Abstract
Background and Aims
Hepatitis G virus/GB virus-C (HGV/GBV-C) is a newly identified member of the Flaviviridae family. Its clinical significance in chronic hepatitis C infection remains controversial. There is a geographical difference in the distribution of GBV-C in the world. The frequency of GBV-C infection among hepatitis C virus (HCV) infected patients varies. The aim of the current study was to determine the prevalence and genotypes of GBV-C among Iranian patients infected with chronic HCV.
Methods
Infection with GBV-C was surveyed in 71 chronic confirmed hepatitis C infected patients. These samples were collected at the Digestive Disease Research Center (DDRC) of Shariati Hospital, Tehran, Iran from January to October 2007. The 5’-UTR region of GBV-C RNA was detected using a novel in-house touchdown nested reverse transcription polymerase chain reaction (RT-PCR), the products were sequenced and the results were aligned and phylogenically analyzed.
Results
Of the 71 HCV-infected patients, 31 (43.6%) were found positive for GBV-C RNA. Sequencing and phylogenic analysis showed that the samples were Genotype 2 of GBV-C.
Conclusions
It seems that there is a high rate of GBV-C infection among Iranian patients infected with chronic HCV. In comparison with the six reference genotypes, it was observed that all the samples were categorized in Genotype 2 of GBV-C, prevalent in North America, Africa and in European countries.
Keywords: GBV-C, HCV, Prevalence, Genotyping, Iranian, 5’-UTR
Introduction
The GB virus C (GBV-C) and hepatitis G virus (HGV) were independently discovered (1, 2), but it was later determined that they were two isolates of the same virus [3][4]. Since subsequent studies did not find an association between these viruses and hepatitis, most researchers refer to the virus as GBV-C [5]. The GBV-C is a single-stranded, positive-sense RNA virus, a member of the Flaviviridae [1][2]. Based on a comparison between genome organization and sequence homologies, GBV-C is most closely related to human hepatitis C virus (HCV), another member of the Flaviviridae [1][6] However, in contrast to HCV, GBV-C does not appear to be hepatotrophic [7][8]. In fact, GBV-C replicates within the cells of the hemopoietic lineage and lymphocytes [9]. Based on sequence and phylogenetic analysis, it was observed that different genotypes of HCV differ by more than 30%, while the most extreme GBV-C variants differ by only 14% [10]. GBV-C is transmitted through blood transfusion and components [11][12][13].
Epidemiological data suggest that the virus is also spread by sexual and vertical modes of transmission [14][15][16]. GBV-C infection is relatively common and prevalent all over the world. Between 1% and 4% of healthy blood donors have GBV-C RNA detected in their sera [7][17][18]. It is prevalent in high-risk groups, such as people with hemodialysis, hemophiliacs, HCV, human immunodeficiency virus (HIV), and hepatitis B virus (HBV) infected patients, as well as in intravenous drug users.
A number of studies have shown that GBV-C has a profound “protective” influence on HIV and inhibits replication of this virus in vitro, and has been associated with a decreased risk of death and better response to treatment among HIV-positive persons as far as co-infection is concerned[19][20][21][22][23]. Co-infection of GBV-C and HCV is common. Among newly diagnosed cases of blood-borne viral hepatitis in the United States, 18% were found positive for GBV-C, and 80% of these patients were also infected with HCV [24]. Other studies have reported the incidence of GBV-C co-infection with HCV, varying from 11–31.5% [3][25][26][27][28]. The high prevalence of HCV infection and its morbidity, as well as its relation to acute and chronic hepatitis, cirrhosis, and hepatocellular carcinoma is very well documented. As for GBV-C, there is little evidence of morbidity and many doubts about its etiological role in liver diseases in situations of co-infection with HCV or HBV [5][29][30][31]. Most of the studies on GBV-C in Iran have been done on HIV co-infected patients, and the GBV-C infection-rate was reported to be 10.97% [32], 11.3% [33] and 15.5% [34]. In another study, 13% of Iranian hemodialysis patients were also found to be infected with GBV-C [35].
The aim of the current study was to determine the rate of co-infection of GBV-C and HCV, as well as the genotype distribution of GBV-C among Iranian patients infected with chronic HCV.
Based on a variation in the nucleotide sequence of 5’-untranslated region (5’-UTR), GBV-C can be classified into six major genotypes [36][37][38] .The GBV-C genotypes were described as originating in West Africa (Genotype 1), North America and Europe (Genotype 2), East Asia and Japan (Genotype 3), Southeast Asia (Genotype 4), South Africa (Genotype 5), and a recently discovered genotype in Indonesia (Genotype 6). It was also shown that different genotypes have different geographical distribution [36][38][39][40]. Apart from the six genotypes of human GBV-C, a closely related chimpanzee virus “GBV-C tro” was also discovered [41].
