Abstract
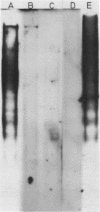
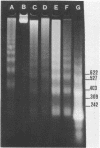
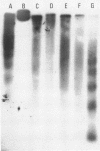
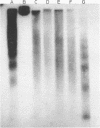
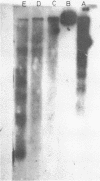
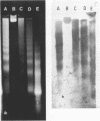
The nucleosomal organization of active and repressed alpha subtype histone genes has been investigated by micrococcal nuclease digestion of P. lividus sperm, 32-64 cell embryo and mesenchyme blastula nuclei, followed by hybridization with 32P-labeled specific DNA probes. In sperms, fully repressed histone genes are regularly folded in nucleosomes, and exhibit a greater resistance to micrococcal nuclease cleavage than bulk chromatin. In contrast, both coding and spacer alpha subtype histone DNA sequences acquire an altered conformation in nuclei from early cleavage stage embryos, i.e., when these genes are maximally expressed. Switching off of the alpha subtype histone genes, in mesenchyme blastulae, restores the typical nucleosomal organization on this chromatin region. As probed by hybridization to D.melanogaster actin cDNA, actin genes retain a regular nucleosomal structure in all the investigated stages.
Full text
PDF














Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Albanese I., Di Liegro I., Cognetti G. Unusual properties of sea urchin unfertilized egg chromatin. Cell Biol Int Rep. 1980 Feb;4(2):201–210. doi: 10.1016/0309-1651(80)90075-2. [DOI] [PubMed] [Google Scholar]
- Arceci R. J., Gross P. R. Histone variants and chromatin structure during sea urchin development. Dev Biol. 1980 Nov;80(1):186–209. doi: 10.1016/0012-1606(80)90508-4. [DOI] [PubMed] [Google Scholar]
- Bellard M., Dretzen G., Bellard F., Oudet P., Chambon P. Disruption of the typical chromatin structure in a 2500 base-pair region at the 5' end of the actively transcribed ovalbumin gene. EMBO J. 1982;1(2):223–230. doi: 10.1002/j.1460-2075.1982.tb01151.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellard M., Kuo M. T., Dretzen G., Chambon P. Differential nuclease sensitivity of the ovalbumin and beta-globin chromatin regions in erythrocytes and oviduct cells of laying hen. Nucleic Acids Res. 1980 Jun 25;8(12):2737–2750. doi: 10.1093/nar/8.12.2737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryan P. N., Hofstetter H., Birnstiel M. L. Nucleosome arrangement on tRNA genes of Xenopus laevis. Cell. 1981 Dec;27(3 Pt 2):459–466. doi: 10.1016/0092-8674(81)90387-1. [DOI] [PubMed] [Google Scholar]
- Camerini-Otero R. D., Zasloff M. A. Nucleosomal packaging of the thymidine kinase gene of herpes simplex virus transferred into mouse cells: an actively expressed single-copy gene. Proc Natl Acad Sci U S A. 1980 Sep;77(9):5079–5083. doi: 10.1073/pnas.77.9.5079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Childs G., Maxson R., Cohn R. H., Kedes L. Orphons: dispersed genetic elements derived from tandem repetitive genes of eucaryotes. Cell. 1981 Mar;23(3):651–663. doi: 10.1016/0092-8674(81)90428-1. [DOI] [PubMed] [Google Scholar]
- Childs G., Maxson R., Kedes L. H. Histone gene expression during sea urchin embryogenesis: isolation and characterization of early and late messenger RNAs of Strongylocentrotus purpuratus by gene-specific hybridization and template activity. Dev Biol. 1979 Nov;73(1):153–173. doi: 10.1016/0012-1606(79)90144-1. [DOI] [PubMed] [Google Scholar]
- Crain W. R., Jr, Durica D. S., Van Doren K. Actin gene expression in developing sea urchin embryos. Mol Cell Biol. 1981 Aug;1(8):711–720. doi: 10.1128/mcb.1.8.711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denhardt D. T. A membrane-filter technique for the detection of complementary DNA. Biochem Biophys Res Commun. 1966 Jun 13;23(5):641–646. doi: 10.1016/0006-291x(66)90447-5. [DOI] [PubMed] [Google Scholar]
- Durica D. S., Schloss J. A., Crain W. R., Jr Organization of actin gene sequences in the sea urchin: molecular cloning of an intron-containing DNA sequence coding for a cytoplasmic actin. Proc Natl Acad Sci U S A. 1980 Oct;77(10):5683–5687. doi: 10.1073/pnas.77.10.5683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felsenfeld G., McGhee J. Methylation and gene control. Nature. 1982 Apr 15;296(5858):602–603. doi: 10.1038/296602a0. [DOI] [PubMed] [Google Scholar]
- Grunstein M. Hatching in the sea urchin Lytechinus pictus is accompanied by a shift in histone H4 gene activity. Proc Natl Acad Sci U S A. 1978 Sep;75(9):4135–4139. doi: 10.1073/pnas.75.9.4135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hentschel C. C., Birnstiel M. L. The organization and expression of histone gene families. Cell. 1981 Aug;25(2):301–313. doi: 10.1016/0092-8674(81)90048-9. [DOI] [PubMed] [Google Scholar]
