Abstract
This study sought to determine whether the quality of enzyme preparations can be determined from their melting curves, which may easily be obtained using a fluorescent probe and a standard RT-PCR machine. Thermal melt data on 31 recombinant enzymes from Plasmodium parasites were acquired by incrementally heating them to 90 °C and measuring unfolding with a fluorescent dye; activity assays specific to each enzyme were also performed. Four of the enzymes were denatured to varying degrees with heat and SDS prior to the thermal melt and activity assays. In general, melting curve quality correlated with enzyme activity; enzymes with high-quality curves were found almost uniformly to be active, while those with lower-quality curves were more varied in their catalytic performance. Inspection of melting curves of bovine xanthine oxidase and Entamoeba histolytica cysteine protease 1 allowed active stocks to be distinguished from inactive stocks, implying that a relationship between melting curve quality and activity persists over a wide range of experimental conditions and species. Our data suggest that melting curves can help to distinguish properly folded proteins from denatured ones and therefore may be useful in selecting stocks for further study and in optimizing purification procedures for specific proteins.
Keywords: thermal melting, malaria, protein denaturation
Thorough characterization of a purified protein requires that it be in its naturally folded (native) state. Functional assays such as enzyme activity assays can indicate whether or not a protein is well-folded; for any given protein, however, an assay may not exist or may require expensive ingredients, extensive sample processing, and/or complex instrumentation. Moreover, as numerous organisms undergo genome-wide characterization, there will be increasing interest in previously obscure (and therefore difficult-to-assay) proteins, including those whose functions are not yet known.
Given the limitations of functional assays as indicators of protein folding, an alternative method for assessing folding status could be quite useful. Thermal melting with fluorescent dye that binds to a protein’s exposed hydrophobic regions [1] might be such a method; a large heat-induced increase in fluorescence suggests that the protein was well folded prior to heating [2]. To our knowledge, though, it is not yet clear whether, among a variety of proteins, melting curve properties can be correlated with an independent readout of folding status.
In the present study, stocks of more than 30 enzymes – mostly from Plasmodium parasites, which cause malaria – were characterized via thermal melt and activity assays. Our goal was to determine whether enzymes’ melting curves are predictive of activity, the latter being a proxy for folding state. Our results suggest that melting curves are indeed a convenient indicator of whether enzymes – and other proteins, presumably – are properly folded.
MATERIALS AND METHODS
Enzyme sources, production, and purification
Recombinant histidine-tagged enzymes from Plasmodium berghei, P. falciparum, P. knowlesi, and P. vivax were expressed in E. coli and purified by immobilized metal affinity chromatography essentially as described previously [3]. GTP cyclohydrolase from E. coli [4] was obtained in a similar manner, while procedures for purification and refolding of cysteine protease 1 from Entamoeba histolytica were based on those of a previous study [5]. Xanthine oxidase from Bos taurus and other non-Plasmodium enzymes used in coupled activity assays (glucose-6-phosphate dehydrogenase, lactate dehydrogenase, phosphogluconate dehydrogenase, pyrophosphatase, and pyruvate kinase) were purchased from Sigma-Aldrich.
Acquisition and quantitation of thermal melt data
Melting curves of enzyme stocks were obtained from samples in 96-well plates using a DNA Engine Opticon 2 (manufactured by MJ Research, now part of Bio-Rad) and the fluorescent probe SYPRO Orange (Invitrogen), as described previously [3]. In brief, a heating rate of ~1.2 °C/minute was used, and fluorescence readings (excitation at 530±30 nm, emission at 575±20 nm) were taken after each 0.2-degree increase. Plasmodium enzymes were diluted both in a standard thermal melt buffer (100 mM HEPES, 150 mM NaCl, pH 7.5) and in the enzyme-specific activity assay buffers listed in Table 1. Xanthine oxidase from B. taurus was heated in the buffers also used for superoxide dismutase, while cysteine protease 1 from E. histolytica was heated in a buffer of 20 mM Tris, 250 mM NaCl, 5% Glycerol, and 125 mM L-Arginine (pH 8.0). Each melting curve was assigned a quality score (Q) calculated as Q = ΔFmelt/ΔFtotal, where ΔFmelt is the melting-associated increase in fluorescence and ΔFtotal is the total range in fluorescence observed between 20 and 90 °C (i.e., the difference between the minimum and maximum values recorded over the 20°-to-90° span). The range of possible Q’s is 0 to 1, with 0 designating an absence of discernable melting behavior and 1 representing a high-quality melting curve.
Table 1.
Plasmodium enzymes studied by thermal melt and activity assays
| Enzymea | Gene IDb | EC number | Buffer for activity assayc | Q (standard buffer/enzyme-specific buffer) | Specific activity (present study/literature) in μmol/(min*mg)d |
|---|---|---|---|---|---|
| 6-phosphogluconolactonase | PF14_0511 | 3.1.1.31 | 25 mM HEPES, 2 mM MgCl2, pH 7.1 [9] | 1.0/1.0 | 145/10.2–60 [8,25] |
| 6-pyruvoyltetrahydropterin synthase | PFF1360w | 4.2.3.12 | 50 mM Tris, 100 mM KCl, 100 μM MgCl2, 2 mM DTT, pH 8.0 [26] | 1.0/0.97 | >0/>0 [26] |
| 6-pyruvoyltetrahydropterin synthase (P. vivax) | PVX_114505 | 4.2.3.12 | 50 mM Tris, 100 mM KCl, 10 mM MgCl2, 2 mM DTT, pH 8.0 [26] | 1.0/1.0 | >0/>0 [26] |
| Adenosine deaminase | PF10_0289 | 3.5.4.4 | 20 mM potassium phosphate, 1 μM EDTA, pH 7.0 [27] | 0.05/0.15 | 11/>0 [28] |
