Abstract
It has been proposed that retinoic acid receptors (RARs) and thyroid hormone receptors (TRs) both bind to AGGTCA “half-site” sequences, but distinguish their different target genes by recognizing different half-site spacings. We report here that artificial DNA binding sites based on these AGGTCA half-sites confer high affinity, but poor specificity, and that spacing alone does not account for the divergent DNA recognition properties of TRs and RARs. Instead, we have determined that the non-consensus half-sites that are present in naturally occurring RAR and TR target genes play a crucial role in defining receptor DNA recognition specificity, and work together with flanking sequences and half-site spacing to produce receptor-specific DNA binding in vitro. We also provide evidence that auxiliary proteins in cells generate an additional layer of receptor-specific target gene recognition, in part by destabilizing the binding of nuclear receptors to the “wrong” response elements.
Keywords: TRs, RARs, DNA recognition, half-site, DNA binding, nuclear receptor
1. INTRODUCTION
Nuclear receptors are ligand-regulated transcription factors that bind to specific DNA sequences (denoted hormone response elements) and modulate the expression of adjacent target genes [Escriva et al., 2004, Flamant et al., 2006, Germain et al., 2006, McEwan, 2009]. The nuclear receptor family includes endocrine receptors, such as the thyroid hormone receptors (TRs), retinoic acid receptors (RARs), estrogen receptors (ERs), vitamin D3 receptors (VDRs), glucocorticoid receptors (GRs) and androgen receptors (ARs), as well as receptors that sense metabolic lipids, respond to xenobiotics, or have no known ligand. Many nuclear receptors can both activate and repress target gene expression by alternatively tethering accessory proteins denoted coactivators and corepressors [McKenna and O’Malley, 2002, Privalsky, 2008, Tsai and Fondell, 2004]. Coactivators are typically recruited in response to ligand agonists, and include acetyltransferases, methyltransferases, chromatin remodelers, and components of the general transcriptional machinery [McKenna and O’Malley, 2002, Privalsky, 2008, Tsai and Fondell, 2004]. Corepressors typically associate with unliganded nuclear receptors or in response to ligand antagonists, and mediate effects opposite to those of the coactivators [McKenna and O’Malley, 2002, Privalsky, 2008, Tsai and Fondell, 2004].
Despite the diversity of their ligands, virtually all members of the nuclear receptor family possess a common zinc-finger DNA binding domain comprised of two α-helical domains that are folded and stabilized through interactions with coordinated zinc ions (reviewed in [Claessens and Gewirth, 2004, Khorasanizadeh and Rastinejad, 2001]). The “P-box” amino acids in the first α-helical domain make major groove contacts with a hexanucleotide sequence, denoted a half-site, on the target DNA. Most nuclear receptors contact their DNA binding sites as protein dimers; therefore two half-sites are required to create a functional hormone response element (e.g. [Brent et al., 1992, Forman et al., 1992, Hirst et al., 1992, Kurokawa et al., 1993, Lazar et al., 1991, Näär et al., 1991, Perlmann et al., 1993, Umesono et al., 1991, Wahlstrom et al., 1992]). The second α-helical domain in the zinc-finger region stabilizes the overall structure and can also contribute to receptor dimerization. In several nuclear receptors a third α-helical domain (the “A/T box”) C-terminal of the zinc-finger domain interacts with the minor groove of the DNA to recognize nucleotides flanking the core half-site sequence [Claessens and Gewirth, 2004, Khorasanizadeh and Rastinejad, 2001].
Idealized half-sites have been elucidated experimentally for many nuclear receptors, but surprisingly tend to distribute into just two generic groupings: AGGTCA (e.g. for RARs, TRs, and ERs) and AGAACA (e.g. for ARs and GRs) [Claessens and Gewirth, 2004, Khorasanizadeh and Rastinejad, 2001]. Similarly, the P-box motifs within the nuclear receptors parallel this binary grouping of the DNA sequences they contact, and are typically comprised of EGCKG or GSCKV sequences, respectively [Claessens and Gewirth, 2004, Escriva et al., 2004, Flamant et al., 2006, Germain et al., 2006, Khorasanizadeh and Rastinejad, 2001, McEwan, 2009]. Given vertebrates encode approximately 50 different nuclear receptors, each with its own characteristic panel of target genes, this raises the question of how adequate target gene specificity is achieved by the different nuclear receptors. A paradigm has emerged proposing that it is the spacing of the half-sites in a response element that defines its specificity [Forman et al., 1992, Kurokawa et al., 1993, Näär et al., 1991, Perlmann et al., 1993, Umesono et al., 1991, Vivanco Ruiz et al., 1991]. In this model, VDRs, TRs, and RARs recognize direct repeats of the same AGGTCA half-sites, but require different spacings between the half-site (a DR3, DR4, or DR5 respectively). Orientation of the half-sites can also play a role, ERs, for example, binds to inverted AGGTCA repeats with a 3 base spacer [Claessens and Gewirth, 2004].
Although this “3, 4, 5 rule” provides an extremely important insight, it falls short of fully explaining the specificity of DNA recognition and target gene regulation by the nuclear receptors. Naturally-occurring hormone response elements can contain half-sites that diverge significantly from the AGGTCA consensus sequences that helped define the spacing rule (e.g. [Williams and Brent, 1995]). Reciprocally, many nuclear receptors are unexpectedly promiscuous in their abilities to recognize half-sites displayed in a variety of spacings or orientations (e.g. [Forman et al., 1992, Hua et al., 2009, Lazar et al., 1991]). Further, as additional nuclear receptors have been identified, the number of possible half-site topologies has proven inadequate to conceptually assign each to a specific receptor.
We therefore performed a detailed study on the DNA recognition properties of TRs and RARs, two receptors with different physiological functions and distinct target gene repertoires in cells, yet with closely related response elements as defined by the 3,4,5 rule [Forman et al., 1992, Kurokawa et al., 1993, Näär et al., 1991, Perlmann et al., 1993, Umesono et al., 1991, Vivanco Ruiz et al., 1991]. We report that the experimentally derived AGGTCA half-site creates high affinity, but low specificity DNA binding sites that fail to adequately discriminate between TRs and RARs whether presented as a DR4 or a DR5. Instead, it is the non-consensus half-sites found in physiologically-relevant hormone response elements that are essential for receptor specificity, with TRs preferentially binding response elements comprised of AGGACA, and RARs preferentially binding response elements comprised of AGTTCAs. DNA sequences flanking these half-sites, and half-site spacing, further contribute to the specificity of DNA binding by these receptors in a combinatorial fashion. Interestingly, only a subset of the DNA elements that bind a given receptor in vitro mediate transcriptional regulation in vivo, and we provide evidence for the ability of auxiliary cellular proteins to destabilize the binding of nuclear receptors to the “wrong” DNA response elements. Our results help clarify how different nuclear receptors mediate their different biological roles.
2. MATERIALS AND METHODS
2.1. Oligonucleotides and molecular clones
Annealed oligonucleotides representing either single (for EMSA) or double (for luciferase assays) copies of each response elements were designed to incorporate either a 5′ Xho1/Sal1 (5′-tcga-3′) or HinDIII (5′-agct-3′) overhang to facilitate both radiolabeling with Klenow polymerase and cloning into reporter vectors. The original M-pTK-Luciferase reporter was digested with HinDIII and the direct-repeat-4 element was discarded prior to introduction of the response elements described here [Vivanco Ruiz et al., 1991]. The expression vectors for wild-type human TRα and RARα were described previously [Hauksdottir and Privalsky, 2001, Rosen and Privalsky, 2009]. All constructs were verified by DNA sequencing.
2.2. Protein Expression
Native, full-length human TRα1, RARα, RXRα were expressed using a recombinant baculovirus system [Chen and Privalsky, 1993]. Briefly, 1.0 × 107 Sf9 cells were infected at high multiplicity with the recombinant baculovirus of interest. After 48 h incubation, infected Sf9 cells were washed in Dulbecco’s Phosphate-Buffered Saline (Mg++ and Ca++ free, Gibco Inc, Grand Island NY) and harvested. The infected Sf9 cells were incubated on ice for 15 min. in 40 μl Buffer A [10 mM HEPES, pH 7.9, 10 mM KCl, 0.1 mM EDTA (pH 8.0), 0.1 mM EGTA (pH 8.0), 1mM DTT and 1x Complete protease inhibitor cocktail (Roche Applied Science, Indianapolis, IN)]. NP-40 was added to 0.6% to lyse the cells, the nuclei were pelleted and were resuspended in 20 mM HEPES (pH 7.9), 400 mM KCl, 0.1 mM EDTA (pH 8.0), 0.1 mM EGTA (pH 8.0), 10% glycerol, 1mM DTT and 1x Complete protease inhibitor cocktail. After 15 min of incubation on ice with shaking, the nuclear extracts were cleared by centrifugation for 10 min at 20,800 x g at 4° C. Receptor yield was assessed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) followed by western blot as described previously [Hauksdottir et al., 2003], using bacterially expressed GST-TRα or GST-RARα as concentration standards. CV-1 cell nuclear extracts were prepared in a similar manner, except they were resuspended in Buffer A and were lysed by Dounce homogenization instead of NP-40. The lysates were incubated on ice for 25 min before centrifugation at 5000 rpm for 5 min. to isolate the nuclei. The pellet was resuspended in 20 mM HEPES (pH 7.9), 420 mM KCl, 25% glycerol, 1.0 mM EDTA (pH 8.0), 1mM DTT and 1x Complete protease inhibitor cocktail and incubated on ice for 45 min. The nuclear extracts were then cleared by centrifugation at 20,800 x g for 20 min at 4° C prior to use.
