Abstract
Tissue-specific alternative splicing in the second half of Ig-like domain 3 (D3) of fibroblast growth factor receptors 1–3 (FGFR1 to -3) generates epithelial FGFR1b-FGFR3b and mesenchymal FGFR1c-FGFR3c splice isoforms. This splicing event establishes a selectivity filter to restrict the ligand binding specificity of FGFRb and FGFRc isoforms to mesenchymally and epithelially derived fibroblast growth factors (FGFs), respectively. FGF1 is termed the “universal FGFR ligand” because it overrides this specificity barrier. To elucidate the molecular basis for FGF1 cross-reactivity with the “b” and “c” splice isoforms of FGFRs, we determined the first crystal structure of FGF1 in complex with an FGFRb isoform, FGFR2b, at 2.1 Å resolution. Comparison of the FGF1-FGFR2b structure with the three previously published FGF1-FGFRc structures reveals that plasticity in the interactions of the N-terminal region of FGF1 with FGFR D3 is the main determinant of FGF1 cross-reactivity with both isoforms of FGFRs. In support of our structural data, we demonstrate that substitution of three N-terminal residues (Gly-19, His-25, and Phe-26) of FGF2 (a ligand that does not bind FGFR2b) for the corresponding residues of FGF1 (Phe-16, Asn-22, and Tyr-23) enables the FGF2 triple mutant to bind and activate FGFR2b. These findings taken together with our previous structural data on receptor binding specificity of FGF2, FGF8, and FGF10 conclusively show that sequence divergence at the N termini of FGFs is the primary regulator of the receptor binding specificity and promiscuity of FGFs.
Keywords: Crystal Structure, Fibroblast Growth Factor (FGF), Fibroblast Growth Factor Receptor (FGFR), Receptor Regulation, Surface Plasmon Resonance (SPR), Ligand-Receptor Specificity
Introduction
Fibroblast growth factor (FGF) signaling plays pleiotropic roles in mammalian development and metabolism (1–3). The mammalian FGF family comprises 18 members (FGF1–FGF10 and FGF16–FGF23), which are divided into six subfamilies based on sequence homology and phylogeny. The FGF1 subfamily comprises FGF1 and FGF2; the FGF7 subfamily comprises FGF3, FGF7, FGF10, and FGF22; the FGF4 subfamily comprises FGF4, FGF5, and FGF6; the FGF8 subfamily comprises FGF8, FGF17, and FGF18; the FGF9 subfamily comprises FGF9, FGF16, and FGF20; and the FGF19 subfamily consists of FGF19, FGF21, and FGF23 (4, 5). In some nomenclatures, FGF homologous factors (FHF1–FHF4)6 are considered to form an additional FGF subfamily, namely the FGF11 subfamily, because these proteins share strong sequence homology to other FGFs. Biochemical and structural analyses of FHFs, however, have shown that these proteins are functionally distinct from FGFs (6–8). The FGF1, FGF4, FGF7, FGF8, and FGF9 subfamilies act in a paracrine fashion to direct tissue patterning and organogenesis during embryogenesis. In contrast, the FGF19 subfamily members have very low affinity for heparan sulfate (HS) and hence act in an endocrine fashion (9) to regulate important metabolic activities, including bile acid and lipid metabolism (10–13), glucose homeostasis (14–17), and phosphate and vitamin D homeostasis (18–20).
The paracrine FGFs carry out their diverse functions by binding and activating the FGF receptor (FGFR) family of tyrosine kinase receptors in an HS-dependent fashion (21–23). The endocrine FGF19 subfamily members require klotho co-receptors to bind and activate their cognate FGFRs (24–26). Whether these endocrine FGFs still require HS for signaling remains to be determined. There are four FGFR genes (FGFR1 to FGFR4) that encode single-pass transmembrane receptors composed of an extracellular ligand-binding region consisting of three immunoglobulin (Ig)-like domains (D1, D2, and D3) connected by flexible linker sequences and a cytoplasmic region that harbors the conserved tyrosine kinase domain (27–29). The D2-D3 segment of the ectodomain is necessary and sufficient for ligand binding (30–34). The D1 and D1-D2 linker regions, albeit dispensable for ligand binding, are implicated in receptor autoinhibition because loss of these regions enhances the FGF and HS binding affinity of the D2-D3 region (35, 36). Upon binding of ligand and HS, FGFRs assemble into a 2:2:2 FGF-FGFR-HS symmetric dimer (32, 37, 38) in which the juxtaposed cytoplasmic kinase domains gain sufficient opportunity to transphosphorylate each other on A-loop tyrosines and become activated (39, 40). FGFR kinase activation triggers activation of various downstream signaling pathways, including the RAS-MAPK (41), PI hydrolysis/PKC/Ca2+ (42, 43), PI3K-AKT (44–46), and RAC1/CDC42 (47) signaling pathways (48, 49).
FGF signaling is tightly regulated by spatial and temporal expression of FGFs, FGFRs, and HS cofactors and, most importantly, by means of FGF-FGFR binding specificity. The tissue-specific alternative splicing in the D3 domain of FGFR1–FGFR3 is the main mechanism in the regulation of FGF-FGFR binding specificity (30, 34, 50, 51). In FGFR1–FGFR3, two alternative exons (IIIb and IIIc) code for the second half of D3 and are spliced in a mutually exclusive fashion to the common exon IIIa that encodes the first half of D3 (52–55). This splicing event results in the expression of epithelial “b” isoforms (FGFR1b–FGFR3b) or mesenchymal “c” isoforms (FGFR1c–FGFR3c), increasing the number of principal FGFRs to seven, namely FGFR1c, FGFR1b, FGFR2c, FGFR2b, FGFR3c, FGFR3b, and FGFR4. The D3 alternative splicing restricts the ligand binding specificity of FGFR1b–FGFR3b isoforms to mesenchymally expressed FGFs and that of FGFR1c–FGFR3c isoforms to epithelially expressed FGFs (30, 34). For example, FGF2 binds with comparably high affinity to both FGFR1c and FGFR2c but does not bind the remaining five FGFRs (50, 56–58). FGF8b binds FGFR1c–FGFR3c and FGFR4 but does not recognize the “b” isoforms (30, 50, 51, 56, 59). FGF10 and FGF7 are the most specific FGF ligands and activate only FGFR2b (50, 51). FGF1 is termed the “universal FGFR ligand” because it overrides the specificity barrier set by alternative splicing and binds equally well to both “b” and “c” isoforms of FGFRs (50, 51).
To date, crystal structures of seven different FGF-FGFR complexes have been solved. These structures include three featuring FGF1 (FGF1-FGFR1c (PDB code 1EVT), FGF1-FGFR2c (PDB code 1DJS), and FGF1-FGFR3c (PDB code 1RY7) (31, 33, 35)), two featuring FGF2 (FGF2-FGFR1c (PDB codes 1CVS and 1FQ9) and FGF2-FGFR2c (PDB code 1EV2) (31, 32)), one featuring FGF8b (FGF8b-FGFR2c (PDB code 2FDB) (30)), and one with FGF10 (FGF10-FGFR2b (PDB code 1NUN) (34)). Analysis of these crystal structures has revealed that alternative splicing controls FGF-FGFR binding specificity by switching the primary sequences of key FGF binding sites in the second half of D3, including the βC′-βE and βF-βG loops and βF and βG strands (30, 31, 34). More specifically, analysis of the FGF2-FGFR1c and FGF2-FGFR2c structures has shown that specific hydrogen bonds between Gln-65 in the β4 strand of FGF2, a residue that is unique to FGF2, and an aspartic acid in the βC′-βE loops of FGFR1c and FGFR2c (Asp-320 in FGFR1c and Asp-321 in FGFR2c) restrict binding of FGF2 to these two FGFRc isoforms (31, 32). Similarly, analysis of the FGF10-FGFR2b structure has identified the highly specific hydrogen bonding between Asp-76 in the N terminus of FGF10 with Ser-315 in the βC′-βE loop of FGFR2b as the main determinant of the tight specificity of FGF10 for the FGFR2b isoform (34). Finally, analysis of the FGF8b-FGFR2c structure has shown that the hydrophobic contacts between Phe-93 (in the β4-β5 loop of FGF8b), Val-36 and Phe-32 (in the N terminus of FGF8b), and residues in the βF and βG strands of receptor narrow the binding specificity/promiscuity of FGF8b to FGFR1c-FGFR3c and FGFR4 (30).
In contrast, the structural basis for indiscriminate binding of FGF1 to both “b” and “c” isoforms of a given FGFR has remained elusive. In the FGF1-FGFR1c structure, the alternatively spliced βC′-βE loop in D3 had poor electron density and could not be modeled. This observation was attributed to the lack of contacts between this inherently flexible loop and FGF1 (31). Because contacts between FGF and the alternatively spliced βC′-βE loop of receptor have been established to comprise one of the key determinants of FGF-FGFR specificity, the observation that FGF1 did not engage this loop to bind FGFR was harmonious with the promiscuity of this ligand toward both isoforms. Hence, it was proposed that indiscriminate binding of FGF1 to both alternative splice isoforms of FGFRs is due to the fact that FGF1 does not rely on the βC′-βE loop for FGFR binding (31). In the FGF1-FGFR2c structure (Protein Data Bank code 1DJS), however, the βC′-βE loop is ordered and makes specific contacts with the core region of FGF1 (33), arguing against the validity of our proposed model.