Although some studies have reported that Genotypes 3 and 4 are prevalent in most parts of Asia [10][42][43], our study showed that Genotype 2 of GBV-C is most prevalent in HCV-infected patients in Iran [33][35][44], as previously reported in the United Arab of Emirates [45] and Turkey [46]. In the current study, 27 GBV-C isolates from Iranian individuals infected with chronic HCV were sequenced and analyzed to determine the GBV-C genotype.
Materials and Methods
Patients
Seventy one Iranian chronic HCV infected patients, in total, who had all been referred to the Digestive Disease Research Center (DDRC) of Shariati Hospital, Tehran, Iran for treatment, were randomly selected for the study during the period January to October 2007. These patients were from different parts of Iran. The study and sampling was approved by Tarbiat Modares University’s Ethics Approval Committee, and informed consent was obtained from the patients. All of the samples were analyzed with Amplicor HCV test v2.0 (Roche Diagnostics, Germany), according to the manufacturer’s instructions, and HCV genotypes were determined based on the HCV genome sequencing of 5’-UTR regions by the restriction fragment length polymorphism (RFLP) method at Keivan Laboratory. The collected sera were stored at -70ºC in an HCV sample bank. The samples consisted of 16 females and 55 males in age ranging from 19 to 57 years old. All of the sera samples were used for RNA extraction, reverse transcription, polymerase chain reaction, and sequencing.
Amplification of GBV-C sequence
GBV-C RNA was detected by an in-house developed touchdown nested reverse transcription polymerase chain reaction (RT-PCR), using nested primers targeting the 5’-UTR region (Table 1).
Table1. Primer sequences and product length for 5'-UTR region of GBV-C.
| Primer | Primer Sequence | Product length |
| GBV-C-G1 (sense, outer) | 5’-GGTCGTAAATCCCGGTCACC-3’ | 262bp |
| GBV-C-G2 (anti-sense, outer) | 5’-CCCACTGGTCCTTGTCAACT-3’ | |
| GBV-C-G3 (sense, inner) | 5’-TAGCCACTATAGGTGGGTCT-3’ | 188bp |
| GBV-C-G4 (anti-sense, inner) | 5’-ATTGAAGGGCGACGTGGACC- |
Briefly, the total viral RNA was extracted from 140 µl of serum using the QIAamp Viral RNA Kit (Qiagen, Germany), according to the manufacturer’s instructions.Complementary DNA (cDNA) was synthesized from 2 µl of extracted RNA at 25°C for 5 min, at 42°C for 1 h, and at 72°C for 10 min, using Moloney Murine Leukaemia virus reverse transcriptase (M-MuLV-RT), and random hexamers. Each 20 µl RT master mixture contained 1 mmol dNTP, 0.01 mg/ml hexanucleotide, 7.5 U/ml M-MuLV-reverse transcriptase, 1 U/ml RNase inhibitor, and 4 µl 5X RT buffer.
Based on the reference sequences from GenBank, oligonucleotide-specific nested primer pairs were designed to amplify the 5’-untranslated region (5’-UTR), and the final expected product was 188 bp in length Table 1. Two microliters of produced cDNA were used as a template for the first round, and one microliter of the first- round PCR product was used for the second.
For the first PCR round, the PCR master mixture contained: 0.15 pmol/ul primers, 0.2 mmol dNTPs, 1.25U Taq DNA polymerase, 2.5 µl 10× PCR buffer, and 1.5 mmol MgCl2. The reagents for the second PCR round were the same as the ones used in the first round. The total volume per reaction in the two rounds was 25 µl.
Amplification was done in 25 cycles for the first round (94°C for 50 s, 55°C for 40 s, 72°C for 50 s with a final extension at 72°C for 3 min followed by a hold at 4°C) and 30 cycles for the second rounds of PCR (94°C for 40 s, 53°C for 35 s, 72°C for 40 s with a final extension at 72°C for 3 min followed by a hold at 4°C).
All PCR contamination precautions were observed; and negative controls using sera from subjects with no GBV-C markers was obtained from the Iranian Blood Transfusion Organization Research Center. PCR procedure was carefully optimized.
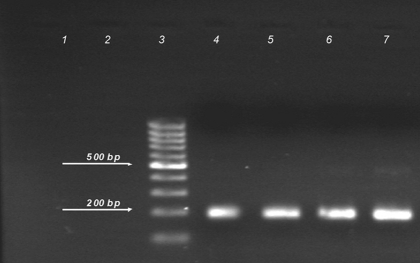
To remove small traces of non-specific bands, a touchdown procedure was adopted for the second round (94°C for 50 s, 72°C for 70 s, 8 cycles; 94°C for 40 s, 69°C for 35 s, 72°C for 40 s, 5 cycles; 94°C for 40 s, 67°C for 35 s, 72°C for 40 s, 5 cycles; 94°C for 40 s, 64°C for 35 s, 72°C for 40 s, 8 cycles; 94°C for 40 s, 62°C for 35 s, 72°C for 40 s, 4 cycles; and the final extension 72°C for 2 min followed by a hold at 4°C). The PCR products were analyzed by electrophoresis in a 2% agarose gel and stained with ethidium bromide (Fig. 1). Finally, the product band was selected and purified with the AccuPrep Gel Purification Kit (Bioneer, Korea), and the optical density was measured.