GST proteins employed as protein standards (GST-TRα, GST-RARα) or for EMSA supershift (a GST-SRC1 representing codons 568-891, or a GST-CBP representing codons 1-451) were isolated from transformed Escherichia coli strain BL-21 by a modification of [Guan and Dixon, 1991]. Protein yield was quantified by SDS-PAGE and Coomassie Brilliant Blue staining, using a Fluorochem 8900 densitometer and AlphaEaseFC software (Alpha Innotech Corp., San Leandro, CA). Defined domains of these coactivators were used in these experiments because the high molecular weight of the native SRC1 and CBP proteins makes obtaining full-length products problematic in most recombinant expression systems.
2.3. Electrophoretic Mobility Shift Assays (EMSA)
Oligonucleotides representing the different response elements were annealed to form double-stranded DNA and were radiolabeled using Klenow polymerase with 32P-α-deoxy-GTP (3000 Ci/mMol). EMSAs were performed by combining 2 μl nuclear extract and 4 μl radiolabeled probe (200,000 cpm, 4 pmol total DNA) in binding buffer and incubating at 25° C for 15 min. For supershift experiments, recombinant SRC1 or CBP proteins were added after the DNA probe and receptor preparations were mixed. Binding buffer for receptor homodimers contained 7.5 mM Tris-HCl (pH 7.5), 2.3% glycerol, 10 mg/ml bovine serum albumin, 200 mM KCl, 1.5 mM MgCl2, 0.66 μg poly(deoxyinosine-deoxycytosine), and 2 μM dithiothreitol. Binding buffer for receptor heterodimers contained 20mM HEPES, 80 mM KCl, 2.5 μg poly(deoxyinosine-deoxycytosine), 1 μM dithiothreitol, 0.1% NP-40, and 10% glycerol as previously described [Umesono et al., 1991]. This buffer was employed to allow a better comparison of heterodimer formation in our study with heterodimer formation analyzed in the Umezono et al. publication; homodimer formation was essentially the same in either buffer system. The resulting DNA-protein complexes were resolved by electrophoresis in a 5% polyacrylamide (29:1 acrylamide/bisacrylamide) gel buffered with 0.5x TBE (45mM Tris-borate, 1mM EDTA) at 180V for 90 min (5% glycerol was included in the polyacrylamide gel for the receptor heterodimer assays). Free and bound DNA probe were visualized by STORM PhosphorImager analysis and quantified using ImageQuant software (Molecular Dynamics, Inc., Sunnyvale, CA). Probe input for each experiment was arbitrarily defined as 1 unit. For EMSA supershifts, the coactivator of interest was added to the binding buffer and nuclear extracts and was incubated for 15 min at 25° C prior to gel electrophoresis; 1 μM triiodothyronine (T3), all-trans retinoic acid (ATRA), or LG69, or an equivalent volume of ethanol carrier, were added to the binding reaction as indicated.
2.4. Reporter gene assays
CV-1 cells were plated in 24-well culture plates (3 × 105/well) in Dulbecco’s modified Eagle medium formulated with high glucose, L-glutamine (DMEM; Invitrogen, Carlsbad, CA), and 5% heat-inactivated fetal bovine serum (HI-FBS; Hyclone, Logan, UT). Cells were maintained at 37° C in a humidified 5% CO2 atmosphere. Twenty-four hrs after plating, the medium was replaced with DMEM containing 5% hormone stripped HI-FBS; 0.5 ng of pSG5-TRα, pSG5-RARα, or an empty pSG5 control were introduced per well together with 200 ng of luciferase reporter and 50 ng of a pCH110-β-galactosidase vector using Effectene and the manufacturer’s protocol (QIAGEN, Valencia, CA). The medium was replaced 24 hrs later with fresh medium containing 1 μM T3-thyronine, all-trans retinoic acid (ATRA), or ethanol-vehicle alone and the incubation continued for an addition 24 h before harvesting. Absolute luciferase activity was measured using a Promega luciferase assay system (Promega, Madison, WI) and a TD 20/20 luminometer (Turner Design, Sunnyvale, CA). Relative luciferase was calculated by normalization to β-galactosidase activity, measured using chlorophenol-red-β-D-galactopyranoside (CPRG) substrate (Roche, Indianapolis, IN) and a Spectramax 250 microplate reader (Molecular Devices, Sunnyvale CA).
3. RESULTS
3.1. AGGTCA half-sites confer high affinity but low specificity receptor binding, whereas naturally-occurring half-sites confer much higher specificity
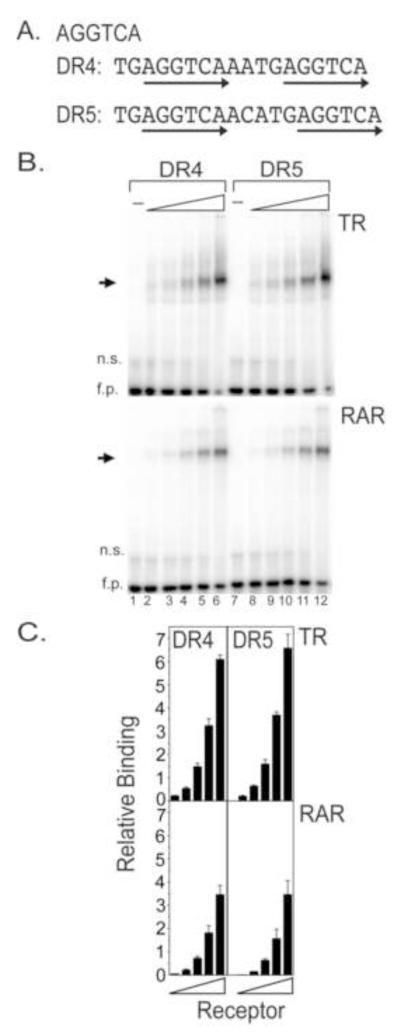
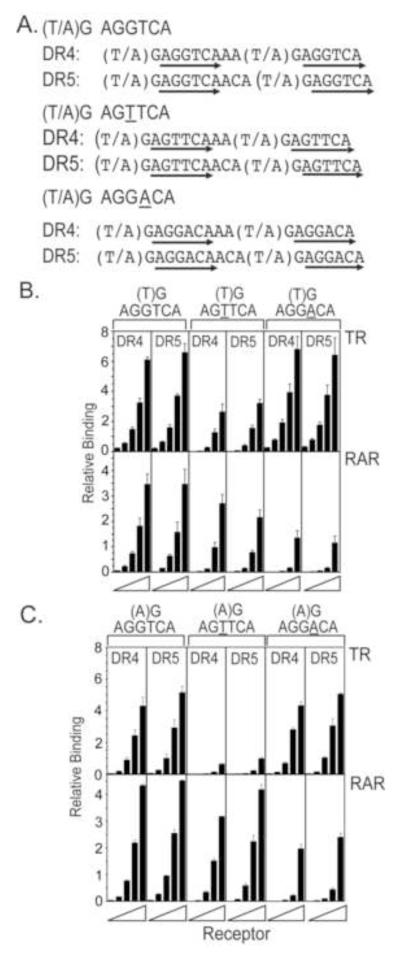
We first explored the ability of TRα or RARα to bind to artificial DR4 and DR5 response elements comprised of AGGTCA consensus half-sites (Fig. 1A). We used an electrophoretic mobility shift assay (EMSA) and a range of TRα and RARα concentrations; the DR4 and DR5 DNA elements were identical except for their spacing (Fig. 1B). Both TRα and RARα bound strongly, but near equally, as homodimers to both the DR4 and DR5 elements (Fig. 1B and quantified in Fig. 1C; the identity of these complexes was previously confirmed by antibody supershift). A similar lack of specificity was observed whether TRα and RARα were isolated from recombinant baculovirus-infected Sf9 cells or from transfected mammalian cells, and over a series of buffer conditions, running temperatures, and poly dI-dC concentrations (data not shown). Neither TRα nor RARα bound to a GR response element (an AGAATA inverted repeat 0) and no mobility shifts were observed using Sf9 lysates infected by a non-recombinant baculovirus vector (Figure 1B and data not shown).
Figure 1. TRα and RARα bind with high affinity, but low specificity to DR4 and DR5 spacings of AGGTCA consensus half-sites.
A. The nucleotide sequences of the consensus half-site DR4 and DR5 response elements are shown. Arrows indicate the hexanucleotide half-sites. B. TRα and RARα bind near equally to both DR4 and DR5 spacings of the AGGTCA consensus half-sites. A representative EMSA gel is presented. Identical two-fold dilution series (25-400 ng protein per pane) of each nuclear receptor, obtained from a recombinant baculovirus/Sf9 expression system, were mixed with a fixed amount of DR4 or DR5 radiolabeled DNA probe. The resulting receptor/DNA complexes (arrows) and free DNA probe (fp) were resolved by native polyacrylamide gel electrophoresis and were visualized by phosphorimager scanning. Extracts of Sf9 cells infected with non-recombinant virus was used as a negative control (lanes 1, 7). n.s. refers to a non-specific band. C. A quantification of the results from panel B is presented. Three EMSA experiments were performed and quantified; the mean and standard error are presented.
