Abstract
The structures and interactions among the macromolecules in the enamel extracellular matrix play vital roles in regulating hydroxyapatite crystal nucleation, growth and maturation. We used dynamic light scattering, circular dichroism, fluorescence spectroscopy and transmission electron microscopy to investigate association of amelogenin and the 32-kDa enamelin, at physiological pH 7.4, in phosphate buffered saline (PBS). Amelogenin (rP148) self-assembly behavior was altered following addition of the 32-kDa enamelin. Dynamic light scattering revealed a trend of decrease in aggregate size in the solution following the addition of enamelin to amelogenin. A blue-shift and intensity increase of the ellipticity minima of rP148 in the circular dichroism spectra, upon the addition of the 32-kDa enamelin, suggest a direct interaction between the two proteins. In the fluorescence spectra, the maximum emission of rP148 was red-shifted from 335 to 341 nm with a marked intensity increase in the presence of enamelin as a result of complexation of the two proteins. In agreement with DLS data, TEM imaging showed that the 32-kDa enamelin dispersed the amelogenin aggregates into oligomeric particles and stabilizing them. Our study provides novel insights into understanding possible cooperation between enamelin and amelogenin in macromolecular co-assembly and in controlling enamel mineral formation.
Keywords: 32-kDa enamelin, assembly, circular dichroism, transmission electron microscopy, fluorescence spectroscopy
Enamel formation is a complex process regulated by ameloblasts that secrete amelogenin (1), ameloblastin (2), enamelin (3,4) and proteinses (5) in a highly controlled and precisely timed manner. All the above components have been shown to be critical for normal enamel formation and interactions between them might be a key to their function in controlling enamel mineral deposition (6–8). Experimental evidence has been accumulating for protein-protein interactions leading to amelogenin self-assembly and processing of the organic matrix. However, knowledge of specific interactions between the dominant protein, amelogenin, and non-amelogenin proteins (i.e. enamelin and ameloblastin) is still limited.
Enamelin is a minor constituent (<5%) of the extracellular matrix. It is the largest enamel protein and is rapidly degraded, following its secretion, into a number of intermediate proteolytic products, among which the 32-kDa enamelin is the most stable. The 32-kDa cleavage product accumulates throughout the developing enamel layer appearing in the inner secretory enamel (9–11). With two phosphorylated serines and three glycosylated asparagines, this hydrophilic and acidic glycoprotein has high affinity for calcium and apatite crystals (10). The 32-kDa enamelin cleavage product can be isolated from developing porcine enamel and has been used for structure-function studies (10–12).
Although the quantity of enamelin in the extracellular matrix of developing enamel is much lower than that of amelogenin, the lack of enamelin has a much more sever effect on enamel formation than does the lack of amelogenin (6, 7). In the absence of amelogenin, enamel is formed but is hypoplastic, and the prisms are disorganized (7). Without enamelin, enamel crystals do not form. Instead, only a pathologically mineralized layer is formed within a layer of accumulated enamel protein in the intercellular spaces between secretory ameloblasts (6). Mutations in the ENAM gene result in formation of abnormal enamel, especially the hypoplastic enamel seen in autosomal dominant amelogenesis imperfecta (AI) (13). Importantly, some mutations in the ENAM gene causing AI have been reported to be within the 32-kDa enamelin fragment (14).
We have recently reported that the 32-kDa enamelin, in cooperation with amelogenin, increased the length to width ratio (aspect ratio) of octacalcium phosphate (OCP) crystals (15). The presence of enamelin in the amelogenin “gel-like matrix” not only increased the aspect ratio of OCP but also enhanced the stability of the transient amorphous calcium phosphate (ACP) phase. Prior to this finding, we reported that enamelin promoted the kinetics of nucleation of apatite crystals in a dose dependent manner (16). We have further shown that enamelin undergoes a conformational change with a structural preference for ®-sheet with addition of its potential target, calcium ions (12). These in vitro studies have revealed that the 32-kDa enamelin has properties consistent with roles in crystal nucleation and regulation of crystal habit (6, 12, 15, 16). Based on the lectin-like properties of amelogenin, namely its affinity for to N-acetyl-D-glucosamine, amelogenin has the potential to bind to the 32-kDa enamelin through its glycosylated functional groups (11, 17). Indeed, our most recent investigations provide evidence that, at least in vitro, amelogenin and enamelin interact to possibly co-assemble (18).
Our recent biophysical studies focused on the pH conditions under which OCP crystal growth experiments are performed (i.e. pH 6.5) (19). In the present manuscript, we have expanded our studies to investigate the interactions between amelogenin and the 32-kDa enamelin at physiological pH 7.4 using phosphate buffered saline (PBS). We have used dynamic light scattering, circular dichroism, fluorescence spectroscopy and transmission electron microscopy to evaluate the effect of enamelin on amelogenin self-association under conditions that better represent the in vivo microenvironment.
