Abstract
Functional data are increasingly encountered in scientific studies, and their high dimensionality and complexity lead to many analytical challenges. Various methods for functional data analysis have been developed, including functional response regression methods that involve regression of a functional response on univariate/multivariate predictors with nonparametrically represented functional coefficients. In existing methods, however, the functional regression can be sensitive to outlying curves and outlying regions of curves, so is not robust. In this paper, we introduce a new Bayesian method, robust functional mixed models (R-FMM), for performing robust functional regression within the general functional mixed model framework, which includes multiple continuous or categorical predictors and random effect functions accommodating potential between-function correlation induced by the experimental design. The underlying model involves a hierarchical scale mixture model for the fixed effects, random effect and residual error functions. These modeling assumptions across curves result in robust nonparametric estimators of the fixed and random effect functions which down-weight outlying curves and regions of curves, and produce statistics that can be used to flag global and local outliers. These assumptions also lead to distributions across wavelet coefficients that have outstanding sparsity and adaptive shrinkage properties, with great flexibility for the data to determine the sparsity and the heaviness of the tails. Together with the down-weighting of outliers, these within-curve properties lead to fixed and random effect function estimates that appear in our simulations to be remarkably adaptive in their ability to remove spurious features yet retain true features of the functions. We have developed general code to implement this fully Bayesian method that is automatic, requiring the user to only provide the functional data and design matrices. It is efficient enough to handle large data sets, and yields posterior samples of all model parameters that can be used to perform desired Bayesian estimation and inference. Although we present details for a specific implementation of the R-FMM using specific distributional choices in the hierarchical model, 1D functions, and wavelet transforms, the method can be applied more generally using other heavy-tailed distributions, higher dimensional functions (e.g. images), and using other invertible transformations as alternatives to wavelets.
Keywords: Adaptive LASSO, Bayesian methods, False discovery rate, Functional Data Analysis, Mixed models, Robust regression, Scale mixtures of normals, Sparsity Priors, Variable Selection, Wavelets
1. INTRODUCTION
An ever-growing number of technologies take automated measurements over fine grids of time, space, or some other domain, and yield functional data, for which the ideal units are curves and the observed data consist of curves sampled on fine grids. Examples include EEG signals, proteomic mass spectra, array CGH copy number arrays, and quantitative image data such as fMRI. These and other functional data have motivated the development of new methodology for functional data analysis (FDA), some of which are reviewed by Ramsay and Silverman (2005), Ferraty and Vieu (2006), and Ruppert, Wand and Carroll (2009).
One class of methods involves functional response regression, an extension of linear regression to functional data whereby a functional response is regressed on a set of predictors, each with its own nonparametrically represented functional coefficient. Early work focused on longitudinal data or functional data on a sparse grid, and involved functional ANOVA with categorical predictors and iid curves (Staniswallis and Lee 1998; Brumback and Rice 1998; Wang 1998; Wu and Zhang 2002; Guo 2002). Larger and more complex functional data sets have increasingly been encountered, with multilevel designs, correlated functions, and functional data sampled on a fine grid. Many of these methods do not scale up to these settings, but recent work attempts to accommodate these complexities and scales up to these larger data sets (Morris, et al. 2003; Morris and Carroll 2006; Morris, et al. 2006; Baladandayuthapani, et al. 2008; Morris, et al. 2008; Di, et al. 2009; Zhou, et al. 2010; Staicu, Crainceanu and Carroll 2010; Morris, et al. 2010; Grevin, et al. 2010). The models underlying many of these methods can be considered variations of a functional mixed effects model (FMM), which adds random effect functions of non-specified functional form to the functional response regression. Methods developed within this general FMM framework have great utility, given their ability to accommodate multiple continuous or categorical fixed effect predictors and random effect predictors to model between-function correlation induced by various experimental designs.
In linear regression, it is well known that outlying values can strongly impact regression coefficient estimators, artificially inflating their standard errors and sometimes leading to bias (Huber 1981). In response, robust regression techniques have been developed that effectively down-weight the influence of the outliers and as a result lead to much improved regression coefficient estimators. For examples of such methods, see Huber (1981) and Hampel, et al. (2005). Outliers are frequently encountered in functional data, as well, including entire outlying curves (global outliers) as well as curves with local outlying features, which can be localized in either the time or frequency domain (local outliers). Analogously, these outliers can have a strong influence on the functional coefficients estimated in functional response regression models. To our knowledge, there are currently no methods in the existing statistical literature for performing robust functional response regression. The limited work we have encountered in robust FDA includes robust estimation of functional principal components (Locantore, et al. 1999; Huber 2002, Gervini 2008, Gervini 2010) and functional predictors of scalar responses (Crambes, Delsol and Laksaci 2008).
In this paper, we introduce Bayesian methods for robust functional regression within the FMM framework, which we refer to as robust functional mixed models (R-FMM).We believe this is the first method in the statistical literature for robust functional response regression, and has great practical utility given it is developed within the general FMM framework, can be applied to functional and image data, is computationally efficient enough to handle large data sets, can be fit in an automated fashion given just the functional responses and design matrices, and yields posterior samples of all model parameters that can be used to perform a wide array of potential Bayesian estimation and inference. The novel model we present involves hierarchical scale mixture distributions for the fixed effect, random effect and residual error functions in the wavelet space. These modeling assumptions across curves result in robust nonparametric estimators of the fixed and random effect functions which down-weight outlying curves and regions of curves, and produce statistics that can be used to flag global and local outliers. This hierarchical model also induces distributions across wavelet coefficients that have connections with some of the best sparsity distributions in current literature and yield desirable adaptive shrinkage properties, which together with the down-weighting of outliers leads to fixed and random effect function estimates that in our simulations demonstrate a remarkable ability to remove spurious features yet retain true features of the functions. While we focus on 1D functions, wavelet modeling, and using particular distributional assumptions in the hierarchical model, the method can also be applied to higher dimensional functions, using basis functions and transformations other than wavelets, and using other specific distributional assumptions.
The outline for the rest of the paper is as follows: Section 2 presents the method, first introducing functional mixed models in Section 2.1, then outlining the robust model in Section 2.2 and summarizing computational model fitting details in Section 2.3. In Section 2.4, we explain how to detect and examine global and local outliers using this method, and then we briefly discuss how to adapt the method use other distributions, basis functions and transformations other than wavelets, and to apply to higher dimensional functions in Section 2.5. In Section 3, we present results from a simulation study to evaluate the performance of the method relative to an existing non-robust method for fitting FMM, and in Section 4, we apply both robust and non-robust methods to real mass spectrometry proteomics data set. Section 5 contains a discussion and some conclusions, and online supplementary materials contain numerous derivations, computational details, and further results beyond what is presented in the text of this paper.
2. METHODS
2.1 Background: Functional Mixed Models and Gaussian Basis Space Modeling
The functional mixed model (FMM) relates functional responses to a set of scalar predictors through functional coefficients, with random effect functions included to account for correlation between functions that may be induced by the experimental design. A general FMM is given by:
| (1) |
where Y(t) = (Y1(t), …, Yn(t))T is a vector of functional responses defined on the same interval 𝒯. B(t) = (B1(t), …, Bp(t))T is a vector of fixed effect functions associated with an n × p design matrix X, with Bj(t) representing the partial effect of covariate j on the function at position t. The U(t) = (U1(t), …, Um(t))T is a vector of mean zero random effect functions associated with an n × m design matrix Z, and E(t) = (E1(t), …, En(t))T is a vector of mean zero residual error functions. A key flexibility of this model is the unspecified forms of its functional quantities.
Before fitting the FMM, assumptions must be specified on the distributions and the structure of the random effect and residual error covariances. Morris and Carroll (2006) describe a Gaussian functional mixed model with separable between- and within-function covariance matrices and a Bayesian, wavelet-based method for fitting it (G-WFMM). They assume the random effects U(t) follow a mean-zero multivariate Gaussian process with an m × m between-function covariance matrix P and a within-function covariance surface Q(t1, t2) ∈ 𝒯 × 𝒯, denoted by U(t) ~ 𝒩 (P, Q), implying that Cov{Ul(t1), Uk(t2)} = PlkQ(t1, t2). The residual error is assumed to be E(t) ~ 𝒩 (R, S), independent of U(t). If the functional responses Yi(t) are all measured on the same equally-spaced fine grid t of length T, the discrete version of model (1) can be represented as
| (2) |
with Y, B, U, and E each having T columns, each corresponding to one of the positions on the grid. The random effects and residual error matrices are mean-zero matrix normals (Dawid 1981): U ~ 𝒩(P, Q), E ~ 𝒩(R, S), with Q and S as T × T matrices. A common special case of this model involves conditionally independent random effect functions and residuals, P = R = I.
