Background: Calsequestrin serves as a calcium storage/buffer protein in sarcoplasmic reticulum and undergoes a posttranslational modification.
Results: The specific site, degree, structure, and effects of glycosylation were determined.
Conclusion: The glycosylation prevented premature polymerization of calsequestrin ensuring mobility to the SR.
Significance: The glycosylation establishes a functional high capacity calcium binding polymer and allows calsequestrin to be retained in SR.
Keywords: Calcium, Crystal Structure, Glycosylation, Post-translational Modification, Sarcoplasmic Reticulum (SR), Skeletal Muscle, Calsequestrin
Abstract
Calsequestrin (CASQ) serves as a major Ca2+ storage/buffer protein in the sarcoplasmic reticulum (SR). When purified from skeletal muscle, CASQ1 is obtained in its glycosylated form. Here, we have confirmed the specific site and degree of glycosylation of native rabbit CASQ1 and have investigated its effect on critical properties of CASQ by comparison with the non-glycosylated recombinant form. Based on our comparative approach utilizing crystal structures, Ca2+ binding capacities, analytical ultracentrifugation, and light-scattering profiles of the native and recombinant rabbit CASQ1, we propose a novel and dynamic role for glycosylation in CASQ. CASQ undergoes a unique degree of mannose trimming as it is trafficked from the proximal endoplasmic reticulum to the SR. The major glycoform of CASQ (GlcNAc2Man9) found in the proximal endoplasmic reticulum can severely hinder formation of the back-to-back interface, potentially preventing premature Ca2+-dependent polymerization of CASQ and ensuring its continuous mobility to the SR. Only trimmed glycans can stabilize both front-to-front and the back-to-back interfaces of CASQ through extensive hydrogen bonding and electrostatic interactions. Therefore, the mature glycoform of CASQ (GlcNAc2Man1–4) within the SR can be retained upon establishing a functional high capacity Ca2+ binding polymer. In addition, based on the high resolution structures, we propose a molecular mechanism for the catecholaminergic polymorphic ventricular tachycardia (CPVT2) mutation, K206N.
Introduction
The sarcoplasmic reticulum (SR)2 in muscle cells serves as a Ca2+ storage/release system that controls the state of the actin-myosin fibrils by regulating cytosolic Ca2+ concentrations (1). Release of Ca2+ from the SR stimulates muscle contraction, and reuptake of cytosolic Ca2+ into the SR returns muscles to a relaxed state. In this pump-storage-release of Ca2+ by skeletal and cardiac SR, calsequestrin (CASQ) plays a major role through buffering free Ca2+ levels within the lumen of the SR together with other luminal acidic proteins (2). Both cardiac CASQ (CASQ2) and skeletal CASQ (CASQ1) do not act as a passive Ca2+ buffer but rather play an active role in regulating Ca2+ levels and facilitating its release from the SR lumen (1–3). Through localizing CASQ and its bound Ca2+ proximal to the ryanodine receptor, diffusion times for Ca2+ release can be drastically reduced (4).
Each CASQ molecule binds large numbers of Ca2+ ions with low affinity (Kd = ∼1 mm) over the physiological range of Ca2+ concentrations and releases it with a high off-rate (5–11). High capacity Ca2+ binding by CASQ is largely known to be nonspecific (12, 13). This high capacity and low affinity Ca2+ binding by CASQ has been directly linked to its unique Ca2+-dependent precipitation (4, 10, 14, 15). According to our widely accepted model of dynamic polymerization (4, 10, 16, 17), Ca2+ fills the electronegative pockets formed within the front-to-front and back-to-back intermolecular interfaces of CASQ, which cross-bridges CASQ molecules through intermolecular cooperative binding of Ca2+ and eventually leads to polymer formation. Evidence of dynamic polymerization associated with physiologic Ca2+ signaling has been confirmed in muscle in vivo (18).
Both CASQ1 and CASQ2 are often purified as glycosylated and/or phosphorylated isoforms. Phosphorylation of CASQ2 results in a drastic increase of its helical content and Ca2+ binding by providing a highly ordered polyanionic network (19). However, the consequence of glycosylation of CASQ is not clearly understood. Although glycosylation is considered the most abundant protein modification found in nature, occurring across all kingdoms of life (20), there is no general model or consensus as to how glycosylation affects protein function. Inhibition and/or suppression of glycosylation often results in protein aggregation or misfolding, leading to non-functional final states (21). Likewise, in a canine tachycardia-induced model of hypertrophy that leads to heart failure, CASQ2 glycosylation and phosphorylation are significantly altered (22). The cellular processes underlying these changes have remained uncertain due to the lack of understanding of CASQ trafficking (23).
Although CASQ does not contain any known targeting sequences, such as the C-terminal KDEL tetrapeptide, CASQ uniquely evades secretion and is retained in junctional SR (24–26). It has been proposed that the unique degree of glycan trimming could regulate its intracellular trafficking and retention in the junctional SR (26). Understanding the molecular mechanism for the unique trafficking mechanism and high capacity Ca2+ binding of CASQ is critical for proper understanding of Ca2+ handling in myocytes and its genetic abnormalities such as malignant hyperthermia or catecholaminergic polymorphic ventricular tachycardia (CPVT) (3, 27–29). Of particular note is a recent report on a new CPVT2 genotype, K206N, that implicates a complex interplay between glycosylation, polymerization, and Ca2+ binding capacity in causing its lethal phenotype (30). In this work we have extensively characterized the structural and functional significance of glycosylation of CASQ by comparing the glycosylated native rCASQ1 to its non-glycosylated recombinant form.
EXPERIMENTAL PROCEDURES
Preparation of Native Rabbit CASQ1
Isolation of SR microsomal membranes and extraction and purification of rCASQ1 contained within were performed as previously described (16, 31).
Preparation of Recombinant Rabbit CASQ1
The open reading frame for Oryctolagus cuniculus fast skeletal muscle CASQ without the N-terminal signal peptide (GenBankTM accession number M15747.1) was commercially synthesized (GenScript) and inserted into pET28a using NcoI and XhoI sites. Plasmid was then transformed into Escherichia coli DH5α cells. After sequence confirmation, the plasmid was transformed into BL21 (DE3) E. coli. Purification of rCASQ1 was performed following the same procedures as described before (10).
