Abstract
The amelogenin proteins are required for normal enamel development; the most abundant amelogenins expressed from alternatively spliced mRNAs are M180 and leucine rich amelogenin protein (LRAP). The Amelx null (KO) mouse has an enamel defect similar to human X-linked amelogenesis imperfecta. The disorganized enamel layer in KO mice is 10–20% the thickness of wild-type (WT) enamel and lacks prismatic structures. When the KO mice were mated with mice that express TgM180-87, partial rescue of the phenotype was observed such that enamel thickness, volume and density increased. A second transgene was introduced by mating the TgM180KO mice with TgLRAP mice, and male offspring were characterized for genotype and tooth phenotype was evaluated by SEM. TgM180LRAPKO molar enamel thickness further increased, and the structure was improved, with a more defined decussation pattern compared to singly rescued mice. We conclude that TgM180 provides significant rescue of the KO phenotype. Although the effectiveness of TgLRAP to rescue by itself is less obvious, the addition of TgLRAP to TgM180 in KO enamel leads to added improvement in both amount and structure and thus these transgenes function in a complementary manner. Together the two most abundant amelogenins lead to formation of obvious enamel decussation patterns.
Keywords: amelogenin, transgenic mice, amelogenesis imperfecta, phenotypic rescue
The composition of the dental enamel that covers the crown of the tooth is greater than 95% mineral when the tooth erupts into the oral cavity. The enamel organic matrix that is secreted by ameloblast cells at the beginning of enamel development is comprised primarily of amelogenins plus lesser amounts of other secreted proteins, including enamelin and ameloblastin (1–3). These proteins form an extracellular matrix that begins to mineralize shortly after secretion begins. Proteases are also released by the ameloblasts, and these enzymes break down the extracellular matrix in an organized way so that the mineral crystals grow as the peptides are removed from the matrix (4,5).
Each secretory stage ameloblast releases enamel proteins from a distal cellular extension known as Tomes’ process, and crystals develop and begin to organize into enamel rods or prisms. Ameloblasts are interconnected by junctional complexes, and alternating rows of ameloblasts are thought to move past each other as the enamel rods are formed (6). In this way, groups of ameloblasts can direct the organization of their secreted product into a unique mineralized structure with a decussating pattern, including rows of interwoven enamel rods.
At least 15 Amelx mRNAs have been described in murine enamel organs due to extensive alternative splicing of the Amelx primary RNA transcript (7–9). Because exons 1 and 2 are present in all mRNAs for this 7 exon gene (9 in rodents), the alternative splicing occurs between the remaining exons 3, 4, and 5 or within exon 6. In order to better understand the function of the amelogenin proteins translated from these mRNAs, an Amelx null (KO) mouse was generated that made none of the amelogenins (10). This mouse had defective dental enamel with an appearance similar to that of humans from kindreds with X-linked amelogenesis imperfecta with AMELX gene mutations (11). Enamel in the Amelx KO mouse was hypoplastic and disorganized, and lacked discernable enamel rod formation and normal decussation patterns (10).
A transgenic mouse was generated with ameloblast-specific expression of TgM180, the most abundant 26 kDa amelogenin protein. These mice developed enamel that lacked obvious defects as expected (12). When this mouse was mated with the Amelx KO mouse, partial rescue of molar enamel thickness was observed, and volume and density were improved (13).
The second most abundant amelogenin, LRAP or leucine rich amelogenin protein, is the product of an alternatively spliced Amelx message (14,15). LRAP mRNA lacks a large segment of the exon 6 coding region, but includes both the N- and C-termini of the most abundant M180 amelogenin protein. When Amelx KO and TgLRAP mice were mated and transgene positive male pups were analyzed, it was found that TgLRAP was unable to substantially rescue the Amelx KO phenotype, although LRAP improved enamel rod organization somewhat when etched samples were observed by scanning electron microscopy (SEM;16). We sequentially mated these mice to generate a new mouse model that was KO for Amelx but expressed both theTgM180 and the TgLRAP transgenes, and evaluated the enamel for phenotypic rescue.