Flexible structures are allowed on Q and S, as induced by the underlying wavelet-space modeling approach. Some early overviews of statistical work using wavelets can be found in Ogden (1997) and Vidakovic (1999). First, the discrete wavelet transform (DWT) is applied to the rows of Y, represented here as D = YWT, with WT an orthonormal wavelet transform matrix. This transform projects the observed functions into the wavelet space, inducing a wavelet-space version of model (2),
| (3) |
where rows of D, B*, U*, and E* correspond to the DWT of the rows of Y, B, U, and E, respectively, and the columns correspond to wavelet coefficients double-indexed by wavelet scale j and location k rather than the location within the function. The induced distributional assumptions are U* ~ N(P, Q*) and E* ~ N(R, S*), with Q* = WQWT and S* = WSWT. The whitening property of the wavelet transform (e.g. Vidakovic 1999) tends to induce decorrelation of the wavelet coefficients in the wavelet domain, so that one might make reasonable independence assumptions , with the induced data space covariance matrices Q = WTQ*W and S = WTS*W. By indexing these wavelet-space variance components by both scale j and location k, this assumption is parsimonious yet flexible enough to model many important types of nonstationarities in Q and S, including different variances and different degrees of autocorrelation at different parts of the curves (Morris and Carroll 2006).
A spike Gaussian-slab prior is assumed for the fixed effects in the wavelet space , the ath component in the (j, k)th column of B*. That is, let , where πaj and τaj are regularization parameters that can be estimated using an empirical Bayes approach or given hyperpriors themselves. When applied to wavelet coefficients, this type of prior induces a nonlinear shrinkage or threshold-like effect which leads to adaptive regularization, or denoising in a way that tends to preserve dominant local features of the corresponding function (Vidakovic 1999). An MCMC method is used to obtain posterior samples for the quantities in model (3), which are then projected back to the data space using the inverse discrete wavelet transform (IDWT) to perform Bayesian inference on the quantities of model (2). Morris, et al. (2010) demonstrate how this method can be extended up to higher dimensional functional data like images, and describe how this three-step approach can be used with basis functions and transformations other than wavelets.
2.2 Robust Wavelet-Based Functional Mixed Models (R-WFMM)
The nonparametric functional regression underlying the fixed and random effect function estimation in the G-WFMM is subject to strong influence by outlying curves or regions of curves, and in this way is not robust. These outliers can be constructive or destructive, i.e. they can either induce spurious artifacts or can attenuate true features of the functional effects. Here, we introduce a new hierarchical modeling framework for functional mixed models that can achieve robustness to global and local outliers and improved adaptive regularization of fixed and random effect functions, leading to a method we believe is the first robust method for functional response regression. In this section, we will present this method for 1D functions, using wavelet transforms, and assuming conditionally independent random effect and residual functions (P = R = I in model (2)) and then later in Section 2.5, we will discuss how the method can be adapted to handle higher dimensional functions like images, general P and R, and using bases and transforms other than wavelets.
Given n observed curves Yi(t), i = 1 …, n, each sampled on equally-spaced grid t of size T, we assume the observed functions and observations on the grid follow the general FMM presented in (1) and (2), respectively. Rather than directly specifying distributional assumptions for these models, we instead specify our distributional assumptions in the wavelet space model (3), and then discuss the distributions these induce in the data space. Like the G-WFMM, our robust method will use a three-step wavelet-based modeling approach, first applying a specified DWT to each curve i to obtain the corresponding set of wavelet coefficients dijk, with j = 1, …, J indexing the wavelet scale (frequency) and k = 1, …, Kj the location. Second, we fit the robust wavelet-space version of the functional mixed model specified below and third, we project our results back to the original data space using the IDWT, obtaining inference on the fixed and random effect functions in model (2). The key novelty in our robust method is the hierarchical modeling assumptions we make on the wavelet coefficients for the residuals, random effect functions, and fixed effect functions, which are completely different from those used in the G-WFMM, and possess the desired robustness properties and improved adaptive regularization.
General Wavelet-Space Hierarchical Model for Robust FMM
Working with the basic wavelet-space FMM (3), we denote the (j, k)th column as . We specify the following hierarchical model on these parameters:
| (4) |
| (5) |
| (6) |
where δ0 is a point mass at 0, are mutually independent. The individual scale parameters λijk, ϕljk, ψajk are mutually independent with specified mixing distributions indexed by population scale parameter vectors , which are also mutually independent with prior distributions indexed by specified hyperparameter vectors ΘE, ΘU, and ΘB, respectively. Note that the G-WFMM is a special case of this model, with a degenerate distribution for g1(•), λijk ~ δsjk, ϕljk ~ δqjk and ψajk = δτaj. This model is fit using a blocked Gibbs sampler, as summarized in Section 2.3.
Robustness Properties
Consider the hierarchical model for the residuals with non-degenerate . For each wavelet coefficient (j, k), each curve i has its own individual scale parameter λijk, which is drawn from a mixture distribution indexed by a population scale parameter , which in turn is given a prior distribution . The individual scale parameters λijk serve as wavelet-space outlier weights. A relatively large λijk (across i) suggests curve i is an outlier with respect to a feature of the curve corresponding to the wavelet basis function (j, k), and will result in a downweighting of observation dijk in estimating the corresponding fixed and random effects , respectively. Similarly, relatively large ϕljk (across l) indicate random effect unit l is an outlier for feature (j, k), and will result in some downweighting of the dijk corresponding to random effect unit l, which are those with Zil ≠ 0. This can be seen by the fact that . The choice of mixing distribution g1 impacts the estimation of the individual scale parameters, and thus the robustness properties of the method. If we marginalize the model by integrating out the individual scale parameters, combining levels (4) and (5), we are left with a heavy-tailed scale mixture distribution Normal ◦ g1 indexed by population scale parameters νjk for each of E, U, and B. We call these population scale parameters because they summarize the overall variability in the population, across i for , across l for , and across k for . These population scale parameters also play a crucial role in the adaptive regularization of the fixed and random effect functions, as we elaborate below.
Normal-Exponential-Gamma Hierarchical Model for Robust FMM
While many different choices can be considered for g1(•) and g2(•), for our calculations in this paper we will assume for each model component E, U, and B, and choose g2(•) to be such that are Gamma distributions, with their parameters determined using the empirical Bayes approach outlined below in Section 2.3. We have found this particular choice to be appealing for several reasons: (1) computations are tractable and efficient, (2) the marginal distributions have good robustness properties, (3) similar models in single-function wavelet regression have robustness properties, and (4) it has connections to various sparsity priors known to be good choices for variable selection, which in the wavelet space should lead to good adaptive regularization for the fixed and random effect functions.
Integrating over the individual scale parameters, this corresponds to double exponential (DE) distributions for the residuals, random effects, and the slab part of the mixture for fixed effects in the wavelet space. The heavier-than-normal exponential tails lead to downweighting of outliers, as described above, and robustness properties. Supplementary materials include a proposition (Proposition 1) and proof that describes some of this model’s robustness properties in a mean+error special case of the FMM. The proposition shows that as the norm of an outlying functional observation approaches infinity, the posterior distribution of the mean function B(t) approaches to a posterior that either depends only on the non-outlying observations, or depends on the non-outlying observations and a part of the outlying observation with bounded norm.
Various researchers have pointed out that the DE distribution is a compelling choice for wavelet-space modeling, since its spike at zero and heavier-than-normal tails match typically encountered empirical characteristics of wavelet coefficients (e.g., Mallat 1989; Kokoszka, et al. 2006; Vidakovic and Ruggeri 2001). In single function wavelet regression, the use of double-exponential likelihoods has been shown to lead to adaptive regularization and efficient function estimation, even when the true noise distribution is Gaussian (Vidakovic 1999; Clyde and George 2000; Vidakovic and Ruggeri 2001; Cutillo et al. 2008). Inspired by Clyde and George (2000), Pensky (2006) examined the theoretical frequentist properties of various choices of likelihoods and priors in Bayesian wavelet regression, and found the combination of double-exponential prior and double-exponential likelihood to have outstanding properties. That combination leads to optimal functional estimators for both spatially homogeneous and spatially heterogeneous functions when the errors are normally distributed; it is robust to heavy-tailed distributions; and it is able to flexibly represent functions in Besov spaces with the full range of potential smoothness. While the FMM setting is more involved than single-function wavelet regression and our model is not quite the same, these optimality results are still compelling and suggest models involving DE distributions might be a good choice in this context. An interesting theoretical exercise beyond the scope of this paper would be to evaluate similar properties for our hierarchical model for estimation of fixed and random effect functions in the FMM framework.
This choice also has connections with distributions commonly used in variable selection. The concept of variable selection is relevant here since effective variable selection across the wavelet coefficients for the random effects and fixed effects leads to effective adaptive regularization of the random effect functions Ul(t) and fixed effect functions Ba(t). The LASSO (Tibshirani 1996) is equivalent to the maximum a posteriori estimator assuming a DE prior, and Bayesian modeling using this prior has also been studied (Park and Casella 2008). While our model behaves like a DE across i for the residuals and across l for the random effects, the fact that the corresponding population scale parameters are indexed by wavelet coefficient (j, k) with their squares having Gamma hyperpriors implies this model behaves like the Normal-Exponential-Gamma (NEG) distribution discussed by Griffin and Brown (2005) across wavelet coefficients, which by mixing over different scale parameters actually has heavier-than-exponential tails. This distribution has better variable selection properties than the LASSO (Griffin and Brown 2005; Carvalho, Polson and Scott 2010), and according to the analysis of Ayers and Cordell (2006), is the best of a range of estimators. This NEG type prior can have quasi Cauchy tail behavior (when shape=0.5) or can have thinner tails (when shape > 0.5) (see Supplementary Material). In our context, this should lead to better nonlinear shrinkage of the random effects’ wavelet coefficients, and in turn improved adaptive regularization of the random effect functions. There are connections between this prior and the adaptive LASSO (Zou 2006) involving coefficient-specific scale parameters, which for our random effects are the corresponding population scale parameters , that unlike the classic regression setting of Zou (2006) can actually be well estimated from the data because of the replication over l.