Crystallization and Structure Determination of Native and Recombinant Rabbit CASQ1
Native and recombinant CASQ protein crystals were grown at 4 °C by the vapor diffusion method using 2-methyl-2,4-pentanediol as the precipitating agent and protein at a concentration of 10 mg ml−1 in 20 mm HEPES, pH 7.0, and 0.5 m NaCl. All crystals belonged to the orthorhombic space group C2221 with one molecule in the asymmetric unit, and the diffraction data were collected to a resolution of 1.8 and 1.9 Å for native and recombinant rCASQ1, respectively. Intensity data were collected at the Advanced Light Source (BL8.2.1 and 8.2.2) for native and recombinant crystals. All data were reduced and scaled using HKL2000 (32). Iterative model adjustment and refinement were completed using the programs COOT (33) and PHENIX (34) using the coordinates of rCASQ1 (PDB code 1A8Y). All coordinates and diffraction data have been deposited in the Protein Data Bank as codes 3US3 (recombinant rCASQ1) and 3TRQ (native rCASQ1).
Equilibrium Dialysis/Atomic Absorption Spectrophotometer
To estimate the fractional occupancy (Y = [bound Ca2+]/[total rCASQ1]) for recombinant and native rCASQ1, equilibrium dialysis and atomic absorption spectrophotometry were as described previously in Sanchez et al. (19).
MALDI Mass Spectrometry of rCASQ1
Native rCASQ1 protein at 1 mg ml−1 was chemically cleaved by the addition of 1 m CNBr in the presence of 4 mm CsI and 70% TFA at 25 °C for 2 h. This chemical cleavage allowed for the isolation of both the C-terminal peptide and the peptide containing the N-linked glycoside. After treatment with CNBr, the sample was dried and then dissolved in 1:1 water:sinopinic acid. The sample was then dried on a MALDI plate and analyzed in negative mode with an Applied Biosystem 4800 mass analyzer. For whole protein mass spectrometry, native rCASQ1 and peptide N-glycosidase F-treated samples at a concentration of 0.5 mg ml−1 in 50 mm NaPi, pH 7.0 were dissolved in 1:1 sinopinic acid with 0.1% TFA and analyzed in positive mode with an Applied Biosystem 4800 mass analyzer. The peptide N-glycosidase F treatment for native rCASQ1 was done at a concentration of 0.5 mg ml−1 in 50 mm NaPi, pH 7.0, and was treated with 500 units of peptide N-glycosidase F (glycerol-free, New England Biolabs) in a reaction volume of 20 μl overnight at 37 °C. Sample was then analyzed with SDS-PAGE electrophoresis to determine completeness of cleavage.
Electrospray Ionization Mass Spectrometry of rCASQ Peptide Fragments
Recombinant and native rCASQ1 preparations were digested with either CNBr or trypsin to produce unique desired peptides. These samples were chromatographically separated and analyzed by a Bruker Esquire HCT mass spectrometer as described before (35). Peptides were then subjected to a MASCOT search (Matrix Science). MASCOT search parameters included tryptophan modification to include both of the dioxindolyalanines as well as asparagine modifications for glycosylation. Other parameters included were chemical alterations to methionine and included oxidative products generated by CNBr treatment. These modifications were both the sulfoxide and sulfone forms of methionine as well as homoserine and homoserine lactone. Serine/threonine phosphorylation was also searched as a variable.
Circular Dichroism (CD)
CD spectra of recombinant and native rCASQ1 were measured using an AVIV 202SF spectropolarimeter (AVIV Biomedical, Inc.) at 25 °C. Spectra of each protein at a concentration of 0.25 mg ml−1 in 20 mm MOPS (pH 7.2) and 300 mm KCl was recorded from 200 to 260 nm.
Thermostability of Recombinant and Native rCASQ1
The thermostability of both recombinant and native rCASQ1 was monitored as a function of CD signal at 222 nm using an AVIV 202SF spectropolarimeter (AVIV Biomedical, Inc.) and temperature. Briefly, samples containing 5 μm concentrations of either native or recombinant rCASQ1 in 20 mm KPi, pH 7.2, and 300 mm KCl were heated in a stepwise fashion by 2.5 °C per step with a 4-min equilibration time and a 3-s scan time. Temperature was regulated with a Peltier device (AVIV Biomedical), and samples were heated from 25 to 85 °C. To determine if the samples were refolded properly, CD spectra were taken after samples returned to 25 °C.
Analytical Ultracentrifugation
Both recombinant and native rCASQ1 (1 mg ml−1 in 20 mm MOPS, 300 mm KCl, pH 7.2) containing various concentrations of mm Ca2+ (0.5, 1, 2, 5, 10) were used to determine sedimentation velocity coefficients using a Beckman Coulter XL-I analytical ultracentrifuge with an An50-Ti rotor spun at 50,000 rpm at 25 °C for 14–16 h. Radial scans were recorded measuring the absorbance of the samples at 280 nm. The sedimentation coefficient of each sedimenting boundary was determined using Sedfit and DCDT+. Each analysis incorporated 100 scans, and the values for the density and viscosity of the buffer relative to water were 1.0038 and 1.0227, respectively.
Molecular Mass Determination by Multiangle Light Scattering
The chromatography and light-scattering experiments were performed as previously described (19).
RESULTS
CD of rCASQ1
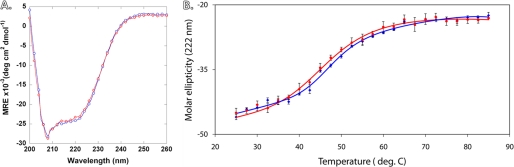
As previously reported (4, 10), a buffer containing 300 mm KCl was chosen for the CD spectra because at this concentration of K+ ion, CASQ1 is driven to a fully folded monomeric structure. Under these conditions both recombinant and native rCASQ1 exhibited identical far-UV CD spectra (Fig. 1A), indicating that there is no overall change in secondary structure between glycosylated and unglycosylated rCASQ1 monomers. The estimated Tm values for the native and the recombinant rCASQ were very similar, 49.25 and 48.75 °C, respectively (Fig. 1B).
FIGURE 1.
CD spectra of rCASQ1. A, shown are wavelength scans of both native and recombinant rCASQ1 from 260 to 200 nm. The blue line represents recombinant rCASQ1, whereas the red line represents native rCASQ1. B, thermostability of rCASQ1 is shown. Molar ellipticity measurements of native and recombinant rCASQ1 at 222 nm are shown. Recombinant measurements are shown in blue, and native measurements are shown in red.