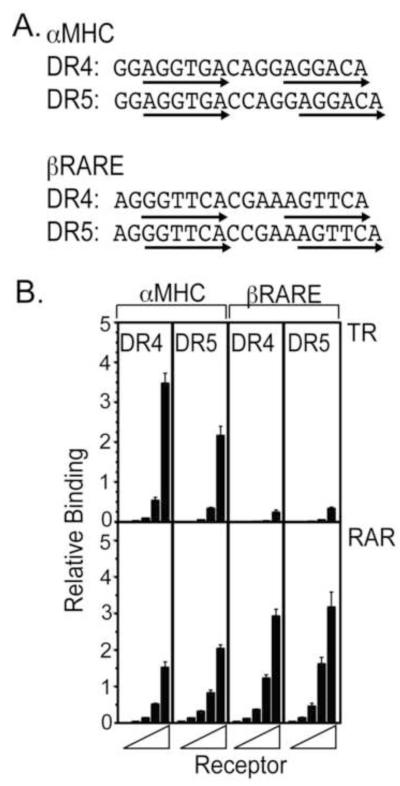
We next turned to two naturally-occurring response elements, a TR binding site in the αMHC promoter (a DR4) and an RAR binding site in the βRAR promoter (a DR5) [Brent et al., 1992, de The et al., 1990] (Fig. 2A). Although these elements were bound by their cognate receptors slightly less strongly than were the artificial AGGTCA elements (e.g. compare TRα1 in Fig. 2B and Fig. 1C), they were bound with much greater specificity (compare Fig. 2B to Fig. 1C). To explore the basis of this specificity, we also created a derivative of the MHC site with a 5 base spacer, and a derivative of the βRARE with a 4 base spacer (Fig. 2A). Interestingly, both the DR4 and DR5 αMHC derivatives retained preferential binding by TRα, and both the DR4 and DR5 βRARE derivatives retained preferentially binding by RARα (Fig. 2B). However, when comparing the DR4 and DR5 versions of the same element, TRα exhibited a moderate preference for the DR4 αMHC element, and RARα displayed a slight preference for the DR5 (Fig. 2B). We conclude that half-site spacing contributes to DNA recognition by these receptors, but is not the dominant discriminatory factor under these conditions.
Figure 2. TRα and RARα bind with lower affinity, but higher specificity, to native αMHC and βRARE elements.
A. The nucleotide sequences of the native MHC (a DR4) and βRARE (a DR5) elements are presented, as are an artificial DR5 version of the MHC element and an artificial DR4 version of the βRARE element. B. TRα and RARα bind specifically to their cognate response elements. The EMSA protocol in Figure 1B was repeated using the native αMHC or βRARE sequences in panel 2A as DNA probes. Three EMSA experiments were performed and quantified; the mean and standard error are presented.
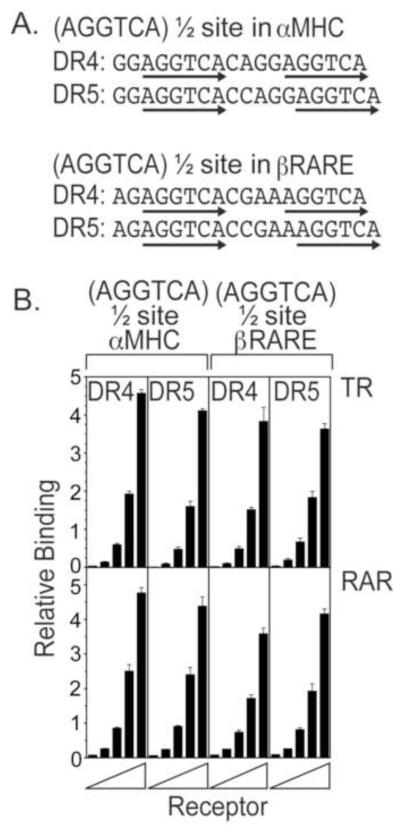
Both the MHC and the βRARE natural elements contain non-consensus hexanucleotide half-sites, as well as additional DNA sequence divergences in the regions flanking the half-sites themselves (Fig. 2A). To test the contributions of these different sequences to specificity, we next replaced the non-consensus half-sites in these elements with AGGTCA sequences, but retained the αMHC and the βRARE flanking sequences (Fig. 3A). Virtually all of the specificity observed for the native elements was lost by these AGGTCA substitutions: both the TRα and RARα bound near equally to both the αMHC (AGGTCA) and the βRARE (AGGTCA) elements, with spacing having only a modest (or no) effect (Fig. 3B).
Figure 3. Substitution of AGGTCA consensus half-sites into the αMHC and βRARE sequences abolished specific recognition by TRα and RARα.
A. Consensus AGGTCA sequences were introduced into the αMHC and βRARE elements in place of the native half-sites, using both a DR4 and a DR5 spacing. Arrows indicate the hexanucleotide half-sites. B. TRα and RARα bind with little or no specificity to the AGGTCA consensus versions of the αMHC and βRARE sequences. The EMSA protocol in Figure 1B was repeated using the AGGTCA versions of the αMHC or βRARE DNA probes indicated in panel 3A. Three EMSA experiments were performed and quantified; the mean and standard error are presented.
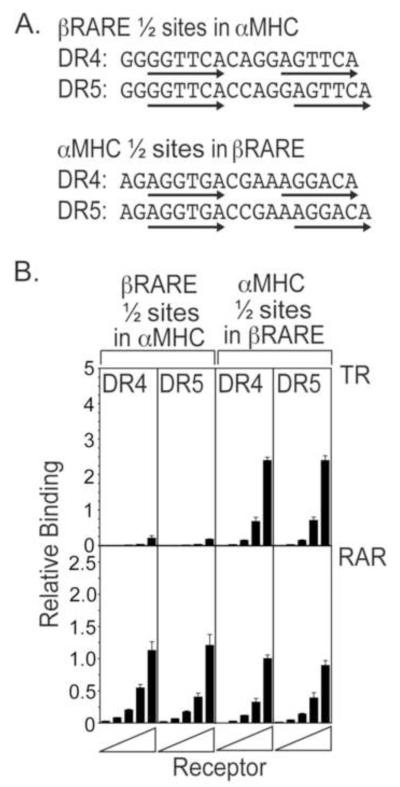
We next exchanged the native non-consensus half-sites between the αMHC and βRARE elements, testing both the DR4 and DR5 spacings (Fig. 4A). The αMHC half-sites conferred strong TR recognition when transferred to the βRARE background whether presented as a DR4 or a DR5, whereas the βRARE half-sites severely destabilized binding of TRα when inserted into the αMHC background, again whether presented as a DR4 or DR5 (Fig. 4B). The reversal of DNA recognition by TRα through swapping of the half-sites alone indicates that these non-consensus half-sites are the primary arbiter of TRα specific DNA recognition in these native response elements, with the spacing and flanking sequences playing secondary roles. Similarly, inserting the αMHC half-sites into the βRARE background greatly decreased RARα binding (compare Fig. 4B to Fig. 3B and Fig. 2B). Unlike the case with TRα, however, this exchange of non-consensus half-sites did not fully reverse RARα recognition, but instead produced response elements near equally recognized by RARα (Fig. 4B). Changing the spacing of the half-sites did not further influence RARα recognition of these chimeric response elements (Fig. 4B), indicating that the flanking sequences (the only other difference between the less-specific αMHC element possessing βRARE half-sites and the more specific native βRARE) must also contribute to the specificity of DNA recognition by RARα.
Figure 4. Exchanging the native half-sites between the αMHC and βRARE elements partially exchanges their recognition by TRα and by RARα.
A. The native αMHC half-sites were introduced into the βRARE sequence background, and the native βRARE half-sites were introduced into the αMHC sequence background as shown, using both a DR4 and a DR5 spacing. Arrows indicate the hexanucleotide half-sites. B. Exchanging the native half-sites between the αMHC and βRARE elements results in a partial exchange in their recognition by TRα and RARα. The EMSA protocol in Figure 1B was repeated using the interchanged half-site versions of the αMHC or βRARE DNA probes indicated in panel 4A. Three EMSA experiments were performed and quantified; the mean and standard error are presented.
3.2. The naturally-occurring, non-consensus half-sites work together with flanking DNA sequences and spacing to confer receptor-specific DNA recognition
To further explore the contributions of the different components of the DNA response element to receptor recognition, we returned to the study of artificial response elements based on AGGTCA repeats and manipulated the half-site, flanking sequences, and spacing individually (Fig. 5A). Substitution of the two AGGTCA consensus half-sites in these elements with two AGGACA half sites (a reiterated form of the non-consensus half-site found in the MHC element) maintained TRα binding and destabilized RARα recognition (Fig. 5B). Reciprocally, substitution of two AGTTCA half sites into the artificial element (a reiterated form of one of the non-consensus half-sites found in the βRARE element) slightly reduced RARα binding but more severely destabilized TRα binding (Fig. 5B). The effect of these half-site substitutions were observed in both the DR4 and DR5 contexts (Fig. 5B). These results confirm that the third and fourth positions in the hexanucleotide half-site play a dominant role in defining the divergent DNA recognition properties of TRα and RARα, can do so in a different flanking sequence context than that observed in the native αMHC and βRARE elements, and can operate largely independent of half-site spacing.
Figure 5. Half-site sequence, flanking sequence, and half-site spacing all contribute to specific DNA recognition by TRα and by RARα.
A. The nucleotide sequences of the DNA response elements used in the EMSA experiments are presented. Different half-sites, flanking sequences (at the −2 position), and different half-site spacings, as indicated, were tested. Arrows indicate the hexanucleotide half-sites. B, C. Specific DNA recognition by TRα and by RARα depends on half-site sequence, half-site spacing, and the identity of the nucleotide at the (−2) position in the flanking region of the response element. The EMSA protocol in Figure 1B was repeated using the different DNA probes indicated in panel 5A with either a T (panel B) or an A (panel C) at the −2 position. Three EMSA experiments were performed and quantified; the mean and standard error are presented.
