Abstract
Recent genome-wide interrogations of transcribed RNA have yielded compelling evidence for pervasive and complex transcription throughout a large majority of the human genome. Tens of thousands of noncoding RNA transcripts have been identified, most of which have yet to be functionally characterized. Along with the revelation that noncoding RNAs in the human genome are surprisingly abundant, there has been a surge in molecular and genetic data showing important and diverse regulatory roles for noncoding RNA. In this report, we summarize the potential roles that noncoding RNAs may play in the molecular pathogenesis of different mental retardation disorders. We suspect that these findings are just the tip of the iceberg, with noncoding RNAs possibly being involved in disease pathogenesis at different levels and through multiple distinct mechanisms.
Keywords: 22q11 deletion/DiGeorge syndrome, Angelman syndrome, Down syndrome, Fragile X syndrome, mental retardation, microRNA, noncoding RNA, Prader-Willi syndrome, Rett syndrome
Recent genome-wide interrogations of transcribed RNA have yielded compelling evidence for pervasive and complex transcription throughout a large majority of the human genome. Nevertheless, a significant portion of this transcribed RNA appears to be non-protein coding and is currently uncharacterized. In-depth analysis of 1% of the human genome (~30 Mb) performed by the Encyclopedia of DNA Elements project revealed that 92.6% of the interrogated bases could be detected as primary transcripts and that, among these, many novel non-protein-coding transcripts could be identified (1). Similarly, another study found that the majority (64%) of polyadenylated (poly-A+) transcripts ≥200 nucleotides (nt) in length lay outside annotated protein-coding regions (2). These and other genome-wide analyses have led to the identification of tens of thousands of noncoding RNA transcripts expressed from the human genome, most of which have yet to be functionally characterized. Along with the revelation that noncoding RNAs in the human genome are surprisingly abundant, there has been a surge in molecular and genetic data showing important and diverse regulatory roles for noncoding RNA. The various classifications of noncoding RNA and their associated functions are summarized in Table 1. In this report, we will discuss the involvement and function of some of these noncoding regulatory RNAs in the context of mental retardation disorders to which each has been linked.
Table 1.
Different types of noncoding RNAs
| Noncoding RNA | Function |
|---|---|
| Ribosomal RNA (rRNA)a | RNA components of ribosome, large and small subunits |
| Transfer RNA (tRNA)a | Amino acid transfer during translation elongation |
| Small nuclear RNA (snRNA)a | Nuclear RNA processing, including splicing |
| Small nucleolar RNAa | Direct necessary chemical modifications of rRNAs, also tRNAs and snRNAs |
| microRNAb | 18- to 24-nt posttranscriptional regulators of mRNA translation; known regulatory roles in developmental timing |
| Piwi-interacting RNAb | Testis-specific, 25- to 32-nt small RNA; ~30% are associated with repeat/transposon sequences; regulatory role in controlling transposition |
| Endogenous small interfering RNAb | 21-nt small RNA derived from long double-strand RNA; regulate transcript stability through the RNA interference pathway, also associated with repeat/transposon sequences |
| Y RNAa | RNA component of Ro ribonucleoprotein complexes involved in 5S rRNA quality control; also implicated in DNA replication |
| SRP RNAa | RNA component of the signal recognition particle (SRP) required for cotranslational targeting of proteins to membranes |
| Long noncoding RNAb | Generally noncoding RNAs ≥200 nt, including Xist, TSIX, AIR, HOTAIR, H19 Evf, and others. Commonly associated with imprinted regions and DNA methylation and may have regulatory function in establishing or maintaining chromatin structure |
Housekeeping.
Regulatory.
FMRP-mediated translational regulation and the microRNA pathway
Fragile X syndrome is a common form of inherited mental retardation, with an estimated prevalence of about 1 in 4000 males and 1 in 8000 females (3). Syndromic phenotypes arise due to loss-of-function mutations in the FMR1 gene, generally due to expansion of an unstable ‘CGG’ trinucleotide repeat in its 5′ untranslated region. Large expansions of the ‘CGG’ tract are associated with dense CpG dinucleotide methylation, transcriptional silencing of the FMR1 gene, and loss of the functional protein, fragile X mental retardation protein (FMRP) (3).
Initial biochemical characterizations of FMRP indicated the involvement of noncoding RNA-based mechanisms in the molecular etiology of Fragile X syndrome. Proteins of the small, highly conserved family to which FMRP belongs (fragile X-related or FXR proteins) harbor two K homology (KH) domains and a cluster of arginine and glycine residues (an RGG box) (4, 5). KH domains and RGG boxes are common among RNA-binding proteins. Indeed, the KH domains and RGG box of FMRP have been found to mediate FMRP–RNA interactions both in vitro and in vivo (6–10). Both domains contribute to the role of FMRP as a suppressor of target messenger RNA (mRNA) translation through binding of noncoding RNA structures, including G-quartets and ‘kissing complexes’ (also known as loop–loop–pseudoknots), within the untranslated regions of target mRNAs (6, 8–15). The ability of FMRP to bind RNA and suppress translation has definite clinical relevance, as evidenced by a severely affected individual with an I304N missense mutation in the KH2 domain of FMRP (10).
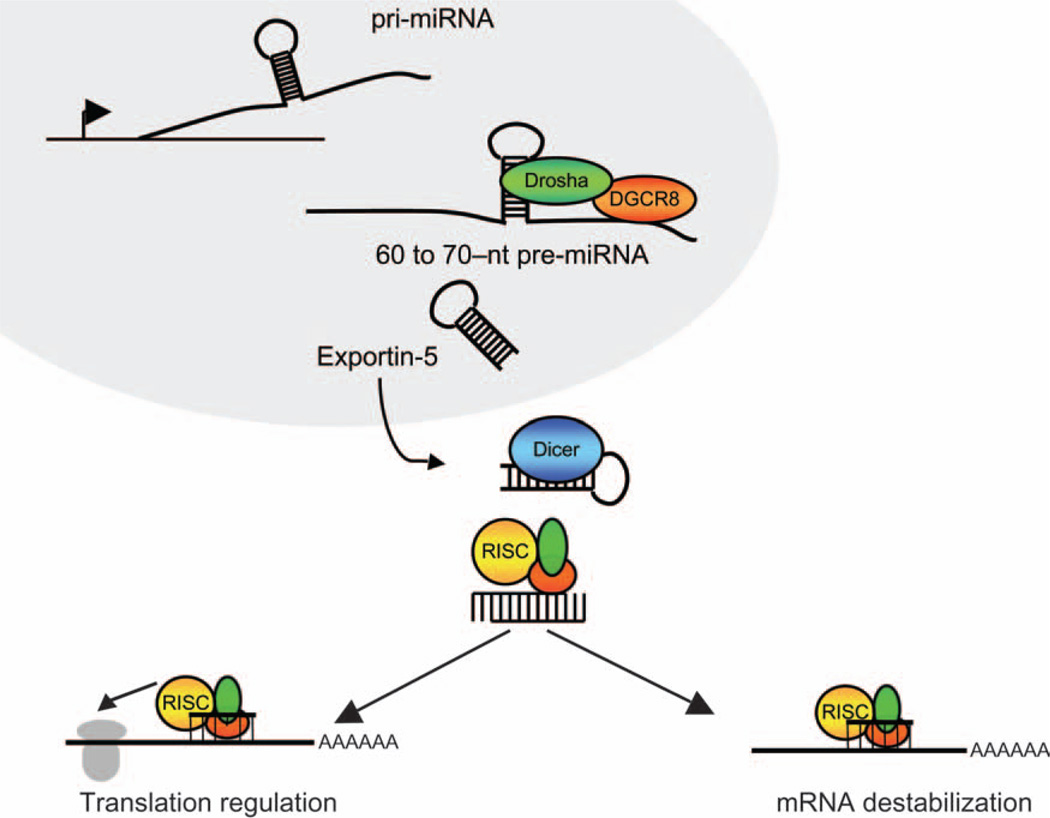