The lack of a crystal structure of FGF1 bound to an FGFRb isoform has been a critical barrier to our comprehension of the molecular basis for the unique ability of FGF1 to break the specificity barrier set by alternative splicing. Hence, in this report, we describe the first crystal structure of FGF1 complexed with an FGFRb isoform, FGFR2b, at 2.1 Å resolution. We also revisit the role of the disordering of the alternatively spliced βC′-βE loop of receptor in the promiscuity of FGF1 by solving the crystal structure of FGF1-FGFR1c in a new crystal lattice at 2.6 Å resolution. By comparing the newly solved FGF1-FGFR2b structure with FGF1-FGFRc structures, we show that the remarkable ability of N-terminal residues of FGF1 to adapt to D3 of both “b” and “c” splice isoforms of FGFRs underlies the ability of FGF1 to cross-react with all isoforms of FGFR.
EXPERIMENTAL PROCEDURES
Protein Expression and Purification
The cDNA fragment encoding full-length wild type human FGF1 (residues 1–155) was subcloned into the expression vector pET30a using the cloning sites NdeI and HindIII. The cDNA for full-length wild type human FGF2 (residues 1–155) was subcloned into pET30a using the NcoI and XhoI cloning sites such that a 47-residue-long N-terminal tag, including a hexahistidine tag, was fused in frame to the N terminus of FGF2. The cDNA fragments for the ligand-binding region (D2-D3 domains) of wild type human FGFR1c (residues 142–365), wild type human FGFR2b (residues 140–369), and wild type human FGFR2c (residues 149–368) were subcloned into pET28a using the NcoI and HindIII cloning sites. The QuikChange XL site-directed mutagenesis kit (Stratagene) was used to sequentially introduce the F26Y, H25N, and G19F mutations into the hexahistidine-tagged wild type FGF2 expression construct; the A172F or P253R gain-of-function mutations, or the E323A or I257A/Y281A loss-of-function mutations into the expression construct for the ligand-binding region of wild type FGFR2b; and the R251Q loss-of-function mutation into the expression construct for wild type FGFR2c ligand-binding region (supplemental Fig. 1). FHF1b and the N-terminally truncated FGF1 (FGF1(21–155)) were expressed and purified as described previously (6, 8, 31, 35).
Competent BL21 DE3 Escherichia coli cells were transformed with expression constructs for FGFs and FGFR ligand-binding regions and were cultured at 37 °C to an A600 of ∼0.5 (Beckman DU530 UV-visible spectrometer) and induced with 1 mm isopropyl 1-thio-β-d-galactopyranoside for 4 h. Cell pellets were lysed in 150 mm NaCl, 10% glycerol, 25 mm Hepes-NaOH, pH 7.5, 5 mm EDTA, and 625 μm PMSF using a French press (Thermo Spectronic). The soluble lysate fraction containing FGF1 or FGF2 protein was loaded onto a heparin affinity column, and ligands were eluted with a step gradient ranging from 500 mm to 2 m NaCl in 25 mm Hepes-NaOH, pH 7.5. Fractions containing FGF1 or FGF2 were pooled, diluted to 200 mm NaCl, loaded onto a cation exchange column (Source S, Amersham Biosciences), and eluted with a continuous gradient ranging from 200 mm NaCl to 700 mm NaCl in 25 mm Hepes-NaOH, pH 7.5. Purified FGF1 and FGF2 proteins were aliquoted and then stored at −80 °C until use.
In contrast to FGFs, the FGFR ligand-binding regions were expressed as insoluble proteins. Inclusion bodies containing the misfolded FGFR ligand-binding regions were isolated and solubilized in 6 m guanidinium hydrochloride, 20 mm EDTA, 20 mm DTT, and either 50 mm Hepes-NaOH, pH 7.5 (for FGFR2c and FGFR2b) or 50 mm Tris-HCl, pH 7.5 (for FGFR1c). The solubilized FGFR proteins were dialyzed at 4 °C against a 22.5 mm Hepes-NaOH or Tris-HCl, pH 7.5, buffer containing 150 mm NaCl, 7.5% glycerol, 1 mm l-cysteine. The dialysis buffer was then exchanged, and the FGFR proteins were dialyzed for a further 48 h at 4 °C against 100 mm NaCl, 5% glycerol, and 22.5 mm Hepes-NaOH or Tris-HCl, pH 7.5. Refolded FGFRs were loaded onto a heparin affinity column, eluted with 1 m NaCl in 25 mm Hepes-NaOH, pH 7.5, and then further purified by size exclusion chromatography (HiLoad 16/60 Superdex 75, Amersham Biosciences) using 25 mm Hepes-NaOH, pH 7.5, buffer containing 1 m NaCl. FGFRs were also aliquoted and stored at −80 °C until use. The expression and purification of mutated FGFs and FGFR ligand-binding regions were carried out as for the wild type counterparts. Protein concentrations were measured under denaturing conditions at a wavelength of 280 nm using the NanoDrop ND-1000 spectrophotometer (Nanodrop Technologies, Wilmington DE). Extinction coefficients for proteins were derived using the ProtParam software (ExPASy Proteomics Server).
Crystallization, Data Collection, and Structure Determination
Purified FGFR1c and FGFR2b ligand-binding regions were mixed with a slight molar excess of FGF1(21–155) or full-length FGF1, respectively. The resulting complexes were concentrated to about 4 mg/ml using a Centricon 10 concentrator and applied onto a Superdex 200 size exclusion column equilibrated in 25 mm Hepes-NaOH, pH 7.5, containing 1 m NaCl. Fractions containing 1:1 FGF-FGFR complexes were pooled, concentrated to ∼60–70 mg/ml, and stored at 4 °C until use. Prior to crystallization, the complexes were diluted to 10 mg/ml using 25 mm HEPES, pH 7.5, which brought down the salt concentrations to ∼150 mm NaCl. The FGF1(21–155)-FGFR1c complex was supplemented with an equimolar amount of fully sulfated heparin octasaccharide prior to crystallization. The FGF-FGFR complexes were crystallized by mixing 1.5 μl of the complex with 1.5 μl of crystallization buffer using the hanging drop vapor diffusion method. The FGF1-FGFR2bP253R complex crystallized in 0.1 m Hepes, pH 7.5, 22% monomethyl ether PEG 5000, and 0.2 m ammonium sulfate. The FGF1-FGFR2bA172F complex crystallized in 0.1 m Hepes, pH 7.5, 20% PEG 4000, and 0.2 m ammonium sulfate. The FGF1-FGFR1c complex crystallized in 0.1 m Tris, pH 8.5, 15% PEG 4000, and 0.1 m ammonium sulfate. Crystals were transferred into cryoprotectant solution composed of the mother liquor and 15% glycerol and were flash-frozen under a nitrogen stream at −196 °C. Diffraction data were collected at NSLS beamline X4A and processed using HKL2000 (60).
The FGF1-FGFR1c complex crystals belong to space group P1 with unit cell dimensions a = 53.324, b = 53.421, c = 80.463, α = 106.441, β = 106.396, and γ = 94.436. There are two FGF1-FGFR1c complexes per asymmetric unit in the crystal with a solvent content of 50.4%. The dimensions of the “a” and “b” axes as well as the α and β angles of this P1 unit cell are nearly equal, implying that the crystal may be in monoclinic space group C2 rather than P1. However, the distortion index for the C2 space group was 1.2%, and accordingly the diffraction data could not be integrated and scaled in C2.
Crystals of the FGF1-FGFR2bWT complex diffracted poorly, and attempts to improve their diffraction failed. In contrast, the crystals of the FGF1-FGFR2bA172F and FGF1-FGFR2bP253R mutant complexes diffracted to 2.2 and 2.1 Å, respectively. The FGF1-FGFR2bA172F and FGF1-FGFR2bP253R crystal structures are in space groups P32 and P212121 and have two and one FGF1-FGFR2b complex per asymmetric unit, respectively. The solvent contents of the FGF1-FGFR2bA172F and FGF1-FGFR2bP253R crystals are 55.8 and 51.9%, respectively. All three crystal structures were solved by the molecular replacement program AMORE (61) using the published FGF1-FGFR1c (PDB code 1EVT) (31) and FGF10-FGFR2b (PDB code 1NUN) (34) structures as search models. The program O (62) was used for model building into the 2Fo − Fc and Fo − Fc electron density maps, and CNS was used for rigid body, positional, and B factor refinements (63). For the FGF1-FGFR1c and FGF1-FGFR2bA172F structures, tight noncrystallographic symmetry restraints were imposed throughout the refinement for the backbone atoms of FGF1, FGFR D2, and FGFR D3 domains. The refined model for the FGF1-FGFR1c structure consists of two FGF1 molecules (residues 21–153), two FGFR1c molecules (residues 147–359), one heparin octasaccharide (of which only six monosaccharide units are modeled), and 32 water molecules. The FGF1-FGFR2bP253R structure in space group P212121 contains one FGF1 molecule (residues 10–154), one FGFR2bP253R molecule (residues 151–361), 161 water molecules, and three sulfate ions. The FGF1-FGFR2bA172F structure in space group P32 contains two FGF1 molecules (residues 19–153), two FGFR2bA172F molecules (residues 151–359), 120 water molecules, and five sulfate ions. The FGF1-FGFR2bP253R structure in the P212121 space group was used to make the structural figures.