Figure 1.
Agarose gel electrophoresis of PCR products. From left: lane 1 and 2 negative samples; lane 3 DNA ladder (100 bp); lane 4 and 5 positive samples (Touchdown RT-PCR); lane 6 and 7 positive samples (Conventional RT-PCR).
PCR products were directly and bidirectionally sequenced using a BigDye Terminator cycle sequencing kit (Applied Biosystems, CA, USA) with an ABI PRISM 3700 DNA analyzer automated sequencer at Sequence Laboratories Göttingen GmbH (SEQLAB), Germany.
Randomization to either the TACE with gemcitabine plus oxaliplatin combination group (GO group) or the TACE with floxuridine plus oxaliplatin combination group (FO group) was performed without stratification by drawing consecutively numbered sealed envelopes. The protocol was approved by the ethics committee of Fujian Provincial Tumor Hospital. Written informed consent was obtained.
Reference sequences from the database
In total, 30 reference sequences of GBV-C Genotype 1–6 were obtained from GenBank and were used to compare the sequences of the isolates in the study. The accession numbers of the reported sequences and country of the reported sequences were as following: Genotype 1, U36380 (USA), AB013500 (Ghana),AB003291 (Japan); Genotype 2, U44402 (USA), AF121950 (USA), AX338086 (USA), AF081782 (China), AF309966 (Germany), D87255 (Japan), AF031827 (USA), AF031829 (USA), U45966 (USA), AF104403 (France), AB003289 (Japan), D90600 (Japan), AB013501 (Bolivia), U63715 (East Africa), AF172543 (South Africa); Genotype 3, D90601 (Japan), D87262 (Japan), AF006500 (Hong Kong), AB013501 (Japan), AB003293 (Japan), AB003288 (Japan), D87253 (Japan), D87252 (Japan); Genotype 4, AB018667 (Vietnam), AB021287 (Myanmar); Genotype 5, AF131112 (South Africa); Genotype 6, AB003292 (Japan); AF070476 (GBV-C tro).
Phylogenetic analysis
Phylogenetic analysis was performed, based on GBV-C 5’-UTR nucleotide sequence and related reference sequences. All of the results were edited and first analyzed with Bioedit (Ibis Biosciences, USA), and ClustalX (EMBL-EBI, UK) software for multiple alignments.
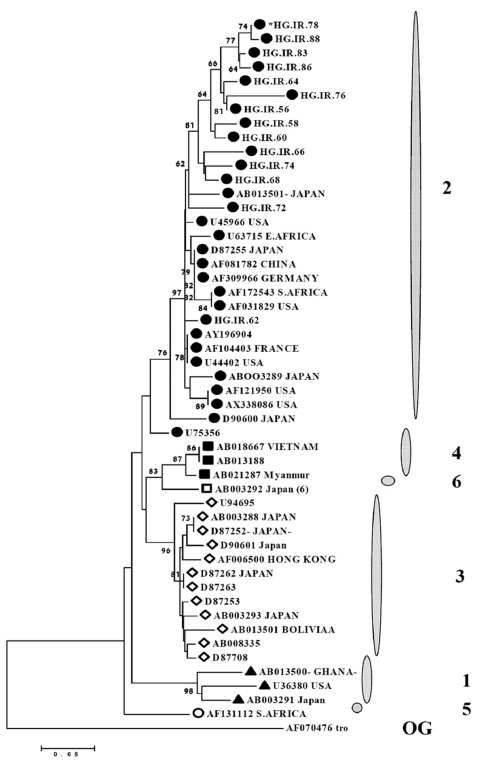
Genetic distance was estimated using the Kimura-two-parameter matrix [47]. Phylogenetic trees were constructed by the neighbor-joining (NJ) method [48]. Bootstrap resampling and reconstruction were carried out on 100 replicates to ensure consistency. The analysis and calculation of nucleotide differences within and between the isolate sequences were carried out using MEGA4 for Windows (Biodesign Institute, USA) software (Fig. 2).
Figure 2.
Phylogenetic tree constructed using the Kimura two-parameter matrix and neighbor-joining method, based on 5'-UTR sequence (188nt, residues 163 – 350). References 5’-UTR sequence from GBV-C genomes are identified by their GenBank accession numbers. 12 out of 27 Iranian sequences determined in this study are indicated by the HG.IR prefix. The numbers 1-6 designated GBV-C genotypes.
