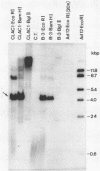
Abstract
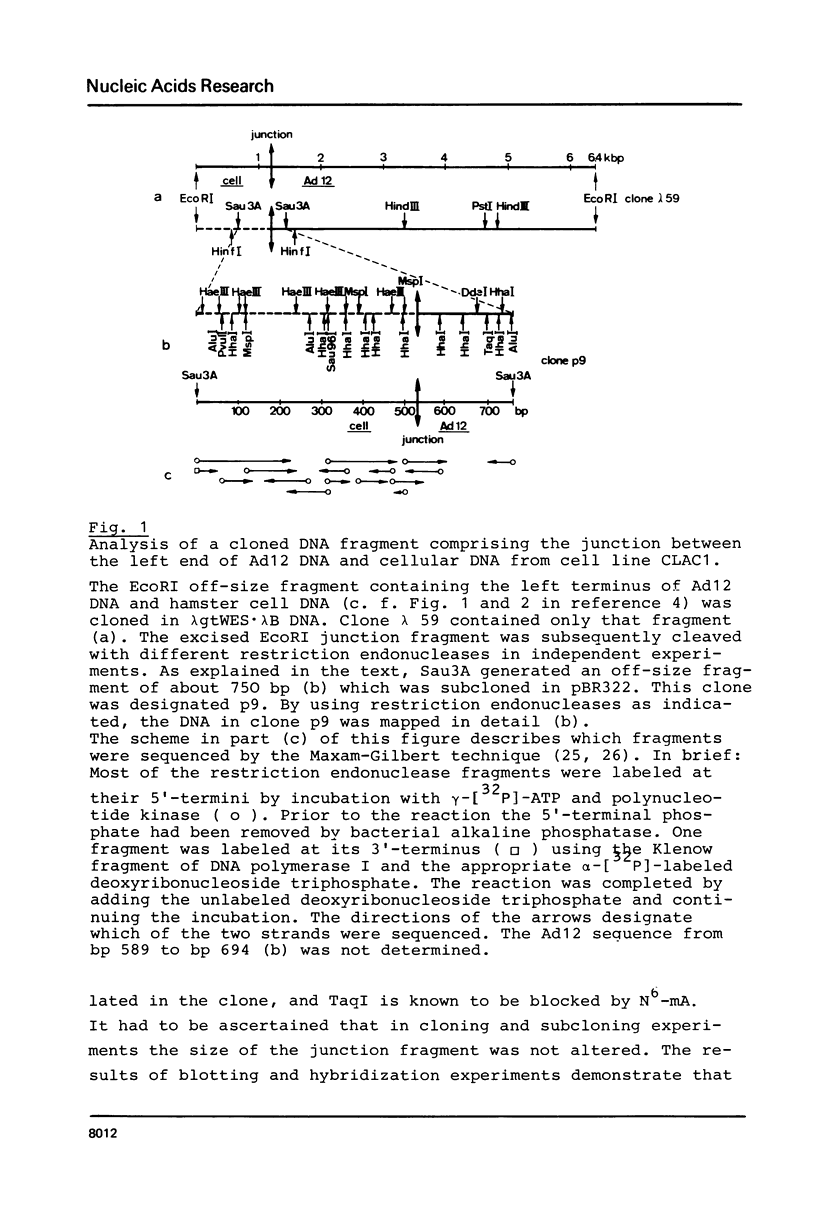
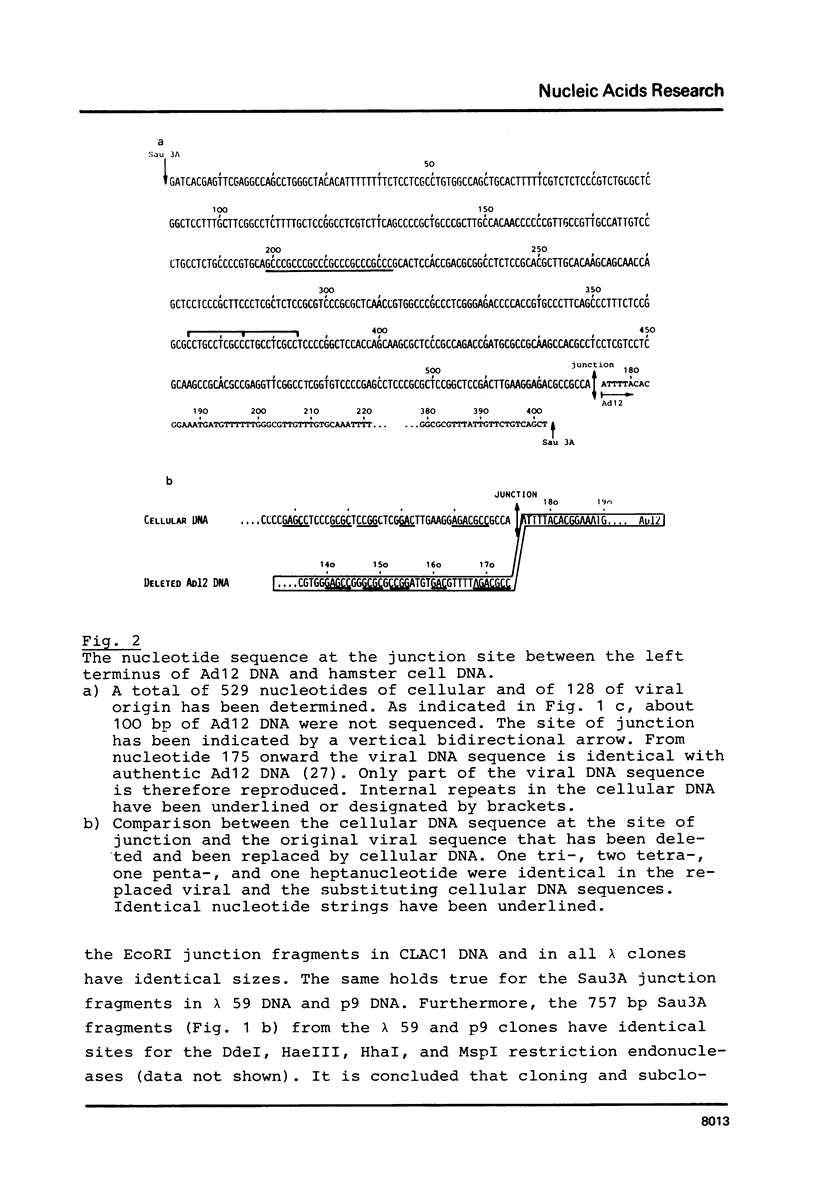
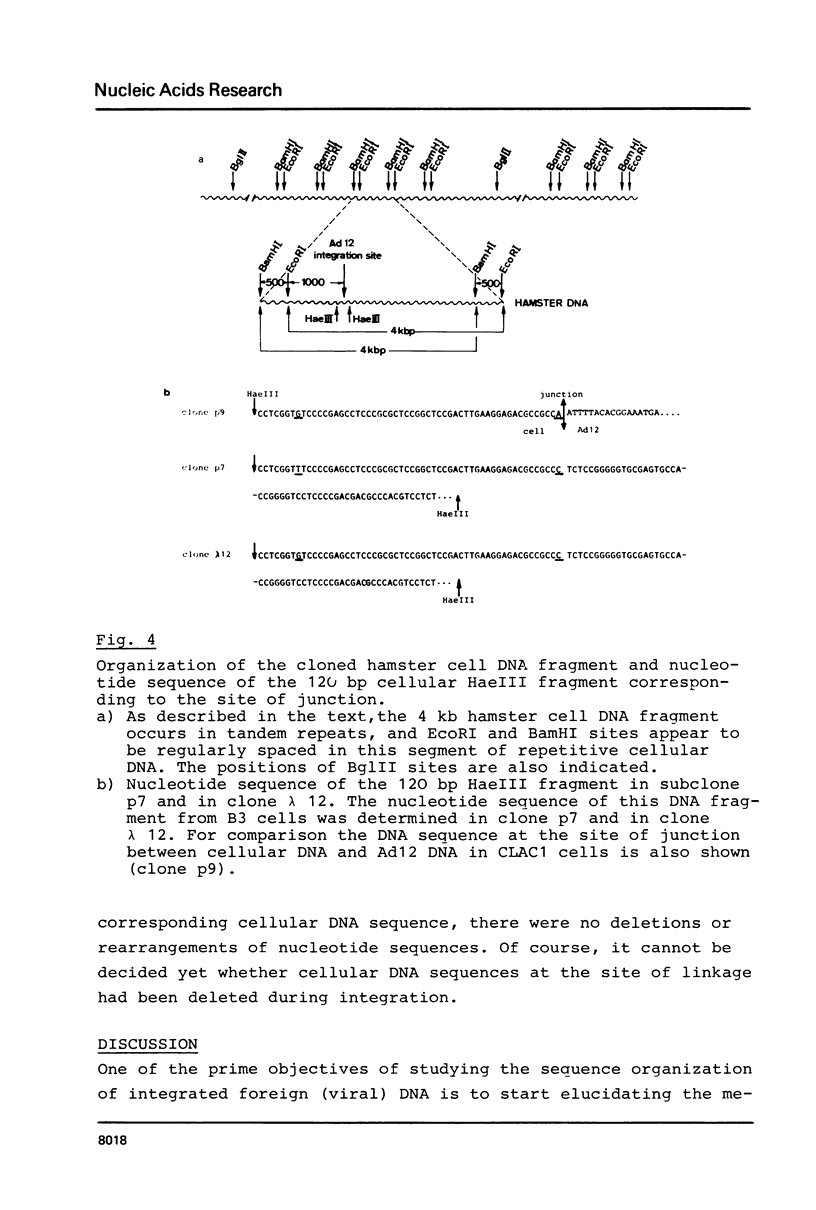
The hamster cell line CLAC1 originated from a tumor induced by injecting human adenovirus type 12 (Ad12) into newborn hamsters. Each cell contained about 12 copies of viral DNA colinearly integrated at two or three different sites. We have cloned and sequenced a DNA fragment comprising the site of junction between the left terminus of Ad12 DNA and cellular DNA. The first 174 nucleotides of Ad12 DNA were deleted at the site of junction. Within 40 nucleotides, there were one tri-, two tetra-, one penta-, and one heptanucleotide which were identical in the 174 deleted viral nucleotides and the cellular sequence replacing them. In addition, there were patch-type homologies ranging from octa- to decanucleotides between viral and cellular sequences. There is no evidence for a model assuming adenovirus DNA to integrate at identical cellular sites. The cellular DNA sequence corresponding to the junction fragment was cloned also from BHK21 (B3) hamster cells and sequenced. Up to the site of linkage with viral DNA, this middle repetitive cellular DNA sequence was almost identical with the equivalent sequence from CLAC1 hamster cells. Taken together with the results of previously published analyses (11, 12), the data suggest a model of viral (foreign) DNA integration by multiple short sequence homologies. Multiple sets of short patch homologies might be recognized as patterns in independent integration events. The model also accounts for the loss of terminal viral DNA sequences.