Materials and Methods
Preparation of the 32-kDa enamelin and amelogenin (rP148)
The 32-kDa enamelin was extracted from the enamel using neutral buffer (25 mM PBS, pH 7.4), purified and isolated in 65% ammonium sulfate precipitation followed by RP-HPLC, and characterized following the method reported previously (12, 17). The purity and homogeneity of the fraction collected from RP-HPLC 10-mm C4 column was verified by a SDS gel and silver-staining (Fig. 1A). The pure 32-kDa enamelin fraction was divided into aliquots of 32μg/tube, lyophilized, and kept at 4 °C. The recombinant porcine amelogenin rP148, which represents amino acids 2–149 of the porcine amelogenin P173 and is an analogue to the major amelogenin proteolytic product (“20k”), was expressed in Escherichia coli, purified using RP-HPLC, and characterized as previously described (20). For all experiments lyophilized proteins were re-suspended in DDW to give the parent solutions of enamelin (0.32 mg/mL) and amelogenin (0.4 mg/mL) before preparing their mixtures.
Fig. 1.
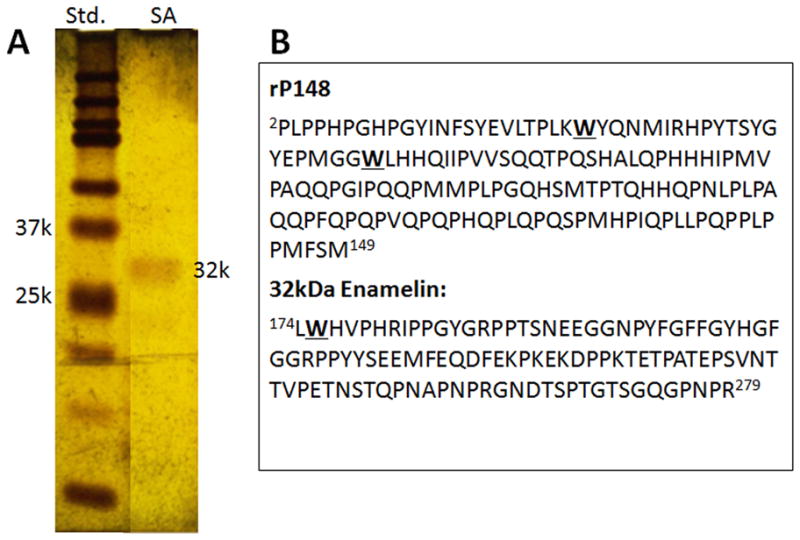
(A) Silver Stained SDS-PAGE of the 32-kDa enamelin fraction after purification with RP-HPLC (B) Amino acid sequence of recombinant rP148 and the 32-kDa enamelin highlighting the positions of Trp (W) residues.
Dynamic light scattering (DLS)
The association between the 32-kDa enamelin and amelogenin was investigated in 25mM PBS (pH 7.4, 0.15M NaCl) by DLS using a DynaPro NanoStar (Wyatt Technologies, Santa Barbara, CA, USA). The final concentrations of rP148 was 0.20 mg/mL and those of enamelin were 0.032 mg/mL, 0.0064 mg/mL, and 0.0032 mg/mL for 1:10, 1:50 and 1:100 enamelin:amelogenin molar ratios respectively. The mixtures were incubated at room temperature for 2 h before measurements and data were analyzed as previously described (12, 21).
Transmission electron microscopy
The effect of the 32-kDa enamelin on amelogenin aggregation or assembly was investigated using a JEOL 1400 TEM with voltage of 100 kV. The mixture of amelogenin with enamelin (final concentrations of 0.20 mg/mL of rP148 with 0.032 mg/mL of enamelin, for the 1:10 molar ratio) was incubated in 25 mM PBS (pH 6.5 or pH 7.4, 0.15 M NaCl) at room temperature for 2 h. The 300 mesh carbon-coated grids were submerged in 30 μl samples of enamelin only (after filtration with Anotop 10 plus 0.1 μm filters, Whatman, City?, England), amelogenin only, and the mixture solution for 30 s, followed by a 30 s immersion in 1% uranyl acetate, and air dried. The diameters of particles were analyzed and measured with ImageJ software.
