Abstract
In most species, each sex produces gametes, usually either sperm or oocytes, from its germline during gametogenesis. The sperm and oocyte subsequently fuse together during fertilization to create the next generation. This review focuses on spermatogenesis and the roles of sperm during fertilization in the nematode Caenorhabditis elegans, where suitable mutants are readily obtained. So far 186 mutants defective in the C. elegans male germline functions have been isolated, and many of these mutations are alleles for one of the ~60 spermatogenesis-defective (spe) genes. Many cloned spe genes are expressed specifically in the male germline, where they play roles during spermatogenesis (spermatid production), spermiogenesis (spermatid activation into spermatozoa), and/or fertilization. Moreover, several spe genes are orthologs of mammalian genes, suggesting that the reproductive processes of the C. elegans and the mammalian male germlines might share common pathways at the molecular level.
Keywords: C. elegans, spe gene, male germline, spermatogenesis, spermiogenesis, fertilization, sperm-oocyte interactions
INTRODUCTION
Germ cells are essential to create the next generation of most multi-cell organisms via gametogenesis followed by fertilization. During gametogenesis, germ cells differentiate into either sperm (spermatogenesis) or oocytes (oogenesis). A mature sperm and oocyte subsequently fuse during fertilization to produce a diploid zygote that is the progenitor of all somatic and germ cells.
The nematode Caenorhabditis elegans is an excellent model system to investigate a variety of biological phenomena, including reproduction (Brenner, 1974). Indeed, many C. elegans genes required for reproduction have been identified, and ~60 genes that can mutate to cause defects in male germline functions, the so-called spermatogenesis-defective (spe) genes, are included among this group (L’Hernault, 1997; L’Hernault and Singson, 2000). In this article, we review how the C. elegans spe genes play roles during spermatogenesis (spermatid production), spermiogenesis (spermatid activation into spermatozoa), and/or fertilization. These C. elegans male germline functions are also reviewed elsewhere (L’Hernault, 1997; L’Hernault, 2006; Singson et al., 2008; L’Hernault, 2009).
OVERVIEW OF C. ELEGANS REPRODUCTION
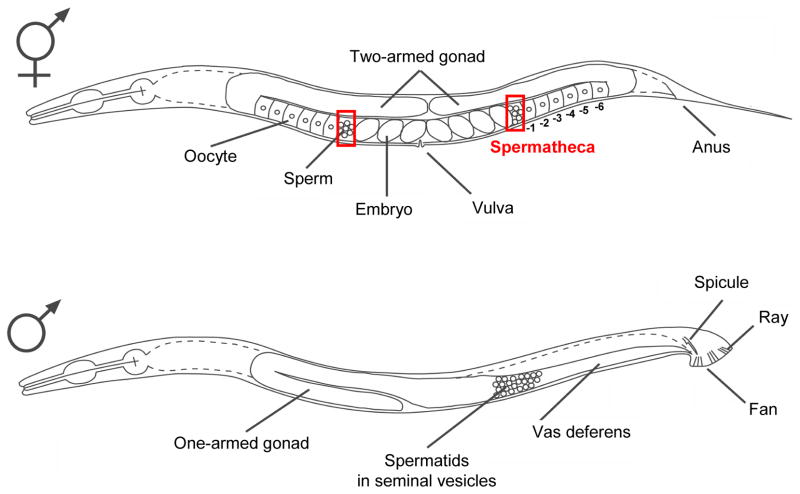
As shown in Fig. 1, C. elegans has a male like most animals, but it lacks a true female and instead has a hermaphrodite where both spermatogenesis and oogenesis occur (Hirsh et al., 1976). Hermaphrodites first undergo spermatogenesis during the fourth larval (L4) stage. When L4 hermaphrodites become young adults, spermatogenesis stops and both arms of the U-shaped gonad completely switch to oogenesis. Hence, adult hermaphrodites are somatically females, although they contain self-sperm. Germ cells that are differentiating into oocytes are aligned in the adult worm gonads, so that immature cells are relatively distal (Fig. 1). The mature, fertilization-ready oocyte resides at the most proximal region (−1 position) of the gonad (Fig. 1). Ovulation of the first −1 oocyte pushes the previously produced spermatids out of the proximal gonad into the spermatheca. Once in the spermatheca, the spermatids are rapidly activated into spermatozoa (sperm), and one of them fertilizes the first oocyte. Prior to onset of embryogenesis, the fertilized oocyte moves into the uterus together with many of the remaining sperm. The sperm subsequently crawl back into the spermatheca to compete again for the next fertilization. An adult hermaphrodite contains ~300 sperm and produces ~300 self-progeny through the entire life of the animal. This indicates that nearly all wild-type sperm, despite being pushed into the uterus, are able to re-establish their position in the spermatheca so that they can be efficiently consumed by fertilization (Ward and Carrel, 1979).
Fig. 1.
The two sexes of the nematode C. elegans. Top: An adult hermaphrodite has a two-armed gonad that exhibits mirror image symmetry around the single vulval opening. The location of each spermatheca is outlined by red squares. Numbers below the gonad indicate the positions of oocytes according to their developmental stages. Bottom: An adult male has a one-armed gonad. Spermatids accumulate in the single vas deferens until they are ejaculated during mating with a hermaphrodite.
In adult males, spermatids are continuously produced and stored in the seminal vesicles of the one-armed gonad (Fig. 1). Upon mating with hermaphrodites, spermatids are ejaculated with the seminal fluid and enter the uterus of hermaphrodites through their vulva. The spermatids mature into spermatozoa in the uterus by exposure to an unknown seminal fluid-derived factor(s), and they crawl into the spermatheca, where they fertilize oocytes.
CYTOLOGY AND PHYSIOLOGY OF MALE GERMLINE FUNCTIONS
Spermatogenesis
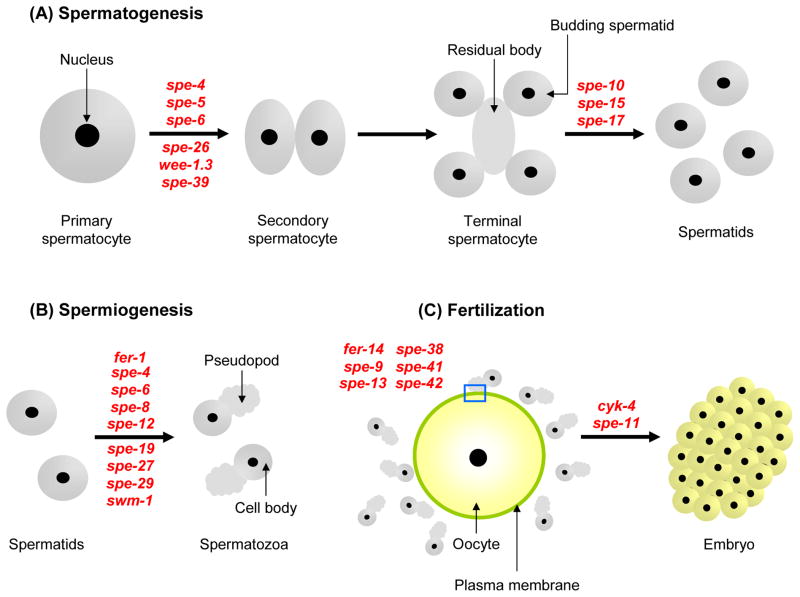
The reproductive processes where the C. elegans male germline is involved are composed of three pivotal steps: spermatogenesis, spermiogenesis, and fertilization (Wolf et al., 1978; Ward et al., 1981; Ward, 1986; Kimble and Ward, 1988). During meiosis I, an early phase of spermatogenesis (Fig. 2A), a primary spermatocyte generates two secondary spermatocytes. Each secondary spermatocyte then undergoes meiosis II, by which two haploid spermatids bud from an acellular residual body (Ward et al., 1981). This second cell division is asymmetric, and the residual body receives many organelles and cytoplasmic proteins, including all ribosomes, the Golgi apparatus, the endoplasmic reticulum (ER), actin, myosin, and most tubulin (except for that contained in the centrioles). The mature spermatid possesses a highly condensed nucleus, the centriole pair embedded in a RNA-enriched, perinuclear layer surrounding the condensed nucleus, mitochondria, and Golgi-derived secretory membranous vesicles (MOs) (Ward, 1986).
Fig. 2.
The three central stages of male germline functions. A: Spermatogenesis. B: Spermiogenesis. C: Fertilization. A blue square shows that a sperm contacts the oocyte plasma membrane through its pseudopod. In these figures, the approximate point where a spe gene is first observed to act (by light microscopy) is indicated in red letters.
