Abstract
The mitogen activated kinases JNK1/2/3 are key enzymes in signaling modules that transduce and integrate extracellular stimuli into coordinated cellular response. Here we report the discovery of the first irreversible inhibitors of JNK1/2/3. We describe two JNK3 co-crystal structures at 2.60 and 2.97 Å resolutions that show the compounds form covalent bonds with a conserved cysteine residue. JNK-IN-8 is a selective JNK inhibitor that inhibits phosphorylation of c-Jun, a direct substrate of JNK kinase, in cells exposed to sub-micromolar drug in a manner that depends on covalent modification of the conserved cysteine residue. Extensive biochemical, cellular and pathway-based profiling establish the selectivity of JNK-IN-8 for JNK and suggest that the compound will be broadly useful as a pharmacological probe of JNK-dependent signal transduction. Potential lead compounds have also been identified for kinases including IRAK1, PIK3C3, PIP4K2C, and PIP5K3.
Introduction
In mammalian cells, the MAPK signaling system is comprised of at least four distinct signaling modules defined by a core of MAP4K, MAP3K, MAP2K and MAPKs that are named after the ‘terminal’ MAPK kinase in each pathway: ERK1/2, JNK1/2/3, p38alpha/beta and ERK5 (Chang et al., 2001; Johnson et al., 2002; Pearson et al., 2001; Raman et al., 2007). JNKs (c-jun NH2-terminal kinase) become highly activated after cells are exposed to stress conditions such as cytokines, osmotic stress, hypoxia and UV light, and are poorly activated by exposure to growth factors or mitogens (Derijard et al., 1994; Pulverer et al., 1991). There are three distinct alternatively spliced genes Jnk1, Jnk2, and Jnk3 that produce approximately ten different proteins. The predominant isoforms JNK1 and JNK2 are ubiquitously expressed but JNK3 is expressed primarily in the nervous system (Derijard et al., 1994; Kallunki et al., 1994; Sluss et al., 1994; Mohit et al., 1995). JNKs are activated by phosphorylation in the activation T-loop at residues Thr183/Tyr185 by the MAP2Ks: MKK4 and MKK7, and are deactivated by MAP kinase phosphatases including MKP1 and MKP5. Signaling through the JNK-pathway is organized through binding to ‘scaffolding’ proteins such as JIP, which assemble signaling complexes containing MAP3K, MAP2K and MAPKs in addition to JNK-phosphorylated transcription factors such as c-Jun, ATF2 and Elk1.
Since JNKs comprise a central node in the inflammatory signaling network, it is not surprising that hyperactivation of JNK signaling is a very common finding in a number of disease states including cancer, inflammatory and neurodegenerative diseases. A significant body of genetic and pharmacological evidence suggests that inhibitors of JNK signaling may provide a promising therapeutic strategy: JNK3 knockout mice exhibit amelioration of neurodegeneration in animal models of Parkinson’s and Alzheimer’s disease (Kyriakis et al., 2001; Zhang et al., 2005; Hunot et al., 2004). JNK1 phosphorylates IRS-1, a key molecule in the insulin-sensing pathway which down-regulates insulin signaling and JNK1 knockout mice are resistant to diet-induced obesity (Aguirre et al., 2000 and 2002; Hirosumi et al., 2002; Sabio et al., 2010); JNK2, often in concert with JNK1, has been implicated in the pathology of autoimmune disorders such as rheumatoid arthritis (Han et al., 2002) and asthma (Wong, W.S., 2005; Pelaia et al., 2005; Blease et al., 2003; Chialda et al., 2005); A recent study suggests that JNK2 may also play a role in vascular disease and atherosclerosis (Osto et al., 2008). However, to date, no inhibitors of JNK have been approved for use in humans.
Numerous small molecules from a variety of scaffolds such as indazoles, aminopyrazoles, aminopyridines, pyridine carboxamides, benzothien-2-ylamides and benzothiazol-2-yl acetonitriles, quinoline derivatives, and aminopyrimidines have been reported to act as selective ATP-competitive JNK inhibitors (LoGrasso and Kamenecka, 2008). Despite this plethora of compounds, many exhibit poor kinase selectivity and/or do not inhibit the phosphorylation of well-characterized substrates of JNK in cells. For example, one of the earliest and still most widely used inhibitors is the anthrapyrazolone, SP-600125 (Bennett et al., 2001; Figure 1A) which exhibits exceptionally low specificity for JNK (Bain et al., 2007) and should only be used in combination with other tools to rule-out a potential role for JNK in a particular process (Inesta-Vaquera et al., 2010). Other reported JNK inhibitors such as AS601245 (Gaillard et al., 2005) only inhibit c-Jun phosphorylation at high concentrations which is likely due to a combination of limited cell penetration, ATP concentration and differences between biochemical and cellular sensitivities to JNK inhibitors.
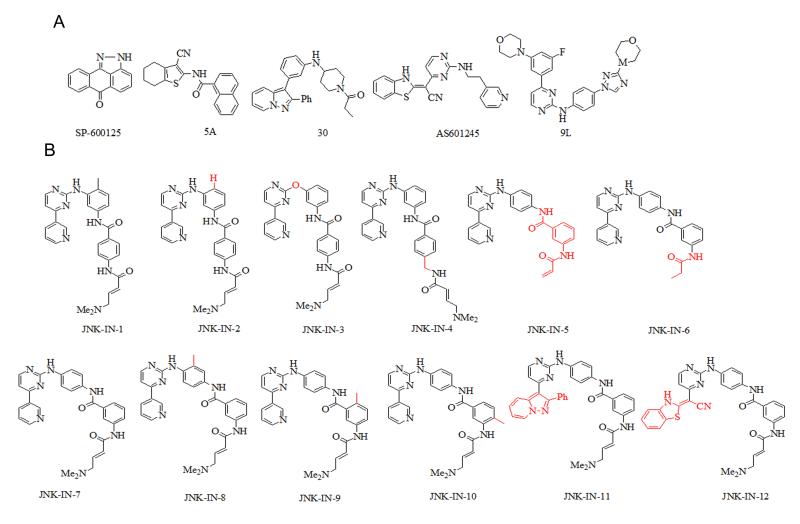
Figure 1. Chemical structures for JNK inhibitors.
(A) Representatives JNK inhibitors (B) Structural modifications relative to JNK-IN-1 for JNK-IN-1 to 6 or relative to JNK-IN-7 for JNK-IN-7 to 12 are highlighted in red.
To address these challenges, we sought to use structure-based drug design to develop ATP-site directed covalent inhibitors of JNK kinases that would target a unique cysteine conserved in all the JNK kinases. Cysteine-directed covalent inhibitors possess a number of potential advantages relative to non covalent inhibitors such as an ability to control kinase selectivity using both non-covalent and covalent recognition of the kinase and the ability to exhibit prolonged pharmacodynamics despite competition with high endogenous intracellular ATP concentrations. Selective cysteine-directed covalent inhibitors have been developed for a number of kinases including Rsk (FMK) (Cohen et al., 2005; Nguyen, T.L., 2008), FGFRs (FIIN-1) (Zhou et al., 2010), Mek (Schirmer et al., 2006), Nek2 (Henise and Taunton, 2011) and other kinases possessing a cysteine immediately proceeding the ‘DFG-motif’ (hypothemycin and analogs) as well as several undergoing clinical investigation as inhibitors of EGFR (HKI-272, BIBW2992, CI1033, EKB569) and BTK (AVL-292, PCI32765) (Singh et al., 2010). Despite these efforts, only four different cysteine positions have been targeted in the ATP-site to date even though at least 180 kinases possess a cysteine that could theoretically be targeted by suitably designed inhibitors (Zhang et al., 2009). Here we report the structure-based design, detailed biochemical and cellular characterization, and crystal structure analysis of JNK3 modified by covalent inhibitors that can irreversibly modify a conserved cysteine residue in JNK.
Serendipitous discovery and rational optimization of a covalent JNK inhibitor
Most currently reported cysteine-directed covalent inhibitors are from the ‘type-1’ (Liu and Gray, 2006) inhibitor class: they bind to the kinase in an ‘active’ conformation with the activation loop in a conformation conducive to substrate binding. We speculated whether ‘type-2’ inhibitors which bind kinases in an ‘inactive’ state with the activation loop in a conformation that blocks substrate from binding might also present a promising platform from which to design a new class of covalent inhibitors. Through an examination of kinases co-crystallized with type-2 inhibitors we noticed that both c-Kit and PDGFR possess a cysteine immediately preceding the ‘DFG-motif’ that marks the beginning of the activation loop and that might be exploited by a suitably designed type-2 inhibitor. We decided to use the phenylaminopyrimidine core of imatinib as a scaffold for elaboration because this compound binds Abl, c-Kit and PDGFR in the type-2 conformation and because it possesses favorable drug properties. Measurement of the distance between the methylpiperazine moiety of imatinib and Cys788 in c-Kit (PDB: 1T46; Mol et al., 2004) inspired us to replace the methylpiperazine moiety with an electrophilic acrylamide bearing a water-solubility enhancing dimethylamino group to generate JNK-IN-1 (Figure 1B). The kinase selectivity of JNK-IN-1 was profiled at a concentration of 10 μM against a 400-kinase panel using KinomeScanTM methodology where, to our surprise, it exhibited significant binding to JNK1/2/3 in addition to the expected imatinib targets of Abl, c-kit, DDR1/2 (Table S1). We confirmed that these binding results by translated into single digit micromolar IC50 for inhibition of JNK kinase activity using the Z’-lyte assay format (Table 1). This result was unanticipated because despite the large number of JNK inhibitors reported in the literature, there are no reports of ‘type-2’ JNK inhibitors and we therefore did not anticipate that imatinib could bind to JNK in an extended ‘type-2’ conformation. However, there are a number of structurally related phenylaminopyrimidines such as 9L (Kamenecka et al., 2010) and 30 (Alam et al., 2007; Figure 1A) that bind to JNK in a type-1 conformation and we speculated that perhaps JNK-IN-1 was binding in an analogous fashion to JNK. In addition, we hypothesized that imatinib might exploit an alternative ‘type-1’ conformation when binding to JNK where the inhibitor assumes an U-shaped configuration as has been observed in a Syk-imatinib co-structure (PDB: 1XBB; Atwell et al., 2004). If JNK-IN-1 were to recognize JNK analogously to how imatinib binds to Syk, the acrylamide moiety of JNK-IN-1 would be placed within covalent bond forming distance of Cys116 of JNK1 and JNK2 and Cys154 of JNK3.
Table 1. JNK inhibitors potency in vitro and in vivo.