Surface Plasmon Resonance (SPR) Spectroscopy
Real-time biomolecular FGF-FGFR interactions were analyzed with a BIAcore 2000 system (GE Healthcare) in HBS-EP buffer (0.01 m Hepes, pH 7.4, 150 mm NaCl, 3 mm EDTA, 0.005% Surfactant P20) at 25 °C. FGF or FGFR proteins were immobilized onto CM5 biosensor chips using an amine coupling kit (GE Healthcare). Briefly, the carboxymethyl groups of a CM5 chip were activated with a freshly prepared mixture of 0.05 m N-hydroxysuccinimide and 0.2 m N-ethyl-N-(dimethyaminopropyl) carbodiimide. Next, FGF or FGFR proteins were immobilized to 0.07 pmol of protein/mm2 density by passing them over the chip in 150 mm NaCl solutions buffered by 50 mm PIPES, pH 6.5 (for FGFR2 ligand-binding regions), 10 mm sodium acetate pH 5.5 (for FGF1), or HBS-EP buffer (for FHF1b). Last, the unreacted sites on the chip surface were blocked using 1 m ethanolamine-HCl, pH 8.5. Next, varying concentrations (0.8–800 nm) of analyte (FGF or FGFR) were prepared in HBS-EP buffer and then passed over the chip at 50 μl/min for 180 s to observe the association phase of the FGF-FGFR interaction. After each analyte injection, HBS-EP buffer was passed over the chip for 180 s to monitor the dissociation phase. Following the dissociation phase, the sensor chip surface was regenerated with 2.5 m NaCl, 5 mm Hepes, pH 7.5.
To evaluate the binding of mutated FGF2 ligands to FGFR2b and FGFR2c, the FGFR2bWT and FGFR2cWT ligand-binding regions were immobilized on a chip, and FGF1, FGF2, and the FGF2 mutants were passed over the chip. To control for nonspecific binding, the FGFR2cR251Q mutant was coupled to the control flow channel of the chip. The R251Q mutation maps to the linker region between D2 and D3 of FGFR2c, and we have previously shown that this mutation results in a major loss of ligand binding because it eliminates key hydrogen bonds between FGFR and FGF (64). To examine binding of mutated FGFR2b ligand-binding regions to FGF1, full-length FGF1 was immobilized on a chip, and FGFR2bWT, FGFR2bI257A/Y281A, and FGFR2bE323A were passed over the chip. As a negative control, FHF1b was immobilized on the chip. FHF1b, despite its high homology to FGFs, does not bind to FGFRs and hence is an ideal negative control for studies of FGF-FGFR interactions (8).
To process the sensorgrams, the reference responses from either the FGFR2cR251Q or FHF1b control flow cells were subtracted from the experimental flow cells for each analyte injection. Equilibrium binding responses were plotted against the concentrations of analyte, and from the fitted saturation binding curves, the KD was derived. The sensorgrams of FGF binding to FGFR have a biphasic nature that is most apparent in the FGF1-FGFR2b interaction. To avoid introducing the error of the second phase into the equilibrium analysis, values for the steady state calculation were taken at t = 10–15 s.
Size Exclusion Chromatography
3 μm solutions of a 1:1 FGF/FGFR mixture and, as controls, FGF and wild type FGFR alone were applied onto a HiLoad 16/60 Superdex 200 size exclusion column (Amersham Biosciences) mounted on an ÄKTAPurifier system (GE Healthcare). Proteins were eluted at 1 ml/min in 1 m NaCl, 25 mm Hepes, pH 7.5, buffer. Ligand-receptor complex formation was judged by the appearance of a new peak with an earlier retention time than receptor alone and by the reduction in the height of the ligand peak. Ligand-receptor complex formation was also confirmed by analyzing fractions by SDS-PAGE. The size exclusion column was calibrated with bovine serum albumin (69 kDa), ovalbumin (44 kDa), carbonic anhydrase (29 kDa), ribonuclease A (14 kDa), and aprotinin (7 kDa). The column volume was determined using acetone.
BaF3 Cell Proliferation Assay
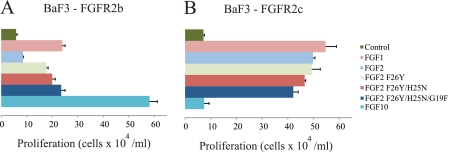
BaF3 cells overexpressing FGFR2b and FGFR2c have been described previously (65). Cells were maintained in RPMI 1640 medium supplemented with 10% fetal bovine serum (Mediatech, Manassas, VA), 5 ng/ml murine recombinant interleukin-3 (R&D Systems, Minneapolis, MN), 2 mm l-glutamine, 100 IU/ml penicillin, 100 μg/ml streptomycin, 50 μm β-mercaptoethanol, and 1200 μg/ml G418. BaF3 cells expressing FGFR2b or FGFR2c were washed three times with RPMI to remove IL-3. Cells were plated at 2 × 104 cells/well in triplicate in a 96-well plate in medium containing 100 ng/ml wild type or mutated FGF ligand, 5 μg/ml heparin, and 1200 μg/ml G418 and cultured for 48 h. Cell counts from triplicate wells were determined using a hemacytometer and averaged. Student's paired t test was used for statistical analysis. The proliferation assay was repeated seven times on different days, and the data presented in Fig. 4, A and B, are representative of one of the seven experiments.
FIGURE 4.
Substitution of three N-terminal residues of FGF2 for the analogous ones in FGF1 confers upon FGF2 the ability to induce proliferation of BaF3 cells ectopically expressing FGFR2b. A, comparison of the ability of FGF1, FGF2, and FGF2 mutants to induce proliferation of FGFR2b-expressing BaF3 cells. FGF10, a FGFR2b-specific ligand, was used as a positive control. Experiments were done in triplicate, and error bars reflect one S.D. The data presented are a representative example of seven independent experiments. As expected, FGF2 induced only minimal proliferation of cells expressing FGFR2b. Compared with wild type FGF2, the FGF2F26Y, FGF2H25N/F26Y, and FGF2G19F/H25N/F26Y mutants elicit a 2.1-, 2.4-, and 2.8-fold greater response, respectively. The increase in cell proliferation induced by FGF2F26Y, FGF2H25N/F26Y, or FGF2G19F/H25N/F26Y relative to FGF2 was statistically significant (p < 0.01). B, comparison of the ability of FGF1, FGF2, and FGF2 mutants to induce proliferation of FGFR2c-expressing BaF3 cells. The FGF2 mutants retain the ability to induce proliferation of FGFR2c-expressing cells. As expected, FGF10 fails to promote proliferation of FGFR2c-expressing cells.
RESULTS
Crystal Structure of FGF1 in Complex with FGFR2b Ligand-binding Region
To elucidate the molecular mechanism underlying the unique ability of FGF1 to override the FGFR specificity barrier set by alternative splicing, we chose to solve the crystal structure of FGF1 complexed with the ligand-binding region of FGFR2b. Crystals of full-length wild type FGF1 (FGF1(1–155)) in complex with the ligand-binding region of wild type FGFR2b encompassing D2, D3, and the D2-D3 linker (residues 158–369) (supplemental Fig. 1) diffracted poorly, and attempts to improve their diffraction failed. To overcome this problem, 1:1 complexes of FGF1 with mutated FGFR2b constructs harboring either the P253R or A172F gain-of-function mutations were generated. The P253R FGFR2 mutation maps to the D2-D3 linker region, and we have previously shown that this mutation enhances FGF-FGFR binding, thereby improving crystal growth/diffraction (30, 66, 67). The A172F mutation maps to the receptor-receptor interface in the FGF-FGFR 2:2 dimer, and it also improves crystal growth/diffraction by introducing FGFR-FGFR contacts (68). The FGF1-FGFR2bP253R and FGF1-FGFR2bA172F complexes crystallized in the orthorhombic (P212121) and hexagonal (P32) space groups, respectively (Table 1). The crystal structures of the complexes were solved by molecular replacement and have been refined to 2.1 Å (in P212121, PDB code 3OJM) and 2.2 Å (in P32, PDB code 3OJ2) resolution. The asymmetric units of the P212121 and P32 crystal forms contain one and two FGF1-FGFR2b complexes, respectively. Data collection and refinement statistics are summarized in Table 1. Superimposition of the Cα traces of residues 20–151 of FGF1 from the FGF1-FGFR2b complexes in the two crystal forms gives a root mean square deviation of only 0.3 Å (supplemental Fig. 2) and shows that the first ordered FGF1 residues in the complex from the P212121 and P32 crystal forms are Thr-10 and Pro-19, respectively (Fig. 1A and supplemental Fig. 2). The observed ordering of the additional nine residues at the FGF1 N-terminal region in the P212121 crystal form is due to the fact that these residues interact with receptor D2 (Fig. 1A). However, because these contacts are not seen in the P32 crystal form, they were deemed crystal-favored and will not be discussed. Aside from the contacts between this distal N-terminal region of FGF1 and the FGFR2b D2 domain, the remaining interactions at the interface between FGF1 and FGFR2b are essentially identical between the two crystal forms. Significantly, in both crystal forms, the alternatively spliced βC′-βE loop in receptor D3, which is a key determinant of the ligand binding specificity of FGFR, is ordered and is engaged by the ligand (Fig. 1A). Hence, our two new FGF1-FGFR2b structures, taken together with our previously published FGF1-FGFR3c structure (PDB code 1RY7) (Fig. 1D) (35) and the FGF1-FGFR2c structure solved by the Hendrickson laboratory (PDB code 1DJS) (Fig. 1C) (33), argue against our initial hypothesis that FGF1 achieves indiscriminate binding to both isoforms by not utilizing the alternatively spliced βC′-βE loop of receptor D3 (31).
TABLE 1.