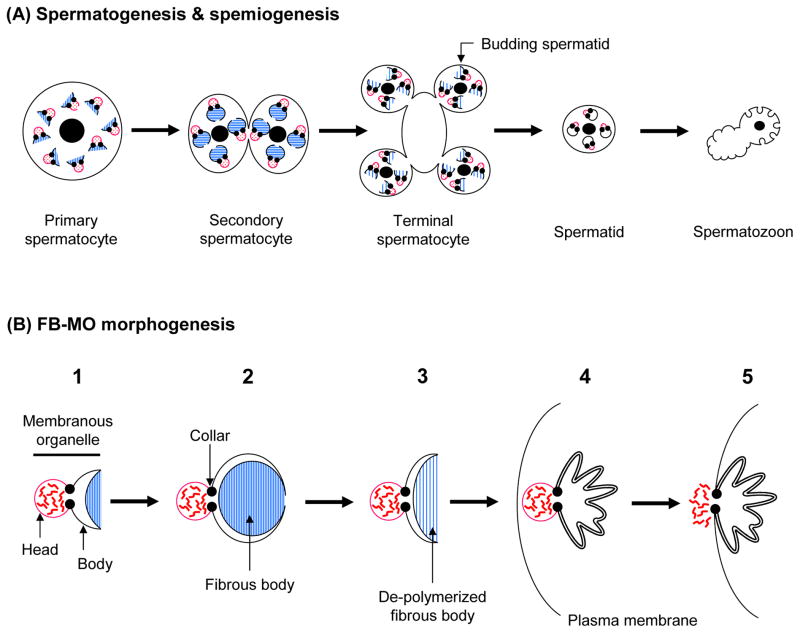
Spermatogenesis involves a dramatic partitioning of the cytoplasm mediated by the fibrous body (FB) and the MO, which form a complex (FB-MO) (Wolf et al., 1978; Ward et al., 1981). One major role of the FB-MO complex is to ensure that sperm proteins segregate into spermatids rather than the residual body. Simultaneous with meiosis I (Fig. 3A), each FB-MO increases in size, and the MO portion forms three compartments: the head, collar, and body regions (Fig. 3B1). The collar constricts the MO like a noose, so that the head region is distinguished from the body. The head is a membrane vesicle, whereas the body membrane folds around and envelopes the developing FB, but never completely seals it off from the rest of the cytoplasm (Fig. 3B2). The FB is mainly composed of the major sperm protein (MSP) fibers, which are hexagonally packed rods within the body membrane. The FB-MO reaches its maximum size in secondary spermatocytes (Fig. 3A), after which the MO’s body membrane surrounding the FB starts to retract. The MSP fibers of the FB are de-polymerized into dimers (Klass and Hirsh, 1981; King et al., 1992; Smith and Ward, 1998), and these disperse throughout the cytoplasm (Fig. 3B3). In mature spermatids (Fig. 3A), the MOs are no longer obviously associated with MSP-fibers, and the head region of the MO localizes near the plasma membrane of spermatids (Fig. 3B4).
Fig. 3.
FB-MO morphogenesis during spermatogenesis and spermiogenesis. A: The cytological stages of spermatogenesis and spermiogenesis, and how they are coordinated with FB-MO morphogenesis. B: The stages of FB-MO morphogenesis. Red, wavy lines represent the contents of the MO vesicles, which are released into the extracellular space upon spermiogenesis. Blue lines show polymerized or de-polymerized MSP filaments. These figures are meant to show the fundamental process of FB-MO morphogenesis, so some details are simplified.
Unlike mammals, C. elegans spermatogenesis will readily occur in vitro; spermatocytes released from dissected males differentiate into spermatids in a simple, chemically defined medium (Nelson and Ward, 1980; Machaca et al., 1996), and accessory cells or hormones are not required for in vitro spermatogenesis. This straightforward and rapid (~90 minutes) in vitro system is a significant advantage of using C. elegans to study spermatogenesis.
Spermiogenesis
C. elegans spermiogenesis is a process that transforms a quiescent, round spermatid into a motile, amoeboid spermatozoon (sperm) (Fig. 2B); nematode spermatozoa do not form a flagellum. Since no ribosomes are present in spermatids, new protein synthesis does not occur during spermiogenesis. C. elegans sperm each have a single pseudopod extending from their cell body, that is used for crawling. Unlike other amoeboid-like cell types, nematode sperm utilize MSP, instead of actin, as their cytoskeletal protein (Ward and Klass, 1982). Most of our knowledge regarding nematode sperm motility has come from Ascaris, since its sperm cells can be isolated in the large quantities required for biochemical analysis (Italiano et al., 2001). Available data suggest that very similar phenomena can occur in C. elegans (Pavalko et al., 1988). Within the pseudopod, MSP forms long polymers, which further associate with each other, leading to a network of MSP-bundles. At the leading edge of the pseudopod, the MSP-bundles are continuously assembled, whereas the MSP-bundle network is disassembled at the back end. The transition of the MSP-bundle network allows sperm to crawl forward (Pavalko et al., 1988; Italiano et al., 2001). Recent data suggest that phosphorylation (Yi et al., 2007) and de-phosphorylation of accessory proteins regulate assembly and disassembly of MSP-bundle, respectively, during sperm movement (Yi et al., 2009).
The MO has a secretory function during spermiogenesis. Upon spermatid activation, the MO head fuses with the plasma membrane and releases its contents extracellularly (Fig. 3B5), while new transmembrane proteins are inserted into the plasma membrane. In this aspect, the MO might be analogous to the acrosome of flagellated sperm (Yanagimachi, 1994). The MO collar leaves a permanent fusion pore in the plasma membrane, and the interior of the MO body is open to the extracellular space (Fig. 3B5).
At present, the physiologically relevant activators of spermatids are not known in either hermaphrodites or males. Male-derived spermatids seem to be activated by a factor(s) that is contained in the seminal fluid (Ward and Carrel, 1979), but how hermaphroditic spermatids are exposed to activator(s) is not yet understood. In vitro, spermatids can be activated into spermatozoa by treatment with either of the cationic ionophore monensin (Nelson and Ward, 1980), the weak base triethanolamine (TEA), the phosphoinositide 3-kinase inhibitor wortmannin (Bae et al., 2009), or proteases, the most effective of which is the serine protease mixture Pronase (Ward et al., 1983). Spermatozoa produced by in vitro activation with TEA, but not Pronase, are competent to fertilize oocytes after they are introduced into hermaphrodites by artificial insemination (LaMunyon and Ward, 1994).
Fertilization
In C. elegans, in vivo fertilization always occurs in the spermatheca of hermaphrodites (Fig. 1). While most aspects of spermatogenesis are highly similar in hermaphrodites and males, the spermatids produced by males are ~50% larger than those produced by hermaphrodites. Furthermore, male-derived spermatozoa can move faster than hermaphrodite-derived spermatozoa (LaMunyon and Ward, 1998). Either or both of these parameters (size and crawling velocity) might explain the superiority of male-derived spermatozoa as compared to hermaphrodite-derived ones in competing to fertilize oocytes (LaMunyon and Ward, 1995; LaMunyon and Ward, 1998). Consequently, after mating, fertilization occurs exclusively between oocytes and male-derived sperm, rather than between oocytes and hermaphroditic sperm (Ward and Carrel, 1979).
C. elegans fertilization differs from mammalian fertilization in several significant ways. Firstly, in mammals, sperm have to bind the zona pellucida (ZP), a glycoprotein matrix surrounding the oocyte, in order to undergo the sperm acrosome reaction, which is a prerequisite for sperm-oocyte fusion (Yanagimachi, 1994). In contrast, C. elegans sperm lack an acrosome, probably due to the absence of any substantial egg coat, like the ZP, on C. elegans oocytes. Secondly, acrosome-reacted sperm of mammals bind to the oocyte plasma membrane at the equatorial region of the sperm head (Yanagimachi, 1994), whereas C. elegans sperm probably first contact the oocyte plasma membrane via the pseudopods (Fig. 2C), which functionally correspond to the flagella of mammalian sperm. Thirdly, unlike mammals, a system for in vitro fertilization (IVF) is not yet available for C. elegans. The major problem is that there is no known method to isolate enough fertilization-competent oocytes to systematically examine and optimize C. elegans IVF.
In certain ways, the reproductive tract of C. elegans adult hermaphrodites is analogous to that of mammalian females; the proximal gonad, spermatheca, and uterus of adult hermaphrodites play similar spatial roles to those of the ovary, oviduct, and uterus of mammalian females. As described above, adult hermaphrodites are somatically females and no longer produce sperm. Consequently, adult hermaphrodites are more accurately thought of “females carrying self-sperm”. Hence, C. elegans might provide insights into certain in vivo aspects of internal fertilization that are analogous to mammalian fertilization.
ROLES OF SPE GENES IN MALE GERMLINE FUNCTIONS
What Are spe Mutants?
spe (spermatogenesis-defective) or fer (fertilization-defective, which is obsolete terminology) mutants produce spermatocytes, spermatids, and/or sperm whose functions are aberrant during spermatogenesis, spermiogenesis, and/or fertilization. spe mutant hermaphrodites produce very few progeny and, instead, lay unfertilized oocytes (L’Hernault et al., 1988; McCarter et al., 1999). However, mating to wild-type males allows spe mutant hermaphrodites to produce outcross progeny. This indicates that sperm, but not oocytes, are functionally defective in spe mutants. In other words, wild-type sperm are necessary and sufficient to rescue the self-sterility of mutant hermaphrodites (Argon and Ward, 1980; L’Hernault et al., 1988). Using these criteria, mutants that define ~60 spe genes have been isolated after treatment of hermaphrodites with chemical mutagens (L’Hernault, 1997; L’Hernault and Singson, 2000), such as ethyl methanesulfonate (EMS) and trimethylpsoralen (TMP) (Anderson, 1995). Table 1 shows a partial list of spe mutants that have been analyzed in some details. While many spe genes are expressed specifically or predominantly in the C. elegans male germline as expected (Reinke et al., 2000), other genes that play important roles in the male germline are also expressed in other tissues.
TABLE 1.