A mixture of point mass at zero and DE prior is the so-called empirical Bayes prior of Johnstone and Silverman (2004), which was shown to have outstanding variable selection properties, equalling the Horseshoe prior in the simulation studies of Carvalho et al. (2010). Our model for the fixed effect wavelet coefficients is like this empirical Bayes prior across k, but across a and j is like a mixture of point mass at zero and NEG prior. This mixture has even more flexibility in modeling heavy tails in the slab and the spike at zero, which provides extra adaptiveness in the variable selection across predictors a and scales j. Various investigators have shown spike heavy-tailed slabs to have better variable selection properties than spike-Gaussian slabs (Vidakovic and Ruggeri 2001; Johnstone and Silverman 2004; Johnstone and Silverman 2005; Nason 2008; Griffin and Brown 2010), since they result in less attenuation of large regression coefficients.
Conditional on the fixed effects B* and population scale parameters for the random effects and residuals , this wavelet-space model with assumptions (4)–(6) induces a data-space FMM (2) for which the random effect and residual error functions Ub(t) and Ei(t) on grid t are mixtures of double-exponentials, with mixing proportions given by the elements of the DWT matrix WT = {Wt(jk)} and component precision parameters given by , respectively. This distribution does not have a simple closed-form expression, but is heavier-tailed than the Gaussian, imbuing it with robustness properties. The distribution is multivariate, and since the weights mix over wavelet coefficients at different frequencies, it is able to account for autocorrelation within the functions in the same manner as the Gaussian model discussed by Morris and Carroll (2006). Since the population scale parameters are double-indexed by both wavelet scale j and location k, it can accommodate nonstationary covariance structures within the random effect and residual curves, e.g. allowing different variances and degrees of smoothness, and thus various borrowing of strength among nearby observations, across different regions of the curves.
2.3 Computational Details of R-WFMM
Here, we outline our computational methods to fit the R-WFMM. We take a fully Bayesian approach, and use a block Gibbs sampler to sample from the joint posterior distribution of the waveletspace FMM (3) with distributional assumptions given by (4)–(6). Here we will briefly summarize the steps; the full details are provided in supplementary materials. For notational convenience, Here we denote .
-
Step 1
For each a, j, k, update the fixed effects from , which is available in closed form as a mixture of point mass at 0 and Gaussian, with γajk the indicator of the Gaussian. Note the random effects are integrated out here, making this a block sampler that mixes more efficiently than a full Gibbs.
-
Step 2
For each j, k, update random effects from , which are MVN.
-
Step 3
For each i, l, a, j, k update the individual scale parameters from , which are Inverse Gaussians, except that when γajk = 0, ψajk is drawn from the exponential prior.
-
Step 4
For each a, j, k, update the population scale parameters from , which are Gamma distributions.
-
Step 5
For each a, j, update the mixture parameter (πaj|γaj), which is a Beta.
These steps are repeated. After a burn-in period, we collect posterior samples from parameters in the wavelet-space FMM (3), and the IDWT can be applied to the posterior samples of B* and U* to obtain posterior samples of B and U in (2) to perform Bayesian inference in the original data space FMM.
If the user is satisfied with default wavelet and the vague proper empirical Bayes hyperpriors at the top hierarchical level, then this method can be run in an automated fashion with no tuning parameters, and the user only required to provide Y, X, and Z. The code is efficient enough to apply to large data sets, and is readily parallelizable when multiple CPU systems are available.
The default hyperparameters for the Gamma priors on , and Beta prior on πaj are chosen using a vague empirical Bayes approach, with modes centered at a moment-matched estimator of the corresponding parameters with the variance large, e.g. 1000. Since the ν are scale parameters, Henderson’s Mixed Model equations (pages 275–286, Searle et al. 1992) are used to get moment-based estimators, and the π are estimated as in Morris and Carroll (2006). Our sensitivity analyses demonstrate our results were not at all sensitive to the vagueness of these prior distributions over a reasonable range. Details of the vague empirical Bayes method, sensitivity, and properties are provided in supplementary materials.
Here, we have chosen to use a fully Bayesian approach to fit our model. In principal, it is possible to fit a similar model using penalized maximum likelihood methods with appropriately chosen penalties and likelihoods, although it is not clear how to proceed on the model fitting, inference, and asymptotics, which are daunting given the complexity of the model and typical size of the data set. It would be interesting to investigate whether such a model could be fit and yield estimation and inference in the frequentist realm, but beyond the scope of this paper. As mentioned above, our Bayesian approach is computationally efficient enough for large data sets, is parallelizable, and can be run automatically and depends only upon vague prior distributions for which we offer automatic choices. Further, our approach does not just yield estimates, but also a wide array of Bayesian inference for all parameters in both the wavelet- and data-space models, and this inference appropriately integrates over the uncertainty of all nuisance parameters in the model.
One type of Bayesian inference that is relevant and interesting here is false discovery rate (FDR)-based pointwise functional inference described by Morris, et al. (2008) that takes both statistical and practical significance into account. Given an effect size of practical interest δ, for each covariate a one can easily compute the posterior probabilities that |Ba(t)| > δ for each t, yielding probability discovery function pa,δ(t). The quantities 1 − pa,δ(t) can be considered pointwise local FDRs for discovering curve regions of at least size δ. A cutpoint on the pa,δ(t) can be determined to flag regions of t as significant based on a specified global FDR α or formal utility considerations. Given this cutpoint, one can use the posterior probabilities to compute false negative rate (FNR), sensitivity, specificity, and to construct ROC curve summaries for detecting significant regions. Details are found in the suppelementary materials.
2.4 Outlier Detection and Characterization Using R-FMM
After fitting the R-FMM, the posterior samples of the individual scale parameters λijk and ϕbjk which can be used to construct global and local outlier diagnostics to identify and characterize outlying curves and individuals. A scalar outlier score for an observed function Yi(t) can be computed by λi‥ = Σj,k λijk. Note that if orthogonal wavelet transforms are used, then this is equivalent to the trace of the covariance of Ei, row i of E in the data space conditional on the scaling parameters, since , and W is the orthogonal linear transformation matrix corresponding to the chosen DWT, with D = YWT and WTW = I. A relatively large value of λi‥ indicates inflated scaling parameters for observation i, thus signifying a possible outlying curve. Posterior samples for these outlier scores can be computed from the MCMC output, and summarized by the posterior mean λ̂i‥ and accompanying posterior credible intervals. If applied to the random effects’ scaling parameters, ϕl‥ = ∑j,k ϕljk, these measures can be used to suggest which individuals may be outliers in their specified populations, i.e., have mean curves that significantly deviate from those of the rest of the population. The posterior statistics and the related inferential values can be combined with traditional box-plots or other testing methods for outlier diagnosis.
For an outlying curve, it is also possible to construct functional summary statistics to characterize which regions of the curve are unusual. An “outlier function” λi(t) can be computed by applying the 2D IDWT to diag{(λijk)}j,k, and then taking the diagonal elements of the resulting matrix. For the orthonormal wavelet-based FMM, this is equivalent to estimating the diagonal elements of Si. By comparing λi(t) across i for each t, we can assess which regions of curve i are unusual for their population, and may be responsible for it being classified as an outlier. Similarly, we can compute and investigate the ϕb(t), for outlying random effect functions. If one suspects outliers in the frequency domain, one can look at the mean individual scale parameters across k, λij. and ϕlj. to flag individuals with outlying activity at scale j.
2.5 Implementing R-FMM for Higher Dimensional Functions, Other Heavy-tailed Distributions, and/or Other Isomorphic Transformations
Sections 2.2 and 2.3 provide modeling and computational details for a specific implementation of the R-FMM assuming P = R = I, double-exponential distributions, 1D functions, and using wavelet transformations. The R-FMM introduced in this paper can be applied much more generally, in some cases with very little additional work, and in other cases requiring some additional derivations and computational work. In this section, we describe how to accommodate general between-function covariance matrices P and R, other heavy-tailed distributions, higher dimensional functions (e.g. images), and transformations other than wavelets.
The FMM of Morris and Carroll (2006) allows correlation between functions through covariance matrices P and R as part of a separable structure, with Var{vec(U*)} = P⊗Q* and Var{vec(E*)} = R ⊗ S*, where vec(•) is the column-stacking vectorizing operator and ⊗ is the kronecker operator. Section 2.2 effectively assumes P = R = I, but the approach can be easily adapted to accommodate general P and R matrices. Given P and R, we can rescale , after which all of the specified steps proceed as described in Section 2.3, with an additional Metropolis-Hastings step to update the (typically very few) covariance parameters in P and R.
Although we focus on exponential-gamma mixtures here, other heavy-tailed distributions could be used as well. Some distributions, such as Student’s t (Andrews and Mallows 1974) and exponential power distributions (West 1987) can be written in ways that lead to tractable Gibbs updating steps. In other cases, alternative modeling strategies can be used, including Metropolis-Hastings steps to update the parameters in heavy-tailed distributions. The observed information matrix can be used to automate the proposal variances of a random walk Metropolis-Hastings, as in the variance component updates in Morris and Carroll (2006).