Full text
PDF










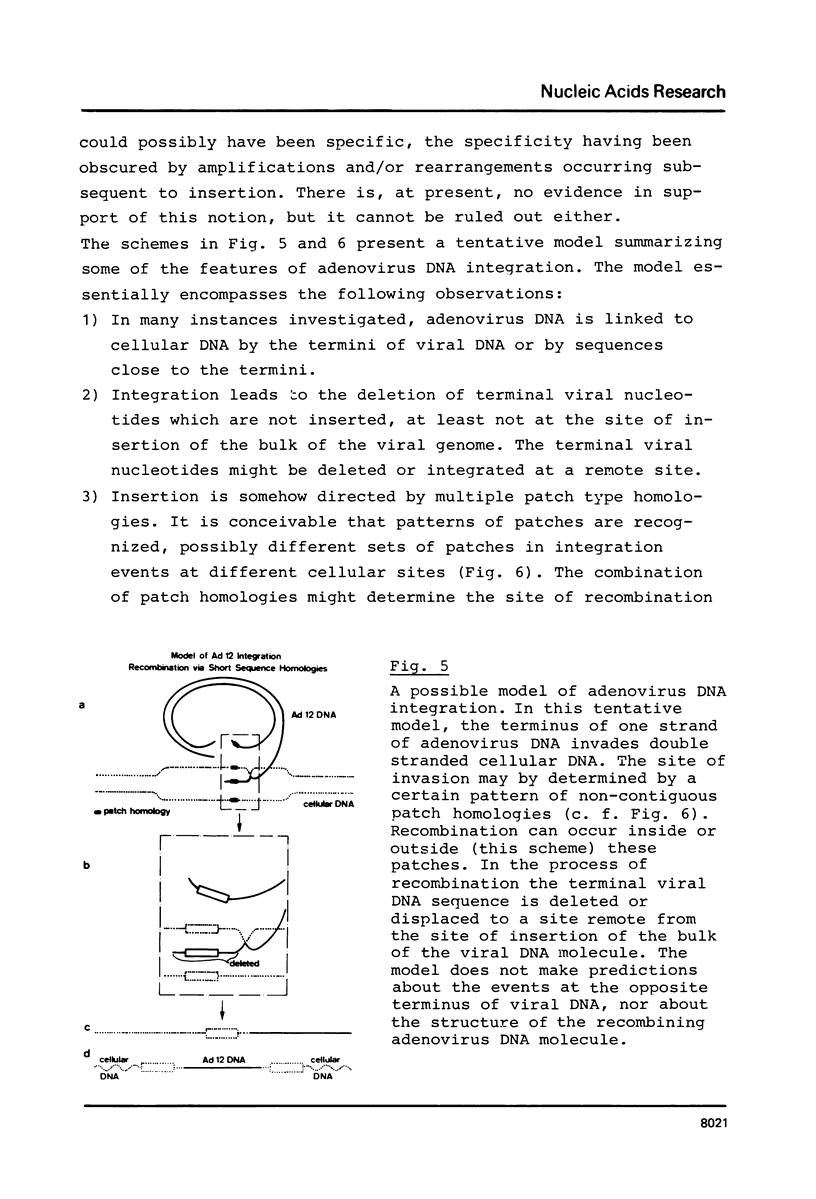
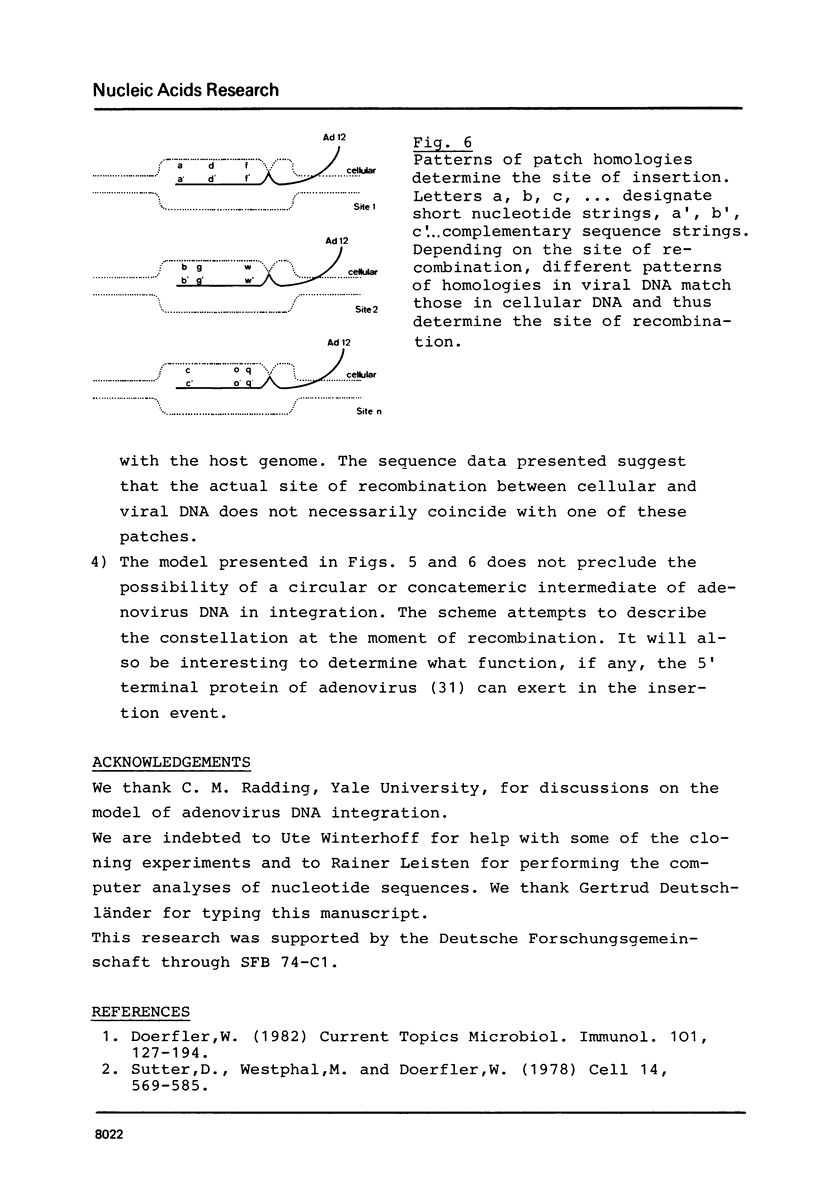

Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Benton W. D., Davis R. W. Screening lambdagt recombinant clones by hybridization to single plaques in situ. Science. 1977 Apr 8;196(4286):180–182. doi: 10.1126/science.322279. [DOI] [PubMed] [Google Scholar]
- Deuring R., Winterhoff U., Tamanoi F., Stabel S., Doerfler W. Site of linkage between adenovirus type 12 and cell DNAs in hamster tumour line CLAC3. Nature. 1981 Sep 3;293(5827):81–84. doi: 10.1038/293081a0. [DOI] [PubMed] [Google Scholar]
- Doerfler W., Lundholm U., Hirsch-Kauffmann M. Intracellular forms of adenovirus deoxyribonucleic acid. I. Evidence for a deoxyribonucleic acid-protein complex in baby hamster kidney cells infected with adenovirus type 12. J Virol. 1972 Feb;9(2):297–308. doi: 10.1128/jvi.9.2.297-308.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doerfler W. Uptake, fixation, and expression of foreign DNA in mammalian cells: the organization of integrated adenovirus DNA sequences. Curr Top Microbiol Immunol. 1982;101:127–194. doi: 10.1007/978-3-642-68654-2_6. [DOI] [PubMed] [Google Scholar]
- Eick D., Doerfler W. Integrated adenovirus type 12 DNA in the transformed hamster cell line T637: sequence arrangements at the termini of viral DNA and mode of amplification. J Virol. 1982 Apr;42(1):317–321. doi: 10.1128/jvi.42.1.317-321.