer containing the AgeI restriction enzyme site (5′-CCGACCGGTGGTTTGACCTTCTTCTTGGGACG-3′). To create the N-terminal deletion mutants (GR-Uba52 Nx), the rat Uba52 gene was PCR-amplified with the primers containing the HindIII or BamHI restriction enzyme sites (forward primer for Uba52 N2, 5′-CTCAAGCTTATGATCTTTGTGAAGACCCTCACT-3′ forward primer for Uba52 N4, 5′-CTCAAGCTTATGGTGAAGACCCTCACTGGCAA-3′ forward primer for Uba52 N6, 5′-CTCAAGCTTATGACCCTCACTGGCAAAACCA-3′ forward primer for Uba52 N10, 5′-CTCAAGCTTATGAAAACCATCACCCTTGAGGTC-3′ forward primer for Uba52 N20, 5′-CTCAAGCTTATGGACACCATTGAGAATGTCAAAG-3′ reverse primer for Uba52 Nx, 5′-GGTGGATCCCGTTTGACCTTCTTCTTGGGAC-3′). The PCR-amplified open-reading frames for Uba52 and Uba52 Nx were subcloned into the pGR vectors. The point mutation was generated using PCR overlap extension technology40 with the mutated PCR primers (5′-GTGTTGCGCCTGCGAGGTGGCAgaAgaGAGCCTTCTCTC-3′ for I77,78R; 5′-GTGTTGCGCCTGCGAGGTGcaAgaATTGAGCCTTCTCTC-3′ for G76A/I77R; 5′-GTGTTGCGCCTGCGAGcaGcaATTATTGAGCCTTCTCTC-3′ for G75,76A; 5′-GTGTTGCGCCTGCGAGGTGGCAgaATTGAGCCTTCTCTC-3′ for I77R; and 5′-GTGTTGCGCCTGCGAGGTGcaATTATTGAGCCTTCTCTC-3′ for G78A). The mutant human Uba52 gene was amplified by PCR with the forward primer containing the EcoRI restriction enzyme site (5′-GGAATTCGCTGACGCAAACATGCAG-3′) and the reverse primer containing the BamHI restriction enzyme site (5′-CGGGATCCTGCCCTTCAACGAAAG-3′). The pGRUba80 expression plasmid tagged at both ends was similarly constructed.
Statistical analysis
The data are presented as the mean±S.E.M. of at least three independent experiments that were each performed in duplicate. All of the data were entered into Microsoft Excel 5.0, and GraphPad Software was used to perform the two-tailed t-tests or analysis of variance where appropriate P-values <0.05 were considered significant.
Acknowledgments
We would like to thank Dr. Ira G Wool (University of Chicago, Chicago, IL, USA) for the generous gift of the rat pUb-S27a-5 and pUb-L40-7 plasmids, Dr. RT Baker (The Australian National University, Canberra, Australia) for kindly providing the DNA fragment (−945 to +84) of the Uba52 promoter, and the Korea Basic Science Institute for use of the laser-scanning microscope. This work was supported by a grant from the Korea Research Foundation funded by the Korean government (Basic Research Promotion Fund, KRF-2008-313-E00434), the National R&D Program for Cancer Control (0620220) and the Korean Health Technology R&D Project (A101834), Ministry for Health, Welfare and Family affairs, Republic of Korea.