Overall Structures of rCASQ1
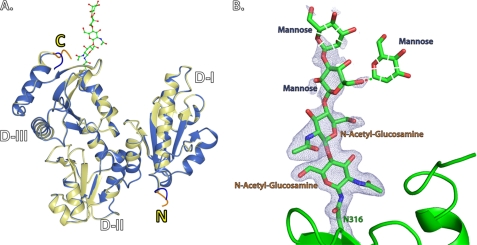
We were also able to crystallize both recombinant and native rCASQ1 under the same conditions, which allowed us a detailed comparison between the native and recombinant form of rCASQ1 (Table 1). The α-carbons of the native and recombinant rCASQ1 were superimposable with a root mean square deviation of 0.16 Å without including two N-terminal and four C-terminal residues, which displayed different conformations between two rCASQ1 structures (Fig. 2A). In addition, the electron density for the triose originating from the carboxyamide side chain of Asn-316, GlcNAc2Man1, was identified in the crystal structure of native rCASQ1 (Fig. 2B). Two structures showed a minor difference in the vicinity of the glycosylation site. The temperature factors of the β-loop near the Asn-316 in native rCASQ1 showed significantly (∼40%) reduced values compared with those in recombinant rCASQ1.
TABLE 1.
Crystallographic data for the native and recombinant rCASQ1
| Native rCASQ1 | Recombinant rCASQ1 | |
|---|---|---|
| Spacegroup | C2221 | C2221 |
| Data collection statistics | ||
| A | 59.271 Å | 59.213 Å |
| B | 144.565 Å | 144.811 Å |
| C | 111.170 Å | 110.471 Å |
| Resolution range | 30.70–1.76 Å | 38.91–1.74 Å |
| No. of reflections (>0) | 46770 | 48166 |
| Completeness | 98.02% (93%) | 97.9% (90%) |
| Refinement statistics | ||
| Rworka | 0.18 | 0.19 |
| Rfreeb | 0.21 | 0.23 |
| r.m.s.d.c | ||
| Bond lengths | 0.012 Å | 0.015Å |
| Bond angles | 0.81 | 0.91 |
| Wilson B-factor | 22.0 | 25.3 |
| Average B-value (Å) | 28.51 | 31.1 |
| No. of atoms | ||
| Nonsolvent | 2972 | 2922 |
| Solvent | 558 | 430 |
| Ramachandran plot | ||
| Favored | 98.30% | 98.6% |
| Allowed | 100.00% | 100.00% |
a Rcryst = Σ‖Fobs| − |Fcal‖/Fobs|.
b Rwork is calculated with removal of ≅5% of the data as the test set at the beginning of refinement.
c Root mean square deviations (r.m.s.d.) for main chain atoms are the root mean-squared deviations of the bond lengths and bond angles from their respective ideal values as implemented in PHENIX.
FIGURE 2.
Structural representations of native and recombinant rCASQ1. A, superimposed views of native rCASQ1 (yellow) and recombinant rCASQ1 (blue) with N and C termini are labeled as N and C, respectively. Attached glycans in native rCASQ1 are represented as ball and stick. Two major differences in N and C termini are highlighted in blue (recombinant rCASQ1) and orange (native rCASQ1). B, shown is a representation of the N-linked glycan, GlcNAc2Man2, present in native rCASQ1. Electron density was shown at 1σ. The expected mannose at the second position is shown in both the β1–3 and β1–6 linkage due to ambiguity. These figures were generated using Open-Source PyMOL™ (Version 1.4).
Multiangle Light Scattering
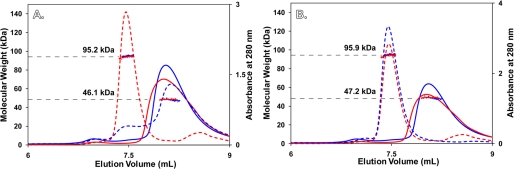
Both native and recombinant rCASQ1 were studied by multiangle light scattering to determine their tendencies to form oligomers. The results clearly indicated the monomeric nature of both molecules in the solution containing 300 mm KCl without any Ca2+ (Fig. 3). With an increase to 1 mm Ca2+, native rCASQ1 was driven to a predominantly dimeric state, whereas recombinant rCASQ1 appeared in an almost equimolar monomer:dimer ratio (Fig. 3). As the Ca2+ concentration increased to 2 mm, recombinant rCASQ1 was driven to a dimeric state similar to native rCASQ1 (Fig. 3). These results indicated a significant difference between native and recombinant rCASQ1 in their oligomeric response to low levels of Ca2+.
FIGURE 3.
Static light scattering of rCASQ1. Static light-scattering profiles of recombinant and native rCASQ1 in the absence and presence of both 1 mm and 2 mm Ca2+ are shown. A280 curves for both recombinant and native rCASQ1 are shown as red and blue lines for both panel A and B. In panel A, the A280 curves in the absence (solid line) and presence (dashed line) of 1 mm Ca2+ are shown. Panel B indicates recombinant and native rCASQ1 behavior in 2 mm Ca2+ following the same convention as panel A. In both panels molecular weights as determined by static light scattering are shown as dots, which are extended to the left axis for easy interpretation. In addition, the average molecular weights for the major peaks are indicated in each panel.
Analytical Ultracentrifugation
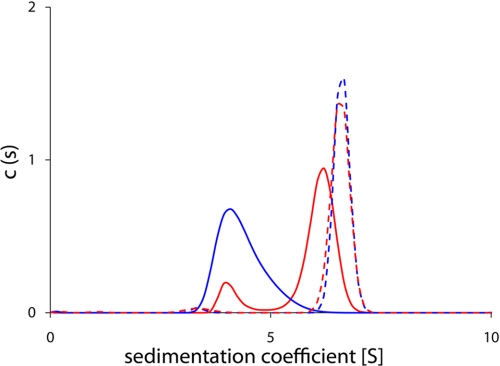
Both CASQ1 proteins precipitated at Ca2+ concentrations higher than ∼3 mm, which disabled any HPLC-based experimental approach. Therefore, to characterize the propensity of oligomerization at higher Ca2+ concentrations, sedimentation velocity experiments were performed using analytical ultracentrifugation. As shown in Fig. 4, the sedimentation coefficient of recombinant rCASQ1 (4.3) was smaller than that of native rCASQ1 (6.0) in the presence of 1 mm Ca2+. This was indicative of a lower oligomeric state, supporting the results of dynamic light-scattering experiments (Fig. 3). However, at 5 mm Ca2+, the sedimentation velocities of recombinant and native rCASQ1 were almost identical, with native slightly higher at 6.6 than that of recombinant at 6.5 (Fig. 4).
FIGURE 4.
Sedimentation equilibrium of rCASQ1. Native rCASQ1 in the presence of 1 mm Ca2+ was shown as a solid red line and in the presence of 5 mm Ca2+ as a dotted red line. Recombinant rCASQ1 in the presence of 1 mm Ca2+ is shown as a solid blue line, and recombinant rCASQ1 in the presence of 5 mm Ca2+ is shown as a dotted blue line.
