Background: Recently, new “targeted” fluorescent probes that react selectively with reactive oxygen and nitrogen species to yield specific products have been discovered.
Results: High-throughput fluorescence and HPLC-based methodology for global profiling of ROS/RNS is described.
Conclusion: This methodology enables real-time monitoring of multiple oxidants in cellular systems.
Significance: The global profiling approach using different ROS/RNS-specific fluorescent probes will help establish the identity of oxidants in redox regulation and signaling.
Keywords: Fluorescence, Nitric Oxide, Reactive Nitrogen Species, Reactive Oxygen Species (ROS), Superoxide Ion, Hydrogen Peroxide, Peroxynitrite
Abstract
Herein we describe a high-throughput fluorescence and HPLC-based methodology for global profiling of reactive oxygen and nitrogen species (ROS/RNS) in biological systems. The combined use of HPLC and fluorescence detection is key to successful implementation and validation of this methodology. Included here are methods to specifically detect and quantitate the products formed from interaction between the ROS/RNS species and the fluorogenic probes, as follows: superoxide using hydroethidine, peroxynitrite using boronate-based probes, nitric oxide-derived nitrosating species with 4,5-diaminofluorescein, and hydrogen peroxide and other oxidants using 10-acetyl-3,7-dihydroxyphenoxazine (Amplex® Red) with and without horseradish peroxidase, respectively. In this study, we demonstrate real-time monitoring of ROS/RNS in activated macrophages using high-throughput fluorescence and HPLC methods. This global profiling approach, simultaneous detection of multiple ROS/RNS products of fluorescent probes, developed in this study will be useful in unraveling the complex role of ROS/RNS in redox regulation, cell signaling, and cellular oxidative processes and in high-throughput screening of anti-inflammatory antioxidants.
Introduction
Recent advances in oxy-radical research reveal multiple roles for reactive oxygen species (ROS)3 (superoxide radical anion, O2˙̄; hydrogen peroxide, H2O2; hypohalous acids such as HOCl and HOBr; lipid hydroperoxides, LOOH; and carbonyl products derived from them) and reactive nitrogen species (RNS) (peroxynitrite, ONOO−; nitrogen dioxide, •NO2; nitrated lipids, LNO2). These species may be involved in redox regulation of cellular proteins, signaling, proliferation and differentiation, senescence, and cell death (1–4). However, in most cases, the identity of the species responsible for the described biologic effects remains to be determined. This is partly due to a lack of suitable methodologies and chemical probes that allow unequivocal detection and distinction between different ROS and RNS and also to the complex intracellular chemistry of these species. Although a considerable amount of work has been done to elucidate reaction mechanisms of ROS/RNS with biological agents, antioxidants, and detection probes (5–7), only recently has real progress been made in uncovering reaction mechanisms (e.g. kinetics, stoichiometry, product analysis) of ROS and RNS with fluorescent probes (8–10). The purpose of this study is to determine the specific products of oxidation of fluorescent probes by ROS and RNS and reveal the strategic use of these probes for real-time detection of these species in a more complex biological setting.
In recent years, new “targeted” chemical probes that are more specific and selective in their reaction with O2˙̄, H2O2, and ONOO− were discovered (10–12). These and the other breakthroughs described below have enabled us to perform global profiling of ROS/RNS in biological systems (8–10). These breakthroughs are highlighted as follows. (i) Hydroethidine (HE) or dihydroethidium and its mitochondria-targeted analog conjugated to a triphenylphosphonium moiety (MitoSOXTM Red) react rapidly with superoxide (k ∼106 m−1 s−1), forming a specific hydroxylated marker product, 2-hydroxyethidium (2-OH-E+) or 2-hydroxymitoethidium (2-OH-Mito-E+). Other oxidants (ONOO−-derived radicals, hydroxyl radical, perferryl iron) react with HE and MitoSOXTM to form the corresponding oxidation/dimeric products (E+, Mito-E+, and dimeric products) but do not yield the same O2˙̄-mediated hydroxylated products (13). Understanding of the oxidation mechanisms of HE and MitoSOXTM has made it possible to use these probes with discretion for detecting intracellular O2˙̄ and other oxidants formed from H2O2 and iron or heme or ONOO−. (ii) Boronate-containing fluorogenic probes react directly and stoichiometrically with H2O2, albeit slowly (k ∼1 m−1 s−1), to form phenolic products that can be monitored in cells. (iii) Furthermore, several aromatic boronates react directly, rapidly, and stoichiometrically with ONOO− (k ∼106 m−1 s−1), forming the corresponding phenols as major products. Other reactive nitrogen species (•NO/O2 and •NO2 formed from myeloperoxidase/NO2−/H2O2) do not react with boronates in a similar manner, thus enabling selective detection of ONOO− (14). Importantly, the chemistry and mechanism of reaction of H2O2 and ONOO− with most boronates are very similar (14). This has prompted the development of several custom-synthesized and targeted boronic acid/ester-based fluorophores that can be used for specific and sensitive detection of ONOO− in chemical and biological systems (12).
The combined use of HPLC and fluorescence detection techniques now enables real-time monitoring and detection with unequivocal characterization of oxidants in biological systems. In this study, we have used a multiwell plate reader as a high-throughput approach to monitor the formation of O2˙̄, H2O2, •NO-derived nitrosating species, and ONOO− in both cell-free and cellular systems. Both newly synthesized and commercially available fluorescent probes were used in the present study for detecting O2˙̄, H2O2, •NO-derived nitrosating species, and ONOO−.
EXPERIMENTAL PROCEDURES
Reagents
HE was purchased from Invitrogen. The standards 2-OH-E+, E+, and E+-E+ were prepared as described previously (15, 16). Coumarin-7-boronic acid (CBA) was synthesized as described earlier (9). Fluorescein dimethylamide boronate (FlAmBE) synthesis is described in supplemental Methods and Figs. S5 and S6. Amplex® Red (10-acetyl-3,7-dihydroxyphenoxazine) and DPTA-NONOate were purchased from Cayman Chemical. Diaminofluorescein (DAF-2), diaminofluorescein–2 diacetate, and DAF-2T and interferon (IFN)-γ were from EMD Calbiochem. Hypoxanthine (HX), xanthine oxidase (XO), superoxide dismutase, catalase, l-NAME, lipopolysaccharide (LPS), and phorbol 12-myristate 13-acetate (PMA) were purchased from Sigma-Aldrich. Peroxynitrite was synthesized as described previously (7–9).
Determination of O2˙̄ and •NO Fluxes
•NO and O2˙̄ fluxes were determined as described previously (9). Briefly, •NO fluxes were determined from the rate of decomposition of DPTA-NONOate measured by following the decrease of its characteristic absorbance at 248 nm (ϵ = 8.1 × 103 m−1 cm−1). The rate of decay of DPTA-NONOate was multiplied by a factor of two to obtain the rate of •NO release, assuming that two molecules of •NO are released during the decomposition of one molecule of DPTA-NONOate. The flux of O2˙̄ generated by XO-catalyzed oxidation of hypoxanthine was determined by monitoring the cytochrome c(Fe3+) reduction and the increase in absorbance at 550 nm (using a difference in the values of the extinction coefficients between reduced and oxidized cytochrome of 2.1 × 104 m−1 cm−1).