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gahlmann R., Leisten R., Vardimon L., Doerfler W. Patch homologies and the integration of adenovirus DNA in mammalian cells. EMBO J. 1982;1(9):1101–1104. doi: 10.1002/j.1460-2075.1982.tb01303.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutai M. W., Nathans D. Evolutionary variants of simian virus 40: Cellular DNA sequences and sequences at recombinant joints of substituted variants. J Mol Biol. 1978 Dec 5;126(2):275–288. doi: 10.1016/0022-2836(78)90363-7. [DOI] [PubMed] [Google Scholar]
- Ibelgaufts H., Doerfler W., Scheidtmann K. H., Wechsler W. Adenovirus type 12-induced rat tumor cells of neuroepithelial origin: persistence and expression of the viral genome. J Virol. 1980 Jan;33(1):423–437. doi: 10.1128/jvi.33.1.423-437.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhlmann I., Achten S., Rudolph R., Doerfler W. Tumor induction by human adenovirus type 12 in hamsters: loss of the viral genome from adenovirus type 12-induced tumor cells is compatible with tumor formation. EMBO J. 1982;1(1):79–86. doi: 10.1002/j.1460-2075.1982.tb01128.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhlmann I., Doerfler W. Shifts in the extent and patterns of DNA methylation upon explanation and subcultivation of adenovirus type 12-induced hamster tumor cells. Virology. 