Abstract
The last several years have seen numerous reports of new chemical modifications for use in RNA. In addition, in that time period, we have seen the discovery of several previously unknown naturally occurring modifications that impart novel properties on the parent RNAs. In this review, we describe recent discoveries in these areas with a focus on RNA modifications that introduce spectroscopic tags, reactive handles, or new recognition properties.
INTRODUCTION
Naturally occurring RNAs are made up primarily of the four common ribonucleosides A, G, C, and U linked via 5′ to 3′ phosphodiesters. However, natural RNAs frequently also contain nucleoside analogs that differ in structure from the four common ribonucleosides (1). These modifications of the typical RNA structure extend the functional properties of the RNA beyond that possible without them. Similarly, chemists have introduced nonnatural nucleosides into RNA that allow it to be manipulated in ways not possible with the native RNA structure alone (2). This has become particularly common recently with the increased focus on the biological function of small RNAs (e.g. siRNAs and miRNAs) that are easily prepared by standard solid phase chemical synthesis of RNA (3). In this review, we describe recent examples of modifications to RNA that introduce new spectroscopic tags, functional groups with reactivity differing from that of native RNA, and novel recognition properties (Figure 1). These modifications have enabled investigators to probe the structure and function of RNAs in new ways. In addition, we also describe newly discovered naturally occurring modifications that impart novel properties on the parent RNAs.
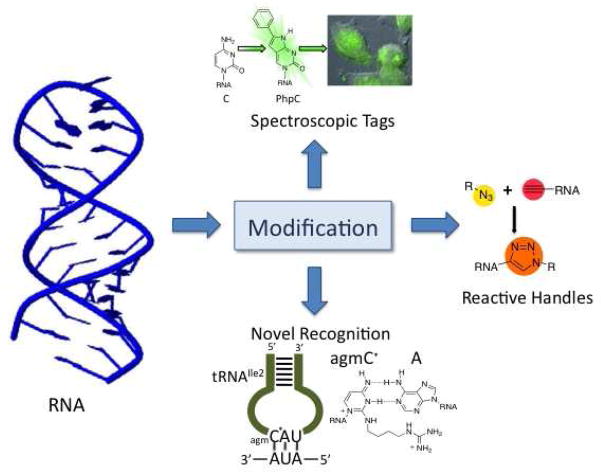
Figure 1.
RNA modification provides access to a wide range of powerful chemical and biochemical tools that enable the study of RNA structure and function.
Spectroscopic Tags
Several recent reports describe new fluorescent base analogs for use in RNA (4–13). The four common nucleobases found in RNA do not have useful fluorescence making it necessary to add fluorophores to RNA for fluorescence-based applications (e.g. FRET, fluorescence microscopy, etc.). For certain applications, minimal structural perturbation to the RNA is preferred when introducing the fluorescent label. In these cases, it is desirable to use an isomorphic fluorophore, or one similar in structure, to the natural nucleobases found in RNA. In the past, this was most frequently accomplished using 2-aminopurine ribonucleoside (1, Figure 2), a constitutional isomer of adenosine. Though its utility and popularity are evident throughout the literature, the shortcomings of 2-aminopurine, such as a short excitation wavelength, severe quenching in a duplex and only functioning as an adenosine analog, have inspired others to expand the scope of fluorescent RNA bases. Recently, Tor et al. reported a single thieno[3,4-d]-pyrimidine heterocycle that, with appropriate modification, can generate emissive isomorphs of each of the four native RNA bases (6). In this report, the guanosine isomorph 2 (Figure 2) was incorporated into a synthetic RNA and shown to maintain pairing specificity. It is interesting to note that emission of this analog remains unquenched in a duplex in contrast to a duplex containing 2-aminopurine. The Tor group has also developed two isomorphic uridines; one that functions as a FRET donor to tryptophan in an RNA binding domain of a protein (11) (3, Figure 2) and one as a FRET donor to a coumarin- linked aminoglycoside used to study binding to the bacterial A-site (4, Figure 2) (10). In both cases, the fluorescent uracil analog provides highly accurate, real-time data for these binding events, regardless of chemical microenvironment.
Figure 2.
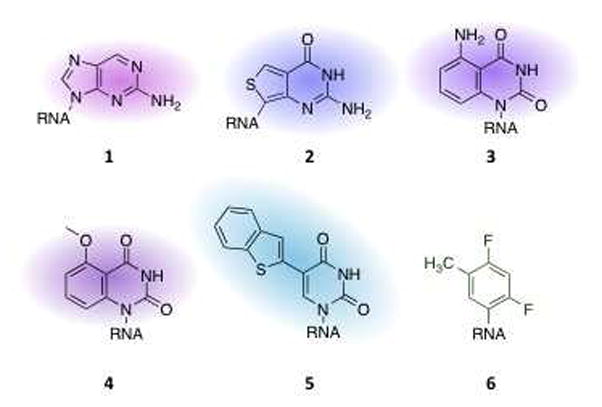
Nucleobase analogs as spectroscopic tags for RNA. 2(6), 3(11), 4(10), 5(5), 6(18)
Damha et al. used the previously synthesized 6-phenylpyrrolocytosine (PhpC, Figure 1) as an emissive cytosine analog to monitor siRNA trafficking inside living cells using fluorescence microscopy (9, 14). These authors showed that the incorporation of multiple PhpC analogs into an siRNA allowed one to image its localization within the cell with very little background. While siRNAs containing a few PhpC analogs show near native levels of silencing activity, the number of analog incorporations required for efficient fluorescence inside of cells did lead to a reduction in activity, suggesting room for improvement for fluorescent base analogs in siRNAs.
Over the past four years, several research groups have developed fluorescent analogs that are sensitive to their microenvironments and have been used for a myriad of different purposes. The Srivatsan lab, for example, has developed a uracil analog (5, Figure 2) to elucidate pairing partners in a duplex by monitoring changes in emission (5). This example marks an exciting new type of fluorescent RNA base that could be applied broadly for the study of RNA structure and function because it reports whether or not the nucleotide is base paired but also its pairing partner.
Recent efforts to develop novel labels for spectroscopy of RNA extend beyond new fluorophores to include labels for NMR of RNA (15–17). Graber, Moroder, and Micura recently reported the use of 2,4-difluorotoluene (6, Figure 2) as a uracil mimetic in one-dimensional 19F NMR (18). 2,4-Difluorotoluene has been used in DNA as a label for 19F NMR (19) and has been used to modify siRNA (20–22) making it a good candidate for labeling RNA for NMR studies. By examining the chemical shifts of the 2-fluoro and 4-fluoro during melting, these authors were able to elucidate secondary structure of RNA (18). This technique is more straightforward than 1H NMR because it is not plagued by severely overlapping signals and is more effective than gel shift assays and UV melting profiles over concentrations not accessible by these alternative methods.
Reactive handles: Alkynes and azides
Since none of the four common RNA nucleosides contain functional groups not shared by at least one of the others, it is generally a challenging task to carry out site-selective chemical modification of a preexisting RNA strand. However, synthesis of the RNA bearing functional groups with reactivity profiles different from those found in the natural RNA structure enables the introduction of a variety of useful modifications at specific positions. These include fluorescent groups for detection or imaging and groups that alter tissue delivery and cellular uptake of the RNA (9, 23). Novel reactive “handles” also allow one to ligate fragments together generating large functional RNAs from smaller synthetic strands or to diversify an RNA structure from a single common intermediate for structure/activity relationship studies (24–26). Early work on this topic included methods to introduce aliphatic amines, thiols, and aldehydes into RNA (27–28). However, over the last few years several research groups have applied the powerful copper-catalyzed azide-alkyne cycloaddition reaction (i.e. CuAAC or click) (23–25, 29–33) and the strain-promoted azide-alkyne cycloaddition reaction (i.e. SPAAC or copper-free click) to the problem of RNA functionalization (34–35).