Material and Methods
Animal Models
Generation of the Amelx KO, TgM180 and TgLRAP mice was described previously (10,12,17). Mice were housed in an AAALAC accredited facility, and procedures were approved by the University of Pennsylvania Institutional Animal Care and Use Committee. Amelx KO females were mated with transgenic males in order to generate offspring, and crossing continued until the desired genotype was generated.
DNA Analysis
High molecular weight genomic DNA was isolated from mouse tails. The Qiagen multiplex PCR kit (Valencia, CA, USA) was used for PCR reactions, and products were analyzed on a 4% NuSieve 3–1 agarose gel (Lonza Rockland, Rockland, ME, USA). PCR primer sequences and conditions for determination of KO, het or WT genotype and transgene status have been described (10,12,13,17)
Microscopic Analysis of Teeth
Mandibles from mice between 6 wk and 3 months of age were fixed in 4% paraformaldehyde overnight, and light microscopy was used for initial characterization of samples. Mandibles were fractured using a razor blade, and SEM analysis of tooth surfaces and fractured internal enamel and dentin surfaces of incisors and molars was completed at 15 kV (FEI Quanta 200 FEG, FEI, Hillboro, OR, USA). Molars and incisors also were examined with ESEM after being embedded in epoxy resin, cut, polished to 0.05 micron finish and etched with 20% vol/vol H3PO4 for 5 to 15 s depending on the thickness of enamel (thin enamel etched for shorter duration).
MicroCT
MicroCT scans were performed using a Skyscan 1074HR portable x-ray microtomograph (Skyscan, Kontich, Belgium) as described previously (13). Fixed samples from 6 wk to 3 month old mice were scanned at a resolution of 20.7 μm/pixel and 40 kV under saline solution in a 600 μl microcentrifuge tube, through 180 degrees of rotation, with exposure time of 420 ms. The specimen was positioned such that the body of the mandible was perpendicular to the sectioning plane with the condyle positioned apically, and the incisor tip pointing up. The resulting images were evaluated using Skyscan CT-analyzer software ver 1.9.3.0 (Skyscan) to determine enamel density and volume. Hydroxyapatite phantoms (250 mg/cc and 750 mg/cc) (CIRS, Norfolk, VA, USA) were used in order to determine densities within molar enamel regions and volumes of interest. The mandibular first molar was analyzed at the position of the mesial root apex to determine enamel volume and density.
Statistical analysis
ANOVA with Bonferroni’s Multiple Comparison Test was performed to compare differences between thickness, volume and density measurements, with significance at P<0.05.
Results
Amelx KO female mice were mated with TgM180-87 or TgLRAP males, and male offspring with the transgene were identified by PCR. Repeated matings were required to generate mice with both TgM180 and TgLRAP on the KO background, and each of the double or single transgene positive mice were also analyzed for genetic background to be certain they were KO. Adult mice were sacrificed and mandibles were dissected and fixed. Mandibles were imaged by light microscopy and subjected to microCT analysis. The mandibles were embedded in epoxy. The sections were etched in phosphoric acid for ESEM analysis. Table 1 gives enamel thickness measurements for molars and incisors from each of the groups of mice, plus WT controls.
Table 1.
Molar and incisor enamel thickness from scanning electron micrographs
| Genotype | Molar Enamel Thickness (μm) | Incisor Enamel Thickness (μm) | ||
|---|---|---|---|---|
| n | Mean +/−SD | n | Mean +/−SD | |
| WT | 10 | 59.5 +/−9.8 | 12 | 124.1 +/−10.5 |
| Amelx het | 5 | 25.3 +/−8.8 * | 4 | 100.7 +/−16.8 * |
| KO | 18 | 13.1 +/−4.0 * | 6 | 13.7 +/−1.2 * |
| LRAPKO | 9 | 18.0 +/−7.6 * | 9 | 18.2 +/−2.3 * |
| M180-87KO | 9 | 30.9 +/−7.0 ** | 5 | 28.2 +/−3.3 * |
| M180-87LRAPKO | 8 | 37.4 +/−8.3 ** | 3 | 25.7 +/−7.2 * |
different from WT P<0.05;
different from WT and from KO P<0.05
Enamel thickness of incisors is greater than twice the value for molars in WT mice. The heterozygous (het) female control mice have only one copy of the Amelx gene and have reduced enamel thickness for both molars and incisors as expected, while the KO mice have the most severe hypoplasia. Improvement in molar and incisor thickness of KO enamel is seen when either transgene is expressed, and additional improvement was observed to 65% of WT molars when both transgenes were present. Transgene amount had previously been measured using Western blot and is greater in molars compared to incisors in 11 experiments using various transgenic strains generated with this vector system (13, and unpublished observations). Greater improvement seems to correlate with the higher level of expression. Although WT molar and incisor enamel thicknesses were significantly different from any of the other genotypes, TgM180 or doubly rescued molar enamel thicknesses were significantly different from KO (P<0.05). Enamel thickness did not vary significantly when samples from 6 wk and 3 month old mice were compared (not shown).