Consistent with its role in translational suppression, FMRP has more recently been linked to the microRNA (miRNA) pathway (14, 16, 17). miRNAs are 18- to 24-nt small noncoding regulatory RNAs that are known to regulate translation of target mRNA molecules in a sequence-specific manner (Table 1; summarized in Fig. 1). These small RNAs are endogenous, evolutionarily conserved genes that are generally transcribed (much like protein-coding mRNA transcripts) by RNA polymerase II, with a subset being expressed by RNA polymerase III. Primary miRNA transcripts are processed to precursor miRNA (pre-miRNA) within the nucleus by a microprocessor complex, which includes an RNAse III-type enzyme (Drosha) and its RNA-binding partner, DGCR8 (Fig. 1). Pre-miRNAs are subsequently exported from the nucleus in a Ran-GTP-dependent manner through an exportin-5-containing nuclear pore complex. Once inside the cytoplasm, pre-miRNAs are further processed into mature 18- to 24-nt miRNA by another RNase III-type enzyme (Dicer) and its associated RNA-binding cofactors. Mature miRNA are quickly loaded as single-stranded RNA onto an Argonaute-containing RNA-induced silencing complex (RISC). Generally, a miRNA then guides the RISC to cognate mRNA through fully or partially complementary sequences within their 3′ untranslated regions (3′-UTRs), thereby repressing translation of the targeted mRNA through inhibition of either translation initiation or elongation (18–21). However, more recent evidence has suggested miRNA function beyond that of translational suppression through targeting of complementary sequences in mRNA 3′-UTRs. miRNAs have now been computationally and experimentally shown to negatively regulate protein expression through targeting of mRNA-coding sequences (22). Conversely, miRNAs have also been found to upregulate translation of target mRNAs in a cell-cycle-dependant manner, switching between translational suppression in proliferating cells and translational activation in quiescent cells (23–25). In these ways, a single miRNA may simultaneously regulate the expression of multiple mRNA targets and thereby act as a rheostat to fine-tune protein expression (26, 27). A central challenge that remains toward the study of miRNA-mediated gene regulation and its relevance to human disease is the development of reliable methods by which miRNA targets may be experimentally determined. Efforts toward this end have and continue to include a combination of sequence-based computational prediction of target sites, immunoprecipitation of miRNA/mRNA-containing Argonaute complexes, and large-scale detection of mRNA destabilization and protein output in response to altered miRNA expression. Methods for the computational and experimental identification of miRNA targets have been reviewed extensively elsewhere, and we refer the reader to these reviews (28–30). Another important consideration relevant to the function of miRNA in human disease is the potential contribution of genetic variation at miRNA target sites. This topic has itself been thoroughly reviewed (31). Although this is a rapidly emerging area of interest in human disease, there are currently no examples of such a mechanism contributing to mental retardation.
Fig. 1.
The miRNA pathway. Within the nucleus, pri-miRNA transcripts are generated by either Pol II or Pol III. The stem-loop structure of the pri-miRNA is recognized and cleaved by the microprocessor complex, including Drosha and its partner, DGCR8. The 60- to 70-nt pre-miRNA is then exported from the nucleus in a Ran-GTP-dependent manner and processed a second time in the cytosol by Dicer. Processing by Dicer is immediately followed by loading of a single-stranded 18- to 24-nt mature miRNA into RISC. The mature miRNA loaded into RISC is referred to as the guide strand as it is what will guide the RISC to a target mRNA in a sequence-specific manner. Once directed to a target mRNA, RISC can mediate translation by inhibiting the initiation or elongation step or through destabilization of the target mRNA. Alternatively, miRNAs may also upregulate translation of target mRNAs in quiescent cells through an AGO2/FXR1-related mechanism, as described in the text. mRNA, messenger RNA; miRNA, microRNA; pri-miRNA, primary miRNA transcripts; RISC, RNA-induced silencing complex.
FMRP interacts biochemically and genetically with known components of the miRNA pathway. Experiments in Drosophila revealed specific biochemical interactions between dFmrp and two functional RISC proteins, dAGO1 and Dicer (16, 17). These interactions were relevant on a genetic level, as well. dAGO1 dominantly interacted with dFmr1 in both dFmr1 overexpression and loss-of-function models. Overexpression of dFmr1 leads to a mild rough eye phenotype due to increased neuronal cell death. Introduction of a recessive lethal allele of AGO1, which contains a P-element insertion that reduced its expression, suppressed the mild rough eye phenotype. The loss-of-function model revealed that dFmr1 regulates synaptic plasticity because the absence of dFmr1 results in pronounced synaptic overgrowth at the neuromuscular junctions (NMJs) of Drosophila larvae; this is reminiscent of the dendritic overgrowth observed in the brains of Fmr1 knockout mice and human patients. While both dFmr1 and dAGO1 heterozygotes had normal NMJs, a trans-heterozygote displayed strong synaptic overgrowth and over-elaboration of synaptic terminals (14). This result suggests that a limiting factor for dFmr1 function at synapses is a functional AGO1 protein, which implicates the miRNA pathway in human disease. Furthermore, dFmr1 also interacts genetically with AGO2, as exemplified by their ability to co-regulate ppk1 mRNA levels (22). Recent studies provide further evidence for the involvement of FMRP in miRNA-containing RISC and P body-like granule in Drosophila neurons (23). Additionally, recombinant human FMRP is able to act as an acceptor for Dicer-derived miRNAs, and, importantly, endogenous miRNAs themselves are associated with FMRP in both flies and mammals (14, 16, 17, 32). In the adult mouse brain, Dicer and eIF2c2 (the mouse homolog of AGO1) interact with FMRP at postsynaptic densities (33). Presumably, this interaction works to regulate translation of target mRNAs in an activity-dependent manner. As a consequence of this localized interaction, FMRP-mediated activity-dependent translational suppression is suspected to occur through the miRNA pathway (summarized in Fig. 2).
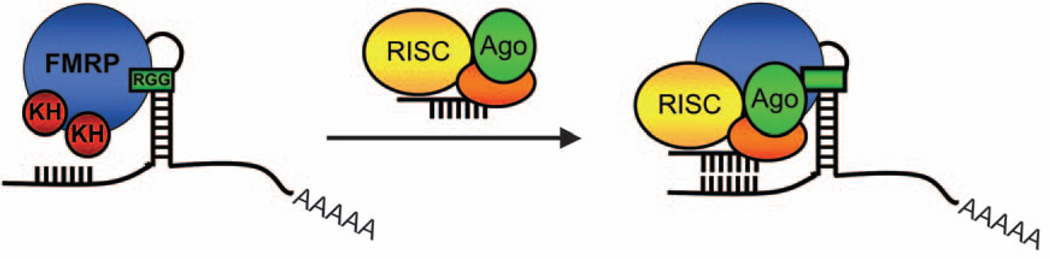
Fig. 2.
Translational regulation by fragile X mental retardation protein (FMRP) mediated through the miRNA pathway. FMRP binds target mRNAs through its two KH domains and single RGG box. RISC proteins, including Argonaute (Ago), may then interact with FMRP and use a loaded guide miRNA to interact with target sequences within the 3′-UTR of RNA bound to FMRP and suppress its translation. In this manner, the KH domains and RGG box of FMRP may help to initially bind target mRNAs, ensure proper targeting of guide miRNA-RISC within 3′-UTRs, and proper translational suppression. 3′-UTR, 3′ untranslated region; KH, K homology; mRNA, messenger RNA; miRNA, microRNA; RISC, RNA-induced silencing complex.
It is also of note that a member of the FXR protein family, FXR1, has been implicated in miRNA-mediated translational upregulation through an association with AGO2 on AU-rich 3′-UTRs in quiescent cells (23–25). However, the relevance of these observations to FMRP-mediated translational regulation and toward involvement of miRNA function in mental retardation disorders and human disease in general remains unexplored.