Results
GBV-C RNA was detected in 31 (43.6%) out of the 71 Iranian patients with chronic hepatitis C infection. Among the 71 patients with chronic hepatitis C, there were no significant differences in sex (P = 0.393), age (P = 0.260), and HCV genotypes in Iran (1a and 3a) (P = 0.471) in those with and without the GBV-C infection. The samples consisted of 16 (22.5%) females, 6 of them positive for GBV-C RNA, and 55 (77.5%) males, 25 of them positive for GBV-C RNA, with an age range of 19–57 years old (Table 2). Four out of the 31 5’-UTR sequenced were excluded because of unacceptable sequencing results.
Table2. Demographic characteristics of HCV-infected and HCV, GBV-C co-infected patients.
| Patients'characteristics | Total patients | Number of HCV infected patients | Number of HCV and GBV-C co-infected patients | |
| Patients’ Sex | Male | 55 | 30 | 25 |
| Female | 16 | 10 | 6 | |
| Patients’ Age Groups (years) | 17-27 | 13 | 7 | 6 |
| 28-37 | 24 | 13 | 11 | |
| 38-47 | 22 | 14 | 8 | |
| 48-57 | 12 | 6 | 6 | |
| HCV Genotype | 1a | 30 | 19 | 11 |
| 3a | 28 | 18 | 10 | |
| 1b | 5 | 1 | 4 | |
| 1a + 1b | 5 | 1 | 4 | |
| 1a + 3a | 2 | - | 2 | |
| 4 | 1 | 1 | - | |
The neighbor- joining method based, using a 1000 bootstrap re-sampling replicate tree, on the 27 5’-UTR nucleotide sequences of patients on whom successful amplification and sequencing were performed. Also 30 different reference GBV-C isolates in the GenBank database were included.
The results also showed that Genotype 2 is the most prevalent in infected patients. Based on the results, the Iranian isolates were classified as Genotype 2 with a bootstrap value of >80% based on clustering with previously characterized Genotype 2 isolates and other genotypes Fig.2.
Discussion
In the current study, GBV-C RNA was detected in 43.6% of Iranian patients infected with chronic HCV. Other studies on GBV-C have reported a relatively lower prevalence. Tanaka et al. demonstrated GBV-C and HCV coinfection in 11% of patients with chronic HCV [27], Alter et al. described this coinfection in 20% [3], Martinol et al. found GBV-C infection in 21% of patients with chronic HCV [26], Feucht et al. found GBV-C infection in 24.4% of HCV-infected patients [25], and Jie Yan et al., Sauleda S et al. and Quiros E et al. described this coinfection, respectively, in 31.5%, 21%, and 19% of patients with chronic HCV [28][49][50]. Al-knawy et al. also demonstrated the GBV-C in 31% of Saudi Arabian HCV-infected patients [51].
The aforementioned studies have shown a wide variation and a difference in prevalence in different geographical areas. The difference between the GBV-C infection-rate, in earlier studies and in the current investigation, may be due to the size of the study group, the methods used to detect GBV-C, the demographic and clinical features of patients, the virulence of different genotypes and strains, and the different patterns of transmission of virus in the world (e.g., blood and blood components, sexual, intravenous injection, etc.). Another preliminary study in Iran reported the prevalence of GBV-C among Iranian chronic HCV-infected patients, about 40% in a smaller study population [52].
The genotypic classification of GBV-C has been extended to six genotypes (1 to 6) based on the 5’-UTR, and/or E2 gene sequences. Earlier studies have suggested that the phylogenetic relationship of GBV-C complete genome sequences can also be reproduced by analysis of the 5’-UTR region [37][40]. The GBV-C genotypes have distinct geographical distributions [40]. Based on the sequence of Residue 163–350 of the 5’-UTR region of Iranian GBV-C isolates, this study found that Genotype 2 seems to be the most common.
Despite the fact that little is known about the GBV-C genotype in the Middle East/Mediterranean region, and despite the lack of significant information on GBV-C sequence(s) from this region, it is reported that Genotype 2 is common. A survey in Turkey has revealed that the predominant genotype is Genotype 2 [46]. In the United Arab Emirates, Abu Odeh et al. have also reported that Genotype 2 was the most prevalent among nationals and non-nationals alike [45].
To date, there is no information about the GBV-C sequences in Iranian patients infected with chronic HCV, and the current study reports the distribution of the GBV-C genotype based on 5’-UTR sequence and phylogenetic analysis in Iran. Twenty-seven GBV-C strains were isolated from Iranian chronic HCV-infected patients and were successfully sequenced. In phylogenetic analysis based on 5’-UTR sequences, Genotype 2 in Iranian isolates was observed with significant bootstrap values. In all phylogenetic trees based on this region, the Iranian isolates were closely related to the strains in North American and European countries.