Circular dichroism spectroscopy
Measurements were conducted on a JASCO J-810 spectropolarimeter calibrated using a 0.06% (+)-10-camphorsulfonic acid solution. The 32-kDa enamelin and the rP148 amelogenin were dissolved in 25 mM PBS buffer (pH 7.4, 0.15 M NaCl) and measurements taken after a 120-min incubation at room temperature. Final concentrations of samples were prepared as described for DLS measurements. The circular dichroism (CD) spectra were recorded at room temperature, as the average of 4 scans, in a 1 mm path-length quartz cell (300 μL) using a scanning speed of 50 nm/min, a time response of 1 s, a bandwidth of 1 nm. CD spectra were expressed as the mean residue ellipticity, [θ]mrw (deg cm2 dmol−1), calculated as previously described (21).
Fluorescence spectroscopy
Both amelogenin and the 32-kDa enamelin fragment have intrinsic fluorescence properties because of the presence of the amino acid tryptophan (Trp). The recombinant rP148 has two Trp residues at position 25 and 45, while the 32-kDa enamelin has only one Trp near the N-terminus (Fig. 1B). The 32-kDa enamelin and the rP148 amelogenin were dissolved in 25 mM PBS buffer (pH 7.4, 0.15 M NaCl) and measurements taken after a 120-min incubation at room temperature. Final concentrations of samples were prepared as described for dynamic light scattering (DLS) measurements. Fluorescence spectra were recorded on a QuantaMaster 4 Spectrofluorometer from PTI (Photon Technology International, Birmingham, NJ, USA). The acquisition interval and the integration time were maintained at 0.5 nm and 0.5 s, respectively. Fluorescence spectra were obtained by measuring the emission spectra in the range from 310 to 390 nm at excitation wavelength 295 nm, spaced by 0.5 nm intervals in the excitation domain. Fully corrected spectra were then concatenated into an excitation-emission matrix. Fluorescence intensities were plotted as a function of the excitation wavelength.
Results
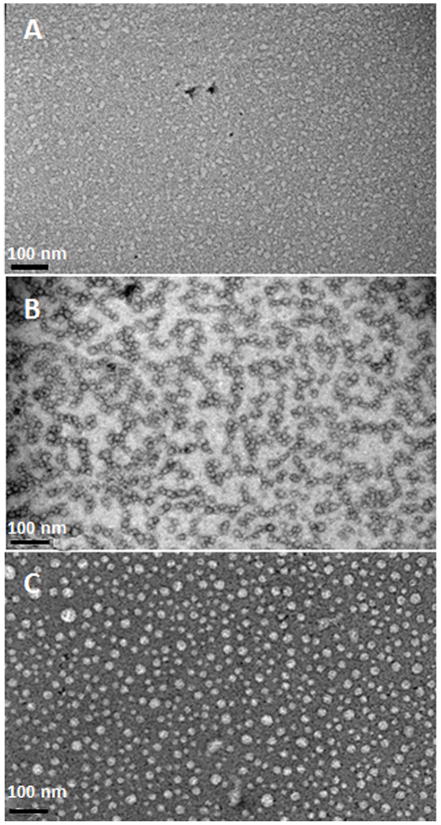
We first examined the effect of the 32-kDa enamelin on amelogenin assembly by using transmission electron microscopy (TEM). TEM images of the 32-kDa emamelin alone in PBS at pH 6.5 at concentration 32 μg/ml revealed irregularly-shaped and lightly-stained nanoparticles (Fig. 2A). Under the same conditions, rP148 alone formed spherical particles with an average particle size of 14.7 ± 3.3 nm (N = 50) in diameter with a tendency to form chain-like structures (Fig. 2B) (Table 1). With the addition of enamelin to the amelogenin solution at a 1:10 enamelin:amelogenin ratio, the size of the spherical particles grew to an average of 18.6 ± 4.2 nm (N = 50), and they remained dispersed without forming chain-like structures (Fig. 2C). The increase in rP148 particle sizes has also been demonstrated using dynamic light scattering (DLS) following addition of enamelin at acidic pH’s (5.5 and 6.5), where the rP148 was more soluble (18–19).
Fig. 2.
Representative TEM images of nanostructures detected in the 32-kDa enamelin and amelogenin solutions and their mixture, resulting from association of rP148 with enamelin in PBS buffer pH 6.5. (A) 32-kDa enamelin (0.032 mg/mL); (B) rP148 (0.2 mg/mL); (C) mixture of rP148 and enamelin at 10:1 ratio.
Table 1.