Ca2+ Binding Capacity Assay
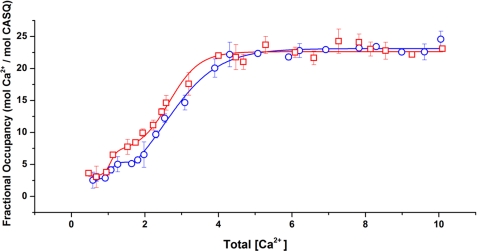
To study the potential effect of glycosylation on the Ca2+ binding capacity of rCASQ1, Ca2+ binding properties of native and recombinant proteins were analyzed by atomic absorption spectroscopy using the same buffer condition as that of multiangle light scattering. As shown in Fig. 5, native and recombinant rCASQ1 showed a multiphasic curve that was indicative of the stepwise formation of dimeric rCASQ1 to tetrameric and then higher-ordered polymeric structures as Ca2+ increases. Consistent with multiangle light scattering results, native rCASQ1 underwent a transition at lower Ca2+ concentrations (0.86 mm Ca2+) relative to recombinant rCASQ1 (1.25 mm Ca2+). However, at the Ca2+ concentrations above ∼5 mm, both recombinant and native CASQ1 bound similar amounts of Ca2+ ions, which indicated that glycosylation did not change Ca2+ binding capacity at those concentrations.
FIGURE 5.
Ca2+-binding capacities of rCASQ1. The number of Ca2+ ions bound to the wild-type rCASQ1 and recombinant rCASQ1 was determined through equilibrium dialysis and atomic absorption spectroscopy. Fractional occupancy (y = [bound Ca2+]/[total protein]) was plotted against [unbound Ca2+]. Capacity traces are as follows; □, native rCASQ1; ○, recombinant rCASQ1.
Mass Spectrometry of rCASQ1
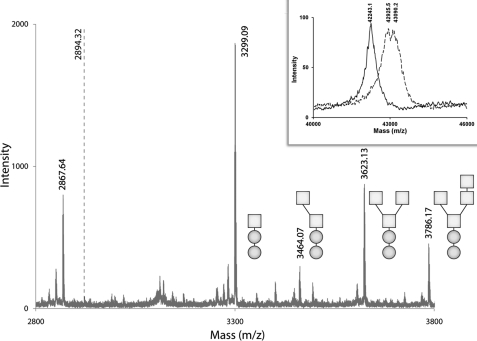
Previously, electrospray ionization mass spectroscopy was used to determine the degree of CASQ1 glycosylation. However, this determination was ambiguous due to the mass of a phosphate ion being almost identical to half the mass of a mannose residue. Our novel approach was to chemically cleave a peptide fragment that contains only Asn-316 and none of the potential phosphorylation sites (Thr-229, Thr-189, and Thr-353). By treating the protein sample with 1 m CNBr with 4 mm CsCl, we were able to selectively cleave a peptide spanning residues 300–324, which contained the N-linked glycan attached to Asn-316. However, due to chemical modification of Trp-324 (36), there was a mass increase of 13.99 Da through its conversion into dioxindolyalanine lactone (33). With this modified mass, the expected molecular mass of a glycan-free peptide would be 2731.31 Da, and with the expected glycan triose core of GlcNAc2Man1, the overall mass would correspond to 3299.54 Da. With the increase of additional mannose residues (22), the mass should increase incrementally by 160.0 Da (Calculated masses for 2, 3, and 4 mannose: 3461.59, 3623.64, and 3785.69, respectively). Therefore, as seen in Fig. 6, the molecular mass peak corresponding to 3299.49 Da was assigned to the GlcNAc2Man1, 3461.5 Da peak to GlcNAc2Man2, 3623.57 Da peak to GlcNAc2Man3 and finally the 3785.59 peak to GlcNAc2Man4.
FIGURE 6.
Mass spectra of rCASQ1. Mass spectra were obtained using the linear high mass negative method supplied by Applied Biosystem/SEIEX. Spectra were obtained from CNBr digestion of native rCASQ1. Mass peaks were fitted according to the glycan composition of the peptide fragment. For clarity, GlcNAc composition is represented by circles, and Man composition is represented by squares. The dotted line at (m/z) of 2894.32 represents the expected m/z for a phosphorylated C-terminal peptide. Inset, shown is the whole protein mass spectra obtained for recombinant (solid line) or native (dashed line) rCASQ1 using the same method as described above. m/z corresponding to the highest intensity peaks are indicated.
We were also able to generate a unique C-terminal peptide under the same cleavage conditions that contains Thr-353, the proposed major phosphorylation site in CASQ1 (37). Although other phosphorylation sites exist within the C-terminus of other CASQs, these sites are not found within the C-terminal of rCASQ1 and were, therefore, excluded as possible outcomes. The expected molecular mass for the unphosphorylated peptide would be seen at 2868.0 Da in negative mode and at 2948.0 Da for its phosphorylated form. As seen in Fig. 6, there was no corresponding peak for the phosphorylated form of this peptide. Instead, two peaks were seen at 2850.0 and 2832.0 Da, which correspond to the loss of one and two waters from the peptide. Those peaks were not due to a loss of phosphate group, as the relative peak intensity increased with increased laser power, and the apparent loss of water has been seen with other acid-rich peptides analyzed in negative mode. We also independently confirmed that CNBr/CsI treatment did not acid-hydrolyze any phosphate from the phosphorylated sites by analyzing casein-derived phosphopeptides using electrospray ionization mass spectroscopy and the same MASCOT search parameters as described under “Experimental Procedures” with the addition of tryptic cut sites (supplemental Fig. 1).
We also used MALDI mass spectrometry to investigate the heterogeneity of rCASQ1 glycosylation by comparing the native rCASQ1 proteins with/without peptide N-glycosidase treatment. The two major peaks of native rCASQ1 were 42,925.5 and 43,090.2, with a broad degree of heterogeneity. After treatment with peptide N-glycosidase F, the determined molecular mass was reduced to 42,243.10 Da, with a much narrower peak indicating a more homogenous sample (Fig. 6, inset).
In addition, tryptic digestion of the same protein preparation followed by a MASCOT search yielded peptides that are unique to rCASQ2, which indicated coexistence of rCAQ1 and rCASQ2 in the skeletal SR. Two unique peptides were found with the expected scores of 0.0057 for a peptide experimentally determined at 1524.96 Da and a calculated mass of 1524.73 Da and an expected value of 1.2E-6 for a peptide of experimental mass 3189.85 Da and a calculated mass of 3189.52. With an adjusted p value of <0.05, both of these peptides are significant and unique for rCASQ2 (supplemental Fig. 1).