Disruption of miRNA biogenesis in 22q11 deletion/DiGeorge syndrome
Individuals with DiGeorge syndrome have behavioral and cognitive deficits that lead to childhood pathologies, including attention-deficit hyperactivity disorder (ADHD), obsessive–compulsive disorder, and autism spectrum disorder (34–36). These manifestations are the result of a common chromosomal abnormality, a 3-Mb hemizygous deletion on chromosome 22 (22q11.2) (37). This region (called the DiGeorge critical region) comprises more than 25 genes, making this syndrome a classic contiguous gene syndrome. Despite numerous human and mouse studies implicating a small subset of these genes (e.g. Tbx1, Comt, Prodh, and Gngl1) as contributors to the morphological or behavioral phenotypes of this syndrome (1, 38–41), the genetic basis of the cognitive impairments have gone largely unexplained. However, Gogos and colleagues recently showed that the heterozygous disruption of a single gene found in the DiGeorge critical region, Dgcr8, results in cognitive delay, specifically in spatial working and memory-based learning (42). DGCR8 was already known to be required for miRNA biogenesis (Fig. 1) (1). Gogos’s group found a reduction of mature miRNAs in the brains of mice containing either the Dgcr8 disruption or the syntenic hemizygous deletion of the DiGeorge critical region. Together, these data argue that the heterozygous loss of DGCR8 causes abnormal miRNA biogenesis and leads to a deficit in cognitive performance. The identity of the downstream targets that are misregulated in these miRNA-deficient mutants may provide further insight into the pathogenesis of DiGeorge syndrome as well as learning and cognition in general.
miRNA regulation by and of MeCP2
MECP2 is the primary gene in which de novo mutations are known to cause the X-linked dominant neurodevelopmental disorder called Rett syndrome (RTT). MECP2 encodes the DNA methyl-CpG-binding protein, MeCP2 (43). The general association of methyl CpG dinucleotides with heterochromatic or transcriptionally silent regions of the genome led to the hypothesis that MeCP2 normally functions as a component of transcriptional repressor complexes (44, 45). MeCP2-null and MeCP2 transgenic mouse models, which, respectively, mimic loss-of-function MECP2 mutations and MECP2 gene duplications, also display RTT-like phenotypes (44). Furthermore, recent clinical observations correlated duplications of MECP2 with Rett-like phenotypes, although overall such duplications result in clinically distinct phenotypes. Together, such observations are consistent with a dose-dependent mechanism for MeCP2-mediated regulation of target transcripts whose misexpression during development is pathogenic. Thus, the molecular etiology of Rett syndrome is thought of as rooted in the transcripts displaying altered expression in the absence or excess of functional MeCP2.
To date, most concerted efforts to identify MeCP2 target transcripts have focused on protein-coding mRNA transcripts. These approaches have revealed a number of direct MeCP2 target genes in specific cell and tissue types [summarized in (44)]. However, as pathogenic target transcripts have yet to be convincingly identified, we will expand the discussion of MeCP2 target transcripts to include noncoding RNAs and highlight recent observations surrounding the involvement of noncoding RNAs in MeCP2 function.
Examination of an imprinted locus on mouse chromosome 9, in which genes are known to be imprinted and expressed specifically in brain, revealed that MeCP2 binds upstream and regulates the paternal expression of a miRNA (miR-184) located within 55 kb of the imprinted locus. Moreover, the induction of miR-184 expression in depolarized cultured neurons is concomitant with a loss of MeCP2 binding upstream of the miR-184 locus. These data indicate that the regulation of miR-184 expression by MeCP2 is activity dependent. It was noted, however, that in whole brain tissue derived from MeCP2-deficient mice, there is a slight decrease in miR-184 expression compared with wild type (46).
Regulation of miRNA expression provides an alternative means by which MeCP2-mediated epigenetic regulation could ultimately influence protein expression and phenotype. Rather than directly influencing the expression of mRNA protein-coding transcripts, MeCP2 may also regulate the transcription of noncoding RNA elements, such as miRNA. Thus, in the absence of MeCP2, some miRNAs might display increased expression, which may result in a negative effect on the translation of mRNAs targeted by that particular miRNA (summarized in Fig. 3).
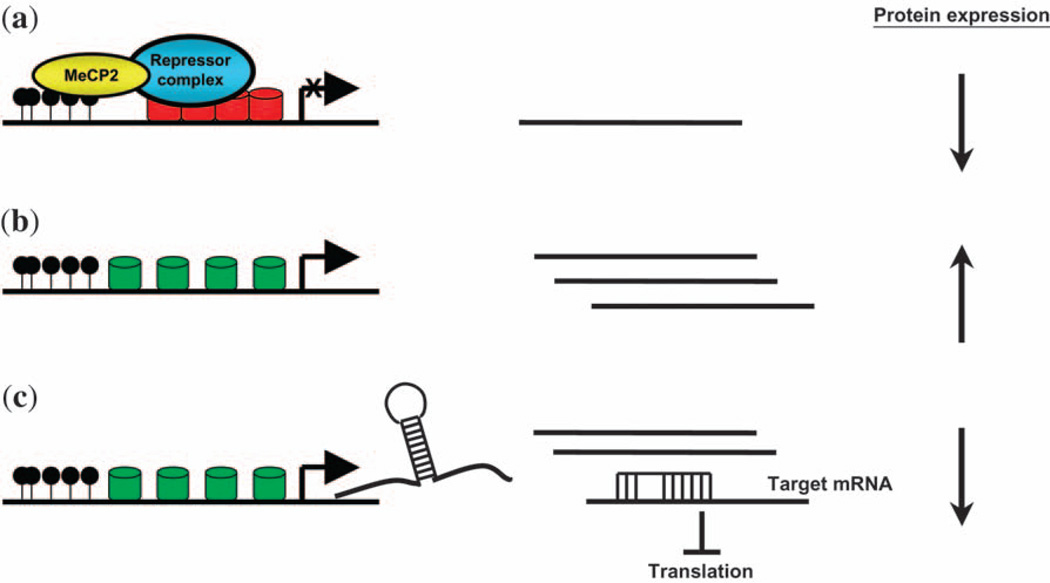
Fig. 3.
The differential effects of MeCP2-mediated transcriptional regulation of mRNA or miRNA on protein expression. (a) In this study, the example of MeCP2-mediated transcriptional repression is shown. Through interactions with methyl-CpG dinucleotides and repressor complexes, MeCP2 is able to associate with an open chromatin context. This ultimately results in the decreased protein expression due to reduced mRNA transcript level. (b) In the absence of functional MeCP2, as is the case in RTT patients, the chromatin to which MeCP2 is normally bound becomes more amenable to transcription, and the mRNA expression may be increased compared with the situation presented in (a). Ultimately, the end result is increased protein expression. (c) Nonetheless, if the target of MeCP2-mediated regulation is a miRNA, then downstream protein expression may be altered in the opposite direction. The absence of MeCP2 proximal to a miRNA results in increased expression of that miRNA. In the example shown, there is an increased translational suppression of mRNAs targeted by the miRNA that is overexpressed, causing a decrease in protein expression. Alternatively, in quiescent cells, such MeCP2-mediated epigenetic regulation of a miRNA may ultimately result in increased translation of the miRNA-targeted mRNA. mRNA, messenger RNA; miRNA, microRNA; RTT, Rett syndrome.
The cAMP response element-binding (CREB) protein is known to be a critical transcription factor regulating neuronal plasticity and activity-dependent refinement of dendritic branching, both of which are defective processes in RTT patients. Initial identification of CREB protein targets identified a miRNA (miR-132) that was predicted to posttranscriptionally regulate MeCP2. In postnatally cultured rat neurons, miR-132 did in fact directly repress the expression of MeCP2. However, by blocking miR-132-mediated regulation of MeCP2, thereby increasing MeCP2 levels, it was found that the expression of brain-derived neurotrophic factor (BDNF) increased (47). These results were contradictory to two previous studies describing the activity-dependent release of MeCP2 from the BDNF locus in embryonic cultured neurons, which indicated that MeCP2 was acting as a negative regulator of BDNF (39, 40). Because BDNF is both a known target of MeCP2 and an activator of CREB, together, these findings led to the hypothesis thatmiR-132 functions within a feedback loop involving homeostatic regulation of MeCP2 expression through BDNF-activated CREB. Homeostatic regulation of MeCP2 by miR-132 may therefore indicate a mechanism by which MeCP2 levels are normally maintained within the narrow range required for proper neuronal development and synaptic maturation in the postnatal brain and highlight the importance of miRNA in these processes (48).