| Adenosine deaminase (P. vivax) | PVX_111245 | 3.5.4.4 | 20 mM potassium phosphate, 1 μM EDTA, pH 7.0 [27] | 0.63/0.75 | 100/>0 [28] |
| Adenylosuccinate lyase (P. vivax) | PVX_114710 | 4.3.2.2 | 50 mM potassium phosphate, pH 7.4 [29] | 1.0/1.0 | 14/5.7–8.0 [29] |
| Adenylosuccinate synthetase | PF13_0287 | 6.3.4.4 | 50 mM sodium phosphate, 5 mM MgCl2, pH 7.5 [30] | 1.0/1.0 | 0.9/1.14 [30] |
| Aspartate carbamoyltransferase (P. vivax) | PVX_083135 | 2.1.3.2 | 50 mM Tris, 1 mM EDTA, 1 mM mercaptoethanol, 400 fM BSA, pH 8.0 [31] | 0.92/1.0 | >0/NA |
| Choline kinase | PF14_0020 | 2.7.1.32 | 100 mM Glycine-NaOH, 150 mM KCl, 6 mM MgCl2, pH 9.2 [6] | 1.0/0.82 | 5/0.8 [6] |
| D-ribulose-5-phosphate 3- epimerase | PFL0960w | 5.1.3.1 | 300 mM triethanolamine, pH 7.4 [32] | 0.91/0.83 | 30/NA |
| Deoxyribose-phosphate aldolase (P. yoelii) | PY02252 | 4.1.2.4 | 100 mM triethanolamine, pH 7.5 [33] | 1.0/1.0 | 4.2/NA |
| Dihydrofolate synthase | PF13_0140 | 6.3.2.12 | 86 mM Tris, 100 mM KCl, 4 mM MgCl2, pH 8.0 [34,35] | 0.05/0.06 | none/NA |
| dUTPase | PF11_0282 | 3.6.1.23 | 2.5 mM MES, 100 mM KCl, 5 mM MgCl2, 2.5 mM DTT, pH 6.2 [7] | 1.0/1.0 | 63/25.8 [7] |
| Farnesyl pyrophosphate synthase (P. vivax) | PVX_092040 | 2.5.1.10 | 25 mM HEPES, 2.5 mM MgCl2, pH 7.4 [36] | 0.78/0.81 | 0.013/>0 [36] |
| Glutamate dehydrogenase, NADP-specific | PF14_0164 | 1.4.1.4 | 100 mM potassium phosphate, 1 mM EDTA, pH 8 [37] | 1.0/0.81 | 4.2/9 [37] |
| Glyceraldehyde-3-phosphate dehydrogenase | PF14_0598 | 1.2.1.12 | 40 mM triethanolamine, 50 mM Na2HPO4, pH 7.6 [38] | 1.0/1.0 | 23/90–120 [38] |
| Glycerol-3-phosphate dehydrogenase | PFL0780w | 1.1.1.8 | 300 mM triethanolamine, pH 7.4 [39] | 1.0/1.0 | 6/NA |
| Guanylate kinase (P. vivax) | PVX_099895 | 2.7.4.8 | 50 mM Tris, 50 mM KCl, 2 mM MgCl2, pH 7.5 [13] | 1.0/1.0 | 350/750 [40] |
| Hydroxymethyl-dihydropterin pyrophosphokinase (P. vivax) | PVX_123230 | 2.7.6.3 | 100 mM Tris, 100 mM mercaptoethanol, 10 mM MgCl2, pH 9.0 [41] | 0.44/0.18 | 0.01/0.01 [41] |
| Methionine adenosyltransferase | PFI1090w | 2.5.1.6 | 100 mM Tris, 150 mM KCl, 20 mM MgCl2, 5 mM mercaptoethanol, pH 8.2 [42] | 0.44/0.49 | none/>0 [42] |
| Methionine aminopeptidase 1 | PF10_0150 | 3.4.11.18 | 25 mM HEPES, 150 mM KCl, pH 7.5 [15] | 1.0/1.0 | 0.02*/0.23 [14] |
| Methionine aminopeptidase 2 | PF14_0327 | 3.4.11.18 | 25 mM HEPES, 150 mM KCl, pH 7.5 [15] | 0.00/0.00 | none/0 [23] |
| N-myristoyltransferase | PF14_0127 | 2.3.1.97 | 20 mM Tris, 100 mM NaCl, 1 mM EDTA, pH 8.0 [43] | 1.0/1.0 | 0.14*/>0 [44] |
| Nucleoside diphosphate kinase B | PF13_0349 | 2.7.4.6 | 50 mM Tris, 40 mM KCl, 2 mM MgCl2, pH 7.2 [45] | 1.0/1.0 | 60*/1880 [45] |
| Orotidine 5′ monophosphate decarboxylase | PF10_0225 | 4.1.1.23 | 25 mM MOPS, 5% glycerol, pH 7.2 [46] | 1.0/1.0 | 11/2.7 [46] |
| Phosphoethanolamine N-methyltransferase | MAL13P1.214 | 2.1.1.103 | 100 mM HEPES, 10% glycerol, 2 mM EDTA, pH 8.6 [47] | 0.38/0.34 | 0.0001/0.0012 [47] |
| Phosphoglycerate kinase | PFI1105w | 2.7.2.3 | 100 mM triethanolamine, 5 mM MgSO4, 1 mM EDTA, 1 mM DTT, pH 7.6 [48] | 1.0/1.0 | none/210[48] |
| Ribose 5-phosphate isomerase | PFE0730c | 5.3.1.6 | 300 mM triethanolamine, pH 7.4 [32] | 1.0/1.0 | 2100/NA |
| S-adenosyl-homocysteine hydrolase | PFE1050w | 3.3.1.1 | 25 mM potassium phosphate, pH 7.2 [49] | 0.45/0.47 | 0.028/0.03 [49] |
| Superoxide dismutase (P. berghei) | PB000490.02.0 | 1.15.1.1 | 50 mM potassium phosphate, 0.1 mM EDTA, pH 7.8 [19] | 1.0/1.0 | 1400*/1631 [19] |
| Superoxide dismutase (P. knowlesi) | PKH_142350 | 1.15.1.1 | 50 mM potassium phosphate, 0.1 mM EDTA, pH 7.8 [19] | 1.0/1.0 | 1400*/1631 [19] |
All enzymes are from P. falciparum unless otherwise noted.
Gene IDs are as assigned by PlasmoDB.org.
Activity assays were performed according to the references given; exceptions and clarifications are noted in the Materials and Methods section.
Literature values for specific activities are taken from previous studies of the same enzyme or an ortholog within the Plasmodium genus. Asterisks in the specific activity column indicate differences in assay substrates between previous studies and the present one (see Materials and Methods). A specific activity of “NA” indicates that a literature value is not available; a value of “>0” means that the enzyme is active, but the specific activity was not reported in the literature (and/or in the present study) could not be determined because of a lack of calibration standards. Approximate assay detection limits for the enzymes with no detectable activity were 0.0002 μmol/(min*mg) for dihydrofolate synthase, 0.003 μmol/(min*mg) for methionine adenosyltransferase, 0.005 μmol/(min*mg) for methionine aminopeptidase 2, and 0.0005 μmol/(min*mg) for phosphoglycerate kinase.
Activity assays
All activity assays were done at room temperature (20–25 °C). Instruments used included a BioSpec-1601 spectrophotometer made by Shimadzu (Kyoto, Japan) and ELx800 and FLx800 microplate readers made by BioTek Instruments (Winooski, VT). Whenever possible, enzymes were assayed according to precedents in the literature (see references in Table 1), using substrate concentrations well above the corresponding Km’s. Enzymes were assayed in the direction corresponding to their names (e.g., with glutamate dehydrogenase, we measured the formation of NADPH corresponding to the dehydrogenation of glutamate, not the consumption of NADPH by the hydrogenation of 2-oxoglutarate) unless specified otherwise. The high bovine serum albumin concentrations (0.5–1.25 mg/mL) used in previous activity assays of choline kinase [6] and dUTPase [7] were omitted from our thermal melt and activity assays, since they interfere with the fluorescent readout of the thermal melt assay. Other exceptions and clarifications are as follows.