The first example of the use of click chemistry to modify RNA came when Jao and Salic metabolically labeled cellular RNA with 5-ethynyluridine (7, Figure 3), which could subsequently be detected via reaction with an azide-bearing fluorophore (29). Using this approach, the authors were able to image sites of transcription in cultured cells as well as in tissues from whole animals. This method provides a sensitive and efficient alternative to monitoring cellular transcription via 5-bromouridine incorporation and, indeed, can now be carried out with a commercially available kit (Click-iT® Nascent RNA Capture Kit, Invitrogen). This pioneering work on click chemistry with RNA was done with cellular RNA in fixed cells using CuSO4 and ascorbic acid for catalysis of the cycloaddition reaction. Unfortunately, these conditions can lead to substantial degradation of RNA and are not suitable as a synthetic protocol for triazole-modified strands. Nevertheless, earlier studies on click reactions with modified DNA suggested that the presence of a copper-binding ligand would reduce degradation observed in the presence of copper salts (36). With this information in hand, we and others have since published protocols for high yielding CuAAC reactions useful for preparing triazole-modified RNA for a variety of applications (23–26, 30–33, 37–38).
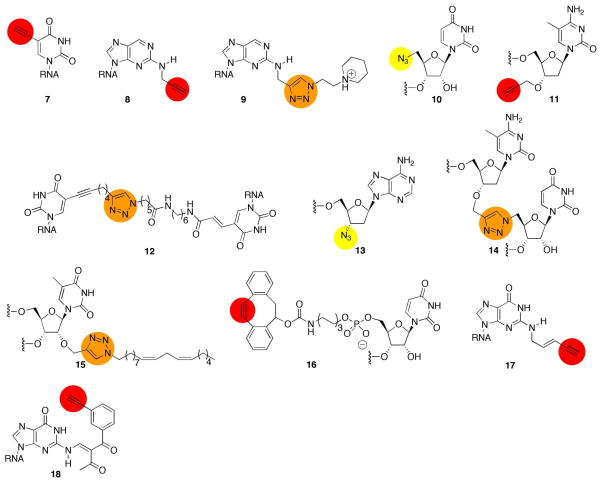
Figure 3.
Structures of alkyne and azide reactive handles recently introduced into RNA. 7(29), 8(30), 9(26), 10(24), 11(24), 12(24), 13(25), 14(24–25), 15(23), 16(34), 17(32), 18(33)
Our lab reported the synthesis of the ribonucleoside phosphoramidite of a purine analog substituted at the 2-position with propargyl amine 8 (Figure 3) (30). This reagent was used to introduce a base-tethered alkyne modification into RNA via solid phase synthesis. Furthermore, triazole formation with the alkyne-bearing RNA strands was efficient with primary azides, copper sulfate, sodium ascorbate and the copper binding ligand tris-(hydroxypropyltriazolylmethyl)amine. Modification of siRNAs with this procedure allowed us to probe the effect of varying minor groove substituents on RNA duplex stability, base pairing specificity, RNA interference, and the binding of known siRNA-binding proteins (26, 30). For instance, the N-ethylpiperidine derivative (9, Figure 3) had miminal effect on RNAi activity at multiple positions in an siRNA but substantially reduced off-pathway protein binding (26). El-Sagheer and Brown described procedures to introduce click reactive groups at several different positions in RNA including an azide at the 5′ end with uridine analog 10 (Figure 3) and an alkyne at the 3′ end with 2′-deoxy-5-methylcytidine analog 11 (Figure 3) (24). They also carried out crosslinking of two strands in a duplex across the major groove with click reactive groups linked to C5 positions of uridines (12, Figure 3). These reagents allowed them to prepare active hairpin ribozymes assembled via a combination of standard synthetic procedures and click reaction ligations. Paredes and Das extended this work by demonstrating that azides could be introduced into RNA enzymatically, for instance, with poly A polymerase and 3′-azido-dATP to give the novel 3′ end modification 13 (Figure 3) (25). Both Brown and Das generated functional ribozymes with a triazole internucleotide linkage prepared from precursors bearing a 5′ azide and 3′ alkyne 14 (Figure 3) (24–25). Rozners has also studied this type of novel RNA backbone modification and reported it to be highly destabilizing in an RNA duplex (~7 °C per modification in a 10 bp duplex) (31). Thus, while the click reaction is useful for ligating short RNA fragments together to generate synthetic RNAs over 100 nt in length, one should chose the ligation site carefully with preference for non essential loop regions.
The efficient and functional group tolerant CuAAC reaction is useful for introducing complex structure into RNA, particularly modifications that would require extensive use of protecting groups or are incompatible with reagents used during automated RNA synthesis, such as carbohydrates, peptides and lipids. These modifications hold promise for altering the tissue delivery and cellular uptake properties of siRNAs, important hurdles to the advancement of RNAi-based therapeutics. Indeed, Alnylam Pharmaceuticals investigators recently described a small library of siRNAs modified with the CuAAC reaction to introduce long lipophilic chains including the linoleyl group (15, Figure 3), cholesterol, oligoamine and a carbohydrate (23). While no novel cellular uptake properties were described for the conjugated siRNAs, initial tests of activity indicated that siRNAs prepared with modified passenger strands effectively silenced a reporter gene with minimal loss of activity.
Although certainly beneficial to the RNA research community, the CuAAC reaction requires millimolar concentrations of copper salts, preventing its use with living cells or with copper-sensitive reagents. However, van der Marel and Filippov, along with investigators at Alnylam, recently incorporated cyclooctynes into RNA (34–35). The Bertozzi and Boons labs had shown that efficient cycloaddition reactions occur with azides and strained cyclooctynes without the requirement for copper catalysis (39–42). Thus, a dibenzocyclooctyne derivative (16, Figure 3) was installed at the 5′ end of an oligoribonucleotide via the corresponding phosphoramidite and used for copper-free click reactions with azides of varying structure including an oligosaccharide and a peptide (34). However, in these initial published examples, it was not obvious that the added benefit of the copper-free reaction conditions justified the additional synthetic effort required to introduce the strained cyclooctyne into the RNA. It will be interesting to see applications for the copper-free click reaction in RNA that fully utilize the power of this novel chemistry (e.g. in living cells or live animals, etc).
A principle benefit of the click reaction in RNA is the site specificity it enables. The site of reaction in the previous examples was determined by the position that the modified nucleosides were incorporated during solid phase synthesis or the enzymatic strategies for 5′ or 3′ end modification. Two labs recently described different approaches to site specifically modify RNA with click reactive handles. Helm and colleagues used the enzyme Trm1 to introduce an alkyne at a specific nucleotide in a tRNA (32). Trm1 is a SAM-dependent tRNA methyl transferase that normally methylates N2 of guanosine at position 26 in tRNAPhe (43). However, these investigators demonstrated that the S-methyl group in the SAM cosubstrate could be replaced with S-pent- 2-en-4-ynyl (32). Trm1 transfers this alkyne-bearing tag from the modified SAM to the N2 of tRNAPhe G26 to give nucleoside analog 17 (Figure 3). The nucleotide was then further modified via CuAAC reaction with a fluorescent azide. In a conceptually similar but potentially more general approach, Sasaki and colleagues synthesized a DNA strand containing a 6-thioguanosine analog, which their lab had previously shown could react to transfer the S6 substitutent to the N2 position of a guanosine in a strand of Watson-Crick complementary RNA (37–38). Thus the sequence selective binding of the DNA directs the transfer reaction to a specific guanosine in the RNA. They showed this selective reaction could be carried out with a 1,3-diketone transfer group bearing an alkyne to give the guanosine analog 18 (Figure 3) (33). RNA so modified was a substrate for a CuAAC reaction with a fluorescent azide.