1982 Apr 15;118(1):169–180. doi: 10.1016/0042-6822(82)90330-0. [DOI] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. A new method for sequencing DNA. Proc Natl Acad Sci U S A. 1977 Feb;74(2):560–564. doi: 10.1073/pnas.74.2.560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- Rigby P. W., Dieckmann M., Rhodes C., Berg P. Labeling deoxyribonucleic acid to high specific activity in vitro by nick translation with DNA polymerase I. J Mol Biol. 1977 Jun 15;113(1):237–251. doi: 10.1016/0022-2836(77)90052-3. [DOI] [PubMed] [Google Scholar]
- Robinson A. J., Younghusband H. B., Bellett A. J. A circula DNA-protein complex from adenoviruses. Virology. 1973 Nov;56(1):54–69. doi: 10.1016/0042-6822(73)90287-0. [DOI] [PubMed] [Google Scholar]
- Sambrook J., Greene R., Stringer J., Mitchison T., Hu S. L., Botchan M. Analysis of the sites of integration of viral DNA sequences in rat cells transformed by adenovirus 2 or SV40. Cold Spring Harb Symp Quant Biol. 1980;44(Pt 1):569–584. doi: 10.1101/sqb.1980.044.01.059. [DOI] [PubMed] [Google Scholar]
- Schmid C. W., Jelinek W. R. The Alu family of dispersed repetitive sequences. Science. 1982 Jun 4;216(4550):1065–1070. doi: 10.1126/science.6281889. [DOI] [PubMed] [Google Scholar]
- Simpson R. B., O'Hara P. J., Kwok W., Montoya A. L., Lichtenstein C., Gordon M. P., Nester E. W. DNA from the A6S/2 crown gall tumor contains scrambled Ti-plasmid sequences near its junctions with plant DNA. Cell. 1982 Jul;29(3):1005–1014. doi: 10.1016/0092-8674(82)90464-0. [DOI] [PubMed] [Google Scholar]