Biochemical IC50 in nanomolar for JNK inhibitors against JNK1/2/3 and cellular EC50 for inhibition of c-Jun phosphorylation in Hela and A375 cells. Values for reference inhibitors 5A (Angell et al., 2007), SP-600125 (Bennett et al., 2001) and AS601245 (Gaillard et al., 2005) are from the literature. ND: not determined. See also Table S1.
| IC50 (nM) | p-c-Jun EC50 (nM) | ||||
|---|---|---|---|---|---|
|
|
|||||
| JNK1 | JNK2 | JNK3 | Hela | A375 | |
| JNK-IN-1 | 7780 | 4230 | 7750 | ND | ND |
| JNK-IN-2 | 809 | 1140 | 709 | 704 | 2400 |
| JNK-IN-3 | >10000 | >10000 | >10000 | ND | ND |
| JNK-IN-4 | 2390 | 5460 | 7220 | ND | ND |
| JNK-IN-5 | 2.11 | 1.93 | 0.96 | 118 | 32 |
| JNK-IN-6 | ND | ND | 148 | 6760 | 1905 |
| JNK-IN-7 | 1.54 | 1.99 | 0.75 | 130 | 244 |
| JNK-IN-8 | 4.67 | 18.7 | 0.98 | 486 | 338 |
| JNK-IN-9 | ND | ND | 0.50 | 104 | 117 |
| JNK-IN-10 | ND | ND | 0.50 | 173 | 141 |
| JNK-IN-11 | 1.34 | 0.50 | 0.50 | 48 | 8.6 |
| JNK-IN-12 | 13 | 11.3 | 11.0 | 605 | 134 |
| 5A | 10000* | ND | 200* | >10000 | >10000 |
| SP-600125 | ND | 110* | 190* | 7450 | 1985 |
| AS601245 | 150* | 220* | 70* | 2025 | 2400 |
To test these hypotheses, a number of analogs of JNK-IN-1 were prepared (Figure 1B). First, the ‘flag methyl’ was removed from JNK-IN-1 to yield JNK-IN-2 since this methyl group is a key driver of selectivity for imatinib to c-kit, Abl and PDGF relative to a number of other kinases (Zimmermann et al., 1996). We also expected JNK-IN-2 to be better able to assume the U-conformation relative to the extended type-2 conformation and thereby increase non-covalent recognition of the JNK ATP-binding site. As shown in Table 1, JNK-IN-2 indeed possessed a 5 to 10-fold improved IC50 for inhibition of JNK1/2/3 kinase activity relative to JNK-IN-1. This encouraged us to obtain direct confirmation of covalent binding between JNK-IN-2 and JNK. Upon incubation of recombinantly produced JNK1 with JNK-IN-2 (5-fold excess), electrospray mass spectrometry revealed that the intact mass of the protein increased by the expected ~493 Da (Figure 2A and 2B), consistent with the covalent addition of one molecule of JNK-IN-2 to the kinase. Subsequent protease digestion and LC/MS2 analysis identified a peptide (residues LMDANLCQVIQME) modified by JNK-IN-2 at Cys 116 as predicted by the molecular modeling (Figure 2C).
Figure 2. Mass spectra for THZ-IN-2 labeled JNK1 protein.
(A) Untreated JNK1 protein (B) JNK-IN-2 treated recombinantly produced JNK1 kinase domain. Inset mass spectra were obtained after deconvolution and show addition of 493 daltons after incubation with JNK-IN-2. (C) HCD MS/MS spectrum of the peptide LMDANLC*QVIQME (JNK residues 110-122; C* indicates labeled cysteine). Identification of ions of type b and y are indicated with lines above and below the sequence, respectively. See also Figure S2.
Despite the confirmation of JNK-IN-2 as a cysteine-directed JNK inhibitor, the approximately 1.0 micromolar IC50 suggests a relatively inefficient labeling of the kinase during the biochemical assay. The molecular modeling of JNK-IN-2 with JNK3 suggested that the amino pyrimidine motif would form the typical bidentate hydrogen bonding interaction with Met149 in the kinase ‘hinge’ segment while the pyridine substituent was situated toward the back of the ATP-pocket adjacent to the gatekeeper Met146 and possibly making a hydrogen bond between the pyridine N and the side chain amino group of Lys93. While the acrylamide of JNK-IN-2 was within covalent bond forming distance of Cys154, the geometry based on the modeling did not appear to be ideal for facilitating nucleophilic addition of the cysteine thiol (Figure S1A). To investigate the functional importance of a potential hydrogen bond between Met149 and JNK-IN-2, the aniline NH was changed to an ether linkage in JNK-IN-3. As expected, this change resulted in more than 100-fold increase in biochemical IC50 against JNK1. Next we explored various changes that might place the acrylamide in a more optimal position for reaction with Cys116 in JNK1. We first attempted to insert an additional methylene spacer in JNK-IN-4 which unfortunately increased IC50 against JNK1 by 3-fold. We investigated different regio-isomers of the 1,3-dianiline and 1,4-benzamide moieties of JNK-IN-2. The most dramatic improvement in IC50 was observed when 1,4-dianiline and 1,3-benzamide were incorporated as the linker segment between the pyrimidine and the acrylamide moiety as exemplified by JNK-IN-5 and JNK-IN-7. These compounds possessed a dramatic 500-fold lower IC50 against JNK1, 2 and 3 when compared with JNK-IN-2. Molecular docking of JNK-IN-7 with JNK3 suggested that this improvement in potency was likely due to a more optimal placement of the acrylamide relative to Cys154 which may result in more efficient covalent bond formation (Figure S1B). Incubation of JNK-IN-7 and JNK3 followed by electrospray mass spectrometry revealed the addition of a single molecule of inhibitor to the protein and labeling of Cys154 (Figure S2).
To investigate the importance of covalent bond formation to the potency of this class of inhibitor, we prepared JNK-IN-6 with an unreactive and approximately isosteric propyl amide group replacing the acrylamide of JNK-IN-5. As expected, this compound exhibited an almost 100-fold less potent biochemical IC50 on JNK1, 2, and 3 (Table 1). We then prepared a small collection of analogs of JNK-IN-7 bearing modifications expected to influence its selectivity relative to other kinases. We prepared three methylated analogs JNK-IN-8, JNK-IN-9 and JNK-IN-10 all of which retained the ability to potently inhibit JNK biochemical activity. We replaced the pyridine ring of JNK-IN-7 with substituents that had previously been described for other JNK inhibitors including a bulky group 2-phenylpyrazolo[1,5-a]pyridine (Alam et al., 2007) and benzothiazol-2-yl acetonitrile (Gaillard et al., 2005). The influence of these changes on kinase selectivity is discussed in detail below.
Co-Crystal structure of JNK-IN-2 and JNK-IN-7 with JNK3
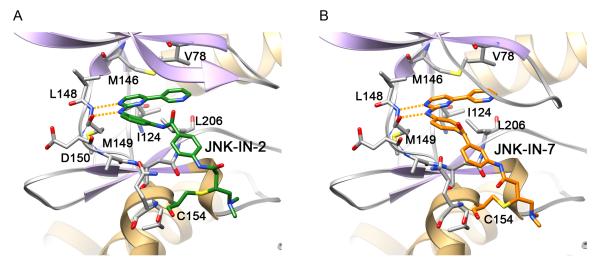
In order to validate the molecular modeling results and to provide a basis for further structure-based optimization efforts, we co-crystallized JNK-IN-2 and JNK-IN-7 with JNK3 de novo using the same JNK3 protein reported previously for 9L (Kamenecka et al., 2010; Figure 3 and Figure S3A). The resulting 2.60 Å and 2.97 Å crystal structures were in good agreement with the docking model described above. Continuous electron density was visible to Cys154 consistent with covalent bond formation (Figure S3B). The inhibitor formed three hydrogen bonds with JNK3, two from the aminopyrimidine motif to the kinase hinge residues Leu148 and Met149 and a third from the amide NH to Asn152. This third hydrogen bond may be important for positioning the terminal ring and orienting the acrylamide moiety proximal to Cys154 thereby facilitating covalent bond formation. The overall kinase conformation of JNK is remarkably similar to the reported 9L crystal structure (average RSMD 2.40 Å; Kamenecka et al., 2010) with the kinase assuming an active conformation. This demonstrates that the covalent inhibitor does not appear to trap an unusual conformation of the kinase. There is a small hydrophobic pocket adjacent to the aniline ortho position which may explain why tolerance exists for the ‘flag’ methyl group in JNK-IN-8, a group that also provided a crucial selectivity determinant. The pyridine moiety binds in a hydrophobic pocket and did not optimally fill this space which was consistent with the potency improvements realized by replacing it with the larger moieties present in JNK-IN-11 and JNK-IN-12. Further modification of the inhibitor in this region would clearly afford significant opportunities for modulating both inhibitor potency and selectivity.
Figure 3. Crystal structure of complex JNK-IN-2 and JNK-IN-7 with JNK3.
Crystal structures of JNK3 residues 39-402 modified at Cys-154 by (A) JNK-IN-2 and (B) JNK-IN-7. The covalent inhibitors (green stick figure for JNK-IN-2 and orange stick figure for JNK-IN-7) and the key residues of JNK3 that are involved in hydrophobic and hydrogen bonding interactions with the covalent inhibitors are labeled and are shown in stick models (grey). The hydrogen bonds between the kinase ‘hinge’ residue Met-149 and the aminopyrimidine-motif of the covalent inhibitors are represented as orange dotted lines (Figure S3). See also Figures S1 and S4.
Inhibition of cellular c-Jun phosphorylation
In parallel with biochemical evaluation, we investigated the ability of the compounds to inhibit JNK activity in cells using two independent assays formats. This is a critical issue because there are several reported JNK inhibitors with nanomolar biochemical potency that translate into micromolar cellular inhibitors. The best characterized direct phosphorylation substrate of JNK is the transcription factor c-Jun. The first assay format is a high-throughput (HTS) compatible cellular assay capable of measuring changes in phosphorylation of c-Jun using the measurement of time resolved fluorescence resonance energy transfer (TR-FRET) between a stably expressed GFP-c-Jun (1-79) fusion protein and a terbium labeled anti pSer73 c-Jun antibody as readout (Robers et al.,2008; Carlson et al., 2009; Stebbins et al., 2008). The second assay format consisted of treating serum starved A375 cells with test compounds followed by stimulation of the JNK kinase pathway with anisomycin and monitoring c-Jun phosphorylation by single-cell microscopy using an anti-phospho Ser73 antibody (Millard et al., 2011). With the exception of a few compounds, both assay formats provided a similar rank-order of potency for this compound series (Table 1). In agreement with the biochemical assays, JNK-IN-5 also provided the break-through in cellular potency and was capable of inhibiting of c-Jun phosphorylation with an IC50 of ~100 nM in HeLa cells and ~30 nM in A375 cells. Introduction of the methylene dimethylamine group to yield JNK-IN-7 resulted in a 2-3-fold loss in potency for cellular JNK inhibition which was not predicted based upon the enzymatic assay. Introduction of methyl groups at the meta-position of the dianiline ring or to the meta and ortho positions of the benzamide resulted in compounds with cellular potency in the hundreds of nanomolar range. JNK-IN-11, the most potent cellular inhibitor of JNK activity in this series, incorporated the phenylpyrazolo[1,5-a]pyridine motif and possessed an IC50 of ~30nM and ~10nM in HeLa and A375 cells respectively. JNK-IN-6, the compound incapable of covalent bond formation, possessed an IC50 50-fold higher than its covalent analog JNK-IN-5, once again underscoring the requirement for the acrylamide moiety to achieve potent cellular inhibition. To allow direct comparison with published JNK inhibitors we tested SP600125, 5A (Angell et al., 2007), and AS601245 in parallel in both assay formats. All these compounds exhibited IC50s in the micromolar range which suggests that covalent inhibition may be required to observe potent JNK inhibition at least under the conditions investigated.
