Abstract
Cell signalling pathways and networks are complex and often non-linear. Signalling pathways can be represented as systems of biochemical reactions that can be modelled using differential equations. Computational modelling of cell signalling pathways is emerging as a tool that facilitates mechanistic understanding of complex biological systems. Mathematical models are also used to generate predictions that may be tested experimentally. In the present chapter, the various steps involved in building models of cell signalling pathways are discussed. Depending on the nature of the process being modelled and the scale of the model, different mathematical formulations, ranging from stochastic representations to ordinary and partial differential equations are discussed. This is followed by a brief summary of some recent modelling successes and the state of future models.
Introduction
The perception of mathematical modelling in cell biology is changing from a theoretical exercise of limited interest to a tool facilitating mechanistic understanding of complex phenomena and better experimental design. As a result, there has been an enhanced incorporation of mathematical modelling in the study of many complex biological processes. A major goal of modelling biological systems is to be able to provide a mechanistic explanation of the underlying processes. More often, models are built to predict behaviours that may be hard to understand because of the multiple variables involved. These predictions can then be tested by experiments and the model can be refined. Modelling is thus an ongoing exercise of analysis and experimental validation, incorporating new biological data as they become available.
Modelling is particularly useful in analysing information flow through cell signalling networks. Cellular signalling pathways regulate the programmes for various cellular functions that change in response to a stimulus. These functions arise from the underlying biochemical reactions and, therefore, lend themselves to quantitative modelling. Modelling of these networks and pathways allows us to organize existing knowledge and explore the signalling pathways for emergent properties.
One of the key features of cell signalling networks is the non-linearity arising from the presence of regulatory loops and branches in the signalling network. This non-linearity makes it challenging to understand and predict responses to a single stimulus; predicting responses is harder still for multiple stimuli, which is the norm in real biological systems. Developing mathematical models for these complex networks allows us to express these non-linearities in the equations that constitute the computational models and explore the models systematically.
Computational models of signalling networks serve two purposes: organization and validation of known experimental data, and prediction of system behaviour that is not intuitive from prior experiments. Predictions made from the numerical simulations can be experimentally verified or nullified and the model can be improved by an iterative process of models→experiments→models. Thus modelling serves as a tool for obtaining systems level understanding of the underlying biological processes.
Complexity of signalling networks
The complexity of biological systems is well-known. Increasing the number of components and the non-linear interactions between them leads to increasing system complexity [1]. This complexity is reflected in the number of interacting species, the number of interactions between them and the parameters involved. One purpose of computational models is to identify behaviours that may not be evident from the experimental data. The number of possible interactions increases combinatorially with the number of components. Assuming each reaction requires at least two parameters (forward and backward rate for mass-action relationships, and kcat and Km for enzymatic relationships), we have at least twice as many kinetic parameters as we have reactions. In addition, we also need the initial concentrations of the components in the system. The heterogeneity and complexity of biological systems makes development of models challenging, but it is this complexity that also makes modelling necessary. Using models it is possible to explain what is happening at different time scales to different modules and components of a network.