The influence of MeCP2 at imprinted loci may also be of significance in RTT and other mental retardation syndromes. In fact, a common feature among imprinted loci is the presence of noncoding RNA and antisense transcription. Although much of the data surrounding MeCP2-mediated transcriptional control at imprinted loci have been controversial, with many groups obtaining conflicting results, few have examined the expression of noncoding RNA derived from these loci. The imprinted loci implicated in RTT have been reviewed in detail elsewhere (49). From these data, it remains possible that MeCP2 mediates allele-specific expression of noncoding RNAs from imprinted loci by directing or establishing a chromatin state in the imprinted region, which in turn affects the transcription of nearby protein-coding genes. In fact, MeCP2 has been proposed to function in establishing ‘chromatin loops’ that allow for proper expression of the included transcripts (49), and it is an intriguing hypothesis that noncoding RNA may play a role in the formation or maintenance of such chromatin structures.
Small nucleolar RNAs and Prader–Willi syndrome
The hallmarks of Prader–Willi syndrome (PWS) appear during infancy as muscle hypotonia, feeding difficulties, and a failure to thrive; around 18 months, these children begin to gain excessive weight, resulting in obesity, and they exhibit hypogonadism and mental retardation (50). The complex genetic etiology of this syndrome includes genomic imprinting and contiguous genes within the 15q11–15q13 genomic region (Fig. 4). The paternal inheritance of a large interstitial deletion within this region is the cause of PWS in ~70% of cases (MIM 176270). The large size of these deletions made it a challenge to identify the gene(s) responsible for this syndrome until recently when a child diagnosed with PWS had a microdeletion within this region that spanned only 175 kb (Fig. 4) (51). Remarkably, C/D box small nucleolar RNAs (snoRNAs) are the only paternal transcripts expressed in this region, effectively demonstrating that a deficiency of these snoRNAs is sufficient to cause PWS.
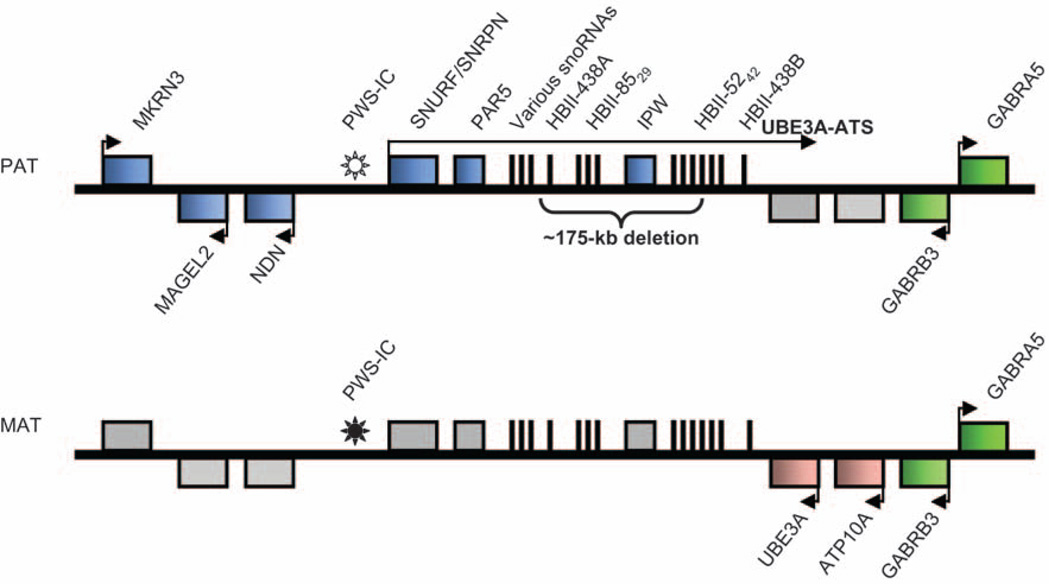
Fig. 4.
Genomic map of the human 15q11-q13 Prader–Willi syndrome/Angelman syndrome genomic region highlighting the allele-specific gene expression found in the brain. The paternally expressed genes (blue boxes) that encode proteins are MKRN3 (makorin, ring finger protein 3), MAGEL2 (MAGE-like protein 2), NDN (Necdin), and SNURF/SNRPN (bicistronic SNURF/small ribonucleoprotein N). Gray boxes indicate silenced alleles. Biallelically expressed genes (green boxes) that encode proteins are GABRB3 (γ-aminobutyric acid receptor β3) and GABRA5 (α5). UBE3A-antisense (UBE3A-ATS) is a noncoding paternally expressed transcript (>460 kb) that initiates within the SNURF/SNRPN gene. UBE3A-ATS is alternatively spliced to generate noncoding transcripts PAR5, IPW, various C/D small nucleolar RNAs (snoRNAs; HBII-13, HBII-436 and HBII-437), HBII-438A, HBII-85, HBII-52, and HBII-438B. There are 29 and 42 tandemly repeated copies of HBII-85 and HBII-52, respectively. The Prader–Willi imprinting center (PWS-IC) is unmethylated on the paternal allele (open sun) and methylated on the maternal allele (closed sun). The maternally expressed genes (pink boxes) are UBE3A (ubiquitin protein ligase E3A) and ATP10A (P-type adenosine triphosphatase). Arrows indicate the transcriptional direction. Each chromosome is depicted by a thick black line; genes shown on top of the line are transcribed from the opposite DNA strand from genes shown on the bottom of the line. The bracket indicates the approximate location of the recently identified ~175-kb deletion.
The C/D box snoRNAs expressed in the Prader–Willi genomic region arise from the introns of the paternally expressed protein-coding gene SNURF-SNRPN, which spans several hundred kilobases (52, 53). The microdeletion resulted in the loss of the HBII-438A snoRNA, the entire HBII-85 cluster (29 snoRNAs), and a portion of the HBII-52 cluster (23 of 42 snoRNAs; Fig. 4) (51). Consistent with the involvement of snoRNAs in proper brain development and mental retardation, the HBII-85 snoRNAs are expressed predominantly in both the human and the mouse brains, and a mouse model containing a deletion of the HBII-85 snoRNA cluster exhibited a deficiency in motor learning and increased anxiety (54). The function of most C/D snoRNAs is to base pair with target sequences (ribosomal RNAs and small nuclear RNAs) and mediate site-specific methylation of a ribose 2′-hydroxyl group (55). To date, the targets of the HBII-85 snoRNAs are unknown, but they are of great interest for the potential development of therapies.
An antisense transcript regulates ubiquitin ligase E3A and Angelman syndrome
Angelman syndrome (AS) is characterized by microcephaly, severe mental retardation, hyperactivity, seizures, abnormal electroencephalogram (EEG), and an ataxic gait with hand flapping (56). AS and PWS will always be linked because they share a genomic region (15q11–15q13) and a reciprocal imprinting regulation mechanism; PWS is caused by a disruption in paternally expressed genes (i.e. expressed from the chromosome contributed by the father) in this region, and AS is caused by a disruption in maternally expressed genes in this same region. Nevertheless, the genes that cause these syndromes are different and are expressed from the opposite DNA strand of their respective chromosomes (paternal vs maternal). The maternal inheritance of a large deletion within this region is the cause of AS in the majority of cases (70%) (57). Mutations in a single maternally inherited gene, ubiquitin ligase E3A (UBE3A; Fig. 4), also causes a slightly less severe AS phenotype, and mutations in other maternally inherited neighboring genes (ATP10A and GABRB3) in combination with UBE3A may contribute to the full severity of the syndrome (27, 58, 59).
