Abstract
AIM: To investigate genetic diversity of Helicobacter pylori (H. pylori) cell division-related gene A (cdrA) and its effect on the host response.
METHODS: Inactivation of H. pylori cdrA, which is involved in cell division and morphological elongation, has a role in chronic persistent infections. Genetic property of H. pylori cdrA was evaluated using polymerase chain reaction and sequencing in 128 (77 American and 51 Japanese) clinical isolates obtained from 48 and 51 patients, respectively. Enzyme-linked immunosorbent assay was performed to measure interleukin-8 (IL-8) secretion with gastric biopsy specimens obtained from American patients colonized with cdrA-positive or -negative strains and AGS cells co-cultured with wild-type HPK5 (cdrA-positive) or its derivative HPKT510 (cdrA-disruptant). Furthermore, the cytotoxin-associated gene A (cagA) status (translocation and phosphorylation) and kinetics of transcription factors [nuclear factor-kappa B (NF-κB) and inhibition kappa B] were investigated in AGS cells co-cultured with HPK5, HPKT510 and its derivative HPK5CA (cagA-disruptant) by western blotting analysis with immunoprecipitation.
RESULTS: Genetic diversity of the H. pylori cdrA gene demonstrated that the cdrA status segregated into two categories including four allele types, cdrA-positive (allele types;Iand II) and cdrA-negative (allele types; III and IV) categories, respectively. Almost all Japanese isolates were cdrA-positive (I: 7.8% and II: 90.2%), whereas 16.9% of American isolates were cdrA-positive (II) and 83.1% were cdrA-negative (III: 37.7% and IV: 45.5%), indicating extended diversity of cdrA in individual American isolates. Comparison of each isolate from different regions (antrum and corpus) in the stomach of 29 Americans revealed that cdrA status was identical in both isolates from different regions in 17 cases. However, 12 cases had a different cdrA allele and 6 of them exhibited a different cdrA category between two regions in the stomach. Furthermore, in 5 of the 6 cases possessing a different cdrA category, cdrA-negative isolate existed in the corpus, suggesting that cdrA-negative strain is more adaptable to colonization in the corpus. IL-8 secretions from AGS revealed that IL-8 levels induced by a cdrA-disrupted HPKT510 was significantly lower (P < 0.01) compared to wild-type HPK5: corresponding to 50%-60% of those of wild-type HPK5. These data coincided with in vivo data that an average value of IL-8 in biopsy specimens from cdrA-positive and cdrA-negative groups was 215.6 and 135.9 pg/mL, respectively. Western blotting analysis documented that HPKT510 had no effect on CagA translocation and phosphorylation, however, nuclear accumulation of NF-κB was lower by HPKT510 compared to HPK5.
CONCLUSION: Colonization by a cdrA-negative or cdrA-dysfunctional strain resulted in decreased IL-8 production and repression of NF-κB, and hence, attenuate the host immunity leading to persistent infection.
Keywords: Helicobacter pylori cell division-related gene A, Genetic diversity, Host immune response, Interleukin-8 secretion, Nuclear factor kappa B, Persistent infection
INTRODUCTION
Helicobacter pylori (H. pylori), a Gram-negative spiral bacterium, colonizes the human stomach and causes chronic inflammation, which may progress to peptic ulceration, atrophic gastritis and gastric cancer[1]. Although the H. pylori population structure appears to be clonal over short periods of time, isolates obtained from different individuals exhibit substantial genetic diversity, consistent with extensive recombination and a panmictic population structure[2-7]. Putative mechanisms for the generation of diversity within H. pylori include frequent horizontal genetic exchange among strains and a high level of spontaneous mutation occurring over a long evolutionary time period within a highly restricted niche[6,8].
Infection by cytotoxin-associated gene A (cagA)-positive H. pylori is a known risk factor for the development of gastroduodenal disease due to major changes in cellular morphology and the release of molecules, including cytokines, from the gastric epithelium. H. pylori infection up-regulates secretion of various inflammatory cytokines, including interleukin (IL)-1, IL-6, IL-8 and tumor necrosis factor α, and contributes to the pronounced inflammatory response[9]. IL-8 production induced by H. pylori in vitro and in vivo is recognized as a host response to microbes[10,11], but the role of CagA on IL-8 production is less clear[12]. IL-8 is a potent pro-inflammatory cytokine which regulates neutrophil infiltration of gastric mucosa in H. pylori gastritis and is released through the signaling pathway concerning nuclear factor-kappa B (NF-κB), which includes extracellular signal-regulated kinase (ERK) activity[12,13] and mitogen-activated protein kinase[14]. Among the strongest transcriptional regulators of H. pylori-induced cytokine expression is the NF-κB family of transcription factors[15]. Activation of NF-κB can affect the expression of several hundred genes, and activation of its signal transduction pathway occurs in response to a wide range of stimuli and results in nuclear accumulation of NF-κB transcription factors, causing changes in the expression of target genes involved in innate and adaptive immunity, and inflammation.
We described previously that the H. pylori cell division-related gene A (cdrA) not only has a repressive role on cell division but is also involved in cell elongation and cell death via cell wall synthesis at the division site[16,17]. The cdrA-disrupted mutant HPKT510 was able to survive for the long-term in liquid medium, even under serum-free and aerobic conditions, and was more resistant to bactericidal of beta-lactam antibiotics than the wild-type HPK5[16]. Inactivation of cdrA during infection in the stomach may have contributed to ensuring persistent infection by altering its ability to adapt to the microenvironment. In fact, the cdrA gene was found to be absent in 3 out of 4 colonies recovered from a mouse infected with H. pylori strain B128[18]. Furthermore, additional isolates of the sequenced H. pylori strain J99 from a patient after a 6-year interval were subjected to microarray analysis, which indicated that the cdrA gene was missing in additional isolates[19]. Thus, we hypothesize that a loss of cdrA in H. pylori during infection might be an evolutionary event to alter its biological characteristics, which affects the host immune response to the microbes and promotes persistent infection.
In this study, the level of IL-8 secretion induced by a cdrA-negative strain was approximately 50% lower than those induced by a cdrA-positive strain in vitro and in vivo. Genetic diversity of cdrA was extended in American isolates and cdrA-negative isolates might be more adaptable to colonize in the corpus. Western blotting analysis documented that the CagA status at the point of its translocation and phosphorylation was not different between HPK5 and HPKT510 strains. However, expression of NF-κB was lower in HPKT510 than that in HPK5, indicating that H. pylori with inactivation of cdrA might escape from rigorous immune clearance and facilitate chronic persistent infection caused by decreased levels of IL-8 and nuclear accumulation of NF-κB.