Data collection and refinement statistics
| FGF1-FGFR2bP253R | FGF1-FGFR2bA172F | FGF1-FGFR1c-heparin octasaccharide | |
|---|---|---|---|
| Data collection | |||
| Space group | P212121 | P32 | P1 |
| Unit cell parameters | |||
| a | 66.740 | 77.945 | 53.324 |
| b | 72.349 | 77.945 | 53.421 |
| c | 91.912 | 137.555 | 80.463 |
| α | 90 | 90 | 106.441 |
| β | 90 | 90 | 106.396 |
| γ | 90 | 120 | 94.436 |
| Solvent content (%) | 51.9 | 55.8 | 50.4 |
| Wavelength (Å) | 0.92018 | 1.277 | 0.97912 |
| Resolution (Å) | 30.0-2.1 | 30.0-2.2 | 50-2.6 |
| Reflections (total/unique) | 330055/27015 | 162990/47189 | 90699/24626 |
| Completeness (%) | 99.5 (98.6)a | 99.3 (99.6)b | 97.8 (91.0)c |
| Rsymd (%) | 5.4 (10.3) | 4.8 (26.5) | 6.4 (32.7) |
| Signal (〈I/σI〉) | 40.5 (35) | 25.4 (5.9) | 16.6 (5.5) |
| Redundancy | 12.2 (10.1) | 3.5 (3.4) | 3.7 (3.5) |
| Refinement | |||
| Resolution (Å) | 25-2.1 | 25-2.2 | 25-2.6 |
| Reflections (total/test) | 26221/2605 | 45665/2287 | 23714/2349 |
| Rcryst/Rfreee (%) | 22.9/26.3 | 26.2/29.4 | 25.1/30.9 |
| Root mean square deviations | |||
| Bond (Å) | 0.005 | 0.008 | 0.008 |
| Angle (degrees) | 1.35 | 1.42 | 1.49 |
| B-factorf (Å2) | 1.05 | 0.888 | 1.2 |
| No. of atoms | |||
| Protein | 2774 | 5372 | 5329 |
| Sulfate | 12 | 20 | 0 |
| Water | 161 | 120 | 32 |
| Heparin octasaccharide | 0 | 0 | 105 |
| Average B-factor (Å2) | |||
| Protein | 25 | 48 | 45 |
| Main chain | 24 | 48 | 45 |
| Side chain | 26 | 48 | 46 |
| Water | 27 | 40 | 27 |
| Sulfate | 44 | 88 | NAg |
| Heparin octasaccharide | NA | NA | 98 |
| Ramachandran plot | |||
| Residues in most favored region | 271 (89.4%) | 516 (88.4%) | 477 (81.1%) |
| Residues in additional allowed region | 31 (9.9%) | 65 (11.1%) | 96 (16.3%) |
| Residues in generously allowed region | 1 (0.3%) | 3 (0.5%) | 8 (1.4%) |
| Residues in disallowed region | 1 (0.3%) | 0 | 7 (1.2%) |
| PDB code | 3OJM | 3OJ2 | 3OJV |
a Values in parentheses in this column are for the higest resoluton shell: 2.18–2.1 Å.
b Values in parentheses in this column are for the highest resoluton shell: 2.28–2.20 Å.
c Values in parentheses in this column are for the highest resoluton shell: 2.69–2.6 Å.
d Rsym = 100 × ΣhklΣI|Ii(hkl) − I(hkl)|/ΣhklΣiIi(hkl).
e Rcryst/Rfree = 100 × Σhkl‖Fo(hkl)| − |Fc(hkl)‖/Σhkl|Fo(hkl)|.
f For bonded protein atoms.
g NA, not applicable.
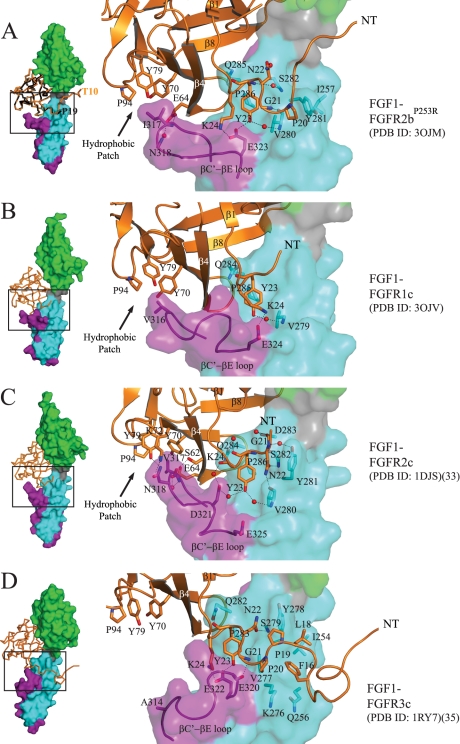
FIGURE 1.
Plasticity at the FGF1-D3 interface. A, left, Cα trace of FGF1 from the FGF1-FGFR2bP253R structure in space group P212121 (PDB code 3OJM) (in orange) is superimposed onto that of FGF1 from the FGF1-FGFR2bA172F structure in space group P32 (PDB code 3OJ2) (in black). The first ordered N-terminal residue in each FGF1 molecule is labeled. FGFR2b is shown as a surface, with D2 colored green, D2-D3 linker in gray, the common region of D3 in cyan, and the alternatively spliced portion of D3 in purple. This color scheme for the receptor is used throughout the paper including in supplemental Figs. 3, 4, 5, and 7. The FGF1-FGFR2bP253R structure in space group P212121 (PDB code 3OJM) is used for comparison with other FGF1-FGFR structures. Right, a close-up view of selected N-terminal and hydrophobic patch contacts of FGF1 with D3 of receptor in the FGF1-FGFR2bP253R crystal structure. FGF1 is shown as a ribbon diagram, and selected residues are rendered as sticks, with oxygen colored red and nitrogen colored blue. Water molecules are indicated as red spheres. This color scheme for atoms is used throughout the paper including in supplemental Figs. 4, 5, and 7. The location of the hydrophobic patch between FGF core and the βC′-βE loop of receptor is indicated by an arrow. B, the new FGF1-FGFR1c structure (PDB code 3OJV) displaying an ordered βC′-βE loop. Selected N-terminal and hydrophobic patch contacts of FGF1 with D3 of receptor are shown. C, selected N-terminal and hydrophobic patch contacts of FGF1 with D3 of receptor in the FGF1-FGFR2c crystal structure (PDB code 1DJS). D, selected N-terminal and hydrophobic patch contacts of FGF1 with D3 of receptor in the FGF1-FGFR3c structure (PDB code 1RY7). Note that Ala-314 in D3 of FGFR3c cannot make a strong hydrophobic contact with Pro-94, Tyr-79, and Tyr-70 in the FGF1 core, and as a result, the βC′-βE loop falls away from the core of the ligand.
In New FGF1-FGFR1c Structure, Alternatively Spliced βC′-βE Loop Is Ordered and Interacts with FGF1
Because in both of our newly solved FGF1-FGFR2b structures FGF1 engages the βC′-βE loop of the receptor, it became imperative to reassess the role of βC′-βE loop disordering in our previous FGF1-FGFR1c structure (PDB code 1EVT) (31). To this end, we decided to solve the crystal structure of FGF1 bound to FGFR1c in a crystal lattice different from that of our first FGF1-FGFR1c structure (PDB code 1EVT) (31). For recrystallization of the FGF1-FGFR1c complex, we used the same N-terminally truncated FGF1 protein (FGF1(21–155)), which we originally used to generate PDB entry 1EVT. FGF1(21–155) and full-length FGF1 bind FGFR1c with identical affinities (69). To encourage crystallization of the FGF1-FGFR1c complex in a crystal lattice different from that of 1EVT, we intentionally supplemented the complex in a 1:1 ratio with a homogeneously sulfated heparin in order to induce formation of a 2:2:2 FGF1-FGFR1c-heparin octasaccharide dimer. The crystal structure of the FGF1-FGFR1c-octasaccharide complex was solved using molecular replacement with 1EVT as the search model. The asymmetric unit of this new FGF1-FGFR1c structure (PDB code 3OJV) contains two complexes, and the structure has been refined to 2.6 Å resolution. Data collection and refinement statistics are given in Table 1. Importantly, although both 3OJV and 1EVT are in triclinic space group P1, the crystal lattice contacts are very different between these two structures (supplemental Fig. 3, A and B). The interface between FGF1 and FGFR1c in 3OJV is similar to that in our previous FGF1-FGFR1c structure (PDB code 1EVT) (31) with the notable difference that the electron density for the alternatively spliced βC′-βE loop in D3 was sufficient to confidently build the entire loop (Fig. 1B). The loop, however, has a high average temperature factor of 80 Å2 (compared with an average temperature factor of 45 Å2 for the whole structure), reflecting the fact that it makes only very few contacts with the ligand (Fig. 1B). In contrast, FGF1 interacts more with the βC′-βE loop in the FGF1-FGFR2c (Fig. 1C) and FGF1-FGFR2b (Fig. 1A) structures, accounting for the lower average temperature factors of the βC′-βE loop in the FGF1-FGFR2c (31 Å2) and FGF1-FGFR2b structures (22 Å2). Based on our new FGF1-FGFR2b and FGF1-FGFR1c structures, we can conclude that the promiscuity of FGF1 toward FGFR isoforms cannot be attributed to the fact that FGF1 does not rely on the alternatively spliced βC′-βE loop of FGFR for binding as we initially proposed (31).