6-phosphogluconolactonase: A previous report on the P. berghei enzyme [8] did not specify the assay buffer used. The buffer listed in Table 1 was taken from a reference [9] cited by that report.
6-pyruvoyltetrahydropterin synthase. Activity was confirmed via a coupled assay in which 7,8-dihydroneopterin triphosphate, the substrate of 6-pyruvoyltetrahydropterin synthase, was generated from GTP by recombinant GTP cyclohydrolase from E. coli. A time-dependent increase in fluorescence (excitation at 360 nm, emission at 460 nm) beyond that seen with GTP and GTP cyclohydrolase alone was observed when 6-pyruvoyltetrahydropterin synthase was added, and the extent of the increase above the baseline rate was proportional to the concentration of 6-pyruvoyltetrahydropterin synthase.
Choline kinase. Choline-dependent ATP consumption was measured with Kinase-Glo [10].
Cysteine protease 1. Measurement of the release of fluorescent 7-amino-4-methyl coumarin (AMC) from the substrate Z–Arg–Arg–AMC was based upon a previous report [11]. The buffer used was 100 mM citric acid-NaHPO4, 5 mM DTT, 2 mM EDTA, pH 6.0.
Dihydrofolate synthase. The dihydropteroate-dependent production of inorganic phosphate was measured with malachite green [12].
dUTPase and farnesyl pyrophosphate synthase. Pyrophosphatase from S. cerevisiae was used to cleave pyrophosphate (a product of both enzymes) into inorganic phosphate, which was detected with malachite green [12].
Glycerol-3-phosphate dehydrogenase. The reaction was studied in the NADH-consuming direction.
Guanylate kinase. The previously published activity assay protocol [13] suggests a buffer including 0.5 mM EDTA and 5.7 mM MgSO4, while the present study used no EDTA and 2 mM MgCl2.
Hydroxymethyldihydropterin pyrophosphokinase. Hydroxymethyldihydropterin-dependent ATP consumption was measured with Kinase-Glo [10].
Methionine adenosyltransferase. Production of inorganic phosphate was measured with malachite green [12].
Methionine aminopeptidase 1. A previous characterization of the Plasmodium enzyme used a dipeptide substrate [14], whereas the present study used L-methionine-p-nitroanilide as the substrate [15].
N-myristoyltransferase. The peptide substrate used was GSSYSRKNK, which was based on the N-terminal sequence of adenylate kinase 2, a substrate of the P. falciparum N-myristoyltransferase in vivo [16]. Production of coenzyme A was measured with ThioGlo [17], which fluoresces (excitation at 379 nm, emission at 513 nm) upon reacting with free sulfhydryl groups.
Nucleoside diphosphate kinase. A previous study of the Plasmodium enzyme used thymidine 5′-diphosphate (TDP) and ATP as substrates, whereas the present study used uridine 5′-diphosphate (UDP) and ATP. Activity was quantified as the rate of UDP-dependent ATP consumption, as measured with Kinase-Glo [10].
Phosphoethanolamine N-methyltransferase. Production of S-adenosylhomocysteine was detected via the coupling enzymes S-adenosylhomocysteine hydrolase (which converts S-adenosylhomocysteine to homocysteine and adenosine) and adenosine deaminase (which converts adenosine to inosine). The concentration of S-adenosylhomocysteine produced was calculated from the slope of the decrease in absorbance at 267 nm representing conversion of adenosine to inosine, based on a standard curve of ΔA267 vs. [S-adenosylhomocysteine].
Phosphoglycerate kinase. Phosphoglycerate-dependent ATP consumption was measured with Kinase-Glo [10].
Superoxide dismutase. A previous study of the Plasmodium enzyme measured inhibition of the autooxidation of pyrrogallol [18], whereas the present study measured inhibition of the reduction of cytochrome c [19].
Xanthine oxidase. Reduction of cytochrome c was tracked as described in the protocol for superoxide dismutase, using the buffer described therein [19].
Pre-assay denaturation of four representative enzymes
Adenosine deaminase (from P. vivax), glyceraldehyde-3-phosphate dehydrogenase, methionine aminopeptidase 1, and orotidine 5′-monophosphate decarboxylase were studied at various levels of denaturation by subjecting them to heating and the detergent sodium dodecyl sulfate (SDS) prior to thermal melt and activity assays. These enzymes were chosen because they cover a range of chemical reactions (oxidoreductase, hydrolase, and lyase, corresponding to EC groups 1, 3, and 4), numbers of subunits (monomer, dimer, and tetrameter), and grand averages of hydropathy (GRAVY; −0.045 to −0.482). The specific conditions (temperature and duration of pre-heating and concentrations of SDS) were empirically adjusted for each enzyme so that a wide range of activities and melting curve qualities could be observed within each dataset. Heating was done at 58 °C for 0, 2, 5, 10 and 20 minutes with adenosine deaminase; 50 °C for 0, 2, 5, 10, and 20 minutes with glyceraldehyde-3-phosphate dehydrogenase; 50 °C for 0, 2, 10, and 20 minutes with methionine aminopeptidase 1; and 61 °C for 1, 10, 20, and 40 minutes with orotidine 5′-monophosphate decarboxylase. SDS concentrations tested were 0%, 0.0125%, 0.02%, 0.03%, and 0.04% for adenosine deaminase; 0%, 0.01%, 0.0125%, 0.02%, and 0.03% for glyceraldehyde-3-phosphate dehydrogenase; 0%, 0.0156%, 0.031%, 0.0625%, and 0.125% for methionine aminopeptidase 1; and 0%, 0.01%, 0.02%, and 0.04% for orotidine 5′-monophosphate decarboxylase. Pre-assay denaturation of orotidine 5′-monophosphate decarboxylase was also attempted through repeated thawing/refreezing and with various concentrations of guanidine hydrochloride and urea.
RESULTS
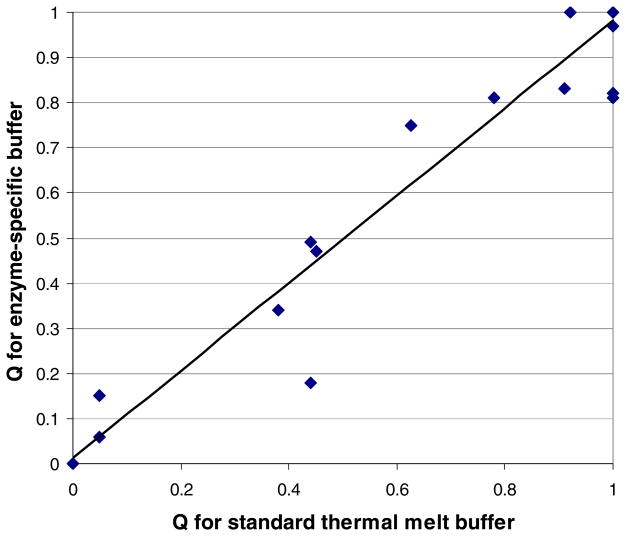
Melting curves were collected for 31 Plasmodium enzymes, both in a standard buffer commonly used for thermal melt assays (100 mM HEPES, 150 mM KCl, pH 7.5) [3,20] and in each enzyme’s activity assay buffer (listed in Table 1). There was a strong correlation (R2 = 0.945) between an enzyme’s Q (which quantifies melting curve quality; see Materials and Methods) in the standard buffer and its Q in activity assay buffer (Figure 1).