Model construction
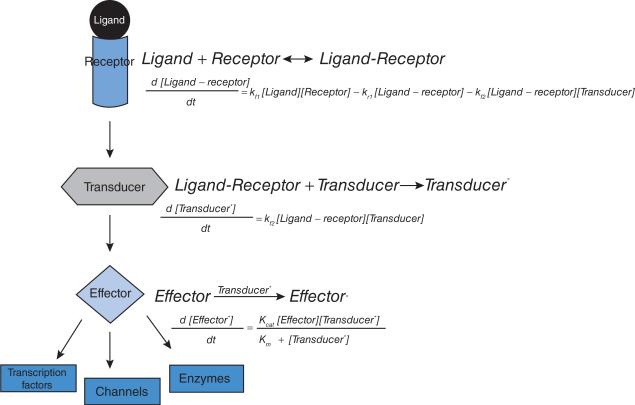
Model construction for signalling networks begins with a wiring diagram that shows the interactions between the system components. The wiring diagram can be translated into a series of coupled biochemical reactions and a system of differential equations (Figure 1). Since we generally model a specific signalling pathway, it becomes necessary to identify the boundaries of the system. Typically, a model for a signalling pathway might begin at the receptor and end at an effector enzyme, transcription factor or channel whose activity is altered by the receptor signal. Such a system can be described by a set of coupled ODEs (ordinary differential equations). In contrast, a network that describes how cell-surface adhesion molecules regulates the actin cytoskeleton requires a nuanced approach. Here, if the focus of the pathway is the remodelling of the actin cytoskeleton, we can model this process in at least two ways: as a biochemical reaction model of coupled ODEs where the signalling interactions from the receptor to the downstream effectors are involved or as a stochastic model where the change in the actin cytoskeleton is described by actin remodelling biochemical reactions (branching, capping, polymerization etc.). The actin remodelling reactions have been considered at various levels of detail [2–4] and, based on the type of information sought, involve different levels of modelling complexity and abstractions. These models range from Brownian ratchet studies of single reactions, such as polymer elongation, to spatio–temporal models of multiple actin remodelling reactions. Therefore the user has to identify and define boundaries of the system to be modelled.
Figure 1. A simple signalling pathway.
A signalling pathway, represented as a wiring diagram. The interactions in the signalling pathway can be rewritten as coupled biochemical reactions and ODEs based on reaction rates that give the temporal dynamics of the activity state of the different components.
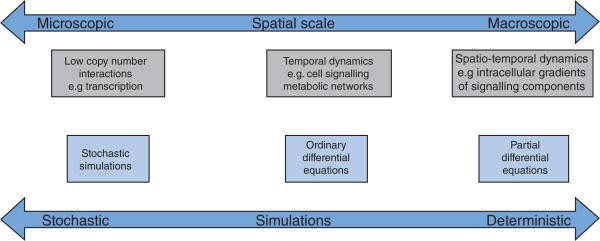
Once the boundaries of the model are well-defined, the next step is the choice of the nature of the mathematical representation; this can vary from fine-grained models for single processes (mostly stochastic) to coarse-grained models for multiple processes or combinations thereof (Figure 2). A stochastic process is one in which the behaviour of the system is non-deterministic and includes some randomness. The behaviour of processes in which the copy number of the species involved is small is often modelled using stochastic calculus, incorporating the randomness and noise in the process. A good example of a stochastic process is the binding of transcription factors to the promoter region of DNA. Deterministic models developed using differential equations are used to model processes in which the concentrations of participating chemical species can be treated as continuous variables because of their abundance. Usually such concentrations are in the nM to mM range. Whereas ODEs capture the temporal dynamics alone, partial differential equations capture both the spatial and temporal dynamics of the signalling pathway.
Figure 2. Different levels of modelling.
Computational modelling can be carried out at different cellular levels, resulting in models of different complexity. Models for processes with a small number of molecules involve stochastic computations. Temporal and spatio–temporal dynamics for cell signalling can be modelled using differential equations.
Parameter estimation
After the biochemical interactions have been identified, the next step in model construction is the estimation of the kinetic parameters and initial concentrations. Kinetic parameters for some biochemical reactions can be found in the literature in studies conducted using purified proteins in vitro. Although it is not clear whether the in vivo reactions occur at the same rate, these parameters are good starting points for conducting simulations using the computational model. The exact value of the amount of a given protein present in a cell is known for very few proteins. Therefore, initial concentrations of proteins and other signalling components are often estimates. In addition to rate constants and initial concentrations, development of a spatial model requires the estimation of diffusion coefficients. Diffusion coefficients are hard to measure experimentally for mixtures of proteins. Because the diffusibility of a molecule depends on its size, we can calculate an approximate diffusion coefficient based on the molecular mass of the component. In some cases, it is possible to measure the diffusion coefficient of a component using FRAP (fluorescence recovery after photobleaching).