In order to evaluate the kinetics with which JNK-IN-5 could covalently modify JNK in cells, we developed a pulse-chase assay. A375 cells were treated with JNK-IN-5 for 1, 2, 3, 4, and 5 hours to allow for cell penetration and labeling of intracellular targets. Cell lysates were then prepared and labeled with ATP-biotin which contains a reactive acyl phosphate anhydride that reacts non-specifically with the catalytic lysine of kinases including JNK (Patricelli et al., 2007). Streptavidin affinity chromatography was then used to isolate all biotinylated proteins and JNK protein was detected following SDS-PAGE and western blotting (Figure S4A). The length of the JNK-IN-5 incubation time required to fully protect JNK from subsequent labeling by ATP-biotin provides a measure of the rate of intracellular covalent bond formation. Three hours were required for JNK-IN-5 to modify JNK to background levels by this assay. As a negative control, the non-covalent inhibitor JNK-IN-6 was subject to the same protocol and was demonstrated to be incapable of protecting JNK from labeling by ATP-biotin. The kinetics of covalent binding between the JNK-IN-5 and JNK3 in vitro was also investigated in a similar way. JNK-IN-5 was capable of completely labeling JNK3 in 45 minutes when introduced at a 27 molar excess (Figure S4B).
Cellular kinase specificity of covalent JNK inhibitors
The kinase selectivity of several key compounds was first evaluated using a chemical proteomic approach KiNativ and which is capable of monitoring 200 kinases in A375 cells (ActivX Biosciences). To probe the intracellular targets of the compounds we incubated A375 cells with the inhibitors and then looked for protection of labeling by an ATP-biotin probe that labels conserved lysines on kinases and other nucleotide-dependent enzymes. This provided an important advantage relative to the in vitro kinase selectivity profiling because in vitro the short incubation times and presence of reactive thiols in the buffers can potentially cause false negatives for acrylamide-modified kinase inhibitors. Treatment of A375 cells with 1 μM of four of the irreversible JNK inhibitors resulted in the identification of JNK as the most potent and common target (Table 2). In contrast, the reversible inhibitor JNK-IN-6 did not inhibit JNK activity in the same live cell treatment. In addition to JNK 1, 2, 3, JNK-IN-7 also bound to IRAK1, PIK3C3, PIP5K3 and PIP4K2C. Since cysteine-directed covalent kinase inhibitors will sometimes cross-react with kinases that contain an equivalently placed cysteine, we performed a sequence alignment to identify all kinases which have a cysteine near JNK1 Cys116 (Table S2). Amongst the 40 kinases revealed through this analysis only IRAK1 exhibited a detectable binding affinity to JNK-IN-7 based upon KinomeScan profiling. Since IRAK1 crystal structure is not available, we examined the IRAK4 crystal structure (PDB: 3CGF). This showed that Cys276 is potentially situated in a similar location relative to the reactive Cys154 of JNK3. Thus, covalent modification of IRAK1 by JNK-IN-7 is a possibility and subsequent biochemical kinase assay revealed an IC50 of ~10 nM against IRAK1. To evaluate whether IRAK1 is a bonafide intracellular target of JNK-IN-7 we also asked whether the compound could inhibit the E3-ligase activity of pellino, which provides an indirect measure of inhibition of IRAK1 kinase activity in cells. JNK-IN-7 inhibited interleukin 1-stimulated Pellino 1 E3 ligase activity but required a relatively high concentration of 10 μM to achieve complete inhibition (Unpublished data courtesy of Philip Cohen group in Dundee University). Sequence alignments did not reveal obvious cysteine residues that could be covalently modified in PIK3C3, PIP4K2C and PIP5K3 but further work will be required to evaluate whether these are indeed functional targets of JNK-IN-7.
Table 2. Cellular kinase profiling with KiNativ™ technology.
Percent inhibition (color coded as indicated in the legend) of kinase labeling by ATP-biotin that results from incubating A375 cells with the inhibitors for 3 hours at a concentration of 1 μM is indicated (larger numbers indicate stronger binding to the kinase). See also Table S2.
| Kinase Family |
Kinase | Labeling Site |
JNK-IN-6 | JNK-IN-7 | JNK-IN-8 | JNK-IN-11 | JNK-IN-12 |
|---|---|---|---|---|---|---|---|
| CMCG | JNK1,JNK2,JNK3 | Lys2 | 27.3 | 99.7 | 98.7 | 99.8 | 98.6 |
| p38a | Lys2 | 32.6 | 16.4 | 13.2 | 95.9 | −28 | |
| p38a | Other | 12.4 | 14.7 | 3.9 | 98.9 | −13.8 | |
| GSK3A | Lys2 | 6.8 | 20.4 | 10.6 | 40.7 | −7.7 | |
| GSK3B | Lys2 | 23.6 | 12.3 | −8.5 | 67.1 | −29.6 | |
| PFTAIRE1 | Lys1 | −6 | 75.3 | 38.1 | 12.8 | 25.7 | |
| CDK9 | Lys2 | 7.6 | 16.6 | −34.1 | −23.4 | −35.3 | |
| AGC | ROCK1 | Other | 24.4 | −11.9 | 10 | −8.2 | −97 |
| ROCK1,ROCK2 | Lys2 | 11.6 | −21.5 | −17.1 | −33.9 | −91 | |
| Lipid | PIP5K3 | ATP | 16.8 | 91.8 | 18.7 | 99.3 | −11.4 |
| PIK3C3 | ATP | 26.5 | 77.5 | 18.1 | 8.9 | 7.5 | |
| PIK3C3 | ATP | 3.1 | 74.2 | 6.4 | −11 | −0.5 | |
| PIP4K2C | ATP | −4.7 | 77.2 | 1.9 | −4.4 | 43.9 | |
| PIP4K2C | ATP | 35.7 | 84.3 | 32.2 | 24.3 | 42.6 | |
| DNAPK | ATP | 51.1 | 17.4 | 34.9 | 23.3 | −65.4 | |
| DNAPK | ATP | 31.7 | 18.4 | 10.5 | −9.4 | −80.7 | |
| ITPK1 | ATP | 27 | −0.2 | 24.6 | −0.7 | 8.6 | |
| CAMK | PKD2 | Lys1 | 12 | 13.6 | 4.9 | 40.9 | −79 |
| PKD1 | Lys1 | 28.7 | 2 | 8.6 | 29.4 | −21.6 | |
| PKD1,PKD2 | Lys2 | 15.9 | 18.2 | −3 | 10.4 | −39.3 | |
| MARK1 | Lys2 | 23.9 | 7.9 | 8.9 | 18.6 | −43.5 | |
| MARK2 | Lys1 | 13.4 | 23.6 | 9.6 | 20.8 | −41.1 | |
| MARK2,MARK3 | Lys2 | 22.9 | 26.5 | 13.9 | 0.4 | −38.7 | |
| MARK3 | Lys1 | −7.3 | −13.5 | −12 | −27.8 | −67.9 | |
| PHKg2 | Lys1 | 28 | −7 | −0.8 | −76 | −94.1 | |
| CK1 | CK1a | Lys2 | 19.5 | −20.2 | −13.6 | 61.1 | −96 |
| CK1d,CK1e | Lys2 | 32.4 | 8.4 | −21.8 | 88.4 | −75 | |
| Others | AurA | Lys2 | 9.9 | 20.9 | −8 | 68.2 | −23 |
| AurA,AurB,AurC | ATP Loop | 4.9 | 19.1 | −11.3 | 53.8 | −20.9 | |
| IKKa | Lys2 | 27.3 | −9.2 | 4.9 | −7.3 | −79.2 | |
| IKKb | Lys2 | 33.4 | −10.1 | −6.5 | −11.2 | −97.5 | |
| STE | ZC2/TNIK,ZC3/MINK | Lys2 | 52.8 | 31 | 45.1 | 75.9 | −36.2 |
| ZC1/HGK,ZC2/TNIK, ZC3/MINK |
Lys2 | 8.8 | 8.3 | −11.3 | 65.1 | −25.5 | |
| ZC2/TNIK | Lys1 | −15.9 | 20.9 | 9.9 | >80 | −8.9 | |
| TAO2 | Lys2 | 22.5 | −3.7 | −21.6 | 42.6 | −13 | |
| MAP2K1,MAP2K2 | Lys2 | 24.3 | 3.2 | 3.2 | −10.4 | −3.7 | |
| TAO1,TAO3 | Lys2 | 35.8 | 13.1 | 2.9 | 34.1 | −10.8 | |
| TK | ABL,ARG | Lys1 | −1.4 | 1.9 | −2.7 | 56.1 | 7.9 |
| TKL | IRAK1 | Lys2 | −4.6 | 81 | 0.8 | 5.4 | −14.9 |
| ZAK | Lys1 | 3 | 15.5 | −7.8 | 84.4 | −40.2 | |
| BRAF | Lys2 | 8.9 | 7 | −23.7 | 27.8 | 6.2 | |
| RAF1 | Lys2 | −20.2 | −7.3 | −41.4 | 25.5 | 8.6 |
Although JNK-IN-7 is a relatively selective JNK inhibitor in cells, introduction of the ‘flag’ methyl to yield JNK-IN-8 resulted in a dramatic improvement in selectivity and eliminated binding to IRAK1, PIK3C3, PIP4K2C and PIP5K3. The dramatic selectivity improvement that results from introduction of this flag-methyl group has been previously reported for imatinib (Zimmermann et al., 1996). Replacement of the pyridine ring with bulkier substituents as exhibited by JNK-IN-11 resulted in a broadening of the selectivity profile as well as further enhancing the potency for inhibition of c-Jun phosphorylation in cells. JNK-IN-11 binds potently to JNKs, p38, PIP5K3, ZAK, ZC2, PIP5K3 and CK1 demonstrating that this compound class might be a valuable lead compound to develop selective inhibitors of these potential alternative targets. In contrast to pyridine in JNK-IN-7, a benzothiazol-2-yl acetonitrile moiety in JNK-IN-12 resulted in enhanced specificity demonstrating the potential to modulate selectivity by the choice of functionality in this region.
In vitro specificity of covalent JNK inhibitors
To complement the KiNativ profiling, the in vitro kinase selectivity of several key compounds was evaluated comprehensively by using two complementary approaches: kinase-binding assays against a panel of 442 distinct kinases using with the KINOMEscan methodology (DiscoverX) and standard radioactivity-based enzymatic assays against a panel of 121 kinases (The National Centre for Protein Kinase Profiling in Dundee). Based upon the KINOMEscan results, JNK-IN-7, JNK-IN-8 and JNK-IN-12 possessed highly selective S (10) scores (defined as the ratio number of kinases inhibited more than 90 percent at screening concentration of 1 μM) of 0.085, 0.031 and 0.025, respectively (Table S1). For example, JNK-IN-7 exhibited binding inhibition of 95% or more to approximately 14 kinases at the concentration of 1.0 μM. We attempted to confirm all these potent binding targets using either an enzymatic kinase assay or through the measurement of a dissociation constant to the kinase in question. JNK-IN-7 was confirmed to have a Kd or IC50 of 100 nM or less against eight additional kinases (Table 3). JNK-IN-7 was next tested for its ability to inhibit the enzymatic activity of a panel of 121 kinases at a concentration of 1.0 μM. This analysis revealed 12 kinases that were inhibited more than 80% relative to the DMSO control and follow-up IC50 determination revealed sub-200 nM IC50 against of IRAK1, ERK8, and NUAK1 (Table S3). JNK-IN-12 bearing a benzothiazol-2-yl acetonitrile in place of the pyridine conferred an improved selectivity relative to JNK-IN-7. The KINOMEscan score for JNK-IN-12 was even smaller than JNK-IN-8 and follow-up enzymatic assays on the potent targets revealed IC50s of 37.6, 57.1, and 89.9 nM for IRAK1, HIPK4 and AKT2 respectively (Table 3). The introduction of phenylpyrazolo[1,5-a]pyridine to JNK-IN-11 resulted in a significant decrease in kinase selectivity as assessed by KINOMEscan (Score = 0.125) and more than 30 additional kinases including different mutants of EGFR, c-Kit, DDR1 and Gsk3b (Tables 3 & Table S3). Consistent with the KiNativ profiling, JNK-IN-8 also exhibited exceptional selectivity based upon KinomeScan and enzymatic profiling. Further biochemical and binding assays failed to identify any target with an IC50 or Kd of less than 1.0 μM. Cumulatively these combined profiling technologies demonstrate that both JNK-IN-8 and JNK-IN-12 are remarkably selective covalent JNK inhibitors and are appropriate for interrogating JNK-dependent biological phenomena.
