1. Introduction
Much of postgenomic biochemistry and all of structural biology are based on the premise that the starting point for both understanding specific biochemical processes, such as affinity, reactivity, or transport, and surveying proteomes is determining the three-dimensional structures of proteins. The two well-established methods for structure determination are highly effective when applied to samples of soluble globular proteins and their complexes: witness the enormous growth of the Protein Data Bank.1 However, the vast majority of biological functions are carried out by proteins associated with supramolecular assemblies, whose samples are problematic for both X-ray crystallography and solution NMR spectroscopy, since they are generally difficult to crystallize and do not reorient rapidly even when soluble. The examples of proteins in supramolecular assemblies whose structures have been determined with atomic resolution are exceptional and highlight the importance of developing new methods of experimental protein structure determination. The essential goals of modern structural biology are to have the capability to select proteins for study based on their biological functions and to perform genuinely unbiased surveys of proteomes unfettered by considerations of the solubility, aggregation state, or other physical properties of the polypeptides. NMR spectroscopy has the potential to accomplish these goals, since it can be applied to molecules in all physical states, including the liquid crystalline environments provided by the lipids associated with membrane proteins.
Determining the atomic resolution structures of membrane proteins is of particular interest in contemporary structural biology.2 Helical membrane proteins constitute one-third of the expressed proteins encoded in a genome.3,4 Furthermore, many drugs have membrane-bound proteins as their receptors, and mutations in membrane proteins result in human diseases. They also provide daunting technical challenges for all methods of protein structure determination, including NMR spectroscopy.5
2. NMR Spectroscopy
NMR is well suited for determining the atomic-resolution structures of proteins. It is possible to obtain a separate signal for each atom in a protein. Not only can these signals be assigned to specific sites, but also they can be characterized by frequencies that provide both distance and orientation constraints as input for structure determination. The potential for determining the structures of proteins with NMR spectroscopy has always been recognized,6,7 and for globular proteins that reorient rapidly in solution, the sample conditions, instrumentation, experiments, and calculations that lead to structure determination are now well established and widely employed.8–11 However, the Achilles’ heel of high-resolution NMR spectroscopy–the correlation time problem–has severely limited its applications to proteins in supramolecular assemblies, especially membrane proteins with their associated lipids.
2.1. The Correlation Time Problem
The rotational correlation time of the polypeptide in the sample is the paramount consideration for NMR spectroscopy. It dictates the sample conditions, instrumentation, experimental methods, and data processing calculations. Dealing with the correlation time problem for membrane proteins is the recurring theme of structural studies by NMR spectroscopy.
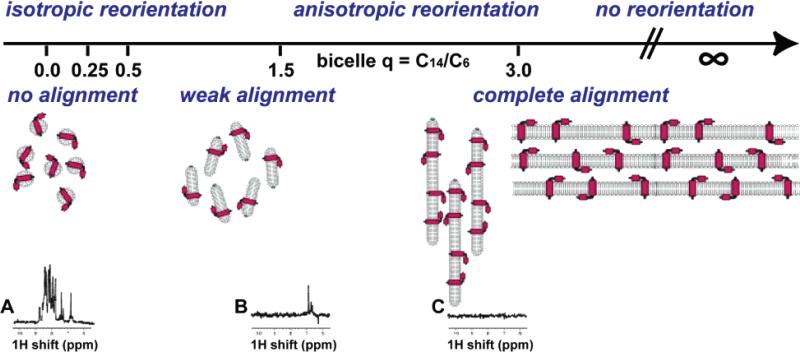
Slow reorientation is the principal reason it is difficult or impossible to obtain high-resolution spectra of proteins that are large, aggregated, or incorporated into supramolecular assemblies using solution NMR methods. This is illustrated with the spectra in Figure 1, where the broad, weak signals from a protein in medium (Figure 1B) and large (Figure 1C) bicelles contrast with those from the same protein in micelles (Figure 1A). For membrane proteins, the correlation time problem can be addressed in two ways. The most fundamental approach is through the application of solid-state NMR methods to membrane proteins in lipid bilayers or large bicelles where they are effectively immobilized by their environment. In this situation, radio frequency irradiations,12,13 and magic angle sample spinning14 or sample alignment,12,15 substitute for molecular motions as the line-narrowing mechanism. Solution NMR methods are also applicable, at least for smaller membrane proteins, since it is possible to prepare samples of protein-containing micelles or small bicelles that reorient rapidly enough to yield well-resolved spectra, as illustrated by the spectrum in Figure 1A, although the range of measurements that can be made is limited compared to what can be done with samples of soluble globular proteins. At present, solid-state NMR studies in bilayer environments and solution NMR studies in micelle environments are complementary approaches to structure determination of membrane proteins.5,16 As the awareness of functional status and the subtleties of interfacial interactions and dynamics becomes more prominent, then studies of the most nativelike bilayer environments where the proteins are immobilized will predominate, and it is likely that the field will rely increasingly on solid-state NMR methodology.
Figure 1.
Linking of correlation times and alignments of the proteins in the samples. From left to right are isotropic reorientation to immobile, and no alignment to complete alignment. The samples are described by the q ratio for bicelles, ranging from 0 for isotropic micelles to 3 for large bicelles to infinity for bilayers, as illustrated schematically. Spectra A–C are one-dimensional 1H NMR spectra of a membrane protein in various samples: (A) q = 0 (isotropic micelle); (B) q = 0.5 (medium sized bicelle); (C) q = 3.0 (large bicelle).
2.2. Sample Alignment
NMR approaches that exploit the properties of aligned samples are crucial for structure determination of proteins that are not compactly folded, for example, helical membrane proteins. As illustrated in Figure 1, the extent of alignment of a protein is inherently linked to its rotational correlation time and is of equal importance to the design and implementation of NMR experiments. Membrane proteins in lipid bilayers are immobile on the millisecond time scale and can be completely aligned for solid-state NMR experiments. It is possible to prepare samples of protein-containing bilayers aligned between glass plates where the extent of alignment is similar to that found in single crystals of peptides.17 In contrast, membranes proteins in micelles or small bicelles have rotational correlation times around 10 ns and can be weakly aligned in samples where the extent of alignment is about 0.1% of that in the completely aligned bilayer samples. Moreover, it is sample alignment that enables seemingly disparate solution NMR and solid-state NMR approaches to structure determination to be unified by the principles of separated local field spectroscopy,18,19 as a way to map protein structure onto the NMR spectra through the orientation-dependent frequencies of the dipole–dipole and chemical shift interactions.16 Oriented lipid bilayer samples are also useful for studying the dynamic properties of small peptides that undergo rotational diffusion in the membrane.20,21
2.3. Local Fields
More than 50 years ago, Pake demonstrated that the dipole–dipole interaction is anisotropic and a source of both distance and angular measurements.15 The development of high-resolution solid-state NMR spectroscopy was initiated 35 years ago so that NMR could be applied to single-crystal and polycrystalline and amorphous samples with molecules that are immobile on laboratory time scales (days). It began with the demonstration that multiple pulse sequences12 could narrow the resonances of abundant nuclei, especially 1H spins in solid samples. It then gained much broader applicability to chemical and biochemical systems by focusing on the multiple roles, including the detection of magnetization under high-resolution conditions, of dilute nuclei, typically 13C, but also 15N.13 These same experimental methods, instruments, and theories work well on samples where the molecules of interest are immobile on the significantly shorter time scales of the dipole–dipole and chemical shift spin interactions (milliseconds). In NMR spectroscopy, the term solid-state refers to the effective rotational correlation time of the molecule rather than the physical state of the sample. Therefore, NMR methods developed for crystalline model peptides can be applied to proteins immobilized by the environment of lipid bilayers.
Both high-resolution spectra and the measurements that provide orientation constraints for structure determination can be obtained from stationary samples of immobile molecules, as long as they are aligned along the direction of the applied magnetic field.22 The archetype of an aligned sample is a single crystal, where any alignment in the magnetic field yields high-resolution NMR spectra. The earliest results15,23 demonstrated two of the most fundamental features of high-resolution solid-state NMR spectroscopy: high spectral resolution can be achieved by sample alignment, and observable spectral parameters, such as the frequencies associated with the dipole–dipole and chemical shift interactions, reflect the orientations of molecular sites with respect to the direction of the magnetic field. Because solid-state NMR procedures can be applied in ways that lead to selective averaging, as well as the temporal separation of the evolution of the anisotropic spin interactions, multidimensional solid-state NMR spectra can be obtained that provide directly interpretable orientation information as input for structure determination.
Separated local field spectroscopy is an extraordinarily powerful approach to structure determination18,19 and is the cornerstone of protein structure determination of aligned samples.22,24 It combines several spectroscopic elements to reduce unwanted broadening and sort the spectra into frequency dimensions from the anisotropic heteronuclear dipolar coupling and chemical shift interactions. Although higher dimensional experiments offer enhanced opportunities for spectral resolution of larger polypeptides and additional frequencies as orientation constraints,17,25–29 the heteronuclear dipole–dipole and chemical shift frequencies in two-dimensional separated local field spectra provide adequate resolution for studies of small and medium sized uniformly labeled polypeptides.30–35
15N can be readily and inexpensively placed in every amide site in the backbone of a protein expressed in bacteria,36 and with somewhat more effort, this is also possible using other types of expression systems. This labeling pattern is ideal for separated local field spectroscopy, since the separation of the nitrogens from each other by the two intervening carbon atoms in the peptide backbone means that they are naturally dilute, obviating the need for homonuclear nitrogen decoupling and enabling the signals from individual sites to be distinguished by their chemical shift frequencies. High-resolution separated local field experiments, such as PISEMA37 or SAMMY,38 associate an orientation-dependent heteronuclear 1H–15N dipolar coupling frequency with each 15N orientation-dependent chemical shift frequency. In solution NMR, IPAP (in-phase–anti-phase) experiments39 are used to measure the residual dipolar couplings associated with each resonance resolved by virtue of the isotropic 1H and 15N chemical shift frequencies. It is also possible to apply this approach to 13C sites in natural abundance or with isotopic labeling and somewhat more complex spectroscopic experiments.
Sample alignment is now an integral part of most NMR structural studies, including solution NMR of weakly aligned soluble proteins39,40 and membrane proteins in micelles and small bicelles,41–44 where motional averaging effectively decouples the interactions and removes line broadening, but there is sufficient alignment for the measurement of residual dipolar couplings and chemical shifts. Thus, both solid-state NMR of completely aligned samples and solution NMR of weakly aligned samples rely on the concept of the separated local field, where individual dipolar couplings, most often 1H–15N in backbone sites, are measured for individual amide sites resolved (separated) by their chemical shift frequencies, which themselves reflect the orientation of the groups in the field. The use of orientation constraints integrates solution NMR and solid-state NMR approaches to protein structure determination through the mapping of structure onto the spectra by means of the anisotropic characteristics of the nuclear spin interactions. This is the key for structure determination of helical membrane proteins, which do not lend themselves to isotropic methods because of the modular nature and topological arrangements of the helical segments. Moreover, the highly hydrophobic amino acid composition of these proteins makes it difficult to resolve among side chain sites on the basis of their isotropic chemical shifts.
3. Structure Determination of Membrane Proteins
The process of determining the structures of membrane proteins can be divided into a number of distinct steps, starting with the selection of target sequences. Many of the preparative steps are similar to those utilized in structure determinations by other methods. However, for NMR spectroscopy, particular attention needs to be paid to the lipid assemblies because of their influence on the global and local motions of the associated polypeptides. Lipids are amphiphilic molecules with one or two hydrocarbon chains and a polar or charged headgroup. Certain lipids assemble into micelles at concentrations above the critical micelle concentration (cmc), while others form bicelles or extended bilayers, all of which can serve as samples for NMR spectroscopy. However, these assemblies of proteins and lipids are the source of the correlation time problems that plague conventional solution NMR studies of membrane proteins.
Micelles are small, roughly spherical aggregates of lipids with their hydrocarbon chains forming an interior hydrophobic core that solubilizes membrane proteins. Protein-containing micelles reorient rapidly enough to give isotropic spectra, as shown in Figure 1A. However, the reorientation occurs slowly compared to that for soluble proteins of similar molecular mass, and as a result, micelle samples are very demanding and require careful sample preparation,45 the use of high-field NMR spectrometers, and elevated temperatures. At that, many of the most informative solution NMR experiments cannot be applied to these samples because the short relaxation times result in the loss of signals during extended pulse sequences. Most critically, the issues with relaxation times also affect measurements of homonuclear 1H–1H NOEs, the principal source of distance constraints in solution NMR structure determination. Very few long-range NOEs can be observed and assigned in helical membrane proteins in micelles. Consequently, the primary source of structural information is residual dipolar couplings (RDCs) measured in weakly aligned samples. Weak alignment can be induced in these samples by the addition of lanthanides41,42 or by their incorporation into stressed polyacrylamide gels.43,46–49 Bicelles are prepared by mixing two different lipids: one type with longer chains forms the extended bilayer portion, and the other with shorter chains forms the caps at the ends of the disks. The sizes of the bicelles can be adjusted through the ratios of the two types of lipids, ranging from small isotropic bicelles to large bicelles that for all practical purposes behave like extended bilayers.50 They provide the experimental connection between solution NMR of micelles on one hand and solid-state NMR of bilayers on the other.
Bilayers (and large bicelles) are the most desirable lipid assemblies for structural studies of membrane proteins because they closely mimic the properties of biological membranes. They require the use of solid-state NMR methods, since the associated polypeptides are immobile on the 104 Hz time scale of the dipole–dipole and chemical shift interactions. Bilayers can be mechanically aligned between glass plates. The line widths of the protein resonances demonstrate that the alignment is complete, with a dispersion similar to that observed in single crystals along the direction of the alignment.17 Bicelles can be aligned magnetically perpendicular to the direction of the magnetic field or flipped to the parallel orientation by the addition of lanthanide ions,51 so that the disks have the same alignment as bilayers on glass plates.
The angular constraints derived from solid-state NMR experiments on completely aligned samples provide reliable and precise structural information. The independent measurement of frequencies associated with the anisotropic heteronuclear dipole–dipole and chemical shift interactions for each backbone amide site in a protein relative to a single reference frame, for example, the magnetic field axis for completely aligned lipid bilayers, means that errors do not propagate in a cumulative manner. This is an extremely important feature of the method that enables the combination of experimental angular constraints from individual residues and the well-established covalent geometry (bond lengths, bond angles, dihedral angles, planarity of the peptide linkages) of proteins to be used as the basis for protein structure determination with atomic resolution. The resonance frequencies, in both the chemical shift and dipolar coupling dimensions, depend on the orientations of the helices, local backbone dihedral angles, the magnitudes and orientations of the principal elements of the 15N chemical shift tensor, and the N–H bond length. Thus, it is possible to calculate spectra for any protein structure.52–59
3.1. Target Selection
The first step is to select a protein for structure determination. A surprise revealed by the analysis of the sequences of many genomes is that the majority of helical membrane proteins are relatively small polypeptides with only one or a few trans-membrane helices.60 Of course, there are larger polypeptides in this category as well, and the structure determination of the 300 residue–400 residue proteins with seven trans-membrane helices is a major goal of structural biology because of the key roles of this class of proteins in signaling and binding of drugs. With one-third of a genome coding for membrane proteins, there are a vast number of potential targets, whether specific questions are being asked about biochemical processes or the structural characteristics of a proteome are being surveyed. Structural studies of membrane proteins are limited by the capabilities of the available technology; therefore, target selection is intimately related to the state of development of NMR spectroscopy.
