Abstract
Recent months have seen rapid advances in the field of transdifferentiation, specifically in the conversion of fibroblasts to neurons. Most surprising is the observation that the ability to drive these transitions is not limited to transcription factors, but that they can be promoted by microRNAs as well. Indeed, in one case, microRNAs alone induced the transdifferentiation of fibroblasts to neuron-like cells, albeit at a low efficiency. Here, we review this rapidly advancing field, discuss possible mechanisms underlying microRNA-induced transdifferentiation and the potential for microRNAs to drive such transitions to any cell type of interest in vitro and in vivo.
Introduction
The holy grail of regenerative medicine is to replace the damaged or lost cells that occur as we age, suffer disease, or are exposed to environmental insults. In theory regeneration could be achieved using the processes of dedifferentiation (reprogramming of a differentiated cell to revert to a stem cell), or transdifferentiation (reprogramming one type of differentiated cell to become another sort of differentiated cell). Recent advances have shown that these phenomena can be induced in adult mammalian cells by modulating the expression of particular genes.
In 1987, Weintraub and colleagues made the pioneering discovery that the addition of a single transcription factor could convert a fibroblast to a muscle cell [1]. In recent years, the number of examples of transcription factor-induced transdifferentiation is growing at a remarkable rate. In 2004, Graf and colleagues induced mouse B-lymphocytes into macrophages [2]. In 2008, Melton and colleagues described the transdifferentiation of mouse pancreatic acinar cells to beta-islet cells [3]. Two years later, Wernig and colleagues induced the conversion of mouse fibroblasts to neurons, showing for the first time that addition of transcription factors can induce transdifferentiation across germ layers – from cell types formed in the mesoderm to those formed in the ectoderm [4]. These efforts were originally performed using mouse cells. A recent litany of papers has taken it a step further directing the conversion of human fibroblasts to neurons as well as the differentiation to specific neuronal subtypes [5-11].
MicroRNAs can change cell fate
While most transdifferentiation experiments have used cocktails of transcription factors ranging from three to as many as eleven, two recent studies have included microRNAs. microRNAs are short non-coding RNAs approximately twenty-one nucleotides in length that bind to complementary pieces of mRNA after transcription and inhibit their translation. Like transcription factors, they can regulate hundreds of targets simultaneously. Also like transcription factors, they recognize short motifs (7-8 nucleotides) in their targets [12]. For microRNAs, these motifs are predominantly in the 3'UTRs (untranslated regions) and occasionally the coding region of mRNAs. In contrast, transcription factors bind DNA motifs within promoter and enhancer regions of the genome's DNA.
Transcription factors have been accepted as potential master regulators of cell fate ever since Weintraub's seminal work. In contrast, microRNAs have rarely been thought of as such. Indeed, they are more typically described as stabilizers of cell fates or buffers of stochastic transcriptional noise and external stressors [13]. This view has been clearly challenged by recent results showing that microRNAs can induce transdifferentiation of fibroblasts to neurons as well as work showing that they can induce the dedifferentiation of fibroblasts to embryonic-like stem cells. In retrospect, hints of this remarkable capacity of microRNAs to drive fates by globally modulating the transcriptome date back to experiments performed by Lim and colleagues [14]. They showed that the introduction of a single microRNA, miR-124, could induce HeLa cells, a cell line derived from a cervical cancer, to partially transition toward a neuronal state as defined by the transcriptional program. However, the treated cells were not shown to have morphological or functional properties of neurons.
Like Lim and colleagues, two recent studies used miR-124 to promote the transition of human fibroblasts to neurons. In particular, Ding and colleagues [5] combined miR-124 with two transcription factors first used by Vierbuchen and colleagues [4], MYT1L and BRN2. In contrast, Crabtree and colleagues showed that miR-124 and miR-9 alone could induce the conversion, albeit at a low efficiency, and functional characterization of the resulting cells was lacking [7]. The addition of the transcription factor NeuroD1 along with miR-124 and miR-9 greatly increased the efficiencies and the resulting cells were shown to have functional properties of neurons. On the surface, the fact that microRNAs alone are able to drive this conversion is puzzling as it requires the microRNAs to somehow activate a new program by suppressing its mRNA targets. How does suppression of targets lead to activation of a new cellular program? One possibility is that the downregulation of non-neural transcripts by the neural-enriched microRNAs creates a permissive atmosphere for stochastic or leaky gene expression to lead to preferential activation of neuronal gene expression programs. These non-neural transcripts would be likely to include mRNAs important for fibroblast identity as well as alternative non-neuronal cell fates. Alternatively, the microRNAs could be situated in a signaling network that directly activates neuron-specific components. That is, the cell fates are driven by double negatives, where the microRNA suppresses a repressor leading to activation of the neuronal cell fate. The answer is likely some combination of the two.