Similar molecular epidemiology surveys have been conducted for HCV genotyping in Iran. In all of these studies, HCV genotypes have been determined based on HCV genome sequencing of 5’-UTR regions. Their data have shown that different HCV genotypes are distributed in Iran. The most prevalent HCV genotypes were 1a and 3a, and the less prevalent ones were 3b and 4 [53].
In summary, our findings indicated that the prevalence of GBV-C in Iranian patients with chronic HCV is notably high. The high prevalence of GBV-C among Iranians with chronic HCV infection probably reflects similar modes of transmission and risk factors, both intravenously and through exposure to blood and blood products, probably since it is not recommended practice to screen blood units for HGV. The study also showed that Genotype 2 of GBV-C is the most common genotype among Iranian chronic HCV-infected patients.
Acknowledgments
The authors acknowledge the support of Tarbiat Modares University and the Shiraz Transplant Research Center for this study.
References
- 1.Leary TP, Muerhoff AS, Simons JN, Pilot-Matias TJ, Erker JC, Chalmers ML, Schlauder GG, Dawson GJ, Desai SM, Mushahwar IK. Sequence and genomic organization of GBV-C: a novel member of the flaviviridae associated with human non-A-E hepatitis. J Med Virol. 1996;48(1):60–7. doi: 10.1002/(SICI)1096-9071(199601)48:1<60::AID-JMV10>3.0.CO;2-A. [DOI] [PubMed] [Google Scholar]
- 2.Linnen J, Wages J, Jr, Zhang-Keck ZY, Fry KE, Krawczynski KZ, Alter H, Koonin E, Gallagher M, Alter M, Hadziyannis S, Karayiannis P, Fung K, Nakatsuji Y, Shih JW, Young L, Piatak M, Jr, Hoover C, Fernandez J, Chen S, Zou JC, Morris T, Hyams KC, Ismay S, Lifson JD, Hess G, Foung SK, Thomas H, Bradley D, Margolis H, Kim JP. Molecular cloning and disease association of hepatitis G virus: a transfusion-transmissible agent. Science. 1996;271(5248):505–8. doi: 10.1126/science.271.5248.505. [DOI] [PubMed] [Google Scholar]
- 3.Alter HJ. The cloning and clinical implications of HGV and HGBV-C. N Engl J Med. 1996;334(23):1536–7. doi: 10.1056/NEJM199606063342310. [DOI] [PubMed] [Google Scholar]
- 4.Polgreen PM, Xiang J, Chang Q, Stapleton JT. GB virus type C/hepatitis G virus: a non-pathogenic flavivirus associated with prolonged survival in HIV-infected individuals. Microbes Infect. 2003;5(13):1255–61. doi: 10.1016/j.micinf.2003.08.006. [DOI] [PubMed] [Google Scholar]
- 5.Berzsenyi MD, Bowden DS, Roberts SK. GB virus C: insights into co-infection. J Clin Virol. 2005;33(4):257–66. doi: 10.1016/j.jcv.2005.04.002. [DOI] [PubMed] [Google Scholar]
- 6.Reed KE, Rice CM. Overview of hepatitis C virus genome structure, polyprotein processing, and protein properties. Curr Top Microbiol Immunol. 2000;242:55-84. 2000;242:55–84. doi: 10.1007/978-3-642-59605-6_4. [DOI] [PubMed] [Google Scholar]
- 7.Alter HJ. G-pers creepers, where’d you get those papers? A reassessment of the literature on the hepatitis G virus. Transfusion. 1997;37(6):569–72. doi: 10.1046/j.1537-2995.1997.37697335149.x. [DOI] [PubMed] [Google Scholar]
- 8.Laskus T, Radkowski M, Wang LF, Vargas H, Rakela J. Lack of evidence for hepatitis G virus replication in the livers of patients coinfected with hepatitis C and G viruses. J Virol. 1997;71(10):7804–6. doi: 10.1128/jvi.71.10.7804-7806.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tucker TJ, Smuts HE, Eedes C, Knobel GD, Eickhaus P, Robson SC, Kirsc RE. Evidence that the GBV-C/hepatitis G virus is primarily a lymphotropic virus. J Med Virol. 2000;61(1):52–8. [PubMed] [Google Scholar]
- 10.Smith DB, Basaras M, Frost S, Haydon D, Cuceanu N, Prescott L, Kamenka C, Millband D, Sathar MA, Simmonds P. Phylogenetic analysis of GBV-C/hepatitis G virus. J Gen Virol. 2000;81(Pt 3):769–80. doi: 10.1099/0022-1317-81-3-769. [DOI] [PubMed] [Google Scholar]