Fifteen years ago, when there were no practical expression systems for membrane proteins, solid-state NMR studies were limited to polypeptides with about 25 residues, that could be synthesized and purified. The two single helix examples provided an in-plane amphipathic helix (magainin)27,62–65 and a trans-membrane hydrophobic helix (AchR M2),34 which are the principal features found in the initial structures of larger membrane proteins determined by X-ray crystallography. Although synthetic methods are now greatly improved,61 only minimal progress could be made with synthetic materials, because of difficulties encountered in preparing well-behaved micelle and bilayers samples and because of the limited or impractical isotopic labeling schemes available for synthetic polypeptides. This motivated the development of bacterial expression systems for membrane proteins.
3.2. Expression and Purification of Isotopically Labeled Membrane Proteins
Expression in bacteria,66–71 and, more recently, in cell-free systems,72 enables milligram amounts of isotopically labeled membrane proteins to be prepared for NMR experiments. A number of factors need to be considered in optimizing the expression system for the preparation of samples for NMR studies. For the expression of smaller membrane proteins in E. coli, synthesizing the DNA encoding the polypeptide sequence makes it possible to optimize the codon usage, while E. coli host strains optimized for the expression of rare E. coli codons are available commercially for larger proteins. In addition, we have found the use of fusion proteins to be essential to express small membrane proteins in E. coli. Depending on the system, large amounts of the expressed fusion proteins can be found in the cell membranes, in inclusion bodies, or, in a few examples, in the cytoplasm. The proteins are isolated through standard methods and then cleaved from the fusion partner with cyanogen bromide or a selective protease. Purification is generally accomplished with reverse-phase HPLC, as well as size-exclusion and ion-exchange chromatography in the presence of lipids.
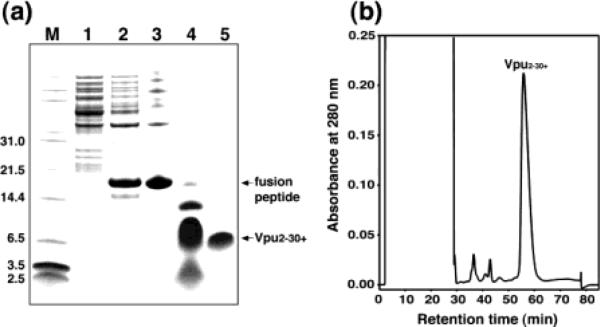
The polypeptide with the amino acid sequence corresponding to residues 2–37 of Vpu was selected as a minimal ion channel-forming domain in order to dissect the structural and functional properties of this HIV-1 accessory protein.33 The ketosteroid isomerase (KSI) fusion system was chosen for the expression of this polypeptide because it can be overproduced in E. coli, purified efficiently as inclusion bodies by nickel chelate chromatography, and subsequently cleaved by cyanogen bromide to yield the polypeptide of interest. This is illustrated with the PAGE patterns in Figure 2a. The major resolved peak in the HPLC trace in Figure 2b corresponds to this polypeptide, as verified by mass spectrometry. The fractions containing the polypeptide are collected, the solvents are removed, and the material is stored as a pure powder that is used to prepare the samples for NMR spectroscopy with the addition of lipids and water.
Figure 2.
(A) SDS-PAGE analysis of the purification procedure for hydrophobic membrane proteins: lane 1, discarded soluble fractions; lane 2, inclusion bodies of the KSI–Vpu fusion peptide; lane 3, fusion peptide purified from Ni chelate chromatography; lane 4, mixture after CNBr cleavage; lane 5, purified polypeptide. (B) Reverse-phase HPLC purification of the polypeptide.
3.3. Micelle Samples
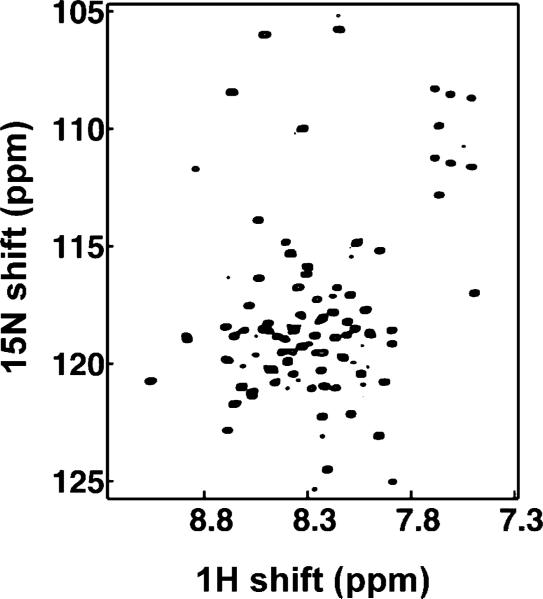
As soon as the polypeptide has been purified, and its sequence verified, samples of protein-containing micelles are prepared for solution NMR spectroscopy. There is a long history to preparing samples of membrane-associated peptides and proteins in micelles for solution NMR studies.45,73–76 For all polypeptides, regardless of length or hydrophobicity, we use four different lipids (SDS (sodium dodecyl sulfate), DPC (dodecyl phosphatidyl choline), DHPC (dihexanoyl phophatidyl choline), LMPC (lyso myristoyl phosphatidyl choline)) in an initial screening of conditions for two-dimensional HSQC spectra of uniformly 15N labeled samples. With the increasing level of activity in the field of solution NMR of membrane proteins, many new suggestions for lipids and combinations of lipids are emerging.68,77 Since the key solution NMR measurements are performed with weakly aligned samples, it is also necessary to simultaneously optimize the conditions for the various gel samples, including the transfer of the protein into the gels. These spectra are highly sensitive monitors of the entire expression, purification, and sample preparation process. Any doubling or anomalous broadening of resonances is a warning sign that the polypeptide is aggregated or improperly folded.45 We do not proceed with structural studies by solution NMR or even with sample preparation in bilayers or bicelles for solid-state NMR until these problems are resolved. This is a crucial checkpoint. Moreover, it is one of the reasons that it is advantageous to study all polypeptides in parallel by solution NMR and solid-state NMR methods. It is generally possible to identify several combinations of lipids, concentration, ionic strength, pH, and temperature that yield solution NMR spectra of membrane proteins in micelles.68–71,77–80 Resolved two-dimensional HSQC spectra, such as that shown in Figure 3, set the stage for structural studies by solution NMR and provide assurance that it is worthwhile to proceed with the preparation of bilayers and bicelle samples for solid-state NMR experiments.
Figure 3.
Two-dimensional HSQC spectrum of uniformly 15N labeled Vpu in micelles.
4. Solution NMR of Micelle and Small Bicelle Samples
Bicelles50 are bilayer disks that self-assemble from mixtures of long chain (DMPC) and short chain (DHPC) phospholipids.81,82 The size of bicelles is controlled by the ratio (q) of DMPC to DHPC. Small isotropic bicelles are of interest as samples in their own right.83,84 They also serve as controls for the use of larger, magnetically alignable bicelles as samples for protein structure determination by solid-state NMR methods.85 The isotropic chemical shift frequency is a highly sensitive qualitative monitor of protein structure. If the resonance frequencies are the same or very similar, except for line widths and intensities, in bicelles of increasing size, then the protein structure is not affected by the differences in the lipid assemblies. Since small bicelles can be weakly aligned in stressed gels,33,43,44,86 and large bicelles can be completely aligned by the magnetic field, these samples provide alternative ways of accessing the orientation information from the anisotropic spin interactions in samples under conditions where the proteins are likely to have very similar structures and local environments. They also provide access to a wide range of dynamics.
The second major class of membrane proteins, β-barrels, has been successfully studied in micelles. Their more compact structures lend themselves to the applications of TROSY-based87 pulse sequences for improved resolution and to NOE-based methods of structure determination.88–90
4.1. Assignment of 1H–15N HSQC Spectra
The two-dimensional 1H–15N HSQC spectra of uniformly 15N labeled membrane proteins in micelles are assigned using a combination of methods, including comparisons to spectra of selectively 15N labeled samples to identify resonances by residue type, the measurement of short-range 1H–1H NOEs in two- and three-dimensional experiments,91 the observation of H/D exchange patterns to confirm the identity of residues in stable helices, as well as triple-resonance experiments92 on uniformly 13C and 15N labeled samples. Even so, the assignment process is challenging because of the extensive spectral overlap resulting from the many hydrophobic residues, the predominantly helical secondary structure, and the broad resonance line widths in these samples. Often it is necessary to reassign the spectra obtained in different micelle or bicelle environments because minor changes in chemical shifts can be problematic due to the crowded nature of the spectra of these highly helical proteins.
In studies of globular proteins in solution by NMR spectroscopy, homonuclear NOE measurements provide the principal input for structure determination in the form of distance constraints.8–11 Unfortunately, this experiment is not nearly as useful in studies of helical membrane proteins in micelles because they typically do not have observable long-range NOEs due to their modular architecture and extensive overlap of side chain resonances. Sufficient NOEs were found between subunits of the trans-membrane helix dimer of glycophorin to describe their interactions and packing.93 In the membrane-bound form of fd coat protein with two helical segments, extraordinary efforts were required to extract a few NOEs in the loop connecting the helices to characterize its fold.94 Although subsequent studies in weakly aligned micelles,95 and completely aligned bilayers,32 have confirmed the NOE-based structure, it is now apparent that fd coat protein is an exception, and for the most part, it will be necessary to determine the structures of helical membrane proteins in micelles without the use of NOEs as distance constraints.
4.2. H/D Exchange
The comparison of HSQC spectra of samples in H2O and D2O solutions gives a useful view of the topology of membrane proteins in micelles by identifying the subset of the most stable helical residues.76,96,97 To extend the range of exchange rates that can be monitored to identify more subtle structural features, we developed hydrogen–deuterium fractionation.98 The experimental data are obtained by lyophilizing the initial NMR sample and redis-solving the protein-containing micelles in solutions with increasing amounts of D2O. Only one sample is needed in order to quantitatively compare the peak intensities. Fractionation factors are derived from the intensities of these resonances as a function of D2O concentration.
4.3. Relaxation Parameters
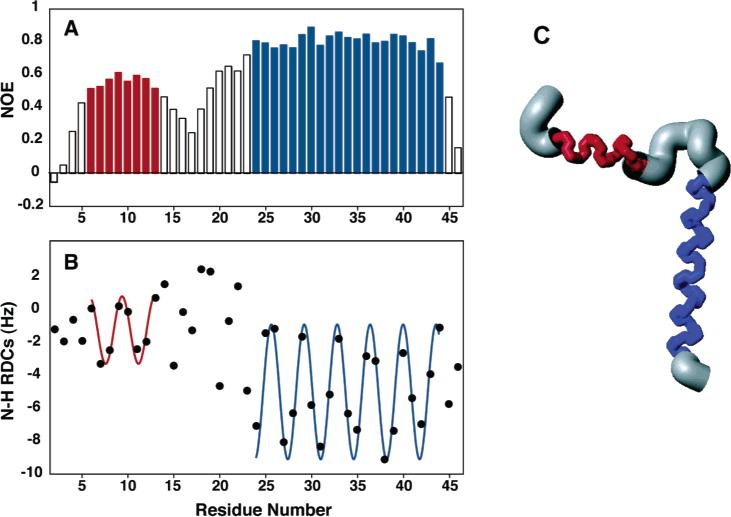
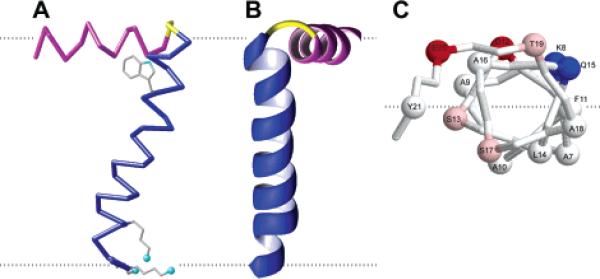
Backbone structure and dynamics are more obviously coupled in helical membrane proteins than they are in most other classes of proteins. There are often dramatic differences between the relaxation parameters for structured helical residues and, for example, those for residues near the N- and C-termini. The heteronuclear 1H–15N NOE, in particular, provides a direct and sensitive indicator of the local dynamics of membrane proteins in micelles.44,99 The experimental heteronuclear NOEs for all backbone amide sites with resolved resonances can be measured using the method of Farrow et al.,100 with and without 1H irradiation to saturate the 1H magnetization. The results shown in Figure 4A give a clear indication of the structural organization of the membrane-bound form of Pf1 coat protein in micelles. The full set of relaxation parameters can be analyzed quantitatively to describe the protein dynamics. Generally, the results for the membrane spanning helices can be fit in a straightforward manner, since the helices pass through the center of the micelle, which itself undergoes effectively isotropic reorientation. The situations for the terminal regions of the proteins, and the loops connecting the helices, are likely to be more complex, since there may be extra modes of motion not well modeled in a simple order parameter analysis.101,102 There is a growing range of interpretation methods capable of dealing with more complex dynamics.103–105
Figure 4.
Structure and dynamics of Pf1 coat protein in micelles: (A) heteronuclear 1H–15N NOE as a function of residue number; (B) RDC as a function of residue number; (C) structure of the membrane-bound form of Pf1 coat protein.