Plasticity at Interface between FGF1 N Terminus and FGFR D3
Comparison of our newly solved FGF1-FGFR2b and FGF1-FGFR1c structures with previous FGF1-FGFR2c (PDB code 1DJS) (33) and FGF1-FGFR3c (PDB code 1RY7) (35) structures reveals that cross-reactivity of FGF1 with both “b” and “c” splice isoforms of FGFRs stems from the fact that the N terminus of FGF1 has a remarkable ability to adapt to D3 of either of the two FGFR splice isoforms (Fig. 1, A–D). Notably, the nine N-terminal residues (Phe-16 to Lys-24) preceding the β-trefoil core region of FGF1 (Fig. 1, A–D) engage in highly variable interactions with D3 of FGFR across different FGF1-FGFR structures. FGF1 employs a different number of residues from this N-terminal region to engage the C-terminal end of the alternatively spliced βC′-βE loop (the most divergent region in D3) and the constant region of D3 in different FGF1-FGFR complexes (Fig. 1, A–D). Moreover, the same residue from this N-terminal region of FGF1 interacts differently with D3 of receptor in different FGF1-FGFR complexes. Starting with Lys-24 and moving N-terminally toward Phe-16, the contacts between FGF1 and D3 are as follows. Lys-24 of FGF1 makes hydrogen bonds with a highly conserved glutamate at the C-terminal end of the βC′-βE loop of the receptor in FGF1-FGFR1c (Glu-324), in FGF1-FGFR2b (Glu-323), and in FGF1-FGFR3c (Glu-322) but not in FGF1-FGFR2c (Glu-325) (Fig. 1, A–D). The hydroxyl group of Tyr-23 directly hydrogen-bonds with the carboxylate side chain of this conserved glutamate of receptor in FGF1-FGFR2b and FGF1-FGFR3c but not in our new FGF1-FGFR1c structure or in FGF1-FGFR2c (Fig. 1, A–D). Tyr-23 of FGF1 also engages in a water-mediated hydrogen bond with a backbone amide of an FGFR-invariant valine (Val-280 in FGFR2b) in the βB′ strand of FGFR in all structures except FGF1-FGFR3c. In addition to these receptor-specific contacts, in all four FGF1-FGFR structures, Tyr-23 engages in conserved hydrophobic contacts with a FGFR-invariant proline in the βB′-βC loop and an FGFR-invariant valine (Val-280 in FGFR2b) in the βB′ strand. Moreover, the backbone atoms of Tyr-23 make hydrogen bonds with a highly conserved glutamine in the βB′-βC loop of the receptor in all four structures (Fig. 1, A–D). As for Asn-22 of FGF1, in FGF1-FGFR2c and FGF1-FGFR3c, its side chain directly hydrogen-bonds to the backbone atoms of D3 of receptor, whereas in FGF1-FGFR2b, Asn-22 makes a water-mediated hydrogen bond with D3. In the FGF1-FGFR1c complex, Asn-22 does not participate in receptor binding at all (Fig. 1, A–D). In the FGF1-FGFR2b structure, the backbone amide of Gly-21 of FGF1 makes a hydrogen bond to the backbone carbonyl oxygen of Val-280 of receptor (Fig. 1A). However, in FGF1-FGFR2c, the carbonyl oxygen of Gly-21 engages in a water-mediated hydrogen bond with the backbone carbonyl oxygen of Asp-283 of receptor (Fig. 1C). In FGF1-FGFR3c, the backbone amide of Gly-21 directly hydrogen-bonds to the backbone of Glu-320 of receptor (Fig. 1D), whereas in our new FGF1-FGFR1c structure, Gly-21 is not involved in receptor binding (Fig. 1B). Last, in the FGF1-FGFR3c structure, Phe-16 and Leu-18 of FGF1 engage in hydrophobic contacts with Ile-254 and Tyr-278 and in van der Waals contacts with Gln-256 and Lys-276 of receptor that are not seen in the other three FGF1-FGFR complexes (Fig. 1D). Hence, based on our detailed structural analysis, plasticity in the interactions of residues Phe-16 to Lys-24 at the FGF1 N terminus with FGFRs imparts receptor binding promiscuity to FGF1.
N-terminal Residues of FGF1 Are Principal Determinants of FGF1 Promiscuity
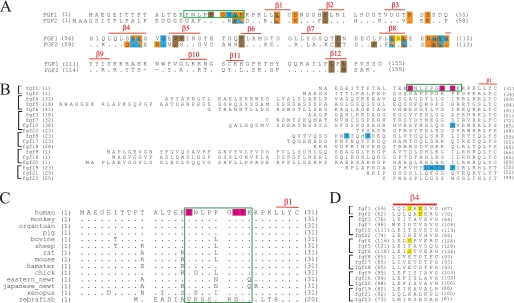
Sequence alignment of the N termini of human FGFs (Fig. 2B) clearly shows that no other FGF would be capable of recapitulating the plastic interactions that this 9-amino acid-long N-terminal region of FGF1 makes with FGFRs in the four different FGF1-FGFR structures. Even FGF2, the other FGF1 subfamily member, shares limited sequence homology with FGF1 at this region. Only four residues in this region are conserved in FGF2 (Fig. 2A). Of the non-conserved residues, Phe-26 of FGF2 cannot provide the direct or water-mediated hydrogen bonds that Tyr-23 of FGF1 makes with Glu-323 of FGFR2b or with Glu-320 of FGFR3c in the FGF1-FGFR2b and FGF1-FGFR3c structures (supplemental Fig. 4 and Fig. 1, A and D). Similarly, Gly-19 of FGF2, which corresponds to Phe-16 of FGF1, is incapable of making the hydrophobic contacts or van der Waals contacts with Ile-254, Gln-256, Lys-276, and Tyr-278 of FGFR3c that are seen in the FGF1-FGFR3c structure (Fig. 1D). His-25 of FGF2 would not be able to make the hydrogen bonds that the corresponding Asn-22 of FGF1 makes with FGFRs in the FGF1-FGFR2b, FGF1-FGFR2c, and FGF1-FGFR3c structures (Fig. 1, A, C, and D). Hence, based on the structural data and sequence analysis, residues 16–24 of FGF1 should play a major role in endowing FGF1 with the ability to bind indiscriminately to either splice isoform of FGFR2.
FIGURE 2.
Sequence and structural comparison of FGF1 and FGF2. A, sequence alignment of human FGF1 and FGF2. FGF1 and FGF2 residues that engage the receptor in the FGF1-FGFR2b (PDB code 3OJM) (see also Fig. 1A) and the FGF2-FGFR2c crystal structures (PDB code 1EV2) (see also supplemental Fig. 4) are colored. Red bars above the sequence indicate the locations of β strands. Residues colored orange participate in direct hydrogen bonds with FGF1 ligand, those colored cyan participate in water-mediated hydrogen bonds, and those colored brown engage in hydrophobic interactions. Yellow indicates a π-cation interaction. Residues participating in more than one type of interaction have multiple colors. Note that although FGF1 and FGF2 share 55% sequence homology, FGF1 is able to bind both “b” and “c” isoforms of FGFR, whereas FGF2 binds only the “c” isoforms of FGFR1 and FGFR2. In A–C, the nine residues at the N terminus of FGF1 that are critical for the indiscriminate binding of FGF1 to both isoforms of FGFR2 are boxed in green. B, sequence alignment of the N termini of the 18 human FGFs. FGFs are grouped into subfamilies, and the location of the β1 strand of FGF1 is indicated. Residues in FGF10, FGF8, and FGF19 that have been shown to be critical to their binding specificity for their cognate receptors are highlighted in cyan. C, a sequence alignment of FGF1 orthologs. In B and C, the three N-terminal residues of FGF1 that when introduced into FGF2 convert FGF2 into a FGF1-like ligand are highlighted in magenta. D, sequence alignment of the β4 strand region of FGFs. Ser-62 and Glu-64 of FGF1, which make plastic contacts with D3 of FGFRs, are highlighted in yellow. Note that Ser-62 and Glu-64 are conserved in FGF4/6 and FGF2, respectively, yet FGF4/6 and FGF2 bind specifically to FGFR1c and FGFR2c.
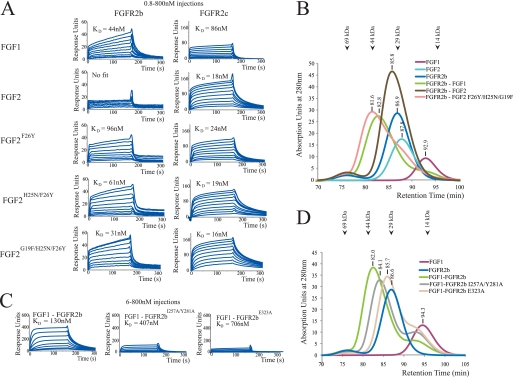
To validate our structural conclusion, we generated three FGF2 mutants in which the N-terminal residues Phe-26, Asn-25, and Gly-19 of FGF2 are progressively replaced with the corresponding residues of FGF1 (Tyr-23, Asn-22, and Phe-16) (supplemental Fig. 1). The ability of these FGF2 mutants, namely FGF2F26Y, FGF2H25N/F26Y, and FGF2G19F/H25N/F26Y, to bind and activate FGFR2b was tested in vitro and in living cells. We reasoned that if these three N-terminal residues of FGF1 were responsible for promiscuous binding of FGF1 to either FGFR2 isoform, then the FGF2 mutants should acquire the ability to bind and activate FGFR2b. SPR spectroscopy and size exclusion chromatography were used to compare binding of FGF1, FGF2, FGF2F26Y, FGF2H25N/F26Y, and FGF2G19F/H25N/F26Y to wild type FGFR2b and FGFR2c ligand-binding regions in vitro. For SPR studies, the ligand-binding regions of wild type FGFR2b and FGFR2c were coupled to biosensor chips, and various concentrations of FGF1, FGF2, and FGF2 mutants were passed over the chips. Consistent with the well known promiscuity of FGF1 toward either of the FGFR isoforms, the SPR data show that FGF1 binds with comparable affinity to both FGFR2c and FGFR2b (Fig. 3A). Similarly, in agreement with our structural data, FGF2 binds with high affinity to FGFR2c and exhibits negligible binding to FGFR2b (Fig. 3A). Interestingly, the affinity of FGF2 for FGFR2c is nearly 5-fold greater than that of FGF1 for FGFR2c (KD of 18 nm versus 86 nm; Fig. 3A), emphasizing the specificity of FGF2 for FGFR2c. The FGF2F26Y, FGF2H25N/F26Y, and FGF2G19F/H25N/F26Y mutants bound to FGFR2b with progressively increased affinity (Fig. 3A). In fact, FGF1 and the FGF2G19F/H25N/F26Y mutant bound FGFR2b with comparable affinity (KD of 44 nm versus 31 nm; Fig. 3A), demonstrating that residues 16, 22, and 23 of FGF1 have imparted to FGF2 the ability to bind FGFR2b (Fig. 3A). All three FGF2 mutants bound to FGFR2c with the same affinity as their parent wild type FGF2 ligand. Size exclusion chromatographic analysis of 1:1 mixtures of FGF1, FGF2, and the FGF2G19F/H25N/F26Y mutant with the ligand-binding region of FGFR2b validates the SPR data (Fig. 3B). The FGF2G19F/H25N/F26Y mutant clearly forms a stable complex with FGFR2b in solution and elutes at a retention time comparable with that of the FGF1-FGFR2b complex (Fig. 3B). In contrast and as expected, wild type FGF2 failed to form a complex with the FGFR2b ligand-binding region. These in vitro data show that residues 16–24 of FGF1 are the main determinants of the ability of FGF1 to override the D3 specificity barrier mediated by alternative splicing (Fig. 3, A and B).