- Southern E. M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975 Nov 5;98(3):503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- Stabel S., Doerfler W., Friis R. R. Integration sites of adenovirus type 12 DNA in transformed hamster cells and hamster tumor cells. J Virol. 1980 Oct;36(1):22–40. doi: 10.1128/jvi.36.1.22-40.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starzinski-Powitz A., Schulz M., Esche H., Mukai N., Doerfler W. The adenovirus type 12 - mouse cell system: permissivity and analysis of integration patterns of viral DNA in tumor cells. EMBO J. 1982;1(4):493–497. doi: 10.1002/j.1460-2075.1982.tb01196.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stringer J. R. DNA sequence homology and chromosomal deletion at a site of SV40 DNA integration. Nature. 1982 Mar 25;296(5855):363–366. doi: 10.1038/296363a0. [DOI] [PubMed] [Google Scholar]
- Stringer J. R. Integrated simian virus 40 DNA: nucleotide sequences at cell-virus recombinant junctions. J Virol. 1981 May;38(2):671–679. doi: 10.1128/jvi.38.2.671-679.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugisaki H., Sugimoto K., Takanami M., Shiroki K., Saito I., Shimojo H., Sawada Y., Uemizu Y., Uesugi S., Fujinaga K. Structure and gene organization in the transformed Hind III-G fragment of Ad12. Cell. 1980 Jul;20(3):777–786. doi: 10.1016/0092-8674(80)90324-4. [DOI] [PubMed] [Google Scholar]
- Sutter D., Westphal M., Doerfler W. Patterns of integration of viral DNA sequences in the genomes of adenovirus type 12-transformed hamster cells. Cell. 1978 Jul;14(3):569–585. doi: 10.1016/0092-8674(78)90243-x. [DOI] [PubMed] [Google Scholar]
- Tiemer D., Enquist L., Leder P. Improved derivative of a phage lambda EK2 vector for cloning recombinant DNA. Nature. 1976 Oct 7;263(5577):526–527. doi: 10.1038/263526a0. [DOI] [PubMed] [Google Scholar]