Recently, Sible and Tyson [4a] have described a step-by-step procedure for the development of ODE models of cell cycle control. A description of parameter choices, parameter optimization and software tools available are also discussed in detail.
Model validation and comparison with experiments
A key step in model validation is comparing experimental results with simulations. It is this step that tells us whether the model recapitulates the known aspects of the biological process. When a model follows observed experimental behaviour closely, the confidence level of the generated predictions increases. For certain signalling pathways [such as EGFR (epidermal growth factor receptor), MAPK (mitogen-activated protein kinase), NF-κB (nuclear factor κB) and cell cycle] extensive information is available [5–9] and models built for these systems are elaborate and match experimental data closely. However, for systems where prior knowledge is limited and experimental studies are ongoing, it is harder to build elaborate models. In these cases, we can generate initial mechanistic models that validate key observations in the system behaviour and make predictions about possible interactions and regulatory mechanisms. Because a number of parameters are estimated, it is not often possible to generate exact matches between experiments and simulation. Parameter space exploration, either by sensitivity analysis [10] or by studying different input conditions [11], can help with understanding the conditions that lead to experimentally observed behaviour. The key point to note in parameter space exploration is that when a large number of parameters influence the system dynamics, a number of combinations of parameters can lead to similar system behaviour. It is therefore essential to choose these parameters in a biologically justifiable manner. The user must validate the model by comparing simulation results to experimental results. It is sometimes difficult to obtain parameters which quantitatively reproduce all the experimental observations. In such cases, the comparison between model and experiment is limited to obtaining good agreement of well-characterized system properties. Often, it is possible to find a middle ground where we can obtain close quantitative matches for some components and qualitatively similar behaviour for others.
Spatio–temporal considerations
Using live-cell imaging techniques, it is increasingly becoming obvious that cell signalling networks are spatially specified and, unlike physico– chemical interactions occurring in vitro, biological interactions are also spatially regulated. Advances in imaging technology and fluorescent probes have resulted in experimental techniques that allow us to capture the spatio– temporal dynamics of cell signalling [12,13]. In addition to the previously described parameters, we then have to consider localization of the component [ER (endoplasmic reticulum), membrane, Golgi, cytoplasm etc.] and rates of transport across these subcellular compartments. Spatial localization of components and interactions across compartments requires the generation of flux mechanisms and diffusive behaviour in addition to kinetic parameters. To accurately model spatial localization, it is important to consider the biophysical properties of the compartment. Consider, for instance, the plasma membrane and synthesis of phosphoinositides [14,15]. Diffusion in the plasma membrane occurs essentially in a two-dimensional plane and is hindered by the presence of other molecules and electrostatic interactions. PtdIns(4,5)P2, in the membrane, has a diffusion coefficient of 1 μm2/s [15]. In contrast, the ubiquitous second messenger cAMP has a diffusion coefficient of 300 μm2/s in the cytoplasm [16]. Thus the cellular localization of the component plays a key role in determining its diffusivity.
An overview of modelling successes
In this section, we discuss a few modelling successes that have led to the identification of emergent properties in signalling networks and enhanced our understanding of certain complex biological phenomena.