Cell Culture
RAW 264.7 cells were grown in DMEM (Invitrogen) medium supplemented with 10% heat-inactivated FBS, 2 mm l-glutamine, 100 units/ml penicillin, and 100 μg/ml streptomycin. Before the experiment, cells were incubated overnight (16–20 h) with LPS (0.5 μg/ml) and IFN-γ (50 units/ml). For monitoring ROS/RNS, the cells were washed and co-treated with the probes and PMA (200 ng/ml) in DPBS supplemented with pyruvic acid and glucose (DPBS-GP). For HPLC analyses, the dishes with cells were incubated for 1 h at 37 °C in a CO2-free incubator. After incubation, an aliquot of the medium was collected and immediately frozen in liquid nitrogen, while the cells were washed twice with ice-cold PBS, scraped in 1 ml of PBS, and centrifuged (1 min, 1000 × g). The supernatant was discarded, and the cell pellet was immediately frozen by immersion of the tube in liquid nitrogen. For measurements using the fluorescence plate reader, the plate with cells was placed immediately after the addition of DPBS-GP containing the probe with or without PMA in the plate reader prewarmed to 37 °C.
Fluorescence Plate Reader
Total fluorescence intensities were acquired using a plate reader (Beckman Coulter DTX-880) equipped with the appropriate excitation and emission filters. The instrument was kept at 37 °C during the measurements, and fluorescence intensity read from the bottom of each well was integrated over 0.5 s every 90 s. The UV-visible absorption and fluorescence parameters of the fluorogenic probes used in this study and their products of interaction with ROS/RNS are listed in Table 1.
TABLE 1.
Spectroscopic properties of ROS/RNS probes and their fluorescent products at pH 7.4
The error of the determined values of extinction coefficients lies within 15%; ND, not determined.
| Probe/product | UV-visible absorption |
Fluorescence |
||
|---|---|---|---|---|
| λmax | ϵ | Excitation λmax | Emission λmax | |
| nm | m−1cm−1 | nm | nm | |
| HE | 265 | 1.8 × 104 | 265, 348 | 400 |
| 345 | 9.75 × 103 | |||
| 2-OH-E+ | 267 | 4.1 × 104 | 369, 470 | 581 |
| 470 | 1.2 × 104 | |||
| 2-OH-E+ + DNA | 325, 383, 503 | ND | 340, 379, 500 | 565 |
| E+ | 286 | 5.6 × 104 | 487 | 601 |
| 480 | 5.8 × 103 | |||
| E+ + DNA | 520 | ND | 330, 523 | 590 |
| DAF-2DA | 289 | 1.0 × 104 | ||
| 325 | 8.1 × 103 | |||
| DAF-2 | 488 | 8.1 × 104 | ||
| DAF-2T | 491 | 8.0 × 104 | 240, 490 | 513 |
| CBA | 287 | 1.2 × 104 | ||
| COH | 324 | 1.3 × 104 | 332 | 450 |
| Amplex® Red | 281 | 5.5 × 103 | ||
| Resorufin | 290 | 7.6 × 103 | 570 | 583 |
| 571 | 6.3 × 104 | |||
HPLC Analyses
HPLC experiments were performed using an Agilent 1100 system equipped with UV-visible absorption and fluorescence detectors. To obtain UHPLC-like conditions, a fused core C18 column (Phenomenex, Kinetex C18, 100 × 4.6 mm, 2.6 μm) was used. Typically, a gradient elution using an aqueous mobile phase with increasing fraction of acetonitrile (from 10–20% to 100% over 5 min) in the presence of 0.1% TFA was used. For the analysis of DAF-2 nitrosation, isocratic elution using an aqueous mobile phase containing 10 mm phosphate buffer, pH 7.4, and 5% acetonitrile was also used. The analytes were eluted using a flow rate of 1.5 ml/min. Mass spectra were obtained using a 7.0-tesla Fourier transform ion cyclotron resonance mass spectrometer, interfaced with an Agilent 1100 HPLC system.
RESULTS
Oxidation of Hydroethidine in Cell-free and Cellular Systems Generating •NO and O2˙̄ as Measured with HPLC and Fluorescence Analyses
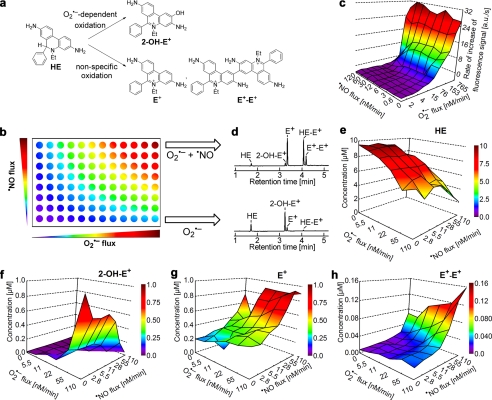
Using an HX and XO system to generate O2˙̄ and H2O2 and a NONOate compound (which releases •NO) (Fig. 1a), we monitored the products formed from oxidation of HE (Fig. 2). The reaction between O2˙̄ and HE generates a fluorescent product, 2-OH-E+. The fluorescence excitation-emission matrix spectrum of DNA-bound 2-OH-E+ is shown in supplemental Fig. S7. Nonspecific oxidation of HE generates another fluorescent product, ethidium (E+), and non-fluorescent dimeric products (e.g. E+-E+) (Fig. 2a). We then monitored the formation of these products under different fluxes of O2˙̄ (derived from hypoxanthine/xanthine oxidase as described in Fig. 1a) and •NO (released slowly from NONOate-based •NO donor) (Fig. 1a). These experiments were performed in a 96-well fluorescence plate containing HE and varying levels of co-generated •NO and O2˙̄ (Fig. 2b), and the rates of increase in the fluorescence intensity were plotted against the fluxes of •NO and O2˙̄ (Fig. 2c). These products were also separated and quantitated using an HPLC technique coupled with absorption and fluorescence detection (Fig. 2, e–h, and supplemental Fig. S1). Upon examining the total profiles of HE oxidation products (Fig. 2, e–h), we conclude the following. (i) 2-OH-E+, the specific marker product of HE reaction with O2˙̄, increased with respect to O2˙̄ flux; •NO inhibited 2-OH-E+ production in a concentration-dependent manner, suggesting the scavenging of O2˙̄ and formation of ONOO−. (ii) The concentration of E+ increased with increasing •NO flux, suggesting the formation of additional oxidants (•NO2 and ONOO− in the presence of superoxide). (iii) Similar to E+, the dimeric oxidation product, E+-E+, increased with increasing •NO flux.