4.4. RDCs from Weakly Aligned Samples
The orientation constraints resulting from the measurement of residual dipolar couplings (RDCs) have had a dramatic impact on structure determination of globular proteins in solution by NMR spectroscopy.39,106 Their importance is even greater for helical membrane proteins in micelles and small bicelles because of the near-total absence of assignable long-range distance constraints from NOEs in these samples. However, until recently it has been difficult to prepare weakly aligned samples of membrane proteins suitable for solution NMR experiments. The well-established methods of obtaining weak alignment of proteins cannot be applied to protein-containing micelles; filamentous bacteriophages are unstable in the presence of lipids, and bicelles or membrane fragments simply merge with the protein-containing micelles. We developed lanthanide-induced alignment of proteins for this purpose.41,42 However, there are limitations to the use of lanthanides, including the extent of alignment and reproducibility. We now utilize an alternative approach that relies on the use of stressed polyacrylamide gels.43,44,47,48,86
Because the dominant type of secondary structure in membrane proteins is the helix, dipolar waves are highly effective at identifying the helical residues and the relative orientations of the helical segments.107 The first step of the analysis consists of plotting the magnitudes of the RDCs as a function of residue number and fitting a sine wave with a period of 3.6 residues to these data.86,95,107,108 The quality of fit is monitored by a scoring function in a four-residue sliding window and the phase of the fit. This is illustrated in Figure 4B. Alignment-induced changes in chemical shift frequencies provide complementary conformational constraints.16,33,44,109
Another useful approach to compensate for insufficient NOEs and to determine global fold in micelle environments involves the combination of site-directed electron spin labeling and NMR. Electron spin labels are incorporated in proteins at Cys residues, and distance constraints are derived from paramagnetic broadening of NMR resonances.110,111 Multiple distance measurements can be made by separately incorporating spin labels into different Cys sites. In addition, spin label probes can be incorporated within the micelles in order to probe protein insertion.75,112,113
4.5. Structure Calculation
We utilize protocols for calculating the structures of membrane proteins that rely primarily on the RDCs measured in weakly aligned samples as input to XPLOR-NIH.114 The structure of the membrane-bound form of the Pf1 coat protein shown in Figure 4 demonstrates the feasibility of determining the three-dimensional structures with this approach. The first step of the calculation is high-temperature equilibration, during which an extended conformational template of the molecule is created with favorable bond lengths and bond angles. Helices are locked into place for regions previously identified as helical by dipolar waves95 with a very strong restraint on their dihedral angles. The second step involves the slow introduction of the residual dipolar coupling restraints using simulated annealing to orient the helices relative to one another and individually refine the helices. The third step is straightforward energy minimization; side chain conformational preferences are applied to adjust the local geometry of the protein. Also included in this protocol is a database term that biases the sampled conformational space on the basis of Ramachandran plot energies and ensures that the packing geometry is reasonable for the under-restrained regions of the protein, which include the N- and C-terminal and loop residues at the current stage of the research. For Pf1 coat protein, four families of solutions were obtained, which is the same result found on the basis of dipolar waves.95 One family could be selected by reference to the solid-state NMR data, assuming that the protein has the same structure in micelles and bilayers. The three-dimensional structure of the membrane-bound form of Pf1 coat protein is shown in Figure 4C.
5. Solid-State NMR of Lipid Bilayer Samples
5.1. Oriented Planar Bilayer Samples
Solid-state NMR spectroscopy is being widely applied to membrane proteins using both aligned sample and magic angle sample spinning approaches.5,30,35,115-136 The basic structural organization of helical membrane proteins, with their well-delineated helical domains that have either trans-membrane or in-plane configurations, means that they are particularly well suited for aligned sample approaches.
Fortunately, it is generally easier to prepare uniaxially aligned samples of membrane proteins in bilayers than it is to crystallize them. Nonetheless, it is still a demanding task to prepare the well-aligned samples of membrane proteins in lipid bilayers that yield well-resolved solid-state NMR spectra. Two different methods for aligning lipid bilayers on glass plates are used (reviewed in refs 66 and 137). These are deposition from organic solvents followed by evaporation and lipid hydration, and fusion of unilamellar reconstituted lipid vesicles with the glass surface. Before insertion into the square coil of the NMR probe, the stacked glass plates are placed in a thin film of polyethylene, which is heat-sealed to maintain sample hydration during the experiments.
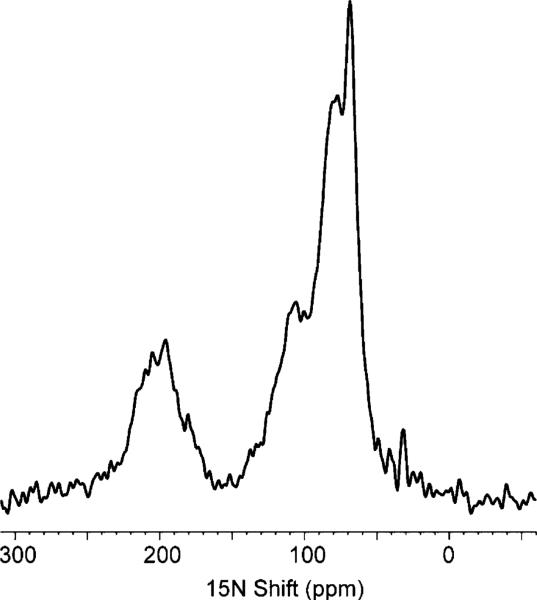
In the early stages of the investigation, one-dimensional solid-state NMR spectra of uniformly 15N labeled samples are used to screen sample conditions. For example, the distinctive dip to the baseline in the middle of the spectrum (near 150 ppm) in Figure 5 is indicative of a well-oriented membrane protein. If the sample contained a significant amount of unoriented protein, then the associated powder pattern intensity would appear in all regions of the spectrum, including in the relatively clear region near 150 ppm. In addition, these spectra give the first view of the dynamics of the protein backbone on the 104 Hz time scale of the 15N amide chemical shift interaction.
Figure 5.
One-dimensional 15N NMR spectrum of uniformly 15N labeled Vpu in bilayers aligned between glass plates.
5.2. Magnetically Aligned Bicelle Samples
The ideal sample for structure determination of membrane proteins can be envisaged as a mixture of lipids, water, and salts that when added to the purified polypeptide self-assembles into bilayers where the protein is in its active, native conformation. The bilayers would then provide the environment that both immobilizes the protein and aligns it magnetically, fulfilling the two prerequisites of structure determination by solid-state NMR of aligned samples. This is remarkably close to the description of large bicelles, which are promising samples for NMR studies of membrane proteins.50,82,138 Since the protein-containing bicelles are in aqueous solution, they ensure that the proteins and lipids are fully hydrated, and they enable the use of a sealed sample tube, which contributes greatly to sample stability. Structure determination by solid-state NMR of aligned samples is predicated on the molecules being im-mobile and uniaxially aligned parallel to the direction of the applied magnetic field.22,139 Large bicelles naturally align perpendicular to the direction of the applied magnetic field; however, they can be flipped to the parallel orientation by the addition of lanthanide ions,51 and it is possible to obtain high-resolution solid-state NMR spectra of membrane proteins aligned in this way.85
5.3. PISEMA Spectra of Uniformly 15N Labeled Samples
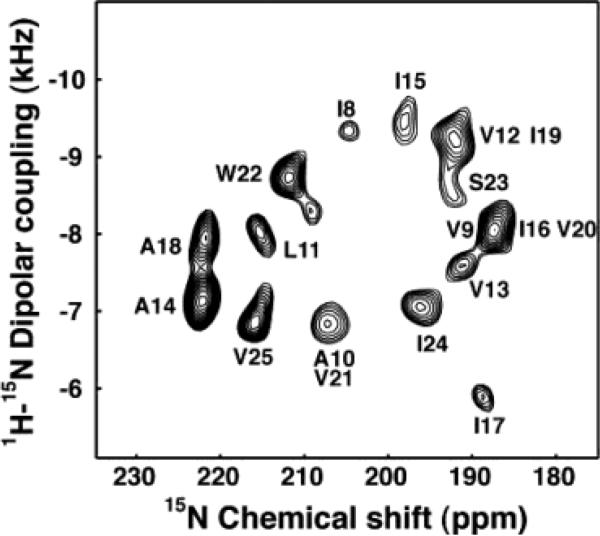
We developed the PISEMA (polarization inversion spin-exchange at the magic angle) experiment37 and its successor, SAMMY,38 to obtain high-resolution separated local field spectra of uniformly labeled samples of aligned proteins. The experimental two-dimensional 1H–15N PISEMA spectrum of the trans-membrane channel-forming domain of Vpu,33 shown in Figure 6, has excellent resolution because the angular dependencies of the heteronuclear dipole–dipole and chemical shift interactions are favorable for helices tilted slightly with respect to the direction of the magnetic field. We have developed,25,28 and applied to membrane proteins,17,27 three-dimensional versions of the experiment as well as a four-dimensional experiment,26 capable of providing additional resolution for larger proteins. The spectra of several uniformly 15N labeled membrane proteins in oriented lipid bilayers have been resolved using these experiments.32–35
Figure 6.
Experimental PISEMA spectrum of uniformly 15N labeled trans-membrane ion-channel domain of Vpu in bilayers aligned on glass plates.
5.4. Analysis and Assignment of PISEMA Spectra
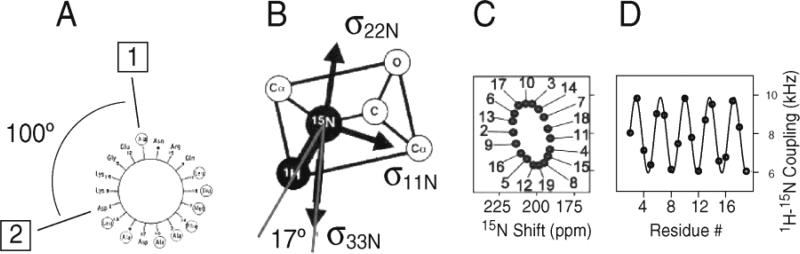
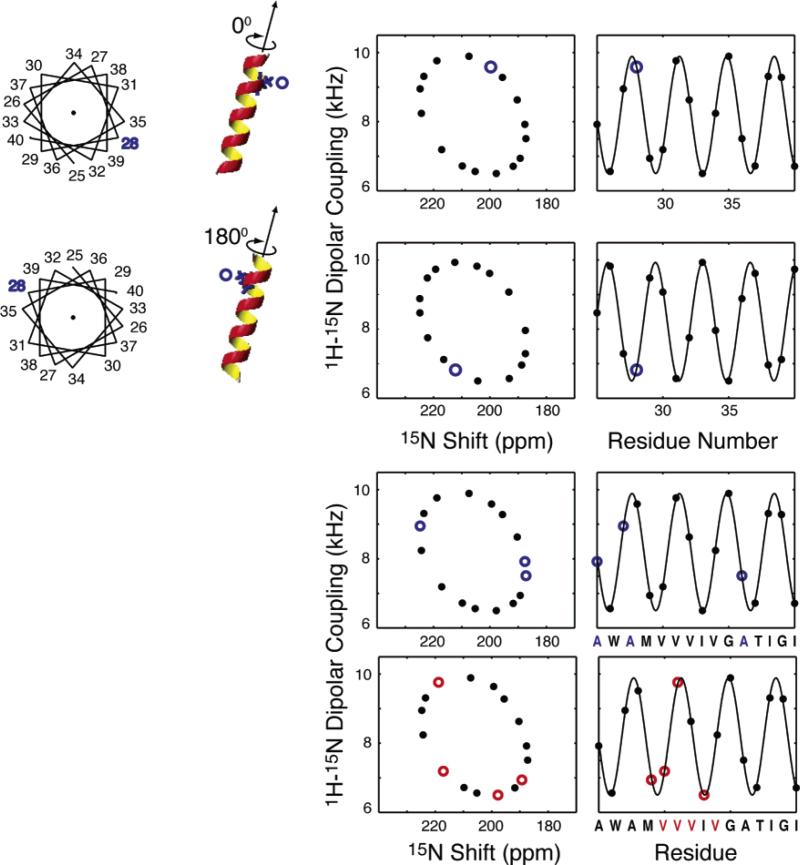
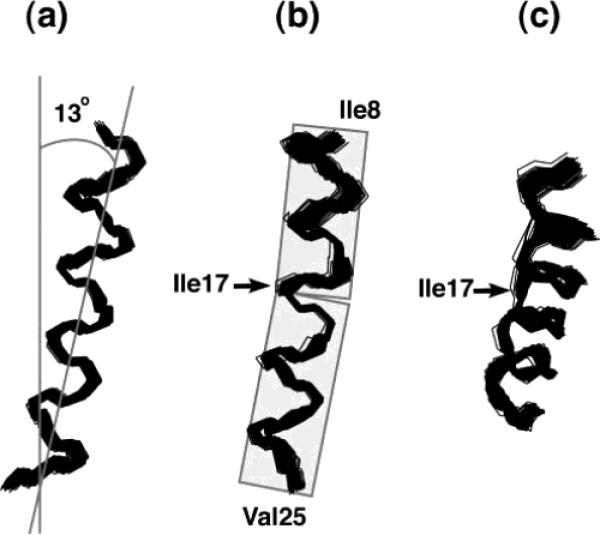
The anisotropy of nuclear spin interactions results in a unique mapping of structure to the resonance frequencies observed in the NMR spectra of completely aligned samples. The wheel-like pattern of resonances in the experimental PISEMA spectrum in Figure 6 is typical of that observed for helical membrane proteins in aligned samples. We refer to these patterns as PISA (polarity index slant angle) wheels.140,141 The principles of protein structure and NMR spectroscopy that result in PISA wheels are illustrated in Figure 7. In Figure 7A, the helical wheel projection down the axis of an α-helix shows that the 3.6 residues per turn periodicity results in an arc of 100° between adjacent residues in the helical wheel, that is mirrored in their resonances in the PISA wheel, shown in Figure 7C. The orientations of the principal axes of the 15N chemical shift and 1H–15N heteronuclear dipole–dipole interactions in the molecular frame of reference are shown in Figure 7B.142 The 17° difference between the alignments of these two interactions, in the plane of the peptide bond, plays a crucial role in the resolution of the spectra, as well as providing orientation constraints for structure determination. The characteristic wheel-like pattern of resonances calculated for a two-dimensional PISEMA spectrum of an ideal helix is shown in Figure 7C, and the corresponding dipolar wave, which is a plot of the dipolar coupling as a function of residue number, is shown in Figure 7D.
Figure 7.
Principles of PISA wheels. (A) Helical wheel showing the 100° arc between adjacent residues that is a consequence of the periodicity of 3.6 residues per turn in an α-helix. (B) Orientations of the principal elements of the spin interaction tensors associated with 15N in a peptide bond. σ11, σ22, and σ33 are the principal elements of the 15N chemical shift interaction tensor, and the 1H–15N dipolar coupling interaction is along the NH bond. σ33 is in the peptide plane and makes an angle of 17° with the NH bond. (C) PISA wheel for an ideal α-helix. (D) Dipolar wave for an ideal α-helix.