The average particle size distribution of enamelin, amelogenin and their complexes was estimated by ImageJ software. Particle sizes in the mixture of enamelin and amelogenin are significantly different that those of enamelin only or amelogenin only (p<0.0001)
| Diameters (nm) (average, N=50) | enamelin (0.032 mg/mL) | rP148 (0.2 mg/mL) | enamelin: rP148 (1:10) |
|---|---|---|---|
| pH 6.5 | 10.9 ± 2.3 | 14.7 ± 3.3 | 18.6 ± 4.2 |
| pH 7.4 | 13.2± 2.9 | 118.5 ± 30.2 | 34.7 ± 10.3 (parent particle*) 11.6 ± 3.7 (oligomeric particle*) |
Parent particle were the particles with the diameters size more than 20 nm; oligomeric particle were the particles with the diameters size less than 20 nm (25).
p = 5.7E-20, (pH6.5; rP148 compare with rP148-enamelin)
p = 5.6E-23, (pH6.5; enamelin compared with rP148-enamelin)
p = 5.3E-23, (pH7.4; rP148 compare with rP148-enamelin)
p = 5.8E-24, (pH7.4;enamelin compare with rP148-enamelin)
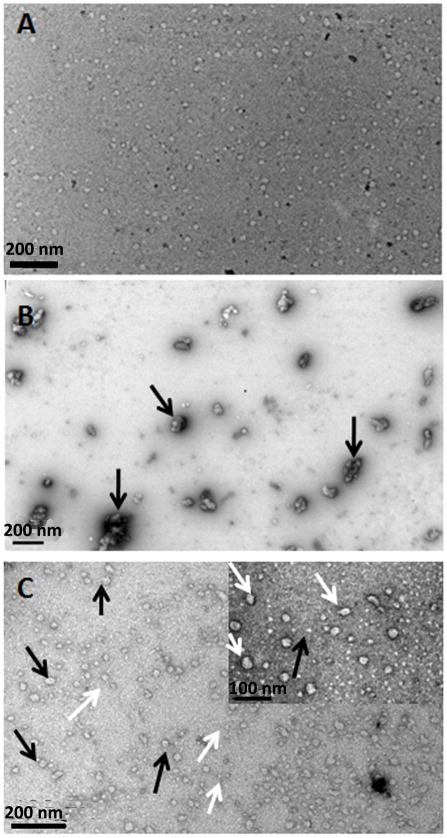
At pH 7.4, the lightly-stained nanoparticles formed by the 32-kDa enamelin had similar size to those formed at pH 6.5 but appeared to be more regularly-shaped (spherical) (Fig. 3A). At this neutral pH rP148 had a tendency to aggregate into large particles ~118.5 ± 30.2 nm in diameter (N = 50) (black arrows in Fig 3B, Table 1). With the addition of enamelin to amelogenin solution at a 1:10 enamelin:amelogenin ratio, the large aggregates dispersed and two populations of particles were observed. The population of larger particles had a diameter of 34.7 ± 10.3 nm (N = 50, counting particles with diameters > 20 nm, black arrows on Fig. 3C, Table 1). These had a tendency to break apart to form a second population of oligomeric nanoparticles with diameters of 11.6 ± 3.7 nm (white arrow, Fig. 3C) (N = 50, counting particles with diameters < 20 nm, white arrows Fig. 3C, Table 1).
Fig. 3.
Representative TEM images of nanostructures detected in the 32-kDa enamelin and amelogenin solutions and their mixture, resulting from association of rP148 with enamelin in PBS buffer pH 7.4. (A) 32-kDa enamelin (0.032 mg/mL); (B) rP148 (0.2 mg/mL); (C) mixture of rP148 and enamelin at 10:1 ratio. Inset is a higher magnification.
The ability of enamelin to break up the large amelogenin aggregates formed at pH 7.4, and stabilize smaller oligomeric particles was also verified by DLS analysis, which provided the estimated hydrodynamic radii (RH), molecular weight (MW), and mass distribution of particles in the rP148-enamelin solutions at pH 7.4 (Table 2). In these experiments, the size of the large aggregates decreased with the addition of enamelin in a dose dependent manner. The rP148 amelogenin particles formed in the absence of enamelin in solution had a predominant population with a mean RH of 673.5 ± 67.4 nm and no minor populations of smaller particles were detected. When the 32-kDa enamelin was added to rP148 at a molar ratio of 1:100, the size of the predominant aggregated particles in the rP148-enamelin solution slightly decreased to 657.9 ± 70 nm while its mass percentage decreased. At the same time, a minor population (2.1% mass percentage) of oligomeric particles with RH = 7.2 ± 0 nm appeared. At a higher 1:50 ratio, the average RH of the aggregates in the major population decreased to 522.6 ± 77 nm and their mass percentage was further reduced, while the mass percentage of the minor population with RH of 5.7 ± 0.5 nm increased to 16.4%. Remarkably, at the highest ratio of enamelin to rP148 used in our study (1:10), the percentage of small oligomeric particles (now RH = 4.5 nm) increased drastically (78.1%) at the expense of the large aggregates, which decreased to 21.6%. The particles were 356.9 nm in radius at the 1:10 enamelin: amelogenin ratio.