C. elegans Genes Involved in Male Germline Functions
| Gene | Chr. | Predicted protein | Phenotype of mutant | Reference |
|---|---|---|---|---|
| cyk-4 | III | Rho GTPase-activating protein (681 aa) |
|
Jantsch-Plunger et al., 2000; Jenkins et al., 2006 |
| fer-1 | I | Transmembrane protein of the ferlin family (1907 and/or 2034 aa) |
|
Ward et al., 1981; Roberts and Ward, 1982; Achanzar and Ward, 1997; Washington and Ward, 2006 |
| fer-14 | I | Transmembrane protein (608 aa) |
|
Kroft et al., unpublished |
| spe-4 | I | 8-Pass transmembrane protein of the presenilin (aspartyl protease) family (465 aa) |
|
L’Hernault and Arduengo, 1992; Arduengo et al., 1998; Gosney et al., 2008 |
| spe-5 | I | B subunit of V-ATPase (501 aa) |
|
L’Hernault et al., 1988; Machaca and L’Hernault, 1997; Gleason et al., unpublished |
| spe-6 | III | Ser/Thr kinase of the casein kinase I family (379 aa) |
|
Varkey et al., 1993; Muhlrad and Ward, 2002 |
| spe-8 | I | Non-receptor tyrosine kinase with the SH2 domain (512 aa) |
|
L’Hernault et al., 1988; Shakes and Ward, 1989a; Muhlrad and Ward, unpublished |
| spe-9 | I | Transmembrane protein with 10 EGF-like domains (661 aa) |
|
L’Hernault et al., 1988; Singson et al., 1998; Zannoni et al., 2003; Putiri et al., 2004 |
| spe-10 | V | 4-Pass transmembrane protein with the DHHC-CRD zinc-finger motif that is a palmitoyl transferase (351 aa) |
|
Shakes and Ward, 1989b; Gleason et al., 2006 |
| spe-11 | I | Soluble protein (299 aa) |
|
L’Hernault et al., 1988; Hill et al., 1989; Browning and Strome, 1996; Royal et al., 1997; McNally and McNally, 2005 |
| spe-12 | I | Transmembrane protein (255 aa) |
|
L’Hernault et al., 1988; Shakes and Ward, 1989a; Nance et al., 1999 |
| spe-13 | I | ND |
|
L’Hernault et al., 1988; Putiri et al., 2004 |
| spe-15 | I | Heavy chain of myosin VI (1219 aa) |
|
L’Hernault at al., 1988; Kelleher et al., 2000 |
| spe-17 | IV | Ser/Thr-rich, highly charged protein (142 aa) |
|
Shakes and Ward, 1989; L’Hernault et al., 1993 |
| spe-19 | V | Transmembrane protein with 11 potential phosphorylation sites in the cytoplasmic tail (300 aa) |
|
Geldziler et al., 2005 |
| spe-26 | IV | Protein similar to the actin-binding protein Kelch and Scruin (570 aa) |
|
Varkey et al., 1995 |
| spe-27 | IV | Soluble protein (131 aa) |
|
Minniti et al., 1996 |
| spe-29 | IV | Transmembrane protein (66 aa) |
|
Nance et al., 2000 |
| wee-1.3 (spe-37) | II | Myt1 (cyclin-dependent Ser/Thr kinase) ortholog (677 aa) |
|
Lamitina and L’Hernault, 2002 |
| spe-38 | I | 4-Pass transmembrane protein (179 aa) |
|
Chatterjee et al., 2005 |
| spe-39 | V | Protein associated with the HOPS complex in vitro (522 aa) |
|
Zhu and L’Hernault, 2003; Zhu et al., 2009 |
| spe-41 (trp-3) | III | Calcium-permeable cation channel of the TRPC family (854 aa) |
|
Xu and Sternberg, 2003 |
| spe-42 | V | 6-Pass transmembrane protein with the DC-STAMP and RING finger domains (774 aa) |
|
Kroft et al., 2005 |
| swm-1 | V | Soluble protein with two trypsin inhibitor-like domains (86 and/or 135 aa) |
|
Stanfield and Villeneuve, 2006 |
Chr., chromosome; aa, amino acid; PM, plasma membrane; SH2, Src homology 2; ND, not determined.
spe Genes Required for Spermatogenesis
Cell cycle during meiosis
wee-1.3
In many higher organisms including humans, the G2/M transition is regulated by activation of a protein complex between cyclin B and Cdk1 (also called Cdc2), a cyclin-dependent kinase (Dunphy et al., 1988; Lindqvist et al., 2009). Phosphorylation of Thr14 and Tyr15 on Cdk1 negatively regulates Cdk1 kinase activity (Coleman and Dunphy, 1994; O’Farrell, 2001; Lindqvist et al., 2009). The Wee1 kinase family Myt1 is a single-pass transmembrane protein whose cytoplasmic tail has unknown function(s). Since the metazoan Myt1 kinases can phosphorylate both of the Thr14 and Tyr15 residues on Cdk1, Myt1 is thought to be a negative regulator for Cdk1 (Booher et al., 1997; O’Farrell, 2001).
In C. elegans, the wee-1.3 gene is widely expressed and encodes the Myt1 kinase ortholog (Lamitina and L’Hernault, 2002). Like other Myt1 orthologs, WEE-1.3 protein controls M-phase entry during oocyte meiosis (Burrows et al., 2006) and, more generally, mitosis. Indeed, a null mutation of wee-1.3 causes embryonic lethality (Lamitina and L’Hernault, 2002). There are also six dominant, missense mutations in the wee-1.3 gene that change amino acids near the C-terminus of WEE-1.3 (Lamitina and L’Hernault, 2002). Interestingly, all these missense mutations prevent primary spermatocytes from entering M-phase, while neither oocyte meiosis nor any mitotic division is affected by these mutations (Lamitina and L’Hernault, 2002). This indicates that the cytoplasmic tail of WEE-1.3 is required for regulation of meiosis during spermatogenesis.
So far wee-1.3 is the only gene that has mutations specifically affecting meiosis during spermatogenesis, although numerous other genes seem to act in the regulation of meiosis. Perhaps, most of the meiosis-regulatory genes are important during oogenesis and/or mitosis, so that mutations of these genes would result in non-Spe-type sterility or embryonic lethality.
FB-MO
fer-1
FER-1 is a transmembrane protein with six C2 (protein kinase C conserved region 2) domains (Achanzar and Ward, 1997), which defines features of the ferlin family in animals (Bashir et al., 1998). The C2 domains usually act in calcium-dependent lipid-processing events, such as vesicle fusion (Lemmon, 2008). FER-1 was demonstrated to regulate the calcium-dependent membrane fusion between the MOs and the plasma membrane during spermatid activation (Washington and Ward, 2006). Indeed, in fer-1 mutant spermatids, MOs do not fuse with the plasma membrane upon spermiogenesis. The resulting fer-1 mutant sperm have short pseudopods and are immotile, because they cannot properly crawl (Ward et al., 1981; Roberts and Ward, 1982; Washington and Ward, 2006).
spe-4
In spe-4 mutants, spermatogenesis arrests as terminal spermatocytes, where four haploid nuclei share a common cytoplasm because cytokinesis is not completed. Unlike wild type, spe-4 mutant cells contain vacuolated MOs that are not near the plasma membrane and are randomly distributed throughout the cytoplasm. Additionally, in mutant spermatocytes, tubulin is present in unusual deposits near the plasma membrane, and FBs are not obviously associated with MOs.
spe-4 encodes an 8-pass transmembrane protein localized within MOs of spermatids and spermatozoa, which belongs to the presenilin family (L’Hernault and Arduengo, 1992; Arduengo et al., 1998). Members of this family are components of the γ-secretase complex, which is an aspartyl protease implicated in intra-membrane proteolysis of single-pass transmembrane proteins, such as β-amyloid precursor protein (APP) and Notch (Iwatsubo, 2004). Domain swapping experiments between SPE-4 and mammalian presenilin 1 revealed that SPE-4 can process APP but not Notch (Yamasaki et al., 2006). While the physiological substrate(s) processed by SPE-4 during spermatogenesis is not yet clear, it is known that a reduction-of-function spe-4 mutant shows genetic suppression of the spe-8 class pathway for spermatid activation (Fig. 4, also see below). Since this pathway includes transmembrane proteins, it will be interesting if they prove to be substrates for the SPE-4 proteolytic activity (Gosney et al., 2008).
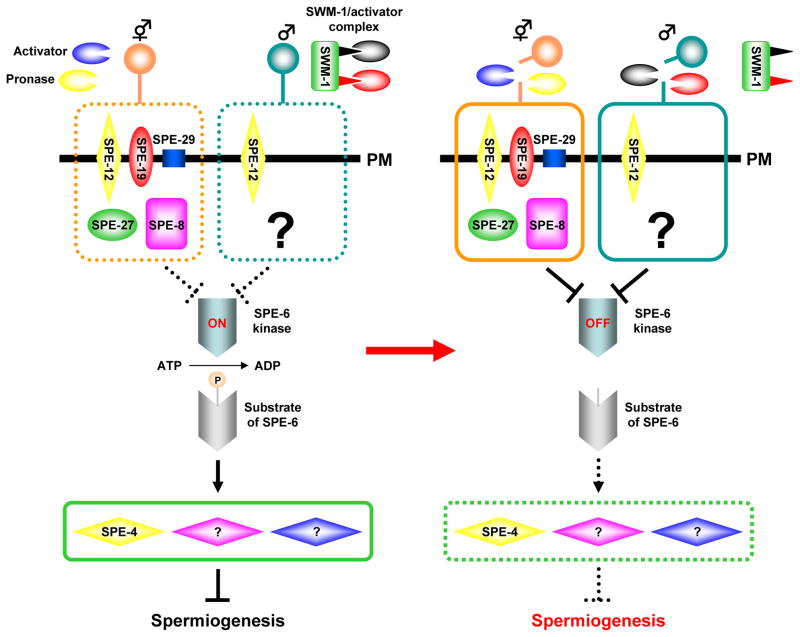
Fig. 4.
