Abstract
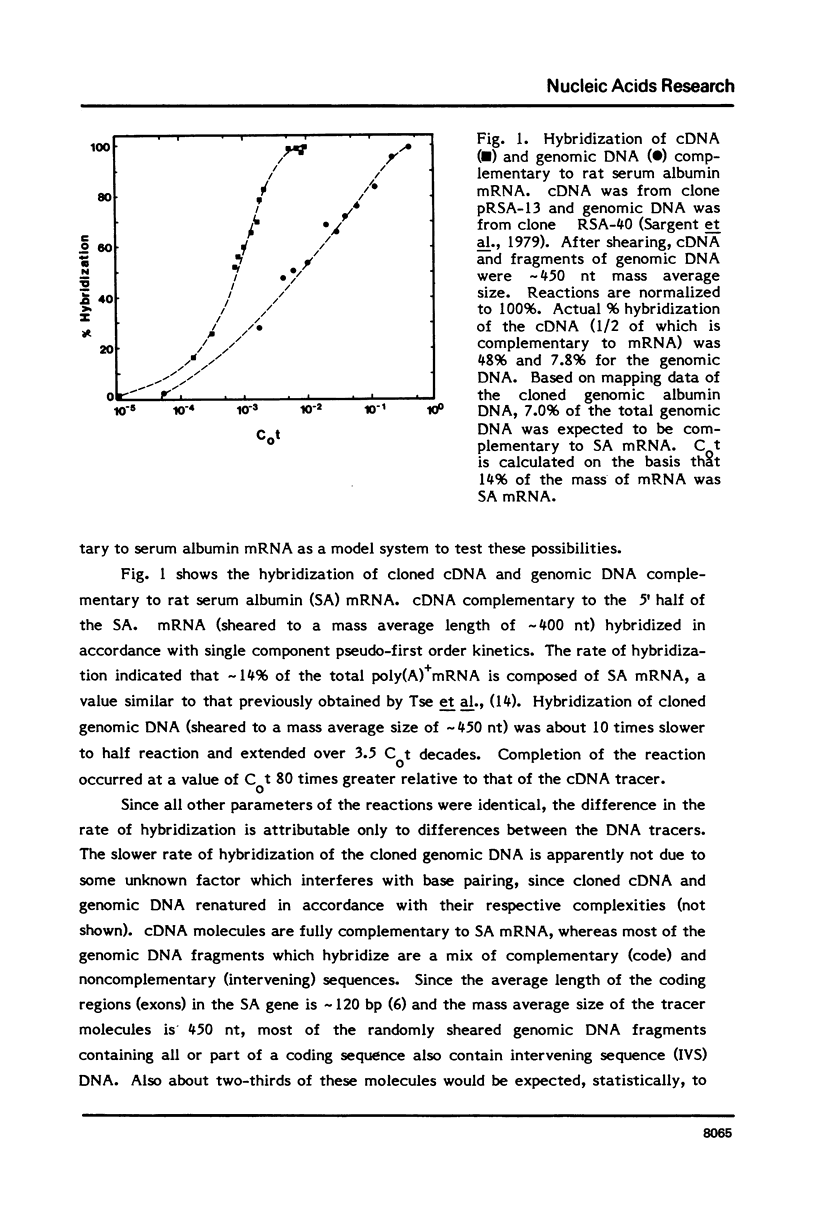
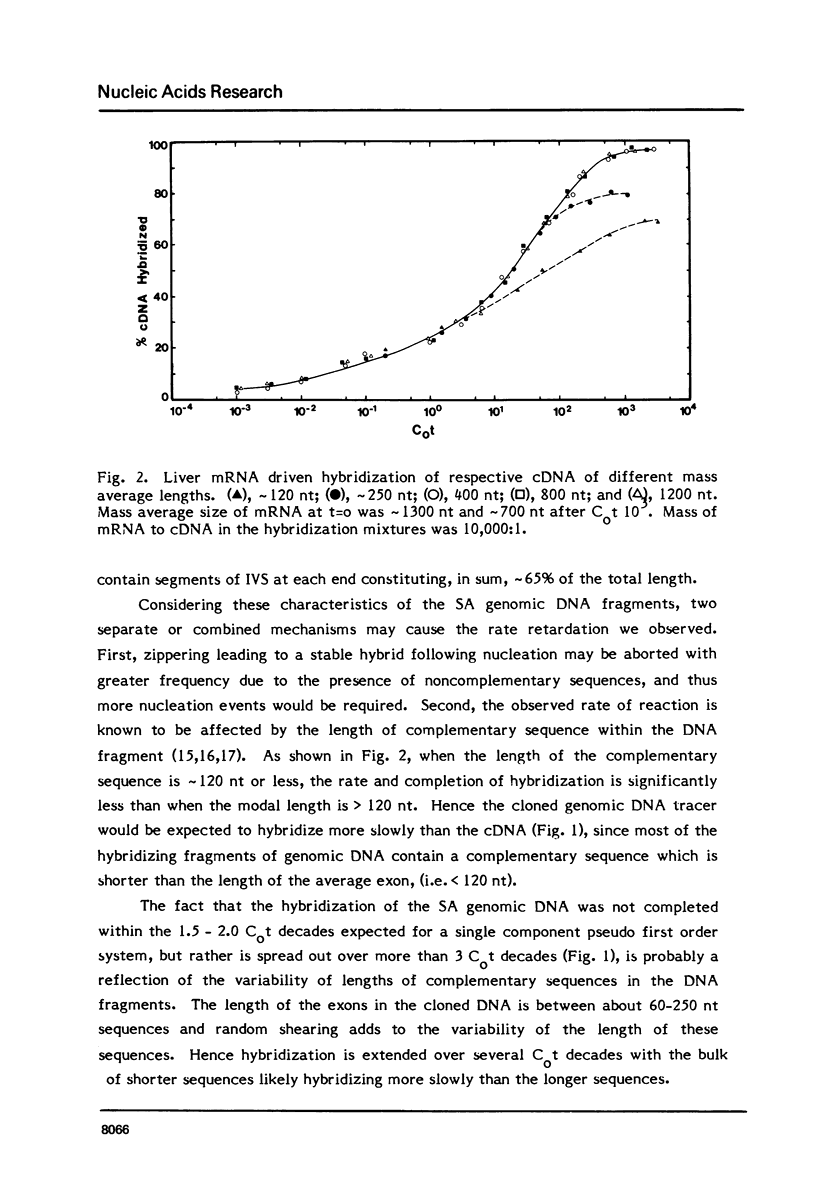
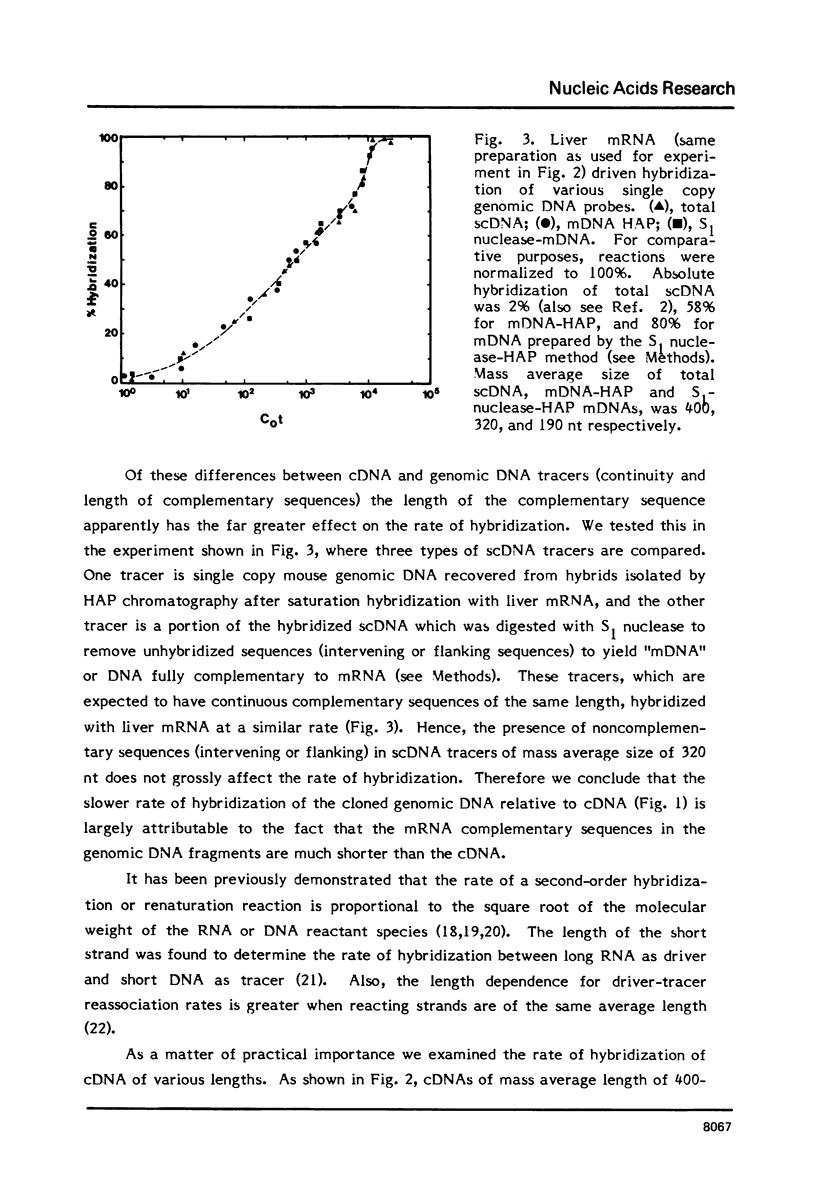
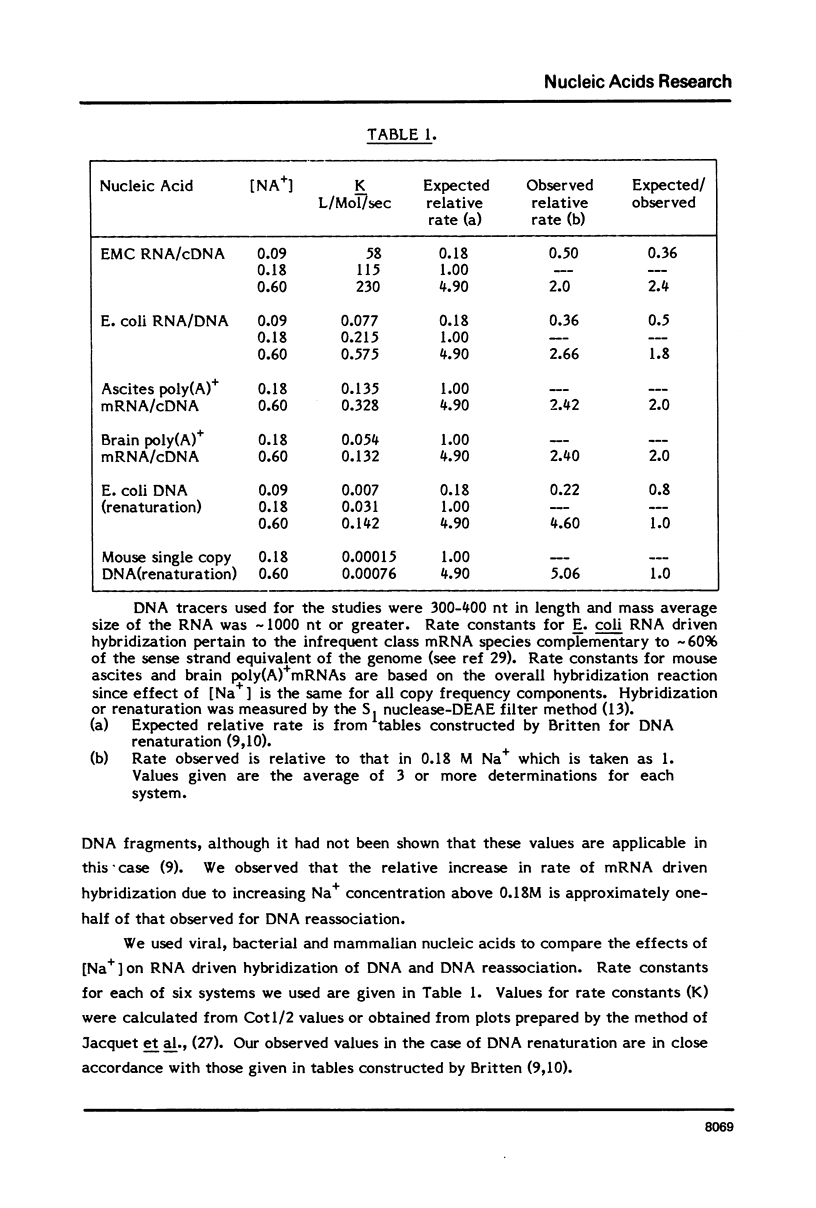
Differences in the RNA-driven hybridization kinetics of genomic DNA and cDNA probes led us to examine physical parameters affecting these reactions. Cloned cDNA complementary to serum albumin (SA) mRNA hybridized in accordance with single component kinetics, whereas cloned SA genomic DNA hybridized more slowly and with multiple component kinetics. This difference is largely attributable to the relatively short and variable lengths of the mRNA complementary regions in the cloned genomic DNA. The rate of mRNA driven hybridization is affected to about half the extent observed for DNA renaturation as Na+ is increased or decreased from 0.18M. In the annealing of nucleic acids of high sequence complexity, after approximately 70% of reaction has been reached, the rate of the reaction is slowed and completion is not reached under "static" conditions. In practical terms, this is not the case for systems of low sequence complexity. This problem can be largely overcome by continuous or frequent mixing of the reactants, so that complex cDNA probes are hybridized essentially to completion, and kinetics can therefore be more readily compared to simple complexity standards.