UBE3A is expressed from the maternal allele in the fetal brain and in the adult frontal cortex in humans and plays a role in catalyzing the transfer of activated ubiquitin to specific target protein substrates (60–64). These findings suggested that UBE3A exhibits a tissue-specific imprint in the brain and led to the discovery of an antisense transcript, UBE3A-ATS, which is paternally expressed in the brain and functions in cis to silence expression of UBE3A from the paternal allele (65, 66). The human UBE3A-ATS is a large (~460 kb) noncoding RNA that initiates near the Prader–Willi imprinting center, extends through the SNURF/SNRPN gene, and overlaps with UBE3A (Fig. 4) (53). While the mechanism of antisense-mediated silencing used by the UBE3A-ATS remains unknown, possibilities include degradation of the sense/antisense RNA double strand, transcriptional interference due to simultaneous occupancy of RNA polymerase on the positive and negative strands of the chromosome, and an antisense-mediated chromatin modification (67–71). A mouse model that deletes the paternal promoter region of Ube3a-ATS does not express Ube3a-ATS in the brain and results in the biallelic expression of Ube3a in these mutant mouse brains (72). Unfortunately, these mutant mice also lack expression of the snoRNAs in the Prader–Willi critical region and cannot be used to determine the effect of the biallelic overexpression of Ube3a in the brain. However, children who inherit a maternal duplication of the 15q11-q13 region, which effectively simulates biallelic expression of UBE3A in the brain, have autism (73). These findings underscore the essential role that the UBE3A-ATS noncoding RNA plays in maintaining the proper amount of UBE3A expression. Further work is necessary to determine the UBE3A-ATS silencing mechanism.
Overexpression of miRNAs contributes to Down syndrome
Down syndrome (DS), which affects 1 in 700 newborns, has a variable phenotype that includes congenital heart defects, craniofacial abnormalities, and cognitive impairment (74). DS is caused by triplication of all or part of human chromosome 21 and is often referred to as trisomy 21. The extra chromosomal segment results in an increase in gene dosage by as much as 50% in multiple genes, perhaps explaining the DS phenotype (75, 76). Genotype and phenotype correlations of partial trisomy cases allowed for the identification of a Down syndrome critical region (DSCR) whose duplication is associated with many of the DS phenotypes, particularly mental retardation (77, 78). To date, we know of more than 30 genes overexpressed in key brain regions within DS individuals; 13 of them reside in the DSCR, including DOPEY2, SIM2, and DYRK1A (reviewed by Rachidi and Lopes) (79). Although these genes are all considered good candidates and need to be characterized further at the proteomic level, explorations of other genetic contributors to mental retardation in DS are underway.
Recently, Feldman and colleagues investigated the potential contribution of miRNAs in DS and computationally found that chromosome 21 encodes five miRNAs (miR-99a, let-7c, miR-125b-2, miR-155, and miR-802), all of which are overexpressed in fetal brain and heart tissues from DS individuals, suggesting that they might contribute, at least in part, to the cognitive and cardiac defects seen in DS (80). Notably, none of these miRNAs are located in the DSCR; however, another finding that supports a role for miRNAs in DS is that miR-155 downregulates a human gene associated with hypertension, angiotensin II type 1 receptor (AGTR1) (81). Indeed, DS individuals do have lower blood pressure and lower AGTR1 protein levels than those without DS.
Associations between a miRNA and a DS phenotype are unlikely to be rare, even for miR-155, because each miRNA has the ability to regulate a large number of protein-coding genes (1, 82). Moreover, improved computational and experimental methods continue to reveal the location of new miRNAs, suggesting that there remain unidentified miRNAs residing on chromosome 21 and in the DSCR. As miRNAs and their interactions with target RNAs become clear, they will make excellent candidates for the development of therapies for individuals with DS.
Other observations and implications about noncoding RNAs in mental retardation
There are additional observations linking noncoding RNAs and mental retardation, as well. Coincident with the transcriptional silencing of the FMR1 gene is also the silencing of a recently identified noncoding transcript, ASFMR1 or FMR4 (83, 84). ASFMR1 overlaps the CGG repeat of FMR1 and is transcribed in the antisense orientation. Similar to FMR1, ASFMR1 expression correlates to CGG repeat length and methylation status displaying increased expression in CGG repeat premutation carriers (55–200 repeats) and absence of expression in full mutation individuals (>200 repeats) with methylated CGG tracts. Additionally, in the presence of premutation range CGG repeats, ASFMR1 was found to display specific alternative splicing (84). Despite the region of overlap in the antisense orientation to FMR1, reduced levels of the antisense transcript were found to have no effect on the expression of FMR1 itself. However, it was noted that a reduction in the ubiquitously expressed antisense transcript resulted in a reduction in cell proliferation of cultured HEK-293T cells (83). Although the function of ASFMR1 remains obscure, the correlation of ASFMR1 expression with CGG repeat length and methylation state warrants further investigation into the contribution of this particular transcript in the etiology of Fragile X syndrome and further implicates the involvement of noncoding RNA in mental retardation.
Other observations linking noncoding RNA and mental retardation have included the detection of a genomic region on chromosome 7, 7q31, in which translocations and deletions have been associated with autism. This particular region holds a complex repertoire of transcripts, including a significant number of noncoding RNAs, collectively known as the ST7 noncoding RNA (85). Although the functions of ST7 noncoding RNAs remain uncharacterized, they are currently considered tumor suppressor genes (86, 87). One genomic region linked to a form of X-linked mental retardation classified as MRX3 and Waisman syndrome (early-onset Parkinsonism with mental retardation) harbors a miRNA, miR-175 (88). Furthermore, Beckwith–Wiedemann syndrome correlates with mild mental retardation in some instances and is linked to the H19 and LIT-1 noncoding RNAs (89, 90). A microdeletion at chromosome Xp11.3 accounts for cosegregation of retinitis pigmentosa and X-linked mental retardation in a large kindred. The region deleted includes the RP2 gene, which presumably accounts for the retinitis pigmentosa phenotype, two annotated protein-coding genes (SLC9A7 and CHST7), a zinc finger protein (FLJ20344), and two highly conserved miRNAs (miR-221 and miR-222) (91). Segmental duplications at breakpoints (BP4–BP5) of chromosome 15q13.2q13.3 result in microdeletions/duplications that are associated with a variety of neuropsychiatric abnormalities including features of autism, ADHD, anxiety disorder, mood disorder, mental retardation, epilepsy, and in some instances EEG abnormality. The ~1.5 Mb region spanning BP4–BP5 include six reference genes and one miRNA, miR-211, while some patients were found to have a smaller ~500-kb deletion that included only three reference genes and miR-211 (92).
Future prospects
Emerging data suggest that RNAs can regulate gene expression at many levels and through an array of mechanisms. Elucidating the functions of these RNAs could vastly improve our understanding and treatment of human diseases. Of particular note, recent discoveries of different types of small RNAs, including miRNAs, Piwi-interacting RNAs, and endogenous small interfering RNAs, revealed a new layer of gene regulation. These ‘small’ regulatory RNAs could play ‘big’ roles in shaping diverse cellular pathways from chromosome architecture, development, and growth control to apoptosis and stem cell maintenance. How these small regulatory RNAs may contribute to human disease pathogenesis is now being studied in earnest.
In this review, we have summarized the potential roles that different noncoding RNAs might play in the molecular pathogenesis of different mental retardation disorders. The alterations could occur at different levels from the transcriptional regulation of noncoding RNAs and the processing of noncoding RNAs (particularly miRNAs) to the potential recognition between noncoding RNAs and protein-coding mRNAs. We suspect that these findings are just the tip of the iceberg, with these and other noncoding RNAs possibly being involved in disease pathogenesis at different levels and by multiple distinct mechanisms. It is therefore important to take noncoding RNAs into consideration when trying to identify disease-causing gene(s) and dissect the biological pathway(s) altered in the pathogenesis of mental retardation.
Acknowledgements
We would like to thank C. Strauss for critical reading of the manuscript. This study was supported in part by National Institutes of Health grant (R01 MH076090). P. J. is a recipient of the Beckman Young Investigator Award and the Basil O’Connor Scholar Research Award as well as an Alfred P Sloan Research Fellow in Neuroscience.