Table 2.
Hydrodynamic radii (RH), molecular weight (MW), and mass distribution of particles in the enamelin-rP148 solutions (rP148, 0.20 mg/ml, 25mM PBS, 0.15M NaCl, at room temperature)
| enamelin: rP148 | RH (nm) | MW (kDa) | Mass (%) |
|---|---|---|---|
| rP148 | 673.5 ± 67.4 | 13960300 | 100.0 |
| - | - | - | |
| 1: 100 | 7.2 ± 0 | 342 | 2.1 |
| 657.9 ± 70 | 13688900 | 97.9 | |
| 1:50 | 5.7 ± 0.5 | 196 | 16.4 |
| 522.6 ± 77 | 7711720 | 83.6 | |
| 1:10 | 4.5 ± 0.1 | 112 | 78.1 |
| 51.3 ± 3 | 33739 | 0.3 | |
| 356.9 ± 48 | 3158770 | 21.6 |
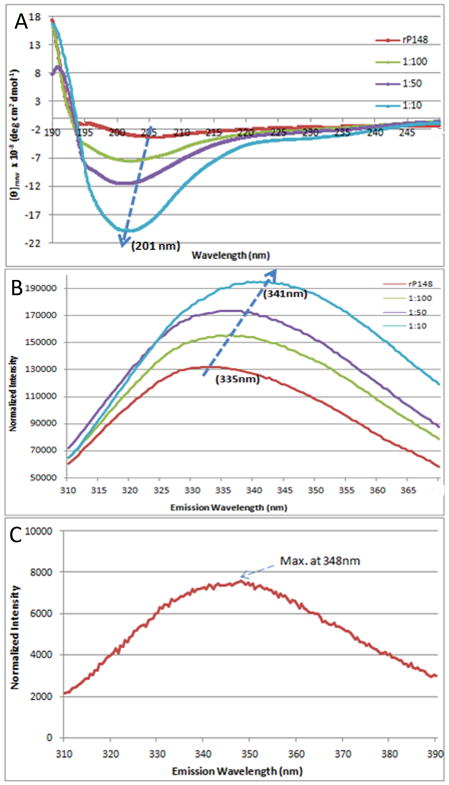
The circuklar dichroism (CD) spectrum of rP148 amelogenin showed a negative ellipticity with a minimum [θ]mrw of −3.75×103 deg cm2 dmol−1 at 205 nm, a characteristic of an unordered polyproline type II structure (22). Upon addition of enamelin to rP148 at molar ratios of 1:100, 1:50, and 1:10, respectively, the intensity of the minimum significantly increased in a dose dependent manner. We subtracted enamelin CD spectra with concentrations equivalent to those in the above ratios from that of the mixed solution and found that the minimum [θ]mrw increased to −20.3×103 deg cm2 dmol−1 in the 1:10 (enamelin:rP148) solution (Fig. 4A). At the same time, the trough slightly shifted from 205 nm to 201 nm upon addition of enamelin to rP148 at molar ratios of 1:100, 1:50, and 1:10.
Fig. 4.
Biophysical studies of the interaction between rP148 and the 32-kDa enamelin at pH 7.4. (A) CD spectra of rP148 in association with the 32-kDa enamelin, (B) and (C) Fluorescence spectra of the rP148/enamelin solutions (B) and that of the 32-kDa enamelin (C). rP148 final concentration was 0.2 mg/ml and those of enamelin were 0.032 mg/mL, 0.0064 mg/mL, and 0.0032 mg/mL for 1:10, 1:50 and 1:100 enamelin:amelogenin molar ratios respectively.
The presence of the intrinsically fluorescent amino acid Trp residues in the N-terminal region of recombinant rP148 allowed us to follow the environment around these Trp residues following the addition of enamelin to the solution. The emission maxima in the fluorescence spectra of the 32-kDa enamelin and the rP148 amelogenin are 348 nm and 335 nm, respectively (Figs. 4B,C). Upon addition of enamelin to the rP148 solution at increasing ratios (1:100, 1:50, and 1:10 of enamelin to rP148), the emission maximum of the solution was red-shifted progressively from 335 to 341 nm. The intensity of the maxima also increased significantly corresponding to the amount of enamelin added (Fig. 4B).