Table 3. JNK inhibitors potency on potential off-targets.
Enzymatic IC50 or dissociation constant (Kd) for the potential additional kinase targets. For JNK-IN-7 and JNK-IN-11, the kinases with the score below 5 were tested and for JNK-IN-8 and JNK-IN-12 kinases with score below 1 were tested (see profiling in table S4). See also Tables S1 and S3.
| JNK-IN-7 | JNK-IN-8 | JNK-IN-11 | JNK-IN-12 | ||||
|---|---|---|---|---|---|---|---|
| kinase | IC50 (nM) | kinase | IC50 (nM) | kinase | IC50 (nM) | kinase | IC50 (nM) |
| (enzymatic) | (enzymatic) | (enzymatic) | (enzymatic) | ||||
| JNK1 | 1.5 | JNK1 | 4.7 | JNK1 | 1.3 | JNK1 | 13 |
| JNK2 | 2 | JNK2 | 18.7 | JNK2 | 0.5 | JNK2 | 11.3 |
| JNK3 | 0.7 | JNK3 | 1 | JNK3 | 0.5 | JNK3 | 11 |
| IRAK1 | 14.1 | MNK2 | 238 | EGFR(L861Q) | 21 | IRAK1 | 37.6 |
| KIT | 2410 | FMS | 287 | EGFR(L858R) | 24.9 | HIPK4 | 57.1 |
| HIPK1 | 5010 | HIPK4 | 8970 | DDR1 | 56.1 | AKT2 | 89.9 |
| Kinase | Kd (nM) | KIT | >10000 | CSNK1E | 82.9 | AKT1 | >370 |
| (KinomeScan™) | MET(M250T) | >10000 | CSNK1G2 | 161 | AKT3 | >370 | |
| YSK4 | 4.8 | PDGFRB | >10000 | PDGFRB | 1030 | SLK | 884 |
| ERK3 | 22 | PRKX | 7500 | KIT | 1320 | ||
| RIOK2 | 30 | Kinase | Kd (nM) | Kinase | Kd (nM) | Kinase | Kd (nM) |
| (KinomeScan™) | (KinomeScan™) | (KinomeScan ™) | |||||
| PIP5K2C | 32 | KIT(V559D) | 92 | EGFR(G719C) | 2.6 | KIT(V559D,T670I) | 160 |
| CDKL5 | 34 | KIT(V559D,T670I) | 56 | EGFR(E746- A750del) |
7 | ||
| KIT(L576P) | 40 | MYLK4 | 4000 | KIT((V559D) | 8.2 | ||
| KIT(V559D) | 48 | RIOK2 | 120 | PFCDPK1 | 14 | ||
| ICK | 54 | DMPK2 | 18 | ||||
| DRAK1 | 100 | CIT | 95 | ||||
| DYRK2 | 120 | ERBB2 | 230 | ||||
| BIKE | 190 | PRKD3 | 240 | ||||
Cellular Pathway Profiling
The profiling above provides an assessment of direct engagement with potential targets, but does not address further perturbations that maybe induced as a consequence of these binding events. We therefore established a microscopy-based assay using phospho-specific antibodies selective for c-Jun phosphorylation, and also sentinel nodes in other signaling pathways such as Erk, p38, JNK, Akt, Stat, NF-κB and Rsk (Millard, et al., 2011; Figure S5). JNK-IN-7, JNK-IN-8 and JNK-IN-12 exhibited only on-pathway activity as monitored by inhibition of c-Jun phosphorylation. JNK-IN-11 was the only compound found to have off-pathway activity as exemplified shown by its ability to potently block phosphorylation of Erk1/2, Rsk1, Msk1 and p38. This finding is consistent with the substantially broadened kinase selectivity profile of this compound. However, JNK-IN-11 also provided the most complete inhibition of c-Jun phosphorylation, a result we interpret as reflecting the ability of the compound inhibit additional kinases involved in phosphorylation of c-Jun.
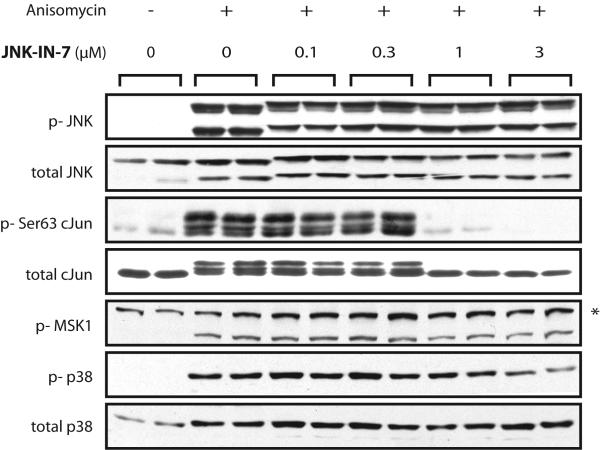
To corroborate these data we also examined the ability of the compounds to inhibit phosphorylation of JNK, c-Jun, MSK1 and p38 in HEK293-ILR1 cells following stimulation by anisomycin by traditional western blotting (Figure 4). All compounds, except the JNK-IN-11, were capable of inhibiting c-Jun phosphorylation without blocking phosphorylation of MSK1 and p38. The inhibition was not reversed by removal of JNK-IN-8 from cell culture medium (Figure S6). The results are in good agreement with the relative compound potencies established using the immunostaining and kinase profiling approaches. A distinct reduction in electrophoretic mobility of JNK protein is apparent upon incubation with the inhibitors presumably as a consequence of covalent modification by the inhibitors. This serves as a simple means to measure kinase modification.
Figure 4. Cellular pathway profiling with Western-blot.
Western blot analysis of inhibition of JNK, cJun, MSK1 and p38 for JNK-IN-7, 8 and 11 following anisomycin stimulation of HEK293-IL1R cells. See also Figures S5 and S6.
Evaluation of the Functional Selectivity
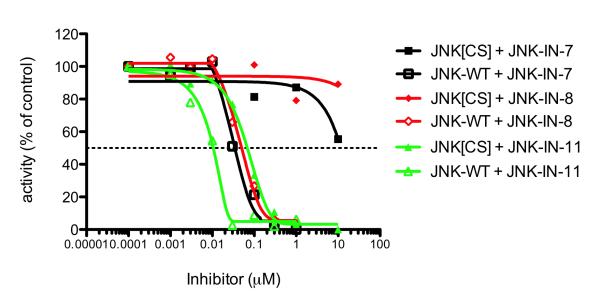
To investigate the extent to which the observed cellular effects resulted from direct covalent modification of JNK1/2/3 cysteine residues versus other potential intracellular targets, we used mutagenesis to engineer a Cys to Ser mutant into JNK2. We purified Cys116Ser JNK2 and confirmed that activated wild type JNK2 and mutant JNK2[Cys116Ser] displayed similar Km and Vmax towards the ATF2 peptide substrate in vitro (Figure 5A). In the presence of inhibitors, the mutation resulted in a 10-fold increase in IC50 for inhibition of JNK activity by JNK-IN-11, and remarkably, at least a 100-fold increase in IC50 for JNK-IN-7 and JNK-IN-8 (Figure 5B). Thus, JNK-IN-7 and JNK-IN-8 require Cys116 for JNK2 inhibition. Overall, our results demonstrate that JNK-IN-8 is an efficient, specific and irreversible intracellular inhibitor of JNK kinase activity by a mechanism that depends on modification of a conserved cysteine in the ATP-binding motif.
Figure 5. Evaluation of the functional selectivity with mutagenesis.
Mutation of the conserved Cys116 to Ser increases the IC50 for inhibition of JNK2 by over 100-fold for JNK-IN-7 and JNK-IN-8 but only by approximately 10-fold for JNK-IN-11.
Discussion
The JNK family of kinases constitutes a central node in the stress-activated MAPK signaling pathway and has been proposed to include drug targets with potential utility in the treatment of cancer, chronic inflammation and neurological disorders. However, with the exception of a recently developed 9L analogue (Crocker et al., 2011), achieving pharmacological inhibition of JNK has been hampered by the lack of potent and selective inhibitors with suitable pharmacokinetic properties for use in proof of concept studies in cells and animals. To address these issues we have pursued the development of irreversible JNK inhibitors that covalently modify a cysteine residue conserved among JNK family members. The major advantage of covalent modification of kinases is that sustained target inhibition can be achieved with only transient exposure of the target to the inhibitor which reduces the need to sustain drug concentration at a level sufficient to achieve complete target inhibition (Singh et al., 2010). From the perspective of pre-clinical research, engineered JNK kinases lacking the cysteine residue (e.g. JNK C116S) that is modified by covalent inhibitors are drug resistant, potentially making it possible to rigorously establish the selectivity of the compounds and thus, the JNK-dependency of various cellular phenotypes.
Our starting point for development of a potent JNK inhibitor was JNK-IN-1 which is an acrylamide-modified phenylaminopyrimidine containing the imatinib backbone that we serendipitously discovered to be capable of binding to JNK based on kinome-wide specificity profiling (Fabian et al., 2005; Goldstein et al., 2008). Recently a similar scaffold was used to develop the first covalent inhibitor of c-Kit, a kinase that possesses a reactive cysteine residue immediately preceding the DFG-motif of the activation loop (Leproult et al., 2011). Molecular docking of JNK-IN-2 into the crystal structures of JNK3 provided a rational basis for structure-guided design of the appropriate linker element that would serve to connect the phenylaminopyrimidine pharmacophore which is predicted to bind to the kinase-hinge region of the protein with a reactive acrylamide moiety. We discovered that the most critical feature for potent inhibition of JNK in vitro and in cellular assays inhibition was for the linker element to contain a 1,4-disposition of the dianiline moiety and a 1,3-disposition of terminal aminobenzoic acid moiety; these features are exemplified by JNK-IN-7 and JNK-IN-8. A 2.97 Å co-structure between JNK-IN-7 and JNK3 showed that our design goals had been made and demonstrated that a covalent bond is indeed formed with residue Cys154 of JNK3.