We next tested the effect of altering the flanking sequences on receptor specificity. We focused on the (−2) position: (a) the sequence at this position differs between the native αMHC and βRARE elements, and (b) prior studies have shown differences in DNA recognition by RARα and TRα at this position [Hauksdottir and Privalsky, 2001, Judelson and Privalsky, 1996]. TRα prefers an T at (−2) [Judelson and Privalsky, 1996], and substitution of this position with an A resulted in slightly reduced binding of TRα to both the AGGTCA half-site elements and the AGGACA half-site elements (Fig. 5C). Notably, however, this unfavorable A at (−2) severely destabilized the binding of TRα to the AGTTCA half-site elements (Fig. 5C); thus two base substitutions, each moderately unfavorable to TRα binding, work together to strongly interfere with TRα recognition. RARα, conversely, favors an A at (−2) [Hauksdottir and Privalsky, 2001]. Response elements bearing an T at this position were bound less efficiently by RARα than were the equivalent response elements bearing a T at (−2), and response elements bearing both the T at (−2) and an AGGACA half-site were particularly unfavorable for RARα binding (compare Fig. 5B and 5C). We conclude that for RARα, as for TRα, the identity of the flanking sequences can further enhance the effects of non-consensus sequences in the half-sites themselves to generate receptor-specific response elements.
The role of half-site spacing was also evaluated in the context of these different half-sites and flanking sequences. For many of the artificial half-site and flanking sequences tested, little or no difference was seen in the ability of a given receptor to bind to the DR4 versus the DR5 version of the corresponding response element (Fig. 5B and C). Intriguingly, however, a subset of combinations of half-site and flanking sequence demonstrated a spacing preference consistent with the 3,4,5 rule. For example, a (−2A) AGTTCA DR5 was preferentially bound by RARα compared to an (−2A) AGTTCA DR4 (Fig. 5B and C). Some of these effects of spacing were enhanced when analyzing TRα or RARα as heterodimers with RXRα, below.
3.3. Heterodimers of TRα or RARα with RXRs display higher affinity, but lower specificity than do the corresponding homodimers
Both TRα and RARα can heterodimerize with other members of the nuclear receptor family, and heterodimer formation with RXRs, in particular, has been proposed to be an important, but not exclusive mode of DNA recognition by these receptors [Glass, 1996, Mangelsdorf and Evans, 1995]. We therefore next extended our studies on DNA binding specificity to RXRα/TRα and RXRα/RARα heterodimers.
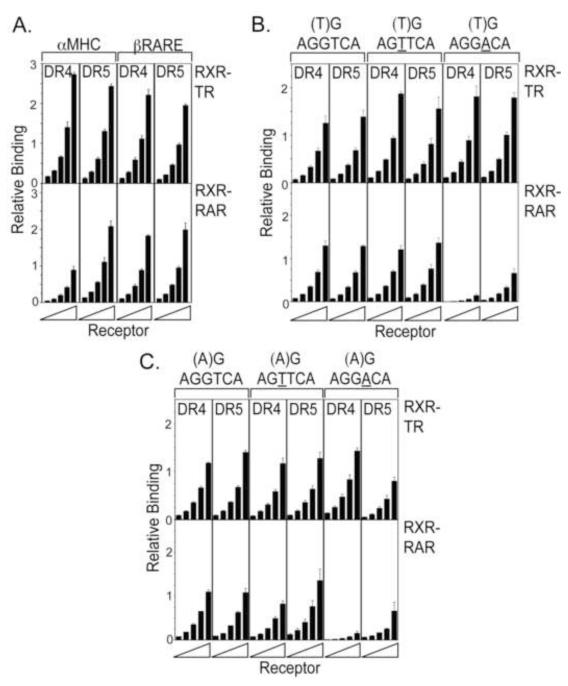
As previously reported (e.g. [Harbers et al., 1996, Marks et al., 1992, Nelson et al., 1996, Olson and Koenig, 1997]), the heterodimeric forms of both TRα and RARα displayed an elevated overall affinity for many of the response elements tested compared to the corresponding TRα or RARα homodimers (note lower concentrations of receptor were employed in Figure 6 than in the prior figures). RXRα itself was able to homodimerize on several of these elements, but at very low efficiencies (data not shown). Somewhat unexpectedly, however, the specificity of DNA binding was reduced in the heterodimers. The RXRα/TRα heterodimers retained a preferential interaction with the naturally occurring DR4 αMHC versus DR5 βRARE elements, but the difference between the two was less than that observed for TRα homodimers (compare Fig. 6A to Fig. 2B). Similarly, RXRα/RARα heterodimers bound the DR5 βRARE better than the DR4 αMHC, but with a lower specificity than did the RARα homodimers (Fig. 6A).
Figure 6. Specific DNA recognition by RXRα heterodimers of TRα and RARα is dependent on all three parameters: half-site sequence, flanking sequence, and half-site spacing.
A. RXRα/TRα and RXRα/RARα heterodimers display higher affinity, but less specificity on the native MHCα and βRARE response elements than do TRα or RARα homodimers. An increasing amount of RXRα and TRα, or RXRα and RARα (approximately 12-200 ng of each nuclear receptor) was mixed with the radioactive DNA probes indicated, and the resulting receptor/DNA complexes were resolved and quantified using the same overall EMSA protocol as in Figure 1B and 1C. Three EMSA experiments were performed; the mean and standard error are presented. B, C. An AGGACA half-site with a T in the (−2) position displays specificity for RXRα/TRα compared to RXRα/RARα. The protocol in panel 6A was repeated using the radioactive DNA probes indicated, containing either a T (panel B) or an A (panel C) at the −2 position.
Notably, all three components of the response element, half-site sequence, flanking sequence, and half-site spacing, worked together to confer response element recognition by these heterodimers. For example, artificial DNA response elements containing a T at the (−2) flanking sequence and consensus AGGTCA half-sites were bound by RXRα/TRα and RXRα/TRα near equally, and this was largely independent of half-site spacing (Fig. 6B). However, the same (−2T) flanking sequence linked to AGGACA half-sites retained binding to RXRα/TRα heterodimers, but significantly destabilized binding to RXRα/RARα heterodimers, an effect that was further enhanced by a DR4 versus a DR5 spacing (Fig. 6B). Conversely, RARα prefers an A at the (−2) position, and RXRα/RARα heterodimers efficiently recognized (−2A) elements containing AGTTCA half-sites with a DR5 spacing (Fig. 6C). Therefore, although half-site sequence alone confers little or no DNA binding discrimination for RARα versus TRα heterodimers, the combining of receptor-selective half-sites with appropriate flanking sequences and half-site spacings generates detectable DNA binding specificity.
We also examined if hormone ligand altered the specificity of DNA recognition by these RXRα/TRα and RXRα/RARα heterodimers. No significant difference was observed in the recognition of the αMHC or βRARE elements in the presence or absence of T3 or all-trans retinoic acid (ATRA), nor was an effect observed if LG68, an RXRα agonist, was added to the binding reactions alone, or together with T3 or ATRA (data not shown). We conclude that in vitro, binding of cognate ligand to RXRα/TRα or to RXRα/RARα does not alter DNA recognition compared to that observed for the corresponding unliganded receptors.
3.4. Half-site spacing contributes an additional level of specificity for transcriptional activation in cells beyond that observed for DNA binding by TRα and RARα in vitro
To evaluate the ability of our different DNA response elements to confer receptor-specific transcriptional regulation in cells, we inserted these elements into a thymidine-kinase promoter-luciferase reporter gene (tk-Luc) and introduced them, together with expression vectors for TRα or RARα, into CV1 cells by transient transfections. We chose CV1 cells because they contain almost no endogenous TRs and relatively low levels of endogenous RARs [Lee and Privalsky, 2005] CV1 cells express endogenous RXRs, and we assume that most of the transcriptional activation we observe in these reporter assays is mediated by RXR/TRα and RXR/RARα heterodimers (based on the higher affinity of heterodimers for DNA, their stability in the presence of hormone, the absence of a further effect of ectopic RXR when co-transfected, and the effects of RXR knockouts in other cell systems) [Andersson et al., 1992, Chiba et al., 1997, Forman et al., 1992, Galli et al., 1998, Lazar et al., 1991, Yen et al., 1992]. Nonetheless we cannot exclude the possibility that TRα homodimers or RARα homodimers may also contribute to reporter gene regulation in this assay (e.g. [Velasco et al., 2007, Diallo et al., 2007]).