Discussion
The evolutionary analyses of mammalian enamelin have revealed conservation in the primary sequence, as well as the phosphorylation and glycosylation positions in the 32-kDa enamelin, supporting critical functional rules for this acidic fragment in all species studied (23). We have recently reported that the 32-kDa enamelin and rP148 amelogenin have a cooperative effect on regulating the morphology of OCP crystals grown at pH 6.5 in a cation selective membrane system (15). It is noteworthy that the presence of enamelin was critical for the control of OCP crystal orientation and stabilization of the transient ACP phase (15). An amelogenin-enamelin interaction has been also demonstrated at pH 6.5, based on a trend of increase in amelogenin oligomers sizes, changes in CD ellipticity minima, and a slight shift in the maximum emission in the fluorescence spectra following addition of enamelin to the amelogenin solution (19). In the present paper, using TEM, we show an increase in the size of amelogenin oligomeric particles following addition of enamelin at pH 6.5. We further investigated association between these two enamel proteins under conditions that better resembles the physiological microenvironment (pH 7.4 and phosphate buffer).
The effect of enamelin on amelogenin assembly/aggregation at pH 7.4, reported in the present study, appeared to be more profound than that at pH 6.5 reported recently (19). When compared to pH 6.5, at pH 7.4 the solubility of rP148 is relatively low and the protein has a tendency to form large aggregates of more than a micron in diameter. Dynamic light scattering (DLS) analysis showed that the size of the aggregates and their mass percentages decreased in a dose dependent manner when enamelin was added to the solution. Interestingly, a growing mass of a minor population of oligomers started to appear simultaneously. The increase in mass percentage of the minor population was presumably due to the formation of complexes between enamelin and rP148 molecules leading to stabilization of oligomeric particles. Enamelin exhibited a similar effect when breaking up rP172 amelogenin nanospheres formed at pH 8, stabilizing oligomeric particles in Tris-HCl buffer (18). Here, we have also demonstrated by TEM a trend of decreasing amelogenin aggregate sizes and increasing mass of the small oligomeric particles in PBS pH 7.4.
The changes in the CD and fluorescence spectra of amelogenin-enamelin mixtures clearly illustrates that there is a direct interaction between rP148 amelogenin and enamelin. Interestingly, the shift of emission maxima and the changes in intensity of fluorescence spectra that resulted from addition of enamelin to amelogenin solutions at pH 6.5 and 7.4 were in opposite directions. In both cases the changes are interpreted to suggest close association and possible co-assembly between these two enamel proteins.
At pH 7.4, addition of enamelin caused a significant red-shift (from λmax = 335 to 341) in the fluorescence spectra, while at pH 6.5 the emission maximum was slightly blue-shifted (λmax = 335 to 333). Five discrete states of Trp in proteins, based on the emission maxima, have been defined for folded and unfolded proteins, that depend on the polarity of the environment of the Trp residue, dictated either by the solvent or the surrounding residues (24). It is noteworthy that porcine amelogenin rP172 monomers stabilized at low pH (3.8) in water have their emission maxima at λmax = 347-346 while amelogenin oligomers stabilized at pH 5.5 have their emission maxima slightly blue-shifted at λmax = 345-343 (25). This emission maximum is close to the maximum observed for the amelogenin-enamelin complex at 1:10 ratio formed at pH 7.4 (Fig 4B). Addition of enamelin to amelogenin at pH 7.4 disintegrates the aggregates, promotes the formation of oligomers and apparently causes exposure of the Trp residues. The tryptophans in the N-terminal region of rP148 could be exposed at the surface of the oligomers and consequently give rise to the increasing intensity and the red-shift of the maximum in the fluorescence spectra of the rP148/enamelin solution. On the other hand, when enamelin is added to amelogenin oligomers formed at pH 6.5, a blue shift is observed, presumably as the result of close association between enamelin and amelogenin making the environment around the N-terminal Trp become more hydrophobic (19).
The increase in ellipticity in the CD spectra of amelogenin following addition of enamelin was more significant at pH 7.4 compared to pH 6.5. The CD trough slightly shifted from 205 nm to 201 nm with the increased ratio of enamelin, indicating a possible change in the conformation of amelogenin.
Our data collectively support the notion that amelognein self-association is affected by addition of enamelin. We propose that such interactions may function to prevent unwanted aggregation of the truncated amelogenin in vivo, stabilize amelogenin oligomers, enhance the hydrophilicity of the amelogenin oligomers, and affect their interactions with the mineral phase. Amelogenin may also function as a chaperone for enamelin. Amelogenin-enamelin interactions may well be affected by local pH changes occurring during enamel maturation. We propose that during the post-secretory stage of enamel formation, amelogenin and enamelin cooperate synchronically to control crystal growth in vivo. Investigations of the role of amelogenin-enamelin association in the extracellular matrix in vivo, however, remains to be explored.