Full text
PDF





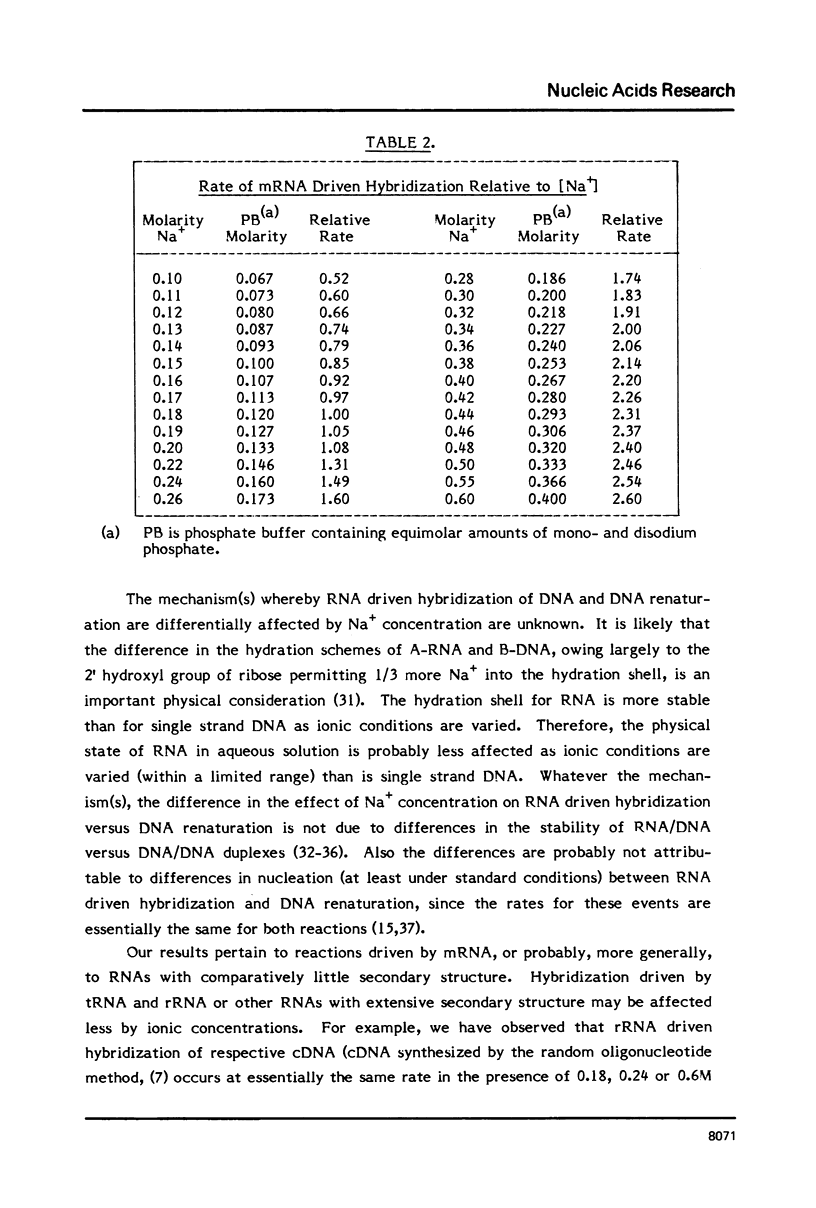

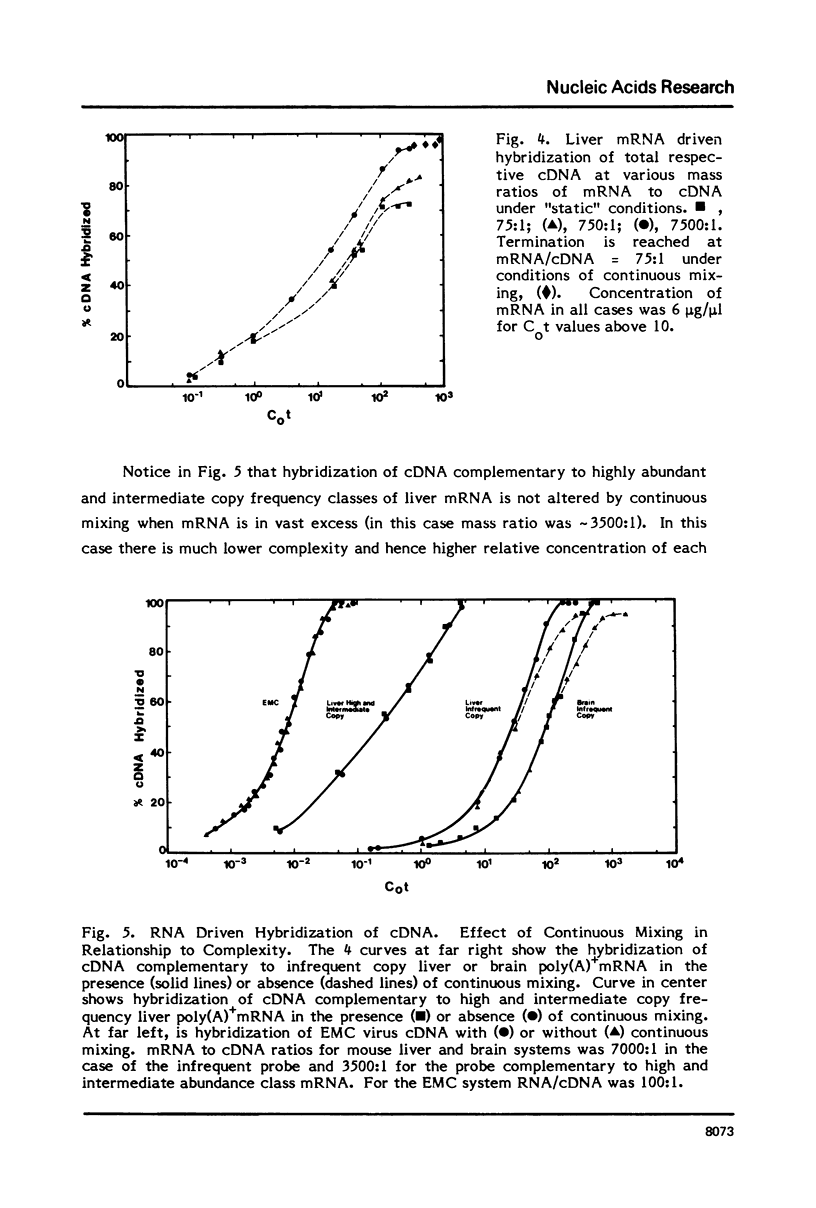
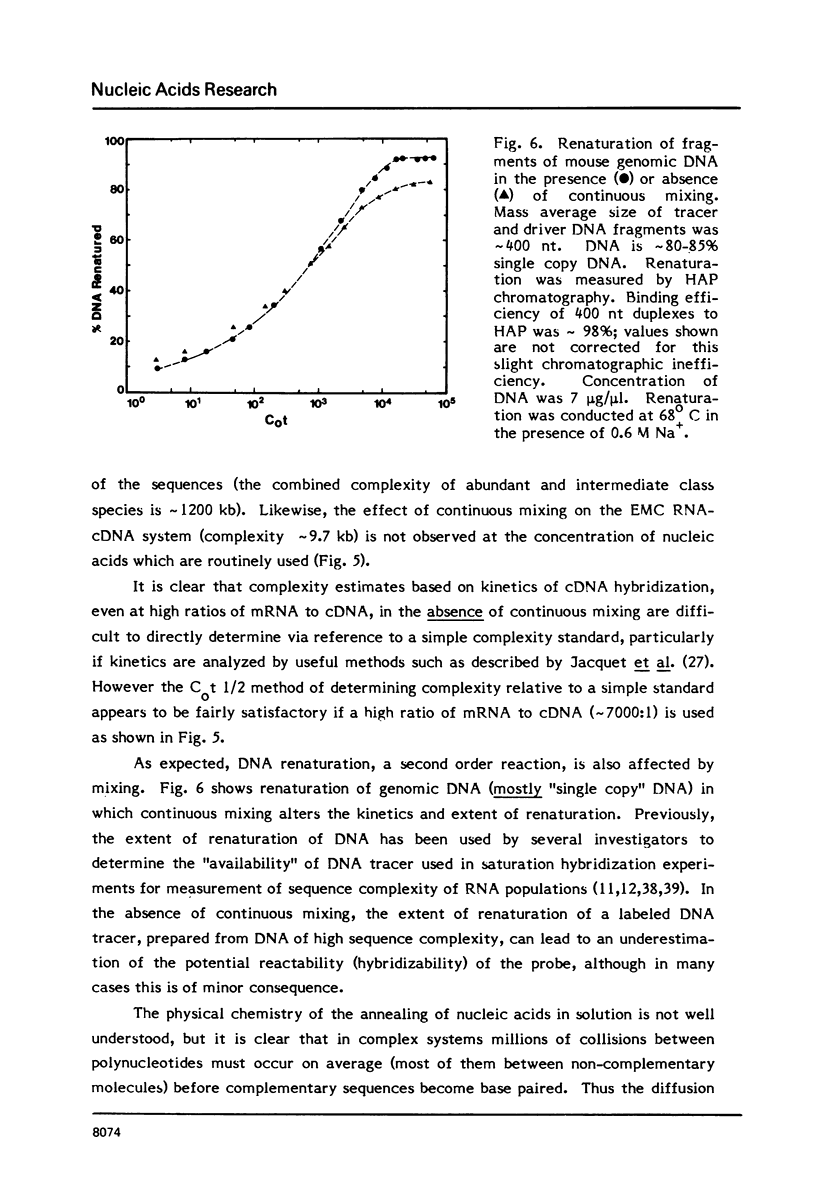



Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- BOLTON E. T., MCCARTHY B. J. FRACTIONATION OF COMPLEMENTARY RNA. J Mol Biol. 1964 Feb;8:201–209. doi: 10.1016/s0022-2836(64)80129-7. [DOI] [PubMed] [Google Scholar]
- Bailey J. M., Davidson N. Methylmercury as a reversible denaturing agent for agarose gel electrophoresis. Anal Biochem. 1976 Jan;70(1):75–85. doi: 10.1016/s0003-2697(76)80049-8. [DOI] [PubMed] [Google Scholar]
- Bantle J. A., Hahn W. E. Complexity and characterization of polyadenylated RNA in the mouse brain. Cell. 1976 May;8(1):139–150. doi: 10.1016/0092-8674(76)90195-1. [DOI] [PubMed] [Google Scholar]
- Bantle J. A., Maxwell I. H., Hahn W. E. Specificity of oligo (dT)-cellulose chromatography in the isolation of polyadenylated RNA. Anal Biochem. 1976 May 7;72:413–427. doi: 10.1016/0003-2697(76)90549-2. [DOI] [PubMed] [Google Scholar]
- Beckmann J. S., Daniel V. Relative stabilities of RNA/DNA hybrids: effect of RNA chain length in competitive hybrization. J Mol Biol. 1974 Oct 25;89(2):355–362. doi: 10.1016/0022-2836(74)90524-5. [DOI] [PubMed] [Google Scholar]
- Bishop J. O., Beckmann J. S., Campo M. S., Hastie N. D., Izquierdo M., Perlman S. DNA-RNA hybridization. Philos Trans R Soc Lond B Biol Sci. 1975 Nov 6;272(915):147–157. doi: 10.1098/rstb.1975.0077. [DOI] [PubMed] [Google Scholar]
- Bonner T. I., Brenner D. J., Neufeld B. R., Britten R. J. Reduction in the rate of DNA reassociation by sequence divergence. J Mol Biol. 1973 Dec 5;81(2):123–135. doi: 10.1016/0022-2836(73)90184-8. [DOI] [PubMed] [Google Scholar]