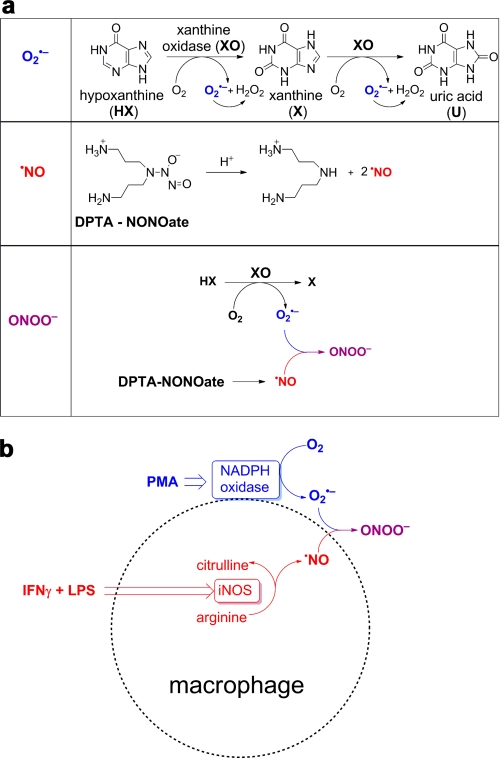
FIGURE 1.
Peroxynitrite generation from co-generated O2˙̄ and •NO. a, schemes showing the formation of O2˙̄ from incubations containing hypoxanthine (100 μm) and xanthine oxidase (0–0.4 milliunits/ml) in a phosphate buffer (pH 7.4; 50 mm) containing DTPA (100 μm), •NO generation from a thermal decomposition of DPTA-NONOate (0–50 μm) and ONOO− formed from co-generated O2˙̄ from HX/XO, and •NO from DPTA-NONOate. b, stimulation of ONOO− from RAW 264.7 macrophages using a mixture of PMA (200 ng/ml, IFN-γ (50 units/ml), and LPS (0.5 μg/ml). •NO is produced from induction and activation of inducible nitric oxide synthase (iNOS), and superoxide is generated from activation of NADPH oxidase by PMA.
FIGURE 2.
Oxidation of HE by O2˙̄, •NO, and co-generated •NO and O2˙̄ and global profiling of products. a, scheme showing HE oxidation by O2˙̄ to the hydroxylated, fluorescent product, 2-OH-E+, and nonspecific oxidation of HE to a red fluorescent product, E+, and non-fluorescent dimeric product, E+-E+, in the presence of •NO and O2˙̄. For the sake of clarity, structures of other dimeric products, HE-HE and HE-E+, are not shown. b, diagrammatic representation of global profiling experiments. Varying fluxes of co-generated •NO and O2˙̄ in a 96-well fluorescence plate containing either HE or CBA in phosphate buffer (50 mm, pH 7.4) containing DTPA (0.1 mm). c, increase in red fluorescence from HE oxidation detected in a 96-well fluorescence plate reader under experimental conditions described in b. a.u., arbitrary units. d, HPLC analyses of incubations with HE from the two corner wells on the right side in b are shown. Oxidation of HE (10 μm) was monitored by HPLC analyses after co-generation of •NO and O2˙̄ in phosphate buffer (50 mm, pH 7.4) containing DTPA (0.1 mm). HE, E+, 2-OH-E+, and E+-E+ were quantified using the appropriate standard. e–h denote global profiling of HE, 2-OH-E+, E+, and E+-E+ levels, respectively, under the experimental conditions described in b.
With increasing O2˙̄ flux alone, the fluorescence intensity due to 2-OH-E+ (independently confirmed by HPLC) increased (Fig. 2, c and f). With increasing •NO flux, the fluorescence intensity was not diminished (Fig. 2c), although the HPLC results indicated a decrease in 2-OH-E+ (Fig. 2f). This is, however, consistent with the HPLC data that showed an increase in E+ that has a similar fluorescence spectrum as 2-OH-E+ under these conditions (cf. Fig. 2g, Table 1, and supplemental Figs. S1 and S7). These results clearly show that the use of the plate reader for high-throughput screening is a feasible approach for detecting O2˙̄- and •NO-derived oxidants. Importantly, the results should be verified by HPLC analysis to determine the mechanism whereby HE is converted to a fluorescent product as E+ and 2-OH-E+ cannot be easily distinguished by using the plate reader alone.
Real-time Monitoring of O2˙̄ Formation from Activated RAW 264.7 Macrophages
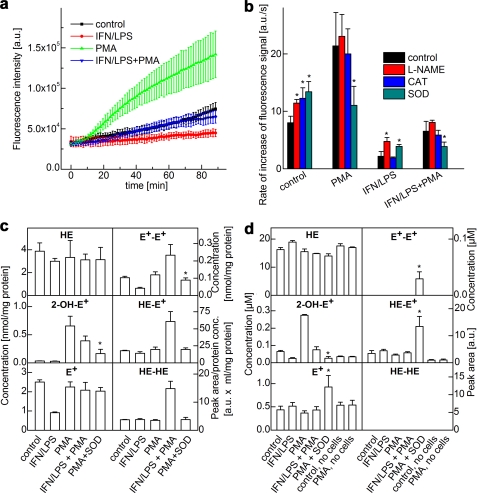
To investigate oxidation products of HE by endogenously generated O2˙̄ and •NO, we used a system consisting of RAW 264.7 macrophages stimulated with a mixture of LPS, IFN-γ, and PMA (Fig. 1b) (17, 18). Oxidation of HE was monitored in a 96-well fluorescence plate reader (Fig. 3, a and b), and products formed in cells (Fig. 3c) and in media (Fig. 3d) were quantitated by HPLC. Macrophages were seeded in 96-well plates, and PMA was added in the presence of HE. This resulted in a steady increase in the fluorescence intensity (Fig. 3a). After examination of the HPLC results obtained from cell lysates and cell culture media (Fig. 3, c and d, and supplemental Fig. S2), the increase in fluorescence intensity was attributed to 2-OH-E+ formation. Consistent with this analysis, the addition of superoxide dismutase, but not catalase, inhibited the rate of increase in fluorescence from macrophages activated by PMA (Fig. 3b). l-NAME did not inhibit the increase in red fluorescence (Fig. 3b). Similar to the cell-free system, co-stimulation of •NO and O2˙̄ production by combined treatment with IFN, LPS, and PMA decreased the yield of 2-OH-E+ with a concomitant increase in the intracellular levels of HE-derived dimeric products. Collectively, these results suggest that although in resting cells the fluorescence intensity reflects the sum of both E+ and 2-OH-E+, the PMA-induced increase resulting from HE-derived fluorescence intensity in incubations containing RAW 264.7 macrophages was mediated predominantly by O2˙̄.
FIGURE 3.