References
- 1.Birney E, Stamatoyannopoulos JA, Dutta A, et al. Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature. 2007;447:799–816. doi: 10.1038/nature05874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kapranov P, Cheng J, Dike S, et al. RNA maps reveal new RNA classes and a possible function for pervasive transcription. Science. 2007;316:1484–1488. doi: 10.1126/science.1138341. [DOI] [PubMed] [Google Scholar]
- 3.Crawford DC, Acuna JM, Sherman SL. FMR1 and the fragile X syndrome: human genome epidemiology review. Genet Med. 2001;3:359–371. doi: 10.1097/00125817-200109000-00006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Siomi MC, Zhang Y, Siomi H, et al. Specific sequences in the fragile X syndrome protein FMR1 and the FXR proteins mediate their binding to 60S ribosomal subunits and the interactions among them. Mol Cell Biol. 1996;16:3825–3832. doi: 10.1128/mcb.16.7.3825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhang Y, O’Connor JP, Siomi MC, et al. The fragile X mental retardation syndrome protein interacts with novel homologs FXR1 and FXR2. Embo J. 1995;14:5358–5366. doi: 10.1002/j.1460-2075.1995.tb00220.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ashley CT, Jr, Wilkinson KD, Reines D, et al. FMR1 protein: conserved RNP family domains and selective RNA binding. Science. 1993;262:563–566. doi: 10.1126/science.7692601. [DOI] [PubMed] [Google Scholar]
- 7.Feng Y, Gutekunst CA, Eberhart DE, et al. Fragile X mental retardation protein: nucleocytoplasmic shuttling and association with somatodendritic ribosomes. J Neurosci. 1997;17:1539–1547. doi: 10.1523/JNEUROSCI.17-05-01539.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Laggerbauer B, Ostareck D, Keidel EM, et al. Evidence that fragile X mental retardation protein is a negative regulator of translation. Hum Mol Genet. 2001;10:329–338. doi: 10.1093/hmg/10.4.329. [DOI] [PubMed] [Google Scholar]
- 9.Li Z, Zhang Y, Ku L, et al. The fragile X mental retardation protein inhibits translation via interacting with mRNA. Nucleic Acids Res. 2001;29:2276–2283. doi: 10.1093/nar/29.11.2276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Feng Y, Absher D, Eberhart DE, et al. FMRP associates with polyribosomes as an mRNP, and the I304N mutation of severe fragile X syndrome abolishes this association. Mol Cell. 1997;1:109–118. doi: 10.1016/s1097-2765(00)80012-x. [DOI] [PubMed] [Google Scholar]
- 11.Schaeffer C, Bardoni B, Mandel JL, et al. The fragile X mental retardation protein binds specifically to its mRNA via a purine quartet motif. Embo J. 2001;20:4803–4813. doi: 10.1093/emboj/20.17.4803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ramos A, Hollingworth D, Pastore A. G-quartet-dependent recognition between the FMRP RGG box and RNA. RNA. 2003;9:1198–1207. doi: 10.1261/rna.5960503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Darnell JC, Fraser CE, Mostovetsky O, et al. Kissing complex RNAs mediate interaction between the fragile-X mental retardation protein KH2 domain and brain polyribosomes. Genes Dev. 2005;19:903–918. doi: 10.1101/gad.1276805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Darnell JC, Jensen KB, Jin P, et al. Fragile X mental retardation protein targets G quartet mRNAs important for neuronal function. Cell. 2001;107:489–499. doi: 10.1016/s0092-8674(01)00566-9. [DOI] [PubMed] [Google Scholar]
- 15.Darnell JC, Warren ST, Darnell RB. The fragile X mental retardation protein, FMRP, recognizes G-quartets. Ment Retard Dev Disabil Res Rev. 2004;10:49–52. doi: 10.1002/mrdd.20008. [DOI] [PubMed] [Google Scholar]
- 16.Caudy AA, Myers M, Hannon GJ, et al. Fragile X-related protein and VIG associate with the RNA interference machinery. Genes Dev. 2002;16:2491–2496. doi: 10.1101/gad.1025202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ishizuka A, Siomi MC, Siomi H. A Drosophila fragile X protein interacts with components of RNAi and ribosomal proteins. Genes Dev. 2002;16:2497–2508. doi: 10.1101/gad.1022002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Du T, Zamore PD. microPrimer: the biogenesis and function of microRNA. Development. 2005;132:4645–4652. doi: 10.1242/dev.02070. [DOI] [PubMed] [Google Scholar]
- 19.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 20.He L, Hannon GJ. MicroRNAs: small RNAs with a big role in gene regulation. Nat Rev Genet. 2004;5:522–531. doi: 10.1038/nrg1379. [DOI] [PubMed] [Google Scholar]
- 21.Kim VN. MicroRNA biogenesis: coordinated cropping and dicing. Nat Rev Mol Cell Biol. 2005;6:376–385. doi: 10.1038/nrm1644. [DOI] [PubMed] [Google Scholar]
- 22.Tay Y, Zhang J, Thomson AM, et al. MicroRNAs to Nanog, Oct4 and Sox2 coding regions modulate embryonic stem cell differentiation. Nature. 2008;455(7216):1124–1128. doi: 10.1038/nature07299. [DOI] [PubMed] [Google Scholar]
- 23.Vasudevan S, Tong Y, Steitz JA. Cell-cycle control of microRNA-mediated translation regulation. Cell Cycle. 2008;7:1545–1549. doi: 10.4161/cc.7.11.6018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vasudevan S, Tong Y, Steitz JA. Switching from repression to activation: microRNAs can up-regulate translation. Science. 2007;318:1931–1934. doi: 10.1126/science.1149460. [DOI] [PubMed] [Google Scholar]
- 25.Vasudevan S, Steitz JA. AU-rich-element-mediated upregulation of translation by FXR1 and Argonaute 2. Cell. 2007;128:1105–1118. doi: 10.1016/j.cell.2007.01.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Baek D, Villen J, Shin C, et al. The impact of microRNAs on protein output. Nature. 2008;455:64–71. doi: 10.1038/nature07242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Selbach M, Schwanhausser B, Thierfelder N, et al. Widespread changes in protein synthesis induced by microRNAs. Nature. 2008;455:58–63. doi: 10.1038/nature07228. [DOI] [PubMed] [Google Scholar]
- 28.Bartel DP, Chen CZ. Micromanagers of gene expression: the potentially widespread influence of metazoan microRNAs. Nat Rev Genet. 2004;5:396–400. doi: 10.1038/nrg1328. [DOI] [PubMed] [Google Scholar]
- 29.Rajewsky N. microRNA target predictions in animals. Nat Genet. 2006;38 Suppl.:S8–S13. doi: 10.1038/ng1798. [DOI] [PubMed] [Google Scholar]
- 30.Sethupathy P, Megraw M, Hatzigeorgiou AG. A guide through present computational approaches for the identification of mammalian microRNA targets. Nat Methods. 2006;3:881–886. doi: 10.1038/nmeth954. [DOI] [PubMed] [Google Scholar]
- 31.Sethupathy P, Collins FS. MicroRNA target site polymorphisms and human disease. Trends Genet. 2008 doi: 10.1016/j.tig.2008.07.004. [DOI] [PubMed] [Google Scholar]
- 32.Plante I, Davidovic L, Ouellet DL, et al. Dicer-derived microRNAs are utilized by the fragile X mental retardation protein for assembly on target RNAs. J Biomed Biotechnol. 2006;2006:64347. doi: 10.1155/JBB/2006/64347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lugli G, Larson J, Martone ME, et al. Dicer and eIF2c are enriched at postsynaptic densities in adult mouse brain and are modified by neuronal activity in a calpain-dependent manner. J Neurochem. 2005;94:896–905. doi: 10.1111/j.1471-4159.2005.03224.x. [DOI] [PubMed] [Google Scholar]