- Britten R. J., Kohne D. E. Repeated sequences in DNA. Hundreds of thousands of copies of DNA sequences have been incorporated into the genomes of higher organisms. Science. 1968 Aug 9;161(3841):529–540. doi: 10.1126/science.161.3841.529. [DOI] [PubMed] [Google Scholar]
- CHAMBERLIN M., BALDWIN R. L., BERG P. AN ENZYMICALLY SYNTHESIZED RNA OF ALTERNATING BASE SEQUENCE: PHYSICAL AND CHEMICAL CHARACTERIZATION. J Mol Biol. 1963 Oct;7:334–349. doi: 10.1016/s0022-2836(63)80028-5. [DOI] [PubMed] [Google Scholar]
- Chamberlin M. J. Comparative properties of DNA, RNA, and hybrid homopolymer pairs. Fed Proc. 1965 Nov-Dec;24(6):1446–1457. [PubMed] [Google Scholar]
- Chikaraishi D. M. Complexity of cytoplasmic polyadenylated and nonpolyadenylated rat brain ribonucleic acids. Biochemistry. 1979 Jul 24;18(15):3249–3256. doi: 10.1021/bi00582a009. [DOI] [PubMed] [Google Scholar]
- Crick F. Split genes and RNA splicing. Science. 1979 Apr 20;204(4390):264–271. doi: 10.1126/science.373120. [DOI] [PubMed] [Google Scholar]
- Dudley J. P., Butel J. S., Socher S. H., Rosen J. M. Detection of mouse mammary tumor virus RNA in BALB/c tumor cell lines of nonviral etiologies. J Virol. 1978 Dec;28(3):743–752. doi: 10.1128/jvi.28.3.743-752.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galau G. A., Britten R. J., Davidson E. H. A measurement of the sequence complexity of polysomal messenger RNA in sea urchin embryos. Cell. 1974 May;2(1):9–20. doi: 10.1016/0092-8674(74)90003-8. [DOI] [PubMed] [Google Scholar]
- Galau G. A., Britten R. J., Davidson E. H. Studies on nucleic acid reassociation kinetics: rate of hybridization of excess RNA with DNA, compared to the rate of DNA renaturation. Proc Natl Acad Sci U S A. 1977 Mar;74(3):1020–1023. doi: 10.1073/pnas.74.3.1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert W. Why genes in pieces? Nature. 1978 Feb 9;271(5645):501–501. doi: 10.1038/271501a0. [DOI] [PubMed] [Google Scholar]