Other reactive handles
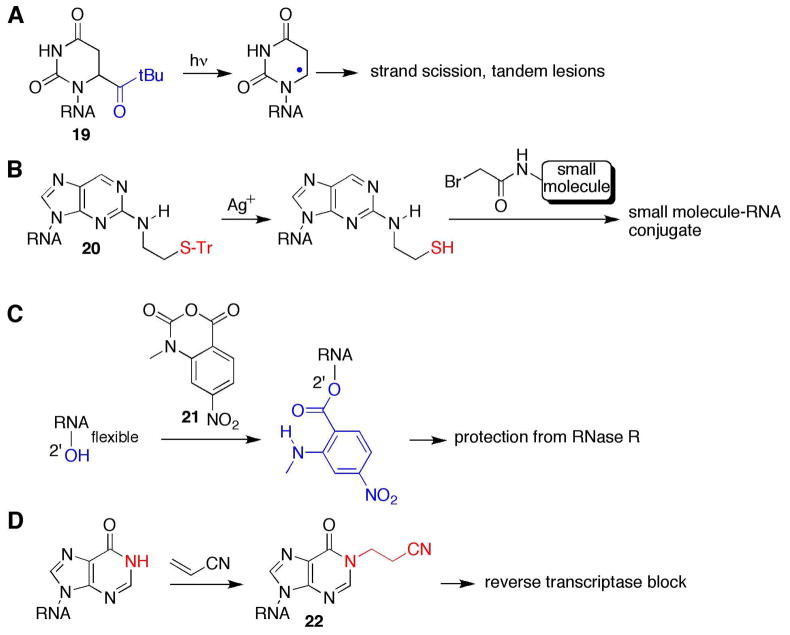
The use of modified phosphoramidites to introduce latent reactivity into RNA continues to be an important research topic. Recent examples include Greenberg’s incorporation of the tert-butyl ketone dihydrouridine analog 19 (Figure 4), which is a precursor to the C6 pyrimidine radical via irradiation of the modified RNA with 350 nm light (Figure 4A) (44). Nucleobase radicals are likely intermediates in the hydroxyl radical cleavage of RNA (45–46), yet prior to this paper, no nucleobase radical had been independently synthesized and studied in RNA. The authors provide convincing evidence that the uridinyl C6 radical produces a direct strand break at the 5′ adjacent nucleotide in RNA (44, 47). Also, the strand scission reaction has an interesting dependence on both the secondary structure of the RNA and the presence or absence of oxygen, with the most efficient cleavage observed under anaerobic conditions in double stranded RNA.
Figure 4.
Recent examples of RNA modifications that impart novel reactivity. A. A precursor to a C6 pyrimidine radical (44); B. A new way to install an RNA thiol (48); C. RNase-detected SHAPE (55); D. Method to detect inosine in RNA (56).
Our laboratory recently described the synthesis and use of a new precursor to thiol-modified RNA (48). The S-trityl protected ethane thiol analog of 2-aminopurine (20, Figure 4) can be incorporated into RNA for subsequent reaction with bromoacetamides (Figure 4B). This derivative is stable in RNA until treated with silver nitrate to reveal the thiol and was used as a replacement for adenosine in a small molecule-binding aptamer near the ligand-binding site. The modification allowed us to stabilize the complex via covalent bond formation between the thiol-containing RNA aptamer and the bromoacetamide-modified small molecule.
Selective acylation of RNA 2′-hydroxyls at flexible nucleotides in folded RNAs followed by detection of those sites as primer extension stops (SHAPE: selective 2′-hydroxyl acylation analyzed by primer extension) is a highly effective method for mapping RNA structure developed by Weeks and colleagues (49–54). However, because of the need to use primer extension in the analysis, structural information cannot be obtained for regions of the RNA close to the 5′ and 3′ ends. Thus, limited information could be obtained from SHAPE analysis for functionally important small RNAs, such as pre-miRNAs or riboswitches. Nevertheless, a recent paper from the Weeks lab has shown that the 2′-O-acylation products from the reaction of flexible nucleotides in RNA and 7-nitroisatoic anhydride (21, Figure 4C) inhibit the reaction of the exoribonuclease RNase R (Figure 4C) (55). Thus, the sites of protection can be directly analyzed with labeled RNA and gel electrophoresis. With the new RNase-detected SHAPE procedure, the authors were able to map the structure of the free thiM thiamine pyrophoshate (TPP) riboswitch from E. coli and characterize the substantial structural reorganization that occurs in the riboswitch upon ligand binding.
In another recent example of a modification reaction in RNA that alters the way enzymes process the modified nucleotide, Suzuki reported the use of the selective reaction of acrylonitrile with inosines in RNA as a method for detecting adenosine to inosine RNA editing events (56). Acrylonitrile reacts with inosine in RNA to cyanoethylate the N1 position (22, Figure 4D), blocking inosine’s Watson-Crick face (Figure 4D). Thus, reverse transcriptase is unable to read through this base analog and stops. This prevents RT-PCR amplification of the inosine containing RNA. Comparing sequencing runs for RT-PCR products from RNA samples with and without prior acrylonitrile treatment identifies the inosines. Suzuki’s approach is useful for distinguishing bona fide RNA editing sites from single nucleotide polymorphisms and led to their discovery of over 4,000 new A to I editing sites in the human transcriptome.
Novel recognition
RNA functions in living systems are dependent on the RNAs’ ability to noncovalently and reversibly bind other cellular components (proteins, other RNA strands, etc.). Alteration of the RNA structure modulates these recognition properties. The following section describes recently discovered naturally occurring modifications that have either been shown to, or have the potential to, substantially alter the way the modified RNAs are recognized by RNA-binding molecules. In addition, we describe a new application of a known naturally occurring modified nucleoside in RNA and new synthetic RNA modifications that impart novel recognition properties.
Newly discovered modified nucleosides in RNA
Three recently reported examples of natural modifications of the RNA are A2503 methylation in 23S rRNA by Cfr (57), agmatidine in tRNAIle (58–59), and NAD linked RNA (60).
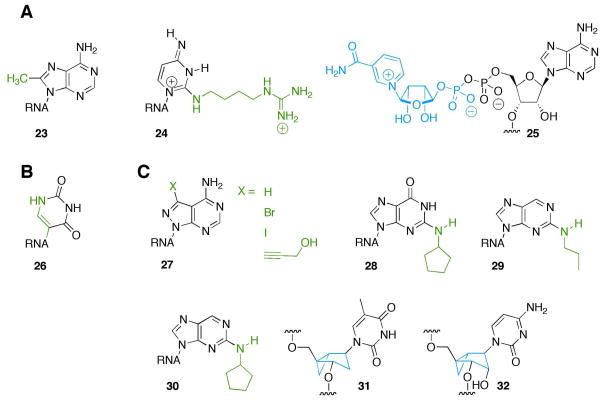
Methylation of the eight-position of A2503 in 23S rRNA by the enzyme Cfr generates C8-methyladenosine (23, Figure 5A) at that position leading to resistance to several ribosome-targeted antibiotics (57). A2503 is located in the peptidyl transferase center of the 50S subunit in the bacterial ribosome, an important target of many antibiotics such as clindamycin and chloramphenicols (61). Adenosine 2503 is methylated by Cfr before or shortly into ribosome assembly (62). According to the model of D. radioduran 50S subunit, this modification points directly into the drug binding site and blocks the binding of antibiotics that target the peptidyl transferase center leading to a loss in recognition of the RNA by those drugs (57). The methylation of A2503 is the first example of a naturally occurring methyl modification at the purine eight-position in RNA. Interestingly, Cfr, along with another methyltransferase that methylates the 2-position of A2503, NlmN, are radical SAM enzymes that do not proceed through the typical SN2 mechanism like other radical SAM enzymes (63–65). Instead, one proposal is that they go through a ping-pong mechanism making this particular modification not only interesting to study from a recognition standpoint but also from a mechanistic standpoint (64).
Figure 5.
A. Studies of recently reported, naturally occurring modified nucleosides in RNA (57–60). B. Known nucleoside analog pseudouridine causes nonsense suppression (73). C. Structures of new synthetic nucleoside analogs that alter RNA recognition (77, 90, 95–96).