Two predicted pathways for spermiogenesis. Spermatids from hermaphrodites and males probably each have a distinct pathway for spermiogenesis. One pathway is spe-8 class-dependent (surrounded by broken or solid orange line) and another is spe-8 class-independent (broken or solid light blue line). A decision to utilize either pathway might depend on activators that are sex-specific. The spe-8 class-dependent pathway seems to be stimulated by a hermaphrodite-derived activator(s) or the serine protease mixture Pronase, whereas male-derived serine proteases that are targets of the trypsin inhibitor-like protein SWM-1 utilize the spe-8 class-independent pathway. To initiate spermiogenesis, these activators might be required to cleave a certain cell-surface protein(s). There seem to be other possible ways to initiate spermiogenesis. For example, serine proteases targeted by SWM-1 might process a precursor protein(s) of the actual activator(s) for spermiogenesis. The SPE-6 kinase is downstream of SPE-8 class proteins and is one of the common points between these two pathways. This kinase phosphorylates (shown as “P” in a light orange ball) unknown substrate(s), and the phosphorylated protein then plays a role in signal transduction to block onset of spermiogenesis. The signal might affect the SPE-4 presenilin and presumably other proteins (purple and blue diamonds; the precise number is unknown). During spermiogenesis, SPE-6 activity is reduced so that negative regulatory proteins such as SPE-4 cannot function to block spermiogenesis. In this figure, the active and inactive status are shown by solid and broken lines, respectively. Thick, black arrows represent positive regulation, whereas negative regulation is expressed by T-shaped lines. PM, plasma membrane.
spe-5
SPE-5 is a B subunit of the vacuolar ATPase (V-ATPase), which is localized in the MOs of spermatids (Gleason et al., unpublished data). spe-5 null mutants produce arrested terminal spermatocytes where the MOs are vacuolated and, unlike spe-4 mutants, associated with the FBs (L’Hernault et al., 1988; Machaca and L’Hernault, 1997). A space between the outer and inner layers of the membrane that surrounds the FBs is greatly expanded in spe-5 mutants, suggesting the V-ATPase participates in this aspect of FB-MO morphology. If SPE-5 is a part of an active V-ATPase, this and other defective aspects of morphogenesis in spe-5 mutants are probably due to failure to acidify the FB-MOs (Cipriano et al., 2008).
spe-6
spe-6 mutants usually produce defective primary spermatocytes that do not properly divide into spermatids. In mutant spermatocytes, MSP fails to assemble into FBs so that unassembled MSP is scattered throughout the cell (Varkey et al., 1993). Since the spe-6 gene encodes a Ser/Thr kinase of the casein kinase I family, the signaling pathway(s) via SPE-6 is probably required for MSP to assemble into FBs (Muhlrad and Ward, 2002). Additionally, the spe-6 gene is involved in spermiogenesis as non-null mutations are suppressors in the spe-8 class pathway that regulates spermatid activation in hermaphrodites (Fig. 4, also see below).
spe-10
In spe-10 mutants, the FBs of terminal spermatocytes are not delivered into budding spermatids and remain in the residual body or bud off as FB cytoplasts (Shakes and Ward, 1989b). The MOs are segregated into spermatids but become vacuolated. spe-10 mutant spermatids are small (~60% of wild-type) and their nuclei are off center. After completing spermiogenesis, the resulting pseudopods are significantly shorter than those found on wild-type spermatozoa. SPE-10 is a four-pass transmembrane protein with a DHHC (Asp-His-His-Cys)-CRD (Cys-rich domain) zinc-finger domain, which represents the catalytic site found in a large family of eukaryotic palmitoyl transferases (Linder and Deschenes, 2007). The presumptive substrate(s) for palmitoylation by SPE-10 and its role in FB-MO morphogenesis are currently unknown.
spe-17
This gene encodes a Ser/Thr-rich, highly charged protein that has no obvious homolog outside nematodes (L’Hernault et al., 1993). spe-17 mutants share several cytological phenotypes with those of spe-10 mutants (Shakes and Ward, 1989b); small spermatids (~60% of wild-type) whose nuclei are off center and are activated into spermatozoa containing short pseudopods. Unlike spe-10 mutants, in spe-17 mutant spermatids, the FBs are successfully segregated from the residual body. However, ribosomes reside on the MO membranes in spe-17 mutant spermatids, and this feature is never observed in wild-type or any other characterized C. elegans mutant that affects spermatogenesis. Despite these abnormalities, some spe-17 spermatids can activate to form spermatozoa that are capable of acting during fertilization, albeit at greatly reduced efficiency (less than 1%) as compared to wild type (L’Hernault et al., 1993).
spe-39
In spe-39 mutants (Zhu and L’Hernault, 2003), spermatogenesis usually arrests as aberrant terminal spermatocytes. Rarely observed spe-39 mutant spermatids are small and they have a nucleus that is located off center. There is no clear MO-like structure (~800 nm) in spe-39 mutants and, instead, numerous small vesicles (~100 nm), that are presumably an earlier stage of the MO biosynthetic pathway, are present in spermatocytes. Although spe-39 mutants fail to form normal FB-MOs, SPE-39 is not a component of these structures; this protein is uniformly distributed in the cytoplasm of wild-type spermatids and spermatozoa (Zhu and L’Hernault, 2003). The SPE-39 protein is widely distributed in C. elegans somatic tissues, and there are orthologs in all animals for which complete genomic sequences are available, including humans. The human spe-39 ortholog FLJ2707 is expressed in more than 20 different tissues, including those derived from embryos, germ cells, testis, ectoderm, mesoderm, and endoderm. Clues to the functions of SPE-39 have come from examination of its role in C. elegans oocytes and coelomocytes. In these diverse cell types, mutation of or RNA interference-induced knockdown of spe-39 alters cargo delivery to lysosomes, probably by affecting the endocytic vesicular pathway. Furthermore, biochemical experiments using cultured human cells revealed that the human SPE-39 ortholog is associated with a subset of the HOPS (homotypic fusion and vacuole protein sorting) complexes, and that this protein is present in sorting, recycling, and late endosomes but absent from lysosomes. These facts, together with other data, imply that the MOs of C. elegans spermatids are specialized lysosomes, and illustrate that proteins important in conserved aspects of eukaryotic vesicular trafficking can be discovered by studying C. elegans spermatogenesis (Zhu et al., 2009).
Cytoskeleton
spe-15
SPE-15 is a heavy chain of myosin VI (L’Hernault et al., 1988; Kelleher et al., 2000). SPE-15 participates in the asymmetric partitioning of organelles and other cellular constituents that occurs during spermatid budding from the residual body. As discussed above, this process typically results in the spermatid receiving MOs, mitochondria, and the nucleus but losing all its ribosomes, many intracellular membranes, and nearly all its conventional cytoskeletal proteins (actin, myosin, and tubulin) except for the paired centrioles (Ward, 1986). While the budding of spermatids does occur in spe-15 mutants, asymmetric partitioning of cellular constituents is abnormal. spe-15 mutant spermatocytes can neither efficiently segregate mitochondria and the FB-MOs into budding spermatids, nor prevent actin filaments and microtubules from segregating to spermatids (L’Hernault et al., 1988; Kelleher et al., 2000). In contrast, nuclei seem to be faithfully segregated to spermatids, despite these other profound sorting problems. The vast majority (~99%) of the resulting spe-15 mutant spermatids are not properly activated into spermatozoa when tested in vitro, but instead acquire a skewered appearance or extend fine spikes. DNA microarray data suggest that gene expression of spe-15 is confined to spermatogenesis (Reinke et al., 2004), which is consistent with the absence of any obvious defect in the somatic and female germline cells of spe-15 mutants.
spe-26
This gene encodes a soluble protein with five tandem repeats, each of ~50 amino acids (Varkey et al., 1995). These repeats are similar to those in the Drosophila protein Kelch (Xue and Cooley, 1993) and the Limulus sperm protein Scruin (Owen and DeRosier, 1993; Schmid et al., 1994; Schmid et al., 2004), that are both actin filament-binding proteins. spe-26 mutants carrying the most severe allele arrest spermatogenesis at the secondary spermatocyte stage and produce almost no spermatids. These arrested secondary spermatocytes sometimes form four nuclei, suggesting that they complete meiosis. However, spe-26 mutant spermatocytes usually show chromosome segregation defects; they can have as many as 12 nuclear DNA masses. While these abnormal spermatocytes make a meiotic apparatus, they later fail to disassemble it and also show other defects including the mislocalization of actin filaments and ER (Varkey et al., 1995). In some ways, spe-26 mutant spermatocytes exhibit morphological features that are reminiscent of spermatids; their FBs at least partly disassemble, the MOs become compact, and nuclei can sometimes have highly condensed chromatin. The spe-26 gene expresses two transcripts but only one is specific to spermatogenesis, consistent with the fact that spe-26 also affects somatic processes, including lifespan (Van Voorhies, 1992) and increased thermotolerance (Lithgow et al., 1995).
spe Genes Required for Sex-specific Spermiogenesis
Hermaphrodite-dependent spermiogenesis
spe-8 class mutants all exhibit the same phenotype; hermaphrodites are self-sterile and males are cross-fertile. These mutant hermaphrodites produce spermatids that are not activated into spermatozoa in vivo. Currently, there are five spe genes known to be in this class [spe-8 (L’Hernault et al., 1988; Shakes and Ward, 1989a), spe-12 (L’Hernault et al., 1988; Shakes and Ward, 1989a; Nance et al., 1999), spe-19 (Geldziler et al., 2005), spe-27 (Minniti et al., 1996), and spe-29 (Nance et al., 2000): for predicted proteins encoded by these genes, see Table 1]. Mating spe-8 class mutant hermaphrodites with males (wild-type or spe-8 class mutants) can result in the appearance of self-progeny. Successful execution of this experiment requires unactivated, hermaphrodite-derived spermatids to be in or near the spermatheca. These spermatids apparently respond to spermatid activator(s) present in male-derived seminal fluids, suggesting that mutations in the spe-8 class genes cause defects in the hermaphrodite-dependent pathway for spermatid activation. Both hermaphrodite-and male-derived spermatids from spe-8 class mutants are morphologically normal and are activated into normal-appearing spermatozoa by in vitro treatment with monensin or TEA. However, in vitro treatment with the activator Pronase causes mutant spermatids from either sex to arrest at an intermediate stage characterized by spiky projections that do not ever transform into a pseudopod. Therefore, spermatids from the spe-8 class mutants of either sex seem to have a defect(s) in those aspects of the spermiogenesis pathway that is affected by Pronase. These data suggest that, in hermaphrodite- and male-derived spermatids, there might be two pathways for spermatid activation that are dependent on or independent of spe-8 class genes (Fig. 4). The spe-8 class-dependent pathway seems to act during spermiogenesis in hermaphrodites, while males probably utilize the spe-8 class-independent pathway for spermatid activation (Fig. 4). Moreover, these two pathways might converge at some common points (Fig. 4). Among spe-8 class genes, spe-12 might be required for both of the spermiogenesis pathways (Fig. 4), since spe-12 mutant males show partially defective spermatid activation into spermatozoa (Nance et al., 1999).