MATERIALS AND METHODS
Bacterial strains and growth conditions
Wild-type H. pylori HPK5 and its derivatives such as HPKT510, a cdrA-disrupted mutant carrying xylE-Km cassette[17], and HPK5CA, a cagA-disrupted mutant carrying xylE-Km cassette[20], were cultivated in Brucella broth (Becton Dickinson, United States) supplemented with 10% horse serum at 37 °C in an atmosphere containing 10% CO2 as previously described[21]. Bacteria grown in 10 mL of Brucella-serum medium in 100-mL conical flasks following sub-culture were subjected to co-culture with AGS cells. Bacterial growth was measured by determining absorbance at 600 nm (A600) with a spectrophotometer (GENEQUANT pro, Amersham Pharmacia Biotech), and colony forming units were determined for bacterial viability. Additional clinical isolates, 77 American and 51 Japanese isolates endoscopically obtained from 48 and 51 patients, respectively, were cultured on Brucella-serum agar at 37 °C in an atmosphere containing 10% CO2. Isolates from different regions in the stomach, such as the antrum and corpus, were also included in 29 of 77 American isolates. The bacterial genomic DNA extracted by a DNA kit (Qiagen, Tokyo, Japan) according to the manufacturer’s instructions were subjected to polymerase chain reaction (PCR) with specific primer pairs to examine cdrA gene diversity.
AGS cell culture conditions
AGS cells (human gastric adenocarcinoma epithelial cell line, CRL1739c) were grown in RPMI 1640 + 7 medium including streptomycin (20 μg/mL) and kanamycin (60 μg/mL) (Nikken bio medical Laboratory, Japan) supplemented with 10% (v/v) heat-inactivated fetal bovine serum (Biowest, France) at 37 °C in an atmosphere containing 5% CO2. AGS cells were seeded at a density of 0.5 × 106 cells in six-well plates (SUMITOMO BAKELIFE, Japan) and grown to about 80% confluence prior to co-culture experiments.
Co-culture of AGS cells with H. pylori strains
Subconfluent AGS cells (1 × 106) cultured in RPMI 1640 + 7 medium supplemented with 10% serum were washed twice by phosphate-buffered saline (PBS) and subsequently cultured with or without each H. pylori strain (HPK5, HPKT510 and HPK5CA) at a multiplicity of infection (MOI) of 150 or 300 in 1.5 mL of RPMI 1640 medium (Gibco BRL, Eggenstein, Germany) alone for 48 h at 37 °C in an atmosphere containing 10% CO2. The supernatant was collected at various times (3-48 h), centrifuged at 7000 r/min for 5 min to pellet bacteria and AGS cells, and subjected to sandwich enzyme-linked immunosorbent assay (ELISA) to measure IL-8 productions. AGS cells after co-culture were collected at the appropriate time and used in the following analyses.
Preparation of whole-cell and nuclear extracts
AGS cells were washed five times in ice-cold PBS, incubated with lysis buffer containing 50 mmol/L NaCl, 1 mmol/L EDTA, 1 mmol/L NaF, 100 μmol/L sodium orthovanadate, 10 μmol/L phenylmethylsulfonyl fluoride (PMSF) and a commercially available protease inhibitor mixture tablet (Complete, Roche Molecular biochemical, Indianapolis, IN), harvested by scraping and transferred to a microcentrifuge tube. Debris was removed by centrifugation at 10 000 g for 10 min (TOMY MRX-150) to collect the total proteins for whole-cell extracts. Nuclear proteins were prepared by Cellytic Nu-CLEAR Extraction Kit (SIGMA, Japan) according to a previous report[22]. Briefly, AGS cells were incubated with lysis buffer (10 mmol/L Hepes, pH 7.9, 1.5 mmol/L MgCl2, 10 mmol/L KCl, 1 mmol/L DTT and 0.25 mmol/L PMSF) on ice for 15 min. The pellet precipitated containing nuclei following centrifugation for 30 s at 10 000 g was resuspended in 100 μL extraction buffer (20 mmol/L Hepes, pH 7.9, 1.5 mmol/L MgCl2, 0.42 mol/L NaCl, 0.2 mmol/L EDTA, 25% glycerol, 1 mmol/L DTT and 0.25 mmol/L PMSF) for 30 min at 4 °C with agitation. After centrifuging for 5 min at 20 000 g, the supernatant (nuclear fraction) was collected for nuclear extracts.
Immunoprecipitation
One milligram of the lysate proteins was incubated with polyclonal rabbit anti-H. pylori CagA antibody (Austral Biologicals, CA, United States) for 1 h at 4 °C, followed by an overnight incubation with a 20 μL aliquot of Protein G Plus-agarose beads (Santa Cruz, United States) at 4 °C, as previously described[20]. Briefly, the beads were washed with lysis buffer and boiled for 10 min in 2 × electrophoresis sample buffer (50 mmol/L Tris (pH 6.8), 10% sodium dodecyl sulfate, 12% 2-mercaptoethanol, 20% (wt/vol) glycine and 1% (v/v) bromophenol blue) to elute the immunoprecipitated proteins.
Western blotting analysis
For detection of CagA and its phosphorylation, equivalent amounts of the immunoprecipitated proteins (100 μg) were resolved by 5% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). Proteins were electro-transferred onto nitrocellulose membranes (Millipore Corporation, United States) by a semidry blotting apparatus KS-8460 (System Instruments Co., Ltd. Tokyo). The blots were blocked overnight with 4% (wt/vol) dried skim milk in Tris-buffered saline with Tween 20 at room temperature and then incubated with monoclonal antibody PY99 (diluted 1/1000; Santa Cruz, United States) for 1 h and anti-mouse IgG peroxidase-linked species-specific whole secondary antibody (diluted 1/1000; GE Healthcare Biosciences, Co. Ltd., United Kingdom) for 1 h to detect phosphorylated tyrosine protein. Immunodetection was performed by enhanced chemiluminescence Plus Western Blotting Detection Reagents (GE Healthcare Biosciences). Next, blots were stripped, reprobed with a specific polyclonal rabbit anti-H. pylori CagA antibody (diluted 1/1000; Austral Biologicals. CA, United States) and incubated with a horseradish peroxidase-conjugated goat anti-rabbit secondary antibody (diluted 1/10 000; Jackson ImmunoResearch, PA, United States)[20]. Each 10 μg of nuclear and total proteins was subjected to 12% SDS-PAGE for detection of NF-κB and inhibition kappa B (IκB), respectively. Procedures of western blot analysis were followed as described above. Rabbit anti-human-NF-κB (p65) (diluted 1/200; Cell Signaling Technology, United States) and rabbit anti-human-IκB antibodies (diluted 1/200; Santa Cruz Biotechnology Inc, United States), and an anti-rabbit IgG antibody-HRP (diluted 1/2000; Santa Cruz Biotechnology Inc, United States) were used as primary and secondary antibodies, respectively. For a standard control, a rabbit anti-human-ERK2 antibody (diluted 1/200; Santa Cruz Biotechnology Inc, United States) was used to detect unphosphorylated ERK2 to confirm equal protein load[23]. The analyses were performed for least three independent experiments.