Extensive biochemical and cellular selectivity profiling allowed us to identify several additional potential kinase targets for JNK-IN-7 including IRAK1, MPSK1, NEK9, PIK3C3, PIP4K2C and PIP5K3. Efficient inhibition of these targets appears to require an acrylamide moiety since they are not inhibited by JNK-IN-6 which lacks the acrylamide group. With the exception of IRAK1, these kinases do not appear to contain a potentially reactive cysteine located in a position analogous to Cys154 on JNK3 suggesting that in binding to MPSK1, NEK9, PIK3C3, PIP4K2C and PIP5K3 JNK-IN-7 may adopt a different conformation than in binding to JNK3 thereby allowing it to access alternative cysteine residues. Alternatively, JNK-IN-7 may form covalent adducts with reactive lysine residues. For example, the natural product Wortmannin undergoes a Michael addition reaction with Lys833 of PI3K, albeit one that involves a non-acrylamide electrophilic moiety. We have validated that JNK-IN-7 can indeed inhibit IRAK-1 dependent E3 ligase activity of pellino, a protein that functions in the Toll-receptor signaling pathway in cells at a relative high compound concentrations (1-10 μM) (unpublished data from Philip Cohen’s group in Dundee). Further compound optimization guided by cell-based assay will be required to establish if more potent cellular inhibition of IRAK-1 can be achieved. We have also initiated chemical and biological experiments to optimize and characterize the potential of compounds such as JNK-IN-11 to inhibit IRAK1, PIK3C3, PIP4K2C, and PIP5K3 in a cellular context. With respect to JNK kinases, we discovered two ways to further enhance the kinase selectivity of JNK-IN-7. The first was to introduce an ortho-methyl group (creating JNK-IN-8) which is analogous to the so-called ‘flag’ methyl group of imatinib or the ortho-methoxy group of the ALK inhibitor TAE684 (Galkin et al., 2007) and of the polo-kinase inhibitor BI-2356 (Kothe et al., 2007). The crystal structure of JNK-IN-7 predicts that the ortho-methyl group may nestle into a small grove along the hinge segment between Asp150 and Ala151 of JNK3. The second was to replace the pyridine moiety with a geometrically more complex benzothiazol-2-yl acetonitrile moiety which was previously shown to represent a favorable pharmacophore for binding to the JNK ATP-site (Gaillard et al., 2005); JNK-IN-12 carries this modification. This portion of the inhibitor is predicted to bind in proximity to the “gatekeeper” methionine and provides a critical selectivity determinant for the compound. In contrast, JNK-IN-11, which contains a large 2-phenylpyrazolo[1,5-a]pyridine group, displays a dramatically broadened inhibition profile in both purified enzyme and cellular assays. JNK-IN-8 and JNK-IN-12 appear to be the most optimal compounds that balance good potency and favorable kinase selectivity profiles. JNK-IN-7 and JNK-IN-11 appear to possess additional targets based upon the KiNativ profiling and these compounds may serve as valuable ‘lead compounds’ to optimize activity against new targets. Our selectivity profiling to date has been limited to kinases and clearly acrylamide containing compounds may also react with other cysteine-containing enzymes, many of which have been cataloged in a recent chemoproteomics study (Weerapana et al., 2010).
Implications for design of covalent kinase inhibitors
Covalent inhibitors are typically designed by rational modification of scaffolds that are already potent non-covalent binders of the desired target protein. For example, the anilinoquinazoline scaffold provided a template for development of highly potent covalent and non covalent inhibitors of EGFR kinase (Smaill et al., 2000). An alternative approach is to start from relatively low affinity non-covalent binders and to allow covalent bond formation to drive potency toward the desired target. For example, the pyrrolopyrimidine Rsk inhibitor FMK (Cohen et al., 2005) and the anilinopyrimidine T790M EGFR inhibitor WZ-4002 (Zhou et al., 2009) both increase approximately 100-fold in potency for their respective targets as a consequence of covalent bond formation. The covalent inhibitors described in this study fall into this second category in that they require covalent bond formation to achieve potent inhibition of JNK kinase activity. One major advantage of this second approach is that it is much easier to identify a relatively selective low affinity non-covalent scaffold as a starting point relative to a selective high affinity scaffold. However, the challenge is that one must identify a scaffold that allows presentation of the electrophile to the kinase with a geometry that allows for efficient covalent bond formation. This is especially true because the residence time for a low affinity non-covalent compound is typically very short. As can be seen from the structure-activity relationship for JNK-IN-1 to 12, relatively minor changes can have dramatic consequences to the potency of inhibition. This is in sharp contrast to the general notion that a covalent inhibitor will always be exceptionally potent. Intracellularly, there is a kinetic competition for modification of the desired target versus ‘off-targets’ which may be other proteins or engagement of cellular pathways that metabolize reactive electrophiles. In addition, proteins are continuously synthesized and degraded with varying kinetics which can allow for regeneration of unmodified protein. Therefore an effective covalent inhibitor must label its target protein rapidly relatively to competing labeling events and protein turn-over.
We have pursued two general approaches to developing potent covalent kinase inhibitors. The first is to generate small, rationally-designed libraries of electrophile modified inhibitors that can be used in cell-based screens to select for compounds with activity against the desired target. Simple molecular modeling based on known ATP-site recognition modes can be used to select where on the scaffold to introduce an electrophilic group. This approach was used to develop WZ-4002 a potent and selective inhibitor of the T790M ‘gatekeeper’ mutation of EGFR. The disadvantage of this approach is that it requires considerable up-front synthetic effort and cell-based screening approach requires a relatively high potency for inhibition to be assayable. The second approach is to search among a larger set of known kinase inhibitor scaffolds lacking electrophiles for low affinity compounds using a biochemical screening approach that allows for screening at high concentrations and then using structure-based drug design to prepare a small library of covalent inhibitors for optimization. The advantage of this approach is that there exist large collections of known kinase inhibitors having established kinase selectivity profiles; the disadvantage is that it can be difficult to predict which scaffolds will be permissive for the correct trajectory for the electrophile relative to the protein nucleophile. Our discovery of JNK-IN-1 as a compound that would enable the second approach was serendipitous, but inspection of published Ambit kinase selectivity data for imatinib (Karaman et. al. 2008) shows that the scaffold had already been annotated as having the ability to bind to JNK non-covalently. We therefore anticipate that it will be possible to create an efficient pipeline for generation of first-in-class covalent inhibitors that target the large number of kinases containing suitably positioned cysteine (and possibly also lysine) residues.
Our study demonstrates that the KiNativ profiling methodology is a powerful tool for discovering and guiding the optimization of new covalent inhibitors. First it allows for an unbiased screen of the majority of available ATP-competitive targets in a cellular system of choice. As discussed above, this enables serendipitous discovery of potential new targets for known compounds. Second by assessing selectivity in a cellular context, the native kinase conformation is accessed and the structure-activity relationships appear to correlate well with functional cellular assays. We anticipate that creation of publically accessible kinase-selectivity profiles for large sets of compounds will further enable the search for low-affinity leads for new kinases of interest.
Use of JNK-IN-8 for studying JNK activities in cellular assays
With respect to enabling analysis of JNK signaling pathways in cells, we have shown that JNK-IN-8 and JNK-IN-11 achieve potent and relatively selective, covalent inhibition of JNK1-3 kinases in cells. We recommend the use of JNK-IN-8 and JNK-IN-12 at concentration of approximately 1.0 μM and we anticipate that transfection of cells with drug-resistant cysteine to serine mutations will make it possible to demonstrate compound selectivity for various cellular phenotypes. Because kinase inhibition appears to reach completion after approximately 3 hours (Figure S4A) we recommend preincubating cells with compound for ~3 hr prior to analyzing JNK activity. A distinct change in the electrophoretic mobility of JNK is observed after exposure to inhibitor (Figure 4 and Figure S6) that may serve as a useful pharmacodynamic marker of JNK inhibition.
Significance
The JNK family of protein kinases are key transducers of extracellular stress signals and inhibition of JNK function may provide a therapeutic strategy to treat a variety of disorders including neurodegeneration, cancer and autoimmune diseases. Here, we report the discovery and characterization of the first irreversible JNK inhibitors that form a covalent bond with a conserved cysteine. Compounds such as JNK-IN-8 and JNK-IN-12 are extremely potent inhibitors of enzymatic and cellular JNK inhibition as monitored by inhibition of c-Jun, a well characterized direct phosphorylation substrate. Extensive biochemical and cellular profiling has been performed to establish the selectivity of these compounds for inhibiting JNK activity. The superior potency and selectivity of JNK-IN-8 and JNK-IN-12 relative to other previously reported JNK inhibitors suggest that these compounds will likely serve as very useful pharmacological probes of JNK-dependent cellular phenomena.
Materials and Methods
Chemistry
All solvents and reagents were used as obtained. 1H NMR spectra were recorded with a Varian Inova 600 NMR spectrometer and referenced to dimethylsulfoxide. Chemical shifts are expressed in ppm. Mass spectra were measured with Waters Micromass ZQ using an ESI source coupled to a Waters 2525 HPLC system operating in reverse mode with a Waters Sunfire C18 5 μm, 4.6 mm x 50 mm column. Purification of compounds was performed with either a Teledyne ISCO CombiFlash Rf system or a Waters Micromass ZQ preparative system. The purity was analyzed on an above-mentioned Waters LC-MS Symmetry (C18 column,4.6 mm x 50 mm, 5 μM) using a gradient of 5-95% methanol in water containing 0.05% trifluoacetic acid (TFA). Detailed synthetic schemes and characterization data are presented in the supplementary data.
JNK1/2/3 specific assay condition (Cascade format) – MAPK8 (JNK1)
The 2X MAPK8 (JNK1)/inactive MAPKAPK3/Ser/Thr 04 Peptide Mixture is prepared in 50 mM HEPES pH 7.5, 0.01% BRIJ-35, 10 mM MgCl2, 1 mM EGTA, 2 mM DTT. The final 10 μL kinase reaction consists of 3.3-13.3 ng MAPK8 (JNK1), 20 ng inactive MAPKAPK3, and 2 μM Ser/Thr 04 Peptide in 50 mM HEPES pH 7.5, 0.01% BRIJ-35, 10 mM MgCl2, 1 mM EGTA, 1 mM DTT. After the 1 hour kinase reaction incubation, 5 μL of a 1:1024 dilution of development reagent A is added. MAPK9 (JNK2) The 2X MAPK9 (JNK2)/inactive MAPKAPK3/Ser/Thr 04 peptide mixture is prepared in 50 mM HEPES pH 7.5, 0.01% BRIJ-35, 10 mM MgCl2, 1 mM EGTA, 2 mM DTT. The final 10 μL kinase reaction consists of 2.4 - 9.8 ng MAPK9 (JNK2), 20 ng inactive MAPKAPK3 and 2 μM Ser/Thr 04 peptide in 50 mM HEPES pH 7.5, 0.01% BRIJ-35, 10 mM MgCl2, 1 mM EGTA, 1 mM DTT. After 1 hour kinase reaction incubation, 5 μL of a 1:1024 dilution of development reagent A is added. MAPK10 (JNK3) The 2X MAPK10 (JNK3)/inactive MAPKAPK3/Ser/Thr 04 peptide mixture is prepared in 50 mM HEPES pH 7.5, 0.01% BRIJ-35, 10 mM MgCl2, 1 mM EGTA, 2 mM DTT. The final 10 μL kinase reaction consists of 1.3-5.3 ng MAPK10 (JNK3), 20 ng inactive MAPKAPK3 and 2 μM Ser/Thr 04 peptide in 50 mM HEPES pH 7.5, 0.01% BRIJ-35, 10 mM MgCl2, 1 mM EGTA, 1 mM DTT. After the 1 hour kinase reaction incubation, 5 μL of a 1:1024 dilution of development reagent A is added.