- 11.Heuft HG, Berg T, Schreier E, Künkel U, Tacke M, Schwella N, Hopf U, Salama A, Huhn D. Epidemiological and clinical aspects of hepatitis G virus infection in blood donors and immunocompromised recipients of HGV-contaminated blood. Vox Sang. 1998;74(3):161–7. [PubMed] [Google Scholar]
- 12.Roth WK, Waschk D, Marx S, Tschauder S, Zeuzem S, Bialleck H, Weber H, Seifried E. Prevalence of hepatitis G virus and its strain variant, the GB agent, in blood donations and their transmission to recipients. Transfusion. 1997;37(6):651–6. doi: 10.1046/j.1537-2995.1997.37697335162.x. [DOI] [PubMed] [Google Scholar]
- 13.Schmidt B, Korn K, Fleckenstein B. Molecular evidence for transmission of hepatitis G virus by blood transfusion. Lancet. 1996;347(9005):909. doi: 10.1016/s0140-6736(96)91396-3. [DOI] [PubMed] [Google Scholar]
- 14.Bourlet T, Guglielminotti C, Evrard M, Berthelot P, Grattard F, Frésard A, Lucht FR, Pozzetto B. Prevalence of GBV-C/hepatitis G virus RNA and E2 antibody among subjects infected with human immunodeficiency virus type 1 after parenteral or sexual exposure. J Med Virol. 1999;58(4):373–7. [PubMed] [Google Scholar]
- 15.Stark K, Doering CD, Bienzle U, Pauli G, Hamouda O, Engel AM, Schreier E. Risk and clearance of GB virus C/hepatitis G virus infection in homosexual men: A longitudinal study. J Med Virol. 1999;59(3):303–6. doi: 10.1002/(sici)1096-9071(199911)59:3<303::aid-jmv7>3.0.co;2-i. [DOI] [PubMed] [Google Scholar]
- 16.Wejstal R, Manson AS, Widell A, Norkrans G. Perinatal transmission of hepatitis G virus (GB virus type C) and hepatitis C virus infections--a comparison. Clin Infect Dis. 1999;28(4):816–21. doi: 10.1086/515187. [DOI] [PubMed] [Google Scholar]
- 17.Moaven LD, Hyland CA, Young IF, Bowden DS, McCaw R, Mison L, Locarnini SA. Prevalence of hepatitis G virus in Queensland blood donors. Med J Aust. 1996;165(7):369–71. doi: 10.5694/j.1326-5377.1996.tb125019.x. [DOI] [PubMed] [Google Scholar]
- 18.Stapleton JT. GB virus type C/Hepatitis G virus. Semin Liver Dis. 2003;23(2):137–48. doi: 10.1055/s-2003-39943. [DOI] [PubMed] [Google Scholar]
- 19.Baggio-Zappia GL, Hernandes Granato CF. HIV-GB virus C co-infection: an overview. Clin Chem Lab Med. 2009;47(1):12–9. doi: 10.1515/CCLM.2009.001. [DOI] [PubMed] [Google Scholar]
- 20.Souza IE, Zhang W, Diaz RS, Chaloner K, Klinzman D, Stapleton JT. Effect of GB virus C on response to antiretroviral therapy in HIV-infected Brazilians. HIV Med. 2006;7(1):25–31. doi: 10.1111/j.1468-1293.2005.00339.x. [DOI] [PubMed] [Google Scholar]
- 21.Xiang J, Wünschmann S, Diekema DJ, Klinzman D, Patrick KD, George SL, Stapleton JT. Effect of coinfection with GB virus C on survival among patients with HIV infection. N Engl J Med. 2001;345(10):707–14. doi: 10.1056/NEJMoa003364. [DOI] [PubMed] [Google Scholar]
- 22.Zhang W, Chaloner K, Tillmann HL, Williams CF, Stapleton JT. Effect of early and late GB virus C viraemia on survival of HIV-infected individuals: a meta-analysis. HIV Med. 2006;7(3):173–80. doi: 10.1111/j.1468-1293.2006.00366.x. [DOI] [PubMed] [Google Scholar]
- 23.Williams CF, Klinzman D, Yamashita TE, Xiang J, Polgreen PM, Rinaldo C, Liu C, Phair J, Margolick JB, Zdunek D, Hess G, Stapleton JT. Persistent GB virus C infection and survival in HIV-infected men. N Engl J Med. 2004;350(10):981–90. doi: 10.1056/NEJMoa030107. [DOI] [PubMed] [Google Scholar]
- 24.Di Bisceglie AM. Hepatitis G virus infection: a work in progress. Ann Intern Med. 1996;125(9):772–3. doi: 10.7326/0003-4819-125-9-199611010-00013. [DOI] [PubMed] [Google Scholar]