Some hints as to how microRNAs may be inducing cell fate transitions that do not normally occur during development comes from recent work in promoting the de-differentiation of fibroblasts to induced pluripotent stem cells (iPSCs). A large family of microRNAs called the ESCC microRNAs, including miR-302 and miR-372 in humans, are strong inducers of iPSCs [15, 16]. Indeed, it has been suggested that these microRNAs together with one or two other microRNAs can convert fibroblasts to iPSCs in the absence of any transcription factors [17, 18]. Recent work has begun to dissect the mRNA targets that underlie the remarkable capacity of the ESCC family. Subramanyam et al. show that microRNAs promote reprogramming by simultaneously targeting several critical cellular pathways including regulators of the G1-S cell cycle checkpoint, the mesenchymal to epithelial transition, and DNA methyl binding proteins [19]. However, these represent only a subset of the mRNAs targeted by these microRNAs. For example, the ESCC microRNAs also target mRNAs that regulate apoptosis and bone morphogenetic protein (BMP) signaling, although the role of these targets in reprogramming has not been studied [20, 21]. Dissection and functional characterization of all of the ESCC targets should provide a detailed and global view of how the microRNAs can promote the transition. Similarly, characterization of all the targets of miR-9 and miR-124 will provide insights on how suppression of targets in a fibroblast eventually results in the activation of the neuronal program.
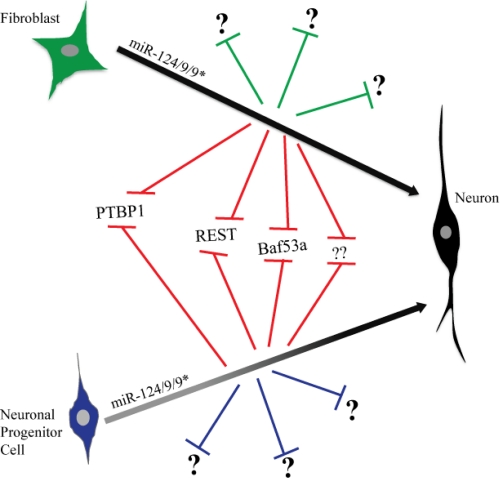
The ability of microRNAs to act in the context of a cell distantly related, in developmental terms, to the cell in which the microRNAs are normally expressed is another surprising feature of microRNA-induced transdifferentiation. That is, while the microRNAs are not normally expressed in the cell, their targets appear to be so. One possibility is that a common set of genes is expressed in the distantly related cell types and then largely regulated through post-transcriptional rather than transcriptional mechanisms. An example of such a target is BAF (Brg/Brm-associated factor)53a. This protein is expressed in both fibroblasts and neural progenitors. Previous work by the Crabtree lab had shown that miR-124 and miR-9* target the BAF53a transcript as neural progenitors differentiate into neurons, thereby repressing protein production [22]. Suppression of BAF53a then leads to the upregulation of BAF53b, an activator of neuronal genes. A similar switch occurs in the microRNA-induced transition of fibroblasts to neurons. While data are not shown or discussed in detail, the authors mention that prolonging expression of Baf53a and other known miR-124 neural progenitor cell targets (PTPB1 [polypyrimidine tract-binding protein 1], REST [RE1-silencing transcription factor], coREST) only partially blocks transdifferentiation, suggesting that these targets play important roles, but do not alone explain the remarkable transition. Of course these are only three of the hundreds of targets of the two microRNAs. It will be interesting to determine how many other targets are shared in the transition of fibroblasts to neurons versus neural progenitors to neurons (see Figure 1).
Figure 1: microRNA-induced differentiation and transdifferentiation to neurons.
Similar microRNAs promote the transition of fibroblasts to neurons and neural progenitors to neurons. The complete set of targets for these microRNAs remain unknown. However, at least a subset including Baf53a, REST, and PTBP1 are common to both fibroblasts and neural progenitors suggesting overlapping pathways in the two transitions. Abbreviations: BAF, Brg/Brm-associated factor; PTBP1, polypyrimidine tract-binding protein 1; REST, RE1-silencing transcription factor.