Glossary
- DAPI
4′-6′-diamidino-z-phenylindole
- 5-FU
5-fluorouracil
- 4HPR
N-(4-hydroxyphenyl)retinamide
- LB
lamin B
- PARP
poly(ADP-ribose)polymerase
- PX
paclitaxel
- TSA
trichostatin A
- RPL40
ribosomal protein L40
- RPS27a
ribosomal protein S27a
- RT-PCR
reverse transcription-PCR
- shRNA
short hairpin RNA
- Ub
ubiquitin
- ubH2A
ubiquitylated histone H2A
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies the paper on Cell Death and Disease website (http://www.nature.com/cddis)
Edited by RA Knight
Supplementary Material
References
- Warner JR. Ubiquitin. A marriage of convenience or necessity. Nature. 1989;338:379. doi: 10.1038/338379a0. [DOI] [PubMed] [Google Scholar]
- Jentsch S, Seufert W, Hauser HP. Genetic analysis of the ubiquitin system. Biochim Biophys Acta. 1991;1089:127–139. doi: 10.1016/0167-4781(91)90001-3. [DOI] [PubMed] [Google Scholar]
- Webb GC, Baker RT, Coggan M, Board PG. Localization of the human UBA52 ubiquitin fusion gene to chromosome band 19p13.1-p12. Genomics. 1994;19:567–569. doi: 10.1006/geno.1994.1108. [DOI] [PubMed] [Google Scholar]
- Lund PK, Moats-Staats BM, Simmons JG, Hoyt E, D'Ercole AJ, Martin F, et al. Nucleotide sequence analysis of a cDNA encoding human ubiquitin reveals that ubiquitin is synthesized as a precursor. J Biol Chem. 1985;260:7609–7613. [PubMed] [Google Scholar]
- Peterson CL, Laniel MA. Histones and histone modifications. Curr Biol. 2004;14:R546–R551. doi: 10.1016/j.cub.2004.07.007. [DOI] [PubMed] [Google Scholar]
- Zhang Y. Transcriptional regulation by histone ubiquitination and deubiquitination. Genes Dev. 2003;17:2733–2740. doi: 10.1101/gad.1156403. [DOI] [PubMed] [Google Scholar]
- Wang H, Wang L, Erdjument-Bromage H, Vidal M, Tempst P, Jones RS, et al. Role of histone H2A ubiquitination in Polycomb silencing. Nature. 2004;431:873–878. doi: 10.1038/nature02985. [DOI] [PubMed] [Google Scholar]
- Bergink S, Salomons FA, Hoogstraten D, Groothuis TA, de Waard H, Wu J, et al. DNA damage triggers nucleotide excision repair-dependent monoubiquitylation of histone H2A. Genes Dev. 2006;20:1343–1352. doi: 10.1101/gad.373706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai SY, Babbitt RW, Marchesi VT. A mutant deubiquitinating enzyme (Ubp-M) associates with mitotic chromosomes and blocks cell division. Proc Natl Acad Sci USA. 1999;96:2828–2833. doi: 10.1073/pnas.96.6.2828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joo HY, Zhai L, Yang C, Nie S, Erdjument-Bromage H, Tempst P, et al. Regulation of cell cycle progression and gene expression by H2A deubiquitination. Nature. 2007;449:1068–1072. doi: 10.1038/nature06256. [DOI] [PubMed] [Google Scholar]
- Nakagawa T, Kajitani T, Togo S, Masuko N, Ohdan H, Hishikawa Y, et al. Deubiquitylation of histone H2A activates transcriptional initiation via trans-histone cross-talk with H3K4 di- and trimethylation. Genes Dev. 2008;22:37–49. doi: 10.1101/gad.1609708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsui SI, Seon BK, Sandberg AA. Disappearance of a structural chromatin protein A24 in mitosis: implications for molecular basis of chromatin condensation. Proc Natl Acad Sci USA. 1979;76:6386–6390. doi: 10.1073/pnas.76.12.6386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller RD, Yasuda H, Hatch CL, Bonner WM, Bradbury EM. Identification of ubiquitinated histones 2A and 2B in Physarum polycephalum. Disappearance of these proteins at metaphase and reappearance at anaphase. J Biol Chem. 1985;260:5147–5153. [PubMed] [Google Scholar]