- Goldberg R. B., Hoschek G., Kamalay J. C., Timberlake W. E. Sequence complexity of nuclear and polysomal RNA in leaves of the tobacco plant. Cell. 1978 May;14(1):123–131. doi: 10.1016/0092-8674(78)90307-0. [DOI] [PubMed] [Google Scholar]
- Hahn W. E., Pettijohn D. E., Van Ness J. One strand equivalent of the Escherichia coli genome is transcribed: complexity and abundance classes of mRNA. Science. 1977 Aug 5;197(4303):582–585. doi: 10.1126/science.327551. [DOI] [PubMed] [Google Scholar]
- Hinnebusch A. G., Clark V. E., Klotz L. C. Length dependence in reassociation kinetics of radioactive tracer DNA. Biochemistry. 1978 Apr 18;17(8):1521–1529. doi: 10.1021/bi00601a026. [DOI] [PubMed] [Google Scholar]
- Hutton J. R., Wetmur J. G. Renaturation of bacteriophage phiX174 DNA-RNA hybrid: RNA length effect and nucleation rate constant. J Mol Biol. 1973 Jul 15;77(4):495–500. doi: 10.1016/0022-2836(73)90218-0. [DOI] [PubMed] [Google Scholar]
- Jacquet M., Affara N. A., Robert B., Jakob H., Jacob F., Gros F. Complexity of nuclear and polysomal polyadenylated RNA in a pluripotent embryonal carcinoma cell line. Biochemistry. 1978 Jan 10;17(1):69–79. doi: 10.1021/bi00594a010. [DOI] [PubMed] [Google Scholar]
- Kim K., Jhon M. S. Theoretical study of hydration of RNA. Biochim Biophys Acta. 1979 Nov 22;565(1):131–147. doi: 10.1016/0005-2787(79)90089-3. [DOI] [PubMed] [Google Scholar]
- Kohne D. E., Levison S. A., Byers M. J. Room temperature method for increasing the rate of DNA reassociation by many thousandfold: the phenol emulsion reassociation technique. Biochemistry. 1977 Nov 29;16(24):5329–5341. doi: 10.1021/bi00643a026. [DOI] [PubMed] [Google Scholar]
- MARMUR J., DOTY P. Determination of the base composition of deoxyribonucleic acid from its thermal denaturation temperature. J Mol Biol. 1962 Jul;5:109–118. doi: 10.1016/s0022-2836(62)80066-7. [DOI] [PubMed] [Google Scholar]