The extension of the R-WFMM to higher dimensional functions such as images is straightforward. For 2D images, the functional quantities of the FMM are indexed by two indices, row t1 and column t2, and higher dimensional wavelet transforms are substituted for the 1D DWT and IDWT used here. If a 2D DWT is used, there are 3 types of wavelet coefficients at each resolution level j, row wavelets (c = 1), column wavelets (c = 2), and diagonal wavelets (c = 3), resulting in wavelet coefficients that are triple-indexed by wavelet resolution level j, type c, and location k. For general r-dimensional data, the r – dimensional DWT has 2r − 1 types of coefficients. This accommodates adaptive smoothing in all dimensions, even when independence among wavelet coefficients is assumed. All modeling and computational details presented in Sections 2.2 and 2.3 remain the same, except that the population scale parameters for the residuals and random effects are triple-indexed by (j, c, k), and the population scale and sparsity parameters for the fixed effects by (a, c, l), yielding additional flexibility in the different functional dimensions. These changes require no additional coding, as our current code already accommodates image data.
Wavelets are a compelling choice of basis representation for irregular functional data, and fit very nicely with the double-exponential distributional assumptions used in the R-WFMM presented in this paper. However, as described by Morris et al. (2010), the FMM can be fit using the same 3-step approach underlying the WFMM but using other basis functions, or more generally using some invertible transformation of the observed functions. Morris, et al. (2010) use the term isomorphic transformation to describe one that preserves all of the information in the original data, i.e., is invertible or lossless. More precisely, given row vector y ∈ ℜ(𝒯), we say a transform f : ℜ(𝒯) → ℜ(𝒯) is isomorphic if there exists a reverse transform f−1 such that f−1{f(y)} = y. The wavelet transform is isomorphic because IDWT(DWT(y))= y, but isomorphic transformations can be constructed in other ways as well, for example, by using other basis functions including Fourier bases, spline bases, and certain empirically determined basis functions like functional principal components (Aston, Chiou and Evans 2010), or even using nonlinear transformations. The same 3-step approach underlying the WFMM can be used to fit the FMM based on other isomorphic transforms, with some of the same computational benefits. That is, apply the transform to each observed function, fit the transformed or basis-space FMM, and then use the reverse transformations to map the estimates (or posterior samples) back to the FMM in the original data space for inference.
In the same way, the R-FMM described here can be applied using isomorphic transformations other than wavelets. Given a choice of transformation, if it is reasonable to use spike-slab or heavy-tailed priors for regularization and to assume independence and the specified heavy-tailed distributions in the transformed space, then the details herein can be straightforwardly applied using our existing code, with the suitable transformations and reverse-transformations substituted for the DWT and IDWT in the first and last steps of the fitting. If other structure is necessary for reasonable modeling in the alternative transformed space, then further work can be done to adapt the modeling to that setting, e.g., by modeling appropriate correlation between coefficients or assuming other types of prior distributions for penalization/regularization.
3. SIMULATION STUDIES
Simulation Setup
We designed a simulation study to compare the performances of R-WFMM and G-WFMM. Since real functional data sets have distinct complex structure in the wavelet space, to make our simulations realistic, we based our simulation upon a real data set: the organ-by-cell line MALDI-MS data of Section 4. We fit the G-WFMM to these data, and then used the fitted values of as the basis for the true distributions from which the data were simulated. To consider the relative performance of G-WFMM and R-WFMM with tails of varying degrees, we considered 5 different random distributions for the random effects and residual errors, with increasing heaviness of tails: Normal, DE, t3, t2, and t1 (Cauchy). We simulated random effects and residuals from these distributions, making the scale parameters approximately the same magnitude as , respectively, and then computed the simulated wavelet space data matrix D according to model (3), using the fixed and random effect design matrices X and Z analogous to those in Section 4. We simulated a total of 50 complete data sets, 10 for each tail type, and each data set consisted of 128 functions, 4 functions from each of 32 “animals”, with each function sampled on an equally spaced grid of 512. Full details are in supplementary materials.
Evaluation Criteria
We used three measures to summarize the methods’ performance in estimating the fixed effect functions Ba(t) and random effect functions Ub(t), the integrated mean squared error (IMSE), the integrated posterior variability (IPVar), and the integrated total variability (ITVar). The IMSE summarizes the variability of the posterior mean estimate about the truth; for a functional parameter θ(t), with true value θ0(t) and posterior mean θ̂(t) it is defined to be IMSE= ∫T {θ̂(t) − θ0(t)}2dt. The IPVar summarizes the posterior variability about the posterior mean; given posterior samples θ(g)(t), g = 1, …, G, it is defined to be . The ITVar summarizes the posterior variability about the true mean; it is defined to be . Note that ITVar=IMSE+IPVar.
For each summary measure, we computed relative efficiency (RE) as the ratio of G-WFMM and R-WFMM, then computing the mean RE across all 10 repetitions, index a for Ba(t) and index b for Ub(t), along with the corresponding 90% intervals. Results are presented in Table 1 and a supplementary figure, with larger numbers indicating greater efficiency for the R-WFMM.
Table 1.
Simulations: Relative efficiency of R-WFMM to G-WFMM (the ratio of G-WFMM/R-WFMM) in terms of integrated mean squared error (IMSE) of the posterior mean, integrated posterior variance about the posterior mean (IPVar), and integrated total variance around the true mean (ITVar), summarized by taking mean and 5% and 95% quantiles of the relative efficiencies
| ITVar | IPVar | IMSE | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Parameter | Tails | mean | Q05 | Q95 | mean | Q05 | Q95 | mean | Q05 | Q95 |
| Normal | 0.98 | 0.84 | 1.17 | 1.11 | 0.88 | 1.33 | 0.87 | 0.58 | 1.20 | |
| DE | 1.36 | 1.05 | 1.72 | 1.50 | 1.17 | 1.92 | 1.27 | 0.85 | 2.16 | |
| B(t) | t3 | 1.52 | 1.161 | 2.00 | 1.63 | 1.29 | 2.10 | 1.49 | 0.88 | 2.53 |
| t2 | 2.61 | 1.57 | 4.08 | 2.54 | 1.65 | 3.49 | 2.81 | 1.15 | 6.39 | |
| t1 | 14.18 | 3.12 | 25.85 | 14.31 | 5.28 | 31.78 | 22.09 | 1.13 | 93.63 | |
| Normal | 0.98 | 0.83 | 1.16 | 1.13 | 0.89 | 1.30 | 0.87 | 0.61 | 1.17 | |
| DE | 1.37 | 1.09 | 1.84 | 1.52 | 1.19 | 1.85 | 1.28 | 0.84 | 2.10 | |
| U(t) | t3 | 1.54 | 1.17 | 2.04 | 1.65 | 1.36 | 2.07 | 1.49 | 0.86 | 2.42 |
| t2 | 2.70 | 1.66 | 4.18 | 2.60 | 1.75 | 3.47 | 2.95 | 1.18 | 5.84 | |
| t1 | 13.05 | 1.87 | 22.17 | 14.04 | 5.00 | 46.72 | 20.64 | 1.11 | 55.50 | |
Simulation Results
For all 3 measures, the R-WFMM performed increasingly better than the G-WFMM as the tails got heavier, while the two methods performed similarly for Gaussian random effects and residual errors. More specifically, for the fixed effect functions, we see that the average improvement in IMSE of the R-WFMM over the G-WFMM is 27%, 49%, 2.81-fold (281%), and 22.09-fold for the DE, t3, t2, and t1, respectively (see Table 1). For Gaussian data, on average the R-WFMM was 13% less efficient than the G-WFMM. The R-WFMM demonstrated a reduction in posterior variation, as measured by the IPVar, for all distributions including the Gaussian, with average improvements of 11.2%, 50%, 63%, 2.54-fold, and 14.31-fold for Gaussian, DE, t3, t2, and t1, respectively. When put together, as measured by ITVar, the R-WFMM demonstrated average improvements of 36%, 52%, 2.61-fold, and 14.18-fold for the heavier-tailed distributions, respectively, while for Gaussian data the R-WFMM and G-WFMM were nearly identical, with estimated mean efficiency loss of just 2% for the R-WFMM. Similar results were obtained for the random effect functions. Thus, we see great improvement in the performance of R-WFMM over that of the G-WFMM, both in terms of estimation (IMSE) and variability (IPVar) for heavier-tailed data. The R-WFMM experienced a slight trade-off (≈ 13%) in estimation accuracy (IMSE) for Gaussian data that was basically offset by a reduction (≈ 11%) in posterior variability (IPVar).