- Marushige Y, Marushige K. Disappearance of ubiquitinated histone H2A during chromatin condensation in TGF beta 1-induced apoptosis. Anticancer Res. 1995;15:267–272. [PubMed] [Google Scholar]
- Tanimoto Y, Onishi Y, Hashimoto S, Kizaki H. Peptidyl aldehyde inhibitors of proteasome induce apoptosis rapidly in mouse lymphoma RVC cells. J Biochem. 1997;121:542–549. doi: 10.1093/oxfordjournals.jbchem.a021620. [DOI] [PubMed] [Google Scholar]
- Mimnaugh EG, Chen HY, Davie JR, Celis JE, Neckers L. Rapid deubiquitination of nucleosomal histones in human tumor cells caused by proteasome inhibitors and stress response inducers: effects on replication, transcription, translation, and the cellular stress response. Biochemistry. 1997;36:14418–14429. doi: 10.1021/bi970998j. [DOI] [PubMed] [Google Scholar]
- Mimnaugh EG, Kayastha G, McGovern NB, Hwang SG, Marcu MG, Trepel J, et al. Caspase-dependent deubiquitination of monoubiquitinated nucleosomal histone H2A induced by diverse apoptogenic stimuli. Cell Death Differ. 2001;8:1182–1196. doi: 10.1038/sj.cdd.4400924. [DOI] [PubMed] [Google Scholar]
- Dantuma NP, Groothuis TA, Salomons FA, Neefjes J. A dynamic ubiquitin equilibrium couples proteasomal activity to chromatin remodeling. J Cell Biol. 2006;173:19–26. doi: 10.1083/jcb.200510071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park JH, Liu L, Kim IH, Kim JH, You KR, Kim DG. Identification of the genes involved in enhanced fenretinide-induced apoptosis by parthenolide in human hepatoma cells. Cancer Res. 2005;65:2804–2814. doi: 10.1158/0008-5472.CAN-04-2221. [DOI] [PubMed] [Google Scholar]
- Kirschner LS, Stratakis CA. Structure of the human ubiquitin fusion gene Uba80 (RPS27a) and one of its pseudogenes. Biochem Biophys Res Commun. 2000;270:1106–1110. doi: 10.1006/bbrc.2000.2568. [DOI] [PubMed] [Google Scholar]
- Baker RT, Board PG. The human ubiquitin-52 amino acid fusion protein gene shares several structural features with mammalian ribosomal protein genes. Nucleic Acids Res. 1991;19:1035–1040. doi: 10.1093/nar/19.5.1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernhard D, Ausserlechner MJ, Tonko M, Löffler M, Hartmann BL, Csordas A, et al. Apoptosis induced by the histone deacetylase inhibitor sodium butyrate in human leukemic lymphoblasts. FASEB J. 1999;13:1991–2001. doi: 10.1096/fasebj.13.14.1991. [DOI] [PubMed] [Google Scholar]
- Nijman SM, Luna-Vargas MP, Velds A, Brummelkamp TR, Dirac AM, Sixma TK, et al. A genomic and functional inventory of deubiquitinating enzymes. Cell. 2005;123:773–786. doi: 10.1016/j.cell.2005.11.007. [DOI] [PubMed] [Google Scholar]
- Qian SB, Ott DE, Schubert U, Bennink JR, Yewdell JW. Fusion proteins with COOH-terminal ubiquitin are stable and maintain dual functionality in vivo. J Biol Chem. 2002;277:38818–38826. doi: 10.1074/jbc.M205547200. [DOI] [PubMed] [Google Scholar]
- Raasi S, Schmidtke G, Groettrup M. The ubiquitin-like protein FAT10 forms covalent conjugates and induces apoptosis. J Biol Chem. 2001;276:35334–35343. doi: 10.1074/jbc.M105139200. [DOI] [PubMed] [Google Scholar]
- Baarends WM, Hoogerbrugge JW, Roest HP, Ooms M, Vreeburg J, Hoeijmakers JH, et al. Histone ubiquitination and chromatin remodeling in mouse spermatogenesis. Dev Biol. 1999;207:322–333. doi: 10.1006/dbio.1998.9155. [DOI] [PubMed] [Google Scholar]