Available from http://tools.invitrogen.com/downloads/CustomerProtocol_AssayConditions.pdf.
Intact protein analysis
For each analysis, ~100 pmol JNK protein +/− inhibitor (JNK-IN-2 or JNK-IN-7) was injected onto a self-packed reversed phase column (1/32” O.D. x 500 μm I.D., 5 cm of POROS 10R2 resin). After desalting, protein was eluted with an HPLC gradient (0-100% B in 4 minutes, A=0.2M acetic acid in water, B=0.2 M acetic acid in acetonitrile, flow rate = 10 μL/min) into a QTRAP mass spectrometer (AB Sciex, Toronto, Canada) or an LTQ Orbitrap mass spectrometer (ThermoFisher Scientific, San Jose, CA). The QTRAP was operated in Q1 MS mode at unit resolution scanning at 2000 amu/sec. LTQ OrbitrapMS spectra were acquired in centroid mode using the electron multipliers for ion detection. Mass spectra were deconvoluted using MagTran1.03b2 software (Zhang et al, 1998).
Protease digestion and nanoLC/MS analysis of peptide fragments
JNK-IN-2 or JNK-IN-7 treated JNK (25 μg, ~620 pmol) was diluted with ammonium bicarbonate buffer, pH 8.0 then reduced for 30 min at 56 °C with 10 mM DTT. After cooling for 5 min, the protein was alkylated with 22.5 mM iodoacetamide for 30 min at room temperature in the dark, and digested overnight with 1.5 μg of trypsin at 37 °C. In the morning, 1 μg of Glu-C was added, and the solution further incubated at 37 °C for 8 hr. Digested peptides (~2 pmol) were injected onto a self-packed pre-column (4 cm POROS10R2) and eluted into the mass spectrometer (LTQ OrbitrapVelos, ThermoFisher Scientific). Peptides were subjected to MS2 by CAD (electron multiplier detection, relative collision energy 35%, q = 0.25) as well as HCD (image current detection, resolution @ m/z 400 = 7500, relative collision energy 35%).
Cell-Based Assays for c-Jun Phosphorylation
The cell based kinase assays for c-Jun phosphorylation carried out by using the LanthaScreen™ c-Jun (1–79) HeLa cell line (Life Technologies, Carlsbad, CA) which stably express GFP-c-Jun 1–79 and GFP-ATF2 19–106, respectively. Phosphorylation was determined by measuring the time resolved FRET (TR-FRET) between a terbium labeled phospho-c-Jun specific antibody and GFP (22). The cells were plated in white tissue culture treated 384 well plates at a density of 10,000 cell per well in 32 μL assay medium (Opti-MEM®, supplemented with 0.5% charcoal/dextran-treated FBS, 100 U/ml penicillin and 100 μg/ml streptomycin, 0.1mM nonessential amino acids, 1mM sodium pyruvate, 25 mM Hepes, pH 7.3, and lacking phenol red). After overnight incubation, cells were pretreated for 90 min with compound (at indicated concentration) diluted in 4 μL assay buffer followed by 30 min of stimulation with 5 ng/ml of TNF-α in 4 μL assay buffer (final assay volume was 40 μl). The medium was then removed by aspiration and the cells were lysed by adding 20 μl of lysis buffer (20 mM Tris-HCl, pH7.6, 5 mM EDTA, 1% Nonidet P-40 substitute, 5 mM NaF, 150 mM NaCl, and 1:100 protease and phosphatase inhibitor mix, SIGMA P8340 and P2850, respectively). The lysis buffer included 2 nM of the terbium-labeled anti-c-Jun (pSer73) detection antibodies (Life Technologies). After allowing the assay to equilibrate for 60 minutes at room temperature, TR-FRET emission ratios were determined on a BMG Pherastar fluorescence plate reader (BMG Labtech, Cary, NC) using the following parameters: excitation at 340 nm, emission 520 nm and 490 nm; 100 μs lag time; 200 μs integration time; emission ratio = Em 520 / Em 490. All data were analyzed and plotted using Graphpad Prism 4.
High Throughput Microscopy
Cells were plated at 7500 cells/well in 96-well microscopy plates (Corning) in recommended media for 24 hours, and then starved in media lacking serum for 16 hours. Cells were pre-treated for 180 minutes with 10-fold stock solutions of JNK inhibitors and for 10 min with control compounds MK2206 (allosteric Akt inhibitor;, Haoyuan Chemexpress co. limited; Hirai et al., 2010), PD0325901 (allosteric Mek inhibitor; Haoyuan Chemexpress co. limited; Barrett et al., 2008), SB239063 (ATP-competitive p38 inhibitor; Haoyuan Chemexpress co. limited; Underwood et al., 2000), KIN001-040 (ATP-competitive JAK1,2,3 inhibitor; Haoyuan Chemexpress co. limited;. Thompson et al., 2002) and KIN001-208 (IKK inhibitor VIII, Haoyuan Chemexpress co. limited., Murata et al., 2004) and treated with 10- fold stock solutions of IGF-1, IL-6, TNF-α (all PeproTech) or anisomycin for 60 minutes. Cells were fixed in 2% paraformaldehyde for 10 min at room temperature and washed with PBS-T (Phosphate Buffered Saline, 0.1% Tween 20). Cells were permeabilized in methanol for 10 min at room temperature, washed with PBS-T, and blocked in Odyssey Blocking Buffer (LI-COR Biosciences) for 1 hour at room temperature. Cells were incubated overnight at 4°C with antibody specific for Erk1/2(pT202/pY204), Akt(pS473), cJUN(pS73), pP38(T180/Y182) and pSTAT3(Y705) (Cell Signaling Technology), pRSK1(S380) and pMSK1(S376) (Epitomics) and NF-κB (Santa Cruz Biotechnology) diluted 1:400 in Odyssey Blocking Buffer. Cells were washed three times in PBS-T and incubated with rabbit-specific secondary antibody labeled with Alexa Fluor 647 (Invitrogen) diluted 1:2000 in Odyssey Blocking Buffer. Cells were washed once in PBS-T, once in PBS and incubated in 250 ng/ml Hoechst 33342 (Invitrogen) and 1:1000 Whole Cell Stain (blue; Thermo Scientific) solution. Cells were washed two times with PBS and imaged in an imageWoRx high-throughput microscope (Applied Precision). Data was plotted using DataPflex (Hendriks et al., 2010).
Binding Kinetics assay
A375 cells (ATCC® CRL-1619™) were pre-treated with 1μM compound for the indicated amounts of time. Remove the medium and wash 3 times with PBS. Resuspend the cell pellet with 1 mL Lysis Buffer (1% NP-40, 1% CHAPS, 25 mM Tris, 150 mM NaCl, Phosphatase Inhibitor Cocktail, Roche 04906845001, and Protease Inhibitor Cocktail, Roche 11836170001). Rotate end-to-end for 30 min at 4°C. Lysates were cleared by centrifugation at 14000 rpm for 15 min in the Eppendorf. The cleared lysates gel filtered into Kinase Buffer (0.1% NP-40, 20 mM HEPES, 150 mM NaCl, Phosphatase Inhibitor Cocktail, Protease Inhibitor Cocktail) using Bio-Rad 10DG colums. The total protein concentration of the gel-filtered lysate should be around 5-15 mg/ml. Cell lysate was labeled with the probe from ActivX® at 5 μM for 1 hour. Samples were reduced with DTT, and cysteines were blocked with iodoacetamide and gel filtered to remove excess reagents and exchange the buffer. Add 1 volume of 2X Binding Buffer (2% Triton-100, 1% NP-40, 2 mM EDTA, 2X PBS) and 50 μL streptavidin bead slurry and rotate end-to-end for 2 hours, centrifuge at 7000 rpm for 2 min. Wash 3 times with 1X Binding Buffer and 3 times with PBS. Add 30 μL 1X sample buffer to beads, heat samples at 95°C for 10 min. Run samples on an SDS-PAGE gel at 110V. After transferred, the membrane was immunoblotted with JNK antibody (Cell signaling 9258).
Incubate 1 μM JNK-IN-5 with purified JNK3 protein for indicated time period, then add the ATP-Biotin probe from ActivX® at 5 μM for 10 min. Denature the protein by adding same volume 8 M urea solution and gel filtered to remove excess reagents and exchange the buffer. Add 1 volume of 2X Binding Buffer (2% Triton-100, 1% NP-40, 2 mM EDTA, 2X PBS) and 50 μL streptavidin bead slurry and rotate end-to-end for 2 hours, centrifuge at 7000 rpm for 2 min. Wash 3 times with 1X Binding Buffer and 3 times with PBS. Add 30 μL 1X sample buffer to beads, heat samples at 95°C for 10 min. Run samples on an SDS-PAGE gel at 110V. After transferred, the membrane was immunoblotted with JNK antibody (Cell signaling 9252).
Buffers
Lysis Buffer contained 50 mM Tris/HCl, pH 7.5, 1 mM EGTA, 1 mM EDTA, 1% (w/v) 1 mM sodium orthovanadate, 10 mM sodium β-glycerophosphate, 50 mM NaF, 5 mM sodium pyrophosphate, 0.27 M sucrose, 1 mM Benzamidine and 2 mM phenylmethanesulphonylfluoride (PMSF) and supplemented with 1% (v/v) Triton X-100. Kinase assay buffer contained 50 mM Tris/HCl, pH 7.5 and 0.1 mM EGTA.
Cell culture, treatments and cell lysis
HEK-293 cells stably expressing Interleukin Receptor 1 (HEK293-IL1R) were cultured in Dulbecco’s Modified Eagle’s medium (DMEM) supplemented with 10% FBS, 2 mM glutamine and 1×antimycotic/antibiotic solution. Cells were serum starved for 18h before incubation with DMSO or different inhibitors, stimulated with 2 μM anisomycin (Sigma) for 1h and lysates were clarified by centrifugation for 10 min at 16000 g and 4°C.
Antibodies
Rabbit polyclonal antibodies against total pan JNK isoforms ((#9252), phospho-pan JNK isoforms (Thr183/Tyr185), (#4668), total p38 (#9212) or phospho-p38 MAPK (Thr180/Tyr182), (4631 resp.), total c-Jun (#9165), phospho-c-Jun (Ser63), (#9261) and phospho-MSK1 (Ser376) (#9591) were from Cell Signalling technology.
SDS-PAGE and western blot
Cell lysates (30 μg) were resolved by electrophoresis on SDS polyacrylamide gels (10%) or Novex 4-12% gradient gels, and electroblotted to nitrocellulose membranes. Membranes were blocked with 5% skimmed milk (w/v) in 50 mM Tris/HCl, pH 7.5, 0.15 M NaCl and 0.1% (v/v) Tween (TBST Buffer). Primary antibodies were used at a concentration of 1 μg/ml, diluted in 5% skimmed milk in TBST and incubated overnight at 4°C. Detection of immune-complexes was performed using horseradish-peroxidase-conjugated secondary antibodies (Pierce) and an enhanced-chemiluminescence reagent (in-house).