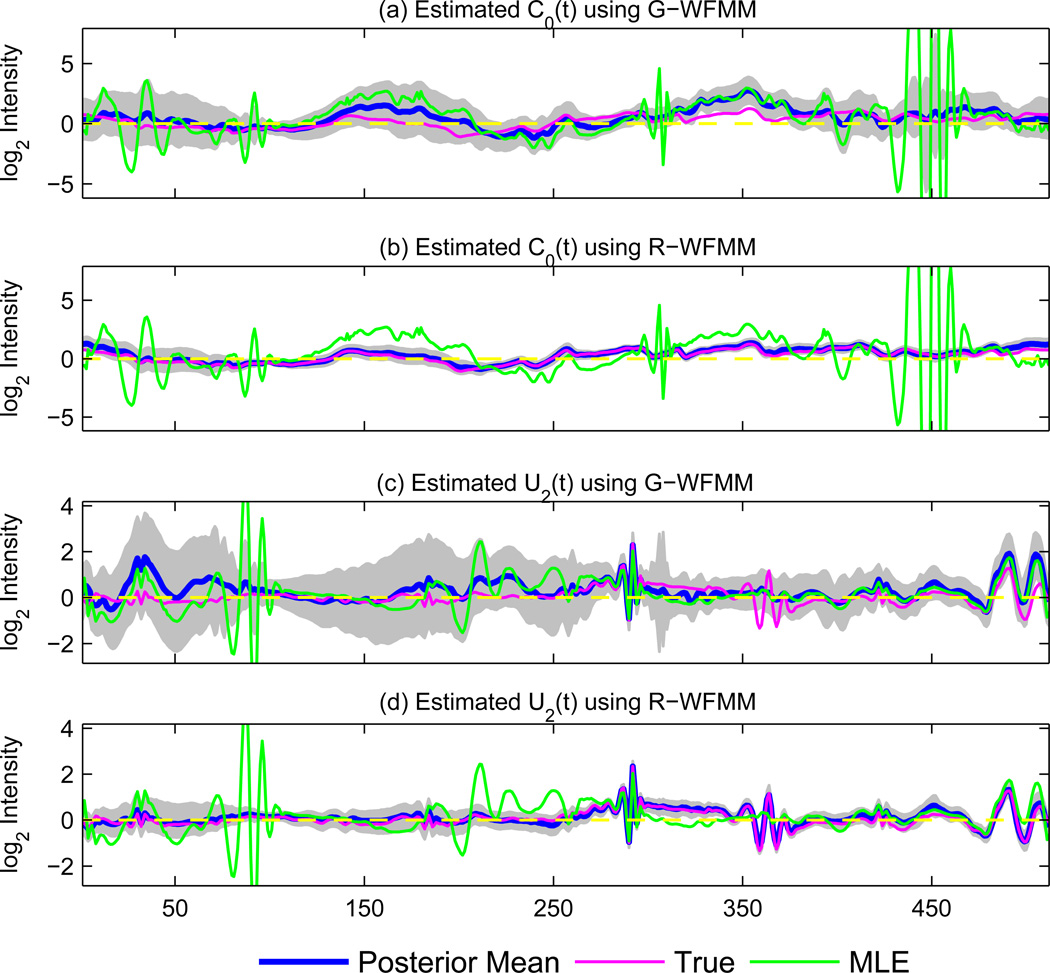
To investigate the nature of this observed improvement, for each data set and distribution simulated we plotted the posterior mean function for the G-WFMM and R-WFMM for each fixed effect function along with the true fixed effect function from which it was simulated. All plots are available as online supplementary materials, but here we present an example in the top two panels (a and b) of Figure 1 involving estimation of the overall mean function , where Ba(t) is the mean function for group a, from one of the Cauchy (t1) simulated data sets. The plot includes the true overall mean function in pink, the posterior mean for the G-WFMM (a) and R-WFMM (b) in blue with grey bands for 95% pointwise credible intervals, and non-regularized maximum likelihood estimators from the Gaussian model in green, obtained by applying the IDWT to the MLE estimates of using Henderson’s mixed model equations (pages 275–286, Searle, Casella and McCulloch 1992). This is the “unshrunken” MLE with no regularization prior and can be considered an unsmoothed non-robust functional estimate. Many of the regions of with large deviations from the true C0(t) correspond to regions with large outliers for some of the observed functions or animals.
Figure 1. Illustrating adaptive estimation of fixed and random effect functions.
The posterior mean (blue line) estimates for C0(t) and U2(t) for data simulated with Cauchy (t1) random effects and residuals and 95% credible intervals (grey bands) for G-WFMM ((a) and (c)) and R-WFMM ((b) and (d)) from one simulation run, along with true C0(t)/U2(t) (pink) and corresponding unregularized maximum likelihood estimates (green).
We see in this plot that the R-WFMM provided much better estimation and more adaptive regularization than the G-WFMM, in the sense that the R-WFMM was able to better capture the “true spikes” in C0(t) while smoothing out more of the “spurious wiggles”, and also providing much tighter pointwise credible intervals. Looking at the simulations with various tail heaviness, we see these results most dramatically for the heavier-tailed simulations (see supplemental plots). Naturally, these effects are most apparent in regions of the curve where the MLE deviates far from the truth (e.g., in intervals [30, 50] and [420, 490]), likely suggesting evidence of some extreme local outliers. In these regions, the G-WFMM is strongly affected by outliers, with relatively poor estimation and wide credible intervals, while the R-WFMM does a much better job, with posterior mean estimates very close to the truth and relatively small credible interval widths. It appears that the re-weighting of observations inherent to the R-WFMM was able to successfully downweight the influence of outliers on estimation, thus leading to improved estimates. The improved performance of the R-WFMM may also be partially due to its use of modern sparsity distributions in the wavelet space with excellent variable selection properties, leading to potential improvements in the adaptive regularization of the functional estimates. These same effects can be seen on analogous plots for the other fixed effect functions and other simulated data sets, which are all available as supplementary web materials (http://odin.mdacc.tmc.edu/~jmorris/papers.html).
We also see greatly improved estimation in the random effect functions Ul(t). The bottom two panels (c and d) of Figure 1 plot the posterior means and posterior credible intervals for G-WFMM and R-WFMM for U2(t) from one of the t1 simulations, again with the true functions and “unregularized MLEs” for U2(t). We once again see that for regions containing outliers, the G-WFMM has poor estimation and large credible intervals, while the R-WFMM does very well. This is most clear in the regions [0, 50], [170, 250], and [350, 400], where the G-WFMM estimate is far from the true U2(t) with very large credible intervals, and the R-WFMM is accurate with tight credible intervals. Notice how outliers appear to induce spurious wiggles in the MLE estimates near 100 and near 200, while attenuating a “true wiggle” near 350. Remarkably, the R-WFMM is able to automatically recognize that the former wiggles are spurious, and regularize them out, yet recognize that the latter wiggle is “real”, estimating it well with tight error bounds, in spite of the fact that it is not even apparent in the MLE. This is an excellent illustration of the interplay among robust estimation, adaptive regularization, and borrowing of strength between curves that we see in robust functional regression. Analogous plots for all random effects from all simulated data are available online at (http://odin.mdacc.tmc.edu/~jmorris/papers.html)
These results show the estimation benefits of R-WFMM. To evaluate the relative inferential performance, we computed posterior samples for the organ, cell line, and organ-by-cell line functional effects Ci(t), i = 1, 2, 3, defined in Section 4, for both the G-WFMM and R-WFMM. We then computed posterior probabilities of 1.5-fold expression changes for all 3 functional effects, and estimated the corresponding thresholds ϕ10 to declare significance based on a global FDR of α = 0.10, as overviewed in Section 2.3 and detailed in supplementary materials. Based on these determinations, we computed both the “realized” and “empirical” FDR, FNR, Sens, and Spec, plus the AUC and AUC10 for the realized and empirical ROC curves. The “realized” statistics are computed based on the true Ba(t), whereas the “empirical” quantities are estimated from the model without knowledge of the true Ba(t). Results are in a supplementary table.
Using the realized AUC to measure performance, we see that the R-WFMM considerably outperformed the G-WFMM for all simulation settings with heavier-than-normal tails, with the magnitude of the difference increasing with the heaviness of the tails. This suggests that the R-WFMM would have better operating characteristics in its detection of significant regions of the curves. This improvement is even more pronounced in the AUC-10, which focuses on the region of the ROC curve with highest specificity, and can also been seen in the individual FDR, FNR, Sens, and Spec statistics. These results were mirrored in the estimated empirical statistics, which did not presume knowledge of the true Ba(t). Note that the G-WFMM yielded slightly higher AUC and AUC-10 than the R-WFMM in the Gaussian simulation. This indicates, as expected, that some inferential price was paid for robust modeling when it was not needed, although the magnitude of this trade-off was not large compared with the improvements seen in setting of heavy-tailed distributions.
Since our primary goal of this simulation study was to compare the R-WFMM and G-WFMM, both of which involve wavelet-space modeling, we simulated the data using heavy tails in the wavelet space. Under the suggestion of a reviewer, we also simulated some data with heavy tails directly in the data space, as detailed in supplementary materials. We again found the R-WFMM performed better than the G-WFMM, with the IMSE approximately 2-fold better for t1 tails in the data space. This is roughly the same order of improvement we saw for the t2 or t3 data in the wavelet space, which is not surprising given wavelet coefficients involve weighted averages of observations in the data space, which may tend to lighten the tails in the wavelet domain.
4. APPLICATION
In this section, we illustrate our new robust R-FMM method by applying it to a cancer proteomics data set and comparing its performance with the G-WFMM. In this study, a tumor from one of two cancer cell lines was implanted into either the brain or lungs of 16 nude mice. The cell lines were A375P, a human melanoma cancer cell line with low metastatic potential, and PC3MM2, a highly metastatic human prostate cancer cell line. The goal was to find blood serum proteins differentially expressed between organ implant sites, implanted cell line types, or the organ-by-cell line interaction. This study was also considered in Morris, et al. (2008).