- 34.Gothelf D, Presburger G, Levy D, et al. Genetic, developmental, and physical factors associated with attention deficit hyperactivity disorder in patients with velocardiofacial syndrome. Am J Med Genet B Neuropsychiatr Genet. 2004;126:116–121. doi: 10.1002/ajmg.b.20144. [DOI] [PubMed] [Google Scholar]
- 35.Marshall CR, Noor A, Vincent JB, et al. Structural variation of chromosomes in autism spectrum disorder. Am J Hum Genet. 2008;82:477–488. doi: 10.1016/j.ajhg.2007.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gothelf D, Presburger G, Zohar AH, et al. Obsessive-compulsive disorder in patients with velocardiofacial (22q11 deletion) syndrome. Am J Med Genet B Neuropsychiatr Genet. 2004;126:99–105. doi: 10.1002/ajmg.b.20124. [DOI] [PubMed] [Google Scholar]
- 37.Edelmann L, Pandita RK, Morrow BE. Low-copy repeats mediate the common 3-Mb deletion in patients with velo-cardio-facial syndrome. Am J Hum Genet. 1999;64:1076–1086. doi: 10.1086/302343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gothelf D, Eliez S, Thompson T, et al. COMT genotype predicts longitudinal cognitive decline and psychosis in 22q11.2 deletion syndrome. Nat Neurosci. 2005;8:1500–1502. doi: 10.1038/nn1572. [DOI] [PubMed] [Google Scholar]
- 39.Torres-Juan L, Rosell J, Morla M, et al. Mutations in TBX1 genocopy the 22q11.2 deletion and duplication syndromes: a new susceptibility factor for mental retardation. Eur J Hum Genet. 2007;15:658–663. doi: 10.1038/sj.ejhg.5201819. [DOI] [PubMed] [Google Scholar]
- 40.Paterlini M, Zakharenko SS, Lai WS, et al. Transcriptional and behavioral interaction between 22q11.2 orthologs modulates schizophrenia-related phenotypes in mice. Nat Neurosci. 2005;8:1586–1594. doi: 10.1038/nn1562. [DOI] [PubMed] [Google Scholar]
- 41.Paylor R, Glaser B, Mupo A, et al. Tbx1 haploinsufficiency is linked to behavioral disorders in mice and humans: implications for 22q11 deletion syndrome. Proc Natl Acad Sci U S A. 2006;103:7729–7734. doi: 10.1073/pnas.0600206103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Stark KL, Xu B, Bagchi A, Lai WS, Liu H, Hsu R, Wan X, Pavlidis P, Mills AA, Karayiorgou M, Gogos JA. Altered brain microRNA biogenesis contributes to phenotypic deficits in a 22q11-deletion mouse model. Nat Genet. 2008;40:751–760. doi: 10.1038/ng.138. [DOI] [PubMed] [Google Scholar]
- 43.Amir RE, Van den Veyver IB, Wan M, et al. Rett syndrome is caused by mutations in X-linkedMECP2, encoding methyl-CpG-binding protein 2. Nat Genet. 1999;23:185–188. doi: 10.1038/13810. [DOI] [PubMed] [Google Scholar]
- 44.Chahrour M, Zoghbi HY. The story of Rett syndrome: from clinic to neurobiology. Neuron. 2007;56:422–437. doi: 10.1016/j.neuron.2007.10.001. [DOI] [PubMed] [Google Scholar]
- 45.Nan X, Ng HH, Johnson CA, et al. Transcriptional repression by the methyl-CpG-binding protein MeCP2 involves a histone deacetylase complex. Nature. 1998;393:386–389. doi: 10.1038/30764. [DOI] [PubMed] [Google Scholar]
- 46.Nomura T, Kimura M, Horii T, et al. MeCP2-dependent repression of an imprinted miR-184 released by depolarization. Hum Mol Genet. 2008;17:1192–1199. doi: 10.1093/hmg/ddn011. [DOI] [PubMed] [Google Scholar]
- 47.Vo N, Klein ME, Varlamova O, et al. A cAMP-response element binding protein-induced microRNA regulates neuronal morphogenesis. Proc Natl Acad Sci U S A. 2005;102:16426–16431. doi: 10.1073/pnas.0508448102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Klein ME, Lioy DT, Ma L, et al. Homeostatic regulation of MeCP2 expression by a CREB-induced microRNA. Nat Neurosci. 2007;10:1513–1514. doi: 10.1038/nn2010. [DOI] [PubMed] [Google Scholar]
- 49.LaSalle JM. The odyssey of MeCP2 and parental imprinting. Epigenetics. 2007;2:5–10. doi: 10.4161/epi.2.1.3697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gunay-Aygun M, Schwartz S, Heeger S, et al. The changing purpose of Prader-Willi syndrome clinical diagnostic criteria and proposed revised criteria. Pediatrics. 2001;108:E92. doi: 10.1542/peds.108.5.e92. [DOI] [PubMed] [Google Scholar]
- 51.Sahoo T, del Gaudio D, German JR, et al. Prader-Willi phenotype caused by paternal deficiency for the HBII-85 C/D box small nucleolar RNA cluster (see comment) Nat Genet. 2008;40:719–721. doi: 10.1038/ng.158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.de los Santos T, Schweizer J, Rees CA, et al. Small evolutionarily conserved RNA, resembling C/D box small nucleolar RNA, is transcribed from PWCR1, a novel imprinted gene in the Prader-Willi deletion region, which Is highly expressed in brain. Am J Hum Genet. 2000;67:1067–1082. doi: 10.1086/303106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Runte M, Huttenhofer A, Gross S, et al. The IC-SNURF-SNRPN transcript serves as a host for multiple small nucleolar RNA species and as an antisense RNA for UBE3A. Hum Mol Genet. 2001;10:2687–2700. doi: 10.1093/hmg/10.23.2687. [DOI] [PubMed] [Google Scholar]
- 54.Ding F, LI H, Zhang S, et al. SnoRNA Snord116 (Pwcr1/MBII-85) deletion causes growth deficiency and hyperphagia in mice. PLoS ONE. 2008;3:e1709, 1–18. doi: 10.1371/journal.pone.0001709. (Epub ahead of print) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kiss T. Small nucleolar RNAs: an abundant group of noncoding RNAs with diverse cellular functions. Cell. 2002;109:145–148. doi: 10.1016/s0092-8674(02)00718-3. [DOI] [PubMed] [Google Scholar]
- 56.Williams CA, Beaudet AL, Clayton-Smith J, et al. Angelman syndrome 2005: updated consensus for diagnostic criteria. Am J Med Genet A. 2006;140:413–418. doi: 10.1002/ajmg.a.31074. [DOI] [PubMed] [Google Scholar]
- 57.Karres JS, Hilgers V, Carrera I, et al. The conserved microRNA miR8 tunes atrophin levels to prevent neuro-degeneration in Drosophila. Cell. 2007;131:136–145. doi: 10.1016/j.cell.2007.09.020. [DOI] [PubMed] [Google Scholar]
- 58.Malzac P, Webber H, Moncla A, et al. Mutation analysis of UBE3A in Angelman syndrome patients. Am J Hum Genet. 1998;62:1353–1360. doi: 10.1086/301877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lossie AC, Whitney MM, Amidon D, et al. Distinct phenotypes distinguish the molecular classes of Angelman syndrome. J Med Genet. 2001;38:834–845. doi: 10.1136/jmg.38.12.834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Scheffner M, Huibregtse JM, Vierstra RD, et al. The HPV-16 E6 and E6-AP complex functions as a ubiquitin-protein ligase in the ubiquitination of p53. Cell. 1993;75:495–505. doi: 10.1016/0092-8674(93)90384-3. [DOI] [PubMed] [Google Scholar]
- 61.Miura K, Kishino T, Li E, et al. Neurobehavioral and electroencephalographic abnormalities in Ube3a maternal-deficient mice. Neurobiol Dis. 2002;9:149–159. doi: 10.1006/nbdi.2001.0463. [DOI] [PubMed] [Google Scholar]