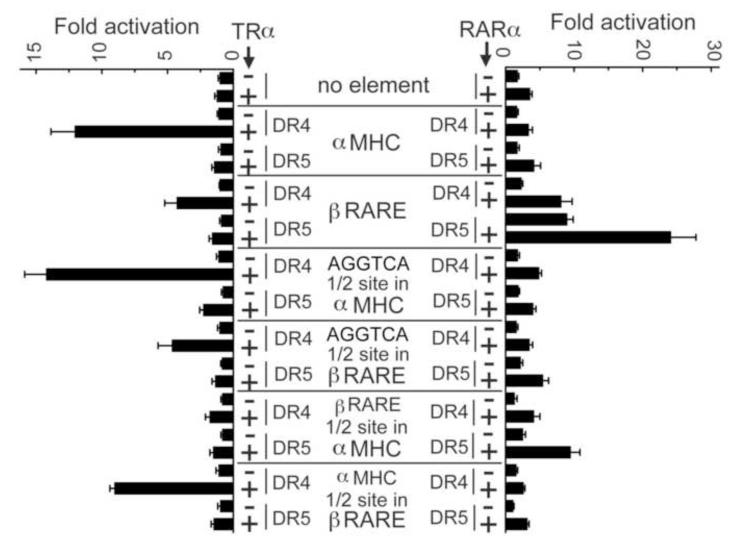
In the absence of a response element, the tk-luciferase reporter displayed a low level of basal expression that was relatively unaltered by introduction of either nuclear receptor or hormone status (Fig. 7 and data not shown). Introduction of the αMHC-DR4 into the tk-Luc reporter resulted in a strong, TRα-mediated response to T3, whereas introduction of the βRARE-DR5 into the tk-Luc reporter resulted in a strong, RARα-mediated response to ATRA (a more modest ATRA response, mediated by endogenous RARs in these cells, was also observed in the absence of the ectopic RARα expression vector) (Fig. 7). As expected, response elements poorly bound by a given receptor in vitro were poor at conferring reporter gene activation in cells (e.g. the αMHC-DR4-TK-Luc reporter unresponsive to RARα, and the βRARE-DR5-TK-Luc reporter was relatively unresponsive to TRα) (Fig. 7). The half-site sequences themselves clearly contributed to this specificity; substitution of the αMHC half-sites into the βRARE-DR4 element permitted significant TRα-mediated reporter expression, and substitution of the βRARE half-sites into the αMHC-DR5 element permitted significant RARα-mediated reporter expression (Fig. 7). In vivo, as in vitro, the flanking sequences also contributed to receptor specificity: (a) the elements with swapped half-sites activated less strongly than did the native elements, and (b) consensus AGGTCA half-sites conferred stronger TRα activation when inserted into the αMHC-DR4 element background (i.e. linked to αMHC flanking sequences) than when inserted into the βRARE-DR4 element background (i.e. linked to βRARE flanking sequences).
Figure 7. The ability of TRα and RARα to activate reporter gene expression in cells reveals an additional layer of DNA recognition specificity.
The response elements indicated (center panel) were engineered into a tk promoter-luciferase reporter and introduced into CV-1 cells by transient transfection, together with an expression vector for TRα (left panel “+”) or for RARα (right panel “+”). A reporter without a response element (“−”) was used as a negative control. Cells were treated with 1 μM T3 (left panel) or 1 μM ATRA (right panel) for 24 hrs, the cells were harvested, and luciferase activity was determined relative to a β-galactosidase internal control. Fold activation was calculated as the ratio of luciferase in the presence versus the absence of hormone. The means and standard error of three independent experiments are shown.
Unexpectedly, not all response elements that bound a given receptor strongly in vitro conferred transcriptional regulation in cells. Most striking was the much greater role of half-site spacing in reporter gene activation in vivo versus receptor binding in vitro. TRα was essentially inactive in the luciferase assay on all of the DR5 elements tested, despite the ability of TRα homo-and heterodimers to bind to both the DR4 and DR5 versions of these element in vitro (Fig. 7). Similarly, RARα displayed much weaker activity in the luciferase assay on the DR4 version of the βRARE element compared to the DR5 version the same was true of the DR4 versus DR5 versions of the response elements bearing βRARE half-sites in the αMHC background (Fig. 7). Parallel results were observed using integrated DR4 and DR5 versions of the βRARE-TK-Luc reporter instead of the transiently transfected reporters (data not shown). Interestingly, neither the DR4 nor DR5 versions of a consensus AGGTCA half-site functioned with RARα in this context; although unexpected, this further highlights the importance of the non-consensus half-sites present in the native βRARE for RARα recognition and function (Fig. 7). We conclude that an additional level of specificity operates in cells to further restrict transcriptional activation to a subset of the response elements that can be recognized by TRα or RARα in vitro, and that the dependence on half-site spacing invoked by the 3,4,5 rule is manifested most strongly at this cellular level.
3.5. Auxiliary proteins in cells can destabilize nuclear receptor binding to the wrong response elements
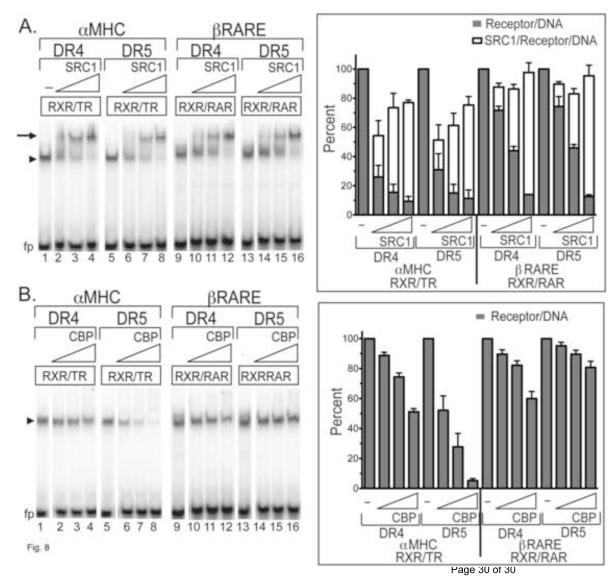
We next tested if the dependence on half-site spacing for reporter gene activation in cells was observable as a difference in coactivator binding in vitro. We employed an EMSA supershift protocol to compare the ability of an SRC1 coactivator construct (representing the receptor-interaction domain of this coactivator; [McKenna and O’Malley, 2002]) to recognize RXRα/TRα or RXRα/RARα heterodimers bound to DR4 or DR5 versions of the αMHC and βRARE elements (all in the presence of cognate agonist) (Fig. 8A). Notably the SRC1 coactivator construct was able to interact with, and supershift, the receptor complexes assembled on these DNA elements independent of half-site spacing (Fig. 8A). We conclude that the DR4/DR5 specificity observed in cell transfections did not reflect the ability of these half-site spacings to recruit our p160 SRC1 coactivator construct in vitro.
Figure 8. SRC1 coactivator is recruited to TRα and RARα independent of half-site spacing, whereas CBP destabilizes nuclear receptor binding to non-cognate half-site spacings.
A. SRC1 coactivator is recruited by nuclear receptors to both DR4 and DR5 response elements. A single concentration of RXRα/TRα or RXRα/RARα heterodimers was mixed with the DNA probes indicated above the panel, together with cognate hormone and increasing amounts of an SRC1 (codons 568-891) construct was then added. The complexes were resolved by native acrylamide gel electrophoresis as in Figure 1B. A representative electrophoretogram is shown (left panel) and the amount of receptor/DNA complex supershifted by SRC1 (arrow), or not (arrowhead), was quantified (right panel). The mean and standard deviation of three independent experiments are presented. B. CBP destabilizes nuclear receptor binding to non-cognate half-site spacings. A single concentration of RXRα/TRα or RXRα/RARα heterodimers was mixed with the radiolabeled DNA probes indicated above the panel, together with cognate hormone and increasing amounts of a CBP (codons 1-451) construct. The complexes were resolved by native acrylamide gel electrophoresis as in Figure 1B. A representative electrophoretogram is shown (left panel) and the amount of receptor/DNA complex formed was quantified (right panel). The means and standard deviation of five independent experiments are presented.
Intriguingly, repeating this experiment with a different transcriptional coactivator, CBP [McKenna and O’Malley, 2002], produced an unexpected result (Fig. 8B). A CBP construct (representing the N-terminal receptor interaction domain) did not generate a dose-dependent supershift with any response elements tested (Fig. 8B). Instead, addition of the CBP construct selectively destabilized the binding of the receptor heterodimers to the elements representing the “wrong” spacings. For example, the CBP construct strongly interfered with the ability of the RXRα/TRα heterodimer to bind to the αMHC-DR5 element, but only modestly interfered with RXRα/TRα binding to the “correctly spaced” αMHC-DR4 element (Fig. 8B). Conversely, the CBP construct interfered with the ability of the RXRα/RARα heterodimer to bind to the βRARE-DR4 element, but displayed little or no ability to interfere with its binding to the “correctly spaced” βRARE-DR5 element (Fig. 8B). A similar spacer-specific destabilization was observed using the CBP construct on RARα homodimers (data not shown). T3 disrupts the ability of TRα homodimers to bind to DNA, preventing us from testing the effect of CBP in the TRα homodimer context [Andersson et al., 1992, Yen et al., 1992]. We suggest that at least one component of the additional selectivity for half-site spacing observed in cell transfections may arise from the presence of auxiliary cell proteins that can destabilize binding to the wrong spacings. CBP may be such a protein, or there may be additional proteins yet to be elucidated that are more relevant to the in vivo context.
4. DISCUSSION
4.1. Half-site sequence, not spacing, dominates in defining the specificity of DNA recognition by RARα and TRα in vitro
Nearly 50 mammalian nuclear receptors have been identified, and yet to function properly, each must be able to specifically recognize its own distinct panel of DNA binding sites and target genes [Escriva et al., 2004, Flamant et al., 2006, Germain et al., 2006, McEwan, 2009]. Complicating any simple explanation for this selectivity, many nuclear receptors share identical P-box amino acid sequences in their zinc-finger domains and recognize identical consensus DNA half-sites [Claessens and Gewirth, 2004, Khorasanizadeh and Rastinejad, 2001]. A major advance in understanding this phenomenon came with the realization that the spacing of the two half-sites in a DNA response element plays an important role in its recognition, an observation subsequently codified as the 3, 4, 5 rule [Forman et al., 1992, Kurokawa et al., 1993, Näär et al., 1991, Perlmann et al., 1993, Umesono et al., 1991, Vivanco Ruiz et al., 1991]. Interestingly, a contribution of half-site sequence was noted in these or in subsequent publications (e.g. [Katz et al., 1995, Mader et al., 1993, Nelson et al., 1996, Umesono et al., 1991, Williams et al., 1992]), but was often not explored in detail and has frequently been overlooked in subsequent interpretations of this pioneering work. We report here a re-examination of these issues by a detailed analysis of the roles of half-site spacing, half-site sequence, and flanking DNA sequence in response element recognition by RARα and TRα, two closely related receptors with different target genes and different biological roles.