- 25.Feucht HH, Zöllner B, Polywka S, Knödler B, Schröter M, Nolte H, Laufs R. Prevalence of hepatitis G viremia among healthy subjects, individuals with liver disease, and persons at risk for parenteral transmission. J Clin Microbiol. 1997;35(3):767–8. doi: 10.1128/jcm.35.3.767-768.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Martinot M, Marcellin P, Boyer N, Detmer J, Pouteau M, Castelnau C, Degott C, Aupérin A, Collins M, Kolberg J, Wilber J, Benhamou JP, Erlinger S. Influence of hepatitis G virus infection on the severity of liver disease and response to interferon-alpha in patients with chronic hepatitis C. Ann Intern Med. 1997;126(11):874–81. doi: 10.7326/0003-4819-126-11-199706010-00004. [DOI] [PubMed] [Google Scholar]
- 27.Tanaka E, Alter HJ, Nakatsuji Y, Shih JW, Kim JP, Matsumoto A, Kobayashi M, Kiyosawa K. Effect of hepatitis G virus infection on chronic hepatitis C. Ann Intern Med. 1996;125(9):740–3. doi: 10.7326/0003-4819-125-9-199611010-00007. [DOI] [PubMed] [Google Scholar]
- 28.Yan J, Dennin RH. A high frequency of GBV-C/HGV coinfection in hepatitis C patients in Germany. World J Gastroenterol. 2000;6(6):833–41. doi: 10.3748/wjg.v6.i6.833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Alter HJ, Seeff LB. Recovery, persistence, and sequelae in hepatitis C virus infection: a perspective on long-term outcome. Semin Liver Dis. 2000;20(1):17–35. doi: 10.1055/s-2000-9505. [DOI] [PubMed] [Google Scholar]
- 30.Berzsenyi MD, Bowden DS, Kelly HA, Watson KM, Mijch AM, Hammond RA, Crowe SM, Roberts SK. Reduction in hepatitis C-related liver disease associated with GB virus C in human immunodeficiency virus coinfection. Gastroenterology. 2007;133(6):1821–30. doi: 10.1053/j.gastro.2007.08.076. [DOI] [PubMed] [Google Scholar]
- 31.Berzsenyi MD, Bowden DS, Roberts SK, Revill PA. GB virus C genotype 2 predominance in a hepatitis C virus/HIV infected population associated with reduced liver disease. J Gastroenterol Hepatol. 2009;24(8):1407–10. doi: 10.1111/j.1440-1746.2009.05920.x. [DOI] [PubMed] [Google Scholar]
- 32.Ramezani A, Mohraz M, Vahabpour R, Jam S, Banifazl M, Eslamifar A, Mahboudi F, Aghakhani A, Edalat R, Hekmat S. Frequency of hepatitis G virus infection among HIV positive subjects with parenteral and sexual exposure. J Gastrointestin Liver Dis. 2008;17(3):269–72. [PubMed] [Google Scholar]
- 33.Hekmat S, Mohraz M, Vahabpour R, Jam S, Bahramali G, Banifazl M, Aghakhani A, Eslamifar A, Mahboudi F, Edalat R, Ramezani A. Frequency and genotype of GB virus C among Iranian patients infected with HIV. J Med Virol. 2008;80(11):1941–6. doi: 10.1002/jmv.21314. [DOI] [PubMed] [Google Scholar]
- 34.Haji Molla Hoseini M, Pourfathollah AA, Mohraz M, Soheili Z, Amini S, Aghaiepour M, Samiee S, Nikoogoftar M, Meshkani R. Evaluation of circulating natural type 1 interferon-producing cells in HIV/GBV-C and HIV/HCV coinfected patients: a preliminary study. Arch Med Res. 2007;38(8):868–75. doi: 10.1016/j.arcmed.2007.05.009. [DOI] [PubMed] [Google Scholar]
- 35.Hosseini-Moghaddam SM, Keyvani H, Samadi M, Alavian SM, Mahdavimazdeh M, Daneshvar S, Razzaghi Z. GB virus type C infection in hemodialysis patients considering co-infection with hepatitis C virus. J Med Virol. 2008;80(7):1260–3. doi: 10.1002/jmv.21161. [DOI] [PubMed] [Google Scholar]
- 36.Muerhoff AS, Dawson GJ, Desai SM. A previously unrecognized sixth genotype of GB virus C revealed by analysis of 5’-untranslated region sequences. J Med Virol. 2006;78(1):105–11. doi: 10.1002/jmv.20510. [DOI] [PubMed] [Google Scholar]
- 37.Muerhoff AS, Smith DB, Leary TP, Erker JC, Desai SM, Mushahwar IK. Identification of GB virus C variants by phylogenetic analysis of 5’-untranslated and coding region sequences. J Virol. 1997;71(9):6501–8. doi: 10.1128/jvi.71.9.6501-6508.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sathar MA, York DF. Group 5: GBV-C/HGV isolates from South Africa. J Med Virol. 2001;65(1):121–2. [PubMed] [Google Scholar]