The physiological role of microRNAs in determining cell fate
Another question is how wide-spread is the capacity of microRNAs to induce cell fate decisions across normal developmental barriers? While there are many examples of transcription factor-induced transdifferentiation across germ layers, including recent studies that have shown conversion of fibroblasts to hepatocytes and cardiomyocytes using the respective lineage-specific transcription factors [23, 24], the variety of cell types using microRNAs is less well known. Are there enough combinations of microRNAs to underlie the remarkably diverse cell types in the human body? Even within the neural lineage there is a remarkable diversity of neural subtypes. It is unclear to what degree each of these subtypes has a unique microRNA signature, which could potentially drive their unique fate. Indeed, there are many less microRNAs than there are transcription factors – hundred versus thousands, respectively. Furthermore, only a small number of microRNAs have been shown to have specific tissue expression. Well-known examples are miR-9 and miR-124 in the brain, miR-1 in muscle, and miR-122 in liver. Both miR-1 and miR-122 are promising candidates for promoting transdifferentiation to their respective cell types. Other microRNAs, such as the let-7 family, are broadly expressed across all differentiated tissues and, hence, are likely general stabilizers of the differentiated adult cell fate [25].
Future Prospects
Together, these recent exciting findings have certainly suggested that microRNAs may play an important role in regenerative medicine. They are not only able to induce cell fate transitions but are also likely to lead to profound insights into the underlying molecular pathways and cellular processes regulating those transitions. From a clinical standpoint, microRNAs are an enticing potential alternative to transcription factors. In particular, they can be introduced relatively easily into cells in their mature form without inducing the cell's innate mature response. In contrast, transcription factors need to be introduced as DNA or as highly modified mRNAs. DNA is risky as it can permanently alter the cell's genome, while microRNAs do not. microRNAs even exist stably within our plasma and are thought to be naturally transferred from one cell to another [26]. Therefore, the introduction of microRNAs directly into patients to induce cell fate conversions does not seem far-fetched. Therefore, both in basic biology and translational medicine, microRNAs should be an explosive field for years to come.
Abbreviations
- BAF
Brg/Brm-associated factor
- BMP
bone morphogenetic protein
- iPSC
induced pluripotent stem cell
- PTBP1
polypyrimidine tract-binding protein 1
- REST
RE1-silencing transcription factor
- UTR
untranslated region
Competing Interests
The authors declare they have no competing interests.
The electronic version of this article is the complete one and can be found at: http://f1000.com/reports/b/4/3
References
- 1.Davis RL, Weintraub H, Lassar AB. Expression of a single transfected cDNA converts fibroblasts to myoblasts. Cell. 1987;51:987–1000. doi: 10.1016/0092-8674(87)90585-X. [DOI] [PubMed] [Google Scholar]; F1000 Factor 8Evaluated by Robert Blelloch 16 Jan 2012
- 2.Xie H, Ye M, Feng R, Graf T. Stepwise reprogramming of B cells into macrophages. Cell. 2004;117:663–76. doi: 10.1016/S0092-8674(04)00419-2. [DOI] [PubMed] [Google Scholar]; F1000 Factor 10Evaluated by Xiaojing Ma 04 Jun 2004, David Serreze 07 Jul 2004
- 3.Zhou Q, Brown J, Kanarek A, Rajagopal J, Melton DA. In vivo reprogramming of adult pancreatic exocrine cells to beta-cells. Nature. 2008;455:627–32. doi: 10.1038/nature07314. [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Factor 25Evaluated by Anthony Means 04 Sep 2008, Phillip Newmark 05 Sep 2008, Linda Jane Mullins and John Mullins 18 Sep 2008, Raghavendra Mirmira 13 Oct 2008, Ray Rodgers 15 Oct 2008, Ivana Novak 29 Oct 2008, Ulf Pettersson 09 Dec 2008, Robert Blelloch 16 Jan 2012