Agmatidine (2-agmatinylcytidine) (24, Figure 5A) is a naturally occurring modification that when present at the 5′ nucleotide in the anticodon of tRNAIle causes the tRNA to recognize codon AUA instead of AUG (58–59). Prior to the discovery of agmatidine from Haloarcula marismortui and other species by Suzuki et al. and Rajbhandary et al., it was unknown how archaea differentiated between codons AUA and AUG (58–59). Agmatidine is a 2-position modified cytosine that is synthesized using ATP and agmatine in the presence of tRNAIle-agm2C synthetase. This modification of cytosine leads to changes in base pairing specificity causing the nucleotide to pair with adenine. Furthermore, the addition of the side chain to the pyrimidine 2-position is believed to prevent wobble pairing with guanosine (66). Indeed, when the modification occurs at position 34 in tRNAIle, the recognition of the anticodon changes from AUG to AUA (58–59).
Another recently discovered naturally occurring RNA modification is nicotinamide adenine dinucleotide (NAD) bound at the 5′ end of small RNAs (25, Figure 5A), which was reported by Liu and colleagues (60). The Lui lab has developed a novel strategy for selecting small molecule ligands to proteins that involves use of DNA-small molecule conjugates (67–68). This research, along with the knowledge that RNA has many biological functions and speculation that early life used nucleic acid enzymes to carry out biochemical processes, led them to hypothesize that small molecule-RNA conjugates may exist in cells today (60, 69–70). To identify covalently linked small molecule-RNA conjugates in bacterial RNA, these investigators used a combination of size-exclusion chromatography, nuclease-catalyzed fragmentation, and mass spectrometry (60). Surprisingly, they observed NAD covalently linked to the 5′ end of small bacterial RNAs. They were able to determine that NAD mostly bound to RNAs less than 200 nucleotides in length and that it is not incorporated through aberrant transcription. Although the role of this modification is unknown, having a NAD at the 5′ end undoubtedly allows these RNAs to bind to NAD-binding proteins and enables new redox chemistry.
Repurposing a known naturally occurring modified nucleoside in RNA
Pseudouridine is a naturally occurring C-nucleoside analog of uridine (26, Figure 5B). Conversion of uridine to pseudouridine in RNA can arise via the action of ribonucleoprotein complexes containing H/ACA snoRNAs that direct the reaction via Watson-Crick binding to target RNAs (71–72). Recently, Karijolich and Yu redirected pseudouridylation to a uridine within a nonsense codon by mutating SNR81, a yeast snoRNA that normally directs pseudouridinylation in a ribosomal RNA (73). Pseudouridinylation within the nonsense codon led to nonsense suppression both in vitro and in vivo. These investigators tested each nonsense codon (UAA, UAG and UGA) to determine the pseudouridinylation effect. Interestingly, ψAA and ψAG led to insertion of serine and threonine into the expressed protein with serine predominately incorporated for ψAG; while, ψGA primarily incorporated tyrosine but also incorporated phenylalanine. The pseudouridylation of uridine is an interesting modification from a recognition standpoint because the modification does not affect the Watson-Crick face but greatly influences how the nucleotide is recognized by both release factors and the tRNA showing that other important interactions are occurring other than Watson-Crick base pairing (74). Ferré-D’Amaré hypothesized that changes in recognition may be caused by the increased energy necessary for release factors to bind to the mRNA and dehydrate hydrated pseudouridine (74). Another hypothesis suggests that pseudouridine increases base stacking stability over uridine that could lead to different recognition properties between the two bases (73–75). While not yet established experimentally, it is possible that pseudouridylation is used naturally for nonsense suppression. If so, this modification could be a previously unrecognized form of RNA editing leading to protein diversity through expansion of the genetic code (73).
Synthetic nucleoside analogs that alter RNA recognition
Our lab has used nucleoside modifications to probe the active site of the RNA editing adenosine deaminase ADAR2 to better understand how this enzyme converts adenosine to inosine in its RNA substrates (76–82). RNA editing by adenosine deamination is an important process for creating new function in RNA transcripts of higher organisms, including by changing the meaning of codons in mRNAs (83–87). A high-resolution crystal structure of the deaminase domain of human ADAR2 has been reported (88). However, no RNA substrate was present in the crystal. Therefore, we used nucleoside-analog containing synthetic RNA substrates, along with active site mutants of the enzyme, to test models for substrate recognition by this important deaminase. We had previously shown that both 7-deazaadenosine and 8-azaadenosine in RNA are good substrates for ADAR2 with 8-azaadenosine significantly enhancing reactivity (81–82). More recently, we used three 7-substituted 8-aza-7-deazaadenosine derivatives (7-iodo, 7-bromo, 7-propargyl alcohol) (27, Figure 5C) along with various active site mutants to probe substrate recognition in the ADAR2 active site (77). Interestingly, while 7-deaza-8-azaadenosine was deaminated almost 8 times faster than adenosine, activity with the three bulky 7-substituted derivatives was lower than expected based on how readily the molecules were predicted to undergo covalent hydration, a key step in the deamination mechanism. Therefore, we suggested the decrease in reactivity was caused by steric factors, which was tested by creating a “hole” in the active site by mutation of nonessential R455 to alanine. This mutation caused an increase in the rate of deamination of the “bumped” adenosine analogs compared to the wild type enzyme. Thus, these new RNA modifications were useful for establishing structure/activity relationships in the ADAR2 reaction and validating a model with the 7-position of the edited purine proximal to the side chain for R455.
The last few years have seen several reports of incorporation of novel nucleoside analogs into siRNAs for a variety of applications, including some described above. Use of siRNA as therapeutics requires modification of the component strands to decrease nuclease sensitivity, enhance delivery and cellular uptake, and to reduce stimulation of the innate immune response (3). Our lab used new base modifications in siRNAs to block binding of human proteins known to confound RNA interference (26, 89) and to evade immune responses (90). The latter example is relevant to the development of new liver cancer therapeutics. Human microRNA-122 (miR122) is downregulated in hepatocellular carcinomas and returning miR122 levels to normal has been shown to reverse tumorgenic properties (91–93). Thus, providing the liver with a source of miR122 in the form of an siRNA guide strand has therapeutic potential (92–93). However, siRNA mimetics of miR122 formulated in lipid nanoparticles for delivery to the liver stimulate the production of cytokines in human immune cells (90). Interestingly, the miR122 sequence contains multiple 5′-(UG)n-3′ motifs, which are found in other immunostimulatory RNAs and believed to be a key feature for RNAs that bind the toll-like receptors 7 and 8 (TLR7/8) (94). Therefore, our lab, in collaboration with investigators at Sirna/Merck, used modifications that change the shapes of the bases while maintaining Watson-Crick pairing to significantly decrease miR122 recognition by TLR7/8 (90). We placed N2-cyclopentylguanosines (28, Figure 5C) in the guide strand and N2-propyl-2-aminopurines or N2-cyclopentyl-2-aminopurines (29 and 30, Figure 5C) in the passenger strand of the miR122 mimetic siRNA. We found these modifications could be used to block cytokine production in human peripheral blood mononuclear cells while maintaining microRNA activity. Importantly, both the gene regulatory and immunostimulatory activities of the modified RNAs showed a profound dependence on the sites and type of modification.
In another example of a novel siRNA modification, Eritja and colleagues have studied the effect North bicyclo[3.1.0]hexane pseudosugars have on target recognition and immune stimulation (95–96). North bicycle methanocarbathymidine (TN) (31, Figure 5C), like a locked nucleic acid (LNA) monomer, is restricted into a northern conformation (2′-exo, 3′ endo) found in A-form helices (96–98). One benefit of this type of modification compared to the well–studied and similarly constrained LNAs is that this structure allows for modifications at the 2′ position not possible for LNA (95–96). TN has been inserted into a DNA/RNA heteroduplex and shown to increase thermal stability (97–98). However, to our knowledge, Eritja and colleagues were the first to incorporate TN into siRNA. The initial study showed that TN substitution could increase RNAi activity and decrease immune stimulation (96). Recently, these authors also incorporated the North ribomethanocarbacytidine (CN) (32, Figure 5C) into siRNA (95). Unlike TN, CN has the 2′-hydroxyl group present. They showed RNAi activity with CN-modified siRNA indicating the cellular RNAi machinery tolerates this analog (95). Thus, North bicyclo[3.1.0]hexane pseudosugars are promising new modifications in siRNA with potential for further structural alterations not possible with LNAs.