Measurement of IL-8 secretion from AGS cells
The amount of IL-8 secreted into culture medium after co-culture with H. pylori strains was determined by ELISA using the CytoSets system (BioSource International) according to the manufacturer’s instructions. Fifty µL aliquots of the supernatant was briefly centrifuged at 7000 r/min for 10 min to remove bacteria and AGS cells, after which supernatants were added to an equal volume of regent on the 96-well ELISA plate. The absorbance of samples was measured at 550 nm using a 96-well microplate reader (Multiskan JX ver1.1, Thermo Labsystems, Finland), and the data was expressed as pg/mL. All samples were measured in triplicate in at least three independent experiments.
Measurement of mucosal IL-8 secretion
Frozen gastric antral biopsy specimens from 25 clinical patients infected with H. pylori were homogenized in 1 mL of PBS, and supernatants obtained by centrifugation were used for determination of IL-8 proteins by ELISA, as described previously[24]. Cytokine concentrations in homogenates were normalized in terms of total protein concentration of the biopsy specimen and were expressed as pg/mL.
PCR for cdrA gene diversity
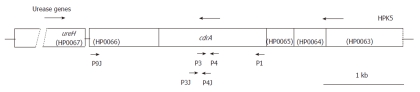
To compare the in vivo IL-8 level in the biopsy specimens between the patients infected with cdrA-positive or -negative strains, PCR was utilized to examine the cdrA status in clinical isolates. The genomic DNA of clinical isolates, including 77 American and 51 Japanese strains, were subjected to PCR with specific primers for the cdrA gene of the HPK5 strain such as P1 (forward), P3 (reverse) and P4 (forward), previously described[17]. Additional new primers, P9J (reverse), P3J (reverse) and P4J (forward), based on the sequence of the cdrA gene of J99 strains, were also utilized in this study. The sequence of primer P1 was identical in strains, HPK5 and J99. The conditions used for PCR to amplify the 194-bp or 336-bp products in the central region of cdrA, which is highly conserved among strains using P3-P4 or P3J-P4J primer pairs, respectively, were as follows: pre-heat for 2 min at 96 °C, followed by 40 cycles of 96 °C for 30 s, 55 °C for 30 s, and 72 °C for 30 s. To amplify the N-terminal region of cdrA using P1-P3 or P1-P3J primer pairs, PCR was performed with the following conditions: pre-heat for 2 min at 96 °C, followed by 40 cycles of 96 °C for 30 s, 52 °C for 30 s, and 72 °C for 2.5 min. PCR was carried out at least two times with all sets of primer pairs, P1-P3, P1-P3J, P3-P4 and P3J-P4J, for determining the standing of the cdrA gene. In particular, for the strains representing no PCR product using various primer pairs mentioned above, P9J-P4 or P9J-P4J primer pairs were used to confirm the existence of the region from central to C-terminal on cdrA corresponding to flanking the downstream region of urease gene cluster. The PCR conditions were as follows: pre-heat for 2 min at 96 °C, followed by 40 cycles of 96 °C for 30 s, 55 °C for 30 s, and 72 °C for 1 min. The location of these primers used for cdrA amplification (Figure 1) and primer sequences are shown (Table 1).
Figure 1.
Location of primers used for cell division-related gene A in the region downstream of the urease gene cluster of Helicobacter pylori HPK5. Arrows above and below the map depict the direction of transcription and primers, respectively. The open reading frames (HP0063 to HP0067) are shown based on strain 26695.
Table 1.
The sequence of primers used and the target region of cell division-related gene A amplified with combination of primers
| Primers | Sequence (5’-3’) | Reference | Combination of primers | Target region of cell division-related gene A |
| P1 (forward) | TGAAGCACACGAAAGGA | 17 | P1-P3 | N-terminal to central |
| P3 (reverse) | CATGCTCTAAAATCGTCG | 17 | P1-P3J | N-terminal to central |
| P3J (reverse) | ATCACCACGATCAGTCTG | This study | P3-P4 | Central |
| P4 (forward) | TTTTGAGATCGGTGAAGC | 17 | P3J-P4J | Central |
| P4J (forward) | AACCACACGCTCATTTGC | This study | P4-P9J | Central to C-terminal |
| P9J (reverse) | GCTGAAAGGCCTGAATTCAG | This study | P4J-P9J | Central to C-terminal |
Statistical analysis
Fischer’s exact test and the χ2 test with Yates’ continuity correction were applied using SPSS version 10.0 for Windows to compare differences on the level of IL-8 productions induced by cdrA-positive and -negative H. pylori strains. P value of < 0.05 were considered significant.
RESULTS
IL-8 secretion from AGS cells
To determine the effect of host response to H. pylori during persistent infection in the human stomach, in vitro levels of IL-8 production from AGS cells co-cultured with HPK5 or HPKT510 strains were examined relative to time (Figure 2). Both strains induced the secretion of IL-8 from AGS cells and the levels increased time-dependently, but HPKT510, the cdrA-disrupted mutant, showed significantly lower levels, corresponding to 50%-60% of levels induced by wild-type HPK5 at MOI of 150 (P < 0.01). Furthermore, the levels of IL-8 were increased dose (MOI)-dependently by HPK5, but not HPKT510, which showed identical levels between MOI of 150 and 300.
Figure 2.
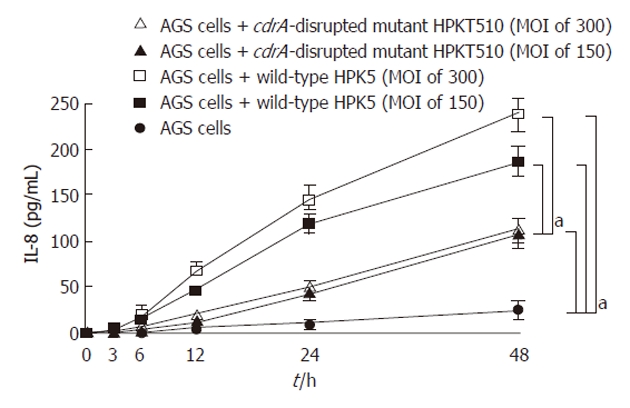
Interleukin-8 production from AGS cells induced by either wild-type or cell division-related gene A-disrupted mutant strains. aP < 0.01.
Western blotting for CagA
Ultrastructural analyses revealed that the HPKT510 strain had a slightly wider periplasmic space between the inner and the outer membrane than that of HPK5[16]. To determine whether such morphological difference affects translocation of CagA into AGS via type IV secretory component system (T4SS) and its phosphorylation, which are considered to be risk factor for development of gastroduodenal disease, western blotting following immunoprecipitation with anti-H. pylori CagA antibody was carried out. CagA was detected in AGS cells co-cultured with HPK5 or HPKT510 strains, but no CagA was observed in AGS alone or when co-cultured with a cagA-disrupted mutant, HPK5CA as a negative control (Figure 3). The intensities of each CagA band are similar between both strains. Phosphorylation of CagA was detected up to 24 h after co-culture with HPK5 or HPKT510 strains and had similar intensity, indicating that there were no differences in the status of CagA between both strains.
Figure 3.