- 4.Vierbuchen T, Ostermeier A, Pang ZP, Kokubu Y, Sudhof TC, Wernig M. Direct conversion of fibroblasts to functional neurons by defined factors. Nature. 2010;463:1035–41. doi: 10.1038/nature08797. [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Factor 13Evaluated by Maripat Corr 26 Feb 2010, James Ellis 26 Mar 2010, Michael G Fehlings 24 Jun 2010
- 5.Ambasudhan R, Talantova M, Coleman R, Yuan X, Zhu S, Lipton SA, Ding S. Direct reprogramming of adult human fibroblasts to functional neurons under defined conditions. Cell Stem Cell. 2011;9:113–8. doi: 10.1016/j.stem.2011.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Factor 6Evaluated by Irit Meivar-Levy and Sarah Ferber 16 Aug 2011
- 6.Son EY, Ichida JK, Wainger BJ, Toma JS, Rafuse VF, Woolf CJ, Eggan K. Conversion of mouse and human fibroblasts into functional spinal motor neurons. Cell Stem Cell. 2011;9:205–18. doi: 10.1016/j.stem.2011.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Factor 6Evaluated by Robert Blelloch 16 Jan 2012
- 7.Yoo AS, Sun AX, Li L, Shcheglovitov A, Portmann T, Li Y, Lee-Messer C, Dolmetsch RE, Tsien RW, Crabtree GR. MicroRNA-mediated conversion of human fibroblasts to neurons. Nature. 2011;476:228–31. doi: 10.1038/nature10323. [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Factor 12Evaluated by Michael Sendtner 26 Jul 2011, Thierry Bertomeu and Roya Khosravi-Far 09 Sep 2011
- 8.Qiang L, Fujita R, Yamashita T, Angulo S, Rhinn H, Rhee D, Doege C, Chau L, Aubry L, Vanti WB, Moreno H, Abeliovich A. Directed conversion of Alzheimer's disease patient skin fibroblasts into functional neurons. Cell. 2011;146:359–71. doi: 10.1016/j.cell.2011.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]; F1000 Factor 6Evaluated by Robert Blelloch 16 Jan 2012
- 9.Pfisterer U, Kirkeby A, Torper O, Wood J, Nelander J, Dufour A, Bjorklund A, Lindvall O, Jakobsson J, Parmar M. Direct conversion of human fibroblasts to dopaminergic neurons. Proc Natl Acad Sci U S A. 2011;108:10343–8. doi: 10.1073/pnas.1105135108. [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Factor 6Evaluated by Oliver Cooper and Ole Isacson 29 Jul 2011
- 10.Pfisterer U, Wood J, Nihlberg K, Hallgren O, Bjermer L, Westergren-Thorsson G, Lindvall O, Parmar M. Efficient induction of functional neurons from adult human fibroblasts. Cell Cycle. 2011;10:3311–6. doi: 10.4161/cc.10.19.17584. [DOI] [PubMed] [Google Scholar]
- 11.Caiazzo M, Dell'Anno MT, Dvoretskova E, Lazarevic D, Taverna S, Leo D, Sotnikova TD, Menegon A, Roncaglia P, Colciago G, Russo G, Carninci P, Pezzoli G, Gainetdinov RR, Gustincich S, Dityatev A, Broccoli V. Direct generation of functional dopaminergic neurons from mouse and human fibroblasts. Nature. 2011;476:224–7. doi: 10.1038/nature10284. [DOI] [PubMed] [Google Scholar]; F1000 Factor 11Evaluated by Alessandra Pasut and Michael Rudnicki 15 Jul 2011, Michael Sendtner 26 Jul 2011, Oliver Cooper and Ole Isacson 29 Jul 2011
- 12.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–33. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hornstein E, Shomron N. Canalization of development by microRNAs. Nat Genet. 2006;38(Suppl):S20–4. doi: 10.1038/ng1803. [DOI] [PubMed] [Google Scholar]
- 14.Lim LP, Lau NC, Garrett-Engele P, Grimson A, Schelter JM, Castle J, Bartel DP, Linsley PS, Johnson JM. Microarray analysis shows that some microRNAs downregulate large numbers of target mRNAs. Nature. 2005;433:769–73. doi: 10.1038/nature03315. [DOI] [PubMed] [Google Scholar]; F1000 Factor 24Evaluated by Isaac Kohane 23 Feb 2005, Harukazu Nakamura 24 Feb 2005, Fritz Eckstein 28 Feb 2005, Ulf Pettersson 28 Feb 2005, Andrew D Sharrocks 02 Mar 2005, Detlef Weigel 02 Mar 2005, Richard Carthew 09 Mar 2005, Alejandro Sanchez-Alvarado 09 Mar 2005, Vishvanath Nene 01 Apr 2005, Miles Wilkinson 19 Apr 2005