To study the proteome, blood serum from each animal was run through a MALDI-TOF mass spectrometer, which produces a proteomic spectrum y(t) that is a function with many peaks, with a peak at location t corresponding to a protein/peptide in the sample with molecular mass of t Daltons, with the spectral intensity y(t) giving a rough estimate of the corresponding protein abundance. In this experiment, we obtained two spectra for each mouse, one using a low laser intensity and once using a high laser intensity. Here we consider the part of the spectrum between t = 2, 000 and t = 14, 000 Daltons, a range that includes T = 7, 985 points per spectrum.
Model Setup
We used the same wavelet transform and FMM design matrices for both the G-WFMM and R-WFMM. After background correction and normalization of the mass spectra (Morris et al. 2005) followed by log2 transformation of the intensities, we applied a DWT to each spectrum, using the Daubechies wavelet with 8 vanishing moments, periodic boundary conditions, decomposed to J = 9 levels. We used the cell mean model for the factorial design with an additional column for the laser intensity effect, so that X in model (1) is a 32 × 5 matrix. Columns one to four indicated the treatment groups: brain-A375P, brain-PC3MM2, lung-A375P, lung-PC3MM2, respectively, while column five indicated whether the observations were from high (coded as 1) or low (coded as −1) laser intensity. The random effect design matrix Z was a 32 × 16 matrix of 0 and 1’s, with Zib = 1 indicating that spectrum i came from the bth animal, accounting for correlation between spectra from the same animal. From the posterior samples of the fixed effect functions, we computed linear transformations of interest, including the overall mean , and three contrast effects: the organ main effect C1(t) = 0.5(B1(t) + B2(t) − B3(t) − B4(t)), the cell-line main effect C2(t) = 0.5(B1(t) − B2(t) + B3(t) − B4(t)), and the organ-by-cell line interaction effect C3(t) = 0.5(B1(t) − B2(t) − B3(t) + B4(t)). Note that these linear combinations differ in scale from what was used by Morris, et al. (2008), which did not have the 0.5 factors.
For the G-WFMM, we specified vague proper beta and inverse gamma hyperpriors for πaj, τaj, qjk, and sjk, centered at the conditional maximum likelihood estimates determined as described in Morris and Carroll (2006), with large variances. For the R-WFMM, we used the vague proper Gamma priors for the population scale parameters and sparsity parameter πaj as described briefly in Section 2.3 and in more detail in supplementary materials. For each method, after a burn-in of 3000, we obtained 2000 posterior samples. Trace plots suggested good mixing. From these, we constructed posterior samples of the organ, cell line, and organ-by-cell line contrast functions Ci(t), i = 1, …, 3, respectively, and computed the posterior probabilities for each to be at least 1.5-fold different (> log2(1.5) in magnitude). The threshold corresponding to FDR of α = 0.10 was computed as described in Morris, et al. (2008), and the corresponding empirical FNR, Sens, Spec and ROC curve summaries were computed as described in supplementary materials. Figure 2 and a supplementary table summarize these results.
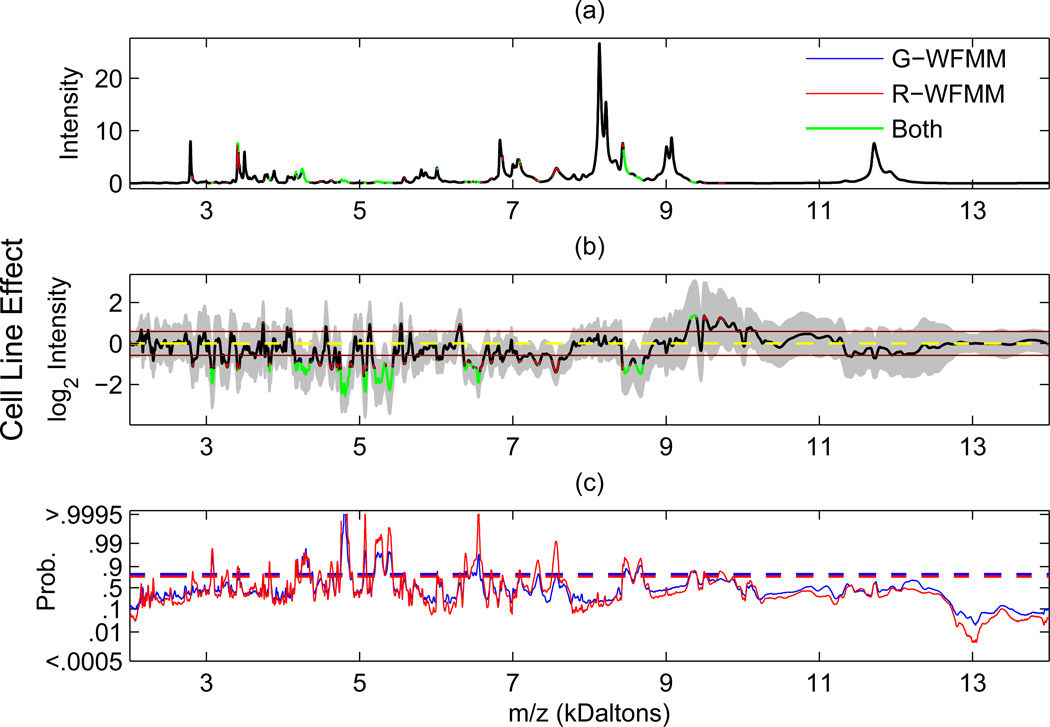
Figure 2. Regions flagged for 1.5-fold cell line effect by G-WFMM and R-WFMM.
(a) The significant regions flagged on the grand mean function C0(t) (defined in Section 3), plotted in the original scale. (b) The same regions flagged on the posterior mean cell line effect function C2(t) with 95% posterior intervals, plotted in log2 scale. In both (a) and (b), blue, red, and green indicate regions flagged by G-WFMM only, R-WFMM only, and by both methods, respectively. (c) The corresponding posterior probability estimates and the thresholds obtained using Bayesian FDR-based inference, with α = 0.10, with blue color for G-WFMM and red color for R-WFMM.
Results
The first two panels of Figure 2 contain for the R-WFMM the posterior mean for the overall mean spectrum C0(t) and the cell line main effect function C2(t). The third panel contains the corresponding posterior probability plots for a 1.5-fold difference, p2(t) = Prob{|C2(t)| > log2(1.5)|D} for the G-WFMM (blue) and R-WFMM (red), along with their respective thresholds determined by constraining the estimated global FDR≤ 0.10. In the first two panels, the colors indicate which methods flagged that region as “significant” in terms of a 1.5-fold difference with a global FDR of α ≤ 0.10: blue=G-WFMM only, red=R-WFMM only, green=both G-WFMM and R-WFMM, and black=neither. Equivalent plots for the organ and organ-by-cell-line interaction effects are available as supplementary material.
In these analyses, the R-WFMM flagged many more regions as significant compared to the G-WFMM. In Figure 2 summarizing the cell line effect function, 20 contiguous regions were flagged by both methods and 10 were flagged by the R-WFMM but not the G-WFMM, including [2815D, 2825D], [3255D, 3285D], [4460D, 4500D], [4610D, 4655D], [4890D, 4910D], [6300D, 6320D], [6705D, 6735D], [7510D, 7610D], [9485D, 9530D], and [9680D, 9770D]. There were no contiguous regions flagged by the G-WFMM for a cell line effect that were not flagged by the R-WFMM. For the organ main effect 28 contiguous regions were flagged by both methods, 10 were flagged by only R-WFMM, and 3 were flagged by only G-WFMM (results shown in supplementary material). For the organ-by-cell-line interaction function, 13 regions were flagged by both methods, 8 were flagged only by R-WFMM, and none were flagged by G-WFMM but not R-WFMM.
Based on the posterior samples for Ci(t), we also computed the empirical estimates of the FNR, Sens and Spec for 1.5 fold change, while specifying the global FDR of α = 0.1. These values are listed in a supplementary table, along with the mean width of the 95% credible intervals averaged across (1 : T), the empirical ROC curves were also computed, and the corresponding AUC and AUC-10. Compared to the performance of the G-WFMM, the R-WFMM model resulted in higher estimated AUC and AUC-10 values, smaller FNR, higher sensitivity, similar levels of specificity, and narrower 95% credible intervals.
Outlier Detection
We used the posterior samples of the scaling parameters to investigate possible outliers in the data as described in Section 2.4. We computed the statistics λi‥ for each individual spectrum, i = 1, …, 32, and ϕl‥ for each individual rat, l = 1, …, 16, and functional outlier statistics {λi(t)} and {ϕl(t)} for all spectra and rats, to check whether regions of certain curves were outliers. Overall and for each t, we computed pointwise medians and IQRs and flagged regions of t that were above median + 1.5 IQR as potential outliers. We found regions of certain spectra and from certain rats were local outliers. For example, spectrum 21 had unusually high levels of protein expression for proteins around 4000D and unusually low levels of expression for several peaks around 5000D and 10,000D. Rat 4 had unusually low levels of some proteins around 5000D, and unusually high levels for some protein around 7000D. These results are readily apparent in the pointwise outlier plots (figure in supplementary material), and serve as useful diagnostics to flag unusual curves or individuals for further investigation.