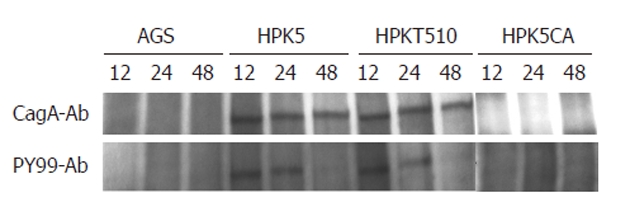
Cytotoxin-associated gene A status in AGS cells co-cultured with or without Helicobacter pylori strains. The immunoprecipitated proteins with anti-Helicobacter pylori (H. pylori) cytotoxin-associated gene A (CagA) antibody (CagA-Ab) were subjected to Western blotting with CagA-Ab (upper) and PY-99 antibody (PY99-Ab) (bottom), respectively. The assay was carried out with strains at MOI of 150. HPK5: Wild-type; HPKT510: Cell division-related gene A-disrupted mutant; HPK5CA: cagA-disrupted mutant.
Western blotting for NF-κB (p65) and IκB
To determine how the expression level of NF-κB transcription factor affects the expression of many genes involved in inflammation and immunities, western blotting was carried out utilizing AGS cells co-cultured with HPK5 or HPKT510 strains for 3, 6 and 12 h. Overall, NF-κB levels in the nuclear extracts from AGS cells with HPKT510 were definitely lower compared to those with HPK5, whereas IκB levels in total extracts were found to be higher in HPKT510 (Figure 4), indicating that activation of NF-κB was relatively repressed in AGS cells stimulated by H. pylori with disruption of cdrA.
Figure 4.
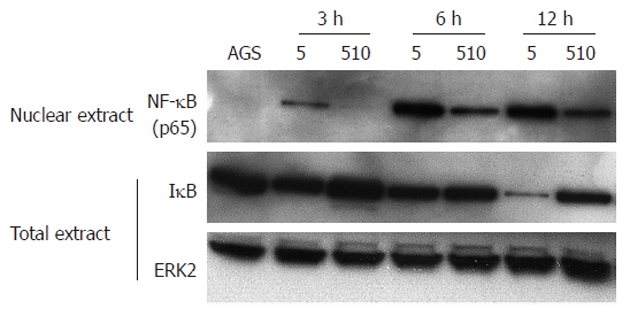
Detection of nuclear factor kappa B (p65) (upper), inhibition kappa B (middle) and extracellular signal-regulated kinase 2 (bottom) in nuclear and total extracts, respectively at 3, 6 and 12 h after being co-cultured with Helicobacter pylori. Unphosphorylated extracellular signal–regulated kinase 2 (ERK2) was detected as the control in this study[23]. Molecular weights of nuclear factor kappa B (NF-κB), inhibition kappa B (IκB) and ERK2 are 65, 35-37 and 42 kDa, respectively. The assay was carried out with strains at a MOI of 150. AGS: AGS cells co-cultured without H. pylori; HPK5: AGS cells co-cultured with wild-type HPK5; HPKT510: AGS cells co-cultured with cdrA-disrupted mutant HPKT510.
Genetic diversity of H. pylori cdrA
To examine the cdrA status [category such as cdrA-positive and -negative, including 4 allele types (Ito IV)] in 128 clinical isolates, PCR was performed with appropriate specific primer pairs. In a previous report, PCR with P1-P3 yielded the 731 bp (short fragment) and approximately 2 kb (long fragment) in HPK5 and other three strains examined respectively, demonstrating that the shorter product of HPK5 was due to partial deletion in the N-terminal region of cdrA. However, cdrA was functionally involved in cell division, morphology, long-term survival and susceptibilities to beta-lactam and its transcript could be detected[16,17]. On the other hand, the 194-bp amplified in the central region of cdrA was detected in all strains, indicating that size diversity exists within the N-terminal region of cdrA among individual strains[17]. In this study, 128 clinical isolates (77 American and 51 Japanese) were subjected to PCR with N-terminal primer pairs (P1-P3 and P1-P3J) which demonstrated that the short fragment (allele typeI) was obtained in only 4 Japanese isolates (7.8%) including HPK5 and the long fragment (allele type II) was detected in 13 American (16.9%) and 46 Japanese (90.2%) isolates. In addition, no product was observed in the 64 American (83.1%) and 1 Japanese (2.0%) isolates by PCR with central region primer pairs (P3-P4 and P3J-P4J). Of the 64 American isolates showing no amplification on the central region, 35 isolates have no product amplified on the C-terminal region with P9J-P4J and P9J-P4 pairs. The results revealed that the downstream region of the urease gene cluster, including cdrA, was absent in the 35 isolates (allele type IV). However, PCR product on the C-terminal region with P9J-P4J or P9J-P4 pairs was detected in remaining 29 isolates, demonstrating that they have partial cdrA sequence loss between the N-terminal and central regions (allele type III). Taken together, the status of the cdrA gene among individual isolates examined was divided into two categories; cdrA-positive including allele typesI(HPK5) and II (J99 and 26695), and cdrA-negative including allele types III and IV (Table 2).
Table 2.
Helicobacter pylori cell division-related gene A status (positive or negative) and genotype (allele type) in clinical isolates n (%)
| H. pylori cdrA | |||
| Status | Genotype | United States (48 patients)1 | Japan (51 patients) |
| Positive | TypeI | 0 (0) | 4 (7.8) |
| Type II | 13 (16.9) | 46 (90.2) | |
| Negative | Type III | 29 (37.7) | 1 (2.0) |
| Type IV | 35 (45.4) | 0 (0) | |
| Total | 772 | 51 | |
H. pylori cdrA: Helicobacter pylori cell division-related gene A.
In 29 out of 48 patients, each isolate was obtained from both antrum and corpus in same stomach;
The total 77 isolates include 44 and 33 isolates obtained from antrum and corpus, respectively.
Comparison of cdrA status between antrum and corpus
The cdrA allele type of each isolate from the antrum and corpus in the stomach of 29 American patients demonstrated that cdrA allele between the regions was identical in 17 patients (59%); however, 9 of the remaining 12 patients possessing a different allele had a isolate with more loss of the cdrA sequence in the corpus compared to the antrum (Table 3). Regarding the status of cdrA, 6 out of 29 patients had different cdrA status between the regions, and of which, 5 patients harbored cdrA-positive and -negative isolates in the antrum and corpus, respectively (Table 3).
Table 3.