FIGURE 1.
Melting curve quality is similar in standard and enzyme-specific buffers. Q’s were calculated as described in Materials and Methods; each score is an average of at least five to eight replicate wells. Each data point represents a separate Plasmodium enzyme; many enzymes are represented by the point at (1,1). The best-fit line is: y = 0.97x + 0.01 (R2 = 0.94).
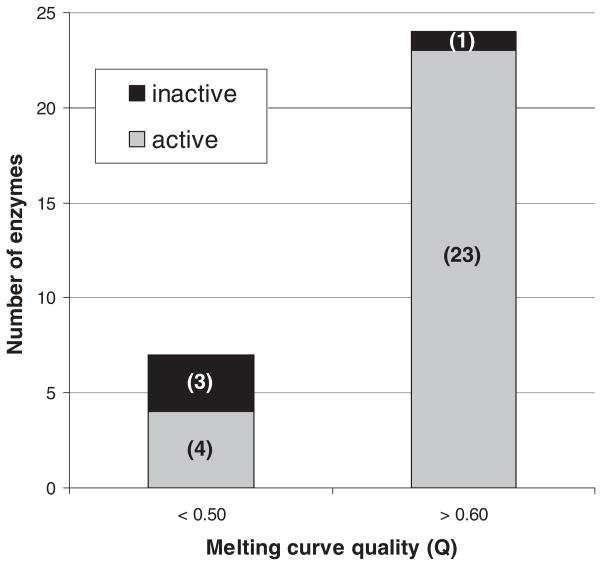
Most enzymes’ melting curves included a temperature span over which fluorescence increased substantially (presumably due to heat-induced unfolding), resulting in Q’s above 0.60. Of these 24 high-Q enzymes, 23 were catalytically active (Figure 2). Fluorescence increases due to melting were small for seven enzymes, leading to Q’s below 0.50. Among these seven, only four were found to be active (Figure 2). Another way of summarizing these results is to say that, of the four inactive enzymes, all but one had Q’s below 0.50. These data suggest that a good melting curve, as indicated by a high Q, is predictive of catalytic activity.
FIGURE 2.
Plasmodium enzymes with high Q’s (>0.60) are almost always active, whereas those with lower Q’s (<0.50) are more variable.
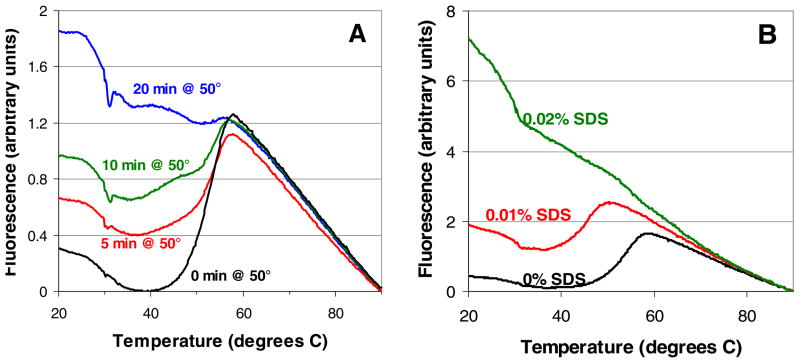
As a further exploration of the relationship between melting curve quality and enzyme activity, we studied how both change in response to varying degrees of pre-assay denaturation. Preliminary experiments gauged the response of orotidine 5′-monophosphate decarboxylase to five possible causes of denaturation: prolonged heating, repeated thawing/refreezing, and exposure to sodium dodecyl sulfate (SDS), guanidine hydrochloride, and urea. Repeated freezing/thawing (5–10 cycles) and exposure to urea (1.8 M and below) did not cause significant changes in enzyme behavior, and guanidine hydrochloride was found to be a competitive inhibitor of the enzyme at sub-denaturing concentrations (data not shown). However, preheating and SDS were found to denature the enzyme effectively and unambiguously, so these perturbations were then used to study three additional Plasmodium enzymes: adenosine deaminase (from P. vivax), glyceraldehyde-3-phosphate dehydrogenase, and methionine aminopeptidase 1. Typical changes in melting curves in response to preheating and SDS are shown in Figure 3. As the [SDS] or duration of preheating increased, the initial fluorescence (at 20 °C) increased and the change in fluorescence associated with melting (ΔFmelt) decreased. The melting temperature (Tm), defined as the temperature at which the fluorescence increases most steeply [3], decreased in response to SDS but not in response to preheating. These trends were similar for all four enzymes.
FIGURE 3.
Melting curves for glyceraldehyde-3-phosphate dehydrogenase in activity assay buffer in response to (A) preheating and (B) SDS. Data shown are typical results from single wells.
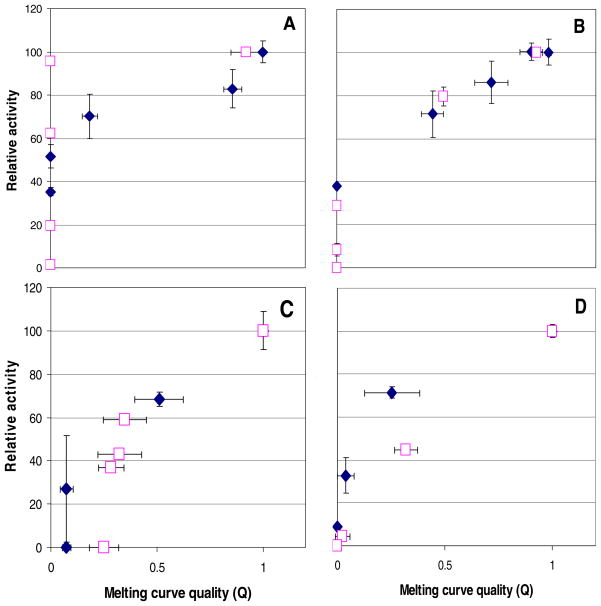
Figure 4 shows the relationships between Q’s and activities of the four enzymes denatured with preheating and with SDS. A full range of conditions was employed, up to and including those producing activities at or near 0%. It is evident that in many instances, a treated enzyme can suffer substantial deterioration of its melting curves, as indicated by Q’s at or near 0, but still retain significant activity. The denaturation of adenosine deaminase by SDS is the clearest example of this somewhat surprising trend. Nevertheless, the general pattern suggested by Figure 2 also applies here: high-Q samples were quite active, whereas the activities of low-Q samples were harder to predict.