Deterministic models
Many signalling pathways have been successfully modelled using ODEs; however, in this section, we will focus on models of MAPK activation to highlight the approaches taken and modelling results. Activation of the MAPK pathway can lead to different cellular responses, including cell survival, cytoskeletal remodelling, motility and differentiation. The MAPK network has been studied extensively from different perspectives using ODE models [6,8,17–23]. Computational analysis of the three-tier cascade of MAPK activation shows that the presence of the multi-layered activation cascade is important for regulation and the feedback loops are necessary for decision making and memory formation [18]. Some of the key results were that signal integration occurs across different time scales and the output depends not only on the duration and strength of input but also on the nature of the feedback loops. Sustained activation of the MAPK pathway after withdrawal of the signal and bistable (with two steady states) steady-state behaviour in response to input signal were observed as a result of feedback mechanisms. Thus the positive-feedback loop acts as a switch through a cascade of biochemical reactions [16,24]. Using a combination of experimental and computational analyses, it has been shown that interactions between pathways in a network lead to adaptation and recovery patterns [25,26]. In the MAPK network, the system exhibits bistable behaviour in response to low concentrations of MKP (MAPK phosphatase) leading to sustained MAPK activation in response to a brief stimulus. An increase in MKP concentrations eliminates this bistability and sustained activation response and instead leads to a monostable, proportional response state [26]. Further feedback effects can result from sequestration effects and competing docking interactions leading to bistability [21]. Thus, even for a simple signalling cascade, modelling allows for insights that are not intuitive.
Delay models
Time delays incorporated in ODEs result in DDEs (delay differential equations). DDEs are used to reduce the number of variables involved and offer a mechanistic insight into regulatory mechanisms, when the entire process details are unknown. Using phosphorylation–dephosphorylation cycles, it has been shown that DDEs can capture observed system behaviour using fewer variables [27,28]. DDEs have been used to mechanistically model transcriptional effects in the feedback regulation of the NF-kB pathway [29] and the effects of pharmacological agents on this pathway [30].
Spatio–temporal models
In order to model the local activation of signalling components, multi-compartment models and spatial models can be used. A compartment-specific model of Ras activation highlights the role of trafficking and compartment-specific feedback loops [31]. Positive and negative regulators of calcium signalling are activated in a context-dependent manner and lead to context-dependent localization of Ras. This study analysed how a signal originating at the plasma membrane propagates to different subcellular locations.
Spatial analysis of signalling from phosphoinositides was carried out in a series of studies that combined experimental and computational modelling methods [32–34]. These studies showed how uniform stimulation can lead to asymmetric phosphoinositide gradients. Furthermore, experiments allowed for parameter estimation for the model and for the analysis of the effect of cell morphology on gradients of phosphoinositides. A partial differential equation model to study the effect of phosphoinositides and Rho-family GTPases on actin cytoskeleton remodelling has been developed [35]. This spatial model explains how Rho-GTPases act as a spatial switch and the phosphoinositide gradients that are important in determining the front from the rear of the cell. Thus spatio–temporal models are necessary for understanding the localization of cellular components and the role played by signal gradients in defining cellular processes.
Stochastic models
Most models of signalling networks are deterministic in nature and use systems of ordinary or partial differential equations. However, not all signalling processes are deterministic in nature and in certain cases, where the number of molecules involved is small, the process is inherently stochastic. Typical examples of stochastic processes include binding of transcription factors to promoter regions and branching of individual actin filaments. Implementation of stochasticity to study temporal dynamics is often carried out using Gillespie's algorithm [36,37]. The core of this algorithm is the chemical master equation that determines the probability that each species will have a defined population at a given future time. The SSA (stochastic simulation algorithm) is a Monte Carlo procedure for computing the dynamics of the population in accordance with the master equation. Stochastic simulation of signalling networks has been used to evaluate the behaviour of the system in many different studies [38–40]. One of the challenges with stochastic approaches is the larger computation time required compared with deterministic systems. Examples of models based on these methods can be found in [38,40]. We have recently developed a spatio–temporal stochastic model for cell spreading based on actin remodelling reactions (Y. Xiong, P. Rangamani, B. Dubin-Thaler, M.P. Sheetz and R. Iyengar, unpublished work). In this model, we capture isotropic cell spreading on a substrate using actin filament elongation, branching and capping regulated by membrane resistance forces. It is anticipated that for large signalling networks, hybrid models will be built where part of the system follows deterministic behaviour and the rest of the system follows stochastic behaviour.