5. CONCLUSIONS AND DISCUSSIONS
We have introduced a novel method, R-FMM, that can be used to perform robust functional regression in the general functional mixed model framework. To our knowledge, this is the first robust functional response regression method in the statistical literature. Our approach involves modeling the functional data on the discrete grid in the wavelet space using a hierarchical scale mixture model that leads to robust modeling and desirable sparsity properties that translate to effective adaptive smoothing of fixed and random effect functions. Our approach leads to tractable calculations and a method that can feasibly be applied to various high-dimensional, complex functional data sets with our automated, efficient software, yielding robust functional inference and providing statistics for outlier detection and investigation. We presented modeling and computational details for a specific implementation of the R-FMM involving double-exponential distributions, 1D functions, and wavelet transforms that we call the R-WFMM, but the method can be applied more generally using other heavy-tailed distributions, higher dimensional functions, and using other invertible transformations as alternatives to wavelets.
Through simulation studies based on real mass spectrometry proteomic data, we demonstrated that the R-WFMM yielded improved estimation and inference over the Gaussian WFMM (G-WFMM) when the random effects and residual errors in the transformed space were heavy-tailed, with the relative improvement increasing with the heaviness of the tails. For both fixed and random effect functions, the R-WFMM demonstrated robustness to outliers, increased precision, and improved adaptive regularization, showing a remarkable ability to distinguish real local features from spurious ones in the functional estimates. This improvement can be explained by the interplay among the robustness properties from heavier-tailed likelihoods, the nonlinear shrinkage induced by the heavier-tailed prior distributions, and the ability to borrow strength across curves to better determine which features in the data are characteristic of the signal and which may be noise. These properties are induced by the specific carefully chosen hierarchical model components of our method that lead to interesting distributional characteristics both across and within the individual curves.
The hierarchical scale mixture distributions for the residuals and random effects in the wavelet space induce heavier-than-normal tails in the distribution across individuals for each wavelet coefficient. This leads to a weighted regression, whereby individuals with outlying values for a given wavelet coefficient are down-weighted in their influence on the regression parameter for that wavelet coefficient. Projected back to the data space, this effectively down-weights corresponding functional features of individual curves and random effect curves that are outliers relative to the rest of the data set. As vividly demonstrated in Figure 1, this robustness can remove outlier-induced spurious features present in naive estimates of the fixed or random effect functions, and can even uncover features truly present in the fixed and random effect functions but obscured by outliers in naive functional regression estimates.
The effective adaptive regularization is related to the model’s distributional assumptions within curves, which is induced by the hierarchical model across wavelet coefficients, for which separate scale parameters are allowed for each wavelet coefficient. In wavelet regression, the key to adaptive regularization is effective variable selection in the wavelet space, which is determined by two aspects of the method: the ability to set nonsignificant coefficients to zero (sparsity) to remove the noise and the ability to estimate the significant regression coefficients with minimal bias (low bias) to preserve the signal features. These shadow the two components of the oracle property studied in the asymptotic variable selection literature, (1) consistent variable selection (sparsity) and (2) optimal estimation (low bias). In Bayesian wavelet regression (and variable selection in general), these two properties depend on two characteristics of the prior distribution across wavelet coefficients: (1) density behavior near zero, and (2) the heaviness of the tails. Effective priors are able to place large amounts of density near zero while retaining heavy tails. The R-WFMM’s distribution for the random effects in the wavelet space across wavelet coefficients (j, k) is like an NEG (Griffin and Brown 2005), known to be an outstanding sparsity prior. It can be viewed as a scale mixture of double exponentials, with separate scale parameters for each wavelet coefficient (j, k) that are estimable because of the replicate random effect functions. This distribution has great flexibility in capturing high density near zero, leading to effective variable selection and shrinkage of the noise coefficients, and yet heavy enough tails to reduce the bias in estimating the large wavelet coefficients corresponding to the signal. Similar effects are seen for the fixed effect functions’ spike double-exponential-slab prior distribution, as well. Together with the down-weighting of outliers induced by the across-curve structure, these properties between wavelet coefficients (i.e.within curves) help explain the astoundingly adaptive properties we observed in our simulation results.
We found it remarkable in our simulations that our method appeared to be sufficiently robust to provide outstanding performance even for data with Cauchy tails, able to down-weight the extreme outliers and obtain accurate functional estimates with reasonably tight pointwise error bounds. Further, the R-WFMM was reasonably competitive with the G-WFMM when the data were truly Gaussian, with some loss in estimation accuracy (≈ 13%), which was partially offset by a realized gain in precision (≈ 12%). One might expect a greater loss of efficiency given the well-known result that the relative efficiency of the median for estimating the location parameter of a normal distribution is 2/π ≈ 0.637, but as described previously, there are other factors at play in our more complex robust FMM framework that may counterbalance the loss of efficiency from misspecified likelihood. There are already documented benefits of using double-exponential likelihoods for wavelet regression even when the true likelihood is Gaussian, and of using double-exponential slabs in variable selection settings. The benefits of these modeling structures are even greater in the multiple function FMM setting, in which we have replicate functions across which we can borrow strength in estimating the parameters regulating the sparsity, regularization, and tails. As a result, it seems reasonable to use the R-WFMM over the G-WFMM by default for functional response regression, given that the R-WFMM can provide the security of excellent robustness properties without trading off too much efficiency even if the data are truly Gaussian.
R-FMM appears to be a promising method for robust functional response regression for the analysis of functional and image data. However, there are some limitations and potential improvements of the method. The independence assumptions across basis coefficients lead to great computational advantages, but for some basis functions and data sets it may be appropriate to consider more general assumptions. The choice of wavelets as a basis space may not be best for all data sets. For example, wavelets have the weakness that individual wavelet coefficients are typically not intuitive to interpret. For a given data set, careful thought should be given to find the most suitable basis spaces or transforms to use. Principal component methods are popular in the FDA literature, and can provide extremely efficient basis representations for many functional data, especially when the functions are simple enough to be well represented by a small number of eigenfunctions. For complex high-dimensional functional data, however, eigenfunction-based analyses may not be ideal, considering recent results suggesting strong inconsistency of eigenvector estimation in high dimension, low sample size settings (Jung and Marron, 2009). As discussed in this paper, our approach to robust FMM does not depend on the choice of wavelets for basis space modeling; the approach could be applied using other bases or transforms, as well. The basis itself need not necessarily be interpretable, but it is sufficient if it is a suitable building block for modeling, since estimation, inference, and interpretation can be done in the original functional or image data space. For a given basis, one would need to consider whether the specific covariance and exponential-gamma assumptions used here make sense, and if not, to adapt the model to have assumptions that make sense.
While our code is automated and efficient enough for large functional and image data sets, for some enormous data sets (e.g. hundreds of GBs in size) the method could not be feasibly applied as described, as memory limitations may prevent the entire data matrix Y from loading into the computer at one time. Some specific calculations regarding computing time and memory requirements for Bayesian FMMs can be found in Herrick and Morris (2006), which provides some information about the feasibility and scalability of these methods to very large data sets. The high degree of parallelizability of these methods help it to scale up and run much quicker for very large data sets, and wavelet compression can be used to reduce the memory requirements by a factor of 20–100 or more (Herrick and Morris 2006; Morris, et al. 2011). However, the method as described will not immediately scale up to extremely large data sets, e.g. fMRI data where the raw data is 100’s of GB in size. For these data, the method would have to be adapted using multi-step approaches or approximations like variational Bayes, but these are topics for future development.
Also, further theoretical studies are needed to study the robustness properties of this hierarchical modeling framework and to explore exactly how we should study robustness in functional data analysis. Under the suggestion of a reviewer, we have performed some preliminary theoretical investigations of robustness, showing the influence of global and local outliers asymptotically go to zero in our hierarchical model applied to a simple mean+error functional model (see supplementary materials). Further investigations in the full FMM setting would be interesting and insightful, but very involved and beyond the scope of this paper.
Supplementary Material
Acknowledgments
Morris’s research is supported by a grant from the National Cancer Institute (CA-107304), and Morris and Zhu’s research is partially supported by the Program on Analysis of Object Data at SAMSI. The authors thank Jim Abbruzzesse, Nancy Shih, Stan Hamilton, Donghui Li, John Koomen, and Ryuji Kobayashi for the data set used in this paper, and thank LeeAnn Chastain for excellent editorial assistance.
Biographies
Zhu is Postdoctoral Fellow, Statistical and Applied Mathematical Sciences Institute, RTP, NC 27709 (E-mail: hzhu@samsi.info).
Brown is Professor, School of Mathematics, Statistics and Actuarial Science, University of Kent, U.K. (E-mail: Philip.J.Brown@kent.ac.uk).
Morris is Professor, Department of Biostatistics, University of Texas MD Anderson Cancer Center, Houston TX 77230 (E-mail: jefmorris@mdanderson.org).