Using microCT analysis, volumes were measured for molars from the different categories of mice, but incisors were not included as most KO or transgeneKO incisor teeth did not have a measurable enamel layer using microCT. Table 2 provides mean measurements from microCT analysis for volume of molars and indicates a volume increase compared to KO when TgM180 was present. Volume measurements were improved to approximately 50% of WT in these transgenic/KO mice. TgM180 singly or doubly rescued enamel differed significantly from KO as well (P<0.05). A thin or undermineralized enamel layer is not easily measured by microCT, yielding 0 values.
Table 2.
Molar enamel volume and density from microCT analysis
| Genotype | n | Volume (voxels)† | Density (gm/cm3) |
|---|---|---|---|
| Mean +/−SD | Mean +/−SD | ||
| WT | 6 | 13,789 +/−3,744 | 2.4 +/−0.10 |
| Amelx het | 2 | 0 | 0 |
| KO | 21 | 0 | 0 |
| LRAPKO | 14 | 0 | 0 |
| M180-87KO | 12 | 6,832 +/−1,864 * | 2.3 +/−0.07 |
| M180-87LRAPKO | 9 | 5,798 +/−987 * | 2.5 +/−0.20 |
Different from WT P<0.05
One voxel is approximately 428.5 μm3
Zero values: below limit of resolution by microCT
Molar enamel density from the microCT analysis is also shown in Table 2. As we previously reported (13), the density is completely rescued as long as TgM180 is expressed in the KO mice. WT molar enamel density was not different from rescued murine enamel when TgM180 or both transgenes were present (P<0.05).
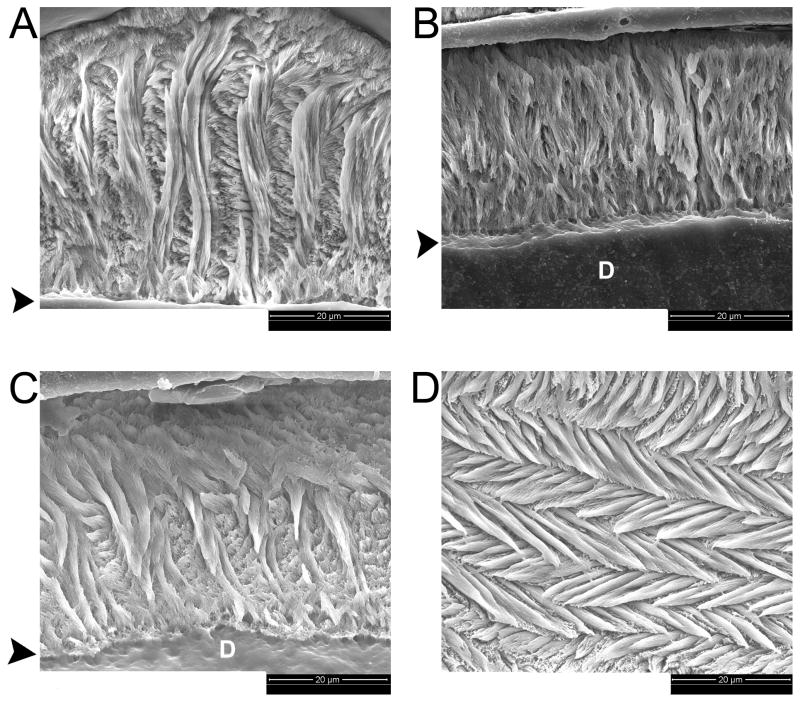
Because TgM180 was able to partially rescue the KO enamel phenotype but TgLRAP by itself provided only minimal change, we were curious to see the structural effect on a KO mouse that expressed both transgenes. Fig. 1 shows SEM images of typical molar teeth from the four different genotypes. Normal WT enamel has the complex interwoven enamel rod pattern (Fig. 1A). The KO mouse has a thin layer of disorganized enamel (Fig. 1B), which was similar to TgLRAPKO (not shown). Rescue is seen in the TgM180-87KO mouse (Fig. 1C), which has a more robust enamel layer. Shown is one of the mice where we were able to detect the decussation patterns within this still hypoplastic enamel layer. The TgM180-87LRAPKO mice developed pronounced decussation patterns in this oblique section through molar enamel (Fig. 1D); this structure was detected in most of the etched double transgenic/KO mouse molars examined.