Real-time monitoring of products of HE oxidation by activated macrophages. a, increase in fluorescence was monitored in a fluorescence plate reader (excitation at 485 nm, emission at 595 nm) from incubations containing RAW 264.7 macrophages activated by different stimulators as shown. a.u., arbitrary units. b, rate of increase in the fluorescence signal intensity from RAW 264.7 macrophages activated by different stimulators in the presence of l-NAME and ROS-detoxifying enzymes. CAT, catalase; SOD, superoxide dismutase. c, same as b except that HE-derived products in cell lysates were analyzed by HPLC after a 1-h incubation. d, same as c except that HE products were detected by HPLC in the media. *, p < 0.05.
Oxidation of Boronate Probes in Cell-free and Cellular Systems Generating •NO and O2˙̄
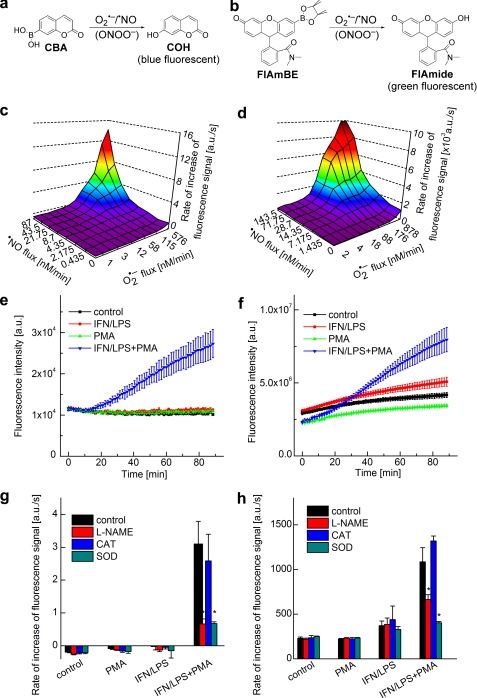
We monitored the profiles of ONOO− formation under conditions of different fluxes of •NO and O2˙̄, using two different boronate-based fluorogenic probes. Kinetic measurements have previously shown that ONOO− reacts rapidly with most boronates (k ∼106 m−1 s−1), yielding the corresponding phenolic products nearly stoichiometrically (9, 14). The fluorogenic probes used in this study were: CBA (9) and newly synthesized FlAmBE (Fig. 4, a and b). The reaction between these probes and ONOO− yields the corresponding phenolic products exhibiting blue and green fluorescence, respectively. The formation of ONOO− was monitored in real time during co-generation of •NO and O2˙̄ in cell-free systems using the same hypoxanthine/xanthine oxidase/DPTA-NONOate system described above (Fig. 1a). As shown in Fig. 4, c and d, the two boronate probes exhibit a similar reaction profile under the same experimental conditions, although the fluorescence intensity formed from FlAmBE was several orders of magnitude higher than that formed from CBA. In the presence of O2˙̄ or •NO alone, there was no detectable fluorescence from the probes. However, with increasing •NO and O2˙̄ fluxes, both probes were oxidized to fluorescent products with rates of oxidation nearly plateauing at a 1:1 flux ratio of •NO and O2˙̄ (Fig. 4, c and d). HPLC analyses of the same incubation mixtures revealed a single fluorescent product with about 80–90% yield (data not shown). These results suggest that the reaction chemistry of ONOO− with the fluorogenic boronate probes is similar under various fluxes of •NO and O2˙̄.
FIGURE 4.
Measurement of ONOO− formation by monitoring oxidation of boronate-based fluorogenic probes. a, scheme showing oxidation of CBA to 7-hydroxycoumarin (COH) (blue fluorescence) by •NO and O2˙̄. b, scheme showing oxidation of FlAmBE to the green fluorescent product, fluorescein N,N-dimethylamide (FlAmide), by •NO and O2˙̄. c, blue fluorescence (excitation at 320 nm, emission at 440 nm) was monitored in a plate reader from incubations containing CBA (20 μm) in phosphate buffer in the presence of varying fluxes of •NO and O2˙̄. a.u., arbitrary units. d, green fluorescence (excitation at 485 nm, emission at 535 nm) was monitored from incubations containing FlAmBE (20 μm) in phosphate buffer and varying fluxes of •NO and O2˙̄. e, increase in the blue fluorescence intensity was monitored using a plate reader, derived from RAW 264.7 macrophages in DPBS-GP buffer containing CBA (20 μM) and different stimulators. f, increase in green fluorescence intensity was monitored using a plate reader from incubations containing RAW 264.7 macrophages and FlAmBE (20μM) in the presence of different stimulators. g, same as e but in the presence of l-NAME and ROS (O2˙̄ and H2O2)-detoxifying enzymes. h, same as f but in the presence of l-NAME and ROS (O2˙̄ and H2O2)-detoxifying enzymes. *, p < 0.05.
Real-time Monitoring of ONOO− from Activated Macrophages
We next compared the oxidation of boronate probes in a cell culture model of ONOO− production with the results observed in a cell-free system. The cell culture conditions were identical to those used to monitor O2˙̄, as in Fig. 3a. Oxidation of CBA and FlAmBE was monitored in a 96-well fluorescence plate reader, and products were characterized by HPLC. Stimulation of macrophages with LPS, IFN-γ, and PMA in the presence of CBA (Fig. 4e) or FlAmBE (Fig. 4f) induced an increase in fluorescence intensity from either ONOO− probe. The addition of LPS and IFN-γ or PMA alone to incubations containing macrophages and CBA or FlAmBE caused little or no increase in fluorescence (Fig. 4, e and f) as compared with the control (non-treated) cells. Concomitant HPLC analysis (Fig. 5, a–c) indicates that the fluorescence intensity was due to the formation of the corresponding phenolic products, COH or fluorescein N,N-dimethylamide (FlAmide). These results are attributed to ONOO− formation and its direct reaction with boronate probes forming the corresponding fluorescent product. As •NO production in response to IFN/LPS is dependent on activation of inducible nitric oxide synthase, we next determined the effect of l-NAME (nonspecific inhibitor of nitric oxide synthase enzymes) on ONOO− formed by activated macrophages. As shown in Fig. 4, g and h, l-NAME inhibited PMA-, LPS-, and IFN-γ-induced fluorescence. Similarly, inhibition of O2˙̄ production using ROS-specific antioxidant enzymes should inhibit ONOO− formation as well. As expected, superoxide dismutase, but not catalase, inhibited CBA and FlAmBE fluorescence derived from ONOO−. Collectively, these results indicate that the boronate-based fluorogenic probes can be used to selectively detect ONOO− formed from macrophages activated to co-generate •NO and O2˙̄.
FIGURE 5.
HPLC analyses of products formed from oxidation of CBA by activated macrophages. Incubations contained CBA (20 μm) and RAW 264.7 macrophages in DPBS-GP buffer in the presence of different stimulators. a, after a 60-min incubation, CBA-derived product was analyzed by HPLC under the same conditions as in the plate reader (Fig. 3b) using the wavelengths as shown in the figure. exc., excitation; emi., emission. b, same as a except that the CBA-derived product was analyzed in the cell culture media. c, conditions same as a except that it gives a quantitative analysis of CBA and COH in cell lysates and media.