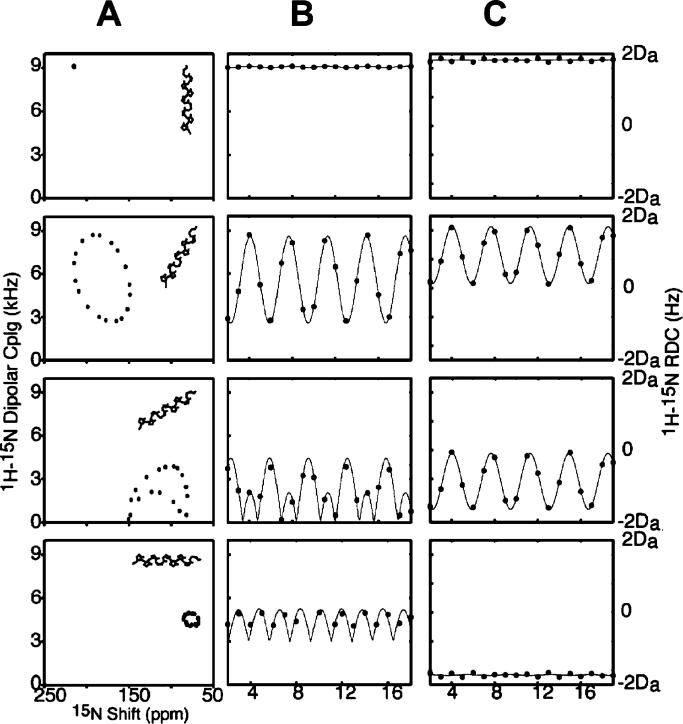
Figure 8A contains PISA wheels for an ideal α-helix at four different tilt angles, ranging from parallel to the field for a trans-membrane helix to perpendicular to field for a helix on the surface of the bilayer. When the helix axis is parallel to the direction of the applied magnetic field, all amide sites have the same orientation relative to the direction of the applied magnetic field; therefore, all of the resonances overlap. Tilting the helix away from parallel breaks the symmetry and introduces variations in the orientations of the amide NH bond vectors relative to the field. This is manifested as dispersions in the chemical shift and dipolar coupling frequencies. All trans-membrane helices studied to date are tilted with respect to the bilayer normal, and it is the combination of the tilt and the difference between the direction of the principal element of the 15N amide chemical shift tensor and the NH bond vector that makes it possible to resolve many resonances in helical membrane proteins. Notably, studies in lipids with different chain lengths show that helix tilt is not appreciably affected by the membrane hydrophobic thickness.109,135
Figure 8.
(A) Ideal α-helix tilted at 0°, 30°, 60°, and 90° relative to the direction of the applied magnetic field. (B) PISA wheels corresponding to the various tilt angles. (C) Dipolar waves derived from the corresponding PISA wheels. (D) Dipolar waves for a weakly aligned sample.
Figure 8B presents the dipolar waves that correspond to the PISA wheels shown in Figure 8A, while dipolar waves from the residual dipolar couplings measured for helices in weakly aligned samples are shown in Figure 8C. The similarities between the wave patterns in Figure 8B and C reflect the convergence of solid-state and solution NMR approaches to protein structure determination that is a consequence of the use of aligned samples. Both PISA wheels and dipolar waves can be used to determine the absolute (solid-state NMR) or relative (solution NMR) tilts of helices. PISA wheels have the advantage that they do not require resonance assignments but the disadvantage that they are susceptible to distortions due to site-to-site variations in the chemical shift tensors. Thus, by utilizing only the dipolar couplings, the analysis of dipolar waves minimizes errors in the determinations of the tilt angles. In practice, the uses of PISA wheels and dipolar waves are complementary, and they are analyzed jointly.
The PISA wheels for two different helix rotations (polarities) are identical, because the dipolar and chemical shift frequencies reflect only the tilt of the helix axis relative to the magnetic field. However, as illustrated in Figure 9, the spectra differ in their resonance assignments, whose patterns mirror exactly the polarity of the corresponding helical wheel. Consequently, the PISA wheel patterns in experimental spectra can be used to assist the assignment process.32,143 The sequential assignment of one residue or the identification of a few residues by type and their position on the rim of the PISA wheel (or the phase of the dipolar wave) directly determines the rotational orientation (polarity) of the α-helix about its long axis. Proteins that are selectively labeled (by residue type) can be readily prepared, and their spectra are instrumental in the identification of the positions of specific residues in experimental PISA wheel spectra and dipolar wave plots. This determines the polarity of the helix in the membrane. Examples are shown in Figure 9 for both single and multiple site labels in a helix. In addition to determining the rotation of the helix in the bilayers, these patterns assist in the assignment of the complete spectrum of a protein because of the direct connection between the structure and its representation in the spectra.
Figure 9.
For an ideal α-helix tilted at 20° relative to the magnetic field, the position of a resonance from a particular residue in the wheel-like pattern is determined by its absolute rotation relative to the long axis of the helix. When a type of residue is present in multiple locations, the pattern of resonances is also uniquely a function of the rotation of the helix. Simulations are shown for a hypothetical Ala selective label and a Val selective label in the amino acid sequence shown on the bottom of the plots for the same helix.
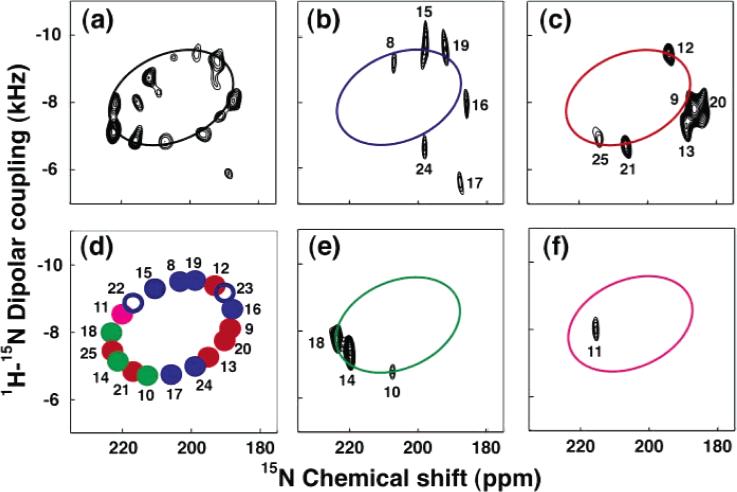
The assignment process begins with the acquisition of PISEMA spectra from one uniformly 15N labeled sample and several selectively 15N labeled samples. NMR spectra of selectively labeled samples are routinely used to assign resonances to types of amino acids; however, in this application they provide additional information. In the experimental PISEMA spectra from one uniformly 15N labeled sample and four selectively 15N labeled samples in Figure 10, a single resonance is observed for each labeled backbone amide site. These spectra illustrate how the wheel-like pattern for a trans-membrane helix emerges from the data. The pattern of the alanine resonances alone is sufficient to index the wheel and determine the helix polarity. The rest of the resonances in the pattern can then be assigned by inspection. The method can be readily extended to three-dimensional spectra if additional resolution is needed. Further, it is possible to select a single peptide plane orientation out of the four symmetry-related possibilities that are consistent with the pair of frequencies associated with each resonance in a PISEMA spectrum because the orientation of each peptide plane in an α-helix can be predicted from the helix tilt and rotation angles, and helical and PISA wheels can be divided into four pie-shaped sections, corresponding to the four symmetry-related peptide plane orientations.32,143 The full application of “shotgun” NMR32 (named for the spread of labeled sites in the sequence and on the PISA wheel target) where the assignment and structure determination processes are performed in parallel, rather than sequentially, yields both complete resonance assignments and unique structures from the NMR data.31-33,143
Figure 10.
PISA wheel analysis of the PISEMA spectrum of Vpu: (a) uniformly 15N labeled; (b) 15N Ile (blue); (c) 15N Val (red); (d) simulated ideal PISA wheel with resonances corresponding to the Ile (blue), Val (red), Ala (green), and Leu (magenta); (e) 15N Ala (green); (f) 15N Leu (magenta).
Inevitably, there will be some resonances that cannot be assigned with the shotgun approach. Certain elements of the mapping of structure onto the spectra are useful in establishing partial assignments and, in combination with many established assignments, can yield unique assignments. A number of spectroscopic approaches for assigning resonances have been developed. Both dilute and abundant homonuclear spin-exchange discriminate strongly toward nuclei in close proximity and have been integrated into multidimensional solid-state NMR experiments. We have utilized spin-exchange as a method for associating resonances from sites in adjacent residues for assignment purposes. In an early example,144 two-dimensional homonuclear 15N spin-exchange experiments were used to identify pairs of nitrogens in adjacent peptide bonds for assignment purposes. In most cases, it is performed as a three-dimensional experiment, and this was used to assign resonances in the trans-membrane helix of the acetylcholine receptor M2 ion channel.34 Abundant spin-exchange relies on homonuclear dipole–dipole couplings among 1H sites. There are several advantages to abundant spin-exchange experiments, including the use of a much shorter mixing time which reduces the overall time of the experiments, the generally shorter internuclear 1H–1H distances, and the well-established relationships among hydrogens in various protein structures as used in solution NMR.8–11 We have also implemented a variety of triple-resonance experiments on oriented protein samples to obtain sequential assignments of 13C and 15N backbone resonances. Initially, resonances were assigned by selective double labeling with 15N amino and 13C carbonyl labeled samples;145 subsequently, this approach has been extended using multidimensional experiments.146 Significantly, this does not depend on the type of structure and, therefore, can be applied equally well to residues in helices and turns. Standard procedures such as comparing spectra in H2O and D2O solutions are also effective in selecting among several possible assignments.31,33,35,147
5.5. Analysis of Orientation-Dependent Results
While the initial motivation for the development of the interpretation methods was the analysis of experimental PISEMA spectra of completely aligned proteins, the emergence of residual dipolar couplings as a structural tool allows the data obtained from weakly aligned soluble proteins or from membrane proteins in micelles to be analyzed in a parallel manner. PISA wheels and dipolar waves involve the analysis of the periodic patterns observed in the spectral data and are applicable to results from both solid-state NMR of completely aligned samples and solution NMR of weakly aligned samples. In the case of dipolar waves, the periodic function is obtained by using a simple distribution on a cone assuming a constant angle between the N–H bond and the helix axis. As a result, the dipolar couplings of helices exhibit wavelike patterns as a function of residue number; the same effects have been noted for the chemical shift interaction. Dipolar waves provide independent validation for the geometry of α-helices. They provide a direct and reliable measure of the regularity of α-helices, since they are independent of the magnitudes and orientations of the 15N chemical shift tensor. Comparisons of experimentally measured dipolar couplings, modeling studies, and bioinformatics have shown that the helices found in proteins typically satisfy this ideal approximation quite well. The frequency due to a dipolar coupling at a 15N labeled backbone amide site is determined by the angle that an N–H bond forms with respect to the direction of the applied magnetic field.
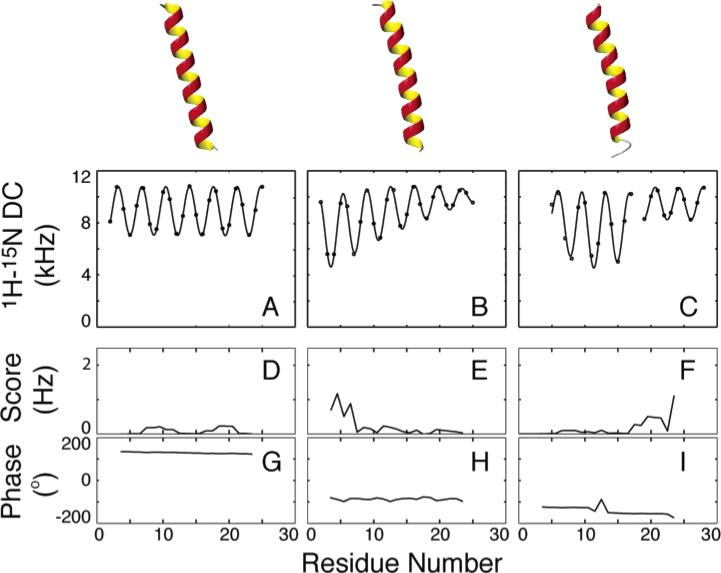
Deviations from ideality have a pronounced effect on the appearance of PISA wheels and dipolar waves. In general, however, the 100° rotations between adjacent residues in the sequence preserve the general wheel-like pattern of resonances from helical residues. Without the influence of chemical shift variability, dipolar waves are highly predictable and reliable indicators of molecular structure. The simulated patterns in Figure 11 demonstrate the effects of deviations from ideal geometry on the appearance of dipolar waves. In general, the periodicity is unaffected, while the changes in average value and amplitude reflect the change in orientation of the local helix axis. Curvature gradually changes these values, and kinks cause abrupt changes.56,95,148
Figure 11.
1H–15N dipolar couplings simulated for (A) a straight ideal α-helix, (B) an α-helix with a 55 Å radius of curvature, and (C) an ideal α-helix with a 20° kink with their average axis tilted 15° relative to the alignment z-axis. (D–F) The average error per point shows that the periodicity in all cases is 3.6 except near the ends, where there is some deviation. (G–I) The phase is also diagnostic, where the kink is evidenced by a slight change in the phase of one sinusoid relative to the other.
Figure 12 illustrates the analysis of the orientation-dependent frequencies measured from the resonances in the spectrum of the uniformly 15N labeled trans-membrane helix of Vpu, shown in Figure 6, and the assignments derived from the spectra of the selectively 15N labeled samples using the “shotgun” approach. To focus on the principal structural features of the domain, the sequence is divided into two segments: residues 8–16 (blue) and residues 17–25 (red). The second column (Figure 12d and e) represents the ideal PISA wheels that correspond to the experimental data, showing that the two segments of the helix have different tilt angles. The magnitudes of the dipolar couplings measured from the PISEMA spectrum of a uniformly 15N labeled sample of Vpu are plotted as a function of residue number in Figure 12f. The magnitudes of the residual dipolar couplings measured from the IPAP spectrum of the weakly aligned micelle sample are shown in Figure 12g.
Figure 12.
(a–c) Representations of the experimental PISEMA spectrum of Vpu2–30+ in completely aligned bilayers: (a) residues 8–25; (b) residues 8–16; (c) residues 17–25. (d and e) Ideal PISA wheels with uniform dihedral angles (ϕ ) –57°; ψ ) –47°) corresponding to the experimental data: (d) residues 8–16 with the tilt angle 12°; (e) residues 17–25 with the tilt angle 15°. (f) Dipolar waves of 1H–15N unaveraged dipolar couplings obtained from completely aligned bilayers. (g) Dipolar waves of 1H–15N residual dipolar couplings obtained from weakly aligned micelles. (h) A tube representation of the trans-membrane helix of Vpu in lipid bilayers.
The measured value of the dipolar coupling of Ile17 deviates markedly from the sinusoidal functions that fit well to the neighboring sites, indicating that there is a deviation from ideality of the helix near Ile17. The different amplitudes and average values of the sine waves show that there is a kink in the helix at residue 17 and that the two helical segments have slightly different orientations. The results for the trans-membrane helix domain of Vpu are consistent in showing a kink at residue 17 in both micelle (Figure 12g) and bilayer (Figure 12f) samples. The tilt angles of the helical segments in the lipid bilayer can only be defined from the solid-state NMR data where the alignment frame is established by the placement of the sample in the magnet. Residues 8–16 have a tilt angle of 12° with a rotation 52°, and residues 18–26 have a tilt angle of 15° with a rotation of 59°. The two distinct components of the trans-membrane helix kinked at Ile17 are represented by tubes in Figure 12h.