Acknowledgments
This study was supported by NIH-NIDCR R01 grants DE-13414 and DE-15644 to JMO. We thank the Nanobiophysics Core Facility at the University of Southern California for use of the CD and fluorescence spectrometers, and Prof. Ralf Langen and the members of his laboratory for assistance in the use of the transmission electron microscope. We thank Dr. Keith M. Bromley and Dr. Sowmya Lokappa for fruitful discussion and assistance in protein purification.
Footnotes
Confllicts of interest. The authors declare no conflicts of interest.
References
- 1.SNEAD ML, LAU EC, ZEICHNER-DAVID M, FINCHAM AG, WOO SL, SLAVKIN HC. DNA sequence for cloned cDNA for murine amelogenin reveal the amino acid sequence for enamel-specific protein. Biochem Biophys Res Commun. 1985;129:812–818. doi: 10.1016/0006-291x(85)91964-3. [DOI] [PubMed] [Google Scholar]
- 2.KREBSBACH PH, LEE SK, MATSUKI Y, KOZAK CA, YAMADA KM, YAMADA Y. Full-length sequence, localization, and chromosomal mapping of ameloblastin. A novel tooth-specific gene. J Biol Chem. 1996;271:4431–4435. doi: 10.1074/jbc.271.8.4431. [DOI] [PubMed] [Google Scholar]
- 3.HU JC, ZHANG CH, YANG Y, KARRMAN-MARDH C, FORSMAN-SEMB K, SIMMER JP. Cloning and characterization of the mouse and human enamelin genes. J Dent Res. 2001;80:898–902. doi: 10.1177/00220345010800031001. [DOI] [PubMed] [Google Scholar]
- 4.HU CC, YAMAKOSHI Y. Enamelin and autosomal-dominant amelogenesis imperfecta. Crit Rev Oral Biol Med. 2003;14:387–398. doi: 10.1177/154411130301400602. [DOI] [PubMed] [Google Scholar]
- 5.BARTLETT JD, SIMMER JP. Proteinases in developing dental enamel. Crit Rev Oral Biol Med. 1999;10:425–441. doi: 10.1177/10454411990100040101. [DOI] [PubMed] [Google Scholar]
- 6.HU CC, SMITH Y, MCKEE MD, WRIGHT JT, YAMAKOSHI Y, PAPAGERAKIS P, HUNTER GK, FENG JQ, SIMMER JP. Enamel defects and ameloblast-specific expression in Enam knockout/lacZ knock-in mice. J Biol Chem. 2008;283:10858–10871. doi: 10.1074/jbc.M710565200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.GIBSON CW, YUAN ZA, HALL B, LONGENECKER G, CHEN E, THYAGARAJAN T, SREENATH T, WRIGHT JT, DECKER S, PIDDINGTON R, HARRISON G, KULKARNI AB. Amelogenin-deficient mice display an amelogenesis imperfecta phenotype. J Biol Chem. 2001;276:31871–31875. doi: 10.1074/jbc.M104624200. [DOI] [PubMed] [Google Scholar]
- 8.FUKUMOTO S, KIBA T, HALL B, IEHARA N, NAKAMURA T, LONGENECKER G, KREBSBACH PH, NANCI A, KULKARNI AB, YAMADA Y. Ameloblastin is a cell adhesion molecule required for maintaining the differentiation state of ameloblasts. J Cell Biol. 2004;167:973–983. doi: 10.1083/jcb.200409077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.TANABE T, AOBA T, MORENO EC, FUKAE M, SHIMUZU M. Properties of phosphorylated 32 kd nonamelogenin proteins isolated from porcine secretory enamel. Calcif Tissue Int. 1990;46:205–215. doi: 10.1007/BF02555046. [DOI] [PubMed] [Google Scholar]
- 10.YAMAKOSHI Y, PINHEIRO FH, TANABE T, FUKAE M, SHIMIZU M. Sites of asparagine-linked oligosaccharides in porcine 32-kDa enamelin. Connect Tissue Res. 1998;9:39–46. doi: 10.3109/03008209809023910. discussion 63–67. [DOI] [PubMed] [Google Scholar]
- 11.YAMAKOSHI Y. Carbohydrate moieties of porcine 32-kDa enamelin. Calcif Tissue Int. 1995;56:323–330. doi: 10.1007/BF00318054. [DOI] [PubMed] [Google Scholar]
- 12.FAN D, LAKSHMINARAYANAN R, MORADIAN-OLDAK J. The 32-kDa enamelin undergoes conformational transitions upon calcium binding. J Struct Biol. 2008;163:109–115. doi: 10.1016/j.jsb.2008.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.KIDA M, SAKIYAMA Y, MATSUDA A, TAKABAYASHI S, OCHI H, SEKIGUCHI H, MINAMITAKE S, ARIGA T. A novel missense mutation (p. P52R) in amelogenin gene causing X-linked amelogenesis imperfecta. J Dent Res. 2007;86:69–72. doi: 10.1177/154405910708600111. [DOI] [PubMed] [Google Scholar]