DISCUSSION
Posttranslational modifications are known to regulate the function of proteins, often by modulating their physical characteristics (38). Among those modifications, phosphorylation and glycosylation are the most common. The attachment of glycans during or after protein synthesis introduces profound consequences on both structure and function of a protein. Inhibition of glycosylation often results in aggregation or misfolding of the corresponding proteins, leading to non-functional and/or disease states (20). Phosphorylation is another essential regulatory mechanism in nearly every aspect of eukaryotic cellular functions, affecting more than 30% of all proteins (39, 40). After phosphorylation, the disordered regions around the modification site of the target protein often become highly ordered (41).
During its transit through the secretory pathway to the SR, CASQ undergoes a yet uncharacterized glycosylation and phosphorylation (37, 42, 43, 45–50). Previously, both glycosylation and phosphorylation have been linked to a junctional SR trafficking mechanism (19, 37, 50–52). Therefore, altered or impaired posttranslational modifications could cause serious pathological symptoms (22, 23). Supporting this, in the animal heart failure models, CASQ2 contains glycan structures that are uncharacteristic of normal junctional SR, indicating altered characteristics or altered trafficking through secretory compartments (22). Previously, we have shown that phosphorylation at the C terminus of human CASQ2 produces a disorder-to-order transition by providing a more stable network of anions, which increases its Ca2+ binding capacity (19, 37). Consequently, phosphorylation could not only affect trafficking due to its effect on solubility but could also regulate a level of Ca2+ binding capacity of CASQ and, hence, a SR Ca2+ release amount in response to physiological needs (19, 37).
Glycosylation
Both CASQ1 and CASQ2 of various species contain an N-glycosylation consensus sequence (Asn-X-Ser/Thr) at their C terminus. Our mass spectometry data showed that the predominant glycan of rabbit CASQ1 is a triose, GlcNAc2Man1, with the presence of lesser, but approximately equal, amounts of GlcNAc2Man2, GlcNAc2Man3, and GlcNAc2Man4 (Fig. 6), which differs from that of canine CASQ (42, 53). Changes in the mannose content of N-linked glycans are often observed during trafficking through distinct ER and Golgi compartments (54, 55). However, trimming of N-linked glycans to Man1, Man3, and Man4, which is the case in rCASQ1, is still poorly understood (23).
In native rCASQ1, GlcNAc2Man1–4 was attached to Asn-316, which is located at the beginning of a short β-turn comprised of residues 316–319. The temperature factors for those residues were substantially reduced upon glycosylation. However, consistent with CD spectra (Fig. 1), those glycans did not cause any significant secondary structural change. On the other hand, consistent with the results of both sedimentation (Fig. 4) and light-scattering studies (Fig. 3), the presence of the glycans significantly stabilizes intermolecular interactions. The comparison of crystal structures between native and recombinant rCASQ1 showed that the interaction with neighboring N-terminal residue of a partner subunit was substantially strengthened by those glycans through both direct and water/ion-mediated indirect interactions (Fig. 7). For example, the carboxyl side chain of Glu-2 of a dimeric partner CASQ1 molecule was within a hydrogen-bonding distance with the N-acetyl group of the second GlcNac (Fig. 7). In addition, one Na+ ion was tightly coordinated by the side-chain oxygens of Glu-1, Asn-316, and Thr-318, the backbone oxygen of Gly-248, and the N-acetyl group of the first GlcNac.
FIGURE 7.
Observed molecular interactions of glycans in front-to-front dimer interfaces. Detailed interaction between the N-terminal residues of one CASQ1 molecule (black) and glycans attached to the other CASQ1 (gray) is shown. The participating Na+ and water molecule are depicted with blue and red spheres. The inset shows a front-to-front dimer that involves N-terminal arm exchange. These figures were generated using CCP4MG.2.5.0.
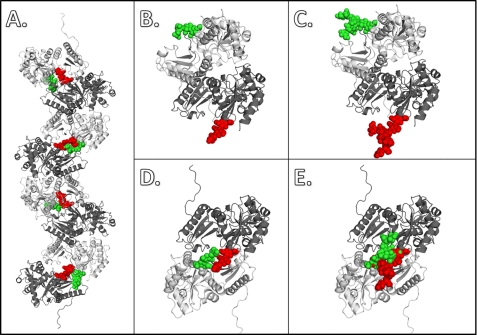
Although the major glycans according to our mass spectometry data, GlcNac2Man1–4, would not cause any steric hindrance in both front-to-front and back-to-back interfaces (Fig. 8, A, B, and D), the expected bulky glycan moiety of CASQ1 in the proximal ER, GlcNac2Man8,9, can certainly interfere with the back-to-back interaction due to severe steric clash (Fig. 8E). However, the same bulky glycans (GlcNac2Man8,9) do not interfere with the formation of the front-to-front interface (Fig. 8C); instead they can accelerate dimer formation through enforced front-to-front interaction between two subunits, as mentioned above. Therefore, it is likely that high mannosylation of CASQ (Man8,9) is a device to prevent premature oligomerization or aggregation while at the same time stabilizing the dimer and keeping CASQ in solution through mimicking the role played by many molecular chaperones (44). As with other proteins, CASQ is progressively modified as it moves from the entry side (cis) to the exit side (trans) of ER. As CASQ is processed, interference in back-to-back interactions on part of the glycan chain is reduced through successive trimming. Glycan moieties in medium levels of glycosylation, such as Man5 or Man6, are no longer inhibitory. Eventually the matured form of glycans (GlcNac2Man1–4) allows CASQ molecules to polymerize in the SR (Fig. 8A). Therefore, through the time- and space-controlled mannose trimming, Ca2+-dependent polymerization causes CASQ to deposit itself at its target site. Considering the similarity of the Ca2+ binding capacity and sedimentation coefficients between native and recombinant CASQ1 in high Ca2+ concentration, it is likely that the major glycan form in mature CASQ1, GlcNac2Man1, might play a passive role once it reaches the SR.
FIGURE 8.
Observed and predicted dimer interaction of native rCASQ1. The front-to-front and back-to-back interactions between two rCASQ1 monomers are represented with a ribbon diagram of protein (gray and black) and space-filing model of glycans (red and green). A, a full octamer with alternating glycan moieties shows their relative positions in polymer packing of CASQ; B. shown is the front-to-front dimer with the trimmed glycan moiety, GlcNAc2Man1. C, shown is the front-to-front dimer with full glycan moieties, GlcNAc2Man9. D, shown is the back-to-back dimer with trimmed glycan moiety, GlcNAc2Man1. E, shown is the back-to-back dimer with full glycan moieties, GlcNAc2Man9. These figures were generated using Open-Source PyMOL™ (v1.4).