CONCLUSION
As the above examples illustrate, major advances have been made recently in our ability to introduce a wide variety of novel modifications into RNA and our understanding of the structure and function of naturally occurring RNA modifications. However, as with any important advance, these have stimulated new questions and created new challenges. Some of these future challenges include the development of fluorophores with high quantum yield that mimic each of the four RNA bases useful for cellular imaging of RNA, copper-free click reactions with RNA applied in ways that take full advantage of this chemistry, modifications to siRNAs that further enhance tissue-specific delivery, cellular uptake, and target specificity and a full understanding, with structural data, of how conversion of cytidine to the C2-modified agmatidine switches the nucleoside’s pairing specificity from G to A. It will be exciting to see how these and other important challenges in RNA modification are addressed in the years to come.
Acknowledgments
P.A.B. acknowledges the National Institutes of Health for financial support in the form of grants R01GM080784 and R01GM061115.
KEY TERMS
- Isomorphic fluorophore
A fluorescent molecule with similar shape and recognition properties to a naturally occurring structure. In this context, the term refers to close structural analogs of nucleosides that are emissive
- siRNA
Short interfering RNA; ~19 bp RNA duplexes used as triggers of RNA interference
- miRNA
microRNA; ~20 nt RNAs encoded in the genome that regulate gene expression through the RNAi pathway
- CuAAC
copper-catalyzed azide-alkyne cycloaddition reaction (click reaction); bioorthogonal reaction of an azide and alkyne to form a triazole, requires copper catalysis
- SPAAC
strain-promoted azide-alkyne cycloaddition reaction (copper-free click reaction) that uses a cyclooctyne
- RNAase-detected SHAPE
a method for identifying flexible nucleotides in a short RNA by selective 2′-hydroxyl acylation
- Cfr
enzyme responsible for introducing a C8 methyl group at A2503 in 23S ribosomal RNA
- Agmatidine
a cytidine analog bearing a C2-substituent found in archea tRNAIle that allows for discrimination between AUA and AUG codons
- ADAR
adenosine deaminase that acts on RNA, an RNA editing enzyme responsible for diversifying RNA sequences in metazoa
- TLR7/8
Toll-like receptors 7 and 8; pathogen-associated molecular pattern receptors that bind RNA, cause of immune stimulation by certain siRNA sequences
References
- 1.Cantara WA, Crain PF, Rozenski J, McCloskey JA, Harris KA, Zhang X, Vendeix FAP, Fabris D, Agris PF. The RNA modification database, RNAMDB: 2011 update. Nucleic Acids Res. 2011;39:D195–D201. doi: 10.1093/nar/gkq1028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Das SR, Fong R, Piccirilli JA. Nucleotide analogues to investigate RNA structure and function. Curr Opin Chem Biol. 2005;9:585–593. doi: 10.1016/j.cbpa.2005.10.009. [DOI] [PubMed] [Google Scholar]
- 3.Shukla S, Sumaria CS, Pradeepkumar PI. Exploring Chemical Modifications for siRNA Therapeutics: A Structural and Functional Outlook. ChemMedChem. 2010;5:328–349. doi: 10.1002/cmdc.200900444. [DOI] [PubMed] [Google Scholar]
- 4.Mitsui T, Kimoto M, Kawai R, Yokoyama S, Hirao I. Characterization of fluorescent, unnatural base pairs. Tetrahedron. 2007;63:3528–3537. [Google Scholar]
- 5.Pawar MG, Srivatsan SG. Synthesis, Photophysical Characterization, and Enzymatic Incorporation of a Microenvironment-Sensitive Fluorescent Uridine Analog. Org Lett. 2011;13:1114–1117. doi: 10.1021/ol103147t. [DOI] [PubMed] [Google Scholar]
- 6.Shin D, Sinkeldam RW, Tor Y. Emissive RNA Alphabet. J Am Chem Soc. 2011;133:14912–14915. doi: 10.1021/ja206095a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Srivatsan SG, Greco NJ, Tor Y. A highly emissive fluorescent nucleoside that signals the activity of toxic ribosome-inactivating proteins. Angew Chem, Int Ed. 2008;47:6661–6665. doi: 10.1002/anie.200802199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Srivatsan SG, Tor Y. Synthesis and enzymatic incorporation of a fluorescent pyrimidine ribonucleotide. Nat Protoc. 2007;2:1547–1555. doi: 10.1038/nprot.2007.222. [DOI] [PubMed] [Google Scholar]
- 9.Wahba AS, Azizi F, Deleavey GF, Brown C, Robert F, Carrier M, Kalota A, Gewirtz AM, Pelletier J, Hudson RHE, Damha MJ. Phenylpyrrolocytosine as an Unobtrusive Base Modification for Monitoring Activity and Cellular Trafficking of siRNA. ACS Chem Biol. 2011;6:912–919. doi: 10.1021/cb200070k. [DOI] [PubMed] [Google Scholar]
- 10.Xie Y, Dix AV, Tor Y. FRET Enabled Real Time Detection of RNA-Small Molecule Binding. J Am Chem Soc. 2009;131:17605–17614. doi: 10.1021/ja905767g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Xie Y, Maxson T, Tor Y. Fluorescent Ribonucleoside as a FRET Acceptor for Tryptophan in Native Proteins. J Am Chem Soc. 2010;132:11896–11897. doi: 10.1021/ja105244t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kimoto M, Mitsui T, Yokoyama S, Hirao I. A Unique Fluorescent Base Analogue for the Expansion of the Genetic Alphabet. J Am Chem Soc. 2010;132:4988–4989. doi: 10.1021/ja100806c. [DOI] [PubMed] [Google Scholar]
- 13.Stengel G, Urban M, Purse BW, Kuchta RD. Incorporation of the Fluorescent Ribonucleotide Analogue tCTP by T7 RNA Polymerase. Anal Chem. 2010;82:1082–1089. doi: 10.1021/ac902456n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hudson RHE, Ghorbani-Choghamarani A. Selective fluorometric detection of guanosine-containing sequences by 6-phenylpyrrolocytidine in DNA. Synlett. 2007;2007:870–873. [Google Scholar]
- 15.Kloiber K, Spitzer R, Tollinger M, Konrat R, Kreutz C. Probing RNA dynamics via longitudinal exchange and CPMG relaxation dispersion NMR spectroscopy using a sensitive (13)C-methyl label. Nucleic Acids Res. 2011;39:4340–4351. doi: 10.1093/nar/gkq1361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kreutz C, Kählig H, Konrat R, Micura R. Ribose 2′-F Labeling: A Simple Tool for the Characterization of RNA Secondary Structure Equilibria by 19F NMR Spectroscopy. J Am Chem Soc. 2005;127:11558–11559. doi: 10.1021/ja052844u. [DOI] [PubMed] [Google Scholar]
- 17.Puffer B, Kreutz C, Rieder U, Ebert M-O, Konrat R, Micura R. 5-Fluoro pyrimidines: labels to probe DNA and RNA secondary structures by 1D 19F NMR spectroscopy. Nucleic Acids Res. 2009;37:7728–7740. doi: 10.1093/nar/gkp862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Graber D, Moroder H, Micura R. (19)F NMR Spectroscopy for the Analysis of RNA Secondary Structure Populations. J Am Chem Soc. 2008;130:17230–17231. doi: 10.1021/ja806716s. [DOI] [PubMed] [Google Scholar]