Oxidation of Amplex® Red in Cell-free and Cellular Systems Generating •NO and O2˙̄ as Measured Using HPLC and Fluorescence Analyses
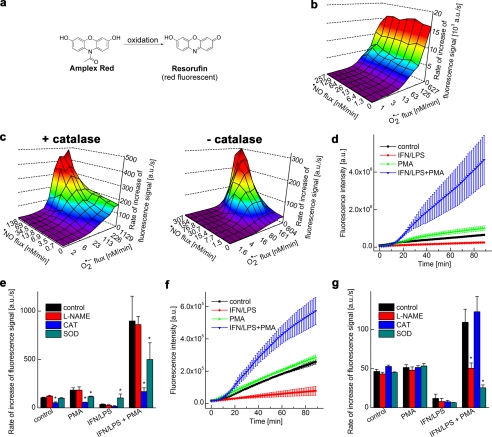
The horseradish peroxidase (HRP)-dependent oxidation of Amplex® Red to a fluorescent product (Fig. 6a) has been used to quantitate H2O2 in biological systems (17). The overall profile of the fluorescent product (i.e. resorufin) formed from Amplex® Red in incubations generating •NO and O2˙̄ in the presence and absence of HRP and catalase are shown (Fig. 6, b and c). HPLC analysis of the incubation mixtures revealed the formation of a single major red fluorescent product, resorufin (supplemental Fig. S3). HRP enhanced the rate of the formation of resorufin by over 30-fold in incubations containing Amplex® Red and HX/XO/NONOate, as observed by real-time monitoring of fluorescence intensity changes. Note that in the absence of HRP, oxidation of Amplex® Red was enhanced during co-generation of •NO and O2˙̄. This indicates that 1-electron oxidants such as radicals derived from ONOO− are able to cause oxidation of Amplex® Red to resorufin, although at a considerably lower yield as compared with HRP-mediated oxidation. Also, catalase increased the rate of Amplex® Red oxidation in the absence of HRP and the •NO donor (Fig. 6c). We attribute this to the peroxidase activity of catalase as H2O2 is produced directly from XO-catalyzed oxidation of HX and xanthine (X) and from the dismutation of O2˙̄ (Fig. 1a).
FIGURE 6.
Global profiling of H2O2 and ONOO−-derived oxidants by monitoring the oxidation of Amplex® Red. a, scheme showing oxidation of Amplex® Red to a red fluorescent product, resorufin. b, increase in the fluorescence intensity (due to resorufin, excitation at 535 nm, emission at 595 nm) caused by HRP-dependent oxidation of Amplex® Red (20 μm) in phosphate buffer in the presence of co-generated •NO and O2˙̄. a.u., arbitrary units. c, same as b except that HRP was not present. Note that the fluorescence intensity was greatly reduced as compared with b. d, oxidation of Amplex® Red (50 μm) by activated macrophages in the presence of HRP was measured. Increase in the fluorescence intensity was monitored in a fluorescence plate reader from incubations containing RAW 264.7 macrophages activated by different stimulators as shown. e, conditions were the same as in d except that the rate of increase in fluorescence intensity was measured in the presence of l-NAME and O2˙̄- and H2O2-detoxifying enzymes. CAT, catalase; SOD, superoxide dismutase. f, same as d but in the absence of HRP. Note the decrease in the fluorescence intensity of the product. g, same as f but in the presence of l-NAME and O2˙̄- and H2O2-detoxifying enzymes. *, p < 0.05.
Real-time Monitoring of H2O2 from Activated Macrophages
Next, we used RAW 264.7 macrophages stimulated with IFN-γ/LPS and PMA. The cell culture conditions were identical to those used to detect O2˙̄ and ONOO−. Oxidation of Amplex® Red was monitored in incubations containing macrophages and the proinflammatory mediators in the presence or absence of HRP (Fig. 6, d–g). The addition of PMA, or PMA plus LPS and IFN-γ to incubations containing macrophages, Amplex® Red, and HRP caused a time-dependent increase in fluorescence intensity (Fig. 6d). In the absence of PMA, no significant increase in fluorescence was detected. Results obtained in the presence of antioxidant enzymes (Fig. 6e) suggest that catalase significantly inhibited PMA-mediated oxidation of Amplex® Red. l-NAME had no effect, indicating that H2O2 was responsible for Amplex® Red oxidation under these conditions. Interestingly, preincubation of macrophages with LPS and IFN-γ caused an increase in the PMA-induced rate of oxidation of Amplex® Red. As this oxidation was inhibited by catalase, but not by l-NAME, we tentatively attribute this effect to the depletion of the intracellular pool of antioxidants during prolonged generation of •NO by LPS/IFN-γ-stimulated macrophages. Similar experiments with activated macrophages were performed in the absence of exogenously added HRP (Fig. 6, f and g). As shown, the fluorescence intensity was considerably diminished; l-NAME and superoxide dismutase but not catalase inhibited Amplex® Red-derived fluorescence (Fig. 6, f and g). These results suggest that it is possible to monitor both H2O2-derived and ONOO−-derived oxidants formed from activated macrophages using the Amplex® Red probe. However, distinguishing between ONOO− and H2O2-dependent mechanisms would require additional studies using several inhibitors (e.g. l-NAME, superoxide dismutase, and catalase).
Oxidative Nitrosation of DAF-2 in Cell-free and Cellular Systems Generating •NO and O2˙̄ as Measured Using HPLC and Fluorescence Analyses
The formation of a green fluorescent product, DAF-2T from DAF-2, has been used to monitor intracellular •NO formation (Fig. 7a) (18). The oxidative nitrosation profile of DAF-2T formed from the probe DAF-2 in incubations generating •NO and O2˙̄ shows that the rate of appearance of green fluorescence reached a maximum at ratios close to 1:1 (Fig. 7b). Independent HPLC analyses revealed that DAF-2T is responsible for the green fluorescence observed under these conditions (supplemental Fig. S4). Superoxide alone did not oxidize DAF-2 to DAF-2T, and •NO alone (in the presence of oxygen) caused only a modest increase in DAF-2T fluorescence (Fig. 7b). Clearly, these results suggest that in the presence of •NO and O2˙̄, DAF-2T is predominantly formed from ONOO−-mediated oxidative nitrosation of DAF-2.
FIGURE 7.
Nitrosation of the probe, DAF-2DA, by co-generated •NO and O2˙̄ and by activated macrophages. a, scheme showing the nitrosation of DAF-2 to the fluorescent product, DAF-2T. b, increase in green fluorescence (excitation at 485 nm, emission at 535 nm) during nitrosation of DAF-2 (5 μm) in phosphate buffer by co-generated •NO and O2˙̄. a.u., arbitrary units. c, increase in green fluorescence due to DAF-2T was monitored in a fluorescence plate reader from incubations containing DAF-2DA (5 μm) and RAW 264.7 macrophages in the presence of different stimulators. d, same as c, but the rate of increase in the fluorescence intensity was monitored in the presence of l-NAME and other O2˙̄- and H2O2-detoxifying enzymes. CAT, catalase; SOD, superoxide dismutase. *, p < 0.05.