One consequence of the sex-specific phenotypic defects in the spe-8 class mutants is that each mutant can be propagated as a homozygous strain where males must mate with hermaphrodites. This requires maintenance on agar plates, where mating is possible, because worms cannot mate in liquid growth media that are shaken. Suppressor mutants that rescue the self-sterility of spe-27 mutant hermaphrodites were isolated by mutagenizing a spe-27 homozygous male/hermaphrodite strain, transferring it to liquid media and selecting for self-fertility (Muhlrad and Ward, 2002). Hermaphrodites with elevated, but not wild-type levels of, self-fertility were identified and, interestingly, some suppressor mutations were non-null alleles of spe-4 (Gosney et al., 2008) or spe-6 (Muhlrad and Ward, 2002).
The suppressor strains that carry a spe-6 allele rescue not only spe-27 mutants but also other spe-8 class mutants (Muhlrad and Ward, 2002). Thus, SPE-6 seems to function downstream of the spe-8 class pathway, and reduction of SPE-6 kinase activity is probably required for onset of spermiogenesis (Fig. 4). Spermatids from males that are homozygous for a spe-6 suppressor allele (other alleles have not been tested yet), but not for a spe-8 class allele, are abnormal in FB-MO morphology and they precociously activate into spermatozoa within the gonads (Muhlrad and Ward, 2002). These data, together with those discussed above, show that the SPE-6 kinase participates in at least two distinct processes: FB-MO morphogenesis in spermatocytes and spermatid activation.
An unusual non-null spe-4 mutant also suppresses the self-sterility of spe-8 class mutants including spe-27 mutants, suggesting that the spe-8 class pathway is bypassed in this spe-4 mutant background (Gosney et al., 2008). This implies that one normal role of the SPE-4 presenilin is to regulate the proteolytic processing of a membrane protein(s) that is a component of the spe-8 class pathway (Fig. 4). In males carrying just the spe-4 suppressor allele, spermatocytes as well as spermatids are stored in the seminal vesicles, as occurs in males carrying spe-4 non-suppressor alleles (L’Hernault and Arduengo, 1992; Arduengo et al., 1998). Moreover, the spe-4 suppressor mutant in a background lacking any spe-8 class gene mutation causes premature spermatid activation within the male gonads. Hence, the spe-4 suppressor mutant, like spe-6 suppressor mutants (see above), causes two distinctive phenotypes; a FB-MO assembly defect in spermatocytes, which is also seen in spe-4 non-suppressor mutants, and defective spermatid activation, which is observed in spe-4 suppressor mutants but not null mutants (Gosney et al., 2008).
Male-dependent spermiogenesis
swm-1
This gene is implicated in male-specific fertility (Stanfield and Villeneuve, 2006). In swm-1 mutant males, spermatids are ectopically activated into spermatozoa within the male-reproductive tract, and these mutant sperm are not transferred normally into hermaphrodites during mating. Since swm-1 mutant spermatozoa are capable of fertilizing oocytes, the sterility of swm-1 mutant males is probably due to this transfer defect. The swm-1 gene encodes a soluble protein with two trypsin inhibitor-like domains, each of which presumably blocks different serine proteases (Fig. 4). Thus, multiple serine proteases seem to act during male-specific spermiogenesis (Fig. 4), and SWM-1 might block serine protease-dependent spermatid activation within the seminal vesicles. After a male ejaculates his spermatids into a hermaphrodite, these serine proteases probably disassociate from SWM-1 in her uterus where they participate in spermatid activation (Fig. 4).
spe Genes Required for Fertilization
Mutations in any of the six spe-9 class genes result in essentially the same phenotype [spe-9 (L’Hernault et al., 1988; Singson et al., 1998; Zannoni et al., 2003; Putiri et al., 2004), spe-13 (L’Hernault et al., 1988; Putiri et al., 2004), spe-38 (Chatterjee et al., 2005), spe-41/trp-3 (Xu and Sternberg, 2003), spe-42 (Kroft et al., 2005), and fer-14 (Kroft et al., unpublished data): also see Table 1]. The spe-9 class mutants produce spermatozoa that cannot fertilize oocytes despite their normal morphology and motility, although these mutant spermatozoa can contact the oocyte surface in the spermatheca. There are at least two interpretations to integrate these phenotypes. Firstly, spe-9 class mutant sperm bind to the oocyte plasma membrane with very low affinities. This results in mutant sperm that are easily detached from the oocyte plasma membrane or cannot proceed to the next step, such as sperm-oocyte fusion. Secondly, the spe-9 class mutant sperm normally bind to the oocyte plasma membrane, but they have a defect in sperm surface-oocyte surface fusion. While we cannot currently distinguish between these two possibilities, it is safe to assume that spe-9 class mutants are probably involved in sperm-oocyte interactions (binding and/or fusion).
spe-9
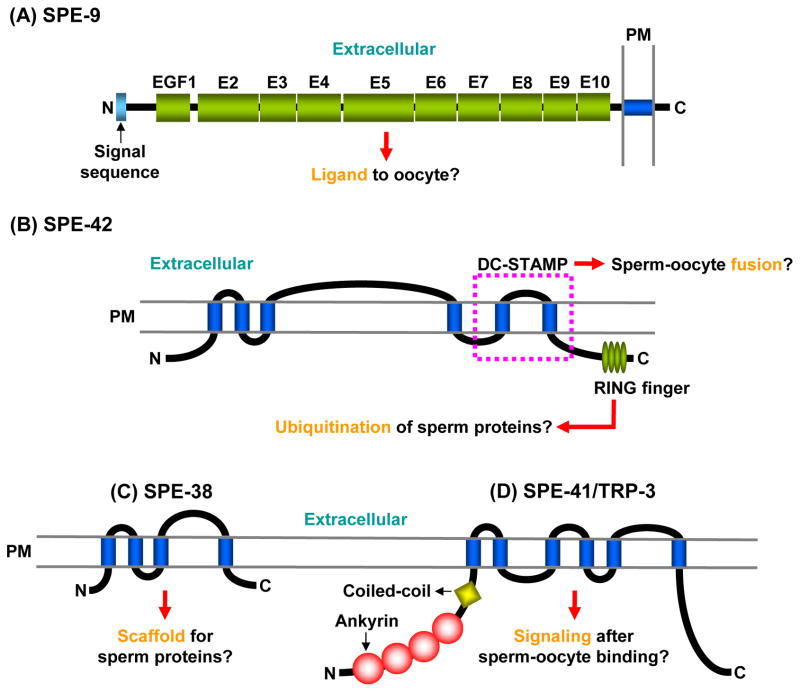
SPE-9 is a transmembrane protein with 10 epidermal growth factor (EGF)-like domains (Fig. 5A), and the overall structure of SPE-9 is similar to those of ligands for the Notch/LIN-12/GLP-1 family: Delta, Serrate, and Jagged1 proteins (Singson et al., 1998; Putiri et al., 2004). Similar to the Notch family ligands, SPE-9 might be a ligand for a putative sperm receptor(s) on the oocyte plasma membrane (Fig. 5A). This hypothesis is consistent with the fact that SPE-9 is localized on the sperm pseudopodial surface (Zannoni et al., 2003), through which sperm interact with oocytes (Fig. 2D). SPE-9 does not have a DSL domain characterizing the Delta/Serrate/LAG-2 family, suggesting that how it functions as a ligand differs significantly from the Delta family (Cordle et al., 2008).
Fig. 5.
SPE-9 class proteins presumably required for sperm-oocyte interactions. A: SPE-9. E, EGF-like domain. B: SPE-42. The region corresponding to the DC-STAMP domain is indicated by a broken purple line. C: SPE-38. D: SPE-41/TRP-3. In these figures, predicted functions are highlighted by orange letters. Blue cylinders represent the transmembrane domains. PM, plasma membrane.
spe-42
SPE-42 is a six-pass transmembrane protein whose localization is not yet clear, and this protein contains two functional domains, the DC-STAMP (dendritic cell-specific transmembrane protein) and the C4C4-type RING finger domains (Kroft et al., 2005) (Fig. 5B). Mammalian DC-STAMPs are known to play a key role in cell-cell fusion of osteoclasts and foreign body giant cells (Miyamoto, 2006). Moreover, the Drosophila gene sneaky, presumably the ortholog of spe-42, is involved in sperm plasma membrane breakdown after fly sperm enter eggs (Wilson et al., 2006). These findings suggest that SPE-42 might play a role(s) in sperm-oocyte fusion through the DC-STAMP domain.
The C4C4-type RING finger domains generally mediate protein-protein interactions (Borden, 2000). Therefore, SPE-42 could associate with another sperm protein(s) through this domain. It is also possible that the SPE-42 RING finger might have a ubiquitin E3 ligase activity, like the human RING finger protein CNOT4 (Hanzawa et al., 2001; Albert et al., 2002), although it is C3HC4-type RING fingers that usually have this enzymatic activity (Deshaies and Joazeiro, 2009). Consequently, SPE-42 possibly catalyzes the ubiquitination of a sperm protein(s) that acts during sperm-oocyte interactions (Fig. 5B), in order to regulate the localization and/or function of that protein(s).
spe-38
This gene encodes a four-pass transmembrane protein with no other significant domains (Chatterjee et al., 2005) (Fig. 5C). SPE-38 is located within MOs in spermatids but it appears on the cell surface after MO fusion with the plasma membrane. SPE-38 is probably involved in sperm-oocyte interactions because it localizes to pseudopods like SPE-9. Although the functional role(s) of SPE-38 is not yet known, this protein might act as a scaffold for another sperm protein(s) that is implicated in sperm-oocyte interactions (Fig. 5C).
spe-41/trp-3
SPE-41/TRP-3 belongs to the TRPC (transient receptor potential (TRP)-canonical) superfamily of cation channels (Xu and Sternberg, 2003) (Fig. 5D). Indeed, SPE-41/TRP-3 was demonstrated to act as a calcium channel in spermatozoa, but not in spermatids. This finding is in good agreement with the SPE-41/TRP-3 localization; SPE-41/TRP-3 is intracellularly localized in the MOs of spermatids, and during spermiogenesis, this protein translocates onto the surface of both the pseudopod and the cell body. Again, the pseudopod is the place where sperm-oocyte interactions are thought to occur. On the C. elegans sperm surface, SPE-41/TRP-3 probably forms a homo- or hetero-tetramer through the ankyrin and coiled-coil domains (Fig. 5D), like other members of the TRP family (Schindl and Romanin, 2007). Sperm-oocyte binding might produce a signal to open the SPE-41/TRP-3 channel, and the ensuing calcium influx would trigger gamete fusion (Fig. 5). Indeed, there is evidence for the TRPC family to play roles in lipid-processing events that might facilitate membrane-membrane fusion (Beech et al., 2009).