- Maxwell I. H., Maxwell F., Hahn W. E. Use of CH3HgOH-agarose gels for the electrophoresis of heterogeneous nuclear RNA and messenger RNA from mammalian cells. Anal Biochem. 1979 Oct 15;99(1):146–160. doi: 10.1016/0003-2697(79)90056-3. [DOI] [PubMed] [Google Scholar]
- Maxwell I. H., Van Ness J., Hahn W. E. Assay of DNA-RNA hybrids by S1 nuclease digestion and adsorption to DEAE-cellulose filters. Nucleic Acids Res. 1978 Jun;5(6):2033–2038. doi: 10.1093/nar/5.6.2033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonell M. W., Simon M. N., Studier F. W. Analysis of restriction fragments of T7 DNA and determination of molecular weights by electrophoresis in neutral and alkaline gels. J Mol Biol. 1977 Feb 15;110(1):119–146. doi: 10.1016/s0022-2836(77)80102-2. [DOI] [PubMed] [Google Scholar]
- Moore R. L., McCarthy B. J. Related base sequences in the DNA of simple and complex organisms. 3. Variability in the base sequence of the reduplicated genes for ribosomal RNA in the rabbit. Biochem Genet. 1968 Jun;2(1):75–86. doi: 10.1007/BF01458452. [DOI] [PubMed] [Google Scholar]
- Sargent T. D., Wu J. R., Sala-Trepat J. M., Wallace R. B., Reyes A. A., Bonner J. The rat serum albumin gene: analysis of cloned sequences. Proc Natl Acad Sci U S A. 1979 Jul;76(7):3256–3260. doi: 10.1073/pnas.76.7.3256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schafer M. P., Boyd C. D., Tolstoshev P., Crystal R. G. Structural organization of a 17 KB segment of the alpha 2 collagen gene: evaluation by R loop mapping. Nucleic Acids Res. 1980 May 24;8(10):2241–2253. doi: 10.1093/nar/8.10.2241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas M., White R. L., Davis R. W. Hybridization of RNA to double-stranded DNA: formation of R-loops. Proc Natl Acad Sci U S A. 1976 Jul;73(7):2294–2298. doi: 