FIGURE 4.
Q’s and activities for (A) adenosine deaminase, (B) glyceraldehyde-3-phosphate dehydrogenase, (C) methionine aminopeptidase 1, and (D) orotidine 5′-monophosphate decarboxylase denatured by heat (solid diamonds) and SDS (open squares). Error bars represent standard deviations.
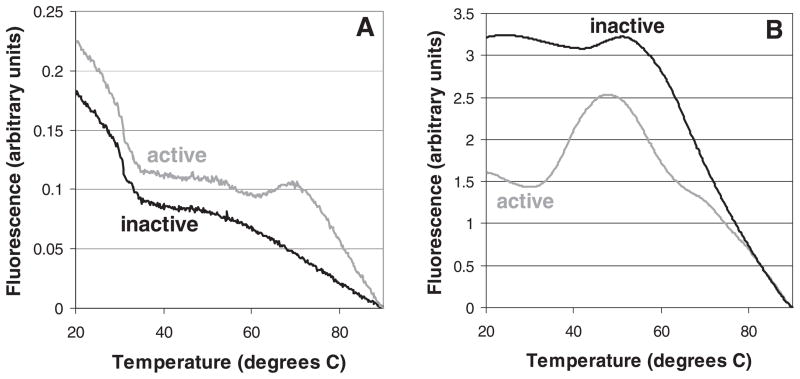
Two limitations of the data presented so far are that (a) only Plasmodium enzymes were studied and (b) the denaturation of enzymes as explored in Figures 3 and 4 was achieved artificially (i.e., with conditions to which valuable proteins are unlikely to be subjected). Can Q’s be used to predict whether enzymes from any organism are active, even when normal measures are taken to preserve their functional 3D shape? As a preliminary look at this question, we studied pairs of stocks of xanthine oxidase from Bos taurus (purchased from Sigma-Aldrich) and cysteine protease 1 from E. histolytica (expressed in E. coli by our lab). Both stocks of xanthine oxidase had been stored at 4 °C as recommended by the manufacturer, but at the time of these experiments, one stock was 14 months old, while the other was only 4 months old. The melting curve of the older stock did not show a melting-related increase in fluorescence (Q=0.00), whereas the newer stock showed a small but distinct increase between 60 and 70 °C (Q=0.05; Figure 5A). When both stocks were tested for catalytic activity, only the newer stock was found to be active (with a specific activity of 0.04 μmol/(min*mg); detection limit ≈ 0.004 μmol/(min*mg)). Similar assays were performed with two stocks of cysteine protease 1, one of which had been refolded with buffers optimized for this particular enzyme. The stock to which optimal refolding buffers had been applied gave a melting curve with a middling Q (0.40) and was active, whereas the other stock had a low Q (0.10; Figure 5B) and was not active (detection limit not known).
FIGURE 5.
Thermal melt curves of catalytically active and inactive stocks of (A) xanthine oxidase from Bos taurus and (B) cysteine protease 1 from Enamoeba histolytica. Data shown are typical results from single wells.
DISCUSSION
This paper presents evidence that enzymes with high-quality thermal melt curves, as indicated by high Q’s, are more likely to be catalytically active than enzymes with poor melting curves. The relationship between melting curves and activity has been examined in three sets of enzymes: 31 recombinant Plasmodium enzymes expressed and purified with standard protocols (Figure 2), a subset of four Plasmodium enzymes artificially denatured with heat and SDS (Figures 3–4), and two pairs of non-Plasmodium enzymes (Figure 5). These datasets collectively indicate that, the higher the Q of an enzyme stock, the more likely it is to be active. The implication is that melting curves are a reasonable indicator of whether a protein is denatured or natively folded, and that thermal melt assays could be used to assess the folding status of noncatalytic proteins as well as enzymes.
Our conclusion that melting curves indicate protein folding status comes with at least four caveats. First, our study focused entirely on enzymes (rather than noncatalytic proteins) and mainly on Plasmodium enzymes, whose amino acid composition differs from that of most organisms due to the AT richness of the Plasmodium genome [21]. Second, melting curves reveal only whether hydrophobic residues are exposed to surrounding solvent and thus cannot distinguish between proteins that are properly folded and proteins that are misfolded in a way that nonetheless shields their hydrophobic regions from solvent. The inactivity of our stock of phosphoglycerate kinase could, in theory, be due to this type of misfolding. Third, an enzyme’s overall 3D structure is only one of several determinants of its activity; for instance, the absence of a cofactor or point mutations in the active site may not significantly alter an enzyme’s gross structure but may render it inactive. As an example, a methionine aminopeptidase 1 stock purified with cobalt rather than manganese gave good melting curves (Q = 1) but was inactive unless manganese, its probable cofactor in vivo [22], was added to the assay buffer (data not shown). Fourth, we have not proven that ΔFmelt/ΔFtotal – the ratio of the melting-associated rise in fluorescence to the total range of fluorescence values observed between 20 and 90 °C – is the best possible measure of melting curve quality. Data such as those in Figure 3 show that, as an enzyme becomes more and more denatured, its ΔFmelt tends to decrease, while its initial fluorescence at 20 °C (and usually ΔFtotal) tends to increase. Calculating Q as the ratio of ΔFmelt to ΔFtotal is a simple and logical way of capturing both trends; however, alternative metrics for rating melting curves may be equally appropriate, depending on the goals of one’s analysis.
In measuring specific activities of our Plasmodium enzymes, we followed precedents reported in the literature whenever possible so that our values could be compared to previously reported values. These comparisons (Table 1) did not reveal any obvious patterns; for example, low-Q enzymes did not consistently have specific activities well below literature values. This may not be surprising, since, aside from possible lab-to-lab differences in assay performance, we do not know how other labs’ enzyme stocks compare to ours in terms of purity and folding state. If a recombinant enzyme expressed in E. coli tends to misfold under standard expression and purification conditions, then it might have a low or nonexistent specific activity both in our lab and elsewhere. Indeed, our failure to detect activity of recombinant methionine aminopeptidase 2 from P. falciparum is consistent with an earlier report [23].
The present study helps clarify previous speculation in the literature regarding melting curves having a high baseline fluorescence and/or lacking a large melting-related increase in fluorescence. It has been proposed that high initial fluorescence is “likely caused by the dye binding to hydrophobic parts of the protein that are exposed even when it is fully folded” and that the lack of a melting transition “can be explained by high protein Tm that exceed the maximum temperature limit of the instruments” [24]. If poor melting curves usually come from properly folded proteins, as implied by the preceding sentence, melting curve quality would not be a useful indicator of an enzyme’s folding status and would not correlate with catalytic activity. On the contrary, our results show that enzymes with poor melting curves are less likely to be active, suggesting that a poor melting curve is often a result of being poorly folded. However, it is possible that well-folded stocks of certain proteins yield poor melting curves for the reasons offered above, and these reasons may account for the relatively high activities of some of our low-Q enzymes.