FIGURE 3.
The plasticity in the interactions of FGF1 N-terminal residues with D3 of FGFR2 underlies the promiscuous binding of FGF1 to either isoform of FGFR2. A, SPR sensorgrams of binding of FGF1, FGF2, and mutated FGF2 proteins to FGFR2b and FGFR2c ligand-binding regions immobilized on a biosensor chip. B, size exclusion chromatography analysis of binding of FGF1, FGF2, and FGF2G19F/H25N/F26Y to FGFR2b ligand-binding region. Retention times of the protein peaks for ligand, receptor, and ligand-receptor complex are provided. The retention times of molecular size standards are also indicated with arrows at the top of the chromatogram overlay. C, SPR sensorgrams of binding of FGFR2b, FGFR2b1257A/Y281A, and FGFR2bE323A ligand-binding regions to FGF1 immobilized on a biosensor chip. D, size exclusion chromatography analysis of FGF1 binding to FGFR2b, FGFR2b1257A/Y281A, and FGFR2bE323A ligand-binding domains.
To provide further biochemical support for our finding that plasticity in the interactions of the N-terminal region of FGF1 with receptor D3 is responsible for the promiscuity of FGF1, we introduced mutations into FGFR2b to specifically ablate the interactions observed in the FGF1-FGFR2b structure between FGFR2b and this N-terminal region. Two FGFR2b ligand-binding region constructs were made, one carrying the E323A single mutation (FGFR2bE323A) and the other carrying the I257A/Y281A double mutation (FGFR2bI257A/Y281A). Based on the FGF1-FGFR2b structure (Fig. 1A), the E323A mutation should cause a reduction in FGF1-FGFR2b binding affinity by eliminating the hydrogen bonding between Tyr-23 of FGF1 and Glu-323 of FGFR2b. The I257A/Y281A double mutation should also reduce the binding affinity of FGFR2b for FGF1 because Tyr-281 engages in a hydrophobic interaction with Pro-20 of FGF1, and Ile-257 helps orient the side chain of Tyr-281 for optimal interaction with Pro-20 of FGF1 (Fig. 1A). Consistent with the FGF1-FGFR2b crystal structure, the SPR data show that relative to the wild type FGFR2b ligand-binding region, the FGFR2bI257A/Y281A and FGFR2bE323A mutants sustained a 3-fold and 5-fold loss in FGF1 binding affinity, respectively (Fig. 3C). Size exclusion chromatography analyses of 1:1 mixtures of wild type and mutated FGFR2b ligand-binding regions with FGF1 support the SPR data. The FGF1-FGFR2bI257A/Y281A and FGF1-FGFR2bE323A complexes eluted two and four fractions later than the wild type FGF1-FGFR2b complex, respectively (Fig. 3D). Taken together, these receptor mutagenesis experiments further confirm that interactions between the FGF1 N terminus and FGFR are crucial for the promiscuity of FGF1 (Fig. 3, C and D).
Last, to validate the biological relevance of our structural and biochemical findings, we examined the ability of FGF1, FGF2, and the three FGF2 mutants to induce proliferation of BaF3 cell lines ectopically expressing FGFR2b or FGFR2c. FGF10, a ligand that is highly selective for FGFR2b, was included as a control in these experiments. BaF3 cells were deprived of interleukin-3 for 3–4 h and then treated with these FGFs (100 ng/ml) in the presence of 5 μg/ml heparin. FGFR2b-expressing cells exhibited a 4-fold and 10-fold increase in cell proliferation in response to FGF1 and FGF10, respectively. In contrast, wild type FGF2 induced a modest (1.5-fold) proliferation of these cells (Fig. 4A). FGFR2c-expressing cells proliferated robustly in response to FGF1 and FGF2 (∼7-fold greater than control), whereas FGF10 had no effect (Fig. 4B). Importantly, relative to the parent FGF2 molecule, the FGF2F26Y, FGF2H25N/F26Y, and FGF2G19F/H25N/F26Y mutants all induced greater proliferation of FGFR2b-expressing cells, and the proliferative activity of these mutants increased as the number of substitutions in the N terminus increased from one (F26Y; 2.1-fold) to two (H25N/F26Y; 2.4-fold) to three (G19F/H25N/F26Y; 2.8-fold). In fact, the FGF2G19F/H25N/F26Y mutant exhibited the same proliferative capacity as wild type FGF1. All three FGF2 mutants retained the ability to induce proliferation of FGFR2c-expressing cells (Fig. 4B). These cell-based data, taken together with our SPR and size exclusion chromatography data, unambiguously demonstrate that residues 16, 22, and 23 of FGF1 confer upon the FGF2 mutant the ability to bind and activate FGFR2b. Hence, we conclude that plasticity in the interactions of N-terminal residues of FGF1 with receptor D3 underlies the cross-reactivity of FGF1 with both alternatively spliced FGFR isoforms. These N-terminal residues of FGF1 are well conserved among orthologs, further supporting our conclusion (Fig. 2C).
DISCUSSION
Our identification of the N-terminal region of FGF1 as the key determinant of the receptor binding promiscuity of FGF1 is harmonious with our published structural data on the determinants of the receptor binding specificity of FGF8b and FGF10. The selectivity of FGF10 for FGFR2b can be accounted for by the highly specific hydrogen bonding between Asp-76 of FGF10 and Ser-315 in the βC′-βE loop of FGFR2b (34) (supplemental Fig. 5A). Asp-76 of FGF10 corresponds to Tyr-23 of FGF1 that participates in degenerate binding of FGF1 to all four FGFRs as observed in the FGF1-FGFR crystal structures (Figs. 1, A–D, and 2B). Similarly, the binding specificity of FGF8b for FGFR1c-FGFR3c and FGFR4 can be traced to the hydrophobic contacts that Phe-32 and Val-36 from the FGF8b N terminus make with a hydrophobic groove in the alternatively spliced region of D3 in these four FGFRs (supplemental Fig. 5B and Fig. 2B) (30). Furthermore, a recent study, in which N-terminal sequences between FGF19 and FGF21 were swapped to map residues in FGF19 that permit binding of FGF19 to FGFR4, led to the identification of residues 38–42 at the N terminus of FGF19 as the key determinants of FGF19-FGFR4 specificity (70). This FGF19 N-terminal region corresponds to the FGF1 region that mediates degenerate binding of FGF1 to both alternative splice isoforms of FGFR2 (Fig. 2B). Hence, collectively our structural data show that the sequence divergence between the N termini of FGFs is the primary determinant of both their receptor binding specificity and promiscuity.
Comparison of the four FGF1-FGFR structures show that FGF1 core also engages in plastic interactions with the alternatively spliced βC′-βE and βF-βG loops in D3, which are the least conserved regions of FGFRs (supplemental Fig. 6D). At the interface between FGF1 and the βC′-βE loop, Glu-64 of FGF1 makes direct and water-mediated hydrogen bonds with the backbone atoms of Ile-317 and Asn-318 of receptor in the FGF1-FGFR2b structure (Fig. 1A), whereas in the FGF1-FGFR2c structure, it only makes a water-mediated hydrogen bond to the backbone of Val-317 of receptor (Fig. 1C). Ser-62 of FGF1 makes a direct hydrogen bond to a backbone atom of Val-317 of receptor in FGF1-FGFR2c (Fig. 1C), whereas in the FGF1-FGFR1c and FGF1-FGFR2b structures, Ser-62 does not contact the loop at all (Fig. 1, A and B). Uniquely in FGF1-FGFR2c, Lys-72 and Tyr-79 of FGF1 directly hydrogen-bond with the side chain of Asn-318 of receptor (Fig. 1C).