- Soldatenkov VA, Dritschilo A. Apoptosis of Ewing's sarcoma cells is accompanied by accumulation of ubiquitinated proteins. Cancer Res. 1997;57:3881–3885. [PubMed] [Google Scholar]
- Gruenbaum Y, Wilson KL, Harel A, Goldberg M, Cohen M. Review: nuclear lamins – structural proteins with fundamental functions. J Struct Biol. 2000;129:313–323. doi: 10.1006/jsbi.2000.4216. [DOI] [PubMed] [Google Scholar]
- Duband-Goulet I, Courvalin JC, Buendia B. LBR, a chromatin and lamin binding protein from the inner nuclear membrane, is proteolyzed at late stages of apoptosis. J Cell Sci. 1998;111:1441–1451. doi: 10.1242/jcs.111.10.1441. [DOI] [PubMed] [Google Scholar]
- Buendia B, Santa-Maria A, Courvalin JC. Caspase-dependent proteolysis of integral and peripheral proteins of nuclear membranes and nuclear pore complex proteins during apoptosis. J Cell Sci. 1999;112:1743–1753. doi: 10.1242/jcs.112.11.1743. [DOI] [PubMed] [Google Scholar]
- Gotzmann J, Vlcek S, Foisner R. Caspase-mediated cleavage of the chromosome-binding domain of lamina-associated polypeptide 2 alpha. J Cell Sci. 2000;113:3769–3780. doi: 10.1242/jcs.113.21.3769. [DOI] [PubMed] [Google Scholar]
- Haas AL, Bright PM. The dynamics of ubiquitin pools within cultured human lung fibroblasts. J Biol Chem. 1987;262:345–351. [PubMed] [Google Scholar]
- Galloway PG, Likavec MJ. Ubiquitin in normal, reactive and neoplastic human astrocytes. Brain Res. 1989;50:343–351. doi: 10.1016/0006-8993(89)90330-2. [DOI] [PubMed] [Google Scholar]
- Kanayama H, Tanaka K, Aki M, Kagawa S, Miyaji H, Satoh M, et al. Changes in expressions of proteosome and ubiquitin genes in human renal cancer cells. Cancer Res. 1991;51:6677–6685. [PubMed] [Google Scholar]
- Ishibashi Y, Takada K, Joh K, Ohkawa K, Aoki T, Matsuda M. Ubiquitin immunoreactivity in human malignant tumors. Br J Cancer. 1991;63:320–322. doi: 10.1038/bjc.1991.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marin F, Cheng Z, Kovacs K. Ubiquitin immunoreactivity in corticotrophs following glucocorticoid treatment and in pituitary adenomas. Arch Pathol Lab Med. 1993;117:254–258. [PubMed] [Google Scholar]
- Shirahashi H, Sakaida I, Terai S, Hironaka K, Kusano N, Okita K. Ubiquitin is a possible new predictive marker for the recurrence of human hepatocellular carcinoma. Liver. 2002;22:413–418. doi: 10.1034/j.1600-0676.2002.01541.x. [DOI] [PubMed] [Google Scholar]
- Bae YC, Ahn HJ, Park KP, Kim HN, Paik SK, Bae JY, et al. The synaptic microcircuitry associated with primary afferent terminals in the interpolaris and caudalis of trigeminal sensory nuclear complex. Brain Res. 2005;1060:118–125. doi: 10.1016/j.brainres.2005.08.042. [DOI] [PubMed] [Google Scholar]
- Lee S, Choi JS, Kang SH. Combination of differential interference contrast with prism-type total internal fluorescence microscope for direct observation of polyamidoamine dendrimer nanoparticle as a gene delivery in living human cells. J Nanosci Nanotechnol. 2007;7:3689–3694. [PubMed] [Google Scholar]
- Ho SN, Hunt HD, Horton RM, Pullen JK, Pease LR. Site-directed mutagenesis by overlap extension using the polymerase chain reaction. Gene. 1989;77:51–49. doi: 10.1016/0378-1119(89)90358-2. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.