Example of a deterministic model
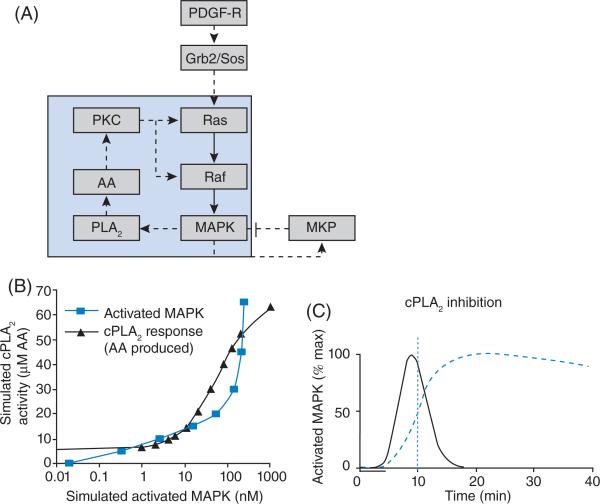
We now discuss an example of a deterministic model to illustrate the concepts discussed in the present chapter. This model studies the role of feedback loops on system dynamics and we can see how analysis of a mathematical model leads to mechanistic understanding of system behaviour [26]. The signalling network is shown in Figure 3(A). Activation of the PDGF (platelet-derived growth factor) receptor by PDGF leads to activation of MAPK via the Grb2/Sos pathway. Negative feedback is present via MKP. MKP is activated by MAPK and inhibits the same. In this case, as more MAPK is activated, more MKP will be activated and leads to the balance of MAPK activity via inhibition. Thus the role of a negative-feedback loop is to attenuate the signal flow in the network. In contrast, positive feedback is present via MAPK activation of PLA2 (phospholipase A2), which leads to further activation of Ras and Raf via arachidonic acid and PKC (protein kinase C). The role of a positive-feedback loop is to enhance signal flow through the network. How does the presence of these loops affect system behaviour? Developing an ODE model for this network allows us to follow the temporal dynamics of the concentrations and study the role of these feedback mechanisms. Simulations showed that a brief stimulus (5 min) could result in sustained MAPK activation for 40 min. When we analyse the steady-state MAPK activation by PLA2 and the activation of PLA2 by MAPK, we see that the concentration–effect curves intersect at three points (Figure 3B). (Each of these defines the system at steady state.) Point 1 is the basal state, point 2 is the metastable state and point 3 is the high stable state of activity. This kind of behaviour is called bistability. Low-level stimulation of the system results in a transient increase in activity with the system coming back to a basal state upon termination of the stimulus. Stimulation by PDGF to levels above the metastable state (point 2) allows the system to attain the activated steady state and stay there even when the stimulus is terminated. If the positive-feedback loop via PLA2, arachidonic acid and PKC is responsible for this sustained activation, then blocking the feedback loop should result in loss of sustained MAPK activation. This prediction was experimentally verified using pharmacological inhibitors of PLA2 (Figure 3C). Thus a computational model was used to provide insight into the role of feedback loops and how the configuration of signalling networks can lead to sustained activity.
Figure 3. Positive-feedback loop and bistability.
(A) The positive-feedback loop between MAPK and arachidonic acid (AA) is shown in the light-blue region. The positive-feedback loop is shown. (B) The concentration–effect curves of MAPK activity and PLA2 intersect at three points resulting in three steady states. Point 1 is the stable basal activity state, point 2 is metastable and point 3 is the highly active stable steady state. AA, arachidonic acid. (C) Disruption of the positive-feedback loop by inhibition of PLA2 results in loss of sustained MAPK activity, confi rming the role of the positive-feedback loop in sustaining MAPK activation. Adapted from [26], with permission. © 2002 American Association for the Advancement of Science.
Perspective
Many of the published works on modelling of biological systems talk about modelling successes and how well the data match experiments and how the predictions were validated by further experiments. This raises the question: what is the value of a model that fails? The failure of a model is equally important. Such failures can be used for gap analysis because we can determine the information about the process that we do not have and that the system can be further explored experimentally to build a refined model. Often by analysing parts of a failed model, it is possible to identify which parts of the system require further experimental study, to better specify the system.