Plasticity is also evident in the interactions of FGF1 core with the alternatively spliced βF-βG loop in D3 of the receptors. In particular, interactions between FGF1 and the βF-βG loop of receptor in the FGF1-FGFR2b complex are markedly different from those observed in FGF1-FGFRc structures (supplemental Fig. 7). At this interface in the FGF1-FGFR2b structure, the phenyl ring of Tyr-345 in the βF-βG loop of receptor engages in a π-cation interaction with Arg-103 of FGF1, whereas the hydroxyl group of this Tyr-345 is directly hydrogen-bonding to the backbone amide of Leu-104 of FGF1 (supplemental Fig. 7A). In FGFRc isoforms, Tyr-345 is replaced by a serine (supplemental Fig. 6D) that is engaged in water-mediated hydrogen bonds with Leu-104 and Glu-102 of ligand in the FGF1-FGFR1c and FGF1-FGFR2c structures (supplemental Fig. 7B).
Structural and sequence-based analyses indicate, however, that plasticity in the interactions of FGF1 core region with D3 of FGFRs plays a minor role in conferring promiscuity upon FGF1. There is significantly greater sequence similarity at the core region between FGFs, suggesting that other FGFs should be capable of recapitulating the contacts that the core region of FGF1 makes with different FGFRs. For example, the Ser-62 in β4 of FGF1 that engages in variable contacts with the βC′-βE loop is conserved in FGF4 and FGF6, which are known to be specific toward FGFR1c and FGFR2c (Fig. 2D). Furthermore, at the position analogous to Glu-64 in β4 of FGF1, FGF2 also possesses a glutamic acid (Glu-67). In the FGF2-FGFR1c and FGF2-FGFR2c structures, Glu-67 of FGF2 makes hydrogen bonds with the βC′-βE loop of FGFR (supplemental Fig. 4) that are reminiscent of those seen between Glu-64 of FGF1 and this loop in the FGF1-FGFR2b and FGF1-FGFR2c structures (Fig. 1, A and C). FGF2 also possesses a tyrosine (Tyr-72) at the position analogous to Tyr-70 of the hydrophobic patch of FGF1 that engages in plastic interactions with the βC′-βE loop of FGFRs (supplemental Fig. 4). In the FGF10-FGFR2b structure (PDB code 1NUN) (34) Arg-155 of FGF10 that corresponds to Arg-103 of FGF1 engages Tyr-345 in the βF-βG loop of D3 in an identical fashion as Arg-103 of FGF1 (supplemental Fig. 7, A and C). As alluded to in the Introduction, FGF10 is highly specific for FGFR2b, and in fact the interactions of Arg-155 of FGF10 with Tyr-345 in the βF-βG loop of D3 further reinforce FGF10-FGFR2b specificity. Notably, FGF2 also has an arginine at the position corresponding to Arg-103 of FGF1, yet FGF2 is highly specific to FGFR1c and FGFR2c. In light of the fact that FGF2 binds specifically to FGFR1c and FGFR2c and does not act on FGFRb isoforms, these observations indicate that plasticity at the interface between FGF1 core and the alternatively spliced βC′-βE and βF-βG loops in D3 of receptor D3 is not a key factor in the promiscuity of FGF1.
FGF1 has been extensively studied for its translational value in treating human diseases, in particular for the treatment of cardiovascular conditions, tissue repair and bioengineering, and wound healing (1). Therefore, understanding the structural determinants of FGF1 promiscuity should aid in the design of more efficacious FGF1 agonists for therapeutic purposes. For example, we can envision that the structural data presented here could facilitate the structure-guided design of FGF1 variants with narrowed specificity toward FGFRs that are expressed in particular tissues, such as the heart. These receptor-specific FGF1 molecules could potentially also aid in determining which FGFR isoforms mediate each of the versatile biological functions of FGF1.
Supplementary Material
Acknowledgments
We thank Dr. Regina Goetz for critically reading and editing the manuscript. We are grateful to Drs. R. Abramowitz and J. Schwanof for synchrotron beamline assistance. Beamlines X-4A and X-4C at the National Synchrotron Light Source, Brookhaven National Laboratory, a Department of Energy facility, are supported by the New York Structural Biology Consortium. The Biacore 2000 SPR instrument is supported by National Institutes of Health Instrument Grant P30 N5050276 (to Dr. Thomas A. Neubert).
This work was supported, in whole or in part, by National Institutes of Health (NIH) Grant DE13686 (to M. M.).

This article contains supplemental Figs. 1–7.
The atomic coordinates and structure factors (codes 3OJ2, 3OJM, and 3OJV) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
- FHF
- FGF homologous factor
- FGFR
- FGF receptor
- HS
- heparan sulfate
- PDB
- Protein Data Bank
- SPR
- surface plasmon resonance.
REFERENCES
- 1. Beenken A., Mohammadi M. (2009) Nat. Rev. Drug Discov. 8, 235–253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Kuro-o M. (2008) Trends Endocrinol. Metab. 19, 239–245 [DOI] [PubMed] [Google Scholar]
- 3. Ornitz D. M. (2005) Cytokine Growth Factor Rev. 16, 205–213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Itoh N., Ornitz D. M. (2004) Trends Genet. 20, 563–569 [DOI] [PubMed] [Google Scholar]
- 5. Popovici C., Roubin R., Coulier F., Birnbaum D. (2005) BioEssays 27, 849–857 [DOI] [PubMed] [Google Scholar]
- 6. Goetz R., Dover K., Laezza F., Shtraizent N., Huang X., Tchetchik D., Eliseenkova A. V., Xu C. F., Neubert T. A., Ornitz D. M., Goldfarb M., Mohammadi M. (2009) J. Biol. Chem. 284, 17883–17896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Goldfarb M. (2005) Cytokine Growth Factor Rev. 16, 215–220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Olsen S. K., Garbi M., Zampieri N., Eliseenkova A. V., Ornitz D. M., Goldfarb M., Mohammadi M. (2003) J. Biol. Chem. 278, 34226–34236 [DOI] [PubMed] [Google Scholar]
- 9. Beenken A., Mohammadi M. (2012) in Versatile Biological Functions of FGF1 (Kuro-o M., ed) Landes Bioscience, Austin, TX [Google Scholar]
- 10. Fu L., John L. M., Adams S. H., Yu X. X., Tomlinson E., Renz M., Williams P. M., Soriano R., Corpuz R., Moffat B., Vandlen R., Simmons L., Foster J., Stephan J. P., Tsai S. P., Stewart T. A. (2004) Endocrinology 145, 2594–2603 [DOI] [PubMed] [Google Scholar]
- 11. Holt J. A., Luo G., Billin A. N., Bisi J., McNeill Y. Y., Kozarsky K. F., Donahee M., Wang D. Y., Mansfield T. A., Kliewer S. A., Goodwin B., Jones S. A. (2003) Genes Dev. 17, 1581–1591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Lundåsen T., Gälman C., Angelin B., Rudling M. (2006) J. Intern. Med. 260, 530–536 [DOI] [PubMed] [Google Scholar]
- 13. Tomlinson E., Fu L., John L., Hultgren B., Huang X., Renz M., Stephan J. P., Tsai S. P., Powell-Braxton L., French D., Stewart T. A. (2002) Endocrinology 143, 1741–1747 [DOI] [PubMed] [Google Scholar]
- 14. Coskun T., Bina H. A., Schneider M. A., Dunbar J. D., Hu C. C., Chen Y., Moller D. E., Kharitonenkov A. (2008) Endocrinology 149, 6018–6027 [DOI] [PubMed] [Google Scholar]
- 15. Kharitonenkov A., Shiyanova T. L., Koester A., Ford A. M., Micanovic R., Galbreath E. J., Sandusky G. E., Hammond L. J., Moyers J. S., Owens R. A., Gromada J., Brozinick J. T., Hawkins E. D., Wroblewski V. J., Li D. S., Mehrbod F., Jaskunas S. R., Shanafelt A. B. (2005) J. Clin. Invest. 115, 1627–1635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Kharitonenkov A., Wroblewski V. J., Koester A., Chen Y. F., Clutinger C. K., Tigno X. T., Hansen B. C., Shanafelt A. B., Etgen G. J. (2007) Endocrinology 148, 774–781 [DOI] [PubMed] [Google Scholar]
- 17. Xu J., Lloyd D. J., Hale C., Stanislaus S., Chen M., Sivits G., Vonderfecht S., Hecht R., Li Y. S., Lindberg R. A., Chen J. L., Jung D. Y., Zhang Z., Ko H. J., Kim J. K., Véniant M. M. (2009) Diabetes 58, 250–259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Inoue Y., Segawa H., Kaneko I., Yamanaka S., Kusano K., Kawakami E., Furutani J., Ito M., Kuwahata M., Saito H., Fukushima N., Kato S., Kanayama H. O., Miyamoto K. (2005) Biochem. J. 390, 325–331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Saito H., Kusano K., Kinosaki M., Ito H., Hirata M., Segawa H., Miyamoto K., Fukushima N. (2003) J. Biol. Chem. 278, 2206–2211 [DOI] [PubMed] [Google Scholar]
- 20. Segawa H., Kawakami E., Kaneko I., Kuwahata M., Ito M., Kusano K., Saito H., Fukushima N., Miyamoto K. (2003) Pflugers Arch. 446, 585–592 [DOI] [PubMed] [Google Scholar]