5.6. Structural Fitting
Structural fitting is a recently developed approach that is complementary to the direct calculation of protein structures from the orientation-dependent frequencies.149 When all of the resonances are assigned to residues in the protein, then results of structural fitting are indistinguishable from direct calculation. In both approaches, the NMR data yield only one solution and a unique structure is obtained. It has been shown to identify kinks and other deviations from ideality in helices, in complete agreement with features from the mapping found in spectra. This is accomplished by the fitting of the spectrum to a model structure assuming a constant peptide plane geometry. When the resonances are assigned, the Ramachandran backbone dihedral angles, ϕ and ψ, are considered the only degrees of freedom. The whole structure is assembled by the sequential walking from one residue to the next, calculating the backbone dihedral angles, ϕ and ψ, directly from two orientation-dependent frequencies for these residues. In addition, it can utilize partial assignment information and has the potential to be an “assignment-free” approach to structure determination.
When a structure is calculated from solid-state NMR data, three sources of error can be anticipated. The first arises from imperfections in the experiments, for example, incomplete decoupling or sample misalignment, which can lead to incorrect measurements of the frequencies. The second source is the residue-to-residue variability of the magnitudes and orientations in the molecular frame of the principal components of the chemical shift tensor. Finally, it is not always clear whether deviations in the spectrum of a helical protein from an ideal PISA wheel are due to the variations in the dihedral angles ϕ and ψ or to a combination of the first two sources of error. Taken together, these three uncertainties determine the accuracy of the structural fit, since multiple solutions for ϕ and ψ, consistent with the experimental measurement (hence for the overall structure), may be possible within the limits of experimental errors and uncertainties in the spin interaction tensors. To assess the error in structure calculations, we perform the following statistical analysis. For every calculation of the Ramachandran angles between two consecutive frequency points, each of the principal values of the chemical shift tensor are allowed to vary within ±5 ppm relative to their canonical values. The experimental accuracy for the determination of the spectral positions is conservatively estimated to be ±100 Hz in each frequency dimension; in other words, a solution for the torsion angles ϕ and ψ is regarded as plausible if the calculated frequency point was lying within a 100 Hz radius relative to the experimental point. The rmsd's of the multiple solutions relative to the average structure are then used to estimate the accuracy and uniqueness of the structural fitting.
A structural fit to a fully assigned spectrum is equivalent to a direct calculation of the protein structure, and this is shown in Figure 13 for Vpu. The structure was calculated sequentially from the N terminus to the C terminus, giving the torsional angles (ϕ and ψ) between each pair of residues. Therefore, there are no ambiguities with respect to the relative orientations of the helical segments. By performing repeated structural fits to the experimental data within its estimated error range, one hundred structural solutions were generated and superimposed. They have a mean rmsd of <1 Å relative to the average structure, which provides an estimate of the precision of the structure determination. The structure of the trans-membrane helix determined by structural fitting has an average tilt angle of 13° with respect to the bilayer normal (Figure 13a) and a slight kink at Ile17 in the middle of the helix (Figure 13b). The average values for the Ramachandran angles between residues 16 and 17, where the kink occurs, are calculated as ϕ = –71° and ψ = –37°. There is complete consistency between the analysis of the PISA wheels and dipolar waves presented in Figure 12 and the results of structural fitting presented in Figure 13. Also, a structure of Vpu was calculated using a simulated annealing protocol in XPLOR-NIH,114 with restraints for helical dihedral angles and dipolar couplings. This is equivalent to refining the structure of an α-helix with an established orientation.
Figure 13.
(a) Superimposition of 100 calculated backbone structures of the trans-membrane helix of Vpu. (b) A 90° rotation of panel part a to the vertical axis. (c) A 30° tilt of panel part b toward the reader. The structures were obtained by structural fitting of the experimental solid-state NMR data and are aligned for the best overlap of residues Ile8–Val25. The average tilt of about 13° relative to the membrane normal and a slight kink near residue Ile17 are apparent when it is noted that the plane of the average tilt of the helix does not coincide with the plane of the kink.
5.7. Three-Dimensional Structures of Proteins from Orientation Constraints
In the most general approach to structure determination of completely aligned samples,24,34,58,139,145,150-154 it is necessary to measure two or more frequencies for each residue of a protein in order to determine the orientation of a peptide plane. Once the orientations of all of the individual peptide planes are determined experimentally, then the planes are assembled into a complete protein structure because they are all related to the common axis defined by the direction of the applied magnetic field. Side chain orientations can be determined in a similar manner.139 A key feature of this approach is that experimental determinations are made relative to an external nonmolecular axis; therefore, the effects of errors and uncertainties in tensors and bond lengths do not accumulate. Previous determinations of protein structures by solid-state NMR relied on orientation constraints derived from the frequencies of independently assigned resonances.30,34,125,145 Newer versions of the approach explicitly exploit the mapping of structure into the spectra and combine the assignment and structure determination steps. In practice, we apply four complementary methods of interpretation to the solid-state NMR data as they are obtained. Once the full data sets are available, these approaches provide important checks on assignments and structural findings. These methods are referred to as PISA wheels,140,141 dipolar waves,95,107 structural fitting,149 and direct structural calculation.24,152
The chemical shift and dipolar coupling frequencies of the PISEMA resonances provided the sole input for structure determination of the membrane-bound form of fd coat protein in lipid bilayers shown in Figure 14. The 16-Å-long in-plane helix (residues 8–18) is amphipathic and rests on the membrane surface, with the boundary separating the polar and nonpolar residues parallel to the lipid bilayer surface and with the nonpolar residues facing the hydrocarbon core of the lipid bilayer (Figure 14C). The aromatic residues, Phe11 in the in-plane helix and Tyr21 in the trans-membrane helix, are near this boundary region and approximately equidistant from the hydrophobic central core of the lipid bilayers. The 35-Å-long trans-membrane helix (residues 21–45) crosses the membrane at an angle of 26° up to residue Lys40, where the helix tilt changes to 16° (Figure 14A and B).
Figure 14.
Structure of the membrane-bound form of the fd coat protein in lipid bilayers with the in-plane helix in magenta, the trans-membrane helix in blue, and the short connecting turn in yellow. The flexible N- and C-termini are not shown. (A) Side view showing the 26° tilt of the TM helix. The dashed gray lines mark the lipid bilayer membrane boundary. The direction of the applied magnetic field is parallel to the arrow. (B) Front view. (C) N-terminal view of the in-plane helix Cα atoms. The dashed gray line marks the boundary between hydrophilic and hydrophobic (gray) residues.
The availability of uniformly 13C and 15N labeled samples will provide spectroscopic access to all of the backbone and side chain sites of proteins. Although we have focused largely on 15N labeled sites because of their strategic locations in the backbone of polypeptides and the ready opportunities for selective and uniform labeling, PISEMA, SAMMY, and related experiments are generally applicable. There are a number of reasons for extending the experiments to 13C sites, including the substantially stronger 1H–13C dipolar coupling resulting from 13C having a higher gyromagnetic ratio. 13C labels provide access to the important 13C backbone site, with its single bonded hydrogen (except for Gly) as well as aliphatic and aromatic side chains. 15N and 13C detected versions of several triple-resonance experiments on single-crystal samples of labeled model peptides,146,155-157 and aligned bacteriophage146 samples, have been described.
5.8. Additional Experimental NMR Constraints
Triple-resonance experiments provide a mechanism for using a combination of heteronuclear correlation experiments, previously assigned 15N resonances, and homonuclear dilute spin-exchange among selectively and uniformly labeled 13C sites to obtain resonance assignments. Significantly, these same experiments can be used to measure additional orientation constraints as further input for structure determination. In addition to the 1H, 13C, and 15N chemical shift frequencies, the 1H–15N, 13C–15N, and 1H–13C dipole–dipole couplings are readily measured orientation-dependent frequencies. While the N–H and Cα–N couplings define the orientation of the peptide plane, the 1Hα–13Cα dipolar interaction occurs along an out-of-plane axis. As an independent orientation constraint, it significantly reduces the number of symmetry-related possibilities for the torsion angles ϕ and ψ between adjacent peptide planes. It directly addresses issues related to ambiguities at loops connecting helices in membrane proteins.
A few distance measurements can resolve among ambiguities associated with the packing of helices in membrane proteins. There are several families of solid-state NMR experiments that measure internuclear distances. The same dilute spin-exchange experiments used to assign resonances provide semi-quantitative distance constraints.144,158 Abundant spin-exchange can also be applied to stationary oriented samples28 or in unoriented samples with magic angle sample spinning.159,160 The distance range is substantially longer. While this experiment can suffer from a lack of specificity due to spin-diffusion, it is possible to tailor the transfer pathways using a combination of deuteration and multiple-pulse sequences.
Rotational echo double resonance (REDOR) spectroscopy developed by Schaefer and co-workers161 is widely applied to structural problems in proteins because it provides very accurate distance measurements between selected sites. It has been used on several membrane-associated polypeptides.64,131,162-164 This experiment relies on the placement of labels in favorable locations, which can be designed on the basis of the initial structural models. In addition, REDOR can provide the limited amount of distance information needed for the structures based on orientation constraints to converge. Proton-driven spin-exchange provides distance constraints in magic angle sample spinning experiments on unoriented samples.165–167 A wide variety of pulse sequences that recouple the dipolar interactions that are averaged out by magic angle sample spinning are being implemented on proteins.117,168,169
As described for solution NMR studies of membrane-associated peptides and proteins,111,112 electron spin labels incorporated into specific sites, for example, on Cys side chains, enable distance-dependent line broadening to be used as a constraint for structure determination that is complementary to the orientation constraints available from the nuclear spin interactions alone.
The incorporation of 19F labeled amino acids in the proteins provides opportunities to measure both orientation and distance parameters that have locations and principal axes that do not overlap with those of the 1H, 13C, and 15N sites in labeled proteins. Thus, this is a method for placing both individual residues and segments of secondary structure in the context of the overall protein structure. 19F has a large chemical shift anisotropy in most environments, and combined with a high gyromagnetic ratio, it has a wide frequency range. In some circumstances, it is possible to design the labeling pattern and experiments to measure internuclear distances between two 19F sites or between 19F and 13C or 15N sites. Prototype experiments have been performed on gram-icidin in aligned bilayers,170 and recently Ulrich and co-workers171 have studied a variety of 19F labeled polypeptides in aligned bilayers. McDowell et al.172 have demonstrated that the combination of 19F with 13C labeling enables the measurement of REDOR experiments over relatively long ranges in unoriented samples with magic angle spinning. The measurement of a few selective distances provides very strong constraints on folding.
5.9. Measurement and Analysis of Motionally Averaged Line Shapes from Backbone and Side Chain Sites
The line shapes of 15N backbone173 and 2H labeled side chain resonances of labeled polypeptides in unoriented lipid bilayer samples are key indicators of local backbone and side chain fluctuations.174-177 It is feasible to extend this approach to 13C labeled backbone and side chain sites.178 This enables NMR to provide a fully integrated description of the structure and dynamics of membrane proteins in bilayer environments over a wide range of time scales.
6. Comparison of Results from Solution NMR and Solid-State NMR
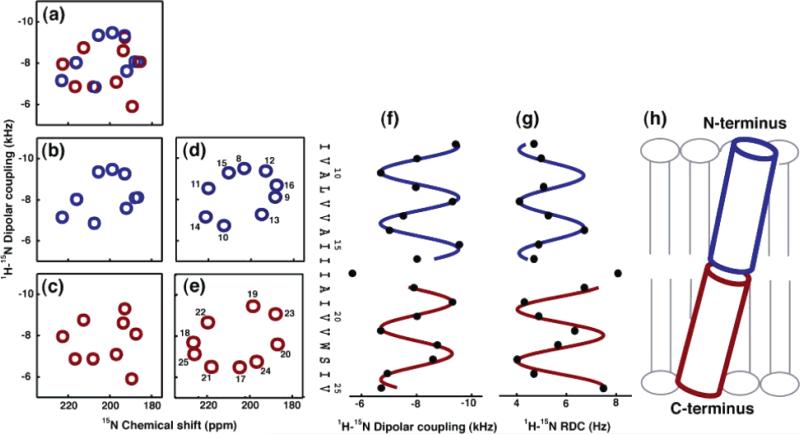
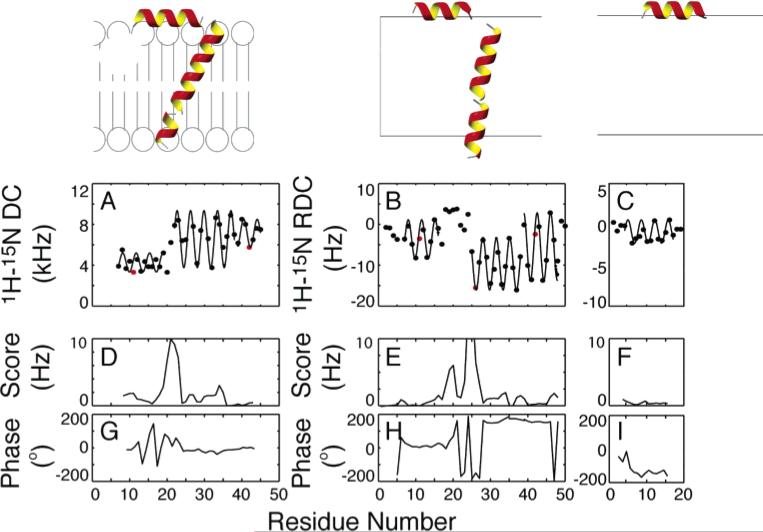
The secondary structures and relative orientations of the helices in the membrane-bound form of fd coat protein can be directly determined from the experimental data and fits to sinusoids shown in Figure 15.86 The results of three experiments on two different polypeptides, full-length fd coat protein and the 20-residue fdN peptide, that corresponds to the N-terminal amphipathic helix of the coat protein, are analyzed in the figure. The dipolar couplings in Figure 15A were measured on a sample of the coat protein in completely aligned bilayers, while those in Figure 15B and C were measured from samples of the 50-residue and 20-residue polypeptides, respectively, in weakly aligned micelles. The protein has very similar properties in bilayer and micelle environments. For example, using the periodicity of the oscillations of the dipolar couplings as a strict criterion, the number of residues in the N-terminal amphipathic helix is well defined and nearly identical in all three samples. Similarly, the length and other properties of the hydrophobic helix in the full-length protein are the same in micelles and bilayers. The average error per measurement for the fit of a four-residue sliding window function is shown in Figure 15D–F and the absolute phases for each window are shown in Figure 15G–I. The large increase in the score between residues Gln15 and Ile22 in Figure 15D and E is evidence of the lack of periodicity in the structures of the residues in the loop connecting the two helices. The helices are straight within experimental error, as evidenced by the low fitting errors for each helix.
Figure 15.
Experimentally measured dipolar couplings are shown for (A) fd coat protein in completely aligned bilayers, (B) fd coat protein in weakly aligned micelles, and (C) fdN (N-terminal 20 residues) in weakly aligned micelles. All datasets are shown with the best-fitting sinusoid and the parametrized expression yielding the tilts and rotations of the helices in the alignment frame. Shown below each dataset is (D–F) the rmsd to an ideal sinusoid and (G–I) the absolute phase of that sinusoid for each point. The positions of residues Phe11, Trp26, and Phe42 are highlighted in red (A, B, C) to characterize the rotations of the helices in the context of dipolar waves.