- 62.Herzing LB, Kim SJ, Cook EH, Jr, et al. The human aminophospholipid-transporting ATPase gene ATP10C maps adjacent to UBE3A and exhibits similar imprinted expression. Am J Hum Genet. 2001;68:1501–1505. doi: 10.1086/320616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Rougeulle C, Glatt H, Lalande M. The Angelman syndrome candidate gene, UBE3A/E6-AP, is imprinted in brain. Nat Genet. 1997;17:14–15. doi: 10.1038/ng0997-14. [DOI] [PubMed] [Google Scholar]
- 64.Vu TH, Hoffman AR. Imprinting of the Angelman syndrome gene, UBE3A, is restricted to brain. Nat Genet. 1997;17:12–13. doi: 10.1038/ng0997-12. [DOI] [PubMed] [Google Scholar]
- 65.Rougeulle C, Cardoso C, Fontes M, et al. An imprinted antisense RNA overlaps UBE3A and a second maternally expressed transcript. Nat Genet. 1998;19:15–16. doi: 10.1038/ng0598-15. [DOI] [PubMed] [Google Scholar]
- 66.Rougeulle C, Lalande M. Angelman syndrome: how many genes to remain silent? Neurogenetics. 1998;1:229–237. doi: 10.1007/s100480050034. [DOI] [PubMed] [Google Scholar]
- 67.Lavorgna G, Dahary D, Lehner B, et al. In search of antisense. Trends Biochem Sci. 2004;29:88–94. doi: 10.1016/j.tibs.2003.12.002. [DOI] [PubMed] [Google Scholar]
- 68.Heard E, Lovell-Badge R, Avner P. Anti-Xistentialism (comment) Nat Genet. 1999;21:343–344. doi: 10.1038/7661. [DOI] [PubMed] [Google Scholar]
- 69.Navarro P, Pichard S, Ciaudo C, et al. Tsix transcription across the Xist gene alters chromatin conformation without affecting Xist transcription: implications for X-chromosome inactivation. Genes Dev. 2005;19:1474–1484. doi: 10.1101/gad.341105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Rougeulle C, Heard E. Antisense RNA in imprinting: spreading silence through air. Trends Genet. 2002;18:434–437. doi: 10.1016/s0168-9525(02)02749-x. [DOI] [PubMed] [Google Scholar]
- 71.Shibata S, Lee JT. Tsix transcription- versus RNA-based mechanisms in Xist repression and epigenetic choice. Curr Biol. 2004;14:1747–1754. doi: 10.1016/j.cub.2004.09.053. [DOI] [PubMed] [Google Scholar]
- 72.Chamberlain SJ, Brannan CI. The Prader-Willi syndrome imprinting center activates the paternally expressed murine Ube3a antisense transcript but represses paternal Ube3a. Genomics. 2001;73:316–322. doi: 10.1006/geno.2001.6543. [DOI] [PubMed] [Google Scholar]
- 73.Cook EH, Jr, Lindgren V, Leventhal BL, et al. Autism or atypical autism in maternally but not paternally derived proximal 15q duplication. Am J Hum Genet. 1997;60:928–934. [PMC free article] [PubMed] [Google Scholar]
- 74.Epstein CJ. Down syndrome (trisomy 21) New York, NY: McGraw-Hill Inc.; 1995. [Google Scholar]
- 75.Gardiner K, Costa AC. The proteins of human chromosome 21. Am J Med Genet C. 2006;142:196–205. doi: 10.1002/ajmg.c.30098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Mao R, Zielke CL, Zielke HR, et al. Global up-regulation of chromosome 21 gene expression in the developing Down syndrome brain. Genomics. 2003;81:457–467. doi: 10.1016/s0888-7543(03)00035-1. [DOI] [PubMed] [Google Scholar]
- 77.Delabar JM, Theophile D, Rahmani Z, et al. Molecular mapping of twenty-four features of Down syndrome on chromosome 21. Eur J Hum Genet. 1993;1:114–124. doi: 10.1159/000472398. [DOI] [PubMed] [Google Scholar]
- 78.Korenberg JR, Chen XN, Schipper R, et al. Down syndrome phenotypes: the consequences of chromosomal imbalance. Proc Natl Acad Sci U S A. 1994;91:4997–5001. doi: 10.1073/pnas.91.11.4997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Rachidi M, Lopes C. Mental retardation in Down syndrome: from gene dosage imbalance to molecular and cellular mechanisms. Neurosci Res. 2007;59:349–369. doi: 10.1016/j.neures.2007.08.007. [DOI] [PubMed] [Google Scholar]
- 80.Kuhn DE, Nuovo GJ, Martin MM, et al. Human chromosome 21-derived miRNAs are overexpressed in down syndrome brains and hearts. Biochem Biophys Res Commun. 2008;370:473–477. doi: 10.1016/j.bbrc.2008.03.120. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 81.Sethupathy P, Borel C, Gagnebin M, et al. Human microRNA-155 on chromosome 21 differentially interacts with its polymorphic target in the AGTR1 3′ untranslated region: a mechanism for functional single-nucleotide polymorphisms related to phenotypes. Am J Hum Genet. 2007;81:405–413. doi: 10.1086/519979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Bushati N, Cohen SM. microRNA functions. Annu Rev Cell Dev Biol. 2007;23:175–205. doi: 10.1146/annurev.cellbio.23.090506.123406. [DOI] [PubMed] [Google Scholar]
- 83.Khalil AM, Faghihi MA, Modarresi F, et al. A novel RNA transcript with antiapoptotic function is silenced in fragile X syndrome. PLoS ONE. 2008;3:e1486. doi: 10.1371/journal.pone.0001486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Ladd PD, Smith LE, Rabaia NA, et al. An antisense transcript spanning the CGG repeat region of FMR1 is upregulated in premutation carriers but silenced in full mutation individuals. Hum Mol Genet. 2007;16:3174–3187. doi: 10.1093/hmg/ddm293. [DOI] [PubMed] [Google Scholar]
- 85.Scherer SW, Cheung J, MacDonald JR, et al. Human chromosome 7: DNA sequence and biology. Science. 2003;300:767–772. doi: 10.1126/science.1083423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Zenklusen JC, Conti CJ, Green ED. Mutational and functional analyses reveal that ST7 is a highly conserved tumor-suppressor gene on human chromosome 7q31. Nat Genet. 2001;27:392–398. doi: 10.1038/86891. [DOI] [PubMed] [Google Scholar]
- 87.Zenklusen JC, Weitzel JN, Ball HG, et al. Allelic loss at 7q31.1 in human primary ovarian carcinomas suggests the existence of a tumor suppressor gene. Oncogene. 1995;11:359–363. [PubMed] [Google Scholar]
- 88.Dostie J, Mourelatos Z, Yang M, et al. Numerous microRNPs in neuronal cells containing novel microRNAs. RNA. 2003;9:180–186. doi: 10.1261/rna.2141503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Sparago A, Cerrato F, Vernucci M, et al. Microdeletions in the human H19 DMR result in loss of IGF2 imprinting and Beckwith-Wiedemann syndrome. Nat Genet. 2004;36:958–960. doi: 10.1038/ng1410. [DOI] [PubMed] [Google Scholar]
- 90.Niemitz EL, DeBaun MR, Fallon J, et al. Microdeletion of LIT1 in familial Beckwith-Wiedemann syndrome. Am J Hum Genet. 2004;75:844–849. doi: 10.1086/425343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Zhang L, Wang T, Wright AF, et al. A microdeletion in Xp11.3 accounts for co-segregation of retinitis pigmentosa and mental retardation in a large kindred. Am J Med Genet A. 2006;140:349–357. doi: 10.1002/ajmg.a.31080. [DOI] [PubMed] [Google Scholar]
- 92.Miller M, Shen Y, Weiss LA, et al. Microdeletion/duplication at 15q13.2q13.3 among individuals with features of autism and other neuropsychiatric disorders. J Med Genet. 2008 doi: 10.1136/jmg.2008.059907. [DOI] [PMC free article] [PubMed] [Google Scholar]