- 19.Pfaff DA, Clarke KM, Parr TA, Cole JM, Geierstanger BH, Tahmassebi DC, Dwyer TJ. Solution structure of a DNA duplex containing a guanine-difluorotoluene pair: A wobble pair without hydrogen bonding? J Am Chem Soc. 2008;130:4869–4878. doi: 10.1021/ja7103608. [DOI] [PubMed] [Google Scholar]
- 20.Xia J, Noronha A, Toudjarska I, Li F, Akinc A, Braich R, Frank-Kamenetsky M, Rajeev KG, Egli M, Manoharan M. Gene sitencing activity of siRNAs with a ribo-difluorotoluyl nucteotide. ACS Chem Biol. 2006;1:176–183. doi: 10.1021/cb600063p. [DOI] [PubMed] [Google Scholar]
- 21.Somoza A, Chelliserrykattil J, Kool ET. The roles of hydrogen bonding and sterics in RNA interference. Angew Chem, Int Ed. 2006;45:4994–4997. doi: 10.1002/anie.200601311. [DOI] [PubMed] [Google Scholar]
- 22.Li F, Pallan PS, Maier MA, Rajeev KG, Mathieu SL, Kreutz C, Fan Y, Sanghvi J, Micura R, Rozners E, Manoharan M, Egli M. Crystal structure, stability and in vitro RNAi activity of oligoribonucleotides containing the ribo-difluorotoluyl nucleotide: insights into substrate requirements by the human RISC Ago2 enzyme. Nucleic Acids Res. 2007;35:6424–6438. doi: 10.1093/nar/gkm664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yamada T, Peng CG, Matsuda S, Addepalli H, Jayaprakash KN, Alam MR, Mills K, Maier MA, Charisse K, Sekine M, Manoharan M, Rajeev KG. Versatile Site-Specific Conjugation of Small Molecules to siRNA Using Click Chemistry. J Org Chem. 2011;76:1198–1211. doi: 10.1021/jo101761g. [DOI] [PubMed] [Google Scholar]
- 24.El-Sagheer AH, Brown T. New strategy for the synthesis of chemically modified RNA constructs exemplified by hairpin and hammerhead ribozymes. Proc Natl Acad Sci USA. 2010;107:15329–15334. doi: 10.1073/pnas.1006447107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Paredes E, Das SR. Click Chemistry for Rapid Labeling and Ligation of RNA. ChemBioChem. 2011;12:125–131. doi: 10.1002/cbic.201000466. [DOI] [PubMed] [Google Scholar]
- 26.Peacock H, Fostvedt E, Beal PA. Minor-Groove-Modulating Adenosine Replacements Control Protein Binding and RNAi Activity in siRNAs. ACS Chem Biol. 2010;5:1115–1124. doi: 10.1021/cb100245u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jin SX, Miduturu CV, McKinney DC, Silverman SK. Synthesis of amine- and thiol-modified nucleoside phosphoramidites for sitespecific introduction of biophysical probes into RNA. J Org Chem. 2005;70:4284–4299. doi: 10.1021/jo050061l. [DOI] [PubMed] [Google Scholar]
- 28.Pfander S, Fiammengo R, Kirin SI, Metzler-Nolte N, Jaschke A. Reversible site-specific tagging of enzymatically synthesized RNAs using aldehyde-hydrazine chemistry and protease-cleavable linkers. Nucleic Acids Res. 2007;35:e25. doi: 10.1093/nar/gkl1110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jao CY, Salic A. Exploring RNA transcription and turnover in vivo by using click chemistry. Proc Natl Acad Sci USA. 2008;105:15779–15784. doi: 10.1073/pnas.0808480105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Peacock H, Maydanovych O, Beal PA. N(2)-Modified 2-aminopurine ribonucleosides as minor-groove-modulating adenosine replacements in duplex RNA. Org Lett. 2010;12:1044–1047. doi: 10.1021/ol100019r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mutisya D, Selvam C, Kennedy SD, Rozners E. Synthesis and properties of triazole-linked RNA. Bioorg Med Chem Lett. 2011;21:3420–3422. doi: 10.1016/j.bmcl.2011.03.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Motorin Y, Burhenne J, Teimer R, Koynov K, Willnow S, Weinhold E, Helm M. Expanding the chemical scope of RNA:methyltransferases to site-specific alkynylation of RNA for click labeling. Nucleic Acids Res. 2011;39:1943–1952. doi: 10.1093/nar/gkq825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Onizuka K, Shibata A, Taniguchi Y, Sasaki S. Pin-point chemical modification of RNA with diverse molecules through the functionality transfer reaction and the copper-catalyzed azide-alkyne cycloaddition reaction. Chem Commun. 2011;47:5004–5006. doi: 10.1039/c1cc10582e. [DOI] [PubMed] [Google Scholar]
- 34.van Delft P, Meeuwenoord NJ, Hoogendoorn S, Dinkelaar J, Overkleeft HS, van der Marel GA, Filippov DV. Synthesis of Oligoribonucleic Acid Conjugates Using a Cyclooctyne Phosphoramidite. Org Lett. 2010;12:5486–5489. doi: 10.1021/ol102357u. [DOI] [PubMed] [Google Scholar]
- 35.van Delft P, van Schie E, Meeuwenoord NJ, Overkleeft HS, van der Marel GA, Filippov DV. Oligonucleotide Conjugates by Means of Copper-Free Click Chemistry - Expanding the Repertoire of Strained Cyclooctyne Phosphoramidites. Synthesis. 2011;2011:2724–2732. [Google Scholar]
- 36.Kočalka P, El-Sagheer AH, Brown T. Rapid and Efficient DNA Strand Cross-Linking by Click Chemistry. ChemBioChem. 2008;9:1280–1285. doi: 10.1002/cbic.200800006. [DOI] [PubMed] [Google Scholar]
- 37.Onizuka K, Taniguchi Y, Sasaki S. Activation and Alteration of Base Selectivity by Metal Cations in the Functionality-Transfer Reaction for RNA Modification. Bioconjugate Chem. 2010;21:1508–1512. doi: 10.1021/bc100131j. [DOI] [PubMed] [Google Scholar]
- 38.Onizuka K, Taniguchi Y, Sasaki S. A new usage of functionalized oligodeoxynucleotide probe for site-specific modification of a guanine base within RNA. Nucleic Acids Res. 2010;38:1760–1766. doi: 10.1093/nar/gkp930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Baskin JM, Prescher JA, Laughlin ST, Agard NJ, Chang PV, Miller IA, Lo A, Codelli JA, Bertozzi CR. Copper-free click chemistry for dynamic in vivo imaging. Proc Natl Acad Sci USA. 2007;104:16793–16797. doi: 10.1073/pnas.0707090104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Agard NJ, Baskin JM, Prescher JA, Lo A, Bertozzi CR. A comparative study of bioorthogonal reactions with azides. ACS Chem Biol. 2006;1:644–648. doi: 10.1021/cb6003228. [DOI] [PubMed] [Google Scholar]
- 41.Agard NJ, Prescher JA, Bertozzi CR. A Strain-Promoted [3 + 2] Azide—Alkyne Cycloaddition for Covalent Modification of Biomolecules in Living Systems. J Am Chem Soc. 2004;126:15046–15047. doi: 10.1021/ja044996f. [DOI] [PubMed] [Google Scholar]
- 42.Ning X, Guo J, Wolfert MA, Boons G-J. Visualizing Metabolically Labeled Glycoconjugates of Living Cells by Copper-Free and Fast Huisgen Cycloadditions. Angew Chem. 2008;120:2285–2287. doi: 10.1002/anie.200705456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Constantinesco F, Motorin Y, Grosjean H. Characterisation and Enzymatic Properties of tRNA(guanine 26, N2,N2)-dimethyltransferase (Trm1p) from Pyrococcus furiosus. J Mol Biol. 1999;291:375–392. doi: 10.1006/jmbi.1999.2976. [DOI] [PubMed] [Google Scholar]