Real-time Monitoring of •NO using DAF-2 from Activated Macrophages
•NO and ONOO− generation from activated macrophages was monitored using the probe DAF-2DA (the diacetate, cell-permeable analog of DAF-2 carboxylate) and DAF-2. The addition of IFN-γ/LPS to macrophages increased the green fluorescence of DAF-2T, which was further enhanced in the presence of PMA (Fig. 7c). HPLC analyses confirmed that the formation of DAF-2T was responsible for the increase in the fluorescence intensity (not shown) (19). l-NAME significantly inhibited the fluorescence intensity due to DAF-2T from macrophages activated by LPS/IFN-γ or LPS/IFN-γ/PMA (Fig. 7d). However, superoxide dismutase did not affect IFN-γ/LPS-induced DAF-2T fluorescence, but significantly inhibited IFN-γ/LPS/PMA-induced DAF-2T fluorescence. Similar results were obtained with DAF-2 probe (not shown). These results strongly suggest that DAF-2 or DAF-2DA is not a suitable probe for measuring •NO selectively under conditions generating both •NO and O2˙̄, although information with regard to RNS generation may be gained with the help of NOS inhibitors and ROS-detoxifying enzymes.
DISCUSSION
As discussed in recent reviews, ROS and RNS do not represent a single reactive species, but encompass a wide range of reactive species that exhibit oxidizing, nitrating, nitrosating, and halogenating properties (20, 21). To better understand the physiological and pathological consequences of ROS and RNS and their inhibitory mechanisms, it is crucial to unequivocally identify and quantify the species that are specifically responsible for a given biologic effect. In this study, we show that the combined use of a multiwell fluorescence plate reader and HPLC will detect all the relevant oxidation products within 5–10 min (UHPLC-like conditions using a fused core C18 column), thus making it feasible to monitor ROS and RNS formation under a variety of experimental conditions. The global profiling of ROS and RNS-derived fluorescent products demonstrated herein gives additional insight into the chemical mechanisms of their formation. For example, the profiles generated from oxidation of boronate-based fluorogenic probes (CBA, FlAmBE) in the presence of •NO and O2˙̄ indicate a direct oxidation of boronates by ONOO− to a fluorescent product that reaches a plateau at nearly a 1:1 ratio of •NO and O2˙̄ (Fig. 4). In contrast, oxidation/hydroxylation of HE under the same conditions shows •NO-mediated inhibition of 2-OH-E+ generated from the reaction of HE with superoxide. This could be explained by competitive scavenging of O2˙̄ by •NO. The concomitant formation of HE-derived dimeric products confirms the generation of a more potent, ONOO−-derived 1-electron oxidant under those conditions.
We have shown recently that ONOO− directly and stoichiometrically reacts with boronates nearly a million times faster than H2O2 and two hundred times faster than HOCl (9, 14). Thus, under conditions that simultaneously induce ONOO− or HOCl formation in a biological setting, boronates cannot be used to measure H2O2. The lack of an effect by catalase on FlAmBE- and CBA-derived fluorescence from activated macrophages (Fig. 4) is in support of this kinetic analysis. However, boronates conjugated to a triphenylphosphonium moiety were used to measure H2O2 production inside mitochondria using tandem LC/MS analysis (21). Under these conditions, it is likely that there is negligible formation of ONOO− or HOCl.
The global profiling of HE and HE-derived oxidation products is essential for proper interpretation of results obtained under different experimental conditions (22). Measurement of intracellular superoxide via monitoring of 2-OH-E+, the diagnostic marker product of O2˙̄/HE reaction, is dependent on HE uptake and its rate of oxidation to ethidium and dimeric products. Although E+ and HE-derived dimeric products are not formed in the presence of O2˙̄ alone, other 1-electron oxidants (perferryl iron or ONOO−-derived oxidants) induce their formation, and consequently, decrease intracellular HE levels (13). This can lead to a decrease in 2-OH-E+ formation. A previous study indicated that the ratio between 2-OH-E+ and HE is a better parameter for assessing O2˙̄ formation in cells (23). However, this approach is based on several kinetic assumptions that have not yet been totally validated (22).
The fluorescent probes, dichlorodihydrofluorescein (DCFH) and dihydrorhodamine 123 (DHR), have been most widely used to measure intracellular oxidants (H2O2, ONOO−). The pitfalls and artifacts of their use in biological systems have been previously discussed (24, 25). These two probes do not directly react with H2O2 or ONOO−. Iron or heme proteins with peroxidase activity are required for H2O2-dependent oxidation of dichlorodihydrofluorescein and dihydrorhodamine. ONOO− oxidizes these probes via a radical mechanism involving •OH, •NO2, or CO3˙̄ in the presence of bicarbonate. Most importantly, the intermediate radicals formed from these probes react with oxygen, forming superoxide and H2O2 and thus artifactually enhancing product formation and fluorescence intensity (25, 26). This is one of the major shortcomings of these probes. In contrast, HE- and Amplex® Red-derived radicals do not react with molecular oxygen, thus preventing artifactual generation of O2˙̄ and H2O2. Boronate reaction with ONOO− is mostly direct (∼85–90%), involving a 2-electron oxidation/reduction. Only a minor fraction (∼10–15%) occurs via a free radical pathway (27).
We have previously shown that the fluorescence spectral overlap between 2-OH-E+ and E+ makes it impossible to assign HE-derived red fluorescence to either 2-OH-E+ or E+ without guidance from HPLC measurements (22). As shown in Fig. 2b, maximal red fluorescence intensity was observed in the presence of O2˙̄ alone and in the presence of equal fluxes of •NO and O2˙̄. However, concomitant HPLC analyses showed the formation of either 2-OH-E+ (marker product of HE and O2˙̄) (15, 28) or E+ (nonspecific oxidation product of HE) under these conditions (Fig. 2d). Extraneous factors (light, sonication, etc.) were shown to induce the formation of 2-OH-E+ and/or E+ (16). Many investigators still have the wrong notion that E+ is the primary product of the HE and O2˙̄ reaction. To properly establish the connection between species and signaling, it is essential to understand the mechanism of oxidation of fluorogenic probes by specific ROS/RNS (29).
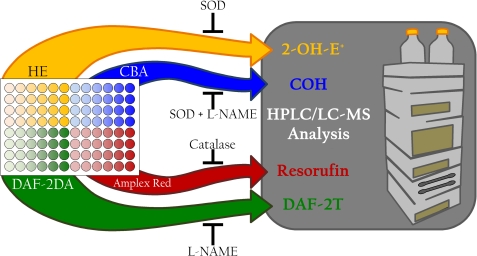
The schematic shown in Fig. 8 describes the approach an investigator may take to determine which reactive oxygen and nitrogen species are produced in their system of interest. In a 96-well plate, cells treated as desired can be assayed using HE, CBA, DAF-2DA, and Amplex® Red. All of these probes are cell-permeable. The fluorescence signal results from both intracellular and extracellular compartments. The use of a fluorescence plate reader for the initial screening, when coupled with inhibitors of specific oxidative products as indicated, allows for real-time simultaneous detection of multiple reactive species. In addition to using exogenous superoxide dismutase and catalase antioxidant enzymes (that are cell-impermeable), the investigator should also use molecular biological approaches that will alter the intracellular levels of these enzymes. Information obtained from the plate reader can then be used to design more selective studies by UHPLC to verify the identity and quantify the amount of intra- and extracellular products. As additional ROS/RNS-specific probes evolve with their oxidation/nitration/nitrosation chemistry properly characterized, the investigator may substitute these probes in this experimental approach.