The amphipathic α-helix begins at Ala7 (which follows Pro6) and ends at Thr19 in bilayers and Ser17 in micelles. There are few discernible differences in the N-terminal helix due to the presence of the hydrophobic helix, demonstrating that the two helices are independent structural entities. In addition, there are no noticeable differences in the properties of this helix in micelle and bilayer samples, indicating that this helix is not affected by the micelle curvature or another property of the lipid assembly. This differs from a recent result on a different, longer polypeptide compared in micelle and bicelle samples.43 The positions of residues Phe11, Trp26, and Phe42 are highlighted in red (Figure 15A, B, and C) to characterize the rotations of the helices in the context of dipolar waves.
A detail of the membrane-bound form of the coat protein structure that may have significance when it is assembled into bacteriophage particles is the change in helix direction after residue Gly38. Remarkably, this same kink is found in the membrane-bound form of the protein, in both micelles (Figure 15A) and bilayers (Figure 15B), and in the structural form of the protein that interacts with DNA but not lipids in the coat of the bacteriophage particles.31 This kink is evident from the rise in the score for that region of the helix in Figure 15D and less dramatically in Figure 15E. In addition, the irregular patterns of the dipolar couplings of the residues connecting the amphipathic and hydrophobic helices demonstrate that there are substantial differences between the short bend in bilayers and the larger, more complex loop structure in micelles. There is evidence from relaxation data that these residues have internal mobility in the micelle samples. In bilayers, the trans-membrane helix begins at residue Tyr21, while in lipid micelles this helix begins at Trp26. This points to the importance of paying particular attention to residues near the bilayer interface in structural studies of membrane proteins. In general, the small size of the linkages in lipid bilayers restricts the possible relative orientations of the two helices and can be used to resolve the ambiguities in possible orientations. The two helical segments are connected by a short turn (Thr19–Glu20) that differs from the longer loop (residues 17–26) determined for the same protein in lipid micelles.94 This may be due to differences between the micelle and bilayer environment or to the limitations with working with the few long-range NOE restraints observed in solution NMR studies of helical membrane proteins. Both reasons argue in favor of the use of lipid bilayer samples for structure determination of membrane proteins.
7. Current Status
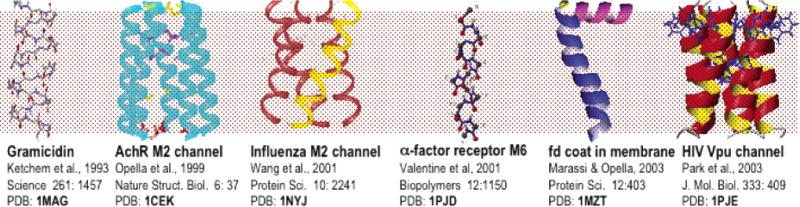
Recent X-ray diffraction structures of channel-forming proteins provide considerable detail about the properties of helices in membrane proteins. Partial145 and complete30–34,125 structure determinations of proteins by solid-state NMR have utilized completely aligned samples. In addition, substantial progress is being made in applying this approach to polycrystalline and other types of disordered protein samples.166,179–182 Recent developments in NMR pulse sequences, methods for data analysis, and methods for protein expression have enabled solid-state NMR spectra,17,148,179,183–191 and structures,30–34,166,180,191,192 to be obtained from several protein and peptide samples. Several atomic-resolution structures have been deposited in the Protein Data Bank (PDB), and those determined in oriented bilayer samples are shown in Figure 16.
Figure 16.
Structures determined by solid-state NMR in oriented bilayers. The PDB file numbers are in bold face.
Figure 16 shows the six structures of membrane proteins that have been determined by solid-state NMR spectroscopy. With the exception of gramicidin, none of the proteins have been crystallized, and none of the proteins in bilayers can be made to reorient rapidly enough to be studied by solution NMR spectroscopy. Therefore, these structures could only be determined by solid-state NMR spectroscopy. Moreover, solid-state NMR yields very accurate representations of the structures, with backbone RMSDs less than 1 Å at this still-early stage of development of the method. Many membrane proteins with between 50 residues and 200 residues are currently under investigation, and the experimental methods and instruments are being further developed so that the structures of substantially larger polypeptides can be determined.
Acknowledgments
We thank A. DeAngelis, D. Jones, and S. Park for preparing figures with their unpublished results. This research was supported by grants from the National Institutes of Health (EB001966, EB002169, GM64676, GM066978, and EB002031 to S.J.O., and CA82864 to F.M.M.), from the Department of the Army (DAMD17-00-1-0506 and DAMD17-02-1-0313 to F.M.M.), and from the California Breast Cancer Research Program (8WB0110 to F.M.M.).
Biography
 Stanley Opella was born in Summit, NJ. He obtained his B.S. in Chemistry from the University of Kentucky in 1969. His Ph.D. was received from Stanford University in 1974, where he studied proteins in solution by NMR spectroscopy with Oleg Jardetzky and Harden McConnell. He was a postdoctoral fellow at MIT 1975–1976 in the group of John Waugh, where he performed research involving solid-state NMR spectroscopy. He was on the faculty of the University of Pennsylvania from 1976 to 2000 and is currently Professor of Chemistry and Biochemistry at the University of California, San Diego, where his research is focused on the development of NMR and its application to proteins.
Stanley Opella was born in Summit, NJ. He obtained his B.S. in Chemistry from the University of Kentucky in 1969. His Ph.D. was received from Stanford University in 1974, where he studied proteins in solution by NMR spectroscopy with Oleg Jardetzky and Harden McConnell. He was a postdoctoral fellow at MIT 1975–1976 in the group of John Waugh, where he performed research involving solid-state NMR spectroscopy. He was on the faculty of the University of Pennsylvania from 1976 to 2000 and is currently Professor of Chemistry and Biochemistry at the University of California, San Diego, where his research is focused on the development of NMR and its application to proteins.
 Francesca Marassi was born in Mantova, Italy. She immigrated to Toronto, Canada, with her family and subsequently obtained her B.Sc., M.Sc., and Ph.D. degrees in Chemistry from the University of Toronto, where she studied NMR of lipids in membranes with Peter MacDonald. She was a postdoctoral fellow at the University of Pennsylvania from 1993 to 1998. She was subsequently an Assistant Professor at the Wistar Institute in Philadelphia from 1998 to 2000 and is currently on the faculty of the Burnham Institute in La Jolla, CA. Her research interests are focused on the application of NMR to proteins involved in the regulation of programmed cell death.
Francesca Marassi was born in Mantova, Italy. She immigrated to Toronto, Canada, with her family and subsequently obtained her B.Sc., M.Sc., and Ph.D. degrees in Chemistry from the University of Toronto, where she studied NMR of lipids in membranes with Peter MacDonald. She was a postdoctoral fellow at the University of Pennsylvania from 1993 to 1998. She was subsequently an Assistant Professor at the Wistar Institute in Philadelphia from 1998 to 2000 and is currently on the faculty of the Burnham Institute in La Jolla, CA. Her research interests are focused on the application of NMR to proteins involved in the regulation of programmed cell death.
References
- 1.Berman H, Henrick K, Nakamura H. Nature Struct. Biol. 2003;10:980. doi: 10.1038/nsb1203-980. [DOI] [PubMed] [Google Scholar]
- 2.Torres J, Stevens TJ, Samso M. Trends Biochem. Sci. 2003;28:137. doi: 10.1016/S0968-0004(03)00026-4. [DOI] [PubMed] [Google Scholar]
- 3.White SH, Wimley WC. Annu. Rev. Biophys. Biomol. Struct. 1999;28:319. doi: 10.1146/annurev.biophys.28.1.319. [DOI] [PubMed] [Google Scholar]
- 4.Fraser CM, Gocayne JD, White O, Adams MD, Clayton RA, Fleischmann RD, Bult CJ, Kerlavage AR, Sutton G, Kelley JM, et al. Science. 1995;270:397. doi: 10.1126/science.270.5235.397. [DOI] [PubMed] [Google Scholar]
- 5.Opella SJ. Nature Struct. Biol. 1997;4(Suppl.):845. [PubMed] [Google Scholar]
- 6.Markley JL, Putter I, Jardetzky O. Science. 1968;161:1249. doi: 10.1126/science.161.3847.1249. [DOI] [PubMed] [Google Scholar]
- 7.Saunders M, Wishnia A, Kirkwood JG. J. Am. Chem. Soc. 1957;79:3289. [Google Scholar]
- 8.Cavanagh J, Fairbrother WJ, Palmer AG, Skelton NJ. Protein NMR Spectroscopy Principles and Practice. Academic Press; New York: 1996. [Google Scholar]
- 9.Wuthrich K. NMR of Proteins and Nucleic Acids. Wiley; New York: 1986. [Google Scholar]
- 10.Ferentz AE, Wagner G. Q. Rev. Biophys. 2000;33:29. doi: 10.1017/s0033583500003589. [DOI] [PubMed] [Google Scholar]
- 11.Clore GM, Gronenborn AM. Crit. Rev. Biochem. Mol. Biol. 1989;24:479. doi: 10.3109/10409238909086962. [DOI] [PubMed] [Google Scholar]
- 12.Waugh JS, Huber LM, Haeberlen U. Phys. Rev. Lett. 1968;20:180. [Google Scholar]
- 13.Pines A, Gibby MG, Waugh JS. J. Chem. Phys. 1973;59:569. [Google Scholar]
- 14.Schaefer J, Stejskal EO. J. Am. Chem. Soc. 1975;98:1031. [Google Scholar]
- 15.Pake GE. J. Chem. Phys. 1948;16:327. [Google Scholar]
- 16.Opella SJ, Nevzorov A, Mesleb MF, Marassi FM. Biochem. Cell Biol. 2002;80:597. doi: 10.1139/o02-154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Marassi FM, Ramamoorthy A, Opella SJ. Proc. Natl. Acad. Sci. U.S.A. 1997;94:8551. doi: 10.1073/pnas.94.16.8551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Waugh JS. Proc. Natl. Acad. Sci. U.S.A. 1976;73:1394. doi: 10.1073/pnas.73.5.1394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hester RK, Ackerman JL, Neff BL, Waugh JS. Phys. Rev. Lett. 1976;36:1081. [Google Scholar]
- 20.Salgado J, Grage SL, Kondejewski LH, Hodges RS, McElhaney RN, Ulrich AS. J. Biomol. NMR. 2001;21:191. doi: 10.1023/a:1012946026231. [DOI] [PubMed] [Google Scholar]
- 21.Smith R, Separovic F, Bennett FC, Cornell BA. Biophys. J. 1992;63:469. doi: 10.1016/S0006-3495(92)81623-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Opella SJ, Waugh JS. J. Chem. Phys. 1977;66:4919. [Google Scholar]
- 23.Pausak S, Pines A, Gibby MG, Waugh JS. J. Chem. Phys. 1973;50:591. [Google Scholar]
- 24.Opella SJ, Stewart PL, Valentine KG. Q. Rev. Biophys. 1987;19:7. doi: 10.1017/s0033583500004017. [DOI] [PubMed] [Google Scholar]
- 25.Ramamoorthy A, Wu CH, Opella SJ. J. Magn. Reson. B. 1995;107:88. doi: 10.1006/jmrb.1995.1063. [DOI] [PubMed] [Google Scholar]
- 26.Ramamoorthy A, Gierasch LM, Opella SJ. J. Magn. Reson. B. 1995;109:112. doi: 10.1006/jmrb.1995.1157. [DOI] [PubMed] [Google Scholar]
- 27.Ramamoorthy A, Marassi FM, Zasloff M, Opella SJ. J. Biomol. NMR. 1995;6:329. doi: 10.1007/BF00197814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ramamoorthy A, Gierasch LM, Opella SJ. J. Magn. Reson. B. 1996;111:81. doi: 10.1006/jmrb.1996.0063. [DOI] [PubMed] [Google Scholar]
- 29.Jelinek R, Ramamoorthy A, Opella SJ. J. Am. Chem. Soc. 1995;117:12348. [Google Scholar]
- 30.Ketchem RR, Hu W, Cross TA. Science. 1993;261:1457. doi: 10.1126/science.7690158. [DOI] [PubMed] [Google Scholar]