JNK2 Kinase assays
Wild type JNK2 or mutant JNK2[Cys116Ser] was activated in a reaction mixture containing 2 μM JNK2, 200 nM MKK4, 200 nM MKK7 in kinase assay buffer containing 0.1 mM ATP and 10 mM magnesium chloride. After incubation at 30 min at 30°C the reaction mixture was snap frozen in aliquots. Activity of JNK2 was assessed in a total reaction volume of 50 μl containing 200 nM activated wild type JNK or mutant JNK2[Cys116Ser], in kinase buffer containing 0.1 mM [γ-32P]ATP (~500-1000 cpm/pmol), 10 mM magnesium chloride and 2 μM ATF2 (residues 19-96) as a substrate. The different inhibitors, or equivalent DMSO volume in controls, were added immediately before to the ATP. Reactions were terminated by adding 20 mM EDTA after 30 min at 30°C incubation 40 μl of the reaction mixture was applied to P81 phosphocellulose paper which were washed in 50 mM phosphoric acid and phosphorylated ATF2 peptide bound to p81 paper quantified by Cerenkov counting.
Supplementary Material
Acknowledgements
Funding was provided by NIH U54 HG006097, NIH 1RC2HG005693, 5 RCD HG005693-02, 5 RC2 CA148164-02, 1 U54 HG 006907-01, NS057153-04 and American Skin Association, Milstein Innovation Award. Thanks to Kendall Nettles for help with the determination of JNK3 inhibitor structures. Thanks to the Medical Research Council UK, the Wellcome Trust and the pharmaceutical companies supporting the Division of Signal Transduction Therapy Unit (AstraZeneca, Boehringer-Ingelheim, GlaxoSmithKline, Merck KgaA and Pfizer) for financial support. Special thanks to Lili Zhou for assistance with high throughput microscopy and Eddy Goh and Philip Cohen for developing the cellular IRAK1 assays and helpful discussions
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Aguirre V, Uchida T, Yenush L, Davis RJ, White MF. The c-Jun NH2-terminal kinase promotes insulin resistance during association with insulin receptor substrate-1 and phosphorylation of Ser307. J. Biol. Chem. 2000;275:9047–9054. doi: 10.1074/jbc.275.12.9047. [DOI] [PubMed] [Google Scholar]
- Aguirre V, Werner ED, Giraud J, Lee Y, Shoelson SE, White MF. Phosphorylation of Ser307 in insulin receptor substrate-1 blocks interactions with the insulin receptor and inhibits insulin action. J. Biol. Chem. 2002;277:1531–1537. doi: 10.1074/jbc.M101521200. [DOI] [PubMed] [Google Scholar]
- Alam M, Beevers RE, Ceska T, Davenport RJ, Dickson KM, Fortunato M, Gowers L, Haughan AF, James LA, Jones MW, et al. Synthesis and SAR of aminopyrimidines as novel c-Jun N-terminal kinase (JNK) inhibitors. Bioorg. Med. Chem. Lett. 2007;17:3463–3467. doi: 10.1016/j.bmcl.2007.03.078. [DOI] [PubMed] [Google Scholar]
- Angell RM, Atkinson FL, Brown MJ, Chuang TT, Christopher JA, Cichy-Knight M, Dunn AK, Hightower KE, Malkakorpi S, Musgrave JR, et al. N-(3-Cyano-4,5,6,7-tetrahydro-1-benzothien-2-yl)amides as potent, selective, inhibitors of JNK2 and JNK3. Bioorg. Med. Chem. Lett. 2007;17:1296–1301. doi: 10.1016/j.bmcl.2006.12.003. [DOI] [PubMed] [Google Scholar]
- Atwell S, Adams JM, Badger J, Buchanan MD, Feil IK, Froning KJ, Gao X, Hendle J, Keegan K, Leon BC, Muller-Deickmann HJ, et al. A novel mode of Gleevec binding is revealed by the structure of spleen tyrosine kinase. J. Biol. Chem. 2004;279:55827–55832. doi: 10.1074/jbc.M409792200. [DOI] [PubMed] [Google Scholar]
- Blease K, Lewis A, Raymon HK. Emerging treatments for asthma. Expert Opin. Emerging Drugs. 2003;8:71–81. doi: 10.1517/14728214.8.1.71. [DOI] [PubMed] [Google Scholar]
- Bennett BL, Sasaki DT, Murray BW, O’Leary EC, Sakata ST, Xu W, Leisten JC, Motiwala A, Pierce S, Satoh Y, et al. SP600125, an anthrapyrazolone inhibitor of Jun N-Terminal kinase. Proc. Natl. Acad. Sci. USA. 2001;98:13681. doi: 10.1073/pnas.251194298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bain J, Later L, Elliot M, Shpiro N, Hastie CJ, Mclauchlan H, Klevernic I, Arthur JSC, Alessi DR, Cohen P. The selectivity of protein kinase inhibitors: a further update. Biochem. J. 2007;408:297–315. doi: 10.1042/BJ20070797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrett SD, Bridges AJ, Dudley DT, Saltiel AR, Fergus JH, Flamme CM, Delaney AM, Kaufman M, LePage S, Leopold WR, et al. The discovery of the benzhydroxamate MEK inhibitors CI-1040 and PD 0325901. Bioorg. Med. Chem. Lett. 2008;18:6501–6504. doi: 10.1016/j.bmcl.2008.10.054. [DOI] [PubMed] [Google Scholar]
- Chang L, Karin M. Mammalian MAP kinase signaling cascades. Nature. 2001;410:37–40. doi: 10.1038/35065000. [DOI] [PubMed] [Google Scholar]
- Chialda L, Zhang M, Brune K, Pahl A. Inhibitors of mitogenactivated protein kinases differentially regulate costimulated T cell cytokine production and mouse airway eosinophilia. Respir. Res. 2005;6:36–54. doi: 10.1186/1465-9921-6-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen MS, Zhang C, Shokat KM, Taunton J. Structural Bioinformatics-Based Design of Selective, Irreversible Kinase Inhibitors. Science. 2005;308:1318–1321. doi: 10.1126/science1108367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlson CB, Robers MB, Vogel KW, Machleidt T. Development of LanthaScreencellular assays for key components within the PI3K/AKT/mTOR pathway. J. Biomol. Screen. 2009;14:121–132. doi: 10.1177/1087057108328132. [DOI] [PubMed] [Google Scholar]
- Crocker CE, Khan S, Cameron MD, Robertson HA, Robertson GS, LoGrasso P. JNK Inhibition Protects Dopamine Neurons and Provides Behavioral Improvement in a Rat 6-Hydroxydopamine Model of Parkinson’s Disease. ACS Chem. Neurosci. 2011;2:207–212. doi: 10.1021/cn1001107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derijard B, Hibi M, Wu IH, Barrett T, Su B, Deng T, Karin M, Davis RJ. JNK1: a protein kinase simulated by UV light and Ha-Ras that binds and phosphorylates the c-Jun activation domain. Cell. 1994;76:1025–1037. doi: 10.1016/0092-8674(94)90380-8. [DOI] [PubMed] [Google Scholar]
- Fabian MA, Biggs WH, III, Treiber DK, Atteridge CE, Azimioara MD, Benedetti1 MG, Carter1 TA, Ciceri1 P, Edeen PT, et al. A small molecule–kinase interaction map for clinical kinase inhibitors. Nature Biotech. 2005;23:329–336. doi: 10.1038/nbt1068. [DOI] [PubMed] [Google Scholar]
- Gaillard P, Jeanclaude-Etter I, Ardissone V, Arkinstall S, Cambet Y, Camps M, Chabert C, Church D, Cirillo R, Gretener D, et al. Design and synthesis of the first generation of novel potent, selective, and in vivo active (benzothiazol-2-yl)-acetonitrile inhibitors of the c-Jun N-terminal kinase. J. Med. Chem. 2005;48:4596–4607. doi: 10.1021/jm0310986. [DOI] [PubMed] [Google Scholar]
- Goldstein DM, Gray NS, Zarrinkar PP. High-throughput kinase profiling as a platform for drug discovery. Nat. Rev. Drug Discov. 2008;7:391–397. doi: 10.1038/nrd2541. [DOI] [PubMed] [Google Scholar]
- Galkin AV, Melnick JS, Kim S, Hood TL, Li N, Li L, Xia G, Steensma R, Chopiuk G, Jiang J, et al. Identification of NVP-TAE684, a potent, selective, and efficacious inhibitor of NPM-ALK. Proc. Natl. Acad. Sci. USA. 2007;104:270–275. doi: 10.1073/pnas.0609412103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunot S, Vila M, Teismann P, Davis RJ, Hirsch EC, Przedborski S, Rakic P, Flavell RA. JNK-mediated induction of cyclooxygenase 2 is required for neurodegeneration in a mouse model of Parkinson’s disease. Proc. Natl. Acad. Sci. U.S.A. 2004;101:665–670. doi: 10.1073/pnas.0307453101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirosumi J, Tuncman G, Chang L, Gorgün CZ, Uysal K.T, ; Maeda, K., Karin M, Hotamisligil GS. A central role for JNK in obesity and insulin resistance. Nature. 2002;420:333–336. doi: 10.1038/nature01137. [DOI] [PubMed] [Google Scholar]
- Han Z, Chang L, Yamanishi Y, Karin M, Firestein GS. Joint damage and inflammation in c-Jun N-terminal kinase 2 knockout mice with passive murine collagen-induced arthritis. Arthritis Rheum. 2002;46:818–823. doi: 10.1002/art.10104. [DOI] [PubMed] [Google Scholar]
- Henise JC, Taunton J. Irreversible Nek2 kinase inhibitor with cellular activity. J. Med. Chem. 2011;54:4133–4146. doi: 10.1021/jm200222m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirail H, Sootome1 H, Nakatsuru Y, Miyama1 K, Taguchi1 S, Tsujioka K, Ueno Y, Hatch H, Majumder PK, Bo-Sheng Pan BS, Kotani H. MK-2206, an Allosteric Akt Inhibitor, Enhances Antitumor Efficacy by Standard Chemotherapeutic Agents or Molecular Targeted Drugs In vitro and In vivo. Mol Cancer Ther. 2010;9:1956–1967. doi: 10.1158/1535-7163.MCT-09-1012. [DOI] [PubMed] [Google Scholar]
- Hendriks BS, Espelin CW. DataPflex: a MATLAB-based tool for the manipulation and visualization of multidimensional datasets. Bioinformatics. 2010;26:432–433. doi: 10.1093/bioinformatics/btp667. [DOI] [PubMed] [Google Scholar]
- Inesta-Vaquera FA, Campbell DG, Arthur JSC, Cuenda A. ERK5 pathway regulates the phosphorylation of tumour suppressor hDlg during mitosis. Biochem. Biophy. Res. Commun. 2010;399:84–90. doi: 10.1016/j.bbrc.2010.07.046. [DOI] [PubMed] [Google Scholar]
- Johnson GL, Lapadat R. Mitogen-activated protein kinase pathways mediated by ERK, JNK, and p38 protein kinases. Science. 2002;298:1911–1912. doi: 10.1126/science.1072682. [DOI] [PubMed] [Google Scholar]