Thermal melt curves have previously been shown useful for optimizing protein purification, concentration, and crystallization, the optimal conditions being those in which the protein’s Tm is maximized [20]. Members of our research group have also found that melting curves help predict whether proteins can be crystallized (F. Zucker et al., unpublished experiments). The present study supports and extends those findings by demonstrating that melting curve quality is predictive of enzyme activity and therefore appears to be a reasonable indicator of folding status. These new results, in turn, hint that thermal melt assays may have additional applications beyond their use in crystallography. For example, a lab that has purified many proteins may wish to prioritize a few of them in developing functional assays for high-throughput screening. Since the functional assays may be difficult and costly to set up, thermal melt assays can be used to identify which proteins are most likely to be well-folded and thus which should be pursued further if resources are limited. (Such assays can probably be conducted with a single common buffer, since generic and enzyme-specific buffers yield similar results [Fig. 1].) Similarly, if a lab has generated many different stocks of the same protein, thermal melt assays may allow rapid assessment of the stocks without the need for functional assays. Finally, if a newly discovered protein appears inactive in an assay of its hypothesized function, thermal melting may help establish whether the apparent inactivity is due to denaturation. If the protein’s melting curves suggest that it is well-folded, the possibility that the protein’s true function is not what was hypothesized should be considered.
The preceding paragraphs should not be taken to mean that thermal melting is the only method conducive to high-throughput assessment of protein folding. The related technique of isothermal denaturation (ITD) also permits scanning of protein samples in 96- or 384-well plates, as does differential static light scattering (DSLS) [24], so those methods may also prove predictive of catalytic activity. The sample and time requirements of the three techniques are similar: 5–25 μL of protein at a concentration of 50–200 μg/mL is needed for each well, and scanning takes ~60 minutes. All three methods are distinct from circular dichroism (CD), another popular way of assessing protein folding, in that the latter requires a CD spectrometer (and therefore greater quantities of protein) and cannot be conducted in a high-throughput manner with commercially available plate readers. However, CD does offer the advantage of being nondestructive to protein samples.
Acknowledgments
We thank Raymond Hui of the University of Toronto’s Structural Genomics Consortium for supplying plasmids for expressing four of the Plasmodium proteins used in this study. We also thank our University of Washington colleagues Erkang Fan, Zhongsheng Zhang, and Frank Zucker for insights on thermal melt assays and analysis, Frederick Buckner for target selection expertise, and Christophe Verlinde for comments on the manuscript.
Footnotes
Funding for this study was provided by the Medicines for Malaria Venture (MMV), the Medical Structural Genomics for Pathogenic Protozoa (MSGPP) program project, the NIH (AI067921 to WGJH, AI080625 to WCVV, AI077822-01 to Sharon L. Reed), and the UNDP/World Bank/WHO Special Programme for Research and Training in Tropical Diseases (05-00508).
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Pantoliano MW, Petrella EC, Kwasnoski JD, Lobanov VS, Myslik J, Graf E, Carver T, Asel E, Springer BA, Lane P, Salemme FR. High-density miniaturized thermal shift assays as a general strategy for drug discovery. J Biomol Screen. 2001;6:429–440. doi: 10.1177/108705710100600609. [DOI] [PubMed] [Google Scholar]
- 2.Mezzasalma TM, Kranz JK, Chan W, Struble GT, Schalk-Hihi C, Deckman IC, Springer BA, Todd MJ. Enhancing recombinant protein quality and yield by protein stability profiling. J Biomol Screen. 2007;12:418–428. doi: 10.1177/1087057106297984. [DOI] [PubMed] [Google Scholar]
- 3.Crowther GJ, Napuli AJ, Thomas AP, Chung DJ, Kovzun KV, Leibly DJ, Castaneda LJ, Bhandari J, Damman CJ, Hui R, Hol WGJ, Buckner FS, Verlinde CLMJ, Zhang Z, Fan E, Van Voorhis WC. Buffer optimization of thermal melt assays of Plasmodium proteins for detection of small-molecule ligands. Journal of Biomolecular Screening. 2009;14 doi: 10.1177/1087057109335749. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bracher A, Eisenreich W, Schramek N, Ritz H, Gotze E, Herrmann A, Gutlich M, Bacher A. Biosynthesis of pteridines. NMR studies on the reaction mechanisms of GTP cyclohydrolase I, pyruvoyltetrahydropterin synthase, and sepiapterin reductase. J Biol Chem. 1998;273:28132–28141. doi: 10.1074/jbc.273.43.28132. [DOI] [PubMed] [Google Scholar]
- 5.Melendez-Lopez SG, Herdman S, Hirata K, Choi MH, Choe Y, Craik C, Caffrey CR, Hansell E, Chavez-Munguia B, Chen YT, Roush WR, McKerrow J, Eckmann L, Guo J, Stanley SL, Jr, Reed SL. Use of recombinant Entamoeba histolytica cysteine proteinase 1 to identify a potent inhibitor of amebic invasion in a human colonic model. Eukaryot Cell. 2007;6:1130–1136. doi: 10.1128/EC.00094-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Choubey V, Guha M, Maity P, Kumar S, Raghunandan R, Maulik PR, Mitra K, Halder UC, Bandyopadhyay U. Molecular characterization and localization of Plasmodium falciparum choline kinase. Biochim Biophys Acta. 2006;1760:1027–1038. doi: 10.1016/j.bbagen.2006.03.003. [DOI] [PubMed] [Google Scholar]
- 7.Whittingham JL, Leal I, Nguyen C, Kasinathan G, Bell E, Jones AF, Berry C, Benito A, Turkenburg JP, Dodson EJ, Ruiz Perez LM, Wilkinson AJ, Johansson NG, Brun R, Gilbert IH, Gonzalez Pacanowska D, Wilson KS. dUTPase as a platform for antimalarial drug design: structural basis for the selectivity of a class of nucleoside inhibitors. Structure. 2005;13:329–338. doi: 10.1016/j.str.2004.11.015. [DOI] [PubMed] [Google Scholar]
- 8.Clarke JL, Scopes DA, Sodeinde O, Mason PJ. Glucose-6-phosphate dehydrogenase-6-phosphogluconolactonase. A novel bifunctional enzyme in malaria parasites. Eur J Biochem. 2001;268:2013–2019. doi: 10.1046/j.1432-1327.2001.02078.x. [DOI] [PubMed] [Google Scholar]
- 9.Collard F, Collet JF, Gerin I, Veiga-da-Cunha M, Van Schaftingen E. Identification of the cDNA encoding human 6-phosphogluconolactonase, the enzyme catalyzing the second step of the pentose phosphate pathway(1) FEBS Lett. 1999;459:223–226. doi: 10.1016/s0014-5793(99)01247-8. [DOI] [PubMed] [Google Scholar]
- 10.Koresawa M, Okabe T. High-throughput screening with quantitation of ATP consumption: a universal non-radioisotope, homogeneous assay for protein kinase. Assay Drug Dev Technol. 2004;2:153–160. doi: 10.1089/154065804323056495. [DOI] [PubMed] [Google Scholar]
- 11.Que X, Brinen LS, Perkins P, Herdman S, Hirata K, Torian BE, Rubin H, McKerrow JH, Reed SL. Cysteine proteinases from distinct cellular compartments are recruited to phagocytic vesicles by Entamoeba histolytica. Mol Biochem Parasitol. 2002;119:23–32. doi: 10.1016/s0166-6851(01)00387-5. [DOI] [PubMed] [Google Scholar]
- 12.Van Veldhoven PP, Mannaerts GP. Inorganic and organic phosphate measurements in the nanomolar range. Anal Biochem. 1987;161:45–48. doi: 10.1016/0003-2697(87)90649-x. [DOI] [PubMed] [Google Scholar]
- 13.Sigma-Aldrich. Enzymatic assay of guanylate kinase (EC 2.7.4.8) 2009 http://www.sigmaaldrich.com/life-science/metabolomics/enzyme-explorer/learning-center/assay-library.html.