- 15.Subramanyam D, Lamouille S, Judson RL, Liu JY, Bucay N, Derynck R, Blelloch R. Multiple targets of miR-302 and miR-372 promote reprogramming of human fibroblasts to induced pluripotent stem cells. Nat Biotechnol. 2011;29:443–8. doi: 10.1038/nbt.1862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Judson RL, Babiarz JE, Venere M, Blelloch R. Embryonic stem cell-specific microRNAs promote induced pluripotency. Nat Biotechnol. 2009;27:459–61. doi: 10.1038/nbt.1535. [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Factor 6Evaluated by Felipe Prosper 11 May 2009
- 17.Anokye-Danso F, Trivedi CM, Juhr D, Gupta M, Cui Z, Tian Y, Zhang Y, Yang W, Gruber PJ, Epstein JA, Morrisey EE. Highly efficient miRNA-mediated reprogramming of mouse and human somatic cells to pluripotency. Cell Stem Cell. 2011;8:376–88. doi: 10.1016/j.stem.2011.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Factor 8Evaluated by Benjamin Resio and David Pellman 04 Aug 2011
- 18.Miyoshi N, Ishii H, Nagano H, Haraguchi N, Dewi DL, Kano Y, Nishikawa S, Tanemura M, Mimori K, Tanaka F, Saito T, Nishimura J, Takemasa I, Mizushima T, Ikeda M, Yamamoto H, Sekimoto M, Doki Y, Mori M. Reprogramming of mouse and human cells to pluripotency using mature microRNAs. Cell Stem Cell. 2011;8:633–8. doi: 10.1016/j.stem.2011.05.001. [DOI] [PubMed] [Google Scholar]
- 19.Subramanyam D, Lamouille S, Judson RL, Liu JY, Bucay N, Derynck R, Blelloch R. Multiple targets of miR-302 and miR-372 promote reprogramming of human fibroblasts to induced pluripotent stem cells. Nat Biotechnol. 29:443–8. doi: 10.1038/nbt.1862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zheng GX, Ravi A, Calabrese JM, Medeiros LA, Kirak O, Dennis LM, Jaenisch R, Burge CB, Sharp PA. A latent pro-survival function for the mir-290-295 cluster in mouse embryonic stem cells. PLoS Genet. 2011;7 doi: 10.1371/journal.pgen.1002054. e1002054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lipchina I, Yechiel E, Hafner M, Sheridan R, Mihailovic A, Tuschl T, Sander C, Studer L, Betel D. Genome-wide identification of microRNA targets in human ES cells reveals a role for miR-302 in modulating BMP response. Genes Dev. 2011;25:2173–86. doi: 10.1101/gad.17221311. [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Factor 6Evaluated by Robert Blelloch 16 Jan 2012
- 22.Yoo AS, Staahl BT, Chen L, Crabtree GR. MicroRNA-mediated switching of chromatin-remodelling complexes in neural development. Nature. 2009;460:642–6. doi: 10.1038/nature08139. [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Factor 11Evaluated by Deyou Zheng 10 Aug 2009, Robert Blelloch 16 Jan 2012
- 23.Sekiya S, Suzuki A. Direct conversion of mouse fibroblasts to hepatocyte-like cells by defined factors. Nature. 2011;475:390–3. doi: 10.1038/nature10263. [DOI] [PubMed] [Google Scholar]; F1000 Factor 6Evaluated by Robert Blelloch 16 Jan 2012
- 24.Ieda M, Fu JD, Delgado-Olguin P, Vedantham V, Hayashi Y, Bruneau BG, Srivastava D. Direct reprogramming of fibroblasts into functional cardiomyocytes by defined factors. Cell. 2010;142:375–86. doi: 10.1016/j.cell.2010.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Factor 13Evaluated by Enrique Lara-Pezzi and Nadia Rosenthal 23 Aug 2010, Sveva Bollini and Paul Riley 16 Sep 2010, Robert Blelloch 16 Jan 2012
- 25.Melton C, Blelloch R. MicroRNA Regulation of Embryonic Stem Cell Self-Renewal and Differentiation. Adv Exp Med Biol. 2010;695:105–17. doi: 10.1007/978-1-4419-7037-4_8. [DOI] [PubMed] [Google Scholar]
- 26.Brase JC, Wuttig D, Kuner R, Sultmann H. Serum microRNAs as non-invasive biomarkers for cancer. Mol Cancer. 2010;9:306. doi: 10.1186/1476-4598-9-306. [DOI] [PMC free article] [PubMed] [Google Scholar]