- Kallunki T, Su B, Tsigelny I, Sluss HK, Derijard B, Moore G, Davis RJ, Karin M. JNK2 contains a specificity-determining region responsible for efficient c-Jun binding and phosphorylation. Genes. Dev. 1994;8:2996–3007. doi: 10.1101/gad.8.24.2996. [DOI] [PubMed] [Google Scholar]
- Kyriakis JM, Avruch J. Mammalian mitogen-activated protein kinase signal transduction pathways activated by stress and inflammation. Physiol. Rev. 2001;81:807–869. doi: 10.1152/physrev.2001.81.2.807. [DOI] [PubMed] [Google Scholar]
- Kamenecka T, Jiang R, Song X, Duckett D, Chen W, Ling YY, Habel J, Laughlin JD, Chambers J, Figuera-Losada M, et al. Synthesis, Biological Evaluation, X-ray Structure, and Pharmacokinetics of Aminopyrimidine c-jun-N-terminal Kinase (JNK) Inhibitors. J. Med. Chem. 2010;53:419–431. doi: 10.1021/jm901351f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kothe M, Kohls D, Low S, Coli R, Rennie GR, Feru F, Kuhn C, Ding YH. Selectivity-determining residues in Plk1. Chem. Biol. Drug Des. 2007;70:540–546. doi: 10.1111/j.1747-0285.2007.00594.x. [DOI] [PubMed] [Google Scholar]
- Karaman MW, Herrgard S, Treiber DK, Gallant P, Atteridge CE, Campbell BT, Chan KW, Ciceri P, Davis MI, Edeen PT, et al. A quantitative analysis of kinase inhibitor selectivity. Nat. Biotechnol. 2008;26:127–132. doi: 10.1038/nbt1358. [DOI] [PubMed] [Google Scholar]
- Liu Y, Gray NS. Rational design of inhibitors that bind to inactive kinase conformations. Nat. Chem. Bio. 2006;2:358–364. doi: 10.1038/nchembio799. [DOI] [PubMed] [Google Scholar]
- Leproult E, Barluenga S, Moras D, Wurtz JM, Winssinger N. Cysteine Mapping in Conformationally Distinct Kinase Nucleotide Binding Sites: Application to the Design of Selective Covalent Inhibitors. J. Med. Chem. 2011;54:1347–1355. doi: 10.1021/jm101396q. [DOI] [PubMed] [Google Scholar]
- LoGrasso P, Kamenecka T. Inhibitors of c-jun-N-Terminal Kinase (JNK) Mini-Rev. in Med. Chem. 2008;8:755–766. doi: 10.2174/138955708784912120. [DOI] [PubMed] [Google Scholar]
- Mohit AA, Martin JH, Miller CA. Ap493F12 kinase: a novel MAPkinase expressed in a subset of neurons in the human nervous system. Neuron. 1995;14:67–78. doi: 10.1016/0896-6273(95)90241-4. [DOI] [PubMed] [Google Scholar]
- Mol CD, Dougan DR, Schneider TR, Skene RJ, Kraus L, Scheibe DN, Snell GP, Zou H, Sang BC, Wilson KP. Structural basis for the autoinhibition and STI-571 inhibition of c-Kit tyrosine kinase. J. Biol. Chem. 2004;279:31655–31663. doi: 10.1074/jbc.M403319200. [DOI] [PubMed] [Google Scholar]
- Millard BJ, Niepel M, Menden MP, Muhlich JL, Sorger PK. Adaptive informatics for multifactorial and high-content biological data. Nat. Meth. 2011;8:487–492. doi: 10.1038/nmeth.1600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murata T, Shimada M, Sakakibara S, Yoshino T, Masuda T, Shintani T, Sato H, Koriyama Y, Fukushima K, Nunami N, et al. Synthesis and structure–activity relationships of novel IKK-β inhibitors. Part 3: Orally active anti-inflammatory agents. Bioorg. Med. Chem. Lett. 2004;14:4019–4022. doi: 10.1016/j.bmcl.2004.05.041. [DOI] [PubMed] [Google Scholar]
- Nguyen TL. Targeting RSK: An Overview of Small Molecule Inhibitors. Anti-Cancer Agents in Med. Chem. 2008;8:710–716. doi: 10.2174/187152008785914770. [DOI] [PubMed] [Google Scholar]
- Osto E, Matter CM, Kouroedov A, Malinski T, Bachschmidt M, Camici GG, Kilic U, Stallmach T, Boren J, Iliceto S, et al. c-Jun N-terminal kinase 2 deficiency protects against hypercholesterolemia-induced endothelial dysfunction and oxidative stress. Circulation. 2008;118:2073–2080. doi: 10.1161/CIRCULATIONAHA.108.765032. [DOI] [PubMed] [Google Scholar]
- Pearson G, Robinson F, Beers GT, Xu BE, Karandikar M, Berman K, Cobb MH. Mitogen-activated protein (MAP) kinase pathways: regulation and physiological functions. Endocr. Rev. 2001;22:153–183. doi: 10.1210/edrv.22.2.0428. [DOI] [PubMed] [Google Scholar]
- Pulverer BJ, Kyriakis JM, Avruch J, Nikolakaki E, Woodgett JR. Phosphorylation of c-Jun mediated by MAP kinases. Nature. 1991;353:670–673. doi: 10.1038/353670a0. [DOI] [PubMed] [Google Scholar]
- Pelaia G, Cuda G, Vatrella A, Gallelli L, Caraglia M, Marra M, Abbruzzese A, Caputi M, Maselli R, Costanzo FS, Marsico SA. Mitogen-activated protein kinases and asthma. J. Cell. Physiol. 2005;202:642–653. doi: 10.1002/jcp.20169. [DOI] [PubMed] [Google Scholar]
- Patricelli MP, Szardenings K, Liyanage M, Nomanbhoy TK, Wu M, Weissig H, Aban A, Doris C, Tanner S, Kozarich JW. Functional Interrogation of the Kinome Using Nucleotide Acyl Phosphates. Biochemistry. 2007;46:350–358. doi: 10.1021/bi062142x. [DOI] [PubMed] [Google Scholar]
- Raman M, Chen W, Cobb MH. Differential regulation and properties of MAPKs. Oncogene. 2007;26:3100–3112. doi: 10.1038/sj.onc.1210392. [DOI] [PubMed] [Google Scholar]
- Robers MB, Horton RA, Bercher MR, Vogel W, Machleidt T. High-throughput cellular assays for regulated posttranslational modifications. Anal. Biochem. 2008;372:189–197. doi: 10.1016/j.ab.2007.09.012. [DOI] [PubMed] [Google Scholar]
- Sluss HK, Barrett T, Derijard B, Davis RJ. Signal transduction by tumor necrosis factor mediated by JNK protein kinases. Mol. Cell. Biol. 1994;14:8376–8384. doi: 10.1128/mcb.14.12.8376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabio J, Davis RJ. cJun NH2-terminal kinase 1 (JNK1): roles in metabolic regulation of insulin resistance. Trends Biochem. Sci. 2010;35:490–496. doi: 10.1016/j.tibs.2010.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schirmer A, Kennedy J, Murli S, Reid R, Santi DV. Targeted covalent inactivation of protein kinases by resorcylic acid lactone polyketides. Proc. Natl. Acad. Sci. USA. 2006;103:4234–4239. doi: 10.1073/pnas.0600445103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh J, Petter RC, Kluge AF. Targeted covalent drugs of the kinase family. Curr. Opin. in Chem. Biol. 2010;14:475–480. doi: 10.1016/j.cbpa.2010.06.168. [DOI] [PubMed] [Google Scholar]
- Stebbins JL, De SK, Machleidt T, Becattini B, Vazquez J, Kuntzen C, Chen LH, Cellitti JF, Riel-Mehan M, Emdadi A, Solinas G, Karin M, Pellecchia M. Identification of a new JNK inhibitor targeting the JNK-JIP interaction site. Proc. Natl. Acad. Sci. USA. 2008;105:16809–16813. doi: 10.1073/pnas.0805677105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smaill JB, Rewcastle GW, Loo JA, Greis KD, Chan OH, Reyner EL, Showalter HD, Vincent PW, Elliott WL, Denny WA. Tyrosine kinase inhibitors. 17. Irreversible inhibitors of the epidermal growth factor receptor: 4-(phenylamino)quinazoline- and 4-(phenylamino)pyrido[3,2-d]pyrimidine-6-acrylamides bearing additional solubilizing functions. J. Med. Chem. 2000;43:1380–1397. doi: 10.1021/jm990482t. [DOI] [PubMed] [Google Scholar]
- Thompson JE, Cubbon RM, Cummings RT, Wicker LS, Frankshun R, Cunningham BR, Cameron PM, Meinke PT, Liverton N, Weng Y, DeMartino JA. Photochemical preparation of a pyridone containing tetracycle: a Jak protein kinase inhibitor. Bioorg. Med. Chem. Lett. 2002;12:1219–1223. doi: 10.1016/s0960-894x(02)00106-3. [DOI] [PubMed] [Google Scholar]
- Underwood DC, Osborn RR, Kotzer CJ, Adams JL, Lee JC, Webb EF, Carpenter DC, Bochnowicz S, Thomas HC, Douglas WP, Hay DWP, et al. SB 239063, a Potent p38 MAP Kinase Inhibitor, Reduces Inflammatory Cytokine Production, Airways Eosinophil Infiltration, and Persistence. J. Pharmacol. Exp. Ther. 2000;293:281–288. [PubMed] [Google Scholar]
- Weerapana E, Wang C, Simon GM, Richter F, Khare S, Dillon MBD, Bachovchin DA, Mowen K, Baker D, Cravatt BF. Quantitative reactivity profiling predicts functional cysteines in proteomes. Nature. 2011;468:790–795. doi: 10.1038/nature09472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong WS. Inhibitors of tyrosine kinase signaling cascade for asthma. Curr. Opin. Pharmacol. 2005;5:264–271. doi: 10.1016/j.coph.2005.01.009. [DOI] [PubMed] [Google Scholar]
- Zhang GY, Zhang QG. Agents targeting c-Jun N-terminal kinase pathway as potential neuroprotectants. Exp. Opin. Invest. Drugs. 2005;14:1373–1383. doi: 10.1517/13543784.14.11.1373. [DOI] [PubMed] [Google Scholar]
- Zhou W, Hur W, McDermott U, Dutt A, Xian W, Ficarro S.B.; Zhang, J., Sharma SV, Brugge J, Meyerson M, Settleman J, Gray NS. A structure-Guided Approach to Creating Covalent FGFR Inhibitors. Chem. Biol. 2010;17:285–295. doi: 10.1016/j.chembiol.2010.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang JM, Yang PL, Gray NS. Targeting cancer with small molecule kinase inhibitors. Nat. Rev. Cancer. 2009;9:28–39. doi: 10.1038/nrc2559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Marshall AG. A universal algorithm for fast and automated charge state deconvolution of electrospray mass-to-charge ratio spectra. Journal of the American Society for Mass Spectrometry. 1998;9:225–233. doi: 10.1016/S1044-0305(97)00284-5. [DOI] [PubMed] [Google Scholar]
- Zimmwermann J, Buchdunger E, Mett H, Meyer T, Lydon NB, Taxler P. a new class of potent and highly selective PDGF-receptor autophosphorylation inhibitors. Bioorg. Med. Chem. Lett. 1996;6:1221–1226. [Google Scholar]
- Zhou WJ, Ercan D, Chen L, Yun CH, Capelletti M, Cortot AB, Chirieac L, Jacob RE, Padera R, Engen JR, Wong KK, Eck MJ, Gray NS, Janne PA. Novel mutant-selective EGFR kinase inhibitors against EGFR T790M. Nature. 2009;462:1070–1074. doi: 10.1038/nature08622. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.