- 14.Chen X, Chong CR, Shi L, Yoshimoto T, Sullivan DJ, Jr, Liu JO. Inhibitors of Plasmodium falciparum methionine aminopeptidase 1b possess antimalarial activity. Proc Natl Acad Sci U S A. 2006;103:14548–14553. doi: 10.1073/pnas.0604101103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mitra S, Dygas-Holz AM, Jiracek J, Zertova M, Zakova L, Holz RC. A new colorimetric assay for methionyl aminopeptidases: examination of the binding of a new class of pseudopeptide analog inhibitors. Anal Biochem. 2006;357:43–49. doi: 10.1016/j.ab.2006.06.012. [DOI] [PubMed] [Google Scholar]
- 16.Rahlfs S, Koncarevic S, Iozef R, Mailu BM, Savvides SN, Schirmer RH, Becker K. Myristoylated adenylate kinase-2 of Plasmodium falciparum forms a heterodimer with myristoyltransferase. Mol Biochem Parasitol. 2009;163:77–84. doi: 10.1016/j.molbiopara.2008.09.008. [DOI] [PubMed] [Google Scholar]
- 17.Wright SK, Viola RE. Evaluation of methods for the quantitation of cysteines in proteins. Anal Biochem. 1998;265:8–14. doi: 10.1006/abio.1998.2858. [DOI] [PubMed] [Google Scholar]
- 18.Gratepanche S, Menage S, Touati D, Wintjens R, Delplace P, Fontecave M, Masset A, Camus D, Dive D. Biochemical and electron paramagnetic resonance study of the iron superoxide dismutase from Plasmodium falciparum. Mol Biochem Parasitol. 2002;120:237–246. doi: 10.1016/s0166-6851(02)00004-x. [DOI] [PubMed] [Google Scholar]
- 19.Sigma-Aldrich. Enzymatic assay of superoxide dismutase (1.15.1.1) 2009 http://www.sigmaaldrich.com/life-science/metabolomics/enzyme-explorer/learning-center/assay-library.html.
- 20.Vedadi M, Niesen FH, Allali-Hassani A, Fedorov OY, Finerty PJ, Jr, Wasney GA, Yeung R, Arrowsmith C, Ball LJ, Berglund H, Hui R, Marsden BD, Nordlund P, Sundstrom M, Weigelt J, Edwards AM. Chemical screening methods to identify ligands that promote protein stability, protein crystallization, and structure determination. Proc Natl Acad Sci U S A. 2006;103:15835–15840. doi: 10.1073/pnas.0605224103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bastien O, Lespinats S, Roy S, Metayer K, Fertil B, Codani JJ, Marechal E. Analysis of the compositional biases in Plasmodium falciparum genome and proteome using Arabidopsis thaliana as a reference. Gene. 2004;336:163–173. doi: 10.1016/j.gene.2004.04.029. [DOI] [PubMed] [Google Scholar]
- 22.Wang J, Sheppard GS, Lou P, Kawai M, Park C, Egan DA, Schneider A, Bouska J, Lesniewski R, Henkin J. Physiologically relevant metal cofactor for methionine aminopeptidase-2 is manganese. Biochemistry. 2003;42:5035–5042. doi: 10.1021/bi020670c. [DOI] [PubMed] [Google Scholar]
- 23.Chen X, Xie S, Bhat S, Kumar N, Shapiro TA, Liu JO. Fumagillin and fumarranol interact with P. falciparum methionine aminopeptidase 2 and inhibit malaria parasite growth in vitro and in vivo. Chem Biol. 2009;16:193–202. doi: 10.1016/j.chembiol.2009.01.006. [DOI] [PubMed] [Google Scholar]
- 24.Senisterra GA, Finerty PJ., Jr High throughput methods of assessing protein stability and aggregation. Mol Biosyst. 2009;5:217–223. doi: 10.1039/b814377c. [DOI] [PubMed] [Google Scholar]
- 25.Miclet E, Stoven V, Michels PA, Opperdoes FR, Lallemand JY, Duffieux F. NMR spectroscopic analysis of the first two steps of the pentose-phosphate pathway elucidates the role of 6-phosphogluconolactonase. J Biol Chem. 2001;276:34840–34846. doi: 10.1074/jbc.M105174200. [DOI] [PubMed] [Google Scholar]
- 26.Dittrich S, Mitchell SL, Blagborough AM, Wang Q, Wang P, Sims PF, Hyde JE. An atypical orthologue of 6-pyruvoyltetrahydropterin synthase can provide the missing link in the folate biosynthesis pathway of malaria parasites. Mol Microbiol. 2008;67:609–618. doi: 10.1111/j.1365-2958.2007.06073.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tyler PC, Taylor EA, Frohlich RF, Schramm VL. Synthesis of 5′-methylthio coformycins: specific inhibitors for malarial adenosine deaminase. J Am Chem Soc. 2007;129:6872–6879. doi: 10.1021/ja0708363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Daddona PE, Wiesmann WP, Lambros C, Kelley WN, Webster HK. Human malaria parasite adenosine deaminase. Characterization in host enzyme-deficient erythrocyte culture. J Biol Chem. 1984;259:1472–1475. [PubMed] [Google Scholar]
- 29.Bulusu V, Srinivasan B, Bopanna MP, Balaram H. Elucidation of the substrate specificity, kinetic and catalytic mechanism of adenylosuccinate lyase from Plasmodium falciparum. Biochim Biophys Acta. 2009;1794:642–654. doi: 10.1016/j.bbapap.2008.11.021. [DOI] [PubMed] [Google Scholar]
- 30.Jayalakshmi R, Sumathy K, Balaram H. Purification and characterization of recombinant Plasmodium falciparum adenylosuccinate synthetase expressed in Escherichia coli. Protein Expr Purif. 2002;25:65–72. doi: 10.1006/prep.2001.1610. [DOI] [PubMed] [Google Scholar]
- 31.Else AJ, Herve G. A microtiter plate assay for aspartate transcarbamylase. Anal Biochem. 1990;186:219–221. doi: 10.1016/0003-2697(90)90069-l. [DOI] [PubMed] [Google Scholar]