FIGURE 8.
Suggested experimental design for identifying the reactive oxygen and nitrogen species generated simultaneously in response to a treatment of interest. The scheme describes the approach an investigator may take to determine which reactive oxygen and nitrogen species are produced in their system of interest. In a 96-well plate, cells treated as desired can be assayed using HE, CBA, DAF-2DA, and Amplex® Red. The use of a fluorescence plate reader for the initial screening, when coupled with inhibitors of specific oxidative products as indicated, allows for simultaneous detection of multiple reactive species. Information obtained from the plate reader can then be used to design more selective studies by UHPLC to verify and quantify the identity, location, and level of the products detected in the plate reader. DAF-2DA, diaminofluorescein–2 diacetate; SOD, superoxide dismutase.
Caveats
There are several important caveats to consider when examining ROS/RNS production in any given system. Chief among these is the tissue-specific expression of antioxidant enzymes. These enzymes will compete for reaction with the oxidants in question, and thus could potentially lead to aberrant quantification of the ROS/RNS involved. For example, the reaction of O2˙̄ with superoxide dismutase (k ∼1 × 109 m−1 s−1) is significantly more rapid than the reaction with HE (k ∼105-106 m−1 s−1) (30, 31). Similarly, performing ROS/RNS quantification studies in complete culture media can also affect the measurements. For example, ascorbate in culture media can react with multiple radical species (e.g. alkoxyl and alkyl peroxyl radicals) (32). Other components (phenol red, pyruvate, etc.) that are present in the culture media may react with ROS/RNS. Thus, it is advisable to maintain consistent culture conditions if comparisons between treatment groups are desired. ROS/RNS also participate in side reactions that can alter their apparent concentration or rate of production. A prime example of this is the reaction of peroxynitrite and carbon dioxide to form nitrosoperoxycarbonate (ONOOCO2−). Another important factor in these studies is the interaction between probe-derived intermediates and products with intra- and extracellular components (e.g. thiols). These side reactions will affect ROS/RNS-dependent formation and quantitation of products formed from the fluorescent probes.
The reactions discussed here are but an example of the considerations that must be made by researchers who wish to examine the profile of ROS/RNS produced in their model system of interest. Alternative methods such as monitoring nitrite/nitrate production or non-mitochondrial oxygen consumption may assist in providing a global view of the levels of ROS/RNS production; however, these methods lack species identification and should be used for gathering general insight into the oxidative biology occurring in a given system.
Future Perspectives
The high-throughput global profiling methodology developed herein for detecting ROS/RNS will be most useful in enzymatic systems regulated by several co-factors and redox modification of proteins in cell signaling. In the future, it is likely that one can monitor real-time changes in intracellular glutathione (GSH:GSSG) redox potential (33), lipid hydroperoxide, lipid aldehyde, isoprostanes, nitrated lipid, and immunospin-trapped protein radical adduct, calcium imbalance, membrane potential, release of pro-inflammatory mediators, and oxidant-induced signaling molecules in response to real-time changes in oxidant generation. Results from such studies will provide a solid foundation for establishing the identity of ROS/RNS species involved in redox signaling, cell proliferation, or cell death.
Supplementary Material
This work was supported, in whole or in part, by National Institutes of Health Grant R01 HL063119 (to B. K.).
This article was selected as a Paper of the Week.

This article contains supplemental Methods and Figs. S1–S7.
- ROS
- reactive oxygen species
- RNS
- reactive nitrogen species
- DTPA
- diethylenetriamine pentaacetic acid
- CBA
- coumarin-7-boronic acid
- DAF-2
- diaminofluorescein
- DAF-2T
- DAF-2 triazole
- E+
- ethidium
- 2-OH-E+
- 2-hydroxyethidium
- HE
- hydroethidine
- HX
- hypoxanthine
- XO
- xanthine oxidase
- DPTA-NONOate
- (Z)-1-[N-(3-aminopropyl)-N-(3-ammoniopropyl)amino]diazen-1-ium-1,2-diolate
- l-NAME
- NG-nitro-l-arginine methyl ester
- PMA
- phorbol 12-myristate 13-acetate
- FlAmBE
- fluorescein dimethylamide boronate
- DPBS
- Dulbecco's PBS
- UHPLC
- ultra high-pressure liquid chromatography.