Figure 1.
Enamel structure in WT, Amelx KO and rescued molars by scanning electron microscopy of etched enamel. A. WT enamel from a female mouse. B. KO enamel from a male mouse. C. A TgM180-87KO male. D. A TgM180-87LRAPKO male seen in oblique section. Arrowheads mark the position of the enamel-dentin junction; D indicates the dentin layer.
Discussion
Using mouse models that express transgenes under control of the Amelx regulatory sequences, we show that TgM180 and TgLRAP transgenes are able to substantially rescue the enamel phenotype of the Amelx KO mice. Although the enamel thickness and volume are not as great as in the WT enamel layer, the addition of the TgLRAP to TgM180 increases enamel thickness and leads to enhanced development of decussating patterns of enamel in molars. These transgenes are expressed at a higher level in molars compared to incisors, but incisors have some improvement nevertheless. It is possible that rescue was limited due to environmental effects, leading to enhanced attrition. However, since 6 wk and 3 month old samples were similar, this does not seem to have a major effect.
The density of the enamel layer is normal in singly or doubly transgenic KO mice as long as TgM180 is present. Previously it was shown that although the density was improved by TgM180, hardness and elastic modulus were not, perhaps because of defective organization of the enamel structure (13).
LRAP protein is abundant in developing enamel and can form structures referred to as nanospheres in vitro (18). While nanospheres generated from M180 can increase mineral formation in vitro, the LRAP nanospheres do not (19). Some proteins are described as bifunctional and LRAP has been proposed, in addition to a functional role in enamel development, to have signaling properties as well (20,21). M180 has also been shown to alter in vitro gene expression (22–24) as well as RNA stability using cell culture (25), and could perhaps have an additional in vivo role as well.
A question to consider is the reason behind the abundant amount of amelogenin alternative splicing, rather than expression of a single protein with a modular design (26) that can provide all required functions. For a protein with both structural and signaling roles, alternative splicing may separate these and other roles both temporally and spatially during development.
At this point it is not possible to resolve whether the improvement in KO enamel mediated by the transgenes is due to contribution to structural properties during enamel development or to signaling to the ameloblasts in order to influence their activity, or both. A question also remains about the role of the other amelogenins as at least 15 mRNAs have been cloned and each has the molecular capability to be translated into amelogenin proteins. Perhaps100 percent rescue requires the expression of one or more additional amelogenin proteins during enamel development.
Acknowledgments
We acknowledge excellent animal care by University of Pennsylvania School of Dental Medicine vivarium personnel. This work was supported by the NIH through NIDCR grant DE011089.