Comparison of cell division-related gene A status and genotype (allete type) between antrum and corpus isolates in 29 American patients
| cdrA status1 | cdrA genetype (allele type) | |||||
| cdrA positive (6 cases) | Same2: 1 | Antrum (+)3 | Antrum | Corpus | ||
| Corpus (+) | Identical4: 1 | Type II | Type II | |||
| Difference: 5 | Antrum (+) | Difference: 5 | 4 | Type II | Type III | |
| Corpus (-) | 1 | Type II | Type IV | |||
| cdrA negative (23 cases) | Same: 22 | Antrum (-) | Identical: 16 | 5 | Type III | Type III |
| Corpus (-) | 11 | Type IV | Type IV | |||
| Difference: 1 | Antrum (-) | Difference: 7 | 4 | Type III | Type IV | |
| Corpus (+) | 2 | Type IV | Type III | |||
| 1 | Type IV | Type II | ||||
| Total (%) | Same: 23 (79) | Same: 17 (59) | ||||
| Difference: 6 (21) | Difference: 12 (41) | |||||
The status of cell division-related gene A (cdrA) determined with isolate from antrum was divided to two category groups (cdrA positive and negative);
Same or difference: cdrA status is same or difference between isolate (antrum and corpus);
+: cdrA positive isolate; -: cdrA negative isolate;
Identical or difference: the genotypes of cdrA is identical or difference between isolate (antrum and corpus). Bold denotes the 6 cases representing differences at cdrA status and genotype between isolates (antrum and corpus).
Measurement of mucosal IL-8 secretion
The cdrA-disrupted mutant HPKT510 showed significantly lower levels of IL-8 secretion in vitro compared to wild-type HPK5. To investigate whether cdrA is associated with IL-8 production in vivo, mucosal IL-8 secretion of biopsy specimens was measured using samples obtained from 20 American patients infected with cdrA-negative and 5 American patients with cdrA-positive H. pylori. Measurement of IL-8 in these specimens demonstrated that the cdrA-negative group exhibited lower IL-8 levels than those of cdrA-positive group. The average value of IL-8 in the cdrA-negative group was 111 pg/mL, corresponding to the average of 60% in the cdrA-positive group (156 pg/mL). This tendency was consisted with in vitro data, revealing that cdrA-negative H. pylori does not induce IL-8 secretion as strongly in the human stomach as cdrA-positive strains.
DISCUSSION
H. pylori colonizes more than half of the world’s population. While it is clear that this organism induces a strong innate and adaptive immune response leading to active inflammation in the gastric mucosa, the ability of H. pylori to establish persistent infection so efficiently has not been fully elucidated. H. pylori possesses a number of virulence factors, and some, such as urease, flagella and lipopolysaccharide (LPS), contribute to its persistence[25-27], whereas others, such as CagA and vacuolating cytotoxin, appear to confer increased virulence[28,29]. Furthermore, the strain-specific genes found outside of the cag pathogenicity island, especially genes in the plasticity regions[30] and genes with variable structures/genotypes, have considerable interest in their contribution to pathogenesis and the host immune response. Nearly half of the strain-specific genes of H. pylori are located in the plasticity regions in strains 26695[31] and J99[30,32]. There is evidence that genetic recombination among H. pylori may occur during the course of infection in the stomach. The diversity or phase variation in H. pylori genes such as babA[33] and fucT1[26] likely contributes to the evasion of the host immune response and thereby has a role in the establishment of persistent infection in the stomach. Based on the genomic information of the sequenced H. pylori strains 26695[31] and J99[32], more than a third of H. pylori genes have unknown function, suggesting that there may be strain-specific genes such as ctkA[34] involved in the mechanisms of persistence as well as the host immune response.
One of unique genes found in the HPK5 strain, cdrA located in the downstream region of urease gene cluster, has not only a inhibitory role for cell division, but is also involved in elongation and death of cells via cell wall synthesis at the site of division[16,17]. Furthermore, the cdrA-disrupted mutant HPKT510 had a longer survival time compared to the wild-type HPK5 in both liquid and solid media, as well as in serum-free medium and aerobic conditions[16]. Loss of the cdrA gene during infection is frequently found in a mouse infected with H. pylori strain B128[18] and in the human stomach infected with J99[19]. The present study found that cdrA-negative strains resulted in lower levels of IL-8 production in vitro and in vivo compared to cdrA-positive strains. In vitro data showed that increased IL-8 production was dose-dependently observed by cdrA-positive HPK5, but not cdrA-disrupted HPKT510. In addition, nuclear accumulation of NF-κB in AGS co-cultured with HPKT510 was lower compared to HPK5, suggesting that activation of the NF-κB signaling pathway was relatively repressed by stimulation of the cdrA-negative strain, which coincided with decreased IL-8 production. These indicated that cdrA-defective H. pylori may evade immune clearance due to limiting bactericidal effects of pro-inflammatory molecules leading to promotion for persistent infection, likely via repression of the NF-κB signaling pathway. Persistent infection was not observed in IL-10-deficient mice with strong inflammation, implying that attenuate host immunity is necessary for H. pylori colonization[35]. Accordingly, it is acceptable that H. pylori infection occurred in the stomach with immature immunity during infantile generation. The proper NF-κB transcriptional response is primarily regulated by post-translational modification of NF-κB signaling components. On the other hand, alternative splicing of NF-κB signaling components is another way to control the NF-κB signaling pathway[36,37]. Alternative splicing of some NF-κB signaling components can be induced by prolonged exposure to a NF-κB-activating signal, such as LPS, suggesting a mechanism for negative feedback to dampen excessive NF-κB signaling. In particular, alternative splicing events in Toll/interleukin-1 NF-κB signaling pathways via Toll like receptors can inhibit the NF-κB response[38].This raises the possibility that changes in bacterial constituents concerned with disruption of cdrA, such as cell envelope components including LPS, may exert on the complicated NF-κB signaling pathway.
As HPKT510 had a slightly wider periplasmic space[16], the effector protein, CagA, was investigated to determine whether this morphological change altered the function of T4SS. The CagA production, its translocation and phosphorylation were indistinguishable between HPK5 and HPKT510 strains, indicating that such morphological differences related with inactivation of cdrA had no effect on CagA-associated pathogenicity, including CagA-related signaling pathway and T4SS. Cytoplasmic nucleotide-binding oligomerization domain (Nod) molecules, Nod1 and Nod2, have been shown to be specific ligands of diaminopimelic acid-containing muropeptides and muramyl dipeptide, respectively[39,40], and were important components of the innate immune response. A peptidoglycan-derived muropeptide possessing a Nod1 motif was translocated through T4SS and affected cell signaling via activation of NF-κB in the host cell, leading to the stimulation of an intracellular pro-inflammatory response[41]. In terms of conserved domains, part of HPK5 cdrA belongs to the SM1-NRK4 family, beta glucan and cell wall synthesis enzyme family, and another belongs to the SpoIIE-FtsK-ATPase family. The properties of penicillin-binding proteins such as PBP1, PBP2, PBP3 and PBP4 varied between HPK5 and HPKT510 strains[16], suggesting that an alteration of such peptidoglycans influences the level of IL-8 production via the Nod1-NF-κB signaling pathway. In fact, the measurement of nod1 transcript level in AGS by real-time RT-PCR documented that HPKT510 induced lower level of nod1 transcript compared to HPK5 (data not shown), which supports the Nod1-NF-κB pathway involved in the difference of IL-8 production.