Thus far, modelling of signalling networks has focused on the behaviour of systems in response to different extracellular stimuli, maintaining the structure of the network itself constant. However, it is well-known that the topology of the network itself is spatio–temporally dynamic [42]. The next advances in realistic modelling of cellular pathways are incorporating interactions that capture the dynamic properties of the network itself. In addition to parameters that define each reaction or interaction in the network, the dynamic network model should then include the conditions for switching of interactions when components move in space. Given the impact modelling has made in a relatively short time on our understanding of cellular processes, it may not be too long before we have hybrid dynamically evolving models that capture both the spatial and temporal characteristics and explain cellular behaviours that currently seem puzzling.
Summary.
Mathematical modelling of signalling networks provides mechanistic understanding of dynamics, of information processing and flow through the system.
Models can be stochastic or deterministic and can also involve spatial specifications.
Parameter estimation is a key step in model development. Constraining a model to experimental data is important for model validation.
Failure of a model to recapitulate experimental observations is often useful and serves as an indicator of the need for more data to specify the system being modelled.
References
- 1.Weng G, Bhalla US, Iyengar R. Complexity in biological signaling systems. Science. 1999;284:92–96. doi: 10.1126/science.284.5411.92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mogilner A, Oster G. Cell motility driven by actin polymerization. Biophys. J. 1996;71:3030–3045. doi: 10.1016/S0006-3495(96)79496-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mogilner A, Edelstein-Keshet L. Regulation of actin dynamics in rapidly moving cells: a quantitative analysis. Biophys. J. 2002;83:1237–1258. doi: 10.1016/S0006-3495(02)73897-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mogilner A, Oster G. Force generation by actin polymerization II: the elastic ratchet and tethered fi laments. Biophys. J. 2003;84:1591–1605. doi: 10.1016/S0006-3495(03)74969-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4a.Sible JC, Tyson JJ. Mathematical modeling as a tool for investigating cell cycle control networks. Methods. 2007;41:238–247. doi: 10.1016/j.ymeth.2006.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bagowski CP, Besser J, Frey CR, Ferrell JE., Jr The JNK cascade as a biochemical switch in mammalian cells: ultrasensitive and all-or-none responses. Curr. Biol. 2003;13:315–320. doi: 10.1016/s0960-9822(03)00083-6. [DOI] [PubMed] [Google Scholar]
- 6.Huang CY, Ferrell JE., Jr Ultrasensitivity in the mitogen-activated protein kinase cascade. Proc. Natl. Acad. Sci. U.S.A. 1996;93:10078–10083. doi: 10.1073/pnas.93.19.10078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sachs K, Perez O, Pe'er D, Lauffenburger DA, Nolan GP. Causal protein-signaling networks derived from multiparameter single-cell data. Science. 2005;308:523–529. doi: 10.1126/science.1105809. [DOI] [PubMed] [Google Scholar]
- 8.Schoeberl B, Eichler-Jonsson C, Gilles ED, Müller G. Computational modeling of the dynamics of the MAP kinase cascade activated by surface and internalized EGF receptors. Nat. Biotechnol. 2002;20:370–375. doi: 10.1038/nbt0402-370. [DOI] [PubMed] [Google Scholar]