References
- 1.TERMINE JD, BELCOURT AB, CHRISTNER PJ, CONN KM, NYLEN MU. Properties of dissociatively extracted fetal tooth matrix proteins. I. Principal molecular species in developing bovine enamel. J Biol Chem. 1980;255:9760–9768. [PubMed] [Google Scholar]
- 2.HU JCC, HU Y, SMITH CE, MCKEE MD, WRIGHT JT, YAMAKOSHI Y, PAPAGERAKIS P, HUNTER GK, FENG JQ, YAMAKOSHI F, SIMMER JP. Enamel defects and ameloblast-specific expression in enamelin knockout/LACZ knockin mice. J Biol Chem. 2008;283:10858–10871. doi: 10.1074/jbc.M710565200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.FUKUMOTO S, KIBA T, HALL B, IEHARA N, NAKAMURA T, LONGENECKER G, KREBSBACH PH, NANCI A, KULKARNI AB, YAMADA Y. Ameloblastin is a cell adhesion molecule required for maintaining the differentiation state of ameloblasts. J Cell Biol. 2004;167:973–983. doi: 10.1083/jcb.200409077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.BARTLETT JD, SIMMER JP. Proteinases in developing dental enamel. Crit Rev Oral Biol Med. 1999;10:425–441. doi: 10.1177/10454411990100040101. [DOI] [PubMed] [Google Scholar]
- 5.SIMMER JP, HU JCC. Expression, structure and function of enamel proteinases. Conn Tiss Res. 2002;43:441–449. doi: 10.1080/03008200290001159. [DOI] [PubMed] [Google Scholar]
- 6.SKOBE Z, STERN D. Scanning and transmission electron microscopy of enamel formation and structure in the molar teeth of rats. Archs Oral Biol. 1978;23:307–316. doi: 10.1016/0003-9969(78)90024-9. [DOI] [PubMed] [Google Scholar]
- 7.SIMMER JP, HU CC, LAU EC, SARTE P, SLAVKIN HC, FINCHAM AG. Alternative splicing of the mouse amelogenin primary RNA transcript. Calcif Tissue Int. 1994;55:302–310. doi: 10.1007/BF00310410. [DOI] [PubMed] [Google Scholar]
- 8.LI W, MATHEWS C, GAO C, DENBESTEN PK. Identification of two additional exons at the 3’ end of the amelogenin gene. Arch Oral Biol. 1998;43:497–504. doi: 10.1016/s0003-9969(98)00013-2. [DOI] [PubMed] [Google Scholar]
- 9.LI Y, YUAN ZA, ARAGON MA, KULKARNI AB, GIBSON CW. Comparison of body weight and gene expression in amelogenin null and wildtype mice. Eur J Oral Sci. 2006;114 (Suppl 1):190–193. doi: 10.1111/j.1600-0722.2006.00286.x. [DOI] [PubMed] [Google Scholar]
- 10.GIBSON CW, YUAN ZA, HALL B, LONGENECKER G, CHEN E, THYAGARAJAN T, SREENATH T, WRIGHT JT, DECKER S, PIDDINGTON R, HARRISON G, KULKARNI AB. Amelogenin-deficient mice display an amelogenesis imperfecta phenotype. J Biol Chem. 2001;276:31871–31875. doi: 10.1074/jbc.M104624200. [DOI] [PubMed] [Google Scholar]
- 11.WRIGHT JT, HART PS, ALDRED MJ, SEOW K, CRAWFORD PJM, HONG SP, GIBSON CW, HART TC. Relationship of phenotype and genotype in X-linked amelogenesis imperfecta. Connect Tissue Res. 2003;44 (Suppl 1):72–78. [PubMed] [Google Scholar]
- 12.GIBSON CW, YUAN ZA, LI Y, DALY B, SUGGS C, ARAGON MA, ALAWI F, KULKARNI AB, WRIGHT JT. Transgenic mice that express normal and mutated amelogenins. J Dent Res. 2007;86:331–335. doi: 10.1177/154405910708600406. [DOI] [PubMed] [Google Scholar]
- 13.LI Y, SUGGS C, WRIGHT JT, YUAN ZA, ARAGON M, FONG H, SIMMONS D, DALY B, GOLUB EE, HARRISON G, KULKARNI AB, GIBSON CW. Partial rescue of the amelogenin null dental enamel phenotype. J Biol Chem. 2008;283:15056–15062. doi: 10.1074/jbc.M707992200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.FINCHAM AG, BELCOURT AB, TERMINE JD, BUTLER WT, COTHRAN WC. Dental enamel matrix: sequences of two amelogenin polypeptides. Biosci Rep. 1981;1:771–778. doi: 10.1007/BF01114799. [DOI] [PubMed] [Google Scholar]