spe Gene Required for Embryogenesis
spe-11
This gene encodes a soluble protein that is localized in the perinuclear region of sperm (Browning and Strome, 1996). Embryos produced by fertilization between spe-11 mutant oocytes and wild-type sperm develop normally (Hill et al., 1989). On the other hand, spe-11 mutant sperm can fertilize wild-type oocytes, but the resulting embryos always die (L’Hernault et al., 1988; Hill et al., 1989; McNally and McNally, 2005). In wild-type sperm, the perinuclear halo surrounding the nucleus and centriolar pair is evenly distributed, while the halo in spe-11 mutant sperm is unevenly distributed and contains extra-granular materials. Additionally, computer-assisted analysis revealed that spe-11 mutant spermatozoa have a defect in the dynamic morphology of their pseudopods, although the defect does not significantly affect the crawling velocity of the mutant sperm (Royal et al., 1997).
cyk-4
CYK-4 is a sperm-enriched, Rho GTPase-activating protein (GAP). CYK-4/GAP targets RHO-1, a C. elegans ortholog of the GTPase RhoA, and cooperation of these two proteins and the kinesin-like protein ZEN-4 are required for assembly of the central spindle in embryos (Jantsch-Plunger et al., 2000). It was recently demonstrated that, after fertilization, the paternally supplied CYK-4/GAP, RHO-1/RhoA, and the guanine nucleotide-exchange factor ECT-2 together regulate an actomyosin network to produce its gradient within a one-cell embryo, which leads to cell polarity (Jenkins et al., 2006).
PERSPECTIVES
Infertility has a diverse series of causes (Matzuk and Lamb, 2008), and this disease/disorder is a serious social problem that seems to be increasing in the developed world (Greil, 1997; Fidler and Bernstein, 1999). However, there has been little progress over the past decade in identifying and treating the underlying causes. This is, at least in part, because the molecular basis of reproduction is poorly understood in animals, including humans.
There are two major reasons why C. elegans is useful for the study of reproductive biology. Firstly, as described above, numerous mutants defective in reproduction can be easily created. These mutants provide access to the molecular basis of reproduction when the affected genes are cloned and analyzed. One area that is especially important and poorly understood in any species is the mechanism of sperm-oocyte binding/fusion. The six C. elegans spe-9 class mutants, where otherwise normal spermatozoa fail to properly interact with oocytes so that fertilization does not occur, are the largest collection of this type known in any organism, and it seems likely that more mutants of this type remain to be identified. Secondly, C. elegans is optically transparent, which allows gamete development and fertilization to be analyzed “in vivo”. Since worms can be treated with small chemicals, peptides, and antibodies by soaking, feeding, and/or microinjection, they can be used as a kind of “in vivo test tube”, which is unique in reproductive research. These important properties of this model animal could allow evaluation of drugs developed for infertile or contraceptive therapies.
While we have superb tools for the in vivo analysis of fertilization in C. elegans, we currently lack an IVF assay. Development of an IVF assay would permit determination of the precise point where mutants (already available and ones obtained in the future) affect fertilization. Hopefully, at some point in the not so distant future, candidate drugs for infertile or contraceptive therapies will be testable by analyzing both in vitro and in vivo C. elegans fertilization.
Acknowledgments
C. elegans studies in our lab have been supported by the National Science Foundation (to S.W.L., IOB-0544180) and National Institute of Health (to S.W.L., GM082932). We thank all past and current members of the L’Hernault lab for valuable discussions and suggestions.
References
- Achanzar WE, Ward S. A nematode gene required for sperm vesicle fusion. J Cell Sci. 1997;110:1073–1081. doi: 10.1242/jcs.110.9.1073. [DOI] [PubMed] [Google Scholar]
- Albert TK, Hanzawa H, Legtenberg YI, de Ruwe MJ, van den Heuvel FA, Collart MA, Boelens R, Timmers HT. Identification of a ubiquitin-protein ligase subunit within the CCR4-NOT transcription repressor complex. EMBO J. 2002;21:355–364. doi: 10.1093/emboj/21.3.355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson P. Mutagenesis. In: Epstein HF, Shakes DC, editors. Caenorhabditis elegans Modern Biological Analysis of an Organism. San Diego: Academic Press, Inc; 1995. pp. 31–58. [Google Scholar]
- Arduengo PM, Appleberry OK, Chuang P, L’Hernault SW. The presenilin protein family member SPE-4 localizes to an ER/Golgi derived organelle and is required for proper cytoplasmic partitioning during Caenorhabditis elegans spermatogenesis. J Cell Sci. 1998;111:3645–3654. doi: 10.1242/jcs.111.24.3645. [DOI] [PubMed] [Google Scholar]
- Argon Y, Ward S. Caenorhabditis elegans fertilization-defective mutants with abnormal sperm. Genetics. 1980;96:413–433. doi: 10.1093/genetics/96.2.413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bae YK, Kim E, L’Hernault SW, Barr MM. The CIL-1 PI 5-phosphatase localizes TRP Polycystins to cilia and activates sperm in C. elegans. Curr Biol. 2009;19:1599–1607. doi: 10.1016/j.cub.2009.08.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bashir R, Britton S, Strachan T, Keers S, Vafiadaki E, Lako M, Richard I, Marchand S, Bourg N, Argov Z, Sadeh M, Mahjneh I, Marconi G, Passos-Bueno MR, Moreira Ede S, Zatz M, Beckmann JS, Bushby K. A gene related to Caenorhabditis elegans spermatogenesis factor fer-1 is mutated in limb-girdle muscular dystrophy type 2B. Nat Genet. 1998;20:37–42. doi: 10.1038/1689. [DOI] [PubMed] [Google Scholar]
- Beech DJ, Bahnasi YM, Dedman AM, Al-Shawaf E. TRPC channel lipid specificity and mechanisms of lipid regulation. Cell Calcium. 2009;45:583–588. doi: 10.1016/j.ceca.2009.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Booher RN, Holman PS, Fattaey A. Human Myt1 is a cell cycle-regulated kinase that inhibits Cdc2 but not Cdk2 activity. J Biol Chem. 1997;272:22300–22306. doi: 10.1074/jbc.272.35.22300. [DOI] [PubMed] [Google Scholar]
- Borden KL. RING domains: master builders of molecular scaffolds? J Mol Biol. 2000;295:1103–1112. doi: 10.1006/jmbi.1999.3429. [DOI] [PubMed] [Google Scholar]
- Brenner S. The genetics of Caenorhabditis elegans. Genetics. 1974;77:71–94. doi: 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Browning H, Strome S. A sperm-supplied factor required for embryogenesis in C. elegans. Development. 1996;122:391–404. doi: 10.1242/dev.122.1.391. [DOI] [PubMed] [Google Scholar]
- Burrows AE, Sceurman BK, Kosinski ME, Richie CT, Sadler PL, Schumacher JM, Golden A. The C. elegans Myt1 ortholog is required for the proper timing of oocyte maturation. Development. 2006;133:697–709. doi: 10.1242/dev.02241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatterjee I, Richmond A, Putiri E, Shakes DC, Singson A. The Caenorhabditis elegans spe-38 gene encodes a novel four-pass integral membrane protein required for sperm function at fertilization. Development. 2005;132:2795–2808. doi: 10.1242/dev.01868. [DOI] [PubMed] [Google Scholar]
- Cipriano DJ, Wang Y, Bond S, Hinton A, Jefferies KC, Qi J, Forgac M. Structure and regulation of the vacuolar ATPases. Biochim Biophys Acta. 2008;1777:599–604. doi: 10.1016/j.bbabio.2008.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coleman TR, Dunphy WG. Cdc2 regulatory factors. Curr Opin Cell Biol. 1994;6:877–882. doi: 10.1016/0955-0674(94)90060-4. [DOI] [PubMed] [Google Scholar]
- Cordle J, Johnson S, Tay JZ, Roversi P, Wilkin MB, de Madrid BH, Shimizu H, Jensen S, Whiteman P, Jin B, Redfield C, Baron M, Lea SM, Handford PA. A conserved face of the Jagged/Serrate DSL domain is involved in Notch trans-activation and cis-inhibition. Nat Struct Mol Biol. 2008;15:849–857. doi: 10.1038/nsmb.1457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deshaies RJ, Joazeiro CA. RING domain E3 ubiquitin ligases. Annu Rev Biochem. 2009;78:399–434. doi: 10.1146/annurev.biochem.78.101807.093809. [DOI] [PubMed] [Google Scholar]
- Dunphy WG, Brizuela L, Beach D, Newport J. The Xenopus cdc2 protein is a component of MPF, a cytoplasmic regulator of mitosis. Cell. 1988;54:423–431. doi: 10.1016/0092-8674(88)90205-x. [DOI] [PubMed] [Google Scholar]
- Fidler AT, Bernstein J. Infertility: from a personal to a public health problem. Public Health Rep. 1999;114:494–511. doi: 10.1093/phr/114.6.494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geldziler B, Chatterjee I, Singson A. The genetic and molecular analysis of spe-19, a gene required for sperm activation in Caenorhabditis elegans. Dev Biol. 2005;283:424–436. doi: 10.1016/j.ydbio.2005.04.036. [DOI] [PubMed] [Google Scholar]