Although canine cardiac CASQ localizes to the proximal ER cisternae of cells, canine skeletal CASQ1 escapes that proximal sites and is found in the distal regions of the ER, which is an ER-Golgi intermediate compartment. A distinct difference has been noticed between CASQ1 and CASQ2 glycoforms, and mannose trimming from the attached glycoside has been proposed as the step responsible for shuttling CASQ2 into the cardiac SR (17). In general, CASQ1 is more negatively charged than CASQ2, although CASQ2 has a more negatively charged C terminus. Previously we have shown CASQ2 forms a polymer at lower Ca2+ levels relative to CASQ1 (10). It is highly plausible that the more negative charge of CASQ1 delays the productive collision between monomers. In addition, the higher degree of glycosylation observed in cardiac CASQ2 (Man5 or Man6) can further stabilize the dimer and polymer, resulting in earlier polymerization at lower Ca2+ concentrations when compared with cardiac CASQ 1 (Man1). Therefore, it is tempting to speculate that differences in both mannose trimming and net charge are the determinant for the observed differential targeting of CASQ1 and CASQ2.
Implication on K206N-CPVT2 Mutation
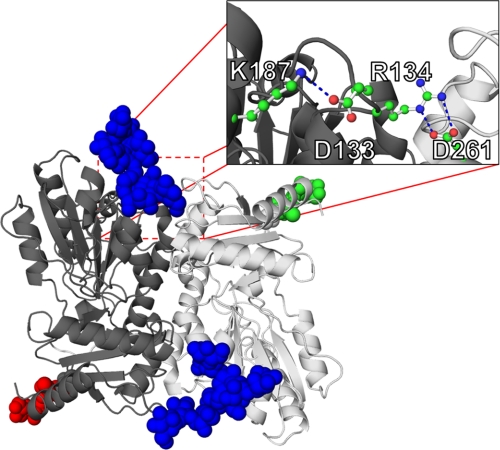
The recently discovered K206N-CPVT2 mutation has been shown to incorporate additional glycans, which results in lower Ca2 binding capacity and altered polymerization in CASQ2 (30). Neumann et al. suggested that the alteration in function is due to the amino acid substitution and not the altered glycosylation. Indeed, as evidenced in the crystal structure, the corresponding Lys-187 in rCASQ1 was critical in stabilizing two salt-bridges (Fig. 9). Those symmetrical salt-bridge pairs stabilized the front-to-front dimer together with N-terminal arm-exchange interactions in the opposite side of the dimer. As shown in Fig. 9 inset, the formation of a salt bridge between Arg-134 and Asp-261 was dependent on the interaction between Glu-133 and Lys-187 (Lys-206). Thus, K206N mutation and consequent glycosylation in this solvent-exposed area will disrupt the network of salt bridges and weaken the front-to-front interaction without introducing much steric hindrance (Fig. 9).
FIGURE 9.
Structural impact of K206N mutation. Shown is a hypothetical model of glycan positioning in K206N mutation. Normal glycosylation moieties positioned at Asp-316 are shown in green and red for each monomer, and expected glycosylation moieties introduced by mutation at K206N are shown in blue. The inset represents the salt bridge that stabilizes the front-to-front interface.
Conclusion
The observed mature forms of oligosaccharide (GlcNAc2Man1–4) did not have any significant effect on the structure and stability of monomeric rCASQ1. Instead, through further stabilizing the N-terminal arm-exchange of a partnering subunit, the glycans significantly enhanced or stabilized formation of a front-to-front dimer. Coupled with the body of evidence in this article, we propose that dynamic mannose-trimming in CASQ is a primary mechanism for intracellular trafficking/targeting. The polymerization of CASQ is a consequence of Ca2+-dependent back-to-back interaction among CASQ dimers (4, 10, 16, 17). The formation of this interface is prevented by the bulky glycan moiety in the proximal ER (GlcNac2Man8,9) but is later enabled by progressive mannose-trimming.
Supplementary Material
Acknowledgments
We thank L. M. Gloss and T. Topping (Washington State University) and J. T. Oxford (Boise State University, M. J. Murdock Charitable Trust, and National Institutes of Health/Center for Research Resources Grant P20RR016454).
This work was supported by American Heart Association Grant 0850084Z, National Science Foundation Grants MCB 1021148 and DBI 0959778, and by the M. J. Murdock Charitable Trust. This work was also supported in part by National Institutes of Health Grant T32GM083864 (NIGMS; to E. J. S.).

This article contains supplemental Fig. 1.
The atomic coordinates and structure factors (codes 3US3 and 3TRQ) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
- SR
- sarcoplasmic reticulum
- ER
- endoplasmic reticulum
- CPVT
- catecholaminergic polymorphic ventricular tachycardia
- rCPVT
- rabbit CPVT
- CASQ
- calsequestrin
- rCASQ
- rabbit CASQ.