- 9.Zaman MH, Kamm RD, Matsudaira P, Lauffenburger DA. Computational model for cell migration in three-dimensional matrices. Biophys. J. 2005;89:1389–1397. doi: 10.1529/biophysj.105.060723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cho KH, Shin SY, Kolch W, Wolkenhauer O. Experimental design in systems biology, based on parameter sensitivity analysis using a Monte Carlo method: a case study for the TNF-α mediated NF-κB signal transduction pathway. Simulation. 2003;79:726–739. [Google Scholar]
- 11.Werner SL, Barken D, Hoffmann A. Stimulus specifi city of gene expression programs determined by temporal control of IKK activity. Science. 2005;309:1857–1861. doi: 10.1126/science.1113319. [DOI] [PubMed] [Google Scholar]
- 12.Tsien RY. Imagining imaging's future. Nat. Rev. Mol. Cell Biol. 2003 Sep;(Suppl):SS16–SS21. [PubMed] [Google Scholar]
- 13.Zacharias DA, Baird GS, Tsien RY. Recent advances in technology for measuring and manipulating cell signals. Curr. Opin. Neurobiol. 2000;10:416–421. doi: 10.1016/s0959-4388(00)00101-x. [DOI] [PubMed] [Google Scholar]
- 14.McLaughlin S, Murray D. Plasma membrane phosphoinositide organization by protein electrostatics. Nature. 2005;438:605–611. doi: 10.1038/nature04398. [DOI] [PubMed] [Google Scholar]
- 15.McLaughlin S, Wang J, Gambhir A, Murray D. PIP(2) and proteins: interactions, organization, and information fl ow. Annu. Rev. Biophys. Biomol. Struct. 2002;31:151–175. doi: 10.1146/annurev.biophys.31.082901.134259. [DOI] [PubMed] [Google Scholar]
- 16.Bhalla US, Iyengar R. Emergent properties of networks of biological signaling pathways. Science. 1999;283:381–387. doi: 10.1126/science.283.5400.381. [DOI] [PubMed] [Google Scholar]
- 17.Orton RJ, Sturm OE, Vyshemirsky V, Calder M, Gilbert DR, Kolch W. Computational modelling of the receptor-tyrosine-kinase-activated MAPK pathway. Biochem. J. 2005;392:249–261. doi: 10.1042/BJ20050908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kolch W, Calder M, Gilbert D. When kinases meet mathematics: the systems biology of MAPK signalling. FEBS Lett. 2005;579:1891–1895. doi: 10.1016/j.febslet.2005.02.002. [DOI] [PubMed] [Google Scholar]
- 19.Mayawala K, Gelmi CA, Edwards JS. MAPK cascade possesses decoupled controllability of signal amplifi cation and duration. Biophys. J. 2004;87:L01–L02. doi: 10.1529/biophysj.104.051888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Asthagiri AR, Lauffenburger DA. A computational study of feedback effects on signal dynamics in a mitogen-activated protein kinase (MAPK) pathway model. Biotechnol. Prog. 2001;17:227–239. doi: 10.1021/bp010009k. [DOI] [PubMed] [Google Scholar]
- 21.Legewie S, Schoeberl B, Blüthgen N, Herzel H. Competing docking interactions can bring about bistability in the MAPK cascade. Biophys. J. 2007;93:2279–2288. doi: 10.1529/biophysj.107.109132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ferrell JE., Jr Tripping the switch fantastic: how a protein kinase cascade can convert graded inputs into switch-like outputs. Trends Biochem. Sci. 1996;21:460–466. doi: 10.1016/s0968-0004(96)20026-x. [DOI] [PubMed] [Google Scholar]
- 23.Ferrell JE., Jr How responses get more switch-like as you move down a protein kinase cascade. Trends Biochem. Sci. 1997;22:288–289. doi: 10.1016/s0968-0004(97)82217-7. [DOI] [PubMed] [Google Scholar]
- 24.Lisman JE, Fallon JR. What maintains memories? Science. 1999;283:339–340. doi: 10.1126/science.283.5400.339. [DOI] [PubMed] [Google Scholar]
- 25.Ingolia NT, Murray AW. Signal transduction. History matters. Science. 2002;297:948–949. doi: 10.1126/science.1075222. [DOI] [PubMed] [Google Scholar]