- Vardimon L., Doerfler W. Patterns of integration of viral DNA in adenovirus type 2-transformed hamster cells. J Mol Biol. 1981 Apr 5;147(2):227–246. doi: 10.1016/0022-2836(81)90439-3. [DOI] [PubMed] [Google Scholar]
- Visser L., Reemst A. C., van Mansfeld A. D., Rozijn T. H. Nucleotide sequence analysis of the linked left and right hand terminal regions of adenovirus type 5 DNA present in the transformed rat cell line 5RK20. Nucleic Acids Res. 1982 Apr 10;10(7):2189–2198. doi: 10.1093/nar/10.7.2189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wahl G. M., Stern M., Stark G. R. Efficient transfer of large DNA fragments from agarose gels to diazobenzyloxymethyl-paper and rapid hybridization by using dextran sulfate. Proc Natl Acad Sci U S A. 1979 Aug;76(8):3683–3687. doi: 10.1073/pnas.76.8.3683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westin G., Visser L., Zabielski J., van Mansfeld A. D., Pettersson U., Rozijn T. H. Sequence organization of a viral DNA insertion present in the adenovirus-type-5-transformed hamster line BHK268-C31. Gene. 1982 Mar;17(3):263–270. doi: 10.1016/0378-1119(82)90142-1. [DOI] [PubMed] [Google Scholar]
- Yasue H., Ishibashi M. The oncogenicity of avian adenoviruses. III. In situ DNA hybridization of tumor line cells localized a large number of a virocellular sequence in few chromosomes. Virology. 1982 Jan 15;116(1):99–115. doi: 10.1016/0042-6822(82)90406-8. [DOI] [PubMed] [Google Scholar]