Significantly, neither RARα nor TRα displayed the ability to discriminate between a DR4 and a DR5 spacing of consensus AGGTCA half-sites, despite this sequence being among the highest affinity DNA binding sites known for these receptors. In contrast, two naturally-occurring response elements, obtained from the T3-regulated αMHC promoter and the ATRA-regulated βRARE promoter, display a strong preference for their cognate receptors. Exchanging the different components of these natural response elements with one another, and with artificial AGGTCA-based DR4 and DR5 elements, demonstrated that it is the non-consensus nature of the half-sites in the αMHC and βRARE elements, not their spacing, that plays the dominant role in their specific recognition in vitro. The αMHC element is comprised of AGGTCA and AGGACA half-sites, whereas the βRARE element is comprised of GGTTCA and AGTTCA half-sites. Significantly, reiteration of these non-consensus half-sites (AGGACA for TRα or AGTTCA for RARα) within an artificial response element conferred or further enhanced the receptor specificity observed with the natural elements. In contrast, changing the spacing in these natural elements had only modest effects on the ability of their cognate receptors to bind to these sequences.
We suggest that AGGTCA sequences generate high affinity, but low specificity receptor recognition and that the “suboptimal” half-site sequences such as are found in the αMHC and βRARE elements (and in many other naturally-occurring response elements) confer slightly lower affinity for their cognate receptors, but are far more disfavorable for binding by non-cognate receptors, thereby helping to confer specificity. Although we analyzed in depth AGTTCA and AGGACA half-sites, additional base changes at other positions in naturally-occurring half-sites may further modify the specificity conferred by these non-consensus position 3 and 4 positions [Williams and Brent, 1995].
The P-box amino acids that mediate the major groove contacts responsible for half-site recognition are identical between TRα and RARα (EGCKG). How then do these receptors preferentially recognize different half-sites, as described here? Differences do exist in the other amino acid sequences that comprise the zinc-finger domains of these two receptors, and we suggest that these differences alter the precise position or conformation of the P box recognition helices themselves. In this fashion receptor amino acids that do not contact the DNA half-site directly can nonetheless influence its recognition. Amino acid sequence differences in the N-terminal domains of TRα and RARα, mapping outside of the zinc-finger domains, may also contribute to this phenomenon, most likely by a similar, indirect effect on P-box conformation [Chen et al., 1993, Wong and Privalsky, 1995]. In support of these proposals, our preliminary studies on chimeric constructs of RARα and TRα demonstrate that the differing recognition properties of these receptors are encoded by multiple determinants mapping within both N-terminal and zinc-finger domains (TQP and MLP, unpublished observations).
4.2. Flanking sequences provide additional response element specificity
Sequences flanking the hexanucleotide half-sites also contributed to receptor-specific DNA recognition in our study. Although observable to some extent when tested with consensus AGGTCA half-sites, the effects of these flanking sequences were particularly evident when used together with the corresponding, receptor-selective half-site sequences. For example, the βRARE half-sites did not fully confer RARα-specific recognition unless the flanking sequences were also derived from the βRARE element. We conclude that flanking and half-site sequences work together to generate receptor-specific DNA recognition. Flanking sequences contribute to DNA binding by other nuclear receptors, and it has been proposed that the “true” nuclear receptor half-site is therefore eight bases long (e.g. [Harbers et al., 1996, Hsu et al., 1998, Olson and Koenig, 1997, Wilson et al., 1992]). However, given the −2 and −1 position are recognized through minor groove contacts mediated by a receptor “AT-box” α-helix lying outside of the zinc finger domain itself [Claessens and Gewirth, 2004, Khorasanizadeh and Rastinejad, 2001], we have treated this aspect of DNA recognition as distinct from the major groove, P-box contacts involved in recognition of the +1 to +6 half-site sequences.
4.3. Heterodimerization with RXRα uncovers a role for half-site spacing in receptor-specific DNA recognition
RARα and TRα can bind to DNA as heterodimers with other nuclear receptors, including RXRs [Glass, 1996], although the relative proportion of homo- and hetero-dimers is likely to vary at different target genes, in different cells, or under different hormone ligand conditions (e.g. [Hsu et al., 1995, Leid et al., 1992, Olson and Koenig, 1997, Schräder et al., 1993]). Notably RXR heterodimers displayed a somewhat greater dependence on half-site spacing than did homodimers, although only when combined with appropriate receptor-selective half-site and flanking DNA sequences. For example, RXRα/TRα heterodimers more strongly recognized agAGGACA half sites when presented as a DR4 than as a DR5. Conversely, RXRα/RARα heterodimers preferred binding to agAGTTCA half-sites presented as a DR5 versus a DR4. In contrast, spacing alone was insufficient to confer selective recognition of AGGTCA half-site elements by either RXRα/TRα or RXRα/RARα heterodimers. We conclude that the spacing of the half-sites does play a role in DNA recognition by TRα and RARα, but that this is most evident for RXR heterodimers and operates together with the half-site and flanking sequence contributions we elucidated for the corresponding receptor homodimers.
Although RXR/TR heterodimers have been reported to bind to certain DNA elements more strongly than do TR homodimers [Harbers et al., 1996, Marks et al., 1992, Nelson et al., 1996, Olson and Koenig, 1997], other studies have indicated that heterodimerization with RXR narrows the DNA binding specificity of these receptors [Yu et al., 1991, Zhang et al., 1992]. One explanation for this apparent contradiction may be the differing sources of receptor used; earlier studies often employed in vitro translated protein, in contrast with our use of baculovirus or mammalian cells expression systems (notably this also permitted us to use a greater range of receptor protein concentrations than those available in these prior studies). The precise nature of the response elements tested may also offer an explanation: the broadening effect of RXR on specificity was most clearly observed on response elements that bound receptor homodimers poorly, whereas less of a difference was observed on elements that bound homodimers strongly.
4.4. The 3, 4, 5 rule resurgent: additional determinants of specificity exist in cells
We examined the ability of our response elements to activate reporter gene expression in cells. As expected, response elements that were bound poorly by a given receptor in vitro also typically mediated poor transcriptional activation in vivo. Significantly, the corollary was not true; several elements that were strongly bound by TRα or RARα in vitro were very poor at mediating reporter gene activation in vivo. Half-site spacing had a particular impact in this context: efficient TRα-mediated reporter regulation required a DR4 and efficient RARα-mediated reporter regulation required a DR5. This dependence on half-site spacing was observed both on transiently introduced and stably integrated reporter. We conclude that an additional layer of specificity is imposed on nuclear receptor-mediated transcriptional regulation in cells beyond that observed for DNA binding in vitro, and may be the predominant basis for the 3,4,5 rule.
Disparities between DNA binding and transcriptional activation has been observed before for various nuclear receptors, although the actual mechanisms underlying these disparities have often remained poorly understood (e.g. [Ikeda et al., 1996, La Vista-Picard et al., 1996, Meijsing et al., 2009, Nagpal et al., 1992, Olson and Koenig, 1997]). In our hands, the ability of RXRα/TRα and RXRα/RARα heterodimers to bind to a p160 coactivator in vitro was the same for DR4 and DR5 elements. We cannot rule out, however, that yet-other coactivators in cells may display a stricter requirement for a given half-site spacing before they can be recruited by their nuclear receptor partners.
Intriguingly, we found that CBP, a coactivator/transcriptional integrator, selectively destabilizes binding of receptors to “wrongly” spaced DNA elements, such as RXRα/TRα to DR5 elements and RXRα/RARα to DR4 elements. We suggest that when CBP associates with these receptors, it imposes a specific topology on the receptor dimer that is incompatible with the incorrect spacing. In this model, DNA binding is mediated by a nuclear receptor/coregulator complex, not by the receptor alone, and DNA specificity is a superposition of the recognition properties of the receptor and the associated coregulator complex. There may also be additional proteins in cells that are not coactivators or corepressors, per se, but that serve primarily to modulate the DNA recognition properties of the nuclear receptors. These specificity-conferring proteins, by defining the recognition of the DNA sequence itself, may operate in additional to factors, such as FoxA1, that help open the chromatin structure and confer access to the DNA [Carroll et al., 2005, Dong et al., 2009, Nielsen et al., 2008]. Interactions between the multiple response elements that often are present within each target gene may further contribute to receptor-specific binding and transcriptional regulation. Conversely, the same mechanisms described here that help explain receptor-specific DNA recognition also help account for how composite response elements, known to permit regulatory crosstalk on genes targeted by multiple members of the nuclear receptor family, can function (e.g. [Adan et al., 1993, Hua et al., 2009, Kato et al., 1995, Nakshatri and Chambon, 1994]).