- 44.Jacobs AC, Resendiz MJE, Greenberg MM. Direct Strand Scission from a Nucleobase Radical in RNA. J Am Chem Soc. 2010;132:3668–3669. doi: 10.1021/ja100281x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Deeble DJ, Schulz D, von Sonntag C. Reactions of OH Radicals with Poly(U) in Deoxygenated Solutions: Sites of OH Radical Attack and the Kinetics of Base Release. Int J Radiat Biol. 1986;49:915–926. doi: 10.1080/09553008514553151. [DOI] [PubMed] [Google Scholar]
- 46.Hildenbrand K, Schulte-Frohlinde D. E.s.r. Studies on the Mechanism of Hydroxyl Radical-induced Strand Breakage of Polyuridylic Acid. Int J Radiat Biol. 1989;55:725–738. doi: 10.1080/09553008914550781. [DOI] [PubMed] [Google Scholar]
- 47.Jacobs AC, Resendiz MJE, Greenberg MM. Product and Mechanistic Analysis of the Reactivity of a C6-Pyrimidine Radical in RNA. J Am Chem Soc. 2011;133:5152–5159. doi: 10.1021/ja200317w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Peacock H, Bachu R, Beal PA. Covalent stabilization of a small molecule-RNA complex. Bioorg Med Chem Lett. 2011;21:5002–5005. doi: 10.1016/j.bmcl.2011.04.136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mortimer SA, Weeks KM. A fast-acting reagent for accurate analysis of RNA secondary and tertiary structure by SHAPE chemistry. J Am Chem Soc. 2007;129:4144–4145. doi: 10.1021/ja0704028. [DOI] [PubMed] [Google Scholar]
- 50.Deigan KE, Li TW, Mathews DH, Weeks KM. Accurate SHAPE-directed RNA structure determination. Proc Natl Acad Sci USA. 2009;106:97–102. doi: 10.1073/pnas.0806929106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gherghe CM, Shajani Z, Wilkinson KA, Varani G, Weeks KM. Strong correlation between SHAPE chemistry and the generalized NMR order parameter (S-2) in RNA. J Am Chem Soc. 2008;130:12244–12245. doi: 10.1021/ja804541s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.McGinnis JL, Duncan CDS, Weeks KM. High-throughput Shape and Hydroxyl Radical Analysis of RNA Structure and Ribonucleoprotein Assembly. In: Herschalag D, editor. Methods in Enzymology, Vol 468: Biophysical, Chemical, and Functional Probes of Rna Structure, Interactions and Folding, Pt A. 2009. pp. 67–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mortimer SA, Weeks KM. Time-Resolved RNA SHAPE Chemistry. J Am Chem Soc. 2008;130:16178–16180. doi: 10.1021/ja8061216. [DOI] [PubMed] [Google Scholar]
- 54.Vasa SM, Guex N, Wilkinson KA, Weeks KM, Giddings MC. ShapeFinder: A software system for high-throughput quantitative analysis of nucleic acid reactivity information resolved by capillary electrophoresis. RNA. 2008;14:1979–1990. doi: 10.1261/rna.1166808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Steen KA, Malhotra A, Weeks KM. Selective 2′-Hydroxyl Acylation Analyzed by Protection from Exoribonuclease. J Am Chem Soc. 2010;132:9940–9943. doi: 10.1021/ja103781u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sakurai M, Yano T, Kawabata H, Ueda H, Suzuki T. Inosine cyanoethylation identifies A-to-I RNA editing sites in the human transcriptome. Nat Chem Biol. 2010;6:733–740. doi: 10.1038/nchembio.434. [DOI] [PubMed] [Google Scholar]
- 57.Giessing AMB, Jensen SS, Rasmussen A, Hansen LH, Gondela A, Long K, Vester B, Kirpekar F. Identification of 8-methyladenosine as the modification catalyzed by the radical SAM methyltransferase Cfr that confers antibiotic resistance in bacteria. RNA. 2009;15:327–336. doi: 10.1261/rna.1371409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ikeuchi Y, Kimura S, Numata T, Nakamura D, Yokogawa T, Ogata T, Wada T, Suzuki T. Agmatine-conjugated cytidine in a tRNA anticodon is essential for AUA decoding in archaea. Nat Chem Biol. 2010;6:277–282. doi: 10.1038/nchembio.323. [DOI] [PubMed] [Google Scholar]
- 59.Mandal D, Kohrer C, Su D, Russell SP, Krivos K, Castleberry CM, Blum P, Limbach PA, Soll D, RajBhandary UL. Agmatidine, a modified cytidine in the anticodon of archaeal tRNA(Ile), base pairs with adenosine but not with guanosine. Proc Natl Acad Sci USA. 2010;107:2872–2877. doi: 10.1073/pnas.0914869107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Chen YG, Kowtoniuk WE, Agarwal I, Shen YH, Liu DR. LC/MS analysis of cellular RNA reveals NAD-linked RNA. Nat Chem Biol. 2009;5:879–881. doi: 10.1038/nchembio.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kehrenberg C, Schwarz S, Jacobsen L, Hansen LH, Vester B. A new mechanism for chloramphenicol, florfenicol and clindamycin resistance: methylation of 23S ribosomal RNA at A2503. Mol Microbiol. 2005;57:1064–1073. doi: 10.1111/j.1365-2958.2005.04754.x. [DOI] [PubMed] [Google Scholar]
- 62.Yan F, LaMarre JM, Rohrich R, Wiesner J, Jomaa H, Mankin AS, Fujimori DG. RImN and Cfr are Radical SAM Enzymes Involved in Methylation of Ribosomal RNA. J Am Chem Soc. 2010;132:3953–3964. doi: 10.1021/ja910850y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Boal AK, Grove TL, McLaughlin MI, Yennawar NH, Booker SJ, Rosenzweig AC. Structural Basis for Methyl Transfer by a Radical SAM Enzyme. Science. 2011;332:1089–1092. doi: 10.1126/science.1205358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Grove TL, Benner JS, Radle MI, Ahlum JH, Landgraf BJ, Krebs C, Booker SJ. A Radically Different Mechanism for SAdenosylmethionine- Dependent Methyltransferases. Science. 2011;332:604–607. doi: 10.1126/science.1200877. [DOI] [PubMed] [Google Scholar]
- 65.Yan F, Fujimori DG. RNA methylation by Radical SAM enzymes RlmN and Cfr proceeds via methylene transfer and hydride shift. Proc Natl Acad Sci USA. 2011;108:3930–3934. doi: 10.1073/pnas.1017781108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Sonawane KD, Tewari R. Conformational preferences of hypermodified nucleoside lysidine (k(2)C) occurring at wobble position in anticodon loop of tRNA(IIe) Nucleosides, Nucleotides Nucleic Acids. 2008;27:1158–1174. doi: 10.1080/15257770802341475. [DOI] [PubMed] [Google Scholar]
- 67.Gartner ZJ, Tse BN, Grubina R, Doyon JB, Snyder TM, Liu DR. DNA-templated organic synthesis and selection of a library of macrocycles. Science. 2004;305:1601–1605. doi: 10.1126/science.1102629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Tse BN, Snyder TM, Shen YH, Liu DR. Translation of DNA into a Library of 13 000 Synthetic Small-Molecule Macrocycles Suitable for in Vitro Selection. J Am Chem Soc. 2008;130:15611–15626. doi: 10.1021/ja805649f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Szostak JW, Bartel DP, Luisi PL. Synthesizing life. Nature. 2001;409:387–390. doi: 10.1038/35053176. [DOI] [PubMed] [Google Scholar]
- 70.White HB. Coenzymes as Fossils of an Earlier Metabolic State. J Mol Evol. 1976;7:101–104. doi: 10.1007/BF01732468. [DOI] [PubMed] [Google Scholar]
- 71.Ni J, Tien AL, Fournier MJ. Small Nucleolar RNAs Direct Site-Specific Synthesis of Pseudouridine in Ribosomal RNA. Cell. 1997;89:565–573. doi: 10.1016/s0092-8674(00)80238-x. [DOI] [PubMed] [Google Scholar]