REFERENCES
- Andrews DF, Mallows CL. Scale mixtures of normal distributions. J. R. Statist. Soc. B. 1974;36:99–102. [Google Scholar]
- Aston J, Chiou J-M, Evans J. Linguistic pitch analysis using functional principal component mixed effect models. Journal of the Royal Statistical Society, Series C. 2010;59:297–317. [Google Scholar]
- Ayers KL, Cordell HJ. SNP selection in Genome-Wide and candidate Gene studies via penalized logistic regression. Genetic Epidemiology. 2006;34:879–891. doi: 10.1002/gepi.20543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baladandayuthapani V, Mallick BK, Hong MY, Lupton JR, Turner ND, Carroll RJ. Bayesian Hierarchical Spatially Correlated Functional Data Analysis with Application to Colon Carcinogenesis. Biometrics. 2008;64:64–73. doi: 10.1111/j.1541-0420.2007.00846.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brumback BA, Rice JA. Smoothing spline models for the analysis of nested and crossed samples of curves. Journal of the American Statistical Association. 1998;93:961–976. [Google Scholar]
- Carvalho CM, Polson NG, Scott JG. The horseshoe estimator for sparse signals. Biometrika. 2010;97(2):465–480. [Google Scholar]
- Clyde M, George EI. Flexible empirical Bayes estimation for wavelets. JRSSB. 2000;62:681–698. [Google Scholar]
- Crambes C, Delsol L, Laksaci A. Robust nonparametric estimation for functional data. Journal of Nonparametric Statistics. 2008;20(7):573–598. [Google Scholar]
- Cutillo L, Jung YY, Ruggeri F, Vidakovic B. Larger posterior mode wavelet thresholding and applications. Journal of Statistical Planning and Inference. 2008;138:3758–3773. [Google Scholar]
- Dawid AP. Some matrix-variate distribution theory: notational considerations and a Bayesian application. Biometrika. 1981;68:265–274. [Google Scholar]
- Di C, Crainiceanu CM, Caffo BS, Punjabi NM. Multilevel Functional Principal Component Analysis. Annals of Applied Statistics. 2009;3(1):458–488. doi: 10.1214/08-AOAS206SUPP. Online access 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferraty F, Vieu P. Nonparametric Functional Data Analysis. New York: Springer-Verlag; 2006. [Google Scholar]
- Gervini D. Robust functional estimation using the median and spherical principal components. Biometrika. 2008;95(3):587–600. [Google Scholar]
- Gervini D. Detecting and handling outlying trajectories in irregularly sampled functional datasets. Annals of Applied Statistics. 2010 (to appear) [Google Scholar]
- Greven S, Crianiceanu CM, Caffo BS, Reich D. Longitudinal functional principal component analysis. Electronic Journal of Statistics. 2010 doi: 10.1214/10-EJS575. to appear. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffin JE, Brown PJ. CRiSM Working Paper No. 05-10. University of Warwick; 2005. Alternative prior distributions for variable selection with very many more variables than observations. [Google Scholar]
- Griffin JE, Brown PJ. Inference with Normal-Gamma prior distributions in regression problems. Bayesian Analysis. 2010;5(1) Posted online 2010–02–16. [Google Scholar]
- Guo W. Functional mixed effects models. Biometrics. 2002;58:121–128. doi: 10.1111/j.0006-341x.2002.00121.x. [DOI] [PubMed] [Google Scholar]
- Hampel FR, Ronchetti EM, Rousseauw PJ, Stahel WA. Robust Statistics: The Approach Based on Inuence Functions. 2 edn. New York: John Wiley & Sons; 2005. [Google Scholar]
- Herrick RC, Morris JS. Wavelet-based functional mixed model analysis:Computational Considerations. Proceedings, Joint Statistical Meetings, ASA Section on Statistical Computing. 2006:2051–2053. [Google Scholar]
- Huber P. Robust Statistics. New York: John Wiley & Sons; 1981. [Google Scholar]
- Hubert M, Rousseeuw PJ, Verboven S. A fast method for robust principal components with applications to chemometrics. Chemometrics and Intelligent Laboratory Systems. 2002;60:101–111. [Google Scholar]
- Johnstone IM, Silverman BW. Needles and straw in haystacks : empirical Bayes estimates of possibly sparse sequences. Ann. Statist. 2004;32:1594–1649. [Google Scholar]
- Johnstone IM, Silverman BW. Empirical Bayes selection of wavelet thresholds. Ann. Statist. 2005;33:1700–1752. [Google Scholar]
- Jung S, Marron JS. PCA Consistency in high dimension, low sample size context. The Annals of Statistics. 2009;37(6B):4104–4130. [Google Scholar]
- Kokoszka P, Maslova I, Sojka J, Zhu L. Probability tails of wavelet coefficients of magnetometer records. J. Geophys. Res. 2006;111 [Google Scholar]
- Locantore N, Marron JS, Simpson DG, Tripoli N, Zhang JT, Cohen KL. Robust principal component analysis for functional data. Test. 1999;8(1):1–73. [Google Scholar]
- Morris JS, Arroyo C, Coull BA, Louise MR, Herrick R, Gortmaker S. Using wavelet-based functional mixed models to characterize population heterogeneity in accelerometer profiles: a case study. J. Am. Statist. Ass. 2006;101:1352–1364. doi: 10.1198/016214506000000465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris JS, Baladandayuthapani V, Herrick RC, Sanna P, Gutstein H. Automated analysis of quantitative image data using isomorphic functional mixed models, with application to proteomics data. Annals of Applied Statistics. 2011 doi: 10.1214/10-aoas407. to appear. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris JS, Brown PJ, Baggerly KA, Coombes KR. Analysis of mass spectrometry data using Bayesian wavelet-based functional mixed models. In: Do K-A, Mueller P, Vannucci M, editors. Bayesian Inference for Gene Expression and Proteomics. New York: Cambridge University Press; 2006. pp. 269–288. [Google Scholar]
- Morris JS, Brown PJ, Herrick RC, Baggerly KA, Coombes KR. Bayesian analysis of mass spectrometry proteomics data using wavelet based functional mixed models. Biometrics. 2008;64:479–489. doi: 10.1111/j.1541-0420.2007.00895.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris JS, Carroll RJ. Wavelet-based functional mixed models. J. R. Statist. Soc. B. 2006;68:179–199. doi: 10.1111/j.1467-9868.2006.00539.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris JS, Coombes KR, Kooman J, Baggerly KA, Kobayashi R. Feature extraction and quantification for mass spectrometry data in biomedical applications using the mean spectrum. Bioinformatics. 2005;21(9):1764–1775. doi: 10.1093/bioinformatics/bti254. [DOI] [PubMed] [Google Scholar]
- Morris JS, Vannucci M, Brown PJ, Carroll RJ. Wavelet-based nonparametric modeling of hierarchical functions in colon carcinogenesis. Journal of the American Statisticsl Association. 2003;98:573–583. [Google Scholar]
- Nason GP, editor. Wavelet Methods in Statistics with R. Springer; 2008. [Google Scholar]
- Ogden R. Essential Wavelets for Statistical Applications and Data Analysis. Boston, USA: Birkhäuser; 1997. [Google Scholar]
- Park T, Casella G. The Bayesian Lasso. J. Am. Statist. Ass. 2008;103:681–686. [Google Scholar]
- Pensky M. Frequentist optimality of Bayesian wavelet shrinkage rules for Gaussian and non-Gaussian noise. Annals of Statistics. 2006;34(2):769–807. [Google Scholar]
- Ramsay JO, Silverman BW. Functional Data Analysis. Second Edition. New York: Springer; 2005. [Google Scholar]
- Ruppert D, Wand MP, Carroll RJ. Semiparametric regression during 2003–2007. Electronic Journal of Statistics. 2009;3:1193–1256. doi: 10.1214/09-EJS525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Searle SR, Casella G, McCulloch CE, editors. Variance Components. New York: John Wiley & Sons; 1992. [Google Scholar]
- Staicu AM, Crainiceanu CM, Carroll RJ. Fast Methods for Spatially Correlated Multilevel Functional Data. Biostatistics. 2010;11(2) doi: 10.1093/biostatistics/kxp058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staniswallis JG, Lee JJ. Nonparametric regression analysis of longitudinal data. Journal of the American Statistical Association. 1998;93:1403–1418. [Google Scholar]
- Tibshirani R. Regression shrinkage and selection via the Lasso. J. R. Statist. Soc. B. 1996;58:267–288. [Google Scholar]
- Vidakovic B. Statistical Modeling by Wavelets. New York: John Wiley & Sons, Inc.; 1999. [Google Scholar]
- Vidakovic B, Ruggeri F. BAMs method: theory and simulations. The Indian Journal of Statistics. 2001;63:234–249. [Google Scholar]
- Wang Y. Mixed effects smoothing spline analysis of variance. Journal of the Royal Statistical Society, Series B. 1998;60:159–174. [Google Scholar]
- West M. On scale mixtures of normal distributions. Biometrika. 1987;74:646–648. [Google Scholar]
- Wu H, Zhang JT. Local polynomial mixed-effect models for longitudinal data. Journal of the American Statistical Association. 2002;97:883–897. [Google Scholar]
- Zhou L, Huang JZ, Martinez JG, Maity A, Baladandayuthapani V, Carroll RJ. Reduced rank mixed effects models for spatially correlated hierarchical functional data. Journal of the American Statistical Association. 2010;105:390–400. doi: 10.1198/jasa.2010.tm08737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zipunnikov V, Caffo BS, Crainiceanu CM, Yousem DM, Davatzikos C, Schwartz BS. Working Paper 219. Johns Hopkins University, Department of Biostatistics Working Papers; 2010. Multilevel functional principal component analysis for high-dimensional data. http://www.bepress.com/jhubiostat/paper219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou H. The adaptive lasso and its oracle properties. Journal of the American Statistical Association. 2006;101:1418–1429. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.