10.1073/pnas.73.7.2294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tonegawa S., Maxam A. M., Tizard R., Bernard O., Gilbert W. Sequence of a mouse germ-line gene for a variable region of an immunoglobulin light chain. Proc Natl Acad Sci U S A. 1978 Mar;75(3):1485–1489. doi: 10.1073/pnas.75.3.1485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tse T. P., Morris H. P., Taylor J. M. Molecular basis of reduced albumin synthesis in Morris hepatoma 7777. Biochemistry. 1978 Jul 25;17(15):3121–3128. doi: 10.1021/bi00608a028. [DOI] [PubMed] [Google Scholar]
- Van Ness J., Hahn W. E. Sequence complexity of cDNA transcribed from a diverse mRNA population. Nucleic Acids Res. 1980 Sep 25;8(18):4259–4270. doi: 10.1093/nar/8.18.4259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Ness J., Maxwell I. H., Hahn W. E. Complex population of nonpolyadenylated messenger RNA in mouse brain. Cell. 1979 Dec;18(4):1341–1349. doi: 10.1016/0092-8674(79)90244-7. [DOI] [PubMed] [Google Scholar]
- Wetmur J. G., Davidson N. Kinetics of renaturation of DNA. J Mol Biol. 1968 Feb 14;31(3):349–370. doi: 10.1016/0022-2836(68)90414-2. [DOI] [PubMed] [Google Scholar]
- Wetmur J. G. Excluded volume effects on the rate of renaturation of DNA. Biopolymers. 1971;10(4):601–613. doi: 10.1002/bip.360100402. [DOI] [PubMed] [Google Scholar]
- Wetmur J. G. Hybridization and renaturation kinetics of nucleic acids. Annu Rev Biophys Bioeng. 1976;5:337–361. doi: 10.1146/annurev.bb.05.060176.002005. [DOI] [PubMed] [Google Scholar]
- Young B. D., Harrison P. R., Gilmour R. S., Birnie G. D., Hell A., Humphries S., Paul J. Kinetic studies of gene frequency. II. Complexity of globin complementary DNA and its hybridization characteristics. J Mol Biol. 1974 Apr 25;84(4):555–568. doi: 10.1016/0022-2836(74)90116-8. [DOI] [PubMed] [Google Scholar]