The cdrA status in almost all Japanese isolates were cdrA-positive (98%), but not in isolates obtained from Americans, demonstrating that American isolates had a greater diversity and higher prevalence of cdrA-negative strains (83.1%). Evaluation of cdrA status in isolates obtained from different regions of the stomach revealed that in 5 of the 6 cases possessing different status between the antrum and corpus of the stomach, the cdrA-negative isolate was from the corpus, suggesting that a cdrA-negative strain might be more adaptable to colonize in the corpus over a longer time period than the cdrA-positive strain. A cdrA-disrupted mutant could survive longer under the stresses, such as presence of beta-lactam antibiotics, serum-free and aerobic conditions, than the wild-type strain[16]. It is possible that changes in biological behavior accompanied by inactivation of cdrA are necessary to stay balanced for the establishment of long-term infection in the human stomach. When the HPK5 cdrA sequence was compared to reference strain 26695[31], the cdrA sequence was a fusion of the ~130 codon hp0064 gene to the middle ~one third of the hp0066 gene, with deletion of the interstitial hp0065 and first part of hp0066. A similar fusion is evident with respect to the reference strain J99 (jhp0059 and jhp0061) or HPAG1 (hpag1_0064 and hpag1_0067)[42]. These two reference strains might represent the majority class in the Japanese population, which gave 2 kb instead of 730 bp PCR products. In contrast, two other sequenced strains, Shi470[43] and G27[44], seem to contain only the ~130 codon ORF, not the longer one, and thus might possibly represent one of the major classes in the United States population. The HPK5 type cdrA stems from gene fusion and synthesizes a partially defective protein that impacts cell wall synthesis or structure and a greater release of peptidoglycan fragments, which can be proinflammatory through the Nod1-NF-κB pathway. More studies are required in the future to dissect the mechanistic role of cdrA and the alteration of comprehensive genomix/proteomix affected by inactivation of cdrA. Analysis for putative deletion alleles might actually provide us with insights to elucidate whether the phenomenon resulted from such sources as large insertions or genome rearrangements.
In histological observation, as far the confined tissue sections of small biopsy specimens were examined, no significant finding was observed in the gastric mucosa colonized with either cdrA-positive or cdrA-negative H. pylori strains. We may need more examinations in detail with extended sections and specimens to confirm the differences in histological findings, including inflammation with neutrophil infiltration.
We concluded that the presence of H. pylori cdrA was associated with effective production of pro-inflammatory cytokine, IL-8, both in vitro and in vivo. Therefore, cdrA-inactivated H. pylori strains may result in attenuated host immunity and evade immune clearance due to repression of the NF-κB signaling pathway in the host cell, leading to persistent infection. Finally, our studies suggest that cdrA-negative H. pylori strains are more likely to colonize in the corpus over long time periods compared with cdrA-positive strains.
COMMENTS
Background
Helicobacter pylori (H. pylori) infection up-regulates secretion of various inflammatory cytokines including interleukin-8 (IL-8), whose production in vitro and in vivo is recognized as a host response to microbes. The levels of IL-8 production differ among individual strains, suggesting the existence of unique genes involved in host response and genetic diversity during infection in the stomach. Furthermore, the effect of fundamental constituents belonging to cell division of H. pylori on host response is unclear.
Research frontiers
H. pylori infection and its persistence in the stomach is the most important event to actually lead a variety of diseases. However, the functions and practices of the cell division of strains colonized in an individual stomach remain unclear. Furthermore, how the bacterial behaviors concerned with the host response is still unknown. In this study, the authors demonstrate that H. pylori cell division-related gene A (cdrA) required for cell division and morphological shape could be a potential role for mediating IL-8 secretion.
Innovations and breakthroughs
Reports have highlighted the importance of H. pylori-host interaction to understand host immunity and pathogenesis. Certain molecules, such as cytotoxin-associated gene A (CagA) and outer membrane proteins, were shown to be involved in these phenomena; however, the relationship between cell division- and morphological shape-related molecules and host response associated with persistent infection in the stomach is unknown. This is the first study to report that cdrA influenced IL-8 secretion (vivo and vitro) and loss of cdrA may attenuate the host immunity due to repression of NF-κB, leading to persistence.
Applications
In this study, how H. pylori cdrA functions and influences bacterium-host interaction was investigated, which provides insights into understanding bacterial fundamental components and their effect on the stomach, including host response, and a future strategy for therapeutic intervention in the treatment of patients with H. pylori infection.
Terminology
H. pylori cdrA, one of the unique genes discovered in strain HPK5, has not only a repressive role on cell division but is also involved in cell elongation and cell death via cell wall synthesis (mainly penicillin binding proteins) at the division site. The cdrA-disrupted mutant is able to survive for long time periods and is more resistant to the bactericidal of beta-lactam antibiotics than the wild-type. The cdrA gene tends to be lost during infection to facilitate adaption in the stomach, resulting in a persistent infection.
Peer review
This interesting manuscript showed that the presence of H. pylori cdrA was associated with the effective production of IL-8 compared to the inactivation of cdrA in vitro and in vivo. Additionally, they observed that cdrA-inactivated H. pylori strains may result in attenuated host immunity and evade immune clearance due to repression of the NF-κB pathway, leading to persistent infection.