- 31.Zeri AC, Mesleh MF, Nevzorov AA, Opella SJ. Proc. Natl. Acad. Sci. U.S.A. 2003;100:6458. doi: 10.1073/pnas.1132059100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Marassi FM, Opella SJ. Protein Sci. 2003;12:403. doi: 10.1110/ps.0211503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Park SH, Mrse AA, Nevzorov AA, Mesleh MF, Oblatt-Montal M, Montal M, Opella SJ. J. Mol. Biol. 2003;333:409. doi: 10.1016/j.jmb.2003.08.048. [DOI] [PubMed] [Google Scholar]
- 34.Opella SJ, Marassi FM, Gesell JJ, Valente AP, Kim Y, Oblatt-Montal M, Montal M. Nature Struct. Biol. 1999;6:374. doi: 10.1038/7610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tian C, Gao PF, Pinto LH, Lamb RA, Cross TA. Protein Sci. 2003;12:2597. doi: 10.1110/ps.03168503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cross TA, DiVerdi JA, Opella SJ. J. Am. Chem. Soc. 1982;104:1759. [Google Scholar]
- 37.Wu CH, Ramamoorthy A, Opella SJ. J. Magn. Reson. 1994;A109:270. [Google Scholar]
- 38.Nevzorov AA, Opella SJ. J. Magn. Reson. 2003;164:182. doi: 10.1016/s1090-7807(03)00240-4. [DOI] [PubMed] [Google Scholar]
- 39.Bax A, Kontaxis G, Tjandra N. Methods Enzymol. 2001;339:127. doi: 10.1016/s0076-6879(01)39313-8. [DOI] [PubMed] [Google Scholar]
- 40.Prestegard JH, Kishore AI. Curr. Opin. Chem. Biol. 2001;5:584. doi: 10.1016/s1367-5931(00)00247-7. [DOI] [PubMed] [Google Scholar]
- 41.Veglia G, Opella SJ. J. Am. Chem. Soc. 2000;122:11733. [Google Scholar]
- 42.Ma C, Opella SJ. J. Magn. Reson. 2000;146:381. doi: 10.1006/jmre.2000.2172. [DOI] [PubMed] [Google Scholar]
- 43.Chou JJ, Kaufman JD, Stahl SJ, Wingfield PT, Bax A. J. Am. Chem. Soc. 2002;124:2450. doi: 10.1021/ja017875d. [DOI] [PubMed] [Google Scholar]
- 44.Lee S, Mesleh MF, Opella SJ. J. Biomol. NMR. 2003;26:327. doi: 10.1023/a:1024047805043. [DOI] [PubMed] [Google Scholar]
- 45.McDonnell PA, Opella SJ. J. Magn. Reson. 1993;102:120. [Google Scholar]
- 46.Tycko R, Blanco FJ, Ishii Y. J. Am. Chem. Soc. 2000;122:9340. [Google Scholar]
- 47.Ishii Y, Markus MA, Tycko R. J. Biomol. NMR. 2001;21:141. doi: 10.1023/a:1012417721455. [DOI] [PubMed] [Google Scholar]
- 48.Sass HJ, Musco G, Stahl SJ, Wingfield PT, Grzesiek S. J. Biomol. NMR. 2000;18:303. doi: 10.1023/a:1026703605147. [DOI] [PubMed] [Google Scholar]
- 49.Meier S, Haussinger D, Grzesiek S. J. BIomol. NMR. 2002;24:351. doi: 10.1023/a:1021609207024. [DOI] [PubMed] [Google Scholar]
- 50.Sanders CR, Prosser RS. Structure. 1998;6:1227. doi: 10.1016/s0969-2126(98)00123-3. [DOI] [PubMed] [Google Scholar]
- 51.Prosser RS, Hunt SA, DiNatale JA, Vold RR. J. Am. Chem. Soc. 1996;118:269. [Google Scholar]
- 52.Sanders CR, 2nd, Schwonek JP. Biophys. J. 1993;65:1460. doi: 10.1016/S0006-3495(93)81215-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bak M, Rasmussen JT, Nielsen NC. J. Magn. Reson. 2000;147:296. doi: 10.1006/jmre.2000.2179. [DOI] [PubMed] [Google Scholar]
- 54.Vosegaard T, Nielsen NC. J. Biomol. NMR. 2002;22:225. doi: 10.1023/a:1014987227285. [DOI] [PubMed] [Google Scholar]
- 55.Bak M, Schultz R, Vosegaard T, Nielsen NC. J. Magn. Reson. 2002;154:28. doi: 10.1006/jmre.2001.2454. [DOI] [PubMed] [Google Scholar]
- 56.Kim S, Cross TA. Biophys. J. 2002;83:2084. doi: 10.1016/S0006-3495(02)73969-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tycko R. J. Biomol. NMR. 1996;8:239. doi: 10.1007/BF00410323. [DOI] [PubMed] [Google Scholar]
- 58.Opella SJ, Stewart PL. Methods Enzymol. 1989;176:242. doi: 10.1016/0076-6879(89)76015-8. [DOI] [PubMed] [Google Scholar]
- 59.Marassi FM. Biophys. J. 2001;80:994. doi: 10.1016/S0006-3495(01)76078-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Arkin IT, Brunger AT, Engelman DM. Proteins. 1997;28:481. doi: 10.1002/(sici)1097-0134(199708)28:4<465::aid-prot1>3.0.co;2-9. [DOI] [PubMed] [Google Scholar]
- 61.Kochendoerfer GG, Jones DH, Lee S, Oblatt-Montal M, Opella SJ, Montal M. J. Am. Chem. Soc. 2004;126:2439. doi: 10.1021/ja038985i. [DOI] [PubMed] [Google Scholar]
- 62.Bechinger B, Zasloff M, Opella SJ. Protein Sci. 1993;2:2077. doi: 10.1002/pro.5560021208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Gesell J, Zasloff M, Opella SJ. J. Biomol. NMR. 1997;9:127. doi: 10.1023/a:1018698002314. [DOI] [PubMed] [Google Scholar]
- 64.Hirsh DJ, Hammer J, Maloy WL, Blazyk J, Schaefer J. Biochemistry. 1996;35:12733. doi: 10.1021/bi961468a. [DOI] [PubMed] [Google Scholar]
- 65.Marassi FM, Ma C, Gesell JJ, Opella SJ. J. Magn. Reson. 2000;144:156. doi: 10.1006/jmre.2000.2036. [DOI] [PubMed] [Google Scholar]
- 66.Opella SJ, Ma C, Marassi FM. Methods Enzymol. 2001;339:285. doi: 10.1016/s0076-6879(01)39319-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Tian C, Tobler K, Lamb RA, Pinto LH, Cross TA. Biochemistry. 2002;41:11294. doi: 10.1021/bi025695q. [DOI] [PubMed] [Google Scholar]
- 68.Krueger-Koplin RD, Sorgen PL, Krueger-Koplin ST, Rivera-Torres IO, Cahill SM, Hicks DB, Grinius L, Krulwich TA, Girvin ME. J. Biomol. NMR. 2004;28:43. doi: 10.1023/B:JNMR.0000012875.80898.8f. [DOI] [PubMed] [Google Scholar]
- 69.Schwaiger M, Lebendiker M, Yerushalmi H, Coles M, Groger A, Schwarz C, Schuldiner S, Kessler H. Eur. J. Biochem. 1998;254:610. doi: 10.1046/j.1432-1327.1998.2540610.x. [DOI] [PubMed] [Google Scholar]
- 70.Schubert M, Kolbe M, Kessler B, Oesterhelt D, Schmieder P. ChemBioChem. 2002;3:1019. doi: 10.1002/1439-7633(20021004)3:10<1019::AID-CBIC1019>3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- 71.Oxenoid K, Sonnichsen FD, Sanders CR. Biochemistry. 2002;41:12876. doi: 10.1021/bi020335o. [DOI] [PubMed] [Google Scholar]
- 72.Klammt C, Lohr F, Schafer B, Haase W, Dotsch V, Ruterjans H, Glaubitz C, Bernhard F. Eur. J. Biochem. 2004;271:568. doi: 10.1111/j.1432-1033.2003.03959.x. [DOI] [PubMed] [Google Scholar]
- 73.Brown LR, Wuthrich K. Biochim. Biophys. Acta. 1981;647:95. doi: 10.1016/0005-2736(81)90298-4. [DOI] [PubMed] [Google Scholar]
- 74.Cross TA, Opella SJ. Biochem. Biophys. Res. Commun. 1980;92:478. doi: 10.1016/0006-291x(80)90358-7. [DOI] [PubMed] [Google Scholar]
- 75.Van Den Hooven HW, Doeland CC, Van De Kamp M, Konings RN, Hilbers CW, Van De Ven FJ. Eur. J. Biochem. 1996;235:382. doi: 10.1111/j.1432-1033.1996.00382.x. [DOI] [PubMed] [Google Scholar]
- 76.Zhang YP, Lewis RN, Henry GD, Sykes BD, Hodges RS, McElhaney RN. Biochemistry. 1995;34:2348. doi: 10.1021/bi00007a031. [DOI] [PubMed] [Google Scholar]
- 77.Vinogradova O, Sonnichsen F, Sanders CR., 2nd J. Biomol. NMR. 1998;11:381. doi: 10.1023/a:1008289624496. [DOI] [PubMed] [Google Scholar]
- 78.Klein-Seetharaman J, Reeves PJ, Loewen MC, Getmanova EV, Chung J, Schwalbe H, Wright PE, Khorana HG. Proc. Natl. Acad. Sci. U.S.A. 2002;99:3452. doi: 10.1073/pnas.052713999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Zamoon J, Mascioni A, Thomas DD, Veglia G. Biophys. J. 2003;85:2589. doi: 10.1016/s0006-3495(03)74681-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Ma C, Marassi FM, Jones DH, Straus SK, Bour S, Strebel K, Schubert U, Oblatt-Montal M, Montal M, Opella SJ. Protein Sci. 2002;11:546. doi: 10.1110/ps.37302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Sanders CR, 2nd, Schwonek JP. Biochemistry. 1992;31:8898. doi: 10.1021/bi00152a029. [DOI] [PubMed] [Google Scholar]
- 82.Sanders CR, Hare B, Howard KP, Prestegard JH. Prog. NMR Spectrosc. 1993;26:421. [Google Scholar]
- 83.Vold RR, Prosser RS, Deese AJ. J. Biomol. NMR. 1997;9:329. doi: 10.1023/a:1018643312309. [DOI] [PubMed] [Google Scholar]
- 84.Luchette PA, Vetman TN, Prosser RS, Hancock RE, Nieh MP, Glinka CJ, Krueger S, Katsaras J. Biochim. Biophys. Acta. 2001;1513:83. doi: 10.1016/s0005-2736(01)00358-3. [DOI] [PubMed] [Google Scholar]
- 85.Howard KP, Opella SJ. J. Magn. Reson. B. 1996;112:91. doi: 10.1006/jmrb.1996.0116. [DOI] [PubMed] [Google Scholar]
- 86.Mesleh MF, Lee S, Veglia G, Thiriot DS, Marassi FM, Opella SJ. J. Am. Chem. Soc. 2003;125:8928. doi: 10.1021/ja034211q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Pervushin K, Riek R, Wider G, Wuthrich K. Proc. Natl. Acad. Sci. U.S.A. 1997;94:12366. doi: 10.1073/pnas.94.23.12366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Arora A, Abildgaard F, Bushweller JH, Tamm LK. Nature Struct. Biol. 2001;8:334. doi: 10.1038/86214. [DOI] [PubMed] [Google Scholar]
- 89.Hwang PM, Choy WY, Lo EI, Chen L, Forman-Kay JD, Raetz CR, Prive GG, Bishop RE, Kay LE. Proc. Natl. Acad. Sci. U.S.A. 2002;99:13560. doi: 10.1073/pnas.212344499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Fernandez C, Adeishvili K, Wutrhich K. Proc. Natl. Acad. Sci. U.S.A. 2001;98:2358. doi: 10.1073/pnas.051629298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Shon K, Opella SJ. J. Magn. Reson. 1989;82:193. [Google Scholar]
- 92.Ikura M, Kay LE, Bax A. Biochemistry. 1990;29:4659. doi: 10.1021/bi00471a022. [DOI] [PubMed] [Google Scholar]
- 93.MacKenzie KR, Prestegard JH, Engelman DM. Science. 1997;276:131. doi: 10.1126/science.276.5309.131. [DOI] [PubMed] [Google Scholar]
- 94.Almeida FC, Opella SJ. J. Mol. Biol. 1997;270:481. doi: 10.1006/jmbi.1997.1114. [DOI] [PubMed] [Google Scholar]
- 95.Mesleh MF, Opella SJ. J. Magn. Reson. 2003;163:288. doi: 10.1016/s1090-7807(03)00119-8. [DOI] [PubMed] [Google Scholar]
- 96.Henry GD, Sykes BD. Biochemistry. 1990;29:6303. doi: 10.1021/bi00478a027. [DOI] [PubMed] [Google Scholar]
- 97.Czerski L, Vinogradova O, Sanders CR. J. Magn. Reson. 2000;142:111. doi: 10.1006/jmre.1999.1920. [DOI] [PubMed] [Google Scholar]
- 98.Veglia G, Zeri AC, Ma C, Opella SJ. Biophys. J. 2002;82:2176. doi: 10.1016/s0006-3495(02)75564-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Bogusky MJ, Schiksnis RA, Leo GC, Opella SJ. J. Magn. Reson. 1987;72:186. [Google Scholar]
- 100.Farrow NA, Muhandiram R, Singer AU, Pascal SM, Kay CM, Gish G, Shoelson SE, Pawson T, Forman-Kay JD, Kay LE. Biochemistry. 1994;33:5984. doi: 10.1021/bi00185a040. [DOI] [PubMed] [Google Scholar]
- 101.Lipari G, Szabo A. J. Am. Chem. Soc. 1982;104:4546. [Google Scholar]
- 102.Clore GM, Driscoll PC, Wingfield PT, Gronenborn AM. Biochemistry. 1990;29:7387. doi: 10.1021/bi00484a006. [DOI] [PubMed] [Google Scholar]
- 103.Kay LE. Biochem. Cell Biol. 1998;76:145. doi: 10.1139/bcb-76-2-3-145. [DOI] [PubMed] [Google Scholar]
- 104.Bruschweiler R. Curr. Opin. Struct. Biol. 2003;13:175. doi: 10.1016/s0959-440x(03)00036-8. [DOI] [PubMed] [Google Scholar]
- 105.Vugmeyster L, Raleigh DP, Palmer AG, 3rd, Vugmeister BE. J. Am. Chem. Soc. 2003;125:8400. doi: 10.1021/ja029480f. [DOI] [PubMed] [Google Scholar]
- 106.Prestegard JH, al-Hashimi HM, Tolman JR. Q. Rev. Biophys. 2000;33:371. doi: 10.1017/s0033583500003656. [DOI] [PubMed] [Google Scholar]
- 107.Mesleh MF, Veglia G, DeSilva TM, Marassi FM, Opella SJ. J. Am. Chem. Soc. 2002;124:4206. doi: 10.1021/ja0178665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Lipsitz RS, Tjandra N. J. Magn. Reson. 2003;164:171. doi: 10.1016/s1090-7807(03)00176-9. [DOI] [PubMed] [Google Scholar]
- 109.Kovacs FA, Denny JK, Song Z, Quine JR, Cross TA. J. Mol. Biol. 2000;295:117. doi: 10.1006/jmbi.1999.3322. [DOI] [PubMed] [Google Scholar]
- 110.Battiste JL, Wagner G. Biochemistry. 2000;39:5355. doi: 10.1021/bi000060h. [DOI] [PubMed] [Google Scholar]
- 111.Girvin ME, Fillingame RH. Biochemistry. 1995;34:1635. doi: 10.1021/bi00005a020. [DOI] [PubMed] [Google Scholar]
- 112.Papavoine CH, Konings RN, Hilbers CW, van de Ven FJ. Biochemistry. 1994;33:12990. doi: 10.1021/bi00248a007. [DOI] [PubMed] [Google Scholar]