- 39.Naito H, Abe K. Genotyping system of GBV-C/HGV type 1 to type 4 by the polymerase chain reaction using type-specific primers and geographical distribution of viral genotypes. J Virol Methods. 2001;91(1):3–9. doi: 10.1016/s0166-0934(00)00207-x. [DOI] [PubMed] [Google Scholar]
- 40.Smith DB, Cuceanu N, Davidson F, Jarvis LM, Mokili JL, Hamid S, Ludlam CA, Simmonds P. Discrimination of hepatitis G virus/GBV-C geographical variants by analysis of the 5’ non-coding region. J Gen Virol. 1997;78(Pt 7):1533–42. doi: 10.1099/0022-1317-78-7-1533. [DOI] [PubMed] [Google Scholar]
- 41.Birkenmeyer LG, Desai SM, Muerhoff AS, Leary TP, Simons JN, Montes CC, Mushahwar IK. Isolation of a GB virus-related genome from a chimpanzee. J Med Virol. 1998;56(1):44–51. doi: 10.1002/(sici)1096-9071(199809)56:1<44::aid-jmv8>3.0.co;2-n. [DOI] [PubMed] [Google Scholar]
- 42.Lu L, Ng MH, Zhou B, Luo H, Nakano T, Robertson BH, Im SW. Detection and genotyping of GBV-C/HGV variants in China. Virus Res. 2001;73(2):131–44. doi: 10.1016/s0168-1702(00)00231-8. [DOI] [PubMed] [Google Scholar]
- 43.Muerhoff AS, Simons JN, Leary TP, Erker JC, Chalmers ML, Pilot-Matias TJ, Dawson GJ, Desai SM, Mushahwar IK. Sequence heterogeneity within the 5’-terminal region of the hepatitis GB virus C genome and evidence for genotypes. J Hepatol. 1996;25(3):379–84. doi: 10.1016/s0168-8278(96)80125-5. [DOI] [PubMed] [Google Scholar]
- 44.Hajikazemi E, Samadi M. Prevalence, risk factors, relation to liver disease, and genotypes of the hepatitis G (HGV) infection in hemodialysis patients in Tehran in 2005. Tehran: Iran University of Medical Sciences. 2007 [Google Scholar]
- 45.Abu Odeh RO, Al-Moslih MI, Al-Jokhdar MW, Ezzeddine SA. Detection and genotyping of GBV-C virus in the United Arab Emirates. J Med Virol. 2005;76(4):534–40. doi: 10.1002/jmv.20394. [DOI] [PubMed] [Google Scholar]
- 46.Kalkan A, Ozdarendeli A, Bulut Y, Saral Y, Ozden M, Keleştimur N, Toraman ZA. Prevalence and genotypic distribution of hepatitis GB-C/HG and TT viruses in blood donors, mentally retarded children and four groups of patients in eastern Anatolia, Turkey. Jpn J Infect Dis. 2005;58(4):222–7. [PubMed] [Google Scholar]
- 47.Kimura M. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J Mol Evol. 1980;16(2):111–20. doi: 10.1007/BF01731581. [DOI] [PubMed] [Google Scholar]
- 48.Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4(4):406–25. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 49.Quiros E, Piedrola G, Maroto MC. GBV-C RNA presence in several high-risk groups of Spain. Eur J Epidemiol. 1998;14(6):617–9. doi: 10.1023/a:1007448501661. [DOI] [PubMed] [Google Scholar]
- 50.Sauleda S, Reesink HJ, Esteban JI, Hess G, Esteban R, Guardia J. Profiles of GBV-C/hepatitis G virus markers in patients coinfected with hepatitis C virus. J Med Virol. 1999;59(1):45–51. [PubMed] [Google Scholar]
- 51.Al-Knawy B, Okamoto H, Ahmed El-Mekki A, Elbagir Khalafalla M, Al Wabel A, Qazi F, Khan A, Shatoor A. Distribution of hepatitis C genotype and co-infection rate with hepatitis G in Saudi Arabia. Hepatol Res. 2002;24(2):95. doi: 10.1016/s1386-6346(02)00080-3. [DOI] [PubMed] [Google Scholar]
- 52.Zali MR, Mayumi M, Raoufi M, Nowroozi A. GBV-C infection among patients with hepatitis C virus in the Islamic Republic of Iran: a preliminary report. East Mediterr Health J. 1999;5(5):1030–4. [PubMed] [Google Scholar]
- 53.Elahi E, Pourmand N, Chaung R, Rofoogaran A, Boisver J, Samimi-Rad K, Davis RW, Ronaghi M. Determination of hepatitis C virus genotype by Pyrosequencing. J Virol Methods. 2003;109(2):171–6. doi: 10.1016/s0166-0934(03)00068-5. [DOI] [PubMed] [Google Scholar]