REFERENCES
- 1. Fruehauf J. P., Meyskens F. L., Jr. (2007) Reactive oxygen species: a breath of life or death? Clin. Cancer Res. 13, 789–794 [DOI] [PubMed] [Google Scholar]
- 2. D'Autréaux B., Toledano M. B. (2007) ROS as signaling molecules: mechanisms that generate specificity in ROS homeostasis. Nat. Rev. Mol. Cell Biol. 8, 813–824 [DOI] [PubMed] [Google Scholar]
- 3. Groeger A. L., Freeman B. A. (2010) Signaling actions of electrophiles: anti-inflammatory therapeutic candidates. Mol. Interv. 10, 39–50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Veal E. A., Day A. M., Morgan B. A. (2007) Hydrogen peroxide sensing and signaling. Mol. Cell 26, 1–14 [DOI] [PubMed] [Google Scholar]
- 5. Rhee S. G. (2006) Cell signaling. H2O2, a necessary evil for cell signaling. Science 312, 1882–1883 [DOI] [PubMed] [Google Scholar]
- 6. Quijano C., Alvarez B., Gatti R. M., Augusto O., Radi R. (1997) Pathways of peroxynitrite oxidation of thiol groups. Biochem. J. 322, 167–173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Hogg N., Joseph J., Kalyanaraman B. (1994) The oxidation of α-tocopherol and trolox by peroxynitrite. Arch. Biochem. Biophys. 314, 153–158 [DOI] [PubMed] [Google Scholar]
- 8. Zhang H., Squadrito G. L., Uppu R. M., Lemercier J. N., Cueto R., Pryor W. A. (1997) Inhibition of peroxynitrite-mediated oxidation of glutathione by carbon dioxide. Arch. Biochem. Biophys. 339, 183–189 [DOI] [PubMed] [Google Scholar]
- 9. Zielonka J., Sikora A., Joseph J., Kalyanaraman B. (2010) Peroxynitrite is the major species formed from different flux ratios of co-generated nitric oxide and superoxide: direct reaction with boronate-based fluorescent probe. J. Biol. Chem. 285, 14210–14216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Zhao H., Joseph J., Fales H. M., Sokoloski E. A., Levine R. L., Vasquez-Vivar J., Kalyanaraman B. (2005) Detection and characterization of the product of hydroethidine and intracellular superoxide by HPLC and limitations of fluorescence. Proc. Natl. Acad. Sci. U.S.A. 102, 5727–5732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Robinson K. M., Janes M. S., Beckman J. S. (2008) The selective detection of mitochondrial superoxide by live cell imaging. Nat. Protoc. 3, 941–947 [DOI] [PubMed] [Google Scholar]
- 12. Miller E. W., Albers A. E., Pralle A., Isacoff E. Y., Chang C. J. (2005) Boronate-based fluorescent probes for imaging cellular hydrogen peroxide. J. Am. Chem. Soc. 127, 16652–16659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Zielonka J., Srinivasan S., Hardy M., Ouari O., Lopez M., Vasquez-Vivar J., Avadhani N. G., Kalyanaraman B. (2008) Cytochrome c-mediated oxidation of hydroethidine and mito-hydroethidine in mitochondria: identification of homo- and heterodimers. Free Radic. Biol. Med. 44, 835–846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Sikora A., Zielonka J., Lopez M., Joseph J., Kalyanaraman B. (2009) Direct oxidation of boronates by peroxynitrite: mechanism and implications in fluorescence imaging of peroxynitrite. Free Radic. Biol. Med. 47, 1401–1407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Zielonka J., Vasquez-Vivar J., Kalyanaraman B. (2008) Detection of 2-hydroxyethidium in cellular systems: a unique marker product of superoxide and hydroethidine. Nat. Protoc. 3, 8–21 [DOI] [PubMed] [Google Scholar]
- 16. Zielonka J., Vasquez-Vivar J., Kalyanaraman B. (2006) The confounding effects of light, sonication, and Mn(III)TBAP on quantitation of superoxide using hydroethidine. Free Radic. Biol. Med. 41, 1050–1057 [DOI] [PubMed] [Google Scholar]
- 17. Mishin V., Gray J. P., Heck D. E., Laskin D. L., Laskin J. D. (2010) Application of the Amplex® Red/horseradish peroxidase assay to measure hydrogen peroxide generation by recombinant microsomal enzymes. Free Radic. Biol. Med. 48, 1485–1491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Ridnour L. A., Thomas D. D., Mancardi D., Espey M. G., Miranda K. M., Paolocci N., Feelisch M., Fukuto J., Wink D. A. (2004) The chemistry of nitrosative stress induced by nitric oxide and reactive nitrogen oxide species. Putting perspective on stressful biological situations. Biol. Chem. 385, 1–10 [DOI] [PubMed] [Google Scholar]
- 19. Thomas S., Kotamraju S., Zielonka J., Harder D. R., Kalyanaraman B. (2007) Hydrogen peroxide induces nitric oxide and proteasome activity in endothelial cells: a bell-shaped signaling response. Free Radic. Biol. Med. 42, 1049–1061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Winterbourn C. C. (2008) Reconciling the chemistry and biology of reactive oxygen species. Nat. Chem. Biol. 4, 278–286 [DOI] [PubMed] [Google Scholar]
- 21. Murphy M. P., Holmgren A., Larsson N. G., Halliwell B., Chang C. J., Kalyanaraman B., Rhee S. G., Thornalley P. J., Partridge L., Gems D., Nyström T., Belousov V., Schumacker P. T., Winterbourn C. C. (2011) Unraveling the biological roles of reactive oxygen species. Cell Metab. 13, 361–366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Zielonka J., Kalyanaraman B. (2010) Hydroethidine- and MitoSOX-derived red fluorescence is not a reliable indicator of intracellular superoxide formation: another inconvenient truth. Free Radic. Biol. Med. 48, 983–1001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Maghzal G. J., Stocker R. (2007) Improved analysis of hydroethidine and 2-hydroxyethidium by HPLC and electrochemical detection. Free Radic. Biol. Med. 43, 1095–1096 [DOI] [PubMed] [Google Scholar]
- 24. Burkitt M. J., Wardman P. (2001) Cytochrome c is a potent catalyst of dichlorofluorescin oxidation: implications for the role of reactive oxygen species in apoptosis. Biochem. Biophys. Res. Commun. 282, 329–333 [DOI] [PubMed] [Google Scholar]
- 25. Bonini M. G., Rota C., Tomasi A., Mason R. P. (2006) The oxidation of 2′,7′-dichlorofluorescin to reactive oxygen species: a self-fulfilling prophecy? Free Radic. Biol. Med. 40, 968–975 [DOI] [PubMed] [Google Scholar]
- 26. Folkes L. K., Patel K. B., Wardman P., Wrona M. (2009) Kinetics of reaction of nitrogen dioxide with dihydrorhodamine and the reaction of the dihydrorhodamine radical with oxygen: implications for quantifying peroxynitrite formation in cells. Arch. Biochem. Biophys. 484, 122–126 [DOI] [PubMed] [Google Scholar]
- 27. Sikora A., Zielonka J., Lopez M., Dybala-Defratyka A., Joseph J., Marcinek A., Kalyanaraman B. (2011) Reaction between peroxynitrite and boronates: EPR spin-trapping, HPLC analyses, and quantum mechanical study of the free radical pathway. Chem. Res. Toxicol. 24, 687–697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Dikalov S., Griendling K. K., Harrison D. G. (2007) Measurement of reactive oxygen species in cardiovascular studies. Hypertension 49, 717–727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Zielonka J., Hardy M., Kalyanaraman B. (2009) HPLC study of oxidation products of hydroethidine in chemical and biological systems: ramifications in superoxide measurements. Free Radic. Biol. Med. 46, 329–338; Correction (2010) Free Radic. Biol. Med. 48, 373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Zhao H., Kalivendi S., Zhang H., Joseph J., Nithipatikom K., Vásquez-Vivar J., Kalyanaraman B. (2003) Superoxide reacts with hydroethidine but forms a fluorescent product that is distinctly different from ethidium: potential implications in intracellular fluorescence detection of superoxide. Free Radic. Biol. Med. 34, 1359–1368 [DOI] [PubMed] [Google Scholar]
- 31. Zielonka J., Sarna T., Roberts J. E., Wishart J. F., Kalyanaraman B. (2006) Pulse radiolysis and steady-state analyses of the reaction between hydroethidine and superoxide and other oxidants. Arch. Biochem. Biophys. 456, 39–47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Buettner G. R., Jurkiewicz B. A. (1996) Catalytic metals, ascorbate, and free radicals: combinations to avoid. Radiat. Res. 145, 532–541 [PubMed] [Google Scholar]
- 33. Gutscher M., Pauleau A. L., Marty L., Brach T., Wabnitz G. H., Samstag Y., Meyer A. J., Dick T. P. (2008) Real-time imaging of the intracellular glutathione redox potential. Nat. Methods 5, 553–559 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.