- 15.GIBSON CW, GOLUB EE, DING W, SHIMOKAWA H, YOUNG M, TERMINE J, ROSENBLOOM J. Identification of the leucine-rich amelogenin peptide (LRAP) as the translation product of an alternatively spliced transcript. Biochem Biophys Res Comm. 1991;174:1306–1312. doi: 10.1016/0006-291x(91)91564-s. [DOI] [PubMed] [Google Scholar]
- 16.GIBSON CW, LI Y, DALY B, SUGGS C, YUAN ZA, FONG H, SIMMONS D, ARAGON M, KUKKARNI AB, WRIGHT JT. The leucine-rich amelogenin peptide alters the amelogenin null enamel phenotype. Cells Tissues Organs. 2009;189:169–174. doi: 10.1159/000151384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.CHEN E, YUAN ZA, WRIGHT JT, HONG SP, LI Y, COLLIER PM, HALL B, D’ANGELO M, DECKER S, PIDDINGTON R, ABRAMS WR, KULKARNI AB, GIBSON CW. The small bovine amelogenin LRAP fails to rescue the amelogenin null phenotype. Calcif Tissue Int. 2003;73:487–495. doi: 10.1007/s00223-002-0036-7. [DOI] [PubMed] [Google Scholar]
- 18.FINCHAM AG, MORADIAN-OLDAK J, DIEKWISCH TG, LYARUU DM, WRIGHT JT, BRINGAS P, JR, SLAVKIN HC. Evidence for amelogenin “nanospheres” as functional components of secretory-stage enamel matrix. J Struct Biol. 1995;115:50–59. doi: 10.1006/jsbi.1995.1029. [DOI] [PubMed] [Google Scholar]
- 19.HABELITZ S, DENBESTEN PK, MARSHALL SJ, MARSHALL GW, LI W. Self-assembly and effect on crystal growth of the leucine-rich amelogenin peptide. Eur J Oral Sci. 2006;114 (Suppl 1):315–319. doi: 10.1111/j.1600-0722.2006.00312.x. [DOI] [PubMed] [Google Scholar]
- 20.VEIS A. Amelogenin gene splice products: potential signaling molecules. Cell Mol Life Sci. 2003;60:38–55. doi: 10.1007/s000180300003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.BOABAID F, GIBSON CW, KUEHL MA, BERRY JE, SNEAD ML, NOCITI FH, JR, KATCHBURIAN E, SOMERMAN MJ. Leucine-rich amelogenin peptide: a candidate signaling molecule during cementogenesis. J Periodontol. 2004;75:1126–1136. doi: 10.1902/jop.2004.75.8.1126. [DOI] [PubMed] [Google Scholar]
- 22.VISWANATHAN HL, BERRY JE, FOSTER BL, GIBSON CW, LI Y, KULKARNI AB, SNEAD ML, SOMERMAN MJ. Amelogenin: a potential regulator of cementum-associated genes. J Periodontol. 2003;74:1423–1431. doi: 10.1902/jop.2003.74.10.1423. [DOI] [PubMed] [Google Scholar]
- 23.SHIMIZU E, SAITO R, NAKAYAMA Y, NAKAJIMA Y, KATO N, TAKAI H, KIM DS, ARAI M, SIMMER J, OGATA Y. Amelogenin stimulates bone sialoprotein (BSP) expression through fibroblast growth factor 2 response element and transforming growth factor-s1 activation element in the promoter of the BSP gene. J Periodontol. 2005;76:1482–1489. doi: 10.1902/jop.2005.76.9.1482. [DOI] [PubMed] [Google Scholar]
- 24.SWANSON EC, FONG HK, FOSTER BL, PAINE ML, GIBSON CW, SNEAD ML, SOMERMAN MJ. Amelogenins regulate expression of genes associated with cementoblasts in vitro. Eur J Oral Sci. 2006;114 (Suppl 1):239–243. doi: 10.1111/j.1600-0722.2006.00321.x. [DOI] [PubMed] [Google Scholar]
- 25.XU L, HARADA H, YOKOHAMA-TAMAKI T, MATSUMOTO S, TANAKA J, TANIGUCHI A. Reuptake of extracellular amelogenin by dental epithelial cells results in increased levels of amelogenin mRNA through enhanced mRNA stabilization. J Biol Chem. 2006;281:2257–2262. doi: 10.1074/jbc.M507695200. [DOI] [PubMed] [Google Scholar]
- 26.SNEAD ML. Amelogenin protein exhibits a modular design: implications for form and function. Connect Tissue Res. 2003;44 (Suppl 1):47–51. [PubMed] [Google Scholar]