- 113.Kutateladze TG, Capelluto DG, Ferguson CG, Cheever ML, Kutateladze AG, Prestwich GD, Overduin M. J. Biol. Chem. 2004;279:3050. doi: 10.1074/jbc.M309007200. [DOI] [PubMed] [Google Scholar]
- 114.Schwieters CD, Kuszewski JJ, Tjandra N, Clore MG. J. Magn. Reson. 2003;160:65. doi: 10.1016/s1090-7807(02)00014-9. [DOI] [PubMed] [Google Scholar]
- 115.Smith R, Separovic F, Milne TJ, Whittaker A, Bennett FM, Cornell BA, Makriyannis A. J. Mol. Biol. 1994;241:456. doi: 10.1006/jmbi.1994.1520. [DOI] [PubMed] [Google Scholar]
- 116.de Planque MR, Bonev BB, Demmers JA, Greathouse DV, Koeppe RE, 2nd, Separovic F, Watts A, Killian JA. Biochemistry. 2003;42:5341. doi: 10.1021/bi027000r. [DOI] [PubMed] [Google Scholar]
- 117.Griffin RG. Nature Struct. Biol. 1998;5(Suppl):508. doi: 10.1038/749. [DOI] [PubMed] [Google Scholar]
- 118.Auger M. Curr. Issues Mol. Biol. 2000;2:119. [PubMed] [Google Scholar]
- 119.de Groot HJ. Curr. Opin. Struct. Biol. 2000;10:593. doi: 10.1016/s0959-440x(00)00136-6. [DOI] [PubMed] [Google Scholar]
- 120.Lam YH, Wassall SR, Morton CJ, Smith R, Separovic F. Biophys. J. 2001;81:2752. doi: 10.1016/S0006-3495(01)75918-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Huster D, Yao X, Hong M. J. Am. Chem. Soc. 2002;124:874. doi: 10.1021/ja017001r. [DOI] [PubMed] [Google Scholar]
- 122.Spooner PJ, Sharples JM, Verhoeven MA, Lugtenburg J, Glaubitz C, Watts A. Biochemistry. 2002;41:7549. doi: 10.1021/bi020007o. [DOI] [PubMed] [Google Scholar]
- 123.Saito H, Yamaguchi S, Okuda H, Shiraishi A, Tuzi S. Solid State Nucl. Magn. Reson. 2004;25:5. doi: 10.1016/j.ssnmr.2003.06.001. [DOI] [PubMed] [Google Scholar]
- 124.Fu R, Cross TA. Annu. Rev. Biophys. Biomol. Struct. 1999;28:235. doi: 10.1146/annurev.biophys.28.1.235. [DOI] [PubMed] [Google Scholar]
- 125.Wang J, Kim S, Kovacs F, Cross TA. Protein Sci. 2001;10:2241. doi: 10.1110/ps.17901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Thompson LK. Curr. Opin. Struct. Biol. 2002;12:661. doi: 10.1016/s0959-440x(02)00374-3. [DOI] [PubMed] [Google Scholar]
- 127.Wang J, Balazs YS, Thompson LK. Biochemistry. 1997;36:1699. doi: 10.1021/bi962578k. [DOI] [PubMed] [Google Scholar]
- 128.Thompson LK, McDermott AE, Raap J, van der Wielen CM, Lugtenburg J, Herzfeld J, Griffin RG. Biochemistry. 1992;31:7931. doi: 10.1021/bi00149a026. [DOI] [PubMed] [Google Scholar]
- 129.Yang J, Weliky DP. Biochemistry. 2003;42:11879. doi: 10.1021/bi0348157. [DOI] [PubMed] [Google Scholar]
- 130.Eilers M, Ying W, Reeves PJ, Khorana HG, Smith SO. Methods Enzymol. 2002;343:212. doi: 10.1016/s0076-6879(02)43137-0. [DOI] [PubMed] [Google Scholar]
- 131.Smith SO, Aschheim K, Groesbeek M. Q. Rev. Biophys. 1996;29:395. doi: 10.1017/s0033583500005898. [DOI] [PubMed] [Google Scholar]
- 132.Mason AJ, Grage SL, Straus SK, Glaubitz C, Watts A. Biophys. J. 2004;86:1610. doi: 10.1016/S0006-3495(04)74228-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Spooner PJ, Sharples JM, Goodall SC, Seedorf H, Verhoeven MA, Lugtenburg J, Bovee-Geurts PH, DeGrip WJ, Watts A. Biochemistry. 2003;42:13371. doi: 10.1021/bi0354029. [DOI] [PubMed] [Google Scholar]
- 134.De Planque MR, Rijkers DT, Liskamp RM, Separovic F. Magn. Reson. Chem. 2004;42:148. doi: 10.1002/mrc.1326. [DOI] [PubMed] [Google Scholar]
- 135.van der Wel PC, Strandberg E, Killian JA, Koeppe RE., 2nd Biophys. J. 2002;83:1479. doi: 10.1016/S0006-3495(02)73918-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Killian JA, Taylor MJ, Koeppe RE., 2nd Biochemistry. 1992;31:11283. doi: 10.1021/bi00161a004. [DOI] [PubMed] [Google Scholar]
- 137.Ketchem R, Roux B, Cross T. Structure. 1997;5:1655. doi: 10.1016/s0969-2126(97)00312-2. [DOI] [PubMed] [Google Scholar]
- 138.Sanders CR., 2nd Chem. Phys. Lipids. 1994;72:41. doi: 10.1016/0009-3084(94)90016-7. [DOI] [PubMed] [Google Scholar]
- 139.Cross TA, Opella SJ. J. Am. Chem. Soc. 1983;105:306. [Google Scholar]
- 140.Marassi FM, Opella SJ. J. Magn. Reson. 2000;144:150. doi: 10.1006/jmre.2000.2035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Wang J, Denny J, Tian C, Kim S, Mo Y, Kovacs F, Song Z, Nishimura K, Gan Z, Fu R, Quine JR, Cross TA. J. Magn. Reson. 2000;144:162. doi: 10.1006/jmre.2000.2037. [DOI] [PubMed] [Google Scholar]
- 142.Wu CH, Ramamoorthy A, Gierasch LM, Opella SJ. J. Am. Chem. Soc. 1995;117:6148. [Google Scholar]
- 143.Marassi FM, Opella SJ. J. Biomol. NMR. 2002;23:239. doi: 10.1023/a:1019887612018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Cross TA, Frey MH, Opella SJ. J. Am. Chem. Soc. 1983;105:7471. [Google Scholar]
- 145.Cross TA, Opella SJ. J. Mol. Biol. 1985;182:367. doi: 10.1016/0022-2836(85)90197-4. [DOI] [PubMed] [Google Scholar]
- 146.Tan WM, Gu Z, Zeri AC, Opella SJ. J. Biomol. NMR. 1999;13:337. doi: 10.1023/a:1008379105545. [DOI] [PubMed] [Google Scholar]
- 147.Franzin CM, Choi J, Zhai D, Reed JC, Marassi FM. Magn. Reson. Chem. 2004;42:172. doi: 10.1002/mrc.1322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Nevzorov AA, Mesleh MF, Opella SJ. Magn. Reson. Chem. 2004;42:162. doi: 10.1002/mrc.1320. [DOI] [PubMed] [Google Scholar]
- 149.Nevzorov AA, Opella SJ. J. Magn. Reson. 2003;160:33. doi: 10.1016/s1090-7807(02)00138-6. [DOI] [PubMed] [Google Scholar]
- 150.Bertram R, Asbury T, Fabiola F, Quine JR, Cross TA, Chapman MS. J. Magn. Reson. 2003;163:300. doi: 10.1016/s1090-7807(03)00147-2. [DOI] [PubMed] [Google Scholar]
- 151.Kim S, Quine JR, Cross TA. J. Am. Chem. Soc. 2001;123:7292. doi: 10.1021/ja003380x. [DOI] [PubMed] [Google Scholar]
- 152.Bertram R, Quine JR, Chapman MS, Cross TA. J. Magn. Reson. 2000;147:9. doi: 10.1006/jmre.2000.2193. [DOI] [PubMed] [Google Scholar]
- 153.Stewart PL, Tycko R, Opella SJ. J. Chem. Soc., Faraday Trans. 1. 1988;84:3803. [Google Scholar]
- 154.Drechsler A, Separovic F. IUBMB Life. 2003;55:515. doi: 10.1080/15216540310001622740. [DOI] [PubMed] [Google Scholar]
- 155.Gu ZT, Opella SJ. J. Magn. Reson. 1999;140:340. doi: 10.1006/jmre.1999.1825. [DOI] [PubMed] [Google Scholar]
- 156.Gu Z, Opella SJ. J. Magn. Reson. 1999;138:193. doi: 10.1006/jmre.1999.1709. [DOI] [PubMed] [Google Scholar]
- 157.Ishii Y, Tycko R. J. Am. Chem. Soc. 2000;122:1443. [Google Scholar]
- 158.Marassi FM, Gesell JJ, Valente AP, Kim Y, Oblatt-Montal M, Montal M, Opella SJ. J. Biomol. NMR. 1999;14:141. doi: 10.1023/a:1008391823293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Reif B, Jaroniec CP, Rienstra CM, Hohwy M, Griffin RG. J. Magn. Reson. 2001;151:320. doi: 10.1006/jmre.2001.2354. [DOI] [PubMed] [Google Scholar]
- 160.Reif B, Griffin RG. J. Magn. Reson. 2003;160:78. doi: 10.1016/s1090-7807(02)00035-6. [DOI] [PubMed] [Google Scholar]
- 161.McDowell LM, Schaefer J. Curr. Opin. Struct. Biol. 1996;6:624. doi: 10.1016/s0959-440x(96)80028-5. [DOI] [PubMed] [Google Scholar]
- 162.Hing AW, Schaefer J. Biochemistry. 1993;32:7593. doi: 10.1021/bi00080a035. [DOI] [PubMed] [Google Scholar]
- 163.Smith SO, Song D, Shekar S, Groesbeek M, Ziliox M, Aimoto S. Biochemistry. 2001;40:6553. doi: 10.1021/bi010357v. [DOI] [PubMed] [Google Scholar]
- 164.Nishimura K, Kim S, Zhang L, Cross TA. Biochemistry. 2002;41:13170. doi: 10.1021/bi0262799. [DOI] [PubMed] [Google Scholar]
- 165.Frey MH, Opella SJ. J. Am. Chem. Soc. 1984;106:4942. [Google Scholar]
- 166.Castellani F, van Rossum B, Diehl A, Schubert M, Rehbein K, Oschkinat H. Nature. 2002;420:98. doi: 10.1038/nature01070. [DOI] [PubMed] [Google Scholar]
- 167.Reif B, Van Rossum BJ, Castellani F, Rehbein K, Diehl A, Oschkinat H. J. Am. Chem. Soc. 2003;125:1488. doi: 10.1021/ja0283697. [DOI] [PubMed] [Google Scholar]
- 168.Brinkmann A, Schmedt auf der Gunne J, Levitt MH. J. Magn. Reson. 2002;156:79. doi: 10.1006/jmre.2002.2525. [DOI] [PubMed] [Google Scholar]
- 169.Karlsson T, Popham JM, Long JR, Oyler N, Drobny GP. J. Am. Chem. Soc. 2003;125:7394. doi: 10.1021/ja0294360. [DOI] [PubMed] [Google Scholar]
- 170.Grage SL, Wang J, Cross TA, Ulrich AS. Biophys. J. 2002;83:3336. doi: 10.1016/S0006-3495(02)75334-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Afonin S, Durr UH, Glaser RW, Ulrich AS. Magn. Reson. Chem. 2004;42:195. doi: 10.1002/mrc.1340. [DOI] [PubMed] [Google Scholar]
- 172.McDowell LM, Lee M, McKay RA, Anderson KS, Schaefer J. Biochemistry. 1996;35:3328. doi: 10.1021/bi9518297. [DOI] [PubMed] [Google Scholar]
- 173.Cross TA, Opella SJ. J. Mol. Biol. 1982;159:543. doi: 10.1016/0022-2836(82)90301-1. [DOI] [PubMed] [Google Scholar]
- 174.Kinsey RA, Kintanar A, Oldfield E. J. Biol. Chem. 1981;256:9028. [PubMed] [Google Scholar]
- 175.Kinsey RA, Kintanar A, Tsai MD, Smith RL, Janes N, Oldfield E. J. Biol. Chem. 1981;256:4146. [PubMed] [Google Scholar]
- 176.Torchia DA. Annu. Rev. Biophys. Bioeng. 1984;13:125. doi: 10.1146/annurev.bb.13.060184.001013. [DOI] [PubMed] [Google Scholar]
- 177.Opella SJ. Methods Enzymol. 1986;131:327. doi: 10.1016/0076-6879(86)31048-6. [DOI] [PubMed] [Google Scholar]
- 178.Frey MH, DiVerdi JA, Opella SJ. J. Am. Chem. Soc. 1985;107:7311. [Google Scholar]
- 179.McDermott A, Polenova T, Bockmann A, Zilm KW, Paulson EK, Martin RW, Montelione GT, Paulsen EK. J. Biomol. NMR. 2000;16:209. doi: 10.1023/a:1008391625633. [DOI] [PubMed] [Google Scholar]
- 180.Rienstra CM, Tucker-Kellogg L, Jaroniec CP, Hohwy M, Reif B, McMahon MT, Tidor B, Lozano-Perez T, Griffin RG. Proc. Natl. Acad. Sci. U.S.A. 2002;99:10260. doi: 10.1073/pnas.152346599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Petkova AT, Ishii Y, Balbach JJ, Antzutkin ON, Leapman RD, Delaglio F, Tycko R. Proc. Natl. Acad. Sci. U.S.A. 2002;99:16742. doi: 10.1073/pnas.262663499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Tycko R, Ishii Y. J. Am. Chem. Soc. 2003;125:6606. doi: 10.1021/ja0342042. [DOI] [PubMed] [Google Scholar]
- 183.Hong M, Jakes K. J. Biomol. NMR. 1999;14:71. doi: 10.1023/a:1008334930603. [DOI] [PubMed] [Google Scholar]
- 184.Petkova AT, Baldus M, Belenky M, Hong M, Griffin RG, Herzfeld J. J. Magn. Reson. 2003;160:1. doi: 10.1016/s1090-7807(02)00137-4. [DOI] [PubMed] [Google Scholar]
- 185.Ernst M, Detken A, Bockmann A, Meier BH. J. Am. Chem. Soc. 2003;125:15807. doi: 10.1021/ja0369966. [DOI] [PubMed] [Google Scholar]
- 186.van Beek JD, Beaulieu L, Schafer H, Demura M, Asakura T, Meier BH. Nature. 2000;405:1077. doi: 10.1038/35016625. [DOI] [PubMed] [Google Scholar]
- 187.Egorova-Zachernyuk TA, Hollander J, Fraser N, Gast P, Hoff AJ, Cogdell R, de Groot HJ, Baldus M. J. Biomol. NMR. 2001;19:243. doi: 10.1023/a:1011235417465. [DOI] [PubMed] [Google Scholar]
- 188.Pauli J, Baldus M, van Rossum B, de Groot H, Oschkinat H. ChemBioChem. 2001;2:272. doi: 10.1002/1439-7633(20010401)2:4<272::AID-CBIC272>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 189.van Rossum BJ, Castellani F, Pauli J, Rehbein K, Hollander J, de Groot HJ, Oschkinat H. J. Biomol. NMR. 2003;25:217. doi: 10.1023/a:1022819921584. [DOI] [PubMed] [Google Scholar]
- 190.Reif B, Hohwy M, Jaroniec CP, Rienstra CM, Griffin RG. J. Magn. Reson. 2000;145:132. doi: 10.1006/jmre.2000.2067. [DOI] [PubMed] [Google Scholar]
- 191.Rienstra CM, Hohwy M, Mueller LJ, Jaroniec CP, Reif B, Griffin RG. J. Am. Chem. Soc. 2002;124:11908. doi: 10.1021/ja020802p. [DOI] [PubMed] [Google Scholar]
- 192.Jaroniec CP, MacPhee CE, Bajaj VS, McMahon MT, Dobson CM, Griffin RG. Proc. Natl. Acad. Sci. U.S.A. 2004;101:711. doi: 10.1073/pnas.0304849101. [DOI] [PMC free article] [PubMed] [Google Scholar]