REFERENCES
- 1. Royer L., Ríos E. (2009) Deconstructing calsequestrin. Complex buffering in the calcium store of skeletal muscle. J. Physiol. 587, 3101–3111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. MacLennan D. H., Abu-Abed M., Kang C. (2002) Structure-function relationships in Ca2+ cycling proteins. J. Mol. Cell. Cardiol. 34, 897–918 [DOI] [PubMed] [Google Scholar]
- 3. MacLennan D. H., Chen S. R. (2009) Store overload-induced Ca2+ release as a triggering mechanism for CPVT and MH episodes caused by mutations in RYR and CASQ genes. J. Physiol. 587, 3113–3115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Park H., Wu S., Dunker A. K., Kang C. (2003) Polymerization of calsequestrin. Implications for Ca2+ regulation. J. Biol. Chem. 278, 16176–16182 [DOI] [PubMed] [Google Scholar]
- 5. Meissner G., Conner G. E., Fleischer S. (1973) Isolation of sarcoplasmic reticulum by zonal centrifugation and purification of Ca2+ pump and Ca2+-binding proteins. Biochim. Biophys. Acta 298, 246–269 [DOI] [PubMed] [Google Scholar]
- 6. Ikemoto N., Nagy B., Bhatnagar G. M., Gergely J. (1974) Studies on a metal-binding protein of the sarcoplasmic reticulum. J. Biol. Chem. 249, 2357–2365 [PubMed] [Google Scholar]
- 7. MacLennan D. H., Wong P. T. (1971) Isolation of a calcium-sequestering protein from sarcoplasmic reticulum. Proc. Natl. Acad. Sci. U.S.A. 68, 1231–1235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Slupsky J. R., Ohnishi M., Carpenter M. R., Reithmeier R. A. (1987) Characterization of cardiac calsequestrin. Biochemistry 26, 6539–6544 [DOI] [PubMed] [Google Scholar]
- 9. Mitchell R. D., Simmerman H. K., Jones L. R. (1988) Ca2+ binding effects on protein conformation and protein interactions of canine cardiac calsequestrin. J. Biol. Chem. 263, 1376–1381 [PubMed] [Google Scholar]
- 10. Park H., Park I. Y., Kim E., Youn B., Fields K., Dunker A. K., Kang C. (2004) Comparing skeletal and cardiac calsequestrin structures and their calcium binding. A proposed mechanism for coupled calcium binding and protein polymerization. J. Biol. Chem. 279, 18026–18033 [DOI] [PubMed] [Google Scholar]
- 11. Kim E., Youn B., Kemper L., Campbell C., Milting H., Varsanyi M., Kang C. (2007) Characterization of human cardiac calsequestrin and its deleterious mutants. J. Mol. Biol. 373, 1047–1057 [DOI] [PubMed] [Google Scholar]
- 12. MacLennan D. H., Reithmeier R. A. (1998) Ion tamers. Nat. Struct. Biol. 5, 409–411 [DOI] [PubMed] [Google Scholar]
- 13. Hidalgo C., Donoso P., Rodriguez P. (1996) Protons induce calsequestrin conformational changes. Biophys. J. 71, 2130–2137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Gatti G., Trifari S., Mesaeli N., Parker J. M., Michalak M., Meldolesi J. (2001) Head-to-tail oligomerization of calsequestrin. A novel mechanism for heterogeneous distribution of endoplasmic reticulum luminal proteins. J. Cell Biol. 154, 525–534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Cho J. H., Ko K. M., Singaruvelu G., Lee W., Kang G. B., Rho S. H., Park B. J., Yu J. R., Kagawa H., Eom S. H., Kim do H., Ahnn J. (2007) Functional importance of polymerization and localization of calsequestrin in C. elegans. J. Cell Sci. 120, 1551–1558 [DOI] [PubMed] [Google Scholar]
- 16. Wang S., Trumble W. R., Liao H., Wesson C. R., Dunker A. K., Kang C. H. (1998) Crystal structure of calsequestrin from rabbit skeletal muscle sarcoplasmic reticulum. Nat. Struct. Biol. 5, 476–483 [DOI] [PubMed] [Google Scholar]
- 17. Milstein M. L., Houle T. D., Cala S. E. (2009) Calsequestrin isoforms localize to different ER subcompartments. Evidence for polymer and heteropolymer-dependent localization. Exp. Cell Res. 315, 523–534 [DOI] [PubMed] [Google Scholar]
- 18. Launikonis B. S., Zhou J., Royer L., Shannon T. R., Brum G., Ríos E. (2006) Depletion “skraps” and dynamic buffering inside the cellular calcium store. Proc. Natl. Acad. Sci. U.S.A. 103, 2982–2987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Sanchez E. J., Munske G. R., Criswell A., Milting H., Dunker A. K., Kang C. (2011) Phosphorylation of human calsequestrin. Implications for calcium regulation. Mol. Cell. Biochem. 353, 195–204 [DOI] [PubMed] [Google Scholar]
- 20. Larkin A., Imperiali B. (2011) The expanding horizons of asparagine-linked glycosylation. Biochemistry 50, 4411–4426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Mitra N., Sinha S., Ramya T. N., Surolia A. (2006) N-Linked oligosaccharides as outfitters for glycoprotein folding, form, and function. Trends Biochem. Sci. 31, 156–163 [DOI] [PubMed] [Google Scholar]
- 22. Kiarash A., Kelly C. E., Phinney B. S., Valdivia H. H., Abrams J., Cala S. E. (2004) Defective glycosylation of calsequestrin in heart failure. Cardiovasc. Res. 63, 264–272 [DOI] [PubMed] [Google Scholar]
- 23. McFarland T. P., Milstein M. L., Cala S. E. (2010) Rough endoplasmic reticulum to junctional sarcoplasmic reticulum trafficking of calsequestrin in adult cardiomyocytes. J. Mol. Cell. Cardiol. 49, 556–564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Campbell K. P., MacLennan D. H., Jorgensen A. O., Mintzer M. C. (1983) Purification and characterization of calsequestrin from canine cardiac sarcoplasmic reticulum and identification of the 53,000-dalton glycoprotein. J. Biol. Chem. 258, 1197–1204 [PubMed] [Google Scholar]
- 25. Franzini-Armstrong C., Kenney L. J., Varriano-Marston E. (1987) The structure of calsequestrin in triads of vertebrate skeletal muscle. A deep-etch study. J. Cell Biol. 105, 49–56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Houle T. D., Ram M. L., McMurray W. J., Cala S. E. (2006) Different endoplasmic reticulum trafficking and processing pathways for calsequestrin (CSQ) and epitope-tagged CSQ. Exp. Cell Res. 312, 4150–4161 [DOI] [PubMed] [Google Scholar]
- 27. Priori S. G., Chen S. R. (2011) Inherited dysfunction of sarcoplasmic reticulum Ca2+ handling and arrhythmogenesis. Circ. Res. 108, 871–883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Chopra N., Kannankeril P. J., Yang T., Hlaing T., Holinstat I., Ettensohn K., Pfeifer K., Akin B., Jones L. R., Franzini-Armstrong C., Knollmann B. C. (2007) Modest reductions of cardiac calsequestrin increase sarcoplasmic reticulum Ca2+ leak independent of luminal Ca2+ and trigger ventricular arrhythmias in mice. Circ. Res. 101, 617–626 [DOI] [PubMed] [Google Scholar]