- 32.Wood T. Spectrophotometric assay for D-ribose-5-phosphateketol-isomerase and for D-ribulose-5-phosphate 3-epimerase. Anal Biochem. 1970;33:297–306. doi: 10.1016/0003-2697(70)90300-3. [DOI] [PubMed] [Google Scholar]
- 33.Sakuraba H, Yoneda K, Yoshihara K, Satoh K, Kawakami R, Uto Y, Tsuge H, Takahashi K, Hori H, Ohshima T. Sequential aldol condensation catalyzed by hyperthermophilic 2-deoxy-d-ribose-5-phosphate aldolase. Appl Environ Microbiol. 2007;73:7427–7434. doi: 10.1128/AEM.01101-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Levin I, Mevarech M, Palfey BA. Characterization of a novel bifunctional dihydropteroate synthase/dihydropteroate reductase enzyme from Helicobacter pylori. J Bacteriol. 2007;189:4062–4069. doi: 10.1128/JB.01878-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bognar AL, Osborne C, Shane B, Singer SC, Ferone R. Folylpoly-gamma-glutamate synthetase-dihydrofolate synthetase. Cloning and high expression of the Escherichia coli folC gene and purification and properties of the gene product. J Biol Chem. 1985;260:5625–5630. [PubMed] [Google Scholar]
- 36.Mukkamala D, No JH, Cass LM, Chang TK, Oldfield E. Bisphosphonate inhibition of a Plasmodium farnesyl diphosphate synthase and a general method for predicting cell-based activity from enzyme data. J Med Chem. 2008;51:7827–7833. doi: 10.1021/jm8009074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wagner JT, Ludemann H, Farber PM, Lottspeich F, Krauth-Siegel RL. Glutamate dehydrogenase, the marker protein of Plasmodium falciparum--cloning, expression and characterization of the malarial enzyme. Eur J Biochem. 1998;258:813–819. doi: 10.1046/j.1432-1327.1998.2580813.x. [DOI] [PubMed] [Google Scholar]
- 38.Campanale N, Nickel C, Daubenberger CA, Wehlan DA, Gorman JJ, Klonis N, Becker K, Tilley L. Identification and characterization of heme-interacting proteins in the malaria parasite, Plasmodium falciparum. J Biol Chem. 2003;278:27354–27361. doi: 10.1074/jbc.M303634200. [DOI] [PubMed] [Google Scholar]
- 39.Sigma-Aldrich. Enzymatic assay of a-glycerophosphate dehydrogenase (EC 1.1.1.8) 2009 http://www.sigmaaldrich.com/life-science/metabolomics/enzyme-explorer/learning-center/assay-library.html.
- 40.Kandeel M, Nakanishi M, Ando T, El-Shazly K, Yosef T, Ueno Y, Kitade Y. Molecular cloning, expression, characterization and mutation of Plasmodium falciparum guanylate kinase. Mol Biochem Parasitol. 2008;159:130–133. doi: 10.1016/j.molbiopara.2008.02.004. [DOI] [PubMed] [Google Scholar]
- 41.Kasekarn W, Sirawaraporn R, Chahomchuen T, Cowman AF, Sirawaraporn W. Molecular characterization of bifunctional hydroxymethyldihydropterin pyrophosphokinase-dihydropteroate synthase from Plasmodium falciparum. Mol Biochem Parasitol. 2004;137:43–53. doi: 10.1016/j.molbiopara.2004.04.012. [DOI] [PubMed] [Google Scholar]
- 42.Chiang PK, Chamberlin ME, Nicholson D, Soubes S, Su X, Subramanian G, Lanar DE, Prigge ST, Scovill JP, Miller LH, Chou JY. Molecular characterization of Plasmodium falciparum S-adenosylmethionine synthetase. Biochem J. 1999;344(Pt 2):571–576. [PMC free article] [PubMed] [Google Scholar]
- 43.Towler DA, Adams SP, Eubanks SR, Towery DS, Jackson-Machelski E, Glaser L, Gordon JI. Purification and characterization of yeast myristoyl CoA:protein N-myristoyltransferase. Proc Natl Acad Sci U S A. 1987;84:2708–2712. doi: 10.1073/pnas.84.9.2708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bowyer PW, Gunaratne RS, Grainger M, Withers-Martinez C, Wickramsinghe SR, Tate EW, Leatherbarrow RJ, Brown KA, Holder AA, Smith DF. Molecules incorporating a benzothiazole core scaffold inhibit the N-myristoyltransferase of Plasmodium falciparum. Biochem J. 2007;408:173–180. doi: 10.1042/BJ20070692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kandeel M, Miyamoto T, Kitade Y. Bioinformatics, enzymologic properties, and comprehensive tracking of Plasmodium falciparum nucleoside diphosphate kinase. Biol Pharm Bull. 2009;32:1321–1327. doi: 10.1248/bpb.32.1321. [DOI] [PubMed] [Google Scholar]
- 46.Langley DB, Shojaei M, Chan C, Lok HC, Mackay JP, Traut TW, Guss JM, Christopherson RI. Structure and inhibition of orotidine 5′-monophosphate decarboxylase from Plasmodium falciparum. Biochemistry. 2008;47:3842–3854. doi: 10.1021/bi702390k. [DOI] [PubMed] [Google Scholar]
- 47.Pessi G, Kociubinski G, Mamoun CB. A pathway for phosphatidylcholine biosynthesis in Plasmodium falciparum involving phosphoethanolamine methylation. Proc Natl Acad Sci U S A. 2004;101:6206–6211. doi: 10.1073/pnas.0307742101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pal B, Pybus B, Muccio DD, Chattopadhyay D. Biochemical characterization and crystallization of recombinant 3-phosphoglycerate kinase of Plasmodium falciparum. Biochim Biophys Acta. 2004;1699:277–280. doi: 10.1016/j.bbapap.2004.01.003. [DOI] [PubMed] [Google Scholar]
- 49.Nakanishi M, Iwata A, Yatome C, Kitade Y. Purification and properties of recombinant Plasmodium falciparum S-adenosyl-L-homocysteine hydrolase. J Biochem. 2001;129:101–105. doi: 10.1093/oxfordjournals.jbchem.a002819. [DOI] [PubMed] [Google Scholar]