- Gosney R, Liau WS, LaMunyon CW. A novel function for the presenilin family member spe-4: inhibition of spermatid activation in Caenorhabditis elegans. BMC Dev Biol. 2008;8:44. doi: 10.1186/1471-213X-8-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greil AL. Infertility and psychological distress: a critical review of the literature. Soc Sci Med. 1997;45:1679–1704. doi: 10.1016/s0277-9536(97)00102-0. [DOI] [PubMed] [Google Scholar]
- Hanzawa H, de Ruwe MJ, Albert TK, van Der Vliet PC, Timmers HT, Boelens R. The structure of the C4C4 ring finger of human NOT4 reveals features distinct from those of C3HC4 RING fingers. J Biol Chem. 2001;276:10185–10190. doi: 10.1074/jbc.M009298200. [DOI] [PubMed] [Google Scholar]
- Hill DP, Shakes DC, Ward S, Strome S. A sperm-supplied product essential for initiation of normal embryogenesis in Caenorhabditis elegans is encoded by the paternal-effect embryonic-lethal gene, spe-11. Dev Biol. 1989;136:154–166. doi: 10.1016/0012-1606(89)90138-3. [DOI] [PubMed] [Google Scholar]
- Hirsh D, Oppenheim D, Klass M. Development of the reproductive system of Caenorhabditis elegans. Dev Biol. 1976;49:200–219. doi: 10.1016/0012-1606(76)90267-0. [DOI] [PubMed] [Google Scholar]
- Italiano JE, Jr, Stewart M, Roberts TM. How the assembly dynamics of the nematode major sperm protein generate amoeboid cell motility. Int Rev Cytol. 2001;202:1–34. doi: 10.1016/s0074-7696(01)02002-2. [DOI] [PubMed] [Google Scholar]
- Iwatsubo T. The γ-secretase complex: machinery for intramembrane proteolysis. Curr Opin Neurobiol. 2004;14:379–383. doi: 10.1016/j.conb.2004.05.010. [DOI] [PubMed] [Google Scholar]
- Jantsch-Plunger V, Gonczy P, Romano A, Schnabel H, Hamill D, Schnabel R, Hyman AA, Glotzer M. CYK-4: a Rho family GTPase activating protein (GAP) required for central spindle formation and cytokinesis. J Cell Biol. 2000;149:1391–1404. doi: 10.1083/jcb.149.7.1391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenkins N, Saam JR, Mango SE. CYK-4/GAP provides a localized cue to initiate anteroposterior polarity upon fertilization. Science. 2006;313:1298–1301. doi: 10.1126/science.1130291. [DOI] [PubMed] [Google Scholar]
- Kelleher JF, Mandell MA, Moulder G, Hill KL, L’Hernault SW, Barstead R, Titus MA. Myosin VI is required for asymmetric segregation of cellular components during C. elegans spermatogenesis. Curr Biol. 2000;10:1489–1496. doi: 10.1016/s0960-9822(00)00828-9. [DOI] [PubMed] [Google Scholar]
- Kimble J, Ward S. Germ-line development and fertilization. In: Wood WB, editor. The Nematode Caenorhabditis elegans. Cold Spring Harbor: Cold Spring Harbor Laboratory Press; 1988. pp. 191–213. [Google Scholar]
- King KL, Stewart M, Roberts TM, Seavy M. Structure and macromolecular assembly of two isoforms of the major sperm protein (MSP) from the amoeboid sperm of the nematode, Ascaris suum. J Cell Sci. 1992;101:847–857. doi: 10.1242/jcs.101.4.847. [DOI] [PubMed] [Google Scholar]
- Klass MR, Hirsh D. Sperm isolation and biochemical analysis of the major sperm protein from Caenorhabditis elegans. Dev Biol. 1981;84:299–312. doi: 10.1016/0012-1606(81)90398-5. [DOI] [PubMed] [Google Scholar]
- Kroft TL, Gleason EJ, L’Hernault SW. The spe-42 gene is required for sperm-egg interactions during C. elegans fertilization and encodes a sperm-specific transmembrane protein. Dev Biol. 2005;286:169–181. doi: 10.1016/j.ydbio.2005.07.020. [DOI] [PubMed] [Google Scholar]
- L’Hernault SW. In: Male germlineC elegans II. Riddle D, Blumenthal R, Meyer BJ, Priess J, editors. Cold Spring Harbor: Cold Spring Harbor Laboratory Press; 1997. pp. 271–294. [Google Scholar]
- L’Hernault SW The C. elegans Research Community. Spermatogenesis. WormBook. 2006 doi: 10.1895/wormbook.1.7.1. http://www.wormbook.org. [DOI]
- L’Hernault SW. The genetics and cell biology of spermatogenesis in the nematode C. elegans. Mol Cell Endocrinol. 2009;306:59–65. doi: 10.1016/j.mce.2009.01.008. [DOI] [PubMed] [Google Scholar]
- L’Hernault SW, Arduengo PM. Mutation of a putative sperm membrane protein in Caenorhabditis elegans prevents sperm differentiation but not its associated meiotic divisions. J Cell Biol. 1992;119:55–68. doi: 10.1083/jcb.119.1.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- L’Hernault SW, Benian GM, Emmons RB. Genetic and molecular characterization of the Caenorhabditis elegans spermatogenesis-defective gene spe-17. Genetics. 1993;134:769–780. doi: 10.1093/genetics/134.3.769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- L’Hernault SW, Shakes DC, Ward S. Developmental genetics of chromosome I spermatogenesis-defective mutants in the nematode Caenorhabditis elegans. Genetics. 1988;120:435–452. doi: 10.1093/genetics/120.2.435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- L’Hernault SW, Singson AW. Developmental genetics of spermatogenesis in the nematode Caenorhabditis elegans. In: Goldberg E, editor. The Testes: From Stem Cell to Sperm Function, Serono Symposium USA. New York: Springer-Verlag; 2000. pp. 109–119. [Google Scholar]
- Lamitina ST, L’Hernault SW. Dominant mutations in the Caenorhabditis elegans Myt1 ortholog wee-1.3 reveal a novel domain that controls M-phase entry during spermatogenesis. Development. 2002;129:5009–5018. doi: 10.1242/dev.129.21.5009. [DOI] [PubMed] [Google Scholar]
- LaMunyon CW, Ward S. Assessing the viability of mutant and manipulated sperm by artificial insemination of Caenorhabditis elegans. Genetics. 1994;138:689–692. doi: 10.1093/genetics/138.3.689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaMunyon CW, Ward S. Sperm precedence in a hermaphroditic nematode (Caenorhabditis elegans) is due to competitive superiority of male sperm. Experientia. 1995;51:817–823. doi: 10.1007/BF01922436. [DOI] [PubMed] [Google Scholar]
- LaMunyon CW, Ward S. Larger sperm outcompete smaller sperm in the nematode Caenorhabditis elegans. Proc R Soc Lond B Biol Sci. 1998;265:1997–2002. doi: 10.1098/rspb.1998.0531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemmon MA. Membrane recognition by phospholipid-binding domains. Nat Rev Mol Cell Biol. 2008;9:99–111. doi: 10.1038/nrm2328. [DOI] [PubMed] [Google Scholar]
- Linder ME, Deschenes RJ. Palmitoylation: policing protein stability and traffic. Nat Rev Mol Cell Biol. 2007;8:74–84. doi: 10.1038/nrm2084. [DOI] [PubMed] [Google Scholar]
- Lindqvist A, Rodriguez-Bravo V, Medema RH. The decision to enter mitosis: feedback and redundancy in the mitotic entry network. J Cell Biol. 2009;185:193–202. doi: 10.1083/jcb.200812045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lithgow GJ, White TM, Melov S, Johnson TE. Thermotolerance and extended life-span conferred by single-gene mutations and induced by thermal stress. Proc Natl Acad Sci U S A. 1995;92:7540–7544. doi: 10.1073/pnas.92.16.7540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machaca K, DeFelice LJ, L’Hernault SW. A novel chloride channel localizes to Caenorhabditis elegans spermatids and chloride channel blockers induce spermatid differentiation. Dev Biol. 1996;176:1–16. doi: 10.1006/dbio.1996.9999. [DOI] [PubMed] [Google Scholar]
- Machaca K, L’Hernault SW. The Caenorhabditis elegans spe-5 gene is required for morphogenesis of a sperm-specific organelle and is associated with an inherent cold-sensitive phenotype. Genetics. 1997;146:567–581. doi: 10.1093/genetics/146.2.567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matzuk MM, Lamb DJ. The biology of infertility: research advances and clinical challenges. Nat Med. 2008;14:1197–1213. doi: 10.1038/nm.f.1895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarter J, Bartlett B, Dang T, Schedl T. On the control of oocyte meiotic maturation and ovulation in Caenorhabditis elegans. Dev Biol. 1999;205:111–128. doi: 10.1006/dbio.1998.9109. [DOI] [PubMed] [Google Scholar]
- McNally KL, McNally FJ. Fertilization initiates the transition from anaphase I to metaphase II during female meiosis in C. elegans. Dev Biol. 2005;282:218–230. doi: 10.1016/j.ydbio.2005.03.009. [DOI] [PubMed] [Google Scholar]
- Minniti AN, Sadler C, Ward S. Genetic and molecular analysis of spe-27, a gene required for spermiogenesis in Caenorhabditis elegans hermaphrodites. Genetics. 1996;143:213–223. doi: 10.1093/genetics/143.1.213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muhlrad PJ, Ward S. Spermiogenesis initiation in Caenorhabditis elegans involves a casein kinase 1 encoded by the spe-6 gene. Genetics. 2002;161:143–155. doi: 10.1093/genetics/161.1.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nance J, Davis EB, Ward S. spe-29 encodes a small predicted membrane protein required for the initiation of sperm activation in Caenorhabditis elegans. Genetics. 2000;156:1623–1633. doi: 10.1093/genetics/156.4.1623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nance J, Minniti AN, Sadler C, Ward S. spe-12 encodes a sperm cell surface protein that promotes spermiogenesis in Caenorhabditis elegans. Genetics. 1999;152:209–220. doi: 10.1093/genetics/152.1.209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson GA, Ward S. Vesicle fusion, pseudopod extension and amoeboid motility are induced in nematode spermatids by the ionophore monensin. Cell. 1980;19:457–464. doi: 10.1016/0092-8674(80)90520-6. [DOI] [PubMed] [Google Scholar]