ACKNOWLEDGEMENTS
The authors thank Liming Liu for superb technical assistance. This work was supported by Public Health Service/National Cancer Institute award RO1DK53528. T.Q.P. was supported in part by a PHS Pre-doctoral Training award, T32-GM007377, from the National Institute of General Medical Sciences.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Adan RA, Cox JJ, Beischlag TV, Burbach JP. A composite hormone response element mediates the transactivation of the rat oxytocin gene by different classes of nuclear hormone receptors. Mol. Endocrinol. 1993;7:47–57. doi: 10.1210/mend.7.1.8383287. [DOI] [PubMed] [Google Scholar]
- Andersson ML, Nordstrom K, Demczuk S, Harbers M, Vennstrom B. Thyroid hormone alters the DNA binding properties of chicken thyroid hormone receptors alpha and beta. Nucleic Acids Res. 1992;20:4803–4810. doi: 10.1093/nar/20.18.4803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brent GA, Williams GR, Harney JW, Forman BM, Samuels HH, Moore DD, Larsen PR. Capacity for cooperative binding of thyroid hormone (T3) receptor dimers defines wild type T3 response elements. Mol. Endocrinol. 1992;6:502–514. doi: 10.1210/mend.6.4.1584220. [DOI] [PubMed] [Google Scholar]
- Carroll JS, Liu XS, Brodsky AS, Li W, Meyer CA, Szary AJ, Eeckhoute J, Shao W, Hestermann EV, Geistlinger TR, Fox EA, Silver PA, Brown M. Chromosome-wide mapping of estrogen receptor binding reveals long-range regulation requiring the forkhead protein FoxA1. Cell. 2005;122:33–43. doi: 10.1016/j.cell.2005.05.008. [DOI] [PubMed] [Google Scholar]
- Chen HW, Privalsky ML. The erbA oncogene represses the actions of both retinoid X and retinoid A receptors but does so by distinct mechanisms. Mol. Cell. Biol. 1993;13:5970–5980. doi: 10.1128/mcb.13.10.5970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen HW, Smitmcbride Z, Lewis S, Sharif M, Privalsky ML. Nuclear hormone receptors involved in neoplasia - Erb-A exhibits a novel DNA sequence specificity determined by amino acids outside of the zinc-finger domain. Mol. Cell. Biol. 1993;13:2366–2376. doi: 10.1128/mcb.13.4.2366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiba H, Clifford J, Metzger D, Chambon P. Distinct retinoid X receptor-retinoic acid receptor heterodimers are differentially involved in the control of expression of retinoid target genes in F9 embryonal carcinoma cells. Mol. Cell. Biol. 1997;17:3013–3020. doi: 10.1128/mcb.17.6.3013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claessens F, Gewirth DT. DNA recognition by nuclear receptors. Essays Biochem. 2004;40:59–72. doi: 10.1042/bse0400059. [DOI] [PubMed] [Google Scholar]
- De the H, Vivanco-Ruiz MM, Tiollais P, Stunnenberg H, Dejean A. Identification of a retinoic acid responsive element in the retinoic acid receptor beta gene. Nature. 1990;343:177–180. doi: 10.1038/343177a0. [DOI] [PubMed] [Google Scholar]
- Diallo EM, Wilhelm KG, Jr., Thompson DL, Koenig RJ. Variable RXR requirements for thyroid hormone responsiveness of endogenous genes. Mol. Cell. Endocrinol. 2007;264:149–156. doi: 10.1016/j.mce.2006.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong H, Yauk CL, Rowan-Carroll A, You SH, Zoeller RT, Lambert I, Wade MG. Identification of thyroid hormone receptor binding sites and target genes using ChIP-on-chip in developing mouse cerebellum. PLoS One. 2009;4:e4610. doi: 10.1371/journal.pone.0004610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Escriva H, Bertrand S, Laudet V. The evolution of the nuclear receptor superfamily. Essays Biochem. 2004;40:11–26. doi: 10.1042/bse0400011. [DOI] [PubMed] [Google Scholar]
- Flamant F, Baxter JD, Forrest D, Refetoff S, Samuels H, Scanlan TS, Vennstrom B, Samarut J. International Union of Pharmacology. LIX. The pharmacology and classification of the nuclear receptor superfamily: thyroid hormone receptors. Pharmacol. Rev. 2006;58:705–711. doi: 10.1124/pr.58.4.3. [DOI] [PubMed] [Google Scholar]
- Forman BM, Casanova J, Raaka BM, Ghysdael J, Samuels HH. Half-site spacing and orientation determines whether thyroid hormone and retinoic acid receptors and related factors bind to DNA response elements as monomers, homodimers, or heterodimers. Mol. Endocrinol. 1992;6:429–442. doi: 10.1210/mend.6.3.1316541. [DOI] [PubMed] [Google Scholar]
- Galli A, Stewart M, Dorris R, Crabb D. High-level expression of RXRalpha and the presence of endogenous ligands contribute to expression of a peroxisome proliferator-activated receptor-responsive gene in hepatoma cells. Arch. Biochem. Biophys. 1998;354:288–294. doi: 10.1006/abbi.1997.0701. [DOI] [PubMed] [Google Scholar]
- Germain P, Chambon P, Eichele G, Evans RM, Lazar MA, Leid M, De Lera AR, Lotan R, Mangelsdorf DJ, Gronemeyer H. International Union of Pharmacology. LX. Retinoic acid receptors. Pharmacol. Rev. 2006;58:712–725. doi: 10.1124/pr.58.4.4. [DOI] [PubMed] [Google Scholar]
- Glass CK. Some new twists in the regulation of gene expression by thyroid hormone and retinoic acid receptors. J. Endocrinol. 1996;150:349–357. doi: 10.1677/joe.0.1500349. [DOI] [PubMed] [Google Scholar]
- Guan KL, Dixon JE. Eukaryotic proteins expressed in Escherichia coli: an improved thrombin cleavage and purification procedure of fusion proteins with glutathione S-transferase. Anal. Biochem. 1991;192:262–267. doi: 10.1016/0003-2697(91)90534-z. [DOI] [PubMed] [Google Scholar]
- Harbers M, Wahlstrom GM, Vennstrom B. Transactivation by the thyroid hormone receptor is dependent on the spacer sequence in hormone response elements containing directly repeated half-sites. Nucleic Acids Res. 1996;24:2252–2259. doi: 10.1093/nar/24.12.2252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hauksdottir H, Farboud B, Privalsky ML. Retinoic acid receptors beta and gamma do not repress, but instead activate target gene transcription in both the absence and presence of hormone ligand. Mol. Endocrinol. 2003;17:373–385. doi: 10.1210/me.2002-0340. [DOI] [PubMed] [Google Scholar]
- Hauksdottir H, Privalsky ML. DNA recognition by the aberrant retinoic acid receptors implicated in human acute promyelocytic leukemia. Cell Growth Differ. 2001;12:85–98. [PMC free article] [PubMed] [Google Scholar]
- Hirst MA, Hinck L, Danielsen M, Ringold GM. Discrimination of DNA response elements for thyroid hormone and estrogen is dependent on dimerization of receptor DNA binding domains. Proc. Natl. Acad. Sci. U.S.A. 1992;89:5527–5531. doi: 10.1073/pnas.89.12.5527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu JH, Zavacki AM, Harney JW, Brent GA. Retinoid-X receptor (RXR) differentially augments thyroid hormone response in cell lines as a function of the response element and endogenous RXR content. Endocrinology. 1995;136:421–30. doi: 10.1210/endo.136.2.7835272. [DOI] [PubMed] [Google Scholar]
- Hsu MH, Palmer CN, Song W, Griffin KJ, Johnson EF. A carboxyl-terminal extension of the zinc finger domain contributes to the specificity and polarity of peroxisome proliferator-activated receptor DNA binding. J. Biol. Chem. 1998;273:27988–27997. doi: 10.1074/jbc.273.43.27988. [DOI] [PubMed] [Google Scholar]
- Hua S, Kittler R, White KP. Genomic antagonism between retinoic acid and estrogen signaling in breast cancer. Cell. 2009;137:1259–1271. doi: 10.1016/j.cell.2009.04.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeda M, Wilcox EC, Chin WW. Different DNA elements can modulate the conformation of thyroid hormone receptor heterodimer and its transcriptional activity. J. Biol. Chem. 1996;271:23096–23104. doi: 10.1074/jbc.271.38.23096. [DOI] [PubMed] [Google Scholar]
- Judelson C, Privalsky ML. DNA recognition by normal and oncogenic thyroid hormone receptors - unexpected diversity in half-site specificity controlled by non-zinc-finger determinants. J. Biol. Chem. 1996;271:10800–10805. doi: 10.1074/jbc.271.18.10800. [DOI] [PubMed] [Google Scholar]
- Kato S, Sasaki H, Suzawa M, Masushige S, Tora L, Chambon P, Gronemeyer H. Widely spaced, directly repeated PuGGTCA elements act as promiscuous enhancers for different classes of nuclear receptors. Mol. Cell. Biol. 1995;15:5858–5867. doi: 10.1128/mcb.15.11.5858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katz RW, Subauste JS, Koenig RJ. The interplay of half-site sequence and spacing on the activity of direct repeat thyroid hormone response elements. J. Biol. Chem. 1995;270:5238–5242. doi: 10.1074/jbc.270.10.5238. [DOI] [PubMed] [Google Scholar]
- Khorasanizadeh S, Rastinejad F. Nuclear-receptor interactions on DNA-response elements. Trends Biochem. Sci. 2001;26:384–390. doi: 10.1016/s0968-0004(01)01800-x. [DOI] [PubMed] [Google Scholar]