- 26.Bhalla US, Ram PT, Iyengar R. MAP kinase phosphatase as a locus of fl exibility in a mitogen-activated protein kinase signaling network. Science. 2002;297:1018–1023. doi: 10.1126/science.1068873. [DOI] [PubMed] [Google Scholar]
- 27.Srividhya J, Gopinathan MS, Schnell S. The effects of time delays in a phosphorylation-dephosphorylation pathway. Biophys. Chem. 2007;125:286–297. doi: 10.1016/j.bpc.2006.09.001. [DOI] [PubMed] [Google Scholar]
- 28.Srividhya J, Gopinathan MS. A simple time delay model for eukaryotic cell cycle. J. Theor. Biol. 2006;241:617–627. doi: 10.1016/j.jtbi.2005.12.020. [DOI] [PubMed] [Google Scholar]
- 29.Rangamani P, Sirovich L. Survival and apoptotic pathways initiated by TNF-α: modeling and predictions. Biotechnol. Bioeng. 2006;97:1216–1229. doi: 10.1002/bit.21307. [DOI] [PubMed] [Google Scholar]
- 30.Sung MH, Simon R. In silico simulation of inhibitor drug effects on nuclear factor-κB pathway dynamics. Mol. Pharmacol. 2004;66:70–75. doi: 10.1124/mol.66.1.70. [DOI] [PubMed] [Google Scholar]
- 31.Eungdamrong NJ, Iyengar R. Compartment-specifi c feedback loop and regulated traffi cking can result in sustained activation of Ras at the Golgi. Biophys. J. 2007;92:808–815. doi: 10.1529/biophysj.106.093104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Haugh JM, Codazzi F, Teruel M, Meyer T. Spatial sensing in fi broblasts mediated by 3′ phosphoinositides. J. Cell Biol. 2000;151:1269–1280. doi: 10.1083/jcb.151.6.1269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Haugh JM, Schneider IC. Spatial analysis of 3′ phosphoinositide signaling in living fi broblasts: I. Uniform stimulation model and bounds on dimensionless groups. Biophys. J. 2004;86:589–598. doi: 10.1016/S0006-3495(04)74137-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schneider IC, Haugh JM. Spatial analysis of 3′ phosphoinositide signaling in living fi broblasts: II. Parameter estimates for individual cells from experiments. Biophys. J. 2004;86:599–608. doi: 10.1016/S0006-3495(04)74138-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dawes AT, Edelstein-Keshet L. Phosphoinositides and Rho proteins spatially regulate actin polymerization to initiate and maintain directed movement in a one-dimensional model of a motile cell. Biophys. J. 2007;92:744–768. doi: 10.1529/biophysj.106.090514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gillespie DT. Stochastic simulation of chemical kinetics. Annu. Rev. Phys. Chem. 2007;58:35–55. doi: 10.1146/annurev.physchem.58.032806.104637. [DOI] [PubMed] [Google Scholar]
- 37.Gillespie DT. Exact stochastic simulation of coupled chemical reactions. J. Phys. Chem. 1977;81:2340–2361. [Google Scholar]
- 38.Vasudeva K, Bhalla US. Adaptive stochastic-deterministic chemical kinetic simulations. Bioinformatics. 2004;20:78–84. doi: 10.1093/bioinformatics/btg376. [DOI] [PubMed] [Google Scholar]
- 39.Samoilov M, Plyasunov S, Arkin AP. Stochastic amplifi cation and signaling in enzymatic futile cycles through noise-induced bistability with oscillations. Proc. Natl. Acad. Sci. U.S.A. 2005;102:2310–2315. doi: 10.1073/pnas.0406841102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Manninen T, Linne ML, Ruohonen K. Developing Ito stochastic differential equation models for neuronal signal transduction pathways. Comp. Biol. Chem. 2006;30:280–291. doi: 10.1016/j.compbiolchem.2006.04.002. [DOI] [PubMed] [Google Scholar]
- 41.Reference deleted
- 42.Zhu H, Huang S, Dhar P. The next step in systems biology: simulating the temporospatial dynamics of molecular networks. BioEssays. 2004;26:68–72. doi: 10.1002/bies.10383. [DOI] [PubMed] [Google Scholar]