- 21. Ornitz D. M., Yayon A., Flanagan J. G., Svahn C. M., Levi E., Leder P. (1992) Mol. Cell Biol. 12, 240–247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Rapraeger A. C., Krufka A., Olwin B. B. (1991) Science 252, 1705–1708 [DOI] [PubMed] [Google Scholar]
- 23. Yayon A., Klagsbrun M., Esko J. D., Leder P., Ornitz D. M. (1991) Cell 64, 841–848 [DOI] [PubMed] [Google Scholar]
- 24. Goetz R., Beenken A., Ibrahimi O. A., Kalinina J., Olsen S. K., Eliseenkova A. V., Xu C., Neubert T. A., Zhang F., Linhardt R. J., Yu X., White K. E., Inagaki T., Kliewer S. A., Yamamoto M., Kurosu H., Ogawa Y., Kuro-o M., Lanske B., Razzaque M. S., Mohammadi M. (2007) Mol. Cell Biol. 27, 3417–3428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Kurosu H., Ogawa Y., Miyoshi M., Yamamoto M., Nandi A., Rosenblatt K. P., Baum M. G., Schiavi S., Hu M. C., Moe O. W., Kuro-o M. (2006) J. Biol. Chem. 281, 6120–6123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Urakawa I., Yamazaki Y., Shimada T., Iijima K., Hasegawa H., Okawa K., Fujita T., Fukumoto S., Yamashita T. (2006) Nature 444, 770–774 [DOI] [PubMed] [Google Scholar]
- 27. Givol D., Yayon A. (1992) FASEB J. 6, 3362–3369 [PubMed] [Google Scholar]
- 28. Johnson D. E., Williams L. T. (1993) Adv. Cancer Res. 60, 1–41 [DOI] [PubMed] [Google Scholar]
- 29. Lee P. L., Johnson D. E., Cousens L. S., Fried V. A., Williams L. T. (1989) Science 245, 57–60 [DOI] [PubMed] [Google Scholar]
- 30. Olsen S. K., Li J. Y., Bromleigh C., Eliseenkova A. V., Ibrahimi O. A., Lao Z., Zhang F., Linhardt R. J., Joyner A. L., Mohammadi M. (2006) Genes Dev. 20, 185–198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Plotnikov A. N., Hubbard S. R., Schlessinger J., Mohammadi M. (2000) Cell 101, 413–424 [DOI] [PubMed] [Google Scholar]
- 32. Plotnikov A. N., Schlessinger J., Hubbard S. R., Mohammadi M. (1999) Cell 98, 641–650 [DOI] [PubMed] [Google Scholar]
- 33. Stauber D. J., DiGabriele A. D., Hendrickson W. A. (2000) Proc. Natl. Acad. Sci. U.S.A. 97, 49–54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Yeh B. K., Igarashi M., Eliseenkova A. V., Plotnikov A. N., Sher I., Ron D., Aaronson S. A., Mohammadi M. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 2266–2271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Olsen S. K., Ibrahimi O. A., Raucci A., Zhang F., Eliseenkova A. V., Yayon A., Basilico C., Linhardt R. J., Schlessinger J., Mohammadi M. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 935–940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Wang F., Kan M., Yan G., Xu J., McKeehan W. L. (1995) J. Biol. Chem. 270, 10231–10235 [DOI] [PubMed] [Google Scholar]
- 37. Mohammadi M., Olsen S. K., Ibrahimi O. A. (2005) Cytokine Growth Factor Rev. 16, 107–137 [DOI] [PubMed] [Google Scholar]
- 38. Schlessinger J., Plotnikov A. N., Ibrahimi O. A., Eliseenkova A. V., Yeh B. K., Yayon A., Linhardt R. J., Mohammadi M. (2000) Mol. Cell 6, 743–750 [DOI] [PubMed] [Google Scholar]
- 39. Chen H., Xu C. F., Ma J., Eliseenkova A. V., Li W., Pollock P. M., Pitteloud N., Miller W. T., Neubert T. A., Mohammadi M. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 19660–19665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Mohammadi M., Dikic I., Sorokin A., Burgess W. H., Jaye M., Schlessinger J. (1996) Mol. Cell Biol. 16, 977–989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Kouhara H., Hadari Y. R., Spivak-Kroizman T., Schilling J., Bar-Sagi D., Lax I., Schlessinger J. (1997) Cell 89, 693–702 [DOI] [PubMed] [Google Scholar]
- 42. Huang J., Mohammadi M., Rodrigues G. A., Schlessinger J. (1995) J. Biol. Chem. 270, 5065–5072 [DOI] [PubMed] [Google Scholar]
- 43. Mohammadi M., Dionne C. A., Li W., Li N., Spivak T., Honegger A. M., Jaye M., Schlessinger J. (1992) Nature 358, 681–684 [DOI] [PubMed] [Google Scholar]
- 44. Carballada R., Yasuo H., Lemaire P. (2001) Development 128, 35–44 [DOI] [PubMed] [Google Scholar]
- 45. Hadari Y. R., Gotoh N., Kouhara H., Lax I., Schlessinger J. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 8578–8583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Ong S. H., Hadari Y. R., Gotoh N., Guy G. R., Schlessinger J., Lax I. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 6074–6079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Seo J. H., Suenaga A., Hatakeyama M., Taiji M., Imamoto A. (2009) Mol. Cell Biol. 29, 3076–3087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Dailey L., Ambrosetti D., Mansukhani A., Basilico C. (2005) Cytokine Growth Factor Rev. 16, 233–247 [DOI] [PubMed] [Google Scholar]
- 49. Eswarakumar V. P., Lax I., Schlessinger J. (2005) Cytokine Growth Factor Rev. 16, 139–149 [DOI] [PubMed] [Google Scholar]
- 50. Ornitz D. M., Xu J., Colvin J. S., McEwen D. G., MacArthur C. A., Coulier F., Gao G., Goldfarb M. (1996) J. Biol. Chem. 271, 15292–15297 [DOI] [PubMed] [Google Scholar]
- 51. Zhang X., Ibrahimi O. A., Olsen S. K., Umemori H., Mohammadi M., Ornitz D. M. (2006) J. Biol. Chem. 281, 15694–15700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Chellaiah A. T., McEwen D. G., Werner S., Xu J., Ornitz D. M. (1994) J. Biol. Chem. 269, 11620–11627 [PubMed] [Google Scholar]
- 53. Johnson D. E., Lu J., Chen H., Werner S., Williams L. T. (1991) Mol. Cell Biol. 11, 4627–4634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Miki T., Bottaro D. P., Fleming T. P., Smith C. L., Burgess W. H., Chan A. M., Aaronson S. A. (1992) Proc. Natl. Acad. Sci. U.S.A. 89, 246–250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Yayon A., Zimmer Y., Shen G. H., Avivi A., Yarden Y., Givol D. (1992) EMBO J. 11, 1885–1890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Chellaiah A., Yuan W., Chellaiah M., Ornitz D. M. (1999) J. Biol. Chem. 274, 34785–34794 [DOI] [PubMed] [Google Scholar]
- 57. Ron D., Reich R., Chedid M., Lengel C., Cohen O. E., Chan A. M., Neufeld G., Miki T., Tronick S. R. (1993) J. Biol. Chem. 268, 5388–5394 [PubMed] [Google Scholar]
- 58. Vainikka S., Partanen J., Bellosta P., Coulier F., Birnbaum D., Basilico C., Jaye M., Alitalo K. (1992) EMBO J. 11, 4273–4280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. MacArthur C. A., Lawshé A., Xu J., Santos-Ocampo S., Heikinheimo M., Chellaiah A. T., Ornitz D. M. (1995) Development 121, 3603–3613 [DOI] [PubMed] [Google Scholar]
- 60. Otwinowski Z., Minor W. (1997) Methods Enzymol. 276, 307–326 [DOI] [PubMed] [Google Scholar]
- 61. Navaza J. (1994) Acta Crystallogr. A 50, 157–163 [Google Scholar]
- 62. Jones T. A., Zou J. Y., Cowan S. W., Kjeldgaard M. (1991) Acta Crystallogr. A 47, 110–119 [DOI] [PubMed] [Google Scholar]
- 63. Brünger A. T., Adams P. D., Clore G. M., DeLano W. L., Gros P., Grosse-Kunstleve R. W., Jiang J. S., Kuszewski J., Nilges M., Pannu N. S., Read R. J., Rice L. M., Simonson T., Warren G. L. (1998) Acta Crystallogr. D Biol. Crystallogr. 54, 905–921 [DOI] [PubMed] [Google Scholar]
- 64. Gartside M. G., Chen H., Ibrahimi O. A., Byron S. A., Curtis A. V., Wellens C. L., Bengston A., Yudt L. M., Eliseenkova A. V., Ma J., Curtin J. A., Hyder P., Harper U. L., Riedesel E., Mann G. J., Trent J. M., Bastian B. C., Meltzer P. S., Mohammadi M., Pollock P. M. (2009) Mol. Cancer Res. 7, 41–54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Byron S. A., Gartside M. G., Wellens C. L., Goodfellow P. J., Birrer M. J., Campbell I. G., Pollock P. M. (2010) Gynecol. Oncol. 117, 125–129 [DOI] [PubMed] [Google Scholar]
- 66. Ibrahimi O. A., Eliseenkova A. V., Plotnikov A. N., Yu K., Ornitz D. M., Mohammadi M. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 7182–7187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Ibrahimi O. A., Zhang F., Eliseenkova A. V., Itoh N., Linhardt R. J., Mohammadi M. (2004) Hum. Mol. Genet. 13, 2313–2324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Ibrahimi O. A., Yeh B. K., Eliseenkova A. V., Zhang F., Olsen S. K., Igarashi M., Aaronson S. A., Linhardt R. J., Mohammadi M. (2005) Mol. Cell Biol. 25, 671–684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Luo Y., Gabriel J. L., Wang F., Zhan X., Maciag T., Kan M., McKeehan W. L. (1996) J. Biol. Chem. 271, 26876–26883 [DOI] [PubMed] [Google Scholar]
- 70. Wu X., Ge H., Lemon B., Vonderfecht S., Baribault H., Weiszmann J., Gupte J., Gardner J., Lindberg R., Wang Z., Li Y. (2010) Proc. Natl. Acad. Sci. U.S.A. 107, 14158–14163 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.