- Kurokawa R, Yu VC, Naar A, Kyakumoto S, Han ZH, Silverman S, Rosenfeld MG, Glass CK. Differential orientations of the DNA-binding domain and carboxy-terminal dimerization interface regulate binding site selection by nuclear receptor heterodimers. Genes and Devel. 1993;7:1423–1435. doi: 10.1101/gad.7.7b.1423. [DOI] [PubMed] [Google Scholar]
- La Vista-Picard N, Hobbs PD, Pfahl M, Dawson MI. The receptor-DNA complex determines the retinoid response: a mechanism for the diversification of the ligand signal. Mol. Cell. Biol. 1996;16:4137–4146. doi: 10.1128/mcb.16.8.4137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazar MA, Berrodin TJ, Harding HP. Differential DNA binding by monomeric, homodimeric, and potentially heteromeric forms of the thyroid hormone receptor. Mol. Cell. Biol. 1991;11:5005–5015. doi: 10.1128/mcb.11.10.5005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S, Privalsky ML. Heterodimers of retinoic acid receptors and thyroid hormone receptors display unique combinatorial regulatory properties. Mol. Endocrinol. 2005;19:863–878. doi: 10.1210/me.2004-0210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leid M, Kastner P, Chambon P. Multiplicity generates diversity in the retinoic acid signalling pathways. Trends Biochem. Sci. 1992;17:427–433. doi: 10.1016/0968-0004(92)90014-z. [DOI] [PubMed] [Google Scholar]
- Mader S, Chen JY, Chen Z, White J, Chambon P, Gronemeyer H. The patterns of binding of RAR, RXR and TR homo- and heterodimers to direct repeats are dictated by the binding specificites of the DNA binding domains. EMBO J. 1993;12:5029–5041. doi: 10.1002/j.1460-2075.1993.tb06196.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mangelsdorf DJ, Evans RM. The RXR heterodimers and orphan receptors. Cell. 1995;83:841–850. doi: 10.1016/0092-8674(95)90200-7. [DOI] [PubMed] [Google Scholar]
- Marks MS, Hallenbeck PL, Nagata T, Segars JH, Appella E, Nikodem VM, Ozato K. H-2RIIBP (RXR beta) heterodimerization provides a mechanism for combinatorial diversity in the regulation of retinoic acid and thyroid hormone responsive genes. EMBO J. 1992;11:1419–1435. doi: 10.1002/j.1460-2075.1992.tb05187.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mcewan IJ. Nuclear receptors: one big family. Methods Mol Biol. 2009;505:3–18. doi: 10.1007/978-1-60327-575-0_1. [DOI] [PubMed] [Google Scholar]
- Mckenna NJ, O’Malley BW. Minireview: nuclear receptor coactivators--an update. Endocrinology. 2002;143:2461–2465. doi: 10.1210/endo.143.7.8892. [DOI] [PubMed] [Google Scholar]
- Meijsing SH, Pufall MA, So AY, Bates DL, Chen L, Yamamoto KR. DNA binding site sequence directs glucocorticoid receptor structure and activity. Science. 2009;324:407–410. doi: 10.1126/science.1164265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Näär AM, Boutin JM, Lipkin SM, Yu VC, Holloway JM, Glass CK, Rosenfeld MG. The orientation and spacing of core DNA-binding motifs dictate selective transcriptional responses to three nuclear receptors. Cell. 1991;65:1267–1279. doi: 10.1016/0092-8674(91)90021-p. [DOI] [PubMed] [Google Scholar]
- Nagpal S, Saunders M, Kastner P, Durand B, Nakshatri H, Chambon P. Promoter context- and response element-dependent specificity of the transcriptional activation and modulating functions of retinoic acid receptors. Cell. 1992;70:1007–1019. doi: 10.1016/0092-8674(92)90250-g. [DOI] [PubMed] [Google Scholar]
- Nakshatri H, Chambon P. The directly repeated RG(G/T)TCA motifs of the rat and mouse cellular retinol-binding protein II genes are promiscuous binding sites for RAR, RXR, HNF-4, and ARP-1 homo- and heterodimers. J. Biol. Chem. 1994;269:890–902. [PubMed] [Google Scholar]
- Nelson CC, Hendy SC, Faris JS, Romaniuk PJ. Retinoid X receptor alters the determination of DNA binding specificity by the P-box amino acids of the thyroid hormone receptor. J. Biol. Chem. 1996;271:19464–19474. doi: 10.1074/jbc.271.32.19464. [DOI] [PubMed] [Google Scholar]
- Nielsen R, Pedersen TA, Hagenbeek D, Moulos P, Siersbaek R, Megens E, Denissov S, Borgesen M, Francoijs KJ, Mandrup S, Stunnenberg HG. Genome-wide profiling of PPARgamma:RXR and RNA polymerase II occupancy reveals temporal activation of distinct metabolic pathways and changes in RXR dimer composition during adipogenesis. Genes and Devel. 2008;22:2953–2967. doi: 10.1101/gad.501108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olson DP, Koenig RJ. 5′-flanking sequences in thyroid hormone response element half-sites determine the requirement of retinoid X receptor for receptor-mediated gene expression. J. Biol. Chem. 1997;272:9907–9914. doi: 10.1074/jbc.272.15.9907. [DOI] [PubMed] [Google Scholar]
- Perlmann T, Rangarajan PN, Umesono K, Evans RM. Determinants for selective RAR and TR recognition of direct repeat HREs. Genes and Devel. 1993;7:1411–1422. doi: 10.1101/gad.7.7b.1411. [DOI] [PubMed] [Google Scholar]
- Privalsky ML. Thyroid hormone receptors, coregulators, and disease. In: KUMAR R, O’MALLEY BW, editors. NR coregulators and human diseases. World Scientific Publishing, LTD.; Singapore: 2008. pp. 243–280. [Google Scholar]
- Rosen MD, Privalsky ML. Thyroid hormone receptor mutations found in renal clear cell carcinomas alter corepressor release and reveal helix 12 as key determinant of corepressor specificity. Mol. Endocrinol. 2009;23:1183–1192. doi: 10.1210/me.2009-0126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schräder M, Wyss A, Sturzenbecker LJ, Grippo JF, Lemotte P, Carlberg C. RXR-dependent and RXR-independent transactivation by retinoic acid receptors. Nucleic Acids Res. 1993;21:1231–1237. doi: 10.1093/nar/21.5.1231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai CC, Fondell JD. Nuclear receptor recruitment of histone-modifying enzymes to target gene promoters. Vitam. Horm. 2004;68:93–122. doi: 10.1016/S0083-6729(04)68003-4. [DOI] [PubMed] [Google Scholar]
- Umesono K, Murakami KK, Thompson CC, Evans RM. Direct repeats as selective response elements for the thyroid hormone, retinoic acid, and vitamin D3 receptors. Cell. 1991;65:1255–1266. doi: 10.1016/0092-8674(91)90020-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velasco LF, Togashi M, Walfish PG, Pessanha RP, Moura FN, Barra GB, Nguyen P, Rebong R, Yuan C, Simeoni LA, Ribeiro RC, Baxter JD, Webb P, Neves FA. Thyroid hormone response element organization dictates the composition of active receptor. J. Biol. Chem. 2007;282:12458–12466. doi: 10.1074/jbc.M610700200. [DOI] [PubMed] [Google Scholar]
- Vivanco Ruiz MM, Bugge TH, Hirschmann P, Stunnenberg HG. Functional characterization of a natural retinoic acid responsive element. EMBO J. 1991;10:3829–3838. doi: 10.1002/j.1460-2075.1991.tb04952.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wahlstrom GM, Sjoberg M, Andersson M, Nordstrom K, Vennstrom B. Binding characteristics of the thyroid hormone receptor homo- and heterodimers to consensus AGGTCA repeat motifs. Mol. Endocrinol. 1992;6:1013–1022. doi: 10.1210/mend.6.7.1324417. [DOI] [PubMed] [Google Scholar]
- Williams GR, Brent GA. Thyroid Hormone Response Elements. In: WEINTRAUB BD, editor. Molecular Endocrinology: Basic Concepts and Clinical Correlations. Raven Press; New York: 1995. pp. 217–239. [Google Scholar]
- Williams GR, Harney JW, Moore DD, Larsen PR, Brent GA. Differential capacity of wild type promoter elements for binding and transactivation by retinoic acid and thyroid hormone receptors. Mol. Endocrinol. 1992;6:1527–1537. doi: 10.1210/mend.6.10.1333048. [DOI] [PubMed] [Google Scholar]
- Wilson TE, Paulsen RE, Padgett KA, Milbrandt J. Participation of non-zinc finger residues in DNA binding by 2 nuclear orphan receptors. Science. 1992;256:107–110. doi: 10.1126/science.1314418. [DOI] [PubMed] [Google Scholar]
- Wong CW, Privalsky ML. Role of the N terminus in DNA recognition by the v-erb A protein, an oncogenic derivative of a thyroid hormone receptor. Mol. Endocrinol. 1995;9:551–562. doi: 10.1210/mend.9.5.7565803. [DOI] [PubMed] [Google Scholar]
- Yen PM, Sugawara A, Chin WW. Triiodothyronine (T3) differentially affects T3-receptor/retinoic acid receptor and T3-receptor/retinoid X receptor heterodimer binding to DNA. J. Biol. Chem. 1992;267:23248–23252. [PubMed] [Google Scholar]
- Yu VC, Delsert C, Andersen B, Holloway JM, Devary OV, Naar AM, Kim SY, Boutin JM, Glass CK, Rosenfeld MG. RXR beta: a coregulator that enhances binding of retinoic acid, thyroid hormone, and vitamin D receptors to their cognate response elements. Cell. 1991;67:1251–1266. doi: 10.1016/0092-8674(91)90301-e. [DOI] [PubMed] [Google Scholar]
- Zhang XK, Hoffmann B, Tran PB, Graupner G, Pfahl M. Retinoid X receptor is an auxiliary protein for thyroid hormone and retinoic acid receptors. Nature. 1992;355:441–446. doi: 10.1038/355441a0. [DOI] [PubMed] [Google Scholar]