Footnotes
Supported by The Project Research Fund from Kochi University, to Takeuchi H; and a Grant-in-Aid for Scientific Research from the Ministry of Education, Science and Culture of Japan, No. 21590631 and 21590629, in part
Peer reviewers: Marcelo Lima Ribeiro, PhD, Clinical Pharma-cology and Gastroenterology Unit, Laboratory of Microbiology and Molecular Biology, Sao Francisco University Medical School, Av Sao Francisco de Assis, 218, Braganca Paulista-SP 12916-900, Brazil; Noriko Nakajima, MD, PhD, Associate Professor, Division of Gastroenterology and Hepatology, Department of Internal Medicine, Nihon University School of Medicine, 1-8-13 Kandasurugadai Chiyoda-ku, Tokyo 101-8309, Japan; Tamara Vorobjova, MD, PhD, Senior Researcher in Immunology, Department of Immunology, Institute of General and Molecular Pathology, University of Tartu, Ravila, 19, Tartu 50411, Estonia
S- Editor Tian L L- Editor Rutherford A E- Editor Li JY
References
- 1.Peek RM, Blaser MJ. Helicobacter pylori and gastrointestinal tract adenocarcinomas. Nat Rev Cancer. 2002;2:28–37. doi: 10.1038/nrc703. [DOI] [PubMed] [Google Scholar]
- 2.Achtman M, Azuma T, Berg DE, Ito Y, Morelli G, Pan ZJ, Suerbaum S, Thompson SA, van der Ende A, van Doorn LJ. Recombination and clonal groupings within Helicobacter pylori from different geographical regions. Mol Microbiol. 1999;32:459–470. doi: 10.1046/j.1365-2958.1999.01382.x. [DOI] [PubMed] [Google Scholar]
- 3.Akopyanz N, Bukanov NO, Westblom TU, Kresovich S, Berg DE. DNA diversity among clinical isolates of Helicobacter pylori detected by PCR-based RAPD fingerprinting. Nucleic Acids Res. 1992;20:5137–5142. doi: 10.1093/nar/20.19.5137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Go MF, Kapur V, Graham DY, Musser JM. Population genetic analysis of Helicobacter pylori by multilocus enzyme electrophoresis: extensive allelic diversity and recombinational population structure. J Bacteriol. 1996;178:3934–3938. doi: 10.1128/jb.178.13.3934-3938.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Salaün L, Audibert C, Le Lay G, Burucoa C, Fauchère JL, Picard B. Panmictic structure of Helicobacter pylori demonstrated by the comparative study of six genetic markers. FEMS Microbiol Lett. 1998;161:231–239. doi: 10.1111/j.1574-6968.1998.tb12953.x. [DOI] [PubMed] [Google Scholar]
- 6.Suerbaum S, Smith JM, Bapumia K, Morelli G, Smith NH, Kunstmann E, Dyrek I, Achtman M. Free recombination within Helicobacter pylori. Proc Natl Acad Sci USA. 1998;95:12619–12624. doi: 10.1073/pnas.95.21.12619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.van Doorn NEM, Namavar F, Kusters JG, van Rees EP, Kuipers EJ, de Graaff J. Genomic DNA fingerprinting of clinical isolates of Helicobacter pylori by REP-PCR and restriction fragment end-labelling. FEMS Microbiol Lett. 1998;160:145–150. doi: 10.1111/j.1574-6968.1998.tb12904.x. [DOI] [PubMed] [Google Scholar]
- 8.Blaser MJ. In Helicobacter pylori. In: Hunt RH, Tytgat GNH, editors. Basic Mechanisms to Clinical Cure. Dordrecht: Kluwer; 1996. pp. 33–39. [Google Scholar]
- 9.Yamaoka Y, Kita M, Kodama T, Sawai N, Kashima K, Imanishi J. Induction of various cytokines and development of severe mucosal inflammation by cagA gene positive Helicobacter pylori strains. Gut. 1997;41:442–451. doi: 10.1136/gut.41.4.442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Crabtree JE, Farmery SM, Lindley IJ, Figura N, Peichl P, Tompkins DS. CagA/cytotoxic strains of Helicobacter pylori and interleukin-8 in gastric epithelial cell lines. J Clin Pathol. 1994;47:945–950. doi: 10.1136/jcp.47.10.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sharma SA, Tummuru MK, Miller GG, Blaser MJ. Interleukin-8 response of gastric epithelial cell lines to Helicobacter pylori stimulation in vitro. Infect Immun. 1995;63:1681–1687. doi: 10.1128/iai.63.5.1681-1687.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nozawa Y, Nishihara K, Peek RM, Nakano M, Uji T, Ajioka H, Matsuura N, Miyake H. Identification of a signaling cascade for interleukin-8 production by Helicobacter pylori in human gastric epithelial cells. Biochem Pharmacol. 2002;64:21–30. doi: 10.1016/s0006-2952(02)01030-4. [DOI] [PubMed] [Google Scholar]
- 13.Sharma SA, Tummuru MK, Blaser MJ, Kerr LD. Activation of IL-8 gene expression by Helicobacter pylori is regulated by transcription factor nuclear factor-kappa B in gastric epithelial cells. J Immunol. 1998;160:2401–2407. [PubMed] [Google Scholar]
- 14.Keates S, Keates AC, Warny M, Peek RM, Murray PG, Kelly CP. Differential activation of mitogen-activated protein kinases in AGS gastric epithelial cells by cag+ and cag- Helicobacter pylori. J Immunol. 1999;163:5552–5559. [PubMed] [Google Scholar]
- 15.Ghosh S, Karin M. Missing pieces in the NF-kappaB puzzle. Cell. 2002;109 Suppl:S81–S96. doi: 10.1016/s0092-8674(02)00703-1. [DOI] [PubMed] [Google Scholar]
- 16.Takeuchi H, Nakazawa T, Okamoto T, Shirai M, Kimoto M, Nishioka M, Con SA, Morimoto N, Sugiura T. Cell elongation and cell death of helicobacter pylori is modulated by the disruption of cdrA (cell division-related gene A) Microbiol Immunol. 2006;50:487–497. doi: 10.1111/j.1348-0421.2006.tb03819.x. [DOI] [PubMed] [Google Scholar]
- 17.Takeuchi H, Shirai M, Akada JK, Tsuda M, Nakazawa T. Nucleotide sequence and characterization of cdrA, a cell division-related gene of Helicobacter pylori. J Bacteriol. 1998;180:5263–5268. doi: 10.1128/jb.180.19.5263-5268.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Israel DA, Salama N, Arnold CN, Moss SF, Ando T, Wirth HP, Tham KT, Camorlinga M, Blaser MJ, Falkow S, et al. Helicobacter pylori strain-specific differences in genetic content, identified by microarray, influence host inflammatory responses. J Clin Invest. 2001;107:611–620. doi: 10.1172/JCI11450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Israel DA, Salama N, Krishna U, Rieger UM, Atherton JC, Falkow S, Peek RM. Helicobacter pylori genetic diversity within the gastric niche of a single human host. Proc Natl Acad Sci USA. 2001;98:14625–14630. doi: 10.1073/pnas.251551698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang Y, Takeuchi H, Nishioka M, Morimoto N, Kamioka M, Kumon Y, Sugiura T. Relationship of IL-8 production and the CagA status in AGS cells infected with Helicobacter pylori exposed to low pH and activating transcription factor 3 (ATF3) Microbiol Res. 2009;164:180–190. doi: 10.1016/j.micres.2006.10.010. [DOI] [PubMed] [Google Scholar]
- 21.Trang VT, Takeuchi H, Kudo H, Aoki A, Katsuno S, Shimamura T, Sugiura T, Ukeda H. Antimicrobial activity of aminoreductone against Helicobacter pylori. J Agric Food Chem. 2009;57:11343–11348. doi: 10.1021/jf9026876. [DOI] [PubMed] [Google Scholar]