- 14.GUTIERREZ SJ, CHAVES M, TORRES DM, BRICEÑO I. Identification of a novel mutation in the enamalin gene in a family with autosomal-dominant amelogenesis imperfecta. Arch Oral Biol. 2007;52:503–506. doi: 10.1016/j.archoralbio.2006.09.014. [DOI] [PubMed] [Google Scholar]
- 15.IIJIMA M, FAN D, BROMLEY KM, SUN Z, MORADIAN-OLDAK J. Tooth enamel proteins enamelin and amelogenin cooperate to regulate the growth morphology of octacacium phosphate crystals. Crystal Growth Des. 2010;10:4815–4822. doi: 10.1021/cg100696r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.BOUROPOOULOS N, MORADIAN-OLDAK J. Induction of apatite by the cooperative effect of amelogenin and the 32-kDa enamelin. J Dent Res. 2004;83:278–282. doi: 10.1177/154405910408300402. [DOI] [PubMed] [Google Scholar]
- 17.RAVINDRANATH RH, MORADIAN-OLDAK J, FINCHAM AG. Tyrosyl motif in amelogenins binds N-acetyl-D-glucosamine. J Biol Chem. 1999;274:2464–2471. doi: 10.1074/jbc.274.4.2464. [DOI] [PubMed] [Google Scholar]
- 18.FAN D, DU C, SUN Z, LAKSHMINARAYANAN R, MORADIAN-OLDAK J. In vitro study on the interaction between the 32-kDa enamelin and amelogenin. J Struct Biol. 2009;166:88–94. doi: 10.1016/j.jsb.2009.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.FAN D, IIJIMA M, BROMLEY KM, YANG X, MATHEW S, MORADIAN-OLDAK J. The cooperation of enamelin and amelogenin in controlling octacalcium phosphate crystal morphology. Cells Tissues Organs. 2011 Apr 28; doi: 10.1159/000324208. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.SUN Z, AHSAN MM, WANG HC, DU C, ABBOTT, MORADIAN-OLDAK J. Assembly and processing of an engineered amelogenin proteolytic product (rP148) Eur J Oral Sci. 2006;114(Suppl 1):59–63. doi: 10.1111/j.1600-0722.2006.00296.x. (discussion 93–95, 379–380) [DOI] [PubMed] [Google Scholar]
- 21.LAKSHMINARAYANAN R, BROMLEY KM, LEI YP, SNEAD ML, MORADIAN-OLDAK J. Perturbed amelogenin secondary structure leads to uncontrolled aggregation in amelogenesis imperfecta mutant proteins. J Biol Chem. 2011;285:40593–40603. doi: 10.1074/jbc.M110.131136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.LAKSHMINARAYANAN R, FAN D, DU C, MORADIAN-OLDAK J. The role of secondary structure in the entropically driven amelogenin self-assembly. Biophys J. 2007;93:3664–3674. doi: 10.1529/biophysj.107.113936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.AL-HASHIMI N, SIRE JY, DELGADO S. Evolutionary analysis of mammalian enamelin, the largest enamel protein, supports a crucial role for the 32-kDa peptide and reveals selective adaptation in rodents and primates. J Mol Evol. 2009;69:635–656. doi: 10.1007/s00239-009-9302-x. [DOI] [PubMed] [Google Scholar]
- 24.RESHETNYAK YK, BURSTEIN EA. Decomposition of protein tryptophan fluorescence spectra into log-normal components. II. The statistical proof of discreteness of tryptophan classes in proteins. Biophys J. 2001;81:1710–1734. doi: 10.1016/S0006-3495(01)75824-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.BROMLEY KM, KISS AS, LOKAPPA SB, LAKSMINARAYANAN R, FAN D, NDAO M, EVANS JS, MORADIAN-OLDAK J. Characterization of amelogenin oligomers as functional components of mineralizing enamel matrix. J Biol Chem. 2001 Aug 11; Epub ahead of print. [Google Scholar]
- 26.YANG X, WANG L, QIN Y, SUN Z, HENNEMAN ZJ, MORADIAN-OLDAK J, NANCOLLAS GH. How Amelogenin Orchestrates the Organization of Hierarchical Elongated Microstructures of Apatite. J Phys Chem B. 2010;114:2293–2300. doi: 10.1021/jp910219s. [DOI] [PMC free article] [PubMed] [Google Scholar]