- 72.Ganot P, Bortolin ML, Kiss T. Site-Specific Pseudouridine Formation in Preribosomal RNA Is Guided by Small Nucleolar RNAs. Cell. 1997;89:799–809. doi: 10.1016/s0092-8674(00)80263-9. [DOI] [PubMed] [Google Scholar]
- 73.Karijolich J, Yu YT. Converting nonsense codons into sense codons by targeted pseudouridylation. Nature. 2011;474:395–398. doi: 10.1038/nature10165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Ferre-D’Amare AR. Protein Synthesis Stop the nonsense. Nature. 2011;474:289–290. doi: 10.1038/474289a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Davis DR. Stabilization of RNA stacking by pseudouridine. Nucleic Acids Res. 1995;23:5020–5026. doi: 10.1093/nar/23.24.5020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Jayalath P, Pokharel S, Veliz E, Beal PA. Synthesis and Evaluation of an RNA Editing Substrate Bearing 2′-Deoxy-2′-Mercaptoadenosine. Nucleosides, Nucleotides Nucleic Acids. 2009;28:78–88. doi: 10.1080/15257770902736459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Pokharel S, Jayalath P, Maydanovych O, Goodman RA, Wang SC, Tantillo DJ, Beal PA. Matching Active Site and Substrate Structures for an RNA Editing Reaction. J Am Chem Soc. 2009;131:11882–11891. doi: 10.1021/ja9034076. [DOI] [PubMed] [Google Scholar]
- 78.Maydanovych O, Beal PA. C6-substituted analogues of 8-azanebularine: Probes of an RNA-editing enzyme active site. Org Lett. 2006;8:3753–3756. doi: 10.1021/ol061354j. [DOI] [PubMed] [Google Scholar]
- 79.Haudenschild BL, Maydanovych O, Veliz EA, Macbeth MR, Bass BL, Beal PA. A transition state analogue for an RNA-editing reaction. J Am Chem Soc. 2004;126:11213–11219. doi: 10.1021/ja0472073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Yi-Brunozzi HY, Easterwood LM, Kamilar GM, Beal PA. Synthetic substrate analogs for the RNA-editing adenosine deaminase ADAR-2. Nucleic Acids Res. 1999;27:2912–2917. doi: 10.1093/nar/27.14.2912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Veliz EA, Easterwood LM, Beal PA. Substrate analogues for an RNA-editing adenosine deaminase: Mechanistic investigation and inhibitor design. J Am Chem Soc. 2003;125:10867–10876. doi: 10.1021/ja029742d. [DOI] [PubMed] [Google Scholar]
- 82.Easterwood LM, Véliz EA, Beal PA. Demethylation of 6-O-Methylinosine by an RNA-Editing Adenosine Deaminase. J Am Chem Soc. 2000;122:11537–11538. [Google Scholar]
- 83.Lomeli H, Mosbacher J, Melcher T, Hoger T, Geiger JRP, Kuner T, Monyer H, Higuchi M, Bach A, Seeburg PH. Control of Kinetic-Properties of AMPA Receptor Channels by Nuclear-RNA Editing. Science. 1994;266:1709–1713. doi: 10.1126/science.7992055. [DOI] [PubMed] [Google Scholar]
- 84.Burns CM, Chu H, Rueter SM, Hutchinson LK, Canton H, SandersBush E, Emeson RB. Regulation of serotonin-2C receptor G-protein coupling by RNA editing. Nature. 1997;387:303–308. doi: 10.1038/387303a0. [DOI] [PubMed] [Google Scholar]
- 85.Higuchi M, Maas S, Single FN, Hartner J, Rozov A, Burnashev N, Feldmeyer D, Sprengel R, Seeburg PH. Point mutation in an AMPA receptor gene rescues lethality in mice deficient in the RNA-editing enzyme ADAR2. Nature. 2000;406:78–81. doi: 10.1038/35017558. [DOI] [PubMed] [Google Scholar]
- 86.Hoopengardner B, Bhalla T, Staber C, Reenan R. Nervous system targets of RNA editing identified by comparative genomics. Science. 2003;301:832–836. doi: 10.1126/science.1086763. [DOI] [PubMed] [Google Scholar]
- 87.Sommer B, Kohler M, Sprengel R, Seeburg PH. RNA Editing in Brain Controls a Determinant of Ion Flow in Glutamate-Gated Channels. Cell. 1991;67:11–19. doi: 10.1016/0092-8674(91)90568-j. [DOI] [PubMed] [Google Scholar]
- 88.Macbeth MR, Schubert HL, VanDemark AP, Lingam AT, Hill CP, Bass BL. Inositol hexakisphosphate is bound in the ADAR2 core and required for RNA editing. Science. 2005;309:1534–1539. doi: 10.1126/science.1113150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Kannan A, Fostvedt E, Beal PA, Burrows CJ. 8-Oxoguanosine Switches Modulate the Activity of Alkylated siRNAs by Controlling Steric Effects in the Major versus Minor Grooves. J Am Chem Soc. 2011;133:6343–6351. doi: 10.1021/ja2003878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Peacock H, Fucini RV, Jayalath P, Ibarra-Soza JM, Haringsma HJ, Flanagan WM, Willingham A, Beal PA. Nucleobase and Ribose Modifications Control Immunostimulation by a MicroRNA-122-mimetic RNA. J Am Chem Soc. 2011;133:9200–9203. doi: 10.1021/ja202492e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Coulouarn C, Factor VM, Andersen JB, Durkin ME, Thorgeirsson SS. Loss of miR-122 expression in liver cancer correlates with suppression of the hepatic phenotype and gain of metastatic properties. Oncogene. 2009;28:3526–3536. doi: 10.1038/onc.2009.211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Bai SM, Nasser MW, Wang B, Hsu SH, Datta J, Kutay H, Yadav A, Nuovo G, Kumar P, Ghoshal K. MicroRNA-122 Inhibits Tumorigenic Properties of Hepatocellular Carcinoma Cells and Sensitizes These Cells to Sorafenib. J Biol Chem. 2009;284:32015–32027. doi: 10.1074/jbc.M109.016774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Tsai WC, Hsu PWC, Lai TC, Chau GY, Lin CW, Chen CM, Lin CD, Liao YL, Wang JL, Chau YP, Hsu MT, Hsiao M, Huang HD, Tsou AP. MicroRNA-122, a Tumor Suppressor MicroRNA that Regulates Intrahepatic Metastasis of Hepatocellular Carcinoma. Hepatology. 2009;49:1571–1582. doi: 10.1002/hep.22806. [DOI] [PubMed] [Google Scholar]
- 94.Judge AD, Sood V, Shaw JR, Fang D, McClintock K, MacLachlan I. Sequence-dependent stimulation of the mammalian innate immune response by synthetic siRNA. Nat Biotechnol. 2005;23:457–462. doi: 10.1038/nbt1081. [DOI] [PubMed] [Google Scholar]
- 95.Terrazas M, Avino A, Siddiqui MA, Marquez VE, Eritja R. A Direct, Efficient Method for the Preparation of siRNAs Containing Ribo-like North Bicyclo 3.1.0 hexane Pseudosugars. Org Lett. 2011;13:2888–2891. doi: 10.1021/ol200909j. [DOI] [PubMed] [Google Scholar]
- 96.Terrazas M, Ocampo SM, Perales JC, Marquez VE, Eritja R. Effect of North Bicyclo 3.1.0 hexane 2′-Deoxy-pseudosugars on RNA Interference: A Novel Class of siRNA Modification. ChemBioChem. 2011;12:1056–1065. doi: 10.1002/cbic.201000791. [DOI] [PubMed] [Google Scholar]
- 97.Altmann KH, Kesselring R, Francotte E, Rihs G. 4′,6′-Methano Carbocyclic Thymidine - a Conformationally Constrained Building-Block for Oligonucleotides. Tetrahedron Lett. 1994;35:2331–2334. [Google Scholar]
- 98.Marquez VE, Siddiqui MA, Ezzitouni A, Russ P, Wang JY, Wagner RW, Matteucci MD. Nucleosides with a twist. Can fixed forms of sugar ring pucker influence biological activity in nucleosides and oligonucleotides? J Med Chem. 1996;39:3739–3747. doi: 10.1021/jm960306+. [DOI] [PubMed] [Google Scholar]