- 22.Mi J, Zhang X, Liu Y, Reddy SK, Rabbani ZN, Sullenger BA, Clary BM. NF-κB inhibition by an adenovirus expressed aptamer sensitizes TNFα-induced apoptosis. Biochem Bioph Res Co. 2007;359:475–480. doi: 10.1016/j.bbrc.2007.05.125. [DOI] [PubMed] [Google Scholar]
- 23.Schmeck B, N’Guessan PD, Ollomang M, Lorenz J, Zahlten J, Opitz B, Flieger A, Suttorp N, Hippenstiel S. Legionella pneumophila-induced NF-kappaB- and MAPK-dependent cytokine release by lung epithelial cells. Eur Respir J. 2007;29:25–33. doi: 10.1183/09031936.00141005. [DOI] [PubMed] [Google Scholar]
- 24.Peek RM, Thompson SA, Donahue JP, Tham KT, Atherton JC, Blaser MJ, Miller GG. Adherence to gastric epithelial cells induces expression of a Helicobacter pylori gene, iceA, that is associated with clinical outcome. Proc Assoc Am Physicians. 1998;110:531–544. [PubMed] [Google Scholar]
- 25.Eaton KA, Morgan DR, Krakowka S. Motility as a factor in the colonisation of gnotobiotic piglets by Helicobacter pylori. J Med Microbiol. 1992;37:123–127. doi: 10.1099/00222615-37-2-123. [DOI] [PubMed] [Google Scholar]
- 26.Khamri W, Moran AP, Worku ML, Karim QN, Walker MM, Annuk H, Ferris JA, Appelmelk BJ, Eggleton P, Reid KB, et al. Variations in Helicobacter pylori lipopolysaccharide to evade the innate immune component surfactant protein D. Infect Immun. 2005;73:7677–7686. doi: 10.1128/IAI.73.11.7677-7686.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Meyer-Rosberg K, Scott DR, Rex D, Melchers K, Sachs G. The effect of environmental pH on the proton motive force of Helicobacter pylori. Gastroenterology. 1996;111:886–900. doi: 10.1016/s0016-5085(96)70056-2. [DOI] [PubMed] [Google Scholar]
- 28.Covacci A, Censini S, Bugnoli M, Petracca R, Burroni D, Macchia G, Massone A, Papini E, Xiang Z, Figura N, et al. Molecular characterization of the 128-kDa immunodominant antigen of Helicobacter pylori associated with cytotoxicity and duodenal ulcer. Proc Natl Acad Sci USA. 1993;90:5791–5795. doi: 10.1073/pnas.90.12.5791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Leunk RD, Johnson PT, David BC, Kraft WG, Morgan DR. Cytotoxic activity in broth-culture filtrates of Campylobacter pylori. J Med Microbiol. 1988;26:93–99. doi: 10.1099/00222615-26-2-93. [DOI] [PubMed] [Google Scholar]
- 30.Yamaoka Y. Roles of the plasticity regions of Helicobacter pylori in gastroduodenal pathogenesis. J Med Microbiol. 2008;57:545–553. doi: 10.1099/jmm.0.2008/000570-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tomb JF, White O, Kerlavage AR, Clayton RA, Sutton GG, Fleischmann RD, Ketchum KA, Klenk HP, Gill S, Dougherty BA, et al. The complete genome sequence of the gastric pathogen Helicobacter pylori. Nature. 1997;388:539–547. doi: 10.1038/41483. [DOI] [PubMed] [Google Scholar]
- 32.Alm RA, Ling LS, Moir DT, King BL, Brown ED, Doig PC, Smith DR, Noonan B, Guild BC, deJonge BL, et al. Genomic-sequence comparison of two unrelated isolates of the human gastric pathogen Helicobacter pylori. Nature. 1999;397:176–180. doi: 10.1038/16495. [DOI] [PubMed] [Google Scholar]
- 33.Aspholm-Hurtig M, Dailide G, Lahmann M, Kalia A, Ilver D, Roche N, Vikström S, Sjöström R, Lindén S, Bäckström A, et al. Functional adaptation of BabA, the H. pylori ABO blood group antigen binding adhesin. Science. 2004;305:519–522. doi: 10.1126/science.1098801. [DOI] [PubMed] [Google Scholar]
- 34.Kim do J, Park KS, Kim JH, Yang SH, Yoon JY, Han BG, Kim HS, Lee SJ, Jang JY, Kim KH, et al. Helicobacter pylori proinflammatory protein up-regulates NF-{kappa} B as a cell-translocating Ser/Thr kinase. Proc Natl Acad Sci USA. 2010;107:21418–21423. doi: 10.1073/pnas.1010153107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Matsumoto Y, Blanchard TG, Drakes ML, Basu M, Redline RW, Levine AD, Czinn SJ. Eradication of Helicobacter pylori and resolution of gastritis in the gastric mucosa of IL-10-deficient mice. Helicobacter. 2005;10:407–415. doi: 10.1111/j.1523-5378.2005.00349.x. [DOI] [PubMed] [Google Scholar]
- 36.Courtois G, Gilmore TD. Mutations in the NF-kappaB signaling pathway: implications for human disease. Oncogene. 2006;25:6831–6843. doi: 10.1038/sj.onc.1209939. [DOI] [PubMed] [Google Scholar]
- 37.Gerondakis S, Grumont R, Gugasyan R, Wong L, Isomura I, Ho W, Banerjee A. Unravelling the complexities of the NF-kappaB signalling pathway using mouse knockout and transgenic models. Oncogene. 2006;25:6781–6799. doi: 10.1038/sj.onc.1209944. [DOI] [PubMed] [Google Scholar]
- 38.Leeman JR, Gilmore TD. Alternative splicing in the NF-kappaB signaling pathway. Gene. 2008;423:97–107. doi: 10.1016/j.gene.2008.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chamaillard M, Hashimoto M, Horie Y, Masumoto J, Qiu S, Saab L, Ogura Y, Kawasaki A, Fukase K, Kusumoto S, et al. An essential role for NOD1 in host recognition of bacterial peptidoglycan containing diaminopimelic acid. Nat Immunol. 2003;4:702–707. doi: 10.1038/ni945. [DOI] [PubMed] [Google Scholar]
- 40.Inohara N, Ogura Y, Fontalba A, Gutierrez O, Pons F, Crespo J, Fukase K, Inamura S, Kusumoto S, Hashimoto M, et al. Host recognition of bacterial muramyl dipeptide mediated through NOD2. Implications for Crohn’s disease. J Biol Chem. 2003;278:5509–5512. doi: 10.1074/jbc.C200673200. [DOI] [PubMed] [Google Scholar]
- 41.Viala J, Chaput C, Boneca IG, Cardona A, Girardin SE, Moran AP, Athman R, Mémet S, Huerre MR, Coyle AJ, et al. Nod1 responds to peptidoglycan delivered by the Helicobacter pylori cag pathogenicity island. Nat Immunol. 2004;5:1166–1174. doi: 10.1038/ni1131. [DOI] [PubMed] [Google Scholar]
- 42.Oh JD, Kling-Bäckhed H, Giannakis M, Xu J, Fulton RS, Fulton LA, Cordum HS, Wang C, Elliott G, Edwards J, et al. The complete genome sequence of a chronic atrophic gastritis Helicobacter pylori strain: evolution during disease progression. Proc Natl Acad Sci USA. 2006;103:9999–10004. doi: 10.1073/pnas.0603784103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Thiberge JM, Boursaux-Eude C, Lehours P, Dillies MA, Creno S, Coppée JY, Rouy Z, Lajus A, Ma L, Burucoa C, et al. From array-based hybridization of Helicobacter pylori isolates to the complete genome sequence of an isolate associated with MALT lymphoma. BMC Genomics. 2010;11:368. doi: 10.1186/1471-2164-11-368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Baltrus DA, Amieva MR, Covacci A, Lowe TM, Merrell DS, Ottemann KM, Stein M, Salama NR, Guillemin K. The complete genome sequence of Helicobacter pylori strain G27. J Bacteriol. 2009;191:447–448. doi: 10.1128/JB.01416-08. [DOI] [PMC free article] [PubMed] [Google Scholar]