- Igo-Kemenes T., Hörz W., Zachau H. G. Chromatin. Annu Rev Biochem. 1982;51:89–121. doi: 10.1146/annurev.bi.51.070182.000513. [DOI] [PubMed] [Google Scholar]
- Kedes L. H. Histone genes and histone messengers. Annu Rev Biochem. 1979;48:837–870. doi: 10.1146/annurev.bi.48.070179.004201. [DOI] [PubMed] [Google Scholar]
- Kornberg R. The location of nucleosomes in chromatin: specific or statistical. Nature. 1981 Aug 13;292(5824):579–580. doi: 10.1038/292579a0. [DOI] [PubMed] [Google Scholar]
- Levy A., Noll M. Chromatin fine structure of active and repressed genes. Nature. 1981 Jan 15;289(5794):198–203. doi: 10.1038/289198a0. [DOI] [PubMed] [Google Scholar]
- Lohr D. E. Detailed analysis of the nucleosomal organization of transcribed DNA in yeast chromatin. Biochemistry. 1981 Oct 13;20(21):5966–5972. doi: 10.1021/bi00524a007. [DOI] [PubMed] [Google Scholar]
- Maniatis T., Jeffrey A., Kleid D. G. Nucleotide sequence of the rightward operator of phage lambda. Proc Natl Acad Sci U S A. 1975 Mar;72(3):1184–1188. doi: 10.1073/pnas.72.3.1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maniatis T., Jeffrey A., van deSande H. Chain length determination of small double- and single-stranded DNA molecules by polyacrylamide gel electrophoresis. Biochemistry. 1975 Aug 26;14(17):3787–3794. doi: 10.1021/bi00688a010. [DOI] [PubMed] [Google Scholar]
- Nedospasov S. A., Georgiev G. P. Non-random cleavage of SV40 DNA in the compact minichromosome and free in solution by micrococcal nuclease. Biochem Biophys Res Commun. 1980 Jan 29;92(2):532–539. doi: 10.1016/0006-291x(80)90366-6. [DOI] [PubMed] [Google Scholar]
- Newrock K. M., Alfageme C. R., Nardi R. V., Cohen L. H. Histone changes during chromatin remodeling in embryogenesis. Cold Spring Harb Symp Quant Biol. 1978;42(Pt 1):421–431. doi: 10.1101/sqb.1978.042.01.045. [DOI] [PubMed] [Google Scholar]
- Newrock K. M., Cohen L. H., Hendricks M. B., Donnelly R. J., Weinberg E. S. Stage-specific mRNAs coding for subtypes of H2A and H2B histones in the sea urchin embryo. Cell. 1978 Jun;14(2):327–336. doi: 10.1016/0092-8674(78)90118-6. [DOI] [PubMed] [Google Scholar]
- Noll M., Kornberg R. D. Action of micrococcal nuclease on chromatin and the location of histone H1. J Mol Biol. 1977 Jan 25;109(3):393–404. doi: 10.1016/s0022-2836(77)80019-3. [DOI] [PubMed] [Google Scholar]
- Palmiter R. D., Mulvihill E. R., McKnight G. S., Senear A. W. Regulation of gene expression in the chick oviduct by steroid hormones. Cold Spring Harb Symp Quant Biol. 1978;42(Pt 2):639–647. doi: 10.1101/sqb.1978.042.01.066. [DOI] [PubMed] [Google Scholar]
- Samal B., Worcel A., Louis C., Schedl P. Chromatin structure of the histone genes of D. melanogaster. Cell. 1981 Feb;23(2):401–409. doi: 10.1016/0092-8674(81)90135-5. [DOI] [PubMed] [Google Scholar]
- Scheller R. H., McAllister L. B., Crain W. R., Jr, Durica D. S., Posakony J. W., Thomas T. L., Britten R. J., Davidson E. H. Organization and expression of multiple actin genes in the sea urchin. Mol Cell Biol. 1981 Jul;1(7):609–628. doi: 10.1128/mcb.1.7.609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spadafora C., Bellard M., Compton J. L., Chambon P. The DNA repeat lengths in chromatins from sea urchin sperm and gastrule cells are markedly different. FEBS Lett. 1976 Oct 15;69(1):281–285. doi: 10.1016/0014-5793(76)80704-1. [DOI] [PubMed] [Google Scholar]
- Spinelli G., Gianguzza F., Casano C., Acierno P., Burckhardt J. Evidences of two different sets of histone genes active during embryogenesis of the sea urchin Paracentrotus lividus. Nucleic Acids Res. 1979 Feb;6(2):545–560. doi: 10.1093/nar/6.2.545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weisbrod S. Active chromatin. Nature. 1982 May 27;297(5864):289–295. doi: 10.1038/297289a0. [DOI] [PubMed] [Google Scholar]
- Wu C. The 5' ends of Drosophila heat shock genes in chromatin are hypersensitive to DNase I. Nature. 1980 Aug 28;286(5776):854–860. doi: 10.1038/286854a0. [DOI] [PubMed] [Google Scholar]
- Wu C., Wong Y. C., Elgin S. C. The chromatin structure of specific genes: II. Disruption of chromatin structure during gene activity. Cell. 1979 Apr;16(4):807–814. doi: 10.1016/0092-8674(79)90096-5. [DOI] [PubMed] [Google Scholar]
- Zachau H. G., Igo-Kemenes T. Face to phase with nucleosomes. Cell. 1981 Jun;24(3):597–598. doi: 10.1016/0092-8674(81)90084-2. [DOI] [PubMed] [Google Scholar]