- O’Farrell PH. Triggering the all-or-nothing switch into mitosis. Trends Cell Biol. 2001;11:512–519. doi: 10.1016/s0962-8924(01)02142-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Owen C, DeRosier D. A 13-Å map of the actin-Scruin filament from the Limulus acrosomal process. J Cell Biol. 1993;123:337–344. doi: 10.1083/jcb.123.2.337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavalko FM, Roberts TM, Holliday LS. Relationship between plasma membrane mobility and substrate attachment in the crawling movement of spermatozoa from Caenorhabditis elegans. Cell Motil Cytoskeleton. 1988;11:16–23. doi: 10.1002/cm.970110103. [DOI] [PubMed] [Google Scholar]
- Putiri E, Zannoni S, Kadandale P, Singson A. Functional domains and temperature-sensitive mutations in SPE-9, an EGF repeat-containing protein required for fertility in Caenorhabditis elegans. Dev Biol. 2004;272:448–459. doi: 10.1016/j.ydbio.2004.05.014. [DOI] [PubMed] [Google Scholar]
- Reinke V, Smith HE, Nance J, Wang J, Van Doren C, Begley R, Jones SJ, Davis EB, Scherer S, Ward S, Kim SK. A global profile of germline gene expression in C. elegans. Mol Cell. 2000;6:605–616. doi: 10.1016/s1097-2765(00)00059-9. [DOI] [PubMed] [Google Scholar]
- Roberts TM, Ward S. Centripetal flow of pseudopodial surface components could propel the amoeboid movement of Caenorhabditis elegans spermatozoa. J Cell Biol. 1982;92:132–138. doi: 10.1083/jcb.92.1.132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Royal DC, Royal MA, Wessels D, L’Hernault S, Soll DR. Quantitative analysis of Caenorhabditis elegans sperm motility and how it is affected by mutants spe11 and unc54. Cell Motil Cytoskeleton. 1997;37:98–110. doi: 10.1002/(SICI)1097-0169(1997)37:2<98::AID-CM2>3.0.CO;2-D. [DOI] [PubMed] [Google Scholar]
- Schindl R, Romanin C. Assembly domains in TRP channels. Biochem Soc Trans. 2007;35:84–85. doi: 10.1042/BST0350084. [DOI] [PubMed] [Google Scholar]
- Schmid MF, Agris JM, Jakana J, Matsudaira P, Chiu W. Three-dimensional structure of a single filament in the Limulus acrosomal bundle: Scruin binds to homologous helix-loop-beta motifs in actin. J Cell Biol. 1994;124:341–350. doi: 10.1083/jcb.124.3.341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid MF, Sherman MB, Matsudaira P, Chiu W. Structure of the acrosomal bundle. Nature. 2004;431:104–107. doi: 10.1038/nature02881. [DOI] [PubMed] [Google Scholar]
- Shakes DC, Ward S. Initiation of spermiogenesis in C. elegans: a pharmacological and genetic analysis. Dev Biol. 1989a;134:189–200. doi: 10.1016/0012-1606(89)90088-2. [DOI] [PubMed] [Google Scholar]
- Shakes DC, Ward S. Mutations that disrupt the morphogenesis and localization of a sperm-specific organelle in Caenorhabditis elegans. Dev Biol. 1989b;134:307–316. doi: 10.1016/0012-1606(89)90103-6. [DOI] [PubMed] [Google Scholar]
- Singson A, Hang JS, Parry JM. Genes required for the common miracle of fertilization in Caenorhabditis elegans. Int J Dev Biol. 2008;52:647–656. doi: 10.1387/ijdb.072512as. [DOI] [PubMed] [Google Scholar]
- Singson A, Mercer KB, L’Hernault SW. The C. elegans spe-9 gene encodes a sperm transmembrane protein that contains EGF-like repeats and is required for fertilization. Cell. 1998;93:71–79. doi: 10.1016/s0092-8674(00)81147-2. [DOI] [PubMed] [Google Scholar]
- Smith HE, Ward S. Identification of protein-protein interactions of the major sperm protein (MSP) of Caenorhabditis elegans. J Mol Biol. 1998;279:605–619. doi: 10.1006/jmbi.1998.1793. [DOI] [PubMed] [Google Scholar]
- Stanfield GM, Villeneuve AM. Regulation of sperm activation by SWM-1 is required for reproductive success of C. elegans males. Curr Biol. 2006;16:252–263. doi: 10.1016/j.cub.2005.12.041. [DOI] [PubMed] [Google Scholar]
- Van Voorhies WA. Production of sperm reduces nematode lifespan. Nature. 1992;360:456–458. doi: 10.1038/360456a0. [DOI] [PubMed] [Google Scholar]
- Varkey JP, Jansma PL, Minniti AN, Ward S. The Caenorhabditis elegans spe-6 gene is required for major sperm protein assembly and shows second site non-complementation with an unlinked deficiency. Genetics. 1993;133:79–86. doi: 10.1093/genetics/133.1.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varkey JP, Muhlrad PJ, Minniti AN, Do B, Ward S. The Caenorhabditis elegans spe-26 gene is necessary to form spermatids and encodes a protein similar to the actin-associated proteins Kelch and Scruin. Genes Dev. 1995;9:1074–1086. doi: 10.1101/gad.9.9.1074. [DOI] [PubMed] [Google Scholar]
- Ward S. Asymmetric localization of gene products during the development of Caenorhaditis elegans spermatozoa. In: Gall JG, editor. Gametogenesis and the Early Embryo. New York: Alan R. Liss, Inc; 1986. pp. 55–75. [Google Scholar]
- Ward S, Argon Y, Nelson GA. Sperm morphogenesis in wild-type and fertilization-defective mutants of Caenorhabditis elegans. J Cell Biol. 1981;91:26–44. doi: 10.1083/jcb.91.1.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward S, Carrel JS. Fertilization and sperm competition in the nematode Caenorhabditis elegans. Dev Biol. 1979;73:304–321. doi: 10.1016/0012-1606(79)90069-1. [DOI] [PubMed] [Google Scholar]
- Ward S, Hogan E, Nelson GA. The initiation of spermiogenesis in the nematode Caenorhabditis elegans. Dev Biol. 1983;98:70–79. doi: 10.1016/0012-1606(83)90336-6. [DOI] [PubMed] [Google Scholar]
- Ward S, Klass M. The location of the major protein in Caenorhabditis elegans sperm and spermatocytes. Dev Biol. 1982;92:203–208. doi: 10.1016/0012-1606(82)90164-6. [DOI] [PubMed] [Google Scholar]
- Washington NL, Ward S. FER-1 regulates Ca2+-mediated membrane fusion during C. elegans spermatogenesis. J Cell Sci. 2006;119:2552–2562. doi: 10.1242/jcs.02980. [DOI] [PubMed] [Google Scholar]
- Wilson KL, Fitch KR, Bafus BT, Wakimoto BT. Sperm plasma membrane breakdown during Drosophila fertilization requires sneaky, an acrosomal membrane protein. Development. 2006;133:4871–4879. doi: 10.1242/dev.02671. [DOI] [PubMed] [Google Scholar]
- Wolf N, Hirsh D, McIntosh JR. Spermatogenesis in males of the free-living nematode, Caenorhabditis elegans. J Ultrastruct Res. 1978;63:155–169. doi: 10.1016/s0022-5320(78)80071-9. [DOI] [PubMed] [Google Scholar]
- Xu XZ, Sternberg PW. A C. elegans sperm TRP protein required for sperm-egg interactions during fertilization. Cell. 2003;114:285–297. doi: 10.1016/s0092-8674(03)00565-8. [DOI] [PubMed] [Google Scholar]
- Xue F, Cooley L. kelch encodes a component of intercellular bridges in Drosophila egg chambers. Cell. 1993;72:681–693. doi: 10.1016/0092-8674(93)90397-9. [DOI] [PubMed] [Google Scholar]
- Yamasaki A, Eimer S, Okochi M, Smialowska A, Kaether C, Baumeister R, Haass C, Steiner H. The GxGD motif of presenilin contributes to catalytic function and substrate identification of γ-secretase. J Neurosci. 2006;26:3821–3828. doi: 10.1523/JNEUROSCI.5354-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanagimachi R. Mammalian fertilization. In: Knobil E, Neill JD, editors. The Physiology of Reproduction. New York: Raven Press, Ltd; 1994. pp. 189–317. [Google Scholar]
- Yi K, Buttery SM, Stewart M, Roberts TM. A Ser/Thr kinase required for membrane-associated assembly of the major sperm protein motility apparatus in the amoeboid sperm of Ascaris. Mol Biol Cell. 2007;18:1816–1825. doi: 10.1091/mbc.E06-08-0741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi K, Wang X, Emmett MR, Marshall AG, Stewart M, Roberts TM. Dephosphorylation of major sperm protein (MSP) fiber protein 3 by protein phosphatase 2A during cell body retraction in the MSP-based amoeboid motility of Ascaris sperm. Mol Biol Cell. 2009;20:3200–3208. doi: 10.1091/mbc.E09-03-0240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zannoni S, L’Hernault SW, Singson AW. Dynamic localization of SPE-9 in sperm: a protein required for sperm-oocyte interactions in Caenorhabditis elegans. BMC Dev Biol. 2003;3:10. doi: 10.1186/1471-213X-3-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu GD, L’Hernault SW. The Caenorhabditis elegans spe-39 gene is required for intracellular membrane reorganization during spermatogenesis. Genetics. 2003;165:145–157. doi: 10.1093/genetics/165.1.145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu GD, Salazar G, Zlatic SA, Fiza B, Doucette MM, Heilman CJ, Levey AI, Faundez V, L’Hernault SW. SPE-39 family proteins interact with the HOPS complex and function in lysosomal delivery. Mol Biol Cell. 2009;20:1223–1240. doi: 10.1091/mbc.E08-07-0728. [DOI] [PMC free article] [PubMed] [Google Scholar]