- 29. Kalyanasundaram A., Bal N. C., Franzini-Armstrong C., Knollmann B. C., Periasamy M. (2010) The calsequestrin mutation CASQ2D307H does not affect protein stability and targeting to the junctional sarcoplasmic reticulum but compromises its dynamic regulation of calcium buffering. J. Biol. Chem. 285, 3076–3083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Kirchhefer U., Wehrmeister D., Postma A. V., Pohlentz G., Mormann M., Kucerova D., Müller F. U., Schmitz W., Schulze-Bahr E., Wilde A. A., Neumann J. (2010) The human CASQ2 mutation K206N is associated with hyperglycosylation and altered cellular calcium handling. J. Mol. Cell. Cardiol. 49, 95–105 [DOI] [PubMed] [Google Scholar]
- 31. He Z., Dunker A. K., Wesson C. R., Trumble W. (1993) Ca2+-induced folding and aggregation of skeletal muscle sarcoplasmic reticulum calsequestrin. The involvement of the trifluoperazine-binding site. J. Biol. Chem. 268, 24635–24641 [PubMed] [Google Scholar]
- 32. Otwinowski Z., Minor W. (1997) Methods Enzymol. 276, 307–326 [DOI] [PubMed] [Google Scholar]
- 33. Emsley P., Cowtan K. (2004) Coot. Model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 [DOI] [PubMed] [Google Scholar]
- 34. Adams P. D., Afonine P. V., Bunkóczi G., Chen V. B., Davis I. W., Echols N., Headd J. J., Hung L. W., Kapral G. J., Grosse-Kunstleve R. W., McCoy A. J., Moriarty N. W., Oeffner R., Read R. J., Richardson D. C., Richardson J. S., Terwilliger T. C., Zwart P. H. (2010) PHENIX. A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66, 213–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Noh S. M., Brayton K. A., Brown W. C., Norimine J., Munske G. R., Davitt C. M., Palmer G. H. (2008) Composition of the surface proteome of Anaplasma marginale and its role in protective immunity induced by outer membrane immunization. Infect. Immun. 76, 2219–2226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Domingues M. R., Domingues P., Reis A., Fonseca C., Amado F. M., Ferrer-Correia A. J. (2003) Identification of oxidation products and free radicals of tryptophan by mass spectrometry. J. Am. Soc. Mass Spectrom. 14, 406–416 [DOI] [PubMed] [Google Scholar]
- 37. Beard N. A., Wei L., Cheung S. N., Kimura T., Varsányi M., Dulhunty A. F. (2008) Phosphorylation of skeletal muscle calsequestrin enhances its Ca2+ binding capacity and promotes its association with junctin. Cell Calcium 44, 363–373 [DOI] [PubMed] [Google Scholar]
- 38. Shental-Bechor D., Levy Y. (2008) Effect of glycosylation on protein folding. A close look at thermodynamic stabilization. Proc. Natl. Acad. Sci. U.S.A. 105, 8256–8261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Hubbard M. J., Cohen P. (1993) On target with a new mechanism for the regulation of protein phosphorylation. Trends Biochem. Sci. 18, 172–177 [DOI] [PubMed] [Google Scholar]
- 40. Johnson L. (2009) The regulation of protein phosphorylation. Biochem. Soc. Trans. 37, 627–641 [DOI] [PubMed] [Google Scholar]
- 41. Iakoucheva L. M., Radivojac P., Brown C. J., O'Connor T. R., Sikes J. G., Obradovic Z., Dunker A. K. (2004) The importance of intrinsic disorder for protein phosphorylation. Nucleic Acids Res. 32, 1037–1049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. O'Brian J. J., Ram M. L., Kiarash A., Cala S. E. (2002) Mass spectrometry of cardiac calsequestrin characterizes microheterogeneity unique to heart and indicative of complex intracellular transit. J. Biol. Chem. 277, 37154–37160 [DOI] [PubMed] [Google Scholar]
- 43. Shoshan-Barmatz V., Ashley R. (1998) The structure, function, and cellular regulation of ryanodine-sensitive Ca2+ release channels. Int. Rev. Cytol. 183, 185–270 [DOI] [PubMed] [Google Scholar]
- 44. Jaenicke R. (1991) Protein folding. Local structures, domains, subunits, and assemblies. Biochemistry. 30, 3147–3161 [DOI] [PubMed] [Google Scholar]
- 45. Cala S. E., Jones L. R. (1991) Phosphorylation of cardiac and skeletal muscle calsequestrin isoforms by casein kinase II. Demonstration of a cluster of unique rapidly phosphorylated sites in cardiac calsequestrin. J. Biol. Chem. 266, 391–398 [PubMed] [Google Scholar]
- 46. Rodriguez M. M., Chen C. H., Smith B. L., Mochly-Rosen D. (1999) Characterization of the binding and phosphorylation of cardiac calsequestrin by ϵ protein kinase C. FEBS Lett. 454, 240–246 [DOI] [PubMed] [Google Scholar]
- 47. Salvatori S., Furlan S., Meggio F. (1994) Dual role of calsequestrin as substrate and inhibitor of casein kinase-1 and casein kinase-2. Biochem. Biophys. Res. Commun. 198, 144–149 [DOI] [PubMed] [Google Scholar]
- 48. Shoshan-Barmatz V., Orr I., Weil S., Meyer H., Varsanyi M., Heilmeyer L. (1996) The identification of the phosphorylated 150/160-kDa proteins of sarcoplasmic reticulum, their kinase, and their association with the ryanodine receptor. Biochim. Biophys. Acta 1283, 89–100 [DOI] [PubMed] [Google Scholar]
- 49. Varsànyi M., Heilmeyer L. (1980) Autocatalytic phosphorylation of calsequestrin. FEBS Lett. 122, 227–230 [DOI] [PubMed] [Google Scholar]
- 50. Ram M. L., Kiarash A., Marsh J. D., Cala S. E. (2004) Phosphorylation and dephosphorylation of calsequestrin on CK2-sensitive sites in heart. Mol. Cell. Biochem. 266, 209–217 [DOI] [PubMed] [Google Scholar]
- 51. Szegedi C., Sárközi S., Herzog A., Jóna I., Varsányi M. (1999) Calsequestrin. More than “only” a luminal Ca2+ buffer inside the sarcoplasmic reticulum. Biochem. J. 337, 19–22 [PMC free article] [PubMed] [Google Scholar]
- 52. Herzog A., Szegedi C., Jona I., Herberg F. W., Varsanyi M. (2000) Surface plasmon resonance studies prove the interaction of skeletal muscle sarcoplasmic reticular Ca2+ release channel/ryanodine receptor with calsequestrin. FEBS Lett. 472, 73–77 [DOI] [PubMed] [Google Scholar]
- 53. Houle T. D., Ram M. L., Cala S. E. (2004) Calsequestrin mutant D307H exhibits depressed binding to its protein targets and a depressed response to calcium. Cardiovasc. Res. 64, 227–233 [DOI] [PubMed] [Google Scholar]
- 54. Helenius A., Aebi M. (2001) Intracellular functions of N-linked glycans. Science 291, 2364–2369 [DOI] [PubMed] [Google Scholar]
- 55. Moremen K. (2002) Golgi α-mannosidase II deficiency in vertebrate systems. Implications for asparagine-linked oligosaccharide processing in mammals. Biochim